Abstract
Pyrimethamine analogs were examined as potential agents against vivax malaria using a bacterial surrogate system carrying Plasmodium vivax dihydrofolate reductase-thymidylate synthase (PvDHFR-TS), in which the PvDHFR complemented chemically knocked out host dihydrofolate reductase. The system was initially tested with P. falciparum dihydrofolate reductase-thymidylate synthase and was found to have good correlation with the parasite-based system. The 50% inhibitory concentrations derived from PvDHFR-TS-dependent bacteria were correlated with their corresponding inhibition constants (Ki) from an enzyme inhibition assay, pointing to the likelihood that the potent enzyme inhibitors will also have potent antimalarial activities. Active compounds against both wild-type and S58R S117N (SP21) double-mutant P. vivax include analogs with structures which can avert a steric clash with the asparagine (S117N) side chain of the mutant, similar to those found for homologous Plasmodium falciparum mutants, raising the possibility that the same compounds can be developed against both types of antifolate-resistant malaria. This rapid and convenient drug screening system should be useful for development of new antifolates against P. vivax, for which a continuous culture system is not yet available.
Malaria caused by Plasmodium vivax is a major public health problem in Asia and South and Central America, where it is most prevalent, with estimates of more than 70 to 80 million cases annually (23). The recent reports on a P. vivax parasite resistant to chloroquine (3, 20), the drug commonly prescribed for P. vivax infection, in addition to the lack of a protective vaccine, highlight the need for new approaches to antimalarial chemotherapy. One promising drug target for the treatment of P. vivax infections is dihydrofolate reductase (DHFR), a key enzyme in folate biosynthesis and utilization. Antifolates, such as pyrimethamine (Pyr), targeting dihydrofolate reductase-thymidylate synthase (DHFR-TS) of the parasite, have been exploited against chloroquine-resistant Plasmodium falciparum, especially as components of combination drugs (2, 9). Nonetheless, these drugs are not recommended for P. vivax treatment due to the preliminary observation that antifolates were ineffective and that the parasite is inherently resistant against them owing to predisposed mutations in the dhfr-ts gene (18, 26). Recently, point mutations of DHFR were revealed to have an association with antifolate resistance in P. vivax in vitro (6, 8, 10, 13), leading to the conclusion that P. vivax is initially sensitive to antifolates, and resistance developed through mutations, similar to the case of P. falciparum. This is an important finding on the molecular basis for drug resistance in P. vivax that gives rise to opportunities for effective drug design for P. vivax therapy.
Several different methods for assessing antimalarial drug sensitivity have been developed (17). These methods mostly rely on culturing malaria parasites (16, 19, 25). Unlike the case for P. falciparum, an in vitro inhibitor susceptibility test for P. vivax is difficult because of the lack of a continuous in vitro culture for this parasite. Although an in vivo assay using rhesus monkeys has been used for drug sensitivity testing for P. vivax, this method is expensive and is not practical (21). In order to facilitate drug screening for P. vivax, we have modified a bacterial complementation system, previously described for selecting drug-resistant P. falciparum DHFR (PfDHFR) mutants generated from error-prone PCR (5), to determine the inhibitor efficacy of a Pyr library against bacteria expressing full-length P. vivax DHFR-TS (PvDHFR-TS) of either wild-type (WT) or S58R S117N (SP21) double mutant enzymes. Furthermore, the results from the bacterial complementation system are compared with the inhibition values obtained from the corresponding target enzyme assay. Highly potent inhibitors are identified as candidates for further lead development and optimization.
MATERIALS AND METHODS
Plasmid construction.
The gene encoding bifunctional PvDHFR-TS was PCR amplified from genomic DNA of P. vivax, generously provided by M. Imwong and S. Pukrittayakamee, in two steps (13). First, oligonucleotide primers, 5′vdrf (5′ATGGAGGACCTTTCAGATGTATTTGACATT3′) and 3′vtsr (5′GGCGGCCATCTCC ATGGTTATTTTATCGTG3′), were used to amplify the pvdhfr-ts sequence. The amplification reaction was set up in a total volume of 50 μl, containing 200 ng genomic template DNA, 2 mM MgSO4, 200 μM (each) deoxynucleoside triphosphates, and 1.5 U of Pfu polymerase. The PCR was performed for 32 cycles: the first cycle at 94°C for 5 min; the subsequent 30 cycles at 94°C for 1 min, 64°C for 2 min, and 72°C for 2 min; and the final cycle at 94°C for 1 min, 64°C for 2 min, and 72°C for 15 min. The obtained product was used as a template for the second PCR step. The primers used in the second PCR were 5′pvdhfr (5′AAGAATTCATATGGAGGACCTTTCAGA3′) and 3′pvdhfrts (5′TATCTCGAGAAGCTTCTTAGGCGGCCATC3′), containing NdeI and HindIII restriction sites, respectively, as underlined. The PCR (50 μl) was performed similarly to the first reaction, but the annealing condition was set at 48°C for 1 min. The obtained 1.8-kb amplified product was cloned into NdeI and HindIII sites of pET17b to yield pETpvDHFR-TS.
