Abstract
The resistance of Candida albicans biofilms to a broad spectrum of antimicrobial agents has been well documented. Biofilms are known to be heterogeneous, consisting of microenvironments that may induce formation of resistant subpopulations. In this study we characterized one such subpopulation. C. albicans biofilms were cultured in a tubular flow cell (TF) for 36 h. The relatively large shear forces imposed by draining the TF removed most of the biofilm, which consisted of a tangled mass of filamentous forms with associated clusters of yeast forms. This portion of the biofilm exhibited the classic architecture and morphological heterogeneity of a C. albicans biofilm and was only slightly more resistant than either exponential- or stationary-phase planktonic cells. A submonolayer fraction of blastospores that remained on the substratum was resistant to 10 times the amphotericin B dose that eliminated the activity of the planktonic populations. A comparison between planktonic and biofilm populations of transcript abundance for genes coding for enzymes in the ergosterol (ERG1, -3, -5, -6, -9, -11, and -25) and β-1,6-glucan (SKN and KRE1, -5, -6, and -9) pathways was performed by quantitative RT-PCR. The results indicate a possible association between the high level of resistance exhibited by the blastospore subpopulation and differential regulation of ERG1, ERG25, SKN1, and KRE1. We hypothesize that the resistance originates from a synergistic effect involving changes in both the cell membrane and the cell wall.
Candida albicans is a commensal opportunistic pathogen that causes both superficial and invasive, disseminated infections (55). Invasive candidiasis is the most prevalent invasive mycosis in humans, with C. albicans being the most common etiological agent (58). Superficial forms of candidiasis that are non-life-threatening but are nevertheless considered serious health concerns include relapsing vaginitis and human immunodeficiency virus-associated oropharyngeal candidiasis (64).
C. albicans colonizes various biomaterials and readily forms dense, complex biofilms under a variety of in vitro conditions (60). A particularly high risk of invasive candidiasis is associated with the use of urinary and vascular catheters and ventricular assistive devices (41). The likelihood of bloodstream infections (BSI) resulting from colonization of intravascular catheters by Candida species ranks second only to that posed by Staphylococcus aureus (19). The prognosis for BSI caused by Candida species is poor, with an attributable mortality for nosocomial BSI of nearly 50% (34).
From a rigorous clinical perspective, in vitro biofilm models are most appropriate for testing hypotheses related to biomaterial-centered infections. However, from a broader, fundamental perspective, their utility for advancing understanding of infective C. albicans communities extends further. The biofilm paradigm is considered an appropriate model for some tissue infections (18), and superficial tissue infections such as oropharyngeal candidiasis resemble, in many respects, C. albicans biofilms that form in vitro (33). Also, some forms of localized invasive candidiasis manifest as a dense pleomorphic community (55), and these infections share conspicuous morphological characteristics with in vitro biofilms.
In general, microbial biofilms exhibit an exceptional ability to survive doses of antimicrobial agents that are many times greater than the dose which is lethal for their planktonic counterparts (18, 22, 45, 68). C. albicans biofilms have been shown to be resistant to a variety of azoles, including fluconazole (36), voriconazole (46), miconazole (44), itraconazole (36), ketoconazole (36), the antiseptic chlorhexidine (15, 16), flucytosine (36), and the polyenes nystatin (15, 16) and amphotericin B (AmB) (15, 16, 36, 61, 62). The reasons for biofilm resistance have not been fully elucidated, but the emerging theme is that biofilm resistance cannot be generally traced to traits characteristic of isolated cells but rather originates from special conditions inherent in the biofilm community lifestyle (18, 22, 60, 68). One possibility is that microenvironments in biofilms accommodate subpopulations of cells with physiological traits that render them less susceptible to antimicrobial agents, a phenomenon sometimes referred to as phenotypic adaptation (48). We present evidence here for the existence of such a subpopulation in C. albicans biofilms cultured in our experimental system.
MATERIALS AND METHODS
C. albicans strain and medium.
C. albicans CA-1 is a clinical isolate obtained from the culture collection of Diane Brawner (Microbiology Department, Montana State University) (35). The biofilm-forming behavior of CA-1 was characterized in previous studies (69, 71). The strain was stored at −80oC. Planktonic cells were cultured in 2% YEPD medium (2% glucose, 1% Bacto yeast extract, and 2% Bacto peptone), and biofilms were cultured in a sixfold dilution of this medium.
Planktonic cultures.
Cultures were grown aerobically in 250-ml Erlenmeyer flasks containing 100 ml growth medium. The flasks were placed in a shaker incubator at 37oC and 160 rpm for the desired period of growth. C. albicans grew as a budding yeast under these conditions. Exponential-phase cells were harvested after a 5-h growth period and stationary-phase cells at 120 h.
Biofilm cultures.
Biofilms were grown in a tubular flow cell (TF). Silicone tubing (Cole-Parmer; catalog no. EW-95802-08) with an inner diameter of 4.78 mm, an outer diameter of 6.35 mm, a wall of 0.79 mm, and a length of 60 cm constituted the biofilm-growing region of the TF. The source of growth medium for the TF was a 2-liter Erlenmeyer flask. A bubble trap was placed between the Erlenmeyer flask and the TF to prevent passage of air bubbles during biofilm growth. Flow rates of 1.17 ml/min (shear rate, 109.5 s−1) were maintained by a peristaltic pump coupled at the effluent end of the TF. The residence time for the volume of liquid contained in the tubular reactor portion of the flow system (20-cm length of tubing) was 3 min. This condition ensured that the contribution to the cell population in the TF from cells in the planktonic mode of growth (doubling time, approximately 80 min) was negligible. After sterilization by autoclaving, the entire setup was placed horizontally on a grilled shelf in an incubator at 37oC. The TF was filled with growth medium before being inoculated with cells. The inoculum was prepared from a 24-h planktonic culture at a concentration of 108 cells/ml in 0.1 M phosphate-buffered saline (PBS, pH 7) buffer. It was fed into the TF from the effluent end by reversing the direction of flow. Flow was then discontinued for 1 h. After the 1-h inoculation period, flow was resumed for 36 h. At the end of this culture period, the section of tubing in which the biofilm grew was clamped at both ends and removed by cutting the tubing with a sterile blade. The liquid column was drained into a petri dish by moving the tubing to a vertical position and releasing the clamps. The tubing was then rinsed with PBS buffer equivalent to one tube volume. The fraction of biofilm collected in the petri dish by this procedure is referred to as the shear-removed biofilm. The cells that remained adhering to the walls of the tube are referred to as the basal blastospores or the basal blastospore subpopulation.
Susceptibility testing.
