Abstract
The molecular mechanisms of reduced susceptibility to cefixime in clinical isolates of Neisseria gonorrhoeae, particularly amino acid substitutions in mosaic penicillin-binding protein 2 (PBP2), were examined. The complete sequence of ponA, penA, and por genes, encoding, respectively, PBP1, PBP2, and porin, were determined for 58 strains isolated in 2002 from Japan. Replacement of leucine 421 by proline in PBP1 and the mosaic-like structure of PBP2 were detected in 48 strains (82.8%) and 28 strains (48.3%), respectively. The presence of mosaic PBP2 was the main cause of the elevated cefixime MIC (4- to 64-fold). In order to identify the mutations responsible for the reduced susceptibility to cefixime in isolates with mosaic PBP2, penA genes with various mutations were transferred to a susceptible strain by genetic transformation. The susceptibility of partial recombinants and site-directed mutants revealed that the replacement of glycine 545 by serine (G545S) was the primary mutation, which led to a two- to fourfold increase in resistance to cephems. Replacement of isoleucine 312 by methionine (I312M) and valine 316 by threonine (V316T), in the presence of the G545S mutation, reduced susceptibility to cefixime, ceftibuten, and cefpodoxime by an additional fourfold. Therefore, three mutations (G545S, I312M, and V316T) in mosaic PBP2 were identified as the amino acid substitutions responsible for reduced susceptibility to cefixime in N. gonorrhoeae.
The increase in the number of Neisseria gonorrhoeae isolates resistant to multiple antimicrobial agents is now a serious problem in Japan. Fluoroquinolones are no longer recommended for the treatment of gonococcal infections in Japan because of a dramatic increase in the incidence of drug-resistant strains (8, 16). Recently, the emergence and spread of strains with reduced susceptibility to oral cephems have been reported (1, 9). Furthermore, clinical failures after treatment of patients with cefixime, which should be a highly effective agent for gonococcal infections, have also been observed (5). Therefore, the treatment guideline in Japan recommends parenteral antibacterials, such as ceftriaxone and spectinomycin, as the first-line treatment for uncomplicated gonococcal infections. Isolates with decreased susceptibility to cefixime have also been reported outside Japan (18).
The mechanisms of chromosomally mediated resistance to β-lactams in N. gonorrhoeae have been studied. One such mechanism involves the mutation of penicillin-binding proteins, PBP1 and PBP2, which decrease the affinity to β-lactams. The replacement of leucine 421 by proline in PBP1, encoded by the ponA gene, is implicated in high-level resistance to penicillin (13). PBP2, encoded by the penA gene, has a greater affinity for penicillin than PBP1. Insertion of an additional amino acid at position 345 in PBP2 is associated with penicillin resistance (4). Recent studies have indicated that in Japan N. gonorrhoeae strains with reduced susceptibility to oral cephems have a mosaic-like structure in the penA gene (2, 7). An alternative mechanism of β-lactam resistance involves mutation of the por gene, which encodes a porin, leading to reduced outer membrane permeability (11). Furthermore, mutation of the mtrR gene can cause overexpression of the MtrCDE efflux pump (17). Overexpression of the pump is required for the porin mutants harboring amino acid substitutions at positions 120 and 121 to confer increased resistance to penicillin (12).
The mosaic-like structure of the penA gene in N. gonorrhoeae has evolved by intragenic recombination with penA genes of commensal Neisseria species (15). Mosaic PBP2 has approximately 60 amino acid alterations from the PBP2 of susceptible strains. Although mosaic PBP2 is known to be associated with reduced susceptibility to cefixime and other cephems, the amino acid substitutions responsible for the development of resistance have not been defined. Moreover, the contribution of other mechanisms to resistance against cephems is unclear. The aim of this study was to identify the mutations essential for the increased MICs of cefixime for clinical isolates of N. gonorrhoeae and, particularly, the mutations in mosaic PBP2.
MATERIALS AND METHODS
Bacterial strains.
Antimicrobial susceptibility testing and nucleotide sequencing of ponA, penA, and por genes were performed on 58 clinical isolates of N. gonorrhoeae collected in Japan during 2002. Isolates were collected at Kotobiken Medical Laboratories Inc. (Tokyo, Japan), from patients with urethritis attending hospitals located in and around Tokyo. β-Lactamase production was negative for all strains. Strains FA1090 (ATCC 700825) and ATCC 49226 were obtained from the American Type Culture Collection. Strain FA1090 was used as the recipient strain for genetic transformation, and strain ATCC 49226 was used as a quality control for susceptibility testing.
Antimicrobial susceptibility testing and antibacterial agents.
MICs were determined by the agar dilution method according to the Clinical and Laboratory Standards Institute (formerly the National Committee for Clinical Laboratory Standards) (10). The following antibiotics were used in this study: penicillin and cephalexin (Sigma-Aldrich Co., St. Louis, MO), cefotiam (Takeda Pharmaceutical Company Limited, Osaka, Japan), ceftriaxone (Nippon Roche Co., Ltd., Tokyo, Japan),cefixime and cefdinir (Astellas Pharma Inc., Tokyo, Japan), ceftibuten (Shionogi and Co., Ltd., Osaka, Japan), and cefditoren (Meiji Seika Kaisha, Ltd., Tokyo, Japan). Cefpodoxime was prepared from the commercial prodrug (Sankyo Co., Ltd., Tokyo, Japan) at the Pharmaceutical Research Center of Meiji Seika Kaisha, Ltd. (Yokohama, Japan). Cefcapene and levofloxacin were synthesized at the Pharmaceutical Research Center of Meiji Seika Kaisha, Ltd.