A similar protocol was adopted for construction of pETpvSP21 with the S58R S117N double mutant.
Complementation.
Plasmids pET17b (Novagen), pETpfTM4 (harboring the WT pfdhfr-ts gene [4]), and pETpfK1 (harboring the C59R S108N mutation [4]) were individually transformed into BL21(DE3) bacteria, while pETpvDHFR-TS and pETpvSP21 were individually transformed into BL21(DE3)pLysS bacteria. BL21(DE3) carrying plasmid was grown on LB agar supplemented with 100 μg ml−1 ampicillin, whereas BL21(DE3)pLysS-transformed cells were grown on LB agar supplemented with 100 μg ml−1 ampicillin and 34 μg ml−1 chloramphenicol. In order to test complementation, cells obtained after transformation were grown on minimal medium (MM) in the absence or presence of 4 μM trimethoprim (Tmp) at 37°C overnight in addition to the antibiotics required to maintain the acquired plasmids.
Inhibitor screening using bacterial system.
Nineteen Pyr analogs were studied for their inhibition activity against cells expressing either WT or SP21 mutant PvDHFR-TS. The structures of these compounds are shown in Table 2. All compounds were maintained at −20°C as 50 mM stock solutions in dimethyl sulfoxide for assay of Escherichia coli bacterial growth in liquid culture. The compounds were diluted to appropriate concentrations in liquid culture media. The assays were conducted with 96-well microplates by monitoring the growth at an optical density of 595 nm (A595). Each compound was assayed in triplicate and tested in at least three separate experiments. Each well contained 180 μl of precultured cells in MM containing 100 μg ml−1 ampicillin, 34 μg ml−1 chloramphenicol, and 4 μM Tmp, precultured cells at the final A595 of 0.07, and 20 μl of drug solution. In the case of BL21(DE3) for expression of PfDHFR-TSs, chloramphenicol was excluded. The final concentration of dimethyl sulfoxide was kept constant at 0.1%. Cultures were grown for approximately 6 h, where the control bacterial cells reached the late log phase. The A595 value for each well was determined by a microplate reader (iEMS reader; Labsystems, Finland). The average A595 of control culture omitting Pyr analogs was scored as 100% growth, and the average readings for the cultures at each drug concentration were divided by this value to obtain relative growth values. The concentrations that inhibited 50% bacterial growth (IC50s) were determined from dose-response curves.
TABLE 2.
Structures of inhibitors and their efficacies against PvDHFR-TSs of WT and SP21 determined by bacterial complementation assay (IC50) and enzyme inhibition assay (Ki)
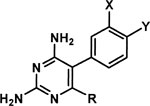
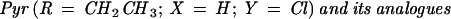
| Compound tested | C-5 substituenta | 6-Ra | Inhibition
|
|||
|---|---|---|---|---|---|---|
| IC50 (μM) for E. coli with PvDHFR-TS | Ki (nM) for WT PvDHFR-TS | IC50 (μM) for E. coli with SP21 | Ki (nM) for SP21 | |||
| Pyr (P1) | p-ClPh | -Et | 0.18 ± 0.05 | 0.21 ± 0.03 | 20.01 ± 5.41 | 3.04 ± 0.44 |
| P25 | p-ClPh | -H | 1.52 ± 0.39 | 3.42 ± 0.28 | >50 | 1,075.93 ± 50 |
| P37 | p-ClPh | -Me | 1.95 ± 0.34 | 0.78 ± 0.15 | >50 | 63.40 ± 6.88 |
| P3 | p-ClPh | -Bui | 0.22 ± 0.05 | 1.40 ± 0.17 | 18.22 ± 3.70 | 28.07 ± 0.58 |
| P8 | p-ClPh | -(CH2)8CH3 | 2.20 ± 0.37 | 4.29 ± 0.20 | >50 | 24.47 ± 2.80 |
| P16 | p-ClPh | -(CH2)3COOMe | 0.20 ± 0.05 | 0.69 ± 0.05 | >50 | 7.43 ± 0.29 |
| P17 | p-MePh | -Et | 0.14 ± 0.03 | 0.39 ± 0.051 | 12.40 ± 3.51 | 2.21 ± 0.21 |
| P18 | p-OMePh | -Et | 0.35 ± 0.03 | 1.01 ± 0.17 | >50 | 37.80 ± 3.33 |
| P21 | p-BrPh | -Et | 0.09 ± 0.02 | 0.78 ± 0.09 | 30.05 ± 7.21 | 6.59 ± 0.49 |