AmB was from Biosource International Inc. (Fungizone with 0.00205% sodium deoxycholate solubilizing agent). A standard broth dilution method was used to assess the AmB MIC of CA-1, with ATCC 24433 used as a reference strain (52). Susceptibility of biofilms and planktonic populations used for comparative analysis were assessed by the alamarBlue metabolic assay (BioSource International Inc.; catalog no. DAL1100) in a 96-well plate format (47, 57, 72). AmB treatment of planktonic cells and shear-removed biofilm was performed in 1.5-ml centrifuge tubes. The tubes contained a total working volume of 450 μl. AmB dissolved in 200 μl of 0.1 M PBS (pH 7.0) and cells resuspended in 250 μl of PBS adjusted to an optical density of 0.05 (A660) were added to the 1.5-ml tube. After more than 10 preliminary experiments, the time between recovery of the shear-removed biofilm in the petri dish and exposure to AmB was reduced to approximately 5 min. The positive controls had no AmB, and the negative controls had no cells. For AmB treatment of the basal blastospore population, each sample consisted of a 0.5-cm long section of tubing cut from the TF. Each tubing sample was positioned in the 96-well plate (Corning Inc.; Costar 3370) such that its outer wall snugly fit along the wall of the well in the 96-well plate. This enabled recording of absorbance data for the alamarBlue dye in real time without perturbing the well contents. The 0.5-cm tubing samples were randomly placed in the 96-well plate in order to randomize the distribution between treated samples and untreated controls. Solution (225 μl) containing AmB in 0.1 M PBS was added to wells to completely submerge the 0.5-cm tubing during the AmB treatment phase. Untreated samples were the positive controls. Three independent TF experiments were run, generating a total of 24 samples per AmB concentration and 61 untreated controls for the basal blastospores. Sterile tubing was used as a negative control. In every experiment, a minimum of four negative controls were used. The AmB treatment period was 1 h across all cell populations. During AmB exposure, the 1.5-ml tubes (containing either the planktonic or shear-removed populations) and 96-well plates (containing the basal blastospores) were placed in a shaker incubator at 37oC and 150 rpm. After AmB treatment, cells in 1.5-ml tubes were centrifuged for 5 min at 4,000 × g, the supernatant was decanted, and the pellet was resuspended in the same volume of fresh PBS buffer. Cells were then transferred into wells of the 96-well plate system for the metabolic assay. For the 96-well plate containing sections of tubing, AmB was carefully aspirated out of the wells and immediately replaced with reagents of the metabolic assay.
The components of the alamarBlue metabolic assay were at the same concentrations for all populations tested. Each well had a total working volume of 230 μl. It contained 25 μl alamarBlue dye, 30 μl 2% YEPD growth medium, 100 μl of cells (or the 0.5-cm tubing section) in PBS, and 75 μl of a PBS solution containing AmB-quenching reagents and antibacterial reagent (penicillin-streptomycin; Invitrogen catalog no. 15140-122; final concentration, 1% [by volume]). The final concentrations of the reagents included in the assay to quench the action of AmB during the posttreatment metabolic assay period were 40 μM ergosterol (Alfa Aesar; catalog no. 57-87-4), 42.5 μM KCl, and 88 μM MgCl2 (26, 27, 38). The metabolic assay in the 96-well format was run for 24 h at 37oC with continuous shaking in a Synergy-HT plate reader (Biotek Inc.). Absorbance data at 570 nm and 600 nm were collected at the 24-h endpoint using KC4 software (Biotek Inc.). The percent reduction of the alamarBlue dye, which is a function of metabolic activity of viable cells, was calculated as per the manufacturer's formula (product literature; BioSource International Inc.; catalog no. DAL1100). The results were expressed as the mean percent reduced for each triplicate culture (except as otherwise stated) ± the standard error.
Scanning electron microscopy.
Biofilm samples from the TF were fixed in 2.5% glutaraldehyde and 1% paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 7.4), overnight, at 4oC. They were rinsed with 0.1 M sodium cacodylate buffer twice for 10 min. Samples were postfixed in 2% osmium tetroxide for 45 min on a rotator. Samples were subsequently dehydrated in a series of ethanol washes (twice for 5 min in 50%, twice for 5 min in 70%, and twice for 5 min in 95%, followed by two washes for 10 min in 100%) and then air dried in an open vial. Biofilms were visualized with a scanning electron microscope in high-vacuum mode at 25 kV after sputter coating with gold. The images were processed using Microsoft Photo Editor software.
Quantitative RT-PCR. (i) Total RNA isolation and cDNA preparation.
Total RNA from planktonic and biofilm populations was isolated using the RiboPure yeast kit (Ambion, Inc.) according to the manufacturer's instructions. Basal blastospores were collected for RNA isolation by treating the tubes with trypsin-EDTA (Invitrogen; catalog no. T25300-120) for 4 min, scraping adherent cells (4 min), concentrating them by centrifugation (2 min), and washing the pellet with PBS (2 min). Total RNA (2.5 μg from biofilm and planktonic samples) was reverse transcribed with oligo(dT) primers using Superscript reverse transcriptase II (Invitrogen) according to the manufacturer's instructions.
(ii) Primer design.
Sequences of C. albicans genes were downloaded from the Candida Genome Database (5) using Omiga (version 2.0) software. Primer3 software (65) was used to design primers for quantitative RT-PCR. The optimal conditions for choosing efficient primers included: biasing the primers toward the 3′ end of the gene, restricting the amplicon size to between 50 and 200 base pairs, maintaining the GC content of the primers at a range of 40 to 60%, and keeping the difference in melting temperature between the two primers below 1oC. Primers were tested to avoid 3′ self-complementarity and self-annealing using the Omiga software. A BLASTn search was performed with the designed primers to ensure that they were unique to the gene of interest within the C. albicans genome. The primer efficiencies for each gene were estimated from standard curves using serial dilutions of cDNA (250, 25, 2.5, and 0.25 ng equivalent of RNA). The PCR primers were considered efficient if the coefficient of determination of the standard curve was >0.98 and the target amplification efficiency (10−slope − 1) was equal to 100% (acceptable error of 10%). Additional information about the genes of interest and their primer sequences are available in Table 1. All primer pairs listed in Table 1 had acceptable efficiencies in the cDNA concentration range tested.
TABLE 1.
