Abstract
Bacterial genes defining intrinsic resistance to antibiotics encode proteins that can be targeted by antibiotic potentiators. To find such genes, a transposon insertion library of Acinetobacter baylyi was screened with subinhibitory concentrations of various antibiotics to find supersusceptible mutants. A DNA microarray printer was used to replica plate 10,000 individual library clones to select mutants unable to grow at 1/10 the MICs of 12 different antibiotics. Transposon insertions in 11 genes were found to cause an eightfold or higher hypersusceptibility to at least one antibiotic. Most of the mutants identified exhibited hypersusceptibility to β-lactam antibiotics. These included mutants with disruptions of genes encoding proteins involved in efflux (acrB and oprM) as well as genes pertaining to peptidoglycan synthesis and modification (ampD, mpl, and pbpG). However, disruptions of genes encoding proteins with seemingly unrelated functions (gph, argH, hisF, and ACIAD0795) can also render cells hypersusceptible to β-lactam antibiotics. A knockout of gshA, involved in glutathione biosynthesis, enhanced the susceptibility to metronidazole, while a knockout of recD, involved in recombination and repair, made the bacteria hypersusceptible to ciprofloxacin. Disruption of acrB in Escherichia coli rendered the cells hypersusceptible to several antibiotics. However, knockout mutants of other homologous genes in E. coli showed no significant changes in antibiotic MICs, indicating that the intrinsic resistance genes are species specific.
The growing problem of antibiotic resistance among bacterial pathogens and the escalating difficulty in finding new antibiotics drive the search for new approaches to antibacterial chemotherapy. One such approach is the development of antibiotic potentiators that can enhance antibiotic efficiency when the antibiotics and the potentiators are used in combination, as well as reduce the chances of the emergence of antibiotic resistance.
Currently, potentiators are available for only one type of antibiotics, β-lactams. The β-lactamase inhibitors currently used in clinics, clavulanate, sulbactam, and tazobactam, have dramatically enhanced the efficacies of important β-lactam antibiotics (6). Other potentiators are being developed, for example, those that target the multidrug resistance efflux pumps (14). However, to date, there are no such potentiators in clinical use.
A number of nonessential bacterial proteins may potentially contribute to the intrinsic antibiotic resistance. The susceptibilities of bacteria to antibiotics depend on many factors, including the structure and the composition of the cell envelope, the presence of inactivating enzymes, and the availability of efflux pumps. The corresponding genes may be present in the genome either because they were selected in the course of evolution to help the organism tolerate antibiotics that it may encounter in the environment or because the encoded enzymes, which have specialized cellular functions, may fortuitously contribute to antibiotic resistance. Conceivably, inactivation of such enzymes by inhibitors may increase the potencies of the antibiotics currently in medical use.
In the present study, we used a genetic approach to identify the putative targets of such antibiotic potentiators. We generated a random transposon gene-knockout library of Acinetobacter baylyi, a close relative of the opportunistic pathogen Acinetobacter baumannii, and selected mutants for the inability to grow at a subinhibitory antibiotic concentration. To identify the A. baylyi genes contributing to the intrinsic resistance to antibiotics, 10,000 bacterial clones carrying random transposon insertions were replica plated in the presence of subinhibitory concentrations of 12 different antibiotics. This otherwise laborious task was facilitated by the use of a microarray-printing robot to “print” the clones for replica plating. Hypersusceptible mutants were identified, and the genes whose disruption increases cell susceptibility to antibiotics were determined by direct genomic DNA sequencing. If a disruption of a certain gene leads to antibiotic hypersusceptibility, inhibition of the encoded protein product is likely to have the same effect. Therefore, an inhibitor can potentially be designed that, when used in combination with the corresponding antibiotic, will enhance its efficacy.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Acinetobacter baylyi strain ADP1 was from the American Type Culture Collection (ATCC 33305). Escherichia coli strain BW25113 gene deletion mutants were obtained from H. Mori, Nara Institute of Science and Technology, Japan (1d). Both A. baylyi and E. coli were grown either in liquid cultures in Luria-Bertani (LB) medium or on LB agar plates at 37°C. When appropriate, overnight cultures were supplemented with kanamycin (KAN; 12.5 μg/ml), spectinomycin (50 μg/ml), and streptomycin (10 μg/ml) for A. baylyi or with KAN (30 μg/ml) for E. coli.
Antibiotics and reagents.