| P23 | p-tBuPh | -Et | 1.76 ± 0.37 | 1.14 ± 0.19 | >50 | 47.49 ± 3.49 |
| P13 | m,p-(Cl)2Ph | -Et | 0.07 ± 0.01 | 0.52 ± 0.11 | 10.45 ± 2.88 | 1.05 ± 0.07 |
| P30 | m-ClPh | -Et | 0.36 ± 0.10 | 0.31 ± 0.07 | 0.30 ± 0.08 | 0.54 ± 0.04 |
| P38 | m-ClPh | -Me | 2.12 ± 0.58 | 0.69 ± 0.07 | 1.98 ± 0.41 | 1.19 ± 0.12 |
| P31 | m-ClPh | -(CH2)3Ph | 2.38 ± 0.49 | 0.77 ± 0.05 | 13.89 ± 1.95 | 1.32 ± 0.36 |
| P40 | m-ClPh | -(CH2)2O(CH2)3OPh | 2.27 ± 0.65 | 0.63 ± 0.06 | 14.18 ± 3.57 | 1.53 ± 0.17 |
| P20 | Ph | -Et | 1.82 ± 0.46 | 0.03 ± 0.006 | 1.52 ± 0.39 | 0.08 ± 0.001 |
| P36 | Ph | -Me | 11.33 ± 3.33 | 4.18 ± 0.29 | 12.85 ± 1.27 | 22.47 ± 2.05 |
| P39 | Ph | -(CH2)5CH3 | 0.23 ± 0.06 | ND | 3.02 ± 0.61 | 0.90 ± 0.21 |
| P33 | Ph | -(CH2)3Ph | 0.80 ± 0.12 | 0.53 ± 0.06 | 2.76 ± 0.34 | 0.81 ± 0.14 |
Ph, phenyl; Et, ethyl; Bui, iso-butyl; tBu, tertiary-butyl; Me, methyl.
Enzyme preparation and antifolate inhibition assay.
The WT and mutant PvDHFR-TS enzymes were expressed in E. coli BL21(DE3)pLysS and purified using a methotrexate-Sepharose column according to previously described methods (5, 13). The methods used for determination of DHFR activities and for the study of inhibition by antifolates were similar to that previously described (13). Inhibition constants (Ki) of antifolates were calculated using a nonlinear least-square fit of the data to the Michaelis-Menten equation, assuming the inhibitor binds competitively to the enzyme active site.
RESULTS
Bacterial complementation by malarial DHFR-TSs.
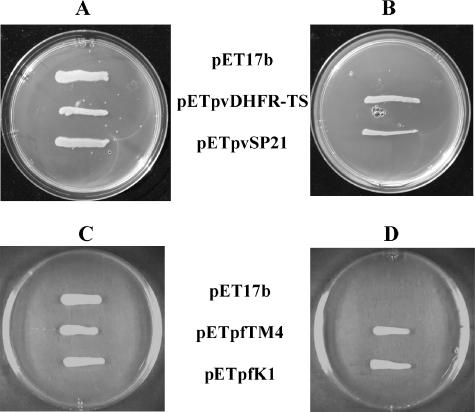
Prior to assessing compounds' efficacies against the bacteria expressing plasmodial DHFR-TSs, the validity of using growth complementation by parasite DHFR-TSs as the basis for inhibitor screening was determined. Besides the PvDHFR-TS wild-type enzyme, the SP21 double mutant carrying S58R and S117N was included, since this mutant is the prevalent mutant found in the field. We also tested the PfDHFR-TS wild type (TM4) and the equivalent C59R S108N double-mutant homolog (K1) for comparison. Since the BL21(DE3)pLysS strain contains its own DHFR, Tmp, a known inhibitor specific to bacterial DHFR, was included to eliminate the endogenous DHFR that can interfere with the testing system. In this scenario, cell survival is based solely on the ability of the expressed plasmodial DHFR-TSs to complement and rescue the growth. The results showed that only transformed BL21(DE3)pLysS cells expressing either WT or SP21 PvDHFR-TS were able to grow (Fig. 1A and B), while host cells carrying the pET17b vector could not survive in the presence of Tmp. In order to complement the growth by PfDHFR-TSs, the BL21(DE3) strain without pLysS encoding the T7 lysozyme was required (Fig. 1C and D). The results indicate that DHFR-TS constructs were fully functioning in the chemically knocked out host cells but PvDHFR-TSs were expressed at higher levels than PfDHFR-TS. The concentration of Tmp at 4 μM was chosen in the screening experiment even though the bacterial DHFR can be inhibited at lower concentrations on MM agar to ensure the need for complementation by parasite DHFR-TS.
FIG. 1.
BL21(DE3)pLysS complemented with pETpvDHFR-TS and pETpvSP21 (A and B) selected in MM containing 100 μg ml−1 ampicillin and 34 μg ml−1 chloramphenicol and BL21(DE3) complemented with pETpfTM4 and pETpfK1 (C and D) grown in MM containing 100 μg ml−1 ampicillin. Cultures were grown in the absence (A and C) or presence (B and D) of 4 μM Tmp. pET17b indicates cell containing vector without the Plasmodium gene.