Nucleotide sequencing of ponA, penA, and por genes.
Full-length ponA, penA, and por genes were amplified from genomic DNA by PCR using Ex Taq polymerase (Takara Bio Inc., Otsu, Japan) with the primers listed in Table 1. In cases where the por gene could not be amplified with the standard primer set, an alternative forward primer was used (shown in parentheses in Table 1). PCR amplifications for the ponA and penA genes were performed as follows: 2 min denaturation at 94°C and 30 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 30 s, and extension at 72°C for 3 min. Amplification of the por gene was performed as follows: 30 s denaturation at 98°C and 30 cycles of denaturation at 98°C for 10 s, followed by annealing and extension at 68°C for 2 min. The cycling reaction was performed with a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA). Sequencing was carried out with a BigDye Terminator v1.1 cycle sequencing kit and 3730 DNA analyzer (Applied Biosystems).
TABLE 1.
Primers used for PCR amplification of the ponA, penA, and por genes and for site-directed mutagenesis
| Use | Forward primer | Reverse primer |
|---|---|---|
| Amplification of complete ponA gene | 5′-ACAAGCGCATGATGAAAGTTC-3′ | 5′-GACAGGCAAAACATCAGCG-3′ |
| Amplification of complete penA gene | 5′-TTTGGCGAATCACGAAGCG-3′ | 5′-CCGGACAGGCAACGAAAATA-3′ |
| Amplification of complete por gene | 5′-GCACATCGGATTCCACACAA-3′ (5′-CTCGGCGGTAAATGCAAAGC-3′) | 5′-TATGGATAGATTCGTCATTCCCGC-3′ |
| Cloning of penA gene | 5′-CGGGATCCAATGTTGATTAAAAGCGAATATAAGC-3′ | 5′-CGGAATTCTTAAGACGGTGTTTTGACGGC-3′ |
| Introduction of F504L in penA gene | 5′-CACGGCGCGCAAGTTGGTCAACGGGCGTTATG-3′ | 5′-CATAACGCCCGTTGACCAACTTGCGCGCCGTG-3′ |
| Introduction of A510V in penA gene | 5′-GTCAACGGGCGTTATGTCGACAACAAACACGTC-3′ | 5′-GACGTGTTTGTTGTCGACATAACGCCCGTTGAC-3′ |
| Introduction of N512Y in penA gene | 5′-GGGCGTTATGCCGACTACAAACACGTCGCTAC-3′ | 5′-GTAGCGACGTGTTTGTAGTCGGCATAACGCCC-3′ |
| Introduction of H541N in penA gene | 5′-GACGAACCGACTGCCAACGGCTATTACGGCG-3′ | 5′-CGCCGTAATAGCCGTTGGCAGTCGGTTCGTC-3′ |
| Introduction of G545S in penA gene | 5′-GCCCACGGCTATTACAGCGGCGTAGTGGCAG-3′ | 5′-CTGCCACTACGCCGCTGTAATAGCCGTGGGC-3′ |
| Introduction of I312M in penA gene | 5′-CCTGGTTCGGCAATGAAACCGTTCGTGATTG-3′ | 5′-CAATCACGAACGGTTTCATTGCCGAACCAGG-3′ |
| Introduction of V316T in penA gene | 5′-GTTCGGCAATCAAACCGTTCACGATTGCGAAGGCATTGG-3′ | 5′-CCAATGCCTTCGCAATCGTGAACGGTTTGATTGCCGAAC-3′ |
Cloning of the penA gene and site-directed mutagenesis.
The penA DNA fragment from the FA1090 strain was amplified by PCR and then cloned into the pCR2.1-TOPO vector (Invitrogen Corp., Carlsbad, CA). Site-directed mutagenesis was carried out with a QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) and the primer sets shown in Table 1.
Genetic transformation.
The penA gene amplified from a strain with reduced susceptibility to cefixime (MSC02236) and recombinant penA genes obtained by site-directed mutagenesis were transformed into a susceptible strain of N. gonorrhoeae (FA1090) as described previously (13). Briefly, cells grown on chocolate II agar (Becton Dickinson, Sparks, MD) were suspended in prewarmed GC broth containing 10 mM MgCl2, 10 mM NaHCO3, and 1% IsoVitaleX (Becton Dickinson) at a cell density of 108 CFU/ml. Following the purification using a MinElute PCR Purification Kit (QIAGEN), donor DNA was added at a final concentration of 1 to 2 μg/ml, and the cells were incubated at 37°C for 5 h in a humidified 5% CO2 atmosphere. Recombinants were selected on GC agar base containing 1% IsoVitaleX and an appropriate concentration of antibiotics (6, 8, or 16 μg/ml cephalexin and 0.09 μg/ml ceftibuten). Nucleotide sequencing of the penA genes from recombinants was performed as indicated above.
RESULTS
Mutations of PBP1, PBP2, and porin in the clinical isolates.