Chloramphenicol, gentamicin, KAN, metronidazole, piperacillin, rifampin, streptomycin, spectinomycin, and vancomycin were from Sigma; ampicillin and tetracycline were from Fisher; azithromycin was from Pfizer; ceftazidime and imipenem (with cilastatin) were from GlaxoWellcome; and ciprofloxacin was from Bayer. All enzymes except DpnI were from Fermentas; DpnI was from Promega.
Determination of MICs.
Logarithmically growing cells were diluted in LB medium to an optical density at 600 nm of 0.002, and 100 μl was placed in the wells of 96-well plates. Antibiotics were added in twofold dilutions, and the bacteria were grown overnight. The MIC was determined as the lowest antibiotic concentration at which no visible growth occurred.
Construction of the transposome insertion library.
A transposome Ω cassette (22) was used to create a random transposon insertion library of A. baylyi, as described previously (11). About 30,000 colonies were obtained on spectinomycin-streptomycin agar plates. A total of 10,000 colonies were picked, individually inoculated into 96-well plates, grown overnight, and stored at −80°C with 15% glycerol.
Selection of hypersusceptible clones of A. baylyi.
Cells were inoculated into 100 μl of LB medium in 96-well plates and grown overnight. Cultures were diluted 10-fold into fresh LB medium and grown for 4 h with shaking at 700 rpm by using a Brinkmann TiterMix100 microtiter plate shaker. The cells were then diluted 20-fold in 50 μl LB broth supplemented with 20% sucrose to prevent drying of the bacterial culture spots during printing. Submicroliter volumes of the cultures were then spotted (in duplicate) on the surfaces of 13 nylon membranes (GE Osmonics) (768 spots per 25-mm by 75-mm membrane with a printing area of 20 mm by 60 mm and a 1-mm distance between spots). Spotting was done by using a LabNext DNA microarray printer. Four pins of the microarray printer, each 0.2 mm in diameter, were dipped into the bacterial cultures; and extra fluid was removed by prespotting onto the first nylon membrane. Replicas of 384 clones were then printed in duplicate on 13 nylon membranes. Printer pins were sterilized by two rounds of dipping into 40% ethanol, followed by air drying. When the printing job was complete, one membrane (a control) was layered on top of an agar plate without antibiotic; the rest of the membranes were placed atop agar plates containing subinhibitory concentrations (1/10 the wild-type MIC) of one of the antibiotics: azithromycin, ceftazidime, chloramphenicol, ciprofloxacin, gentamicin, imipenem, metronidazole, piperacillin, rifampin, or tetracycline. The membranes were also placed on LB agar plates containing ampicillin (3 μg/ml) or vancomycin (10 μg/ml). After 16 to 18 h of incubation at 37°C, the colonies on the filters were analyzed, and clones that grew on the control plate but failed to grow on plates containing antibiotics at subinhibitory concentrations were identified. For such clones, the MICs of the antibiotics were then determined in liquid cultures.
In order to determine the site of the transposon insertion in antibiotic-hypersusceptible clones, genomic DNA was isolated from 3-ml overnight cultures by using a genomic DNA isolation kit (Sigma). Sequencing of the genomic DNA segment adjacent to the site of the transposon insertion was performed by using an outwards-directed transposon-specific primer (5′ AGAGTCGACCTGCAGGCATGC) and the Promega fmol DNA cycle sequencing system. The disrupted gene was identified by using the genome sequence of A. baylyi strain ADP1 (NCBI accession number CR543861) (2).
To validate the causative relation between the transposon insertion and the hypersusceptible phenotype, total DNA was prepared from the hypersusceptible mutants that were identified and was used to directly transform logarithmically growing wild-type A. baylyi (20). Transformants were selected on a spectinomycin-streptomycin agar plate. Individual colonies were picked, the location of the transposon insertion was verified by PCR, and the antibiotic MICs were determined.
Targeted inactivation of A. baylyi genes.