Application of bacterial complementation as an antifolate antimalarial screening.
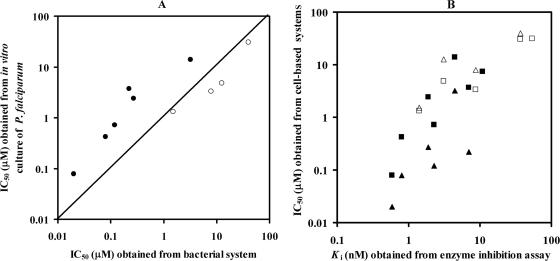
The validity of the above bacterial complementation system for determining the biological activities of potential antifolate inhibitors was assessed through the comparison of the inhibition profile obtained from the bacteria expressing PfDHFR-TS and that from the in vitro P. falciparum assay. Since the dhfr-ts gene from P. falciparum is expressed in bacteria whose endogenous DHFR activity is inhibited by Tmp, the susceptibilities to antifolates displayed from the complemented bacteria reflect the susceptibilities of the Plasmodium dhfr-ts alleles. In this study, Pyr analogs (P1, P30, P38, P20, P36, P34, and P64) with diverse antifolate activities (11, 24) were chosen as model compounds for screening using bacteria expressing WT or C59R S108N PfDHFR-TS enzymes in a 96-well plate format. The response of each bacterial construct to the Pyr analogs was measured by its relative growth in the presenpce of the analogs at 0 to 50 μM concentrations. The results were then compared with their antiplasmodial activity (IC50) and enzyme inhibition constant (Ki) values reported previously by our laboratory (11, 24) (Table 1). As shown in Fig. 2A, IC50s obtained from bacterial and parasite systems were well correlated (r2 = 0.82). The IC50s obtained from both systems were also well correlated with the Ki values against PfDHFR (r2 = 0.89 and 0.85 for the bacterial and parasite systems, respectively) (Fig. 2B), although the IC50s were in micromolar and the Ki values were in nanomolar scales. In detail, the results show that bacteria complemented with WT PfDHFR-TS behaved similarly to the TM4 parasite, which is sensitive to Pyr (parent Pyr, P1). Consistently, bacteria carrying C59R S108N mutant PfDHFR-TS responded poorly to this compound, similarly to the K1 parasite, which carries these double mutations. A similar pattern was also observed for other selected analogs. It should be noted that although the absolute IC50s are different between the two systems, the results are quite consistent, suggesting that the bacterial system reflects the Plasmodium system in general. From the aforementioned observation, we therefore adopted the bacterial complementation system to screen for effective Pyr analogs against DHFR-TS of P. vivax for which the long-term in vitro culture is not available.
TABLE 1.
Comparisons of inhibitor efficacies of pyrimethamine analogs against WT and C59R S108N double-mutant PfDHFR-TS, determined by bacterial complementation (IC50), in vitro parasite culture assay (IC50), and PfDHFR enzyme (Ki) inhibition screening methodsa
| Compound tested | Inhibition of wild type
|
Inhibition of mutant (C59R S108N)
|
||||
|---|---|---|---|---|---|---|
| IC50 (μM) for E. coli | IC50 (μM) for TM4* | Ki (nM) for PfDHFR* | IC50 (μM) for E. coli | IC50 (μM) for K1* | Ki (nM) for PfDHFR* | |
| Pyr (P1) | 0.02 ± 0.004 | 0.08 ± 0.01 | 0.6 ± 0.2 | >50 | 30.9 ± 8.4 | 53.9 ± 6.5 |
| P30 | 0.08 ± 0.02 | 0.4 ± 0.1 | 0.8 ± 0.1 | 1.51 ± 0.45 | 1.3 ± 0.5 | 1.4 ± 0.0 |
| P38 | 0.27 ± 0.06 | 2.4 ± 1.1 | 1.9 ± 0.5 | 12.48 ± 2.56 | 4.8 ± 0.7 | 3.1 ± 0.3 |
| P20 | 0.12 ± 0.04 | 0.7 ± 0.24 | 2.3 ± 0.3 | 7.88 ± 1.67 | 3.3 ± 0.4 | 8.7 ± 0.2 |
| P34 | 0.22 ± 0.04 | 3.7 ± 0.8 | 7.0 ± 1.0 | >50 | 17.4 ± 3.1 | 534.5 ± 65.2 |
| P36 | ND | 7.4 ± 2.6 | 10.9 ± 1.5 | 39.69 ± 8.41 | 30.8 ± 6.5 | 36.7 ± 4.3 |
| P64 | 3.19 ± 0.80 | 13.9 ± 1.9 | 4.5 ± 0.3 | >50 | >50 | 14.2 ± 2.0 |
FIG. 2.
