Abstract
The objectives of these analyses were to assess the feasibility of the latest WHO recommendations (28-day follow-up with PCR genotyping) for the assessment of antimalarial drug efficacy in vivo and to examine how different statistical approaches affect results. We used individual-patient data from 13 studies of uncomplicated pediatric falciparum malaria conducted in sub-Saharan Africa, using chloroquine (CQ), sulfadoxine/pyrimethamine (SP), or amodiaquine (AQ). We assessed the use effectiveness and test performance of PCR genotyping in distinguishing recurrent infections. In analyzing data, we compared (i) the risk of failure on target days (days 14 and 28) by using Kaplan-Meier and per-protocol evaluable patient analyses, (ii) PCR-corrected results allowing (method 1) or excluding (method 2) new infections, (iii) and day 14 versus day 28 results. Of the 2,576 patients treated, 2,287 (89%) were evaluable on day 28. Of the 695 recurrences occurring post-day 14, 650 could be processed and 584 were resolved (PCR use effectiveness, 84%; test performance, 90%). The risks of failure on day 28 with Kaplan-Meier and evaluable-patient analyses tended to be generally close (except in smaller studies) because the numbers of dropouts were minimal, but attrition rates on day 28 were higher with the latter method. Method 2 yielded higher risks of failure than method 1. Extending observation to 28 days produced higher estimated risks of failure for SP and AQ but not for CQ (high failure rates by day 14). Results support the implementation of the current WHO protocol and favor analyzing PCR-corrected outcomes by Kaplan-Meier analysis (which allows for dropouts) and retaining new infections (which minimizes losses).
Malaria is one of the leading causes of morbidity and mortality worldwide (14). The spread of parasite resistance to first-line drugs adds to the burden of the disease (9). A contributing factor has been the continued use of failing drugs, partly because of the difficulty of assessing their efficacy in vivo. Methodological issues still being debated include how long patients should be monitored after treatment and whether clinical or parasitological outcomes should be given greater weight. Methods used so far differ greatly, making comparison and synthesis of data very difficult (16). The WHO has issued different sets of guidelines, shifting emphasis with time from parasitological (17) to clinical (18) assessment and more recently recommending that patients be assessed both parasitologically and clinically and monitored for 28 days if true failures are to be distinguished from new infections; it also makes provision for the use of life table analysis of results (19).
In areas of intense transmission, a second episode of malaria or a recurrent parasitemia during the drug-free follow-up period may be due to the same infection or a different infection (a recrudescence, thus a treatment failure, or a new infection, respectively). To distinguish these two events, polymorphic Plasmodium falciparum genes, such as the merozoite surface protein 1 and 2 genes (msp1 and msp2) and the glutamate-rich protein gene (glurp), can be genotyped by PCR (13). The duration of follow-up required to capture all true treatment failures will depend on a drug's residence time in the body (15) and the level of parasite sensitivity (21).
We analyzed data from 13 antimalarial drug studies with the standard single-agent first- and second-line drugs: chloroquine (CQ), sulfadoxine/pyrimethamine (SP), and amodiaquine (AQ). These studies enrolled 2,576 children in eight African countries from 2001 to 2004. The aims of this analysis were to (i) determine the feasibility of in vivo assessment of antimalarial drugs by using PCR genotyping for resolving recurrent infections (use effectiveness and test performance) and (ii) compare current and alternative methods of data analysis. The current recommendation is that failure rates should be calculated on target days (days 14 and 28) by using PCR-corrected outcomes after excluding both missing values and new infections (19). Specifically, we wanted to compare (i) treatment outcomes expressed as product limit estimates of failure (Kaplan-Meier) versus per-protocol evaluable-patient analyses on target days (days 14 and 28), (ii) analysis of PCR-corrected results allowing or excluding new infections, and (iii) day 14 versus day 28 results.
MATERIALS AND METHODS
In vivo study methods.
All 13 studies included in this analysis adhered to current recommendations (19), with a few minor exceptions (see Results). Briefly, the children, aged 6 to 59 months and with consenting caregivers, were eligible for enrolment if they had P. falciparum mono-infections (density threshold at entry varied with endemicity at the study site; lower threshold, 500, 1,000, or 2,000/μl; upper threshold, 100,000 or 200,000/μl), no signs of severity, no reported hypersensitivities to the study drug, and no serious concomitant febrile illnesses. We directly observed the intake of all doses (days 0, 1, and 2) and reassessed the children clinically and parasitologically on days 2, 3, 7, 14, 21, and 28 or any day in between in the event of illness. When asymptomatic parasitemia occurred after day 3, we monitored the children closely and administered a rescue antimalarial (quinine) to those who had developed fever or were parasitemic on day 28. All studies underwent ethical review and were approved by the relevant national authority.