Selected A. baylyi genes were disrupted by using the method of chromosomal gene replacement (,17). A PCR product containing a kanamycin resistance marker, Tn903, which was amplified from the KAN-2 transposon (Epicenter), flanked by the 600-bp-long upstream and downstream regions of the gene of interest was prepared by consecutive rounds of PCR. First, the kanamycin resistance marker and 300- to 500-bp-long regions flanking the gene of interest were amplified by PCR. During PCR, the NotI and SdaI restriction sites were introduced at the appropriate ends of the upstream and downstream gene-flanking regions of the PCR-amplified kanamycin resistance gene. The PCR products were cut with the restriction enzymes NotI and SdaI, ligated together, treated with DpnI, and then used as the template for the second round of PCR with primers specific to the distal ends of the gene-flanking regions. The PCR products were purified by using a Wizard SV Gel and PCR cleanup system (Promega). In some cases, the second round of PCR produced several bands. In this case, the correct product was extracted from the gel and purified by using a QIAquick gel extraction kit (QIAGEN) or a Wizard SV Gel and PCR cleanup system (Promega). The resulting PCR product was used to transform exponentially growing A. baylyi cells, and transformants were selected on LB medium-KAN plates. The disruption of the appropriate gene in the selected transformants was verified by PCR. The MICs for the generated mutants were determined and compared with the MICs for the original transposon-disrupted clone and wild type.
Identification of E. coli genes homologous to A. baylyi genes.
Eleven A. baylyi genes were used as queries in a BLAST search of the E. coli MG1655 genome (NCBI accession number U00096) (4). The top hits were then used as queries in a BLAST search of the A. baylyi ADP1 genome (NCBI accession number CR543861) (2). The antibiotic MICs for E. coli strains from the Keio collection (1a) were determined as described above for A. baylyi. The MICs of E. coli wild-type strain BW25113 were as follows: ampicillin, 100 μg/ml; azithromycin, 12.5 μg/ml; ceftazidime, 0.3 μg/ml; chloramphenicol, 8 μg/ml; ciprofloxacin, 0.06 μg/ml; gentamicin, 10 μg/ml; imipenem, 1.3 μg/ml; metronidazole, 4,000 μg/ml; piperacillin, 2.5 μg/ml; rifampin, 1 μg/ml; tetracycline, 4 μg/ml; and vancomycin, 325 μg/ml.
RESULTS
Selection and validation of antibiotic-hypersusceptible mutants of A. baylyi.
Previously we described an approach that allows the selection of antibiotic-hypersusceptible gene-knockout mutants of A. baylyi (11), based on the release of DNA from the cells lysed in the presence of subinhibitory concentrations of antibiotics. Although this method identified several genes of interest, its intrinsic limitations left open the possibility that many interesting mutations have not been detected. A more straightforward approach for identification of the genes contributing to the intrinsic resistance of bacteria to antibiotics is the selection of hypersusceptible mutants in a gene-knockout library by direct replica plating of individual mutants on plates containing subinhibitory antibiotic concentrations. In order to accomplish this inherently laborious task, especially when thousands of mutants are to be screened against a number of antibiotics, we applied a variant of the clone microarray printing technique (26). This technique allowed the replication of arrays of clones of the A. baylyi ADP1 transposon library for the simultaneous screening for mutants susceptible to low concentrations of various antibiotics (11). The drugs that were used in our screen included the cell wall synthesis inhibitors ampicillin, imipenem, piperacillin, ceftazidime, and vancomycin; the protein synthesis inhibitors azithromycin, chloramphenicol, gentamicin, and tetracycline; the transcription inhibitor rifampin; and the DNA synthesis inhibitor metronidazole.
After the initial screening of 10,000 clones, we identified 29 clones that exhibited diminished or no growth on plates with low concentrations (1/10 the MICs) of ampicillin, ceftazidime, piperacillin, vancomycin, ciprofloxacin, imipenem, rifampin, and metronidazole. These clones showed at least an eightfold reduction in MICs for at least one antibiotic compared to that for the wild type. (We were unable to find mutants with MIC reductions of eightfold or greater for the translation inhibitors azithromycin, chloramphenicol, gentamicin, and tetracycline). Sequencing of the regions flanking the transposon insertions, performed directly with the genomic DNA prepared from these clones, revealed the identities of the disrupted genes. Independent transpositions were found in 22 distinct genes, with some clones having disruptions within the same gene.