Graphics show inhibition of Pyr analogs. (A) Correlation of IC50s derived from bacterial complementation assay and in vitro culture of P. falciparum assay. (B) Relationship between IC50s obtained from cell-based assay systems (either parasite [square] or bacteria [triangle]) and the Ki values from the in vitro enzyme inhibition assay. Closed and open symbols are data points derived from the wild type and the mutant, respectively (data with Ki values and IC50s greater than 60 nM and 50 μM were excluded from the plot). Each data point is the average from at least three independent experiments (each performed in triplicate).
Enzyme inhibition and antimalarial activity against the surrogate host by the Pyr library.
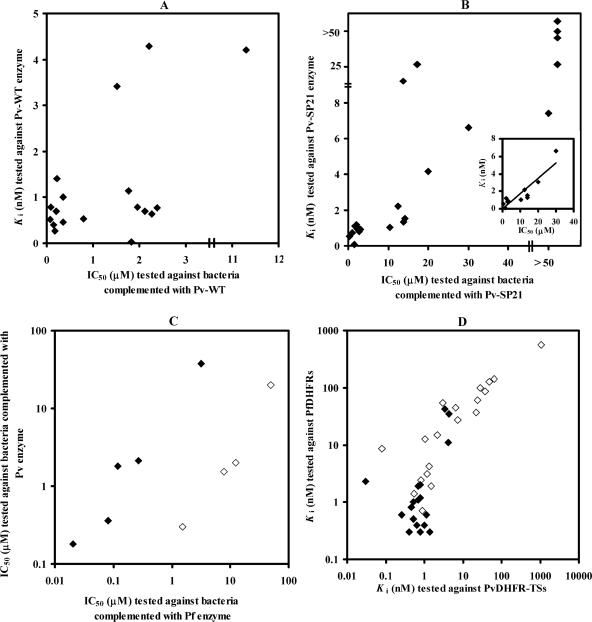
Nineteen analogs from a library of 2,4-diaminopyrimidine (Pyr) derivatives previously developed against P. falciparum (11, 24) were tested for their antimalarial activities (IC50) against bacteria expressing WT and SP21 PvDHFR-TS. At the same time, Ki values of these compounds were determined against corresponding purified recombinant PvDHFR-TS enzymes. Similar to the results from the P. falciparum system, the IC50s of these compounds against P. vivax were in the micromolar range, whereas the compounds inhibited the target enzyme (Ki values) at the nanomolar level (11) (Table 2). The results showed that all except P36 inhibited bacteria carrying WT pvdhfr-ts with IC50s of less than 5 μM. The Ki values of the compounds tested, except P25, P8, and P36, are in the range of ≤2 nM and correspond well with those IC50s. The inhibition profile for the SP21 mutant reveals that P20, P30, P33, P38, and P39 are potent inhibitors for both enzyme- and cell-based assays in which the Ki values and IC50s are lower than 1.2 nM and 3 μM, respectively. In contrast, compounds P8, P16, P18, P23, P25, and P37 yielded Ki values and IC50s greater than 7 nM and 50 μM and are therefore classified as poor inhibitors for the SP21 mutant. It is noted that, unlike the case for the WT, SP21 is less sensitive to analogs with substituents at the para position of the phenyl ring. While compounds P13, P17, P31, and P40 inhibited the PvDHFR-TS-SP21 enzyme strongly, with Ki values of about 2 nM, they only moderately inhibited the bacteria bearing SP21 (IC50, ∼10 μM). When the IC50s and Ki values of each compound were plotted, a linear relationship (r2 = 0.81) was observed for the SP21 mutant (Fig. 3B), whereas in the case of the wild type, most data points were clustered around an IC50 of 3 μM and a Ki value of 2 nM (Fig. 3A). The IC50s for inhibition of growth of bacteria bearing PfDHFR-TSs are correlated with those bearing PvDHFR-TSs for both WTs (r2 = 0.9) and double mutants (r2 = 0.9) (Fig. 3C). Linear correlations were also obtained for Ki values of PfDHFR and PvDHFR-TS enzymes (r2 = 0.9) (Fig. 3D).
FIG. 3.
Inhibition profile of Pyr library. Panels A and B show relationships of IC50s derived from the bacterial assay system and Ki values from the in vitro enzyme assay of WT PvDHFR-TS and the SP21 mutant, respectively. Inset (B) shows the data used for calculating r2 (data with Ki and IC50 greater than 7 nM and 50 μM were excluded from calculation). (C) Relationship of IC50s between bacteria expressing DHFR-TS of P. falciparum or P. vivax. (D) Relationship of Ki values tested against PvDHFR-TS and PfDHFR enzymes. For C and D, closed and open symbols are data points derived from wild types and mutants (SP21 for P. vivax and C59R S108N for P. falciparum), respectively. Each data point is the average from at least three independent experiments (each performed in triplicate). Pf, P. falciparum; Pv, P. vivax.