We applied the WHO 2004 endpoint classification (Table 1). When late-clinical or parasitological failure occurred after day 14, we used PCR (2, 11, 12, 13) to compare the genotypic profiles of msp1 and msp2 for post- and pretreatment parasites (in Bundi Bugyo, Uganda, glurp was also analyzed due to the low allelic heterogeneity of local isolates). PCR analyses were conducted at Prince Leopold Institute of Tropical Medicine, Antwerp, Belgium (four sites, 37% of total samples); University of Glasgow, Glasgow, United Kingdom (three sites, 19% of total samples); Hôpital Bichat, Paris, France (two sites, 15% of total samples); Shoklo Malaria Research Unit, Mae Sod, Thailand (one site, 9% of total samples); Kenya Medical Research Institute, Nairobi, Kenya, and Malaria Research and Training Centre, Bamako, Mali (one site each, 8% of total samples); and Faculté de Médicine-Pharmacie, Rouen, France (one site, 4% of total samples). Methods were not standardized across these laboratories, which used different extraction methods, primers, and reaction conditions.
TABLE 1.
Efficacy endpoints for WHO classification
| Endpoint | Criteria
|
|
|---|---|---|
| Day 14 follow-up | Day 28 follow-up | |
| Early treatment failure | Parasite density on day 2 > that on day 0, parasite density on day 3 ≥ 25% that on day 0, fever in the presence of parasites on day 3, or severe malaria in the presence of parasites on days 1-3 | Parasite density on day 2 > that on day 0, parasite density on day 3 ≥ 25% that on day 0, fever in the presence of parasites on day 3, or severe malaria in the presence of parasites on days 1-3 |
| Late clinical failure | Fever in the presence of parasites between days 4 and 14 or severe malaria in the presence of parasites between days 4 and 14 | Fever in the presence of parasites between days 4 and 28 or severe malaria in the presence of parasites between days 4 and 28 |
| Late parasitological failure | Parasites without fever on day 14 | Parasites without fever on day 28 |
| ACPR | Follow-up completed without meeting any of the above criteria | Follow-up completed without meeting any of the above criteria |
Cases in which pre- and posttreatment genotypes were identical were considered failures; cases in which pre- and posttreatment genotypes were different were considered new infections; cases with mixed genotypes were classified as failures. It is generally agreed that, contrary to epidemiological studies, the persistence of pretreatment alleles after treatment indicates a failure to clear all parasites. In the studies, there was no provision (except in Bundi Bugyo; see above) for establishing the a priori diversity of the parasite population.
PCR genotyping.
For each study, we identified cases requiring PCR genotyping, i.e., children with a recurrence after day 14 (it is generally assumed that, for P. falciparum, recurrences on or before day 14 are recrudescences). These cases were classified as either (i) resolved by PCR and further categorized as recrudescences or new infections or (ii) unresolved by PCR, with the reason recorded (missing sample, failure to extract DNA, or result not interpretable). PCR genotyping was assessed in terms of (i) use effectiveness, defined as the number of cases resolved by PCR divided by the total number of children with recurrences after day 14 (the denominator includes missing samples), and (ii) test performance, defined as the number of cases resolved by PCR out of the total number of paired samples available for genotyping (the denominator does not include missing samples).
Failure rate (day 14 and day 28 evaluable-patient analysis) versus Kaplan-Meier analysis.
Failure rates at day 14 and day 28 were calculated using the method of analysis recommended by the WHO (18) and compared to the product limit estimates of failure obtained by Kaplan-Meier analysis. The WHO method takes into account only patients with an outcome on the target day (day 14 or day 28) (evaluable patients). The Kaplan-Meier method allows for dropouts (losses during follow-up and withdrawals). In our Kaplan-Meier analysis, the event measured was “failure” and the time to the event was the duration of the follow-up (either 28 days for patients generating adequate clinical and parasitological response [ACPR] at day 28, x days for patients who failed at day x of follow-up, or the number of days of follow-up until the last visit for patients lost during follow-up or excluded). For both methods, we calculated the number of patients included in the analysis and the risks of failure and compared these figures between the two methods.
Day 28 PCR-corrected results for two methods of analysis.
The WHO currently recommend calculation of failure rates on target days by using PCR-corrected outcomes after excluding both missing values and new infections (19).