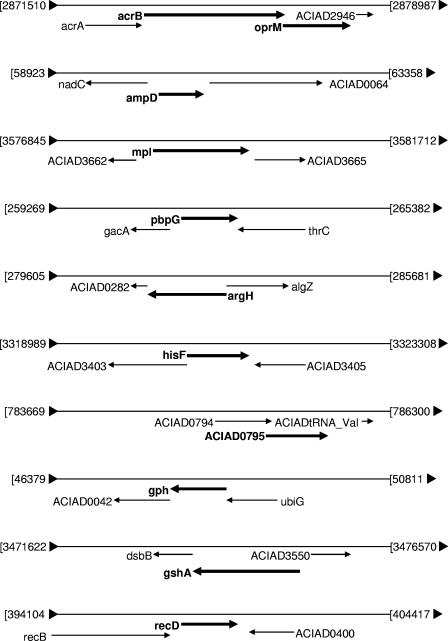
Since A. baylyi has a very high natural competence for transformation and recombination (2), we were able to easily transfer transposon-mediated gene disruptions into the fresh wild-type bacteria to verify whether the hypersusceptibility phenotype was the direct result of the disruption of the identified gene. From the 22 gene disruptions, 11 withstood this test: acrB, oprM, ampD, mpl, pbpG, argH, hisF, gph, ACIAD0795, gshA, and recD (Table 1). We further constructed 11 strains in which these genes were completely deleted and replaced by a kanamycin resistance marker (see Materials and Methods). MIC determination for the deletion mutants showed levels of antibiotic hypersusceptibility similar to those obtained with the transposon disruption mutants. In order to exclude possible polar effects of chromosomal gene replacement, we performed deletions of the genes located immediately downstream from several of the identified genes (Fig. 1). We used the gene replacement technique in an attempt to inactivate the genes ACIAD0042, dsbB, ACIAD2946, ACIAD3665, ACIAD0064, and ACIAD0282, located downstream from gph, gshA, oprM, mpl, ampD, and argH, respectively (the genes recD, pbpG, and ACIAD0795 had no downstream protein genes belonging to the same operon). While we were not able to obtain the knockout mutants for ACIAD0064 and ACIAD0282, the A. baylyi mutants lacking ACIAD0042, dsbB, ACIAD2946, and ACIAD3665 showed no changes in susceptibility compared to that of the wild type. This result indicated that the hypersusceptibilities of the initial mutants were likely the direct result of the disruption of the corresponding genes rather than a polar effect of the transposon insertion. Altogether, these experiments led us to conclude that inactivation of the 11 genes identified in A. baylyi ADP1 (acrB, oprM, ampD, mpl, pbpG, argH, hisF, gph, ACIAD0795, gshA, and recD) led to the antibiotic hypersusceptibility phenotypes.
TABLE 1.
Antibiotic susceptibilities of the gene disruption mutants of Acinetobacter baylyi ADP1 mutants
| Gene product | Gene | Gene identifiera | MIC (μg/ml)b
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | PIP | CAZ | IPM | VAN | AZM | CHL | GEN | TET | RIF | CIP | MTZ | |||
| Wild type | 128.0 | 10.0 | 5.0 | 0.20 | 160.0 | 0.3 | 4.0 | 1.3 | 1.0 | 0.6 | 0.1 | 2000 | ||
| Acriflavin resistance protein B | acrB::kan | 2879610 | 8.0 | 0.3 | 0.3 | 0.10 | 160.0 | 0.2 | 1.0 | 1.3 | 0.1 | 0.3 | 0.006 | 2000 |
| Outer membrane protein OprM | oprM::kan | 2879678 | 8.0 | 0.6 | 0.6 | 0.05 | 160.0 | 0.2 | 2.0 | 1.3 | 0.3 | 0.3 | 0.013 | 2000 |
| Anhydro-N-acetylmuramyl tripeptide amidase | ampD::kan | 2880355 | 4.0 | 0.6 | 0.3 | 0.01 | 80.0 | 0.3 | 4.0 | 0.7 | 1.0 | 0.3 | 0.025 | 2000 |
| UDP-MurNAc-l-Ala-d-Glu-meso-diaminopimelate ligase | mpl::kan | 2878498 | 8.0 | 1.3 | 0.6 | 0.05 | 160.0 | 0.3 | 4.0 | 1.3 | 1.0 | 0.6 | 0.100 | 2000 |
| d-Alanyl-d-alanine endopeptidase | pbpG::kan | 2879431 | 8.0 | 0.6 | 0.6 | 0.10 | 40.0 | 0.1 | 2.0 | 0.7 | 0.5 | 0.08 | 0.050 | 1000 |
| Argininosuccinate lyase | argH::kan | 2877980 | 16.0 | 2.5 | 2.5 | 0.05 | 160.0 | 0.2 | 4.0 | 0.7 | 1.0 | 0.2 | 0.100 | 2000 |
| Imidazole glycerol phosphate synthase | hisF::kan | 2878927 | 16.0 | 1.3 | 1.3 | 0.03 | 80.0 | 0.2 | 4.0 | 0.7 | 1.0 | 0.6 | 0.025 | 2000 |
| Unknown | ACIAD0795::kan | 2877853 | 8.0 | 0.6 | 2.5 | 0.20 | 160.0 | 0.2 | 4.0 | 1.3 | 1.0 | 0.3 | 0.100 | 2000 |