DISCUSSION
The lack of a continuous P. vivax culture and the difficulties associated with the use of an animal model have impeded the development of new antimalarials against P. vivax malaria. It is desirable to have a heterologous system that would simulate the parasite so as to allow convenient inhibitor screening. This study demonstrates the use of a bacterial complementation system as another approach to assess the efficacies of antifolates against P. vivax, which enables us to identify leads to be modified that alter the level of inhibition against PvDHFR-TS.
The bacterial model presented in this study is BL21(DE3)pLysS, a widely used expression host. Arguably, the DHFR-disrupted host, like E. coli PA414 (1), might be a better system, since the endogenous DHFR activity is absent and Tmp can be left out. However, that system did not give dependable results in our laboratory, and a small amount of thymidine is usually required in the culture, affecting the observed IC50. Studies using yeast as a heterologous system for antifolate screening have been described. However, inhibitor responses between the pathogen and yeast systems were not compared in those reports (7, 14, 22). It has been suggested that the proper heterologous host for antifolate screening should express only an amount of enzyme sufficient to support host cell growth, since a high level of expression is likely to increase the inhibitor necessary to inhibit host cell growth. Therefore, pLysS encoding the T7 lysozyme to diminish T7 RNA polymerase was supplied in the case of antifolate screening against vivax DHFR-TS.
As a surveillance tool, bacterial complementation has many advantages. The assay is very simple and rapid, where drug sensitivity determination can be accomplished within a day. In this study, the assay was performed with a 96-well titer plate, which can be automated and processed in a high-throughput format. The assay requires only media and basic laboratory equipment to support and follow cell growth, making the system relatively cheap and suitable for initial screening. The system can be easily adapted for the assessment of the relative sensitivities of various Plasmodium DHFR alleles against an antifolate inhibitor. In addition, the ease of genetic transformation in E. coli makes it suitable for screening DHFR inhibitors of similar pathogens in general.
It should be noted that our method may reflect only the interaction of the inhibitor and the target protein in a surrogate system and may not parallel an assay based on the parasite, since various factors, such as membrane transport, drug metabolism, and access to the target, are different for the two organisms. The differences in the Ki values and IC50s likely reflect the limitations imposed by these factors. However, a linear relationship was observed, indicating that the inhibitors can penetrate the cell, find the PvDHFR target, and exert a killing effect on the cell through disruption of folate metabolism. The fact that different linear Ki-IC50 relationships were obtained for the WTs and double mutants probably reflects the minor differences of the components between the two systems used. However, it is clear that the bacterial surrogate system works for the P. vivax DHFR inhibitors as well as those for P. falciparum and will therefore be useful in primary cell-based screening activities.
In this study, the full-length DHFR-TS construct, rather than the DHFR construct alone, was used, since the inhibitory properties of the compounds might be different for DHFR alone than for DHFR-TS. Since the inhibitors were designed to target malarial DHFR, the relationship between the bacterium- and enzyme-based screenings was examined. The assay results obtained from both enzyme inhibition and bacterial complementation assay systems support the idea of the association of point mutations with antifolate resistance in P. vivax, as indicated by higher Ki values and IC50s for the SP21 mutant tested against parental Pyr (P1). In this study, different derivatives of 2,4-diaminopyrimidine were screened for their inhibitor characteristics in order to identify inhibitors against P. vivax for further optimization. Inhibitors with p-substituent at C-5 of 2,4-diaminopyrimidine (P1, P13, P16, P17, P18, P21, and P23) are effective against the WT enzyme; however, they are less effective against the SP21 mutant. Compounds with bulky substituents at the p position (P18 and P23) also resulted in high Ki values against the double-mutant enzymes. These findings support the conclusion that the active sites of the PvDHFR and PfDHFR domains share similar features and indicate a steric clash between the p-substituent group of the inhibitors and Asn at position 117 in the PvDHFR domain, which was similar to, but stronger than, Asn at position 108 of the PfDHFR mutant (15, 27). Compounds either with no substituent on the phenyl ring (P20, P33, and P39) or with the substituent at other positions (P30 and P38), however, showed improvement in inhibitor efficacy toward the SP21 mutant by a factor of 2 to 38 for Ki values and 6 to 60 for IC50s.
These results are supported by the recently published crystal structures of WT and SP21 PvDHFRs in complex with P1 and P20 (12), where a large protein conformational change and inhibitor displacement were observed for the SP21-P1 complex compared to WT-P1, rendering resistance to the drug. Optimal conformation of the protein and favorable interactions of the inhibitor and protein were recovered upon the binding of SP21 with P20, which retains good activity against this mutant. The nature of the substituent (R) at position six of the pyrimidine ring is anticipated to influence the binding affinity of the inhibitor to the DHFRs. Finally, 5-m-substituted-phenyl analogues with a small 6-substituent (ethyl) showed more favorable binding to the mutant enzyme than longer 6-alkyl substituents, whereas this effect is rather small for 5-phenyl analogues (Table 2).