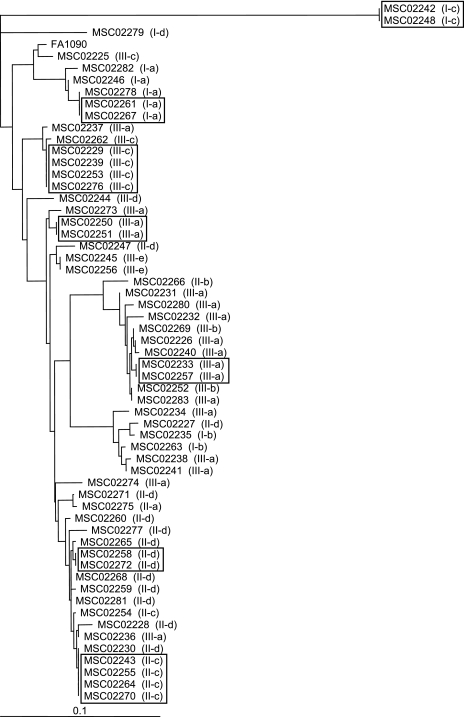
The complete sequences of the ponA, penA, and por genes were determined for the 58 clinical isolates of N. gonorrhoeae. Potentially important amino acid substitutions were identified by comparing the obtained sequences with those from a cefixime-susceptible strain of N. gonorrhoeae (FA1090) (Table 2). Only four alterations were identified in PBP1. A total of 48 out of 58 strains (82.8%) had the point mutation L421P in PBP1. All of the alterations after position 501 in PBP2 are shown in Table 2. Most strains carried some mutations in PBP2, particularly after position 504. Twenty-eight out of 58 strains (48.3%) harbored a mosaic-like structure in the penA gene. Interestingly, every strain with mosaic PBP2 had the L421P mutation in PBP1. Fifty-eight strains were classified into three groups according to the mutations in PBP1 and PBP2. Strains classified into group I (n = 10; 17.2%) had no alteration at amino acid 421 in PBP1 nor a mosaic mutation in PBP2. Isolates in group II (n = 20; 34.5%) possessed the L421P mutation in PBP1, i.e., PBP1(L421P), but no mosaic PBP2. Isolates in group III (n = 28; 48.3%) had both PBP1(L421P) and mosaic PBP2. The nucleotide sequences of the por genes were diverse among the 58 strains under investigation (42 distinct amino acid sequences). The major porin type was PIB and most strains possessed G120K and A121D mutations in putative loop 3. Figure 1 shows the high genetic diversity of the strains based on the sequences of porin, PBP1, and PBP2. Clinical isolates consisted of 47 individual strains typed from the sequences of porin, PBP1, PBP2 and the susceptibility to antibiotics (Table 2).
TABLE 2.
Susceptibility to cefixime and levofloxacin, and amino acid substitutions identified in PBP1, PBP2, and porin of the N. gonorrhoeae strains used in this study
| Group | Subgroupa | Strain no. | MIC (μg/ml)
|
Amino acid substitutions in:b
|
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PBP1c
|
PBP2d
|
Porine
|
|||||||||||||||||||||||||
| Cefixime | Levofloxacin | 375A | 421L | 537Y | 696P | 1 to 500 | 501A | 504F | 510A | 512N | 516A | 541H | 542G | 545G | 549A | 551P | 552P | 555K | 556I | 566I | 574A | Type | 120G | 121A | |||
| I (n = 10) | a | MSC02246 | 0.008 | 0.008 | . | . | . | . | None | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | PIB | . | . |
| a | MSC02282 | 0.008 | 0.03 | . | . | . | . | None | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | PIB | . | . | |
| a | MSC02261, MSC02267 | 0.016 | 0.008 | . | . | . | . | None | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | PIB | . | . | |
| a | MSC02278 | 0.016 | 0.03 | . | . | . | . | None | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | PIB | . | . | |
| b | MSC02235 | 0.008 | 0.008 | T | . | C | . | None | . | L | V | . | G | . | . | . | . | . | . | . | . | . | . | PIB | . | . | |
| b | MSC02263 | 0.008 | 0.06 | T | . | C | . | None | . | L | V | . | G | . | . | . | . | . | . | . | . | . | . | PIB | . | . | |
| c | MSC02242, MSC02248 | 0.008 | 0.004-0.008 | . | . | . | . | None | . | L | V | . | G | . | . | . | . | . | . | . | . | . | NV | PIA | D | G | |
| d | MSC02279 | 0.03 | 0.25 | . | . | . | . | None | V | L | V | . | G | N | . | . | . | . | V | Q | V | V | NV | PIB | D | . | |
| II (n = 20) | a | MSC02275 | 0.03 | 1 | . | P | . | . | None | . | L | V | . | G | . | . | . | . | . | . | . | . | . | . | PIB | K | D |
| b | MSC02266 | 0.008 | 1 | . | P | . | . | None | . | L | V | . | G | . | S | . | . | . | . | . | . | . | . | PIB | K | N | |
| c | MSC02243, MSC02255, MSC02264, MSC02270 | 0.03 | 8 | . | P | . | . | None | . | L | V | . | G | . | . | . | . | S | . | . | . | . | . | PIB | K | D | |
| c | MSC02254 | 0.03 | 16 | . | P | . | . | None | . | L | V | . | G | . | . | . | . | S | . | . | . | . | . | PIB | K | D | |
| d | MSC02268 | 0.008 | 1 | . | P | . | . | None | . | L | V | . | G | . | S | . | . | . | . | . | . | V | NV | PIB | K | D | |
| d | MSC02228 | 0.016 | 4 | . | P | . | . | None | . | L | V | . | G | . | S | . | . | . | . | . | . | V | NV | PIB | K | D | |
| d | MSC02247 | 0.016 | 4 | . | P | . | . | None | . | L | V | . | G | . | S | . | . | . | . | . | . | V | NV | PIB | R | D | |
| d | MSC02259 | 0.016 | 8 | . | P | . | . | None | . | L | V | . | G | . | S | . | . | . | . | . | . | V | NV | PIB | K | D | |
| d | MSC02260 | 0.016 | 8 | . | P | . | . | None | . | L | V | . | G | . | S | . | . | . | . | . | . | V | NV | PIB | K | D | |