We genotyped only recurrences occurring after day 14 and assumed all earlier failures to be recrudescences. Two methods of analysis were used. For method 1, all PCR-confirmed new infections were reclassified as successes (ACPR), with individuals unresolved by PCR excluded. For method 2, individuals with new infections or infections unresolved by PCR were excluded from the analysis, per the current WHO approach (19). For both methods, results are presented by drug and study site as probabilities of failure [1 − St, where St is probability of survival] on day 28 with 95% confidence intervals (CIs). Since recurrent episodes occurring on or before day 14 were not genotyped, for day 14, we report only crude (uncorrected) results (with no comparisons between data for methods 1 and 2).
Day 14 versus day 28 failures.
Estimates of risk of failure by days 14 and 28 were analyzed separately (Table 1), based on results obtained with method 1 and Kaplan-Meier analysis. Comparisons were expressed as absolute differences (summarized across sites as medians and ranges). Correlations between day 14 and 28 results were expressed as coefficients of correlation.
Data analysis was performed using Stata 8.2 (Stata Corp., College Station, Texas).
RESULTS
Study profiles.
The 13 studies analyzed children aged 6 to 59 months enrolled in 28 treatment arms in eight countries in Africa between February 2001 and July 2004. Malaria was hyperendemic in all sites except three (Table 2). All studies were performed according to the latest WHO recommendations, with minor deviations at some sites (the lower limit of parasitemia at entry was 1,000 parasites/μl instead of 2,000 for Bundi Bugyo; the upper limit of parasitemia at entry was 100,000 parasites/μl instead of 200,000 for Bundi Bugyo, Mapel, Caala, and Kuito; fever or history of fever in the previous 24 h instead of fever at entry was noted for Harper, Caala, and Bundi Bugyo; and there was primary exclusion of children with recent histories of antimalarial intake for Bundi Bugyo, Harper, Caala, and Kuito).
TABLE 2.
Profile of PCR genotyping of recurrent episodes of malaria during follow-up
| Site (drug[s])a | No. of recurrences (needing genotyping) | No. of unresolved cases (PCR results not available)
|
No. of resolved cases (PCR results available)
|
|||||
|---|---|---|---|---|---|---|---|---|
| Total (% recurrences) | Missing sample | No DNA | Not interpretable | Total (% recurrences) | Recrudescence | New infection | ||
| Angola, Caala (AQ, CQ, SP) | 70 | 13 (18.6) | 11 | 0 | 2 | 57 (81.4) | 44 | 13 |
| Angola, Kuito (AQ, SP) | 55 | 6 (10.9) | 4 | 2 | 0 | 49 (89.1) | 35 | 14 |
| Chad, Bongorb (Q, CQ, SP) | 38 | 16 (42.1) | 5 | 11 | 0 | 22 (57.9) | 11 | 11 |
| Liberia, Harper (AQ) | 27 | 4 (14.8) | 4 | 0 | 0 | 23 (85.2) | 11 | 12 |
| Mali, Koumantoub (CQ, SP) | 54 | 3 (5.6) | 3 | 0 | 0 | 51 (94.4) | 47 | 4 |
| DRC, Boende (AQ, SP) | 97 | 3 (3.1) | 1 | 1 | 1 | 94 (96.9) | 43 | 51 |
| Sierra L, Freetown (AQ) | 8 | 2 (25.0) | 2 | 0 | 0 | 6 (75.0) | 2 | 4 |
| Sierra L, Kabala (AQ, CQ, SP) | 48 | 4 (8.3) | 0 | 0 | 4 | 44 (91.7) | 19 | 25 |
| Sierra L, Kailahun (AQ, CQ, SP) | 84 | 27 (32.1) | 7 | 6 | 14 | 57 (67.9) | 36 | 21 |
| Sierra L, Makeni (AQ, CQ, SP) | 74 | 14 (18.9) | 4 | 0 | 10 | 60 (81.1) | 25 | 35 |
| Sierra L, Matru (AQ) | 23 | 0 | 0 | 0 | 0 | 23 (100.0) | 10 | 13 |
| S. Sudan, Mapel (CQ, SP) | 53 | 3 (5.7) | 1 | 0 | 2 | 50 (94.3) | 33 | 17 |
| Uganda, B. Bugyob (AQ, SP) | 64 | 16 (25.0) | 3 | 0 | 13 | 48 (75.0) | 18 | 30 |
| Total | 695 | 111 (16.0) | 45 (40.5%) | 20 (18.0%) | 46 (41.4%) | 584 (84.0) | 334 (57.2%) | 250 (42.8%) |
Sierra L, Sierra Leone; S. Sudan, South Sudan; B. Bugyo, Bundi Bugyo.