C. albicans genes chosen for transcript analysis using quantitative RT-PCRa
| Gene | Accession no. | Protein product and/or function | Primer sequencesb |
|---|---|---|---|
| GAP | U72203 | Glyceraldehyde-3-phosphate dehydrogenase; enzyme of glycolysis | TGACCACTGTCCACTCCATC |
| AGCAGTTCTACCACCTCTCCA | |||
| ACT1 | X16377 | Actin | GCTGAACGTATGCAAAAGGAA |
| TGTGGTGAACAATGGATGGA | |||
| EFB1 | X96517 | Translation elongation factor EF-1 beta | GAAGGCTGCTAAAGGTCCAAA |
| ACCCCAAGTCAAACCTTCCA | |||
| ERG1 | U69674 | Squalene epoxidase, catalyzes epoxidation of squalene to 2,3(S)-oxidosqualene in the ergosterol biosynthetic pathway | TCGTTGTTCTCCCTTTCCTC |
| CCTTGCTCAACTCCCATTCT | |||
| ERG3 | AF069752 | C-5 sterol desaturase; introduces C-5(6) double bond into episterol in ergosterol biosynthesis | GCTTGTCACACTGTCCATCAC |
| AATCTTCTTTTCTTCTTCTGCCTTT | |||
| ERG5 | CA4418 | Putative C-22 sterol desaturase; catalyzes formation of the C-22(23) double bond in the sterol side chain in ergosterol biosynthesis | TCTTTGAGATACCGTCCACCA |
| GTCGTGCAAAGCAGGATACA | |||
| ERG6 | AF031941 | Delta(24)-sterol C-methyltransferase; converts zymosterol to fecosterol in ergosterol biosynthesis by methylating position C-24 | GGTGGTCCTGGTAGAGAAATCACA |
| AGCTTCAATGGCATAAACAGCATC | |||
| ERG9 | D89610 | Putative farnesyl-diphosphate farnesyl transferase (squalene synthase) involved in the sterol biosynthesis pathway | AGTAAAAACCCCTGCCAAAGA |
| GCACAAAATGAGAATGAAGAAGG | |||
| ERG11 | X13296 | Lanosterol 14-alpha-demethylase, member of the cytochrome P450 family that functions in ergosterol biosynthesis | GGATACTGCTGCTGCCAAA |
| GCAAATTGTTCCCCAATACATC | |||
| ERG25 | AF051914 | Putative C-4 methyl sterol oxidase with role in C4 demethylation of ergosterol biosynthesis intermediates (based on similarity to S. cerevisiae) | GCAGCAGAATATGCTCATCCA |
| ATCGGAATACCAACCGTACCC | |||
| SKN1 | D88491 | Protein with predicted role in β-1,6-glucan synthesis; probable N-glycosylated type II membrane protein | TCGATCAGGGATGATTCAAAG |
| ACCATAACCCAGGCCAAAA | |||
| KRE1 | M81588 | Predicted GPIc anchor, role in β-1,6-glucan biosynthesis; production in S. cerevisiae increases glucan content and complements killer toxin sensitivity of KRE1 mutant | TTAGCCACCACCCAATCAA |
| ACCCCAATCCAATAGAACCA | |||
| KRE5 | XM_714894 | Predicted UDP-glucose:glycoprotein glucosyltransferase; required for wild-type β-1,6-glucan biosynthesis | TGTCTGCATCGGAAAACAAA |
| AAACTGGCACCTCAATGCTC | |||
| KRE6 | D88490 | Protein of β-1,6-glucan biosynthesis | GTGATGAATTTGATGCTGAAGG |
| GAATGTCAGGAGCGGTGAA | |||
| KRE9 | AF069763 | Protein of β-1,6-glucan biosynthesis | AAAGGCTGGACAACCAAA |
| TGGATATGGAGCAACACTAGCA |
(iii) Quantitative RT-PCR protocol.
Each PCR mixture contained the following (final concentrations): 1× 20 mM Tris-HCl (pH 8.3) buffer, 50 mM KCl, 3 mM MgCl2, 0.3 μM forward primer, 0.3 μM reverse primer, 1× additive reagent (0.2 mg/ml bovine serum albumin, 150 mM trehalose, and 0.2% Tween-20), 0.25× SYBR green, 1.5 U Platinum Taq polymerase, a 200 μM concentration of each deoxynucleoside triphosphate, 2 μl cDNA, and H2O to bring the final volume to 25 μl. The cDNA was amplified according to the following steps: (i) 95oC for 2 min, (ii) 95oC for 12 s, (iii) 58 to 63oC for 15 s, (iv) 72oC for 20 s, and (v) 80oC for 6 s to detect SYBR green (nonspecific products melt at <80oC and therefore were not detected). Steps 2 to 5 were repeated for 40 cycles, and at the end of the last cycle the temperature was increased 0.2oC/s from 60 to 95oC to produce a melting curve. Smart Cycler software calculated corresponding cycle threshold (CT) values for a threshold fluorescence of 30. All samples had RNA obtained from three independent experiments. To avoid variation between runs, samples of all populations were run simultaneously for each gene. Relative quantification of gene expression was performed using the comparative CT method (sequence detection systems chemistry guide; Applied Biosystems). The most stable housekeeping genes, EFB1 and ACT1 (11), across the sample states were chosen using the statistical technique described by Vandesompele et al. (73). The internal normalization factor was the geometric mean of the stable housekeeping genes (73). The propagated standard error of a set of triplicate samples was estimated using the differential equation of Gauss as described by Muller et al. (51). The change in gene expression was further normalized relative to a calibrator state chosen from the pool of populations being tested. In our study, the exponential-phase planktonic population was selected as the calibrator state.
RESULTS
Biofilm subpopulations.
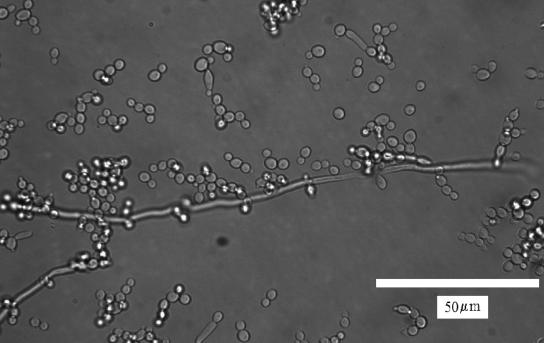
The effluent that was drained from the TF at the end of the 36-h culture period contained masses of material that had the slimy appearance characteristic of many biofilms. Although C. albicans biofilms vary in architecture depending on growth conditions (7, 8), many studies have reported an architecture consisting of a pleomorphic mixture of filamentous and yeast forms in which the filaments are intertwined and interspersed with clusters of blastospores (3, 15, 63). In a previous study, we found that the architecture of CA-1 biofilms conformed to this description (69). In these respects, the material displaced from the TF as the tubing was drained resembled a typical C. albicans biofilm (Fig. 1).
FIG. 1.
Scanning electron microscopy image of the shear-removed C. albicans biofilm displaced by passage of the three-phase boundary layer.