| d | MSC02277 | 0.016 | 4 | . | P | . | . | None | . | L | V | . | G | . | S | . | . | . | . | . | . | V | NV | PIB | K | D | |
| d | MSC02227 | 0.03 | 4 | . | P | . | . | None | . | L | V | . | G | . | S | . | . | . | . | . | . | V | NV | PIB | . | . | |
| d | MSC02230 | 0.03 | 4 | . | P | . | . | None | . | L | V | . | G | . | S | . | . | . | . | . | . | V | NV | PIB | K | D | |
| d | MSC02258, MSC02272 | 0.03 | 8 | . | P | . | . | None | . | L | V | . | G | . | S | . | . | . | . | . | . | V | NV | PIB | K | D | |
| d | MSC02265 | 0.03 | 4 | . | P | . | . | None | . | L | V | . | G | . | S | . | . | . | . | . | . | V | NV | PIB | K | D | |
| d | MSC02271 | 0.03 | 4 | . | P | . | . | None | . | L | V | . | G | . | S | . | . | . | . | . | . | V | NV | PIB | K | D | |
| d | MSC02281 | 0.03 | 8 | . | P | . | . | None | . | L | V | . | G | . | S | . | . | . | . | . | . | V | NV | PIB | K | D | |
| III (n = 28) | a | MSC02231 | 0.12 | 32 | . | P | . | . | Mosaic-1 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | K | D |
| a | MSC02238 | 0.12 | 2 | . | P | . | . | Mosaic-1 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | . | . | |
| a | MSC02240 | 0.12 | 4 | . | P | . | . | Mosaic-1 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | K | D | |
| a | MSC02226 | 0.25 | 8 | . | P | . | . | Mosaic-1 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | K | D | |
| a | MSC02234 | 0.25 | 2 | . | P | . | . | Mosaic-1 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | . | . | |
| a | MSC02237 | 0.25 | 4 | . | P | . | . | Mosaic-1 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | K | D | |
| a | MSC02241 | 0.25 | 2 | . | P | . | . | Mosaic-1 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | . | . | |
| a | MSC02250, MSC02251 | 0.25 | 16 | . | P | . | . | Mosaic-1 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | K | D | |
| a | MSC02273 | 0.25 | 16 | . | P | . | . | Mosaic-1 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | K | D | |
| a | MSC02274 | 0.25 | 8 | . | P | . | . | Mosaic-1 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | N | D | |
| a | MSC02280 | 0.25 | 16 | . | P | . | . | Mosaic-1 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | K | D | |
| a | MSC02283 | 0.25 | 4 | . | P | . | . | Mosaic-1 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | K | D | |
| a | MSC02232 | 0.5 | 16 | . | P | . | . | Mosaic-1 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | K | D | |
| a | MSC02233, MSC02257 | 0.5 | 16 | . | P | . | . | Mosaic-1 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | K | D | |
| a | MSC02236 | 0.5 | 16 | . | P | . | . | Mosaic-1 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | K | D | |
| b | MSC02252 | 0.25 | 16 | . | P | . | S | Mosaic-1 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | K | D | |
| b | MSC02269 | 0.25 | 16 | . | P | . | S | Mosaic-1 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | K | D | |
| c | MSC02225 | 0.25 | 4 | . | P | . | . | Mosaic-2 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | K | D | |
| c | MSC02229, MSC02239, MSC02253, MSC02276 | 0.25 | 1-2 | . | P | . | . | Mosaic-2 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | K | D | |
| c | MSC02262 | 0.25 | 2 | . | P | . | . | Mosaic-2 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | K | D | |
| d | MSC02244 | 0.25 | 4 | . | P | . | . | Mosaic-3 | . | L | V | Y | . | N | . | S | T | . | V | Q | V | V | NV | PIB | N | D | |
| e | MSC02256 | 0.12 | 2 | . | P | . | . | Mosaic-4 | V | L | V | . | . | N | . | . | T | . | V | Q | V | V | NV | PIB | K | D | |
| e | MSC02245 | 0.25 | 8 | . | P | . | . | Mosaic-4 | V | L | V | . | . | N | . | . | T | . | V | Q | V | V | NV | PIB | K | D | |
Groups were subdivided on the basis of the amino acid sequences of PBP1 and PBP2.
Dots, identical amino acids.
All of the mutations found in PBP1 are shown.
All the mutations after position 501 in PBP2 are shown. Every substitution is presented in Fig. 2 for mosaic PBP2.
Only the mutations at position 120 and 121 are shown for porin.
FIG. 1.
Genetic relationship among the 58 clinical isolates. The dendrogram was constructed from the amino acid sequence of porin using the neighbor-joining method. Classification of the strains based on the sequences of PBP1 and PBP2 are shown in parentheses. Strains that were indistinguishable based on the sequences of porin, PBP1, and PBP2 and also on their susceptibilities to antibiotics are boxed. Bar, 0.1 genetic distance.