It is worth noting that although this series of compounds was previously developed against P. falciparum, several compounds exhibited similar action toward PvDHFR-TS (Fig. 3C and D). These compounds are therefore suitable for further selection as candidate agents against mixed infections, which are quite common in many areas of endemicity. In summary, this study introduced the simple bacterial complementation system for antifolate screening against P. vivax. Insight into the strategies for developing new inhibitors effective against the SP21 mutant was provided, which should benefit rational drug design of effective compounds targeted against antifolate-resistant parasites. The system is also useful for P. falciparum and has been validated against parasite-based assays. A similar system may also be generally valid for screening of other similar pathogens.
Acknowledgments
This research was supported by EU, the UNICEF/UNDP/World Bank/WHO Special Programme on Tropical Diseases, The Wellcome Trust, Medicines for Malaria Venture (MMV), and the Target Research Unit Network of Thailand-TDR program.
We thank M. Imwong and S. Pukrittayakamee from Mahidol University for P. vivax's genomic DNA. We are grateful to D. Kongkasuriyachai of BIOTEC for critically reviewing the manuscript. S.K. is a Research Fellow of the Howard Hughes Medical Institute (HHMI).
Footnotes
Published ahead of print on 5 September 2006.
REFERENCES
- 1.Ahrweiler, P. M., and C. Frieden. 1988. Construction of a fol mutant strain of Escherichia coli for use in dihydrofolate reductase mutagenesis experiments. J. Bacteriol. 170:3301-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, A. C. 2005. Targeting DHFR in parasitic protozoa. Drug Discov. Today 10:121-128. [DOI] [PubMed] [Google Scholar]
- 3.Baird, J. K. 2004. Chloroquine resistance in Plasmodium vivax. Antimicrob. Agents Chemother. 48:4075-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chitnumsub, P., J. Yavaniyama, J. Vanichtanankul, S. Kamchonwongpaisan, M. D. Walkinshaw, and Y. Yuthavong. 2004. Characterization, crystallization and preliminary X-ray analysis of bifunctional dihydrofolate reductase-thymidylate synthase from Plasmodium falciparum. Acta Crystallogr. D Biol. Crystallogr. 60:780-783. [DOI] [PubMed] [Google Scholar]
- 5.Chusacultanachai, S., P. Thiensathit, B. Tarnchompoo, W. Sirawaraporn, and Y. Yuthavong. 2002. Novel antifolate resistant mutations of Plasmodium falciparum dihydrofolate reductase selected in Escherichia coli. Mol. Biochem. Parasitol. 120:61-72. [DOI] [PubMed] [Google Scholar]
- 6.de Pécoulas, P. E., R. Tahar, T. Ouatas, A. Mazabraud, and L. K. Basco. 1998. Sequence variations in the Plasmodium vivax dihydrofolate reductase-thymidylate synthase gene and their relationship with pyrimethamine resistance. Mol. Biochem. Parasitol. 92:265-273. [DOI] [PubMed] [Google Scholar]
- 7.Gerum, A. B., J. E. Ulmer, D. P. Jacobus, N. P. Jensen, D. R. Sherman, and C. H. Sibley. 2002. Novel Saccharomyces cerevisiae screen identifies WR99210 analogues that inhibit Mycobacterium tuberculosis dihydrofolate reductase. Antimicrob. Agents Chemother. 46:3362-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hastings, M. D., and C. H. Sibley. 2002. Pyrimethamine and WR99210 exert opposing selection on dihydrofolate reductase from Plasmodium vivax. Proc. Natl. Acad. Sci. USA 99:13137-13141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hyde, J. E. 2002. Mechanisms of resistance of Plasmodium falciparum to antimalarial drugs. Microbes Infect. 4:165-174. [DOI] [PubMed] [Google Scholar]
- 10.Imwong, M., S. Pukrittayakamee, L. Rénia, F. Letourneur, J.-P. Charlieu, U. Leartsakulpanich, S. Looareesuwan, N. J. White, and G. Snounou. 2003. Novel point mutations in the dihydrofolate reductase gene of Plasmodium vivax: evidence for sequential selection by drug pressure. Antimicrob. Agents Chemother. 47:1514-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamchonwongpaisan, S., R. Quarrell, N. Charoensetakul, R. Ponsinet, T. Vilaivan, J. Vanichtanankul, B. Tarnchompoo, W. Sirawaraporn, G. Lowe, and Y. Yuthavong. 2004. Inhibitors of multiple mutants of Plasmodium falciparum dihydrofolate reductase and their antimalarial activities. J. Med. Chem. 47:673-680. [DOI] [PubMed] [Google Scholar]