By holding the tubing vertically and allowing it to drain, we essentially implemented a method previously used to remove biofilms cultured in vitro from biomaterials (32). It was shown that shear forces exerted at the three-phase (solid/liquid/air) interface imparted by passage of an air bubble through a biofilm flow cell are considerable (approximately 2 × 10−7 N) and can peel away biofilms grown under continuous medium flow (32). During growth, our C. albicans biofilms were subjected to a constant shear of 109.5 s−1. During this period there was continual detachment of cells (mostly yeast forms) (Fig. 2). However, we did not observe detachment of large aggregates, as has been reported for bacterial biofilms (76).
FIG. 2.
Transmittance image of C. albicans cells that were present in the effluent of the TF during biofilm growth. These cells resemble typical planktonic-phase cells, with an occasional hypha.
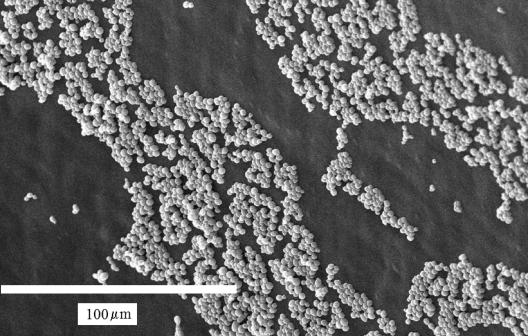
After the bulk of the biofilm had been removed by draining the tubing, there were still cells left on the surface (Fig. 3). This population consisted entirely of blastospores, arranged in small, fairly dense monolayer domains. Thus, by using this relatively simple method, we separated the biofilm into two subpopulations: a complex mixture of different morphological types that comprised the bulk of the biofilm and a submonolayer of basal blastospores that were more tenaciously attached to the surface of the tubing.
FIG. 3.
Scanning electron microscopy image of the C. albicans basal blastospore subpopulation that remained on the silicone tubing surface of the TF after passage of the three-phase boundary layer.
Biofilm and planktonic resistance to AmB.
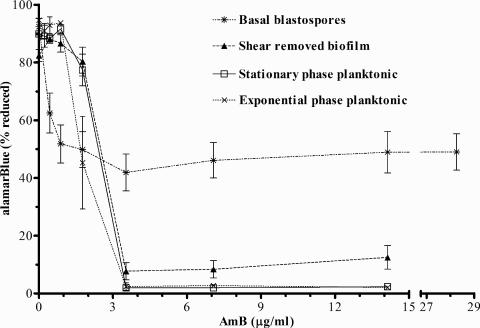
The dose-response curve for the biofilm subpopulations and planktonic populations used for comparative analysis is shown in Fig. 4. At AmB concentrations greater than or equal to 3.7 μg/ml, the basal blastospores were clearly more active than the other three populations. At these AmB concentrations, the planktonic populations showed no statistically significant difference between AmB-treated and negative (no-cell) controls (P < 0.0001, two-tailed Student's t test), and the shear-removed biofilm exhibited significant inhibition of metabolic activity (>90%).
FIG. 4.
AmB dose-response curves for C. albicans biofilm and planktonic populations assessed by an alamarBlue metabolic assay. The data are means ± standard errors for all populations considered in the study and are from three independent experiments.
As assessed by the broth dilution method, CA-1 exhibited an AmB MIC (0.5 μg/ml) within a standard range for susceptible C. albicans strains. Based on previous studies (26, 28), it was anticipated that stationary-phase planktonic cells would exhibit greater AmB resistance than exponential-phase planktonic cells when evaluated by a metabolic assay sensitive to membrane integrity. This difference was evident only at 1.77 μg/ml AmB. Data not shown here indicated that when replication efficiency on solid medium was used to assess AmB action, the greater tolerance of stationary-phase batch cultures than exponential-phase batch cultures was more apparent for a range of AmB concentrations (0.88 to 3.53 μg/ml).
The basal blastospore subpopulation exhibited metabolic activity up to a tested concentration of 28.26 μg/ml AmB. Even at this highest AmB concentration, microscopic observation revealed the appearance of budding planktonic yeast forms during the metabolic assay, suggesting that the basal blastospores produced viable cells. At lower concentrations (<3.53 μg/ml), the basal blastospore subpopulation showed a greater loss of metabolic activity than the other samples. This result would be expected if a portion of the cells in the basal blastospore subpopulation were more susceptible to AmB than the other populations and suggests that this subpopulation is itself not completely homogeneous with respect to its susceptibility to AmB.
Biofilm and planktonic gene expression.
Transcript analysis using quantitative RT-PCR indicated differential gene expression of several genes in the ergosterol (Table 2) and β-1,6-glucan (Table 3) pathways. Data for biofilm and planktonic populations were obtained from three independent culture experiments. GAP, ACT1, and EFB1 were chosen as potential housekeeping candidates to estimate an internal normalization factor. Based on the statistical algorithm proposed by Vandesompele et al. (73), only ACT1 and EFB1 proved to be stable across the four cell populations tested in this study. Their geometric mean was used to estimate the internal normalization control. Gene expression values were calculated relative to the calibrator state, which in our study was the exponential-phase planktonic state.
TABLE 2.
Relative transcript abundance for selected genes of the ergosterol pathway in C. albicans
| Genea | Change (fold) in gene expressionb
|
|||
|---|---|---|---|---|
| Planktonic forms
|
Shear-removed biofilm | Basal blastospore subpopulation | ||
| Exponential phase | Stationary phase | |||
| ACT1 | 1.0 ± 0.05 | 1.61 ± 0.15 | 0.85 ± 0.31 | 0.9 ± 0.04 |
| EFB1 | 1.0 ± 0.10 | 0.62 ± 0.05 | 1.18 ± 0.41 | 1.11 ± 0.11 |
| ERG1 | 1.0 ± 0.72 | 35.55 ± 24.03 | −2.06 ± 1.67 | −11.93 ± 4.69 |
| ERG3 | 1.0 ± 0.06 | 3.67 ± 0.6 | 5.31 ± 1.74 | 0.91 ± 0.22 |
| ERG5 | 1.0 ± 0.06 | 2.1 ± 0.55 | 2.11 ± 0.96 | 1.48 ± 0.27 |
| ERG6 | 1.0 ± 0.12 | 0.72 ± 0.23 | 3.95 ± 1.95 | 1.32 ± 0.55 |
| ERG9 | 1.0 ± 0.15 | 1.05 ± 0.21 | 1.89 ± 0.45 | −2.2 ± 0.78 |
| ERG11 | 1.0 ± 0.06 | 0.98 ± 0.29 | 2.0 ± 0.96 | 0.88 ± 0.27 |
| ERG25 | 1.0 ± 0.18 | 7.1 ± 1.13 | 13.28 ± 3.83 | 3.7 ± 0.73 |
See Table 1 for descriptions of genes. ACT1 and EFB1 were internal normalization controls.