Malaria is hyperendemic at all sites except these, where it is meso-endemic.
Ten studies were compared: five studies (Angola Caala, Chad Bongor, Sierra Leone Kabala, Sierra Leone Kailahun, and Sierra Leone Makeni) compared all three drugs, three studies (Angola Kuito, DRC Boende, and Uganda Bundi Bugyo) compared AQ and SP, and two studies (Mali Koumantou and South Sudan Mapel) compared CQ and SP. Three studies used single-arm AQ treatment (Liberia Harper, Sierra Leone Freetown, and Sierra Leone Matru). All studies but two (Angola Caala and Mali Koumantou) were nonrandomized (patients enrolled in one treatment arm at a time).
Of the 9,610 patients screened (based on data available from 12 studies), 4,087 (42.3%) were slide positive; 2,576 (26.8%) were treated with CQ (n = 593), SP (n = 989), or AQ (n = 994) (see Table 3 for details); 112 (4.3%) patients were lost during follow-up (64 before day 14, 48 on or after day 14); and 177 (6.9%) were withdrawn (127 before day 14, 50 on or after day 14). The characteristics of these patients on inclusion are reported elsewhere (1, 3, 4-8, 10).
TABLE 3.
Estimated risk of failure for CQ, SP, and AQ on day 28, calculated by Kaplan-Meier analysis using method 1 (unresolved PCR excluded from the analysis) and method 2 (both unresolved PCR and new infections excluded)
| Drug | Sitea | % Day 28 failure (95% CI)
|
Differenceb | |
|---|---|---|---|---|
| Method 1 | Method 2 | |||
| CQ | Angola, Caala | 81.2 (70.8-89.6) | 82.8 (72.7-90.8) | +1.6 |
| Chad, Bongor | 23.7 (16.1-34.0) | 26.3 (18.0-37.4) | +2.6 | |
| Mali, Koumantou | 86.9 (79.5-92.6) | 89.6 (82.7-94.7) | +2.7 | |
| S.Leone, Kabala | 44.2 (30.2-61.2) | 52.8 (37.3-70.2) | +8.6 | |
| S.Leone, Kailahun | 87.5 (75.8-95.3) | 90.4 (79.4-96.9) | +2.9 | |
| S.Leone, Makeni | 71.2 (56.2-84.7) | 77.4 (62.6-89.5) | +6.2 | |
| S.Sudan, Mapel | 85.8 (76.3-92.8) | 96.5 (89.5-99.3) | +10.7 | |
| SP | Angola, Caala | 26.7 (18.3-37.9) | 27.8 (19.1-39.4) | +1.1 |
| Angola, Kuito | 34.9 (25.6-46.5) | 35.4 (25.9-47.0) | +0.5 | |
| Chad, Bongor | 20.8 (14.1-30.1) | 21.0 (14.3-30.4) | +0.2 | |
| Mali, Koumantou | 7.0 (3.4-14.1) | 7.0 (3.4-14.1) | 0% | |
| DRC, Boende | 39.0 (29.8-49.8) | 45.2 (35.0-56.8) | +6.2 | |
| S.Leone, Kabala | 20.6 (12.9-32.0) | 21.2 (13.2-32.8) | +0.6 | |
| S.Leone, Kailahun | 41.5 (31.7-53.0) | 42.7 (32.7-54.3) | +1.2 | |
| S.Leone, Makeni | 24.0 (16.5-34.2) | 26.1 (18.0-37.0) | +2.1 | |
| S.Sudan, Mapel | 15.6 (9.7-24.5) | 17.0 (10.6-26.6) | +1.4 | |
| Uganda, Bundi Bugyo | 39.3 (29.6-50.9) | 49.8 (38.3-62.6) | +10.5 | |
| AQ | Angola, Caala | 17.3 (10.4-27.9) | 18.8 (11.4-30.1) | +1.5 |
| Angola, Kuito | 22.2 (14.9-32.3) | 24.4 (16.4-35.2) | +2.2 | |
| Chad, Bongor | 8.4 (4.1-16.8) | 8.5 (4.1-17.0) | +0.1 | |
| Liberia, Harper | 18.3 (11.0-29.6) | 20.9 (12.6-33.3) | +2.6 | |
| DRC, Boende | 16.9 (10.5-26.5) | 22.7 (14.4-34.8) | +5.8 | |
| S.Leone, Freetown | 5.5 (1.8-16.1) | 5.9 (1.9-17.3) | +0.4 | |
| S.Leone, Kabala | 18.1 (10.7-29.9) | 21.1 (12.5-34.3) | +3.0 | |
| S.Leone, Kailahun | 27.9 (18.7-40.4) | 34.5 (23.5-48.8) | +6.6 | |
| S.Leone, Makeni | 4.8 (1.8-12.2) | 5.9 (2.2-14.9) | +1.1 | |
| S.Leone, Matru | 13.2 (7.3-23.2) | 15.6 (8.7-27.0) | +2.4 | |
| Uganda, Bundi Bugyo | 21.4 (13.0-34.0) | 24.8 (15.2-38.9) | +3.4 | |
S. Leone, Sierra Leone; S. Sudan, South Sudan.
Result for method 2 minus result for method 1.