- 12.Kongsaeree, P., P. Khongsuk, U. Leartsakulpanich, P. Chitnumsub, B. Tarnchompoo, M. D. Walkinshaw, and Y. Yuthavong. 2005. Crystal structure of dihydrofolate reductase from Plasmodium vivax: pyrimethamine displacement linked with mutation-induced resistance. Proc. Natl. Acad. Sci. USA 102:13046-13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leartsakulpanich, U., M. Imwong, S. Pukrittayakamee, N. J. White, G. Snounou, W. Sirawaraporn, and Y. Yuthavong. 2002. Molecular characterization of dihydrofolate reductase in relation to antifolate resistance in Plasmodium vivax. Mol. Biochem. Parasitol. 119:63-73. [DOI] [PubMed] [Google Scholar]
- 14.Ma, L., Q. Jia, and J. A. Kovacs. 2002. Development of a yeast assay for rapid screening of inhibitors of human-derived Pneumocystis carinii dihydrofolate reductase. Antimicrob. Agents Chemother. 46:3101-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKie, J. H., K. T. Douglas, C. Chan, S. A. Roser, R. Yates, M. Read, J. E. Hyde, M. J. Dascombe, Y. Yuthavong, and W. Sirawaraporn. 1998. Rational drug design approach for overcoming drug resistance: application to pyrimethamine resistance in malaria. J. Med. Chem. 41:1367-1370. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen-Dinh, P., and W. Trager. 1980. Plasmodium falciparum in vitro: determination of chloroquine sensitivity of three new strains by a modified 48-hour test. Am. J. Trop. Med. Hyg. 29:339-342. [DOI] [PubMed] [Google Scholar]
- 17.Noedl, H., C. Wongsrichanalai, and W. H. Wernsdorfer. 2003. Malaria drug-sensitivity testing: new assays, new perspectives. Trends Parasitol. 19:175-181. [DOI] [PubMed] [Google Scholar]
- 18.Peters, W. 1998. Drug resistance in malaria parasites of animals and man. Adv. Parasitol. 41:1-62. [DOI] [PubMed] [Google Scholar]
- 19.Richards, W. H., and B. K. Maples. 1979. Studies on Plasmodium falciparum in continuous cultivation. I. The effect of chloroquine and pyrimethamine on parasite growth and viability. Ann. Trop. Med. Parasitol. 73:99-108. [PubMed] [Google Scholar]
- 20.Ruebush, T. K., II, J. Zegarra, J. Cairo, E. M. Andersen, M. Green, D. R. Pillai, W. Marquino, M. Huilca, E. Arévalo, C. Garcia, L. Solary, and K. C. Kain. 2003. Chloroquine-resistant Plasmodium vivax malaria in Peru. Am. J. Trop. Med. Hyg. 69:548-552. [PubMed] [Google Scholar]
- 21.Russell, B. M., R. Udomsangpetch, K. H. Rieckmann, B. M. Kotecka, R. E. Coleman, and J. Sattabongkot. 2003. Simple in vitro assay for determining the sensitivity of Plasmodium vivax isolates from fresh human blood to antimalarials in areas where P. vivax is endemic. Antimicrob. Agents Chemother. 47:170-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sibley, C. H., and I. Macreadie. 2001. Novel approaches to tackling malarial drug resistance using yeast. IUBMB Life 52:285-289. [DOI] [PubMed] [Google Scholar]
- 23.Sina, B. 2002. Focus on Plasmodium vivax. Trends Parasitol. 18:287-289. [DOI] [PubMed] [Google Scholar]
- 24.Tarnchompoo, B., C. Sirichaiwat, W. Phupong, C. Intaraudom, W. Sirawaraporn, S. Kamchonwongpaisan, J. Vanichtanankul, Y. Thebtaranonth, and Y. Yuthavong. 2002. Development of 2,4-diaminopyrimidines as antimalarials based on inhibition of the S108N and C59R+S108N mutants of dihydrofolate reductase from pyrimethamine-resistant Plasmodium falciparum. J. Med. Chem. 45:1244-1252. [DOI] [PubMed] [Google Scholar]
- 25.World Health Organization. 1990. In vitro microtest (mark II) for the assessment of the response of Plasmodium falciparum to chloroquine, mefloquine, quinine, sulfadoxin/pyrimethamine and amodiaquine. WHO/MAP/87.2, revision 1. World Health Organization, Geneva, Switzerland.
- 26.Young, M. D., and R. W. Burgess. 1959. Pyrimethamine resistance in Plasmodium vivax malaria. Bull. W. H. O. 20:27-36. [PMC free article] [PubMed] [Google Scholar]
- 27.Yuvaniyama, J., P. Chitnumsub, S. Kamchonwongpaisan, J. Vanichtanankul, W. Sirawaraporn, P. Taylor, M. D. Walkinshaw, and Y. Yuthavong. 2003. Insights into antifolate resistance from malarial DHFR-TS structures. Nat. Struct. Biol. 10:357-365. [DOI] [PubMed] [Google Scholar]