Values were estimated relative to those for the exponential-phase planktonic population (calibrator state) and are given as means ± propagated standard errors.
TABLE 3.
Relative transcript abundance for selected genes of the β-1,6-glucan pathway in C. albicans
| Genea | Change (fold) in gene expressionb
|
|||
|---|---|---|---|---|
| Planktonic forms
|
Shear-removed biofilm | Basal blastospore subpopulation | ||
| Exponential phase | Stationary phase | |||
| SKN1 | 1.0 ± 0.15 | 9.09 ± 4.23 | 2.49 ± 1.63 | 30.70 ± 4.48 |
| KRE1 | 1.0 ± 0.34 | 12.92 ± 2.06 | 17.01 ± 6.03 | 29.86 ± 7.35 |
| KRE5 | 1.0 ± 0.2 | 7.35 ± 2.58 | 1.11 ± 0.42 | −2.22 ± 0.84 |
| KRE6 | 1.0 ± 0.1 | 7.83 ± 2.12 | 4.88 ± 2.24 | 1.9 ± 1.28 |
| KRE9 | 1.0 ± 0.07 | 2.92 ± 0.86 | 2.08 ± 0.7 | 2.62 ± 0.30 |
The basal blastospore subpopulation had a unique transcript profile for both the pathways compared to the other cell states. ERG1 (−11.93 ± 4.68) and ERG25 (+3.70 ± 0.73) were highly differentially regulated in this resistant fraction. For the β-1,6-glucan pathway, the resistant basal blastospores showed a high degree of differential up regulation of the SKN1 (+30.7 ± 4.48) and KRE1 (+29.86 ± 7.39) genes. The KRE1 gene exhibited an interesting trend of increasing transcript abundance for populations exhibiting increased resistance at higher concentrations of AmB.
DISCUSSION
We have demonstrated that C. albicans biofilms cultured in our in vitro system harbored a subpopulation of blastospores that were substantially more resistant to AmB than both planktonic cells and the bulk of the biofilm that had been displaced from the substratum. It is not surprising to find a subpopulation of microbes that exhibit superior tolerance to an antimicrobial challenge in a biofilm. In contrast to suspended cultures, biofilms are not well-mixed systems; thus, any substance that is consumed by the organisms at a substantial rate (such as the carbon source or electron acceptor) will be relatively depleted in certain regions of any given biofilm to some degree, with a consequent effect on metabolic activity. This phenomenon has been well documented for bacterial biofilms (24, 37, 77). The level of metabolic activity is known to influence susceptibility of microbes to antimicrobials (30) as well as other stresses (31), and in situ characterization of bacterial biofilms has confirmed that regions with reduced access to nutrients correspond to regions exhibiting tolerance to an antimicrobial (2).
Our results have broad implications if similar subpopulations of highly resistant blastospores are present in other types of C. albicans biofilms, or in infective C. albicans communities that have salient features in common with C. albicans biofilms. There is sufficient suggestive evidence that our biofilm has some universally relevant characteristics to justify entertaining this hypothesis. The biofilms that formed on the TF had an architecture that resembled that reported by others, including, in particular, the presence of a basal layer of blastospores (9, 15, 42). We showed previously that, in early-stage biofilms, attached blastospores were less susceptible to membrane disruption by the antiseptic chlorhexidine than filamentous forms (70). The basal layer of blastospores was located furthest away from the medium source and thus may have experienced nutrient limitations similar to that of stationary-phase cultures. Similar nutrient-deprived microenvironments are likely to exist in many C. albicans biofilms, as well as in infective C. albicans communities, and it has been demonstrated that resistance to membrane disruption by AmB increases dramatically when yeast forms transition into stationary phase (26, 28).
The transcript analysis provides a starting point for investigating the molecular basis of the resistance exhibited by the basal layer of blastospores. The data indicated possible association of genes involved with both the ergosterol and β-1,6-glucan synthetic pathways (Table 1). The results indicated that the basal blastospore subpopulation was a distinct population at this level of molecular characterization.
Transcript abundance of the ERG1 gene was significantly reduced in the resistant blastospore subpopulation (Table 2). The ergosterol synthetic pathway has been extensively characterized in Saccharomyces cerevisiae and, with the exception of one gene, the genes corresponding to an analogous pathway in C. albicans are probably all orthologs (1). Reduction of the ergosterol content of the plasma membrane is the most common mechanism for C. albicans to acquire resistance to AmB (29). The ERG1 gene product catalyzes epoxidation of squalene to 2,3-oxidosqualene, a step that occurs early in the ergosterol synthetic pathway, just after commitment to sterol synthesis, and thus could serve as a point of down-regulation of ergosterol synthesis (21). In a C. albicans strain in which both copies of ERG1 were disrupted, ergosterol was absent from the membrane and was replaced primarily by squalene (56). It is possible that the basal blastospore subpopulation up-regulated the ERG25 gene in order to shuttle nonergosterol intermediates into the membrane (10).
Transcripts encoded by two genes involved in β-1,6-glucan synthesis (SKN1 and KRE1) were clearly more abundant in the basal blastospore subpopulation than in either the shear-removed biofilm or planktonic populations (Table 3). Along with mannoproteins and chitin, β-1,3-glucans and β-1,6-glucans form the polymeric network that comprises the C. albicans cell wall (40, 66). In C. albicans, β-1,6-glucans constitute 20% (dry weight) of the cell wall (40). In general, the cell wall plays a protective role. Restructuring of the glucan portion of the cell wall has been implicated in conferring transient AmB resistance to C. albicans (26). Although the SKN1 gene has been implicated in β-1,6-glucan synthesis in S. cerevisiae, it has not been shown to be essential for β-1,6-glucan synthesis in C. albicans (49). The role of the KRE1 gene in β-1,6-glucan synthesis was unambiguously demonstrated in S. cerevisiae (13), and closely corresponding structural similarities, including the presence of an amino-terminal signal sequence (12), indicate that the corresponding gene in C. albicans is an ortholog. A more recent study suggests that the KRE1 gene product plays a role in both cell wall assembly and architecture in S. cerevisiae (14). Thus, the up-regulation of KRE1 in the basal blastospores suggests either an increase in or restructuring of the β-1,6-glucan portion of the cell wall. In addition, the trend of increasing KRE1 transcript abundance for populations exhibiting increasing levels of AmB resistance (Table 3; Fig. 4) supports the hypothesis that differential regulation of the KRE1 gene may confer resistance.