β-Lactam susceptibilities and mutations in PBPs.
The MICs of various β-lactams against the 58 strains revealed that resistance was associated with mutations in PBP1 and PBP2 and not with that of the porin. The MIC50s and MIC90s for isolates categorized into three groups on the basis of mutations in PBPs (groups I, II, and III) to various β-lactams are shown in Table 3. While the susceptibility to penicillin was reduced for strains in group II and III, which all possess PBP1(L421P), the MICs of the oral cephems toward isolates in group III were significantly elevated in comparison to those isolates in group II. For example, the MICs of cefixime, ceftibuten, and cefpodoxime were 4- to 64-fold, 16- to 256-fold, and 1- to 256-fold greater for group III isolates compared to group II isolates. Importantly, all strains with reduced susceptibility to cefixime (MIC of ≥0.12 μg/ml) had a mosaic-like structure in PBP2 (Table 2). In contrast to cefixime (16-fold), ceftriaxone and cefditoren had only a four- and twofold increase in the MIC for strains with mosaic PBP2 (MIC90 ratio for group III to group II).
TABLE 3.
Association between mutations in PBPs and antimicrobial susceptibility
| Antibiotics and strain groups testeda | MIC (μg/ml)
|
||
|---|---|---|---|
| 50% | 90% | Range | |
| Penicillin | |||
| I | 0.12 | 0.25 | 0.12-0.25 |
| II | 1 | 2 | 0.25-2 |
| III | 2 | 4 | 0.25-4 |
| Ceftriaxone | |||
| I | 0.004 | 0.008 | 0.004-0.016 |
| II | 0.03 | 0.03 | 0.004-0.06 |
| III | 0.06 | 0.12 | 0.016-0.12 |
| Cefixime | |||
| I | 0.008 | 0.016 | 0.008-0.03 |
| II | 0.03 | 0.03 | 0.008-0.03 |
| III | 0.25 | 0.5 | 0.12-0.5 |
| Ceftibuten | |||
| I | 0.03 | 0.06 | 0.03-0.12 |
| II | 0.06 | 0.12 | 0.03-0.12 |
| III | 8 | 8 | 2-8 |
| Cefpodoxime | |||
| I | 0.03 | 0.06 | 0.016-0.12 |
| II | 0.12 | 0.25 | 0.016-0.5 |
| III | 2 | 4 | 0.5-4 |
| Cefdinir | |||
| I | 0.008 | 0.03 | 0.008-0.03 |
| II | 0.03 | 0.06 | 0.008-0.06 |
| III | 1 | 1 | 0.5-1 |
| Cefcapene | |||
| I | 0.016 | 0.06 | 0.008-0.06 |
| II | 0.12 | 0.25 | 0.008-0.25 |
| III | 1 | 2 | 0.12-4 |
| Cefditoren | |||
| I | 0.03 | 0.06 | 0.008-0.12 |
| II | 0.12 | 0.25 | 0.004-0.5 |
| III | 0.25 | 0.5 | 0.016-0.5 |
Strains were classified into three groups according to the mutations in PBP1 and PBP2 (Table 2). Group I, n = 10; group II, n = 20; group III, n = 28.
Susceptibility of transformants with full and partial recombination of mosaic penA gene.
The mosaic-like structure of PBP2 was classified into four groups (mosaic-1 to mosaic-4) based on their amino acid sequences (Fig. 2). Nineteen out of 28 strains (67.9%) possessed mosaic-1 sequence, followed by 6 strains with mosaic-2 (21.4%). Mosaic-3 and mosaic-4 sequences were detected in 1 and 2 isolates, respectively. In order to identify the mutations responsible for the reduced susceptibility to cefixime among mosaic PBP2 (mosaic-1), the penA gene from MSC02236 (cefixime MIC of 0.5 μg/ml) was transformed into FA1090 (cefixime MIC of 0.008 μg/ml). The full-length recombinant (TF1) had reduced susceptibility to all the cephems tested (Table 4), and the susceptibility to cefixime was reduced by 16-fold (MIC of 0.12 μg/ml). Transformants with partial recombination were also obtained, and their susceptibilities to β-lactam antibiotics were determined. The absence of a mutation downstream of position 545 did not necessarily affect the susceptibility to all the β-lactams (TF1 to TF4). As anticipated, the loss of mutations upstream of the transpeptidase domain had no effect on susceptibility to the antibiotics (TF5 and TF6). Comparison of the susceptibilities of TF7 and TF8 suggests that mutations between amino acids 312 and 322 (i.e., I312M and V316T) may increase resistance to cefixime, ceftibuten, and cefpodoxime by at least fourfold. A comparison of the various transformants with N-terminal reversion of mosaic penA (TF5 to TF11) found that mutations between 504 and 575 were important for the increase in cephem MICs. Moreover, a recombinant with mutations from amino acids 504 to 545 (TF12), including F504L, A510V, N512Y, H541N, and G545S, had a significantly reduced susceptibility to cephems, despite being the shortest recombinant to be isolated.
FIG. 2.
Amino acid sequences of mosaic PBP2 from clinical isolates. The amino acid sequences of the mosaic PBP2 (mosaic-1 to mosaic-4) are aligned with those of penicillin-susceptible strains, LM306 and FA1090. Dashes indicate amino acid residues identical to those of LM306 (GenBank accession no. M32091).