| Phosphoglycolate phosphatase | gph::kan | 2881034 | 8.0 | 2.5 | 1.3 | 0.03 | 160.0 | 0.3 | 4.0 | 0.7 | 1.0 | 0.3 | 0.050 | 2000 |
| Glutamate cysteine ligase | gshA::kan | 2879245 | 128.0 | 10.0 | 5.0 | 0.20 | 80.0 | 0.3 | 4.0 | 1.3 | 1.0 | 0.3 | 0.100 | 250 |
| Exodeoxyribonuclease V α chain | recD::kan | 2879479 | 64.0 | 10.0 | 5.0 | 0.10 | 80.0 | 0.3 | 2.0 | 1.3 | 1.0 | 0.6 | 0.013 | 1000 |
Gene identifiers are from the complete Acinetobacter baylyi genome (NCBI accession number CR543861).
MICs with eightfold or higher decreases are shown in boldface.
FIG. 1.
Gene maps of the Acinetobacter ADP1 chromosome in the vicinity of genes whose inactivation renders cells hypersusceptible to antibiotics. Numbers refer to positions in the complete sequence of the A. baylyi ADP1 genome, and ACIAD numbers correspond to the genome annotation (NCBI accession number CR543861) (2).
Effects of gene disruptions in E. coli on antibiotic sensitivity.
In order to test whether the intrinsic resistance genes discovered in A. baylyi play similar roles in other gram-negative bacteria, we investigated the antibiotic sensitivities of E. coli strains in which the homologs of these genes were deleted. The E. coli homologs of the A. baylyi genes were identified by a reciprocal BLAST search (24). First, the highest-scoring E. coli homolog was identified for each of the 11 A. baylyi intrinsic resistance genes. Then, each of the highest-scoring E. coli genes was used as a query in the BLAST search of the A. baylyi genome. For 10 of the 11 genes (with the exception of ACIAD0795), the reciprocal BLAST search consistently revealed the homologous genes in A. baylyi and E. coli. The similarity scores for the homologous genes were 46% or higher for the aligned sections of the E. coli and A. baylyi proteins (Table 2). Of these 10 genes, all but 1 (oprM) showed the highest similarity to the expected functional homologs in the E. coli genome. E. coli lacks a homolog of the oprM gene (which encodes a component of the tripartite efflux pump in Pseudomonas aeruginosa and several other bacteria) (13). The putative E. coli functional equivalent of oprM is tolC, which encodes a component of the acrAB-tolC tripartite efflux pump whose inactivation is known to lead to antibiotic hypersusceptibility (reviewed in reference 12). However, E. coli tolC shows only limited similarity to A. baylyi oprM. The E. coli gene showing the highest degree of similarity (55%) to the A. baylyi oprM gene was cusC, which encodes an outer membrane component of the copper efflux system (7). Therefore, E. coli cusC was targeted for disruption. Of the 11 genes whose disruption makes A. baylyi hypersensitive to antibiotics, only 1, ACIAD0795, did not have a clear homolog in E. coli. A 48-amino-acid segment of this putative 167-amino-acid A. baylyi protein showed homology to the conserved domains of the YbiS and ErfK proteins of E. coli, whose functions are unknown but which may be related to the outer membrane porins and the stress response, respectively (3, 5). However, the reciprocal BLAST search with either YbiS or ErfK against the A. baylyi genome showed that these proteins have the highest homology to another A. baylyi gene, ACIAD2475. Nevertheless, we did investigate the effects of ybiS and erfK deletions on the sensitivity of E. coli to antibiotics.
TABLE 2.