The transcript analysis of both β-1,6-glucan and ergosterol pathways is consistent with a regulatory mechanism that reduced the susceptibility of the subpopulation of basal blastospores to AmB by a synergistic effect of increasing and/or restructuring the glucan portion of the cell wall and reducing the ergosterol content of the plasma membrane. If this is the case, then in the context of C. albicans biology, the basal blastospore subpopulation was exploiting fairly generic molecular mechanisms to minimize their susceptibility to AmB which were transiently activated by the biofilm environment. There is a precedent for involvement of the glucan portion of the cell wall in conferring the biofilm resistance phenotype, since echinocandins, which interfere with β-1,3-glucan synthesis, are singularly effective against C. albicans biofilms (6, 43). Although strains with mutation-based alterations resulting in a resistant phenotype associated with reduced ergosterol content can be induced to form in the laboratory (10), they are rarely isolated from clinical samples (1, 29, 67, 75). However, this does not preclude the in vivo existence of a similar transient, reversible phenotype in some infective C. albicans populations.
A number of reviews outline the various theories proposed to explain resistance of biofilms (18, 22, 25, 68). Hindered transport of an antimicrobial by interaction with biofilm components (cells and the exopolysaccharide matrix) was one of the first theories posed (4, 53, 54). Since the basal layer of blastospores was exposed to AmB after the removal of the bulk of the biofilm, lack of delivery of AmB to the cells is an unlikely explanation for their enhanced resistance. Although our flow cell design precluded any direct measurements of the microenvironment occupied by the basal layer of blastospores that might have lead to increased resistance, the high oxygen permeability of the silicone tubing should have maintained aerobic conditions at base of the biofilm. It seems likely that the basal blastospores experienced a reduced level of medium nutrients. However, the superior resistance of the basal blastospore subpopulation compared to the stationary-phase cultures, as well as the difference in transcript abundance for genes involved in the ergosterol pathway (ERG1, ERG3, and ERG25) and β-1,6-glucan pathway (SKN1, KRE5, and KRE6) between these two populations (Table 2 and Table 3), suggests that there were additional factors, besides nutrient deprivation, that led to the enhanced resistance of this biofilm subpopulation.
One possibility is that these highly resistant blastospores regulated their genes in direct response to their relatively intimate association with the surface. A similar mechanism was proposed to explain AmB resistance of C. albicans biofilms grown on PVC catheters (8). Bacterial biofilms have been shown to alter genetic programs in response to surface association (23, 39, 50, 59), and even early-stage (monolayer) biofilms can become more resistant than their planktonic counterparts (17, 20). It is not unreasonable to expect that the resistant blastospores used some of the same mechanisms previously identified in C. albicans for sensing surface contact (74).
In summary, we have discovered that C. albicans biofilms cultured under our experimental conditions harbored a subpopulation of basal blastospores that were exceptionally resistant to AmB. We have presented arguments that subpopulations with a similar phenotype may exist in C. albicans biofilms cultured under different conditions, and perhaps even in infective C. albicans communities that share salient features with C. albicans biofilms. The molecular mechanisms underlying the resistance of the basal blastospore subpopulation were probed by performing a comparative analysis of transcript abundance for key genes in the ergosterol and β-1,6-glucan pathways. These results show that at this level of genetic regulation the basal blastospore subpopulation was distinct both from the bulk of the biofilm and from exponential- and stationary-phase planktonic populations and suggest possible involvement of these pathways in conferring the resistant phenotype.
Acknowledgments
This study was supported by a grant from the National Institute of Dental and Craniofacial Research (DE13231-02) (B. J. Tyler), the National Institute of General Medical Sciences (1 R21 GM 079554-01A1) (P. A. Suci), and The University of Utah, Salt Lake City.
Footnotes
Published ahead of print on 11 September 2006.
REFERENCES
- 1.Akins, R. A. 2005. An update on antifungal targets and mechanisms of resistance in Candida albicans. Med. Mycol. 43:285-318. [DOI] [PubMed] [Google Scholar]
- 2.Anderl, J. N., J. Zahller, F. Roe, and P. S. Stewart. 2003. Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 47:1251-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andes, D., J. Nett, P. Oschel, R. Albrecht, K. Marchillo, and A. Pitula. 2004. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect. Immun. 72:6023-6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anwar, H., M. Dasgupta, K. Lam, and J. W. Costerton. 1989. Tobramycin resistance of mucoid Pseudomonas aeruginosa biofilm grown under iron limitation. J. Antimicrob. Chemotherapy 24:647-655. [DOI] [PubMed] [Google Scholar]
- 5.Arnaud, M. B., M. C. Costanzo, M. S. Skrzypek, G. Binkley, C. Lane, S. R. Miyasato, and G. Sherlock. 2005. The Candida Genome Database (CGD), a community resource for Candida albicans gene and protein information. Nucleic Acids Res. 33:D358-D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachmann, S. P., K. VandeWalle, G. Ramage, T. F. Patterson, B. L. Wickes, J. R. Graybill, and J. L. Lopez-Ribot. 2002. In vitro activity of caspofungin against Candida albicans biofilms. Antimicrob. Agents Chemother. 46:3591-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baillie, G. S., and L. J. Douglas. 1998. Effect of growth rate on resistance of Candida albicans biofilms to antifungal agents. Antimicrob. Agents Chemother. 42:1900-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baillie, G. S., and L. J. Douglas. 2000. Matrix polymers of Candida biofilms and their possible role in biofilm resistance to antifungal agents. J. Antimicrob. Chemotherapy 46:397-403. [DOI] [PubMed] [Google Scholar]
- 9.Baillie, G. S., and L. J. Douglas. 1999. Role of dimorphism in the development of Candida albicans biofilms. J. Med. Microbiol. 48:671-679. [DOI] [PubMed] [Google Scholar]
- 10.Barker, K. S., S. Crisp, N. Wiederhold, R. E. Lewis, B. Bareither, J. Eckstein, R. Barbuch, M. Bard, and P. D. Rogers. 2004. Genome-wide expression profiling reveals genes associated with amphotericin B and fluconazole resistance in experimentally induced antifungal resistant isolates of Candida albicans. J. Antimicrob. Chemother. 54:376-385. [DOI] [PubMed] [Google Scholar]
- 11.Beggs, K. T., A. R. Holmes, R. D. Cannon, and A. M. Rich. 2004. Detection of Candida albicans mRNA in archival histopathology samples by reverse transcription-PCR. J. Clin. Microbiol. 42:2275-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boone, C., A. M. Sdicu, M. Laroche, and H. Bussey. 1991. Isolation from Candida albicans of a functional homolog of the Saccharomyces cerevisiae kre1 gene, which is involved in cell wall beta-glucan synthesis. J. Bacteriol. 173:6859-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boone, C., S. S. Sommer, A. Hensel, and H. Bussey. 1990. Yeast Kre genes provide evidence for a pathway of cell-wall beta-glucan assembly. J. Cell Biol. 110:1833-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breinig, F., K. Schleinkofer, and M. J. Schmitt. 2004. Yeast Kre1p is GPI-anchored and involved in both cell wall assembly and architecture. Microbiology SGM 150:3209-3218. [DOI] [PubMed] [Google Scholar]