TABLE 4.
MICs of β-lactams for donor strain (MSC02236), recipient strain (FA1090), and various penA recombinants
| Strain | Substitutions in PBP2 (aa)a | MIC (μg/ml)
|
||||||
|---|---|---|---|---|---|---|---|---|
| Penicillin | Cephalexin | Cefotiam | Ceftriaxone | Cefixime | Ceftibuten | Cefpodoxime | ||
| MSC02236 | 101-575 | 4 | 512 | 16 | 0.12 | 0.5 | 8 | 8 |
| FA1090 | None | 0.12 | 4 | 0.06 | 0.004 | 0.008 | 0.03 | 0.016 |
| TF1 | 101-575 | 0.25 | 128 | 1 | 0.016 | 0.12 | 4 | 0.5 |
| TF2 | 101-556 | 0.25 | 128 | 1 | 0.008 | 0.12 | 2 | 0.25 |
| TF3 | 101-552 | 0.25 | 128 | 1 | 0.016 | 0.12 | 2 | 0.25 |
| TF4 | 101-545 | 0.25 | 64 | 1 | 0.016 | 0.06 | 2 | 0.25 |
| TF5 | 201-575 | 0.25 | 128 | 1 | 0.016 | 0.12 | 4 | 0.5 |
| TF6 | 279-575 | 0.25 | 128 | 1 | 0.016 | 0.12 | 4 | 0.5 |
| TF7 | 312-575 | 0.25 | 256 | 1 | 0.016 | 0.12 | 4 | 0.5 |
| TF8 | 323-575 | 0.25 | 128 | 0.5 | 0.004 | 0.03 | 0.5 | 0.06 |
| TF9 | 406-575 | 0.25 | 64 | 0.5 | 0.008 | 0.03 | 0.5 | 0.06 |
| TF10 | 437-575 | 0.25 | 128 | 0.5 | 0.016 | 0.06 | 1 | 0.12 |
| TF11 | 504-575 | 0.25 | 64 | 0.5 | 0.016 | 0.06 | 1 | 0.12 |
| TF12 | 504-545 | 0.12 | 32 | 0.5 | 0.008 | 0.03 | 0.25 | 0.06 |
MSC02236 had alterations in PBP2 between amino acid (aa) positions 101 and 575 (each substitution is shown in Fig. 2, mosaic-1 sequence). The region that was acquired from MSC02236 through homologous recombination is indicated for recombinants.
Identification of the mutations associated with reduced susceptibility to cefixime.
Five mutations between positions 504 and 545 were individually introduced into penA from the FA1090 strain by site-directed mutagenesis. After transformation, only the recombinant with the G545S mutation in PBP2 was recovered. Two mutations, I312M and V316T, were introduced into penA containing the mutation G545S, because no transformant with a single mutation of either I312M or V316T was obtained. A recombinant with all three of these mutations could not be isolated. We therefore investigated the antibiotic susceptibility of the three recombinants (with amino acid substitutions G545S, G545S and I312M, and G545S and V316T) (Table 5). Acquisition of the G545S mutation resulted in a two- to fourfold increase in the MICs of cephems. In addition to the G545S mutation, the I312M or V316T mutation led to a further fourfold increase in the MICs of cefixime, ceftibuten, cefpodoxime, and cefdinir. Thus, two substitutions in PBP2, G545S plus I312M or V316T, were found to confer an eightfold reduction in susceptibility to cefixime. Ceftriaxone and cefditoren differed from the other cephems in that the susceptibility of the strains to these two antibiotics was not greatly affected by mutations in mosaic PBP2.
TABLE 5.
MICs of β-lactams for transformants with point mutations in PBP2
| Mutations in PBP2 | MIC (μg/ml)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Penicillin | Cephalexin | Cefotiam | Ceftriaxone | Cefixime | Ceftibuten | Cefpodoxime | Cefdinir | Cefcapene | Cefditoren | |
| None (FA1090) | 0.12 | 4 | 0.06 | 0.004 | 0.008 | 0.03 | 0.016 | 0.016 | 0.008 | 0.008 |
| G545S | 0.12 | 16 | 0.25 | 0.008 | 0.016 | 0.12 | 0.03 | 0.03 | 0.016 | 0.016 |
| G545S, I312M | 0.12 | 32 | 0.5 | 0.016 | 0.06 | 0.5 | 0.12 | 0.12 | 0.06 | 0.03 |
| G545S, V316T | 0.06 | 32 | 0.5 | 0.016 | 0.06 | 0.5 | 0.12 | 0.12 | 0.03 | 0.03 |
DISCUSSION
Recently, there has been a marked decrease in the susceptibility of N. gonorrhoeae strains to cefixime throughout Japan. These bacterial strains were previously shown to possess a mosaic-like structure of PBP2. Although earlier investigations have reported mutations of either PBP1 or PBP2 for numerous isolates (7, 14), there has been no systematic study of PBP sequences from various clinical isolates. In this study, the complete sequence of the genes encoding PBP1, PBP2, and porin were determined from 58 isolates. We found that the presence of mosaic PBP2 was strongly correlated with reduced susceptibility to cefixime and other cephems. Interestingly, all the strains with a mosaic PBP2 carried the L421P mutation in PBP1. The L421P substitution in PBP1, together with overexpression of MtrCDE and mutations in porin and PilQ, are reported to be involved in high-level resistance to penicillin (19). Our transformation experiments demonstrate that recombination of full-length mosaic PBP2 could not fully explain the observed resistance of the clinical isolates (MSC02236) to cephem antibiotics, such as cefotiam, ceftriaxone, and cefpodoxime (Table 4). Therefore, the amino acid substitution in PBP1 could be involved not only in the high-level resistance to penicillin (13) but also in resistance to the cephem antibiotics. Indeed, the PBP1(L421P) mutation was found in all the strains with reduced susceptibility to cefixime, suggesting that it might partly contribute to the increase in MIC. However, further study is needed to clarify the association of the PBP1(L421P) mutation and resistance to cephems. As reported previously (6), extensive sequence variability was identified for the porin, with 42 different amino acid patterns determined from 58 strains. Although most of the strains possess mutations at positions 120 and 121 of the porin protein, there is no clear correlation with increased resistance to cefixime.