Homology between homologous portions of the proteins in A. baylyi and E. coli as identified by BLAST search
| A. baylyi (length [no. of amino acids]) | E. coli (length [no. of amino acids]) | % Similarity | Length (amino acids) of the homologous regiona |
|---|---|---|---|
| AcrB (1,059) | AcrB (1,049) | 72 | 1,062 |
| OprM (484) | CusC (457) | 55 | 434 |
| AmpD (196) | AmpD (183) | 66 | 181 |
| Mpl (453) | Mpl (457) | 68 | 452 |
| PbpG (352) | PbpG (310) | 63 | 248 |
| ArgH (477) | ArgH (457) | 61 | 455 |
| HisF (269) | HisF (258) | 61 | 257 |
| ACIAD0795 (167) | YbiS (306) | 65 | 38 |
| ACIAD0795 (167) | ErfK (310) | 58 | 48 |
| Gph (235) | Gph (252) | 46 | 232 |
| GshA (530) | GshA (518) | 57 | 476 |
| RecD (592) | RecD (608) | 50 | 526 |
The total length of the homologous region, including gaps introduced for the optimal alignment.
The E. coli deletion mutants (acrB, cusC, mpl, ampD, pbpG, argH, hisF, gph, gshA, recD, ybiS, and erfK) from the Keio collection (1a), which is composed of E. coli clones in which all nonessential gene are replaced by a kanamycin resistance marker, were used for antibiotic sensitivity testing. The disruption of the acrB gene in E. coli rendered the cells hypersusceptible to several antibiotics: ampicillin (MICs, 12.5 μg/ml versus 100 μg/ml for the wild type), piperacillin (MICs, 0.1 μg/ml versus 2.5 μg/ml for the wild type), chloramphenicol (MICs, 1 μg/ml versus 8 μg/ml for the wild type), and azithromycin (MICs, 0.8 μg/ml versus 12.5 μg/ml for the wild type). Surprisingly however, no significant changes in the MICs for any of the other strains tested were observed, indicating that the newly identified potentiator targets may be specific to A. baylyi.
DISCUSSION
Our goal in this work was to develop a strategy for identifying new targets for antibiotic potentiators. Using a straightforward replica-plating approach, we succeeded in identifying genes whose disruption renders A. baylyi hypersusceptible to several clinical antibiotics. The corresponding gene products could be considered putative targets for antibiotic potentiators. In these studies, we used A. baylyi ADP1 as a model organism. The very high degree of competence of A. baylyi greatly facilitates the transfer of the identified transposon insertion mutations into wild-type cells and the subsequent preparation of strains with chromosomal replacement of the identified genes. In general, however, our approach can be applied to any microorganism for which a gene-knockout library is available. The task of identifying the intrinsic antibiotic resistance genes was significantly aided by the use of a DNA microarray printer, which allowed the fast and easy replica plating of 10,000 individual strains on plates with 12 different antibiotics.
Eleven genes in A. baylyi whose disruption leads to antibiotic hypersusceptibility were identified. Among these genes was the homolog of acrB, which encodes a component of the multidrug resistance pump in E. coli (19). Disruption of such pumps in other bacteria is known to confer multidrug resistance (12). Some of the previous studies also indicated that mutations in the rec genes may increase cell sensitivity to quinolones (18, 25). This result validated our approach and demonstrated that the colony-printing technique can be used effectively to detect mutants hypersusceptible to antibiotics. To the best of our knowledge, the other genes that we identified were not previously described as direct contributors to the intrinsic antibiotic resistance and thus were not previously considered targets for putative antibiotic potentiators.
Several of the genes identified provide intrinsic resistance to one or more β-lactam antibiotics (Table 1). In three cases, ampD, mpl, and pbpG, this conclusion is in generally good agreement with the known functions of the corresponding gene products, since they are involved in biosynthesis or recycling of peptidoglycan and thus belong to the same biochemical pathway that is targeted by β-lactam antibiotics. AmpD is involved in the breakdown of the anhydromuropeptides produced upon degradation of old peptidoglycan (9, 10), whereas Mpl links the tripeptide l-alanyl-γ-d-glutamyl-meso-diaminopimelate, released by AmpD, to one of the main precursors of peptidoglycan synthesis, UDP-N-acetylmuramate (16). Gene pbpG encodes two low-molecular-weight penicillin-binding proteins (PBPs), PBPs 7 and 8, in E. coli, which play a role in the remodeling of peptidoglycan (8, 23). However, beyond the general observation that the protein products identified are involved in the biochemical pathway affected by the antibiotic, it is hard to explain why disruptions of these particular genes and not of multiple others involved in the same pathway render cells hypersusceptible to the drugs. This uncertainty underscores the importance of experimental approaches, similar to the one described in this paper, for identification of putative targets for antibiotic potentiators.