- 15.Chandra, J., D. M. Kuhn, P. K. Mukherjee, L. L. Hoyer, T. McCormick, and M. A. Ghannoum. 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J. Bacteriol. 183:5385-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandra, J., P. K. Mukherjee, S. D. Leidich, F. F. Faddoul, L. L. Hoyer, L. J. Douglas, and M. A. Ghannoum. 2001. Antifungal resistance of candidal biofilms formed on denture acrylic in vitro. J. Dent. Res. 80:903-908. [DOI] [PubMed] [Google Scholar]
- 17.Cochran, W. L., G. A. McFeters, and P. S. Stewart. 2000. Reduced susceptibility of thin Pseudomonas aeruginosa biofilms to hydrogen peroxide and monochloramine. J. Appl. Microbiol. 88:22-30. [DOI] [PubMed] [Google Scholar]
- 18.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 19.Crump, J. A., and P. J. Collignon. 2000. Intravascular catheter-associated infections. Eur. J. Clin. Microbiol. Infect. Dis. 19:1-8. [DOI] [PubMed] [Google Scholar]
- 20.Das, J. R., M. Bhakoo, M. V. Jones, and P. Gilbert. 1998. Changes in the biocide susceptibility of Staphylococcus epidermidis and Escherichia coli cells associated with rapid attachment to plastic surfaces. J. Appl. Microbiol. 84:852-858. [DOI] [PubMed] [Google Scholar]
- 21.Daum, G., N. D. Lees, M. Bard, and R. Dickson. 1998. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast 14:1471-1510. [DOI] [PubMed] [Google Scholar]
- 22.Davies, D. 2003. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discovery 2:114-122. [DOI] [PubMed] [Google Scholar]
- 23.Davies, D. G., and G. G. Geesey. 1995. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl. Environ. Microbiol. 61:860-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Debeer, D., P. Stoodley, F. Roe, and Z. Lewandowski. 1994. Effects of biofilm structures on oxygen distribution and mass transport. Biotechnol. Bioeng. 43:1131-1138. [DOI] [PubMed] [Google Scholar]
- 25.Douglas, L. J. 2003. Candida biofilms and their role in infection. Trends Microbiol. 11:30-36. [DOI] [PubMed] [Google Scholar]
- 26.Gale, E. F. 1986. Nature and development of phenotypic resistance to amphotericin B in Candida albicans. Adv. Microb. Physiol. 27:277-320. [DOI] [PubMed] [Google Scholar]
- 27.Gale, E. F. 1974. The release of potassium ions from Candida albicans in the presence of polyene antibiotics. J. Gen. Microbiol. 80:451-465. [DOI] [PubMed] [Google Scholar]
- 28.Gale, E. F., E. A. Miles, D. Kerridge, and A. M. Johnson. 1978. Phenotypic resistance to amphotericin B in Candida albicans: role of reduction. J. Gen. Microbiol. 109:191-204. [DOI] [PubMed] [Google Scholar]
- 29.Ghannoum, M. A., and L. B. Rice. 1999. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 12:501-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilbert, P., P. J. Collier, and M. R. W. Brown. 1990. Influence of growth rate on susceptibility to antimicrobial agents: biofilms, cell cycle, dormancy, and stringent response. Antimicrob. Agents Chemother. 34:1865-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Givskov, M., L. Eberl, and S. Molin. 1994. Responses to nutrient starvation in Pseudomonas putida Kt2442: two-dimensional electrophoretic analysis of starvation-induced and stress-induced proteins. J. Bacteriol. 176:4816-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomez-Suarez, C., H. J. Busscher, and H. C. van der Mei. 2001. Analysis of bacterial detachment from substratum surfaces by the passage of air-liquid interfaces. Appl. Environ Microbiol. 67:2531-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Green, C. B., G. Cheng, J. Chandra, P. Mukherjee, M. A. Ghannoum, and L. L. Hoyer. 2004. RT-PCR detection of Candida albicans ALS gene expression in the reconstituted human epithelium (RHE) model of oral candidiasis and in model biofilms. Microbiology SGM 150:267-275. [DOI] [PubMed] [Google Scholar]
- 34.Gudlaugsson, O., S. Gillespie, K. Lee, J. V. Berg, J. F. Hu, S. Messer, L. Herwaldt, M. Pfaller, and D. Diekema. 2003. Attributable mortality of nosocomial candidemia, revisited. Clin. Infect. Dis. 37:1172-1177. [DOI] [PubMed] [Google Scholar]
- 35.Han, Y. M., R. P. Morrison, and J. E. Cutler. 1998. A vaccine and monoclonal antibodies that enhance mouse resistance to Candida albicans vaginal infection. Infect. Immun. 66:5771-5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hawser, S. P., and L. J. Douglas. 1995. Resistance of Candida albicans biofilms to antifungal agents in vitro. Antimicrob. Agents Chemother. 39:2128-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang, C. T., K. D. Xu, G. A. McFeters, and P. S. Stewart. 1998. Spatial patterns of alkaline phosphatase expression within bacterial colonies and biofilms in response to phosphate starvation. Appl. Environ Microbiol. 64:1526-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kerridge, D., T. Y. Koh, and A. M. Johnson. 1976. The interaction of amphotericin B methyl ester with protoplasts of Candida albicans. J. Gen. Microbiol. 96:117-123. [DOI] [PubMed] [Google Scholar]
- 39.Kilic, A. O., L. Tao, Y. S. Zhang, Y. Lei, A. Khammanivong, and M. C. Herzberg. 2004. Involvement of Streptococcus gordonii beta-glucoside metabolism systems in adhesion, biofilm formation, and in vivo gene expression. J. Bacteriol. 186:4246-4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klis, F. M., P. De Groot, and K. Hellingwerf. 2001. Molecular organization of the cell wall of Candida albicans. Med. Mycol. 39:1-8. [PubMed] [Google Scholar]
- 41.Kojic, E. M., and R. O. Darouiche. 2004. Candida infections of medical devices. Clin. Microbiol. Rev. 17:255-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuhn, D. M., J. Chandra, P. K. Mukherjee, and M. A. Ghannoum. 2002. A comparison of biofilms formed by Candida albicans and Candida parapsilosis on bioprosthetic surfaces. Infect. Immun. 70:878-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuhn, D. M., T. George, J. Chandra, P. K. Mukherjee, and M. A. Ghannoum. 2002. Antifungal susceptibility of Candida biofilms: unique efficacy of amphotericin B lipid formulations and echinocandins. Antimicrob. Agents Chemother. 46:1773-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamfon, H., S. R. Porter, M. McCullough, and J. Pratten. 2004. Susceptibility of Candida albicans biofilms grown in a constant depth film fermentor to chlorhexidine, fluconazole and miconazole: a longitudinal study. J. Antimicrob. Chemother. 53:383-385. [DOI] [PubMed] [Google Scholar]
- 45.Lewis, K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis, R. E., D. P. Kontoyiannis, R. O. Darouiche, Raad, I. I., and R. A. Prince. 2002. Antifungal activity of amphotericin B, fluconazole, and voriconazole in an in vitro model of Candida catheter-related bloodstream infection. Antimicrob. Agents Chemother. 46:3499-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McBride, J., P. R. Ingram, F. L. Henriquez, and C. W. Roberts. 2005. Development of colorimetric microtiter plate assay for assessment of antimicrobials against Acanthamoeba. J. Clin. Microbiol. 43:629-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McDonnell, G., and A. D. Russell. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12:147-179. (Erratum, 14:227-228, 2001.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mio, T., T. Yamada-Okabe, T. Yabe, T. Nakajima, M. Arisawa, and H. Yamada-Okabe. 1997. Isolation of the Candida albicans homologs of Saccharomyces cerevisiae KRE6 and SKN1: expression and physiological function. J. Bacteriol. 179:2363-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moorthy, S., and P. I. Watnick. 2004. Genetic evidence that the Vibrio cholerae monolayer is a distinct stage in biofilm development. Mol. Microbiol. 52:573-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muller, P. Y., H. Janovjak, A. R. Miserez, and Z. Dobbie. 2002. Processing of gene expression data generated by quantitative real-time RT-PCR. BioTechniques 32:1372-1379. [PubMed] [Google Scholar]
- 52.NCCLS. 1997. Reference method for broth dilution antifungal susceptibility testing of yeasts. NCCLS document M27-A. NCCLS, Wayne, Pa.