Four different mosaic PBP2 sequences, designated mosaic-1 to mosaic-4, were detected from 28 isolates. Mosaic-1 sequence, the most frequently identified in this study, was identical in the isolates from Kitakyushu, Japan (strains SNG32, SNG33, SNG46 and SNG50; GenBank accession no. AY146782 to 146785), and also from the central region of Japan (47 strains of pattern X) (7, 9). Twelve out of 22 isolates with reduced susceptibility to cefixime (MIC of ≥0.25 μg/ml) from the Kinki area of Japan also possessed the mosaic-1 mutation in PBP2 (unpublished data). These observations suggest that isolates with mosaic-1 are spreading nationwide and comprise the strains with reduced susceptibility to cefixime.
We have identified three amino acid substitutions in mosaic-1 (G545S, I312M, and V316T) associated with reduced susceptibility to cefixime and other oral cephems. While full-length recombination of mosaic-1 led to a 16-fold reduction in susceptibility to cefixime, the mutation G545S plus either I312M or V316T conferred an eightfold reduction, suggesting that these substitutions are important for the resistance mechanism. The primary mutation G545S is located downstream of the conserved K497TG motif. The mutations conferring additional resistance to cephems, I312M and V316T, are associated with the conserved S310AIK motif. Mosaic-2 and mosaic-3 sequences differ from the sequence of mosaic-1 by a single amino acid change at position 101 or 214, respectively. Because these amino acid substitutions are not located in the transpeptidase domain, which constitutes the β-lactam binding site, the same three mutations found in mosaic-1 are likely to be crucial for conferring resistance to the mosaic-2 and -3 isolates. By contrast, the mosaic-4 sequence does not contain the G545S alteration. However, during the transformation experiments a spontaneous mutant with a single A501V mutation in PBP2, which was found in mosaic-4 sequence, was selected with cephalexin. Analysis of this transformant revealed that the A501V mutation in PBP2 led to a two- to fourfold increase in the MICs of cefixime and other cephems. This increase in resistance is similar to that observed for the PBP2(G545S) mutation. We assume that A501V complements the G545S substitution and is the primary mutation in mosaic-4 isolates.
It has been proposed that horizontal genetic exchange of the penA genes between commensal Neisseria species, such as Neisseria cinerea, Neisseria perflava and Neisseria flavescens, resulted in the mosaic-like structure of PBP2 in N. gonorrhoeae and Neisseria meningitidis (3, 15). Analysis of the amino acid sequences of PBP2 from various Neisseria species indicated that PBP2 from N. perflava/sicca 1654/1659 (GenBank accession no. X76422) and N. flavescens NCTC8263 (GenBank accession no. M26645) had methionine 312 and threonine 316, as does mosaic PBP2 in N. gonorrhoeae. These observations suggest that the I312M and V316T substitutions originated from N. perflava/sicca or N. flavescens through horizontal gene transfer. However, no commensal Neisseria species was found to possess serine 545. Therefore, the G545S alteration is probably the result of antibiotic selective pressure.
The results of our study suggest that the extensive use of cefixime, or other cephems with reduced affinity to mosaic PBP2, will select the resistant N. gonorrhoeae strains harboring mosaic PBP2. The antibacterial activity of ceftriaxone and cefditoren was only slightly reduced for strains with mosaic PBP2. Both these cephalosporins possess a long side chain at the C-3 position of the cephem skeleton, which might result in a strong affinity for the altered PBP2. Continued surveillance of antimicrobial susceptibility and genetic studies to identify the mechanisms of resistance will be needed to establish appropriate treatments. The results from such studies may also assist in the development of new antibiotics of therapeutic utility.
Footnotes
Published ahead of print on 28 August 2006.