Four other genes, argH, hisF, gph, and ACIAD0795, whose knockout increases the susceptibility of A. baylyi to β-lactam antibiotics, encode proteins with functions seemingly unrelated to the biosynthesis of the bacterial cell wall. ArgH and HisF are involved in the biosynthesis of arginine and histidine, respectively, while gph encodes the housekeeping enzyme 2-phosphoglycolate phosphatase, which is induced during oxidative stress (21). The presumed function of Gph is to metabolize the 2-phosphoglycolate produced in the repair of DNA lesions (21). A hypothetical 167-amino-acid protein is encoded by an A. baylyi gene, ACIAD0795, that contains a domain that exhibits similarity to the conserved domain of the erfK family of proteins in E. coli, whose function is obscure. At the moment, it is unclear why disruption of any of these four genes in A. baylyi causes hypersusceptibility to β-lactam antibiotics.
Finally, genetic knockouts of gshA and recD increased the sensitivity of A. baylyi to metronidazole and ciprofloxacin, respectively. Metronidazole is a prodrug which, upon activation, forms highly active radical species that cause DNA damage (15). Therefore, it makes sense that the disruption of gshA, the gene whose product is involved in the biosynthesis of the reducing agent glutathione, increased the sensitivity of A. baylyi to metronidazole. Similarly, it is not surprising that the genetic knockout of recD, a component of the recBCD complex that plays a major role in DNA repair and recombination (1), increases bacterial sensitivity to the DNA-damaging drug ciprofloxacin. Nevertheless, as mentioned earlier, without the experimental data obtained in this study, it would be difficult to “handpick” these particular genes as targets for antibiotics potentiators.
Although in our experiments a number of genes were found to contribute to the intrinsic antibiotic resistance of A. baylyi, we cannot accurately estimate how exhaustive our screening was and whether we identified all or even the majority of the A. baylyi intrinsic resistance genes. On the one hand, our screen revealed five independent insertions into recD. This result may indicate that either mutagenesis was close to saturation or that recD represents a transposon insertion hot spot. On the other hand, the random nature of colony picking and the testing of a fairly limited number of clones (which was of the same order as the number of genes in A. baylyi) could have left some mutants with the hypersusceptible phenotype untested.
One of the unexpected findings of this work was that disruption of many genes which contribute to the intrinsic resistance of A. baylyi to antibiotics had very little or no effect on the susceptibility of E. coli to these drugs. Although this result might be viewed as discouraging for the development of broad-range antibiotic potentiators, it opens the possibility of expansion of the spectrum of available drugs to specific classes of pathogens as well as the development of narrow-spectrum potentiators fine-tuned to combat particular infections.
Acknowledgments
We are grateful to Alexander Mankin for his help with preparing the manuscript and to Dennis Kaznadzey (LabNext) for help in modifying the microarray printing routine. Special thanks are extended to H. Mori (Nara Institute, Japan) and his Japanese and American collaborators for unparalleled generosity in providing the Keio collection of E. coli gene knockout clones.
The work was supported by NIH grants AI49214 and AI56575.
Footnotes
Published ahead of print on 28 August 2006.
This paper is dedicated to the memory of Alexander A. Neyfakh, deceased 20 April 2006.