PCR findings.
Among the total of 695 children with recurrent parasitemia after day 14, the question of recrudescence versus new infection could not be resolved for 111 (16.0%), either because at least one sample was missing (41%), because DNA could not be extracted (18%), or because the result was not interpretable (41%) (Table 2). Among the 584 children for whom this question could be resolved, 334 (57%) had recrudescences and 250 (43%) new infections. With the exception of those for Bundi Bugyo (Uganda), infections were polyclonal (six to eight alleles for msp1 and msp2). The use effectiveness of PCR was 84% (584/695), and its test performance was 90% (584/650). The median value for failure to genotype was 4% (range, 0 to 33). There was no obvious difference between laboratories in terms of test performance.
Risk of failure: per-protocol versus Kaplan-Meier analysis.
By day 14, 8% of the 2,576 children had been lost during follow-up or withdrawn and are not included in the estimation of risk of failure based on evaluable children, while with the Kaplan-Meier method, only children with no follow-up (i.e., lost at day 0) are not considered (1%) (Table 4) . By day 28, 11% of enrolled children had been lost during follow-up or withdrawn. After PCR correction, the loss from the initial sample size was 6% for Kaplan-Meier and 15% for day 28 evaluable-patient analysis (method 1); losses were 15 and 25%, respectively, after removing new infections (method 2).
TABLE 4.
Patient attrition with the various analysesa
| Day | Total no. of patients enrolled (% lost) | No. of patients enrolled (% lost) for indicated analysis and method
|
|||||
|---|---|---|---|---|---|---|---|
| Kaplan-Meier
|
Per-protocol
|
||||||
| Method 1 | Method 2 | No PCR genotyping | Method 1 | Method 2 | No PCR genotyping | ||
| 0 | 2,576 | ||||||
| 14 | 2,364 (8) | 2,544 (1) | 2,364 (8) | ||||
| 28 | 2,287 (11) | 2,433 (6) | 2,183 (15) | 2,176 (15) | 1,926 (25) | ||
Method 1 (cases unresolved by PCR excluded) and method 2 (cases unresolved by PCR and new infections both excluded) were applied to Kaplan-Meier and evaluability (per-protocol) data sets after PCR genotyping. No genotyping was done until day 14.
Figure 1 shows the differences in sample size (percent differences between data sets for Kaplan-Meier and evaluability analyses) and estimated risk of failure (risk difference) on day 28 (PCR corrected, with unresolved genotypes removed [method 1] and unresolved genotypes and new infections both removed [method 2]) for the two analyses. There were 256 more patients in the data set for the Kaplan-Meier analysis than in that for the evaluable-patient analysis (6%); proportionally, with the larger sample used for method 1, the difference between the results for the Kaplan-Meier and evaluable-patient analyses was smaller with method 1 than with method 2.
FIG. 1.
Comparison of Kaplan-Meier and evaluability (per-protocol) analyses (failure rates on day 28) using method 1 (PCR corrected, with cases unresolved by PCR excluded) (open symbols) and method 2 (PCR corrected, with cases unresolved by PCR and new infections both excluded) (closed symbols) by drug treatment.
Failure rates were lower with the Kaplan-Meier analysis and with method 2 in all studies but one (DRC Boende). Risk differences ranged from −2.3% to +0.9% for CQ, −1.8% to +2.3% for SP, and −1.8% to +1.2% for AQ with method 1 and −9.7% to 0% for CQ, −2.2% to +0.2% for SP, and −1.5% to 0% for AQ with method 2.
Day 28 results for two methods of PCR analysis.
Table 3 shows failure rates at day 28 estimated by Kaplan-Meier analysis with the two methods for analysis of PCR-corrected outcomes. Method 2 (unresolved genotypes and new infections removed) yields higher failure rates than method 1: +2.9 (range, +1.6 to +10.7) for CQ, +1.15 (range, 0 to +10.5) for SP, and +2.4 (range, +0.1 to +6.6) for AQ.
Day 14 and day 28 Kaplan-Meier failure estimates.