REFERENCES
- 1.Akasaka, S., T. Muratani, Y. Yamada, H. Inatomi, K. Takahashi, and T. Matsumoto. 2001. Emergence of cephem- and aztreonam-high-resistant Neisseria gonorrhoeae that does not produce β-lactamase. J. Infect. Chemother. 7:49-50. [DOI] [PubMed] [Google Scholar]
- 2.Ameyama, S., S. Onodera, M. Takahata, S. Minami, N. Maki, K. Endo, H. Goto, H. Suzuki, and Y. Oishi. 2002. Mosaic-like structure of penicillin-binding protein 2 gene (penA) in clinical isolates of Neisseria gonorrhoeae with reduced susceptibility to cefixime. Antimicrob. Agents Chemother. 46:3744-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowler, L. D., Q. Y. Zhang, J. Y. Riou, and B. G. Spratt. 1994. Interspecies recombination between the penA genes of Neisseria meningitidis and commensal Neisseria species during the emergence of penicillin resistance in N. meningitidis: natural events and laboratory simulation. J. Bacteriol. 176:333-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brannigan, J. A., I. A. Tirodimos, Q. Y. Zhang, C. G. Dowson, and B. G. Spratt. 1990. Insertion of an extra amino acid is the main cause of the low affinity of penicillin-binding protein 2 in penicillin-resistant strains of Neisseria gonorrhoeae. Mol. Microbiol. 4:913-919. [DOI] [PubMed] [Google Scholar]
- 5.Deguchi, T., M. Yasuda, S. Yokoi, K. Ishida, M. Ito, S. Ishihara, K. Minamidate, Y. Harada, K. Tei, K. Kojima, M. Tamaki, and S. Maeda. 2003. Treatment of uncomplicated gonococcal urethritis by double-dosing of 200 mg cefixime at a 6-h interval. J. Infect. Chemother. 9:35-39. [DOI] [PubMed] [Google Scholar]
- 6.Fudyk, T. C., I. W. Maclean, J. N. Simonsen, E. N. Njagi, J. Kimani, R. C. Brunham, and F. A. Plummer. 1999. Genetic diversity and mosaicism at the por locus of Neisseria gonorrhoeae. J. Bacteriol. 181:5591-5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito, M., T. Deguchi, K. Mizutani, M. Yasuda, S. Yokoi, S. Ito, Y. Takahashi, S. Ishihara, Y. Kawamura, and T. Ezaki. 2005. Emergence and spread of Neisseria gonorrhoeae clinical isolates harboring mosaic-like structure of penicillin-binding protein 2 in central Japan. Antimicrob. Agents Chemother. 49:137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito, M., M. Yasuda, S. Yokoi, S. Ito, Y. Takahashi, S. Ishihara, S. Maeda, and T. Deguchi. 2004. Remarkable increase in central Japan in 2001-2002 of Neisseria gonorrhoeae isolates with decreased susceptibility to penicillin, tetracycline, oral cephalosporins, and fluoroquinolones. Antimicrob. Agents Chemother. 48:3185-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muratani, T., S. Akasaka, T. Kobayashi, Y. Yamada, H. Inatomi, K. Takahashi, and T. Matsumoto. 2001. Outbreak of cefozopran (penicillin, oral cephems, and aztreonam)-resistant Neisseria gonorrhoeae in Japan. Antimicrob. Agents Chemother. 45:3603-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 11.Olesky, M., M. Hobbs, and R. A. Nicholas. 2002. Identification and analysis of amino acid mutations in porin IB that mediate intermediate-level resistance to penicillin and tetracycline in Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 46:2811-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olesky, M., S. Zhao, R. L. Rosenberg, and R. A. Nicholas. 2006. Porin-mediated antibiotic resistance in Neisseria gonorrhoeae: ion, solute, and antibiotic permeation through PIB proteins with penB mutations. J. Bacteriol. 188:2300-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ropp, P. A., M. Hu, M. Olesky, and R. A. Nicholas. 2002. Mutations in ponA, the gene encoding penicillin-binding protein 1, and a novel locus, penC, are required for high-level chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 46:769-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shigemura, K., T. Shirakawa, N. Massi, K. Tanaka, S. Arakawa, A. Gotoh, and M. Fujisawa. 2005. Presence of a mutation in ponA1 of Neisseria gonorrhoeae in numerous clinical samples resistant to various β-lactams and other, structurally unrelated, antimicrobials. J. Infect. Chemother. 11:226-230. [DOI] [PubMed] [Google Scholar]
- 15.Spratt, B. G., L. D. Bowler, Q. Y. Zhang, J. Zhou, and J. M. Smith. 1992. Role of interspecies transfer of chromosomal genes in the evolution of penicillin resistance in pathogenic and commensal Neisseria species. J. Mol. Evol. 34:115-125. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka, M., H. Nakayama, T. Notomi, S. Irie, Y. Tsunoda, A. Okadome, T. Saika, and I. Kobayashi. 2004. Antimicrobial resistance of Neisseria gonorrhoeae in Japan, 1993-2002: continuous increasing of ciprofloxacin-resistant isolates. Int. J. Antimicrob. Agents 24S:S15-S22. [DOI] [PubMed] [Google Scholar]
- 17.Veal, W. L., R. A. Nicholas, and W. M. Shafer. 2002. Overexpression of the MtrC-MtrD-MtrE efflux pump due to an mtrR mutation is required for chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. J. Bacteriol. 184:5619-5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, S. A., M. V. Lee, N. O'Connor, C. J. Iverson, R. G. Ohye, P. M. Whiticar, J. A. Hale, D. L. Trees, J. S. Knapp, P. V. Effler, and H. S. Weinstock. 2003. Multidrug-resistant Neisseria gonorrhoeae with decreased susceptibility to cefixime—Hawaii, 2001. Clin. Infect. Dis. 37:849-852. [DOI] [PubMed] [Google Scholar]
- 19.Zhao, S., D. M. Tobiason, M. Hu, H. S. Seifert, and R. A. Nicholas. 2005. The penC mutation conferring antibiotic resistance in Neisseria gonorrhoeae arises from a mutation in the PilQ secretin that interferes with multimer stability. Mol. Microbiol. 57:1238-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]