REFERENCES
- 1.Amundsen, S. K., A. F. Taylor, A. M. Chaudhury, and G. M. Smith. 1986. recD: the gene for an essential third subunit of exonuclease V. Proc. Natl. Acad. Sci. USA 83:5558-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:msb4100050-E1-msb4100050-E11. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbe, V., D. Vallenet, N. Fonknechten, A. Kreimeyer, S. Oztas, L. Labarre, S. Cruveiller, C. Robert, S. Duprat, P. Wincker, L. N. Ornston, J. Weissenbach, P. Marliere, G. N. Cohen, and C. Medigue. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 32:5766-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennik, M. H., P. J. Pomposiello, D. F. Thorne, and B. Demple. 2000. Defining a rob regulon in Escherichia coli by using transposon mutagenesis. J. Bacteriol. 182:3794-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Bochkareva, E. S., A. S. Girshovich, and E. Bibi. 2002. Identification and characterization of the Escherichia coli stress protein UP12, a putative in vivo substrate of GroEL. Eur. J. Biochem. 269:3032-3040. [DOI] [PubMed] [Google Scholar]
- 6.Buynak, J. D. 2006. Understanding the longevity of the beta-lactam antibiotics and of antibiotic/beta-lactamase inhibitor combinations. Biochem. Pharmacol. 71:930-940. [DOI] [PubMed] [Google Scholar]
- 7.Franke, S., G. Grass, C. Rensing, and D. H. Nies. 2003. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 185:3804-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henderson, T. A., M. Templin, and K. D. Young. 1995. Identification and cloning of the gene encoding penicillin-binding protein 7 of Escherichia coli. J. Bacteriol. 177:2074-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holtje, J. V., U. Kopp, A. Ursinus, and B. Wiedemann. 1994. The negative regulator of beta-lactamase induction AmpD is a N-acetyl-anhydromuramyl-l-alanine amidase. FEMS Microbiol. Lett. 122:159-164. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs, C., B. Joris, M. Jamin, K. Klarsov, J. Van Beeumen, D. Mengin-Lecreulx, J. van Heijenoort, J. T. Park, S. Normark, and J. M. Frere. 1995. AmpD, essential for both beta-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-l-alanine amidase. Mol. Microbiol. 15:553-559. [DOI] [PubMed] [Google Scholar]
- 11.Lee, H., N. Vazquez-Laslop, K. A. Klyachko, and A. A. Neyfakh. 2003. Isolation of antibiotic hypersusceptibility mutants of Acinetobacter spp. by selection for DNA release. Antimicrob. Agents Chemother. 47:1267-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, X. Z., and H. Nikaido. 2004. Efflux-mediated drug resistance in bacteria. Drugs 64:159-204. [DOI] [PubMed] [Google Scholar]
- 13.Li, X. Z., H. Nikaido, and K. Poole. 1995. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lomovskaya, O., M. S. Warren, A. Lee, J. Galazzo, R. Fronko, M. Lee, J. Blais, D. Cho, S. Chamberland, T. Renau, R. Leger, S. Hecker, W. Watkins, K. Hoshino, H. Ishida, and V. J. Lee. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendz, G. L., and F. Megraud. 2002. Is the molecular basis of metronidazole resistance in microaerophilic organisms understood? Trends Microbiol. 10:370-375. [DOI] [PubMed] [Google Scholar]
- 16.Mengin-Lecreulx, D., J. van Heijenoort, and J. T. Park. 1996. Identification of the mpl gene encoding UDP-N-acetylmuramate: l-alanyl-gamma-d-glutamyl-meso-diaminopimelate ligase in Escherichia coli and its role in recycling of cell wall peptidoglycan. J. Bacteriol. 178:5347-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metzgar, D., J. M. Bacher, V. Pezo, J. Reader, V. Doring, P. Schimmel, P. Marliere, and V. de Crecy-Lagard. 2004. Acinetobacter sp. ADP1: an ideal model organism for genetic analysis and genome engineering. Nucleic Acids Res. 32:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niga, T., H. Yoshida, H. Hattori, S. Nakamura, and H. Ito. 1997. Cloning and sequencing of a novel gene (recG) that affects the quinolone susceptibility of Staphylococcus aureus. Antimicrob. Agents Chemother. 41:1770-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmen, R., B. Vosman, P. Buijsman, C. K. Breek, and K. J. Hellingwerf. 1993. Physiological characterization of natural transformation in Acinetobacter calcoaceticus. J. Gen. Microbiol. 139:295-305. [DOI] [PubMed] [Google Scholar]
- 21.Pellicer, M. T., M. F. Nunez, J. Aguilar, J. Badia, and L. Baldoma. 2003. Role of 2-phosphoglycolate phosphatase of Escherichia coli in metabolism of the 2-phosphoglycolate formed in DNA repair. J. Bacteriol. 185:5815-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 23.Romeis, T., and J. V. Holtje. 1994. Penicillin-binding protein 7/8 of Escherichia coli is a dd-endopeptidase. Eur. J. Biochem. 224:597-604. [DOI] [PubMed] [Google Scholar]
- 24.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 25.Urios, A., G. Herrera, V. Aleixandre, and M. Blanco. 1991. Influence of recA mutations on gyrA dependent quinolone resistance. Biochimie 73:519-521. [DOI] [PubMed] [Google Scholar]
- 26.Van Dyk, T. K., E. J. DeRose, and G. E. Gonye. 2001. LuxArray, a high-density, genomewide transcription analysis of Escherichia coli using bioluminescent reporter strains. J. Bacteriol. 183:5496-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]