- 53.Nichols, W. W., S. M. Dorrington, M. P. E. Slack, and H. L. Walmsley. 1988. Inhibition of tobramycin diffusion by binding to alginate. Antimicrob. Agents Chemother. 32:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nichols, W. W., M. J. Evans, M. P. E. Slack, and H. L. Walmsley. 1989. The penetration of antibiotics into aggregates of mucoid and non-mucoid Pseudomonas aeruginosa. J. Gen. Microbiol. 135:1291-1303. [DOI] [PubMed] [Google Scholar]
- 55.Odds, F. C. 1988. Candida and candidosis. Baillière Tindall, London, England.
- 56.Pasrija, R., S. Krishnamurthy, T. Prasad, J. F. Ernst, and R. Prasad. 2005. Squalene epoxidase encoded by ERG1 affects morphogenesis and drug susceptibilities of Candida albicans. J. Antimicrob. Chemother. 55:905-913. [DOI] [PubMed] [Google Scholar]
- 57.Pfaller, M. A., and A. L. Barry. 1994. Evaluation of a novel colorimetric broth microdilution method for antifungal susceptibility testing of yeast isolates. J. Clin. Microbiol. 32:1992-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pfaller, M. A., and D. J. Diekema. 2002. Role of sentinel surveillance of candidemia: trends in species distribution and antifungal susceptibility. J. Clin. Microbiol. 40:3551-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prigent-Combaret, C., O. Vidal, C. Dorel, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181:5993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramage, G., S. P. Saville, D. P. Thomas, and J. L. Lopez-Ribot. 2005. Candida biofilms: an update. Eukaryot. Cell 4:633-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramage, G., K. vande Walle, B. L. Wickes, and J. L. Lopez-Ribot. 2001. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob. Agents Chemother. 45:2475-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramage, G., K. VandeWalle, S. P. Bachmann, B. L. Wickes, and J. L. Lopez-Ribot. 2002. In vitro pharmacodynamic properties of three antifungal agents against preformed Candida albicans biofilms determined by time-kill studies. Antimicrob. Agents Chemother. 46:3634-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramage, G., K. Vandewalle, B. L. Wickes, and J. L. Lopez-Ribot. 2001. Characteristics of biofilm formation by Candida albicans. Rev. Iberoam. Micol. 18:163-170. [PubMed] [Google Scholar]
- 64.Rex, J. H., M. A. Pfaller, T. J. Walsh, V. Chaturvedi, A. Espinel-Ingroff, M. A. Ghannoum, L. L. Gosey, F. C. Odds, M. G. Rinaldi, D. J. Sheehan, and D. W. Warnock. 2001. Antifungal susceptibility testing: practical aspects and current challenges. Clin. Microbiol. Rev. 14:643-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 66.Ruiz-Herrera, J., M. V. Elorza, E. Valentin, and R. Sentandreu. 2006. Molecular organization of the cell wall of Candida albicans and its relation to pathogenicity. FEMS Yeast Res. 6:14-29. [DOI] [PubMed] [Google Scholar]
- 67.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73-85. [DOI] [PubMed] [Google Scholar]
- 68.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 69.Suci, P. A., G. G. Geesey, and B. J. Tyler. 2001. Integration of Raman microscopy, differential interference contrast microscopy, and attenuated total reflection Fourier transform infrared spectroscopy to investigate chlorhexidine spatial and temporal distribution in Candida albicans biofilms. J. Microbiol. Methods 46:193-208. [DOI] [PubMed] [Google Scholar]
- 70.Suci, P. A., and B. J. Tyler. 2002. Action of chlorhexidine digluconate against yeast and filamentous forms in an early-stage Candida albicans biofilm. Antimicrob. Agents Chemother. 46:3522-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suci, P. A., and B. J. Tyler. 2003. A method for discrimination of subpopulations of Candida albicans biofilm cells that exhibit relative levels of phenotypic resistance to chlorhexidine. J. Microbiol. Methods 53:313-325. [DOI] [PubMed] [Google Scholar]
- 72.Tiballi, R. N., X. He, L. T. Zarins, S. G. Revankar, and C. A. Kauffman. 1995. Use of a colorimetric system for yeast susceptibility testing. J. Clin. Microbiol. 33:915-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Watts, H. J., A. A. Very, T. H. S. Perera, J. M. Davies, and N. A. R. Gow. 1998. Thigmotropism and stretch-activated channels in the pathogenic fungus Candida albicans. Microbiology UK 144:689-695. [DOI] [PubMed] [Google Scholar]
- 75.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilson, S., M. A. Hamilton, G. C. Hamilton, M. R. Schumann, and P. Stoodley. 2004. Statistical quantification of detachment rates and size distributions of cell clumps from wild-type (PAO1) and cell signaling mutant (JP1) Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 70:5847-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu, K. D., P. S. Stewart, F. Xia, C. T. Huang, and G. A. McFeters. 1998. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 64:4035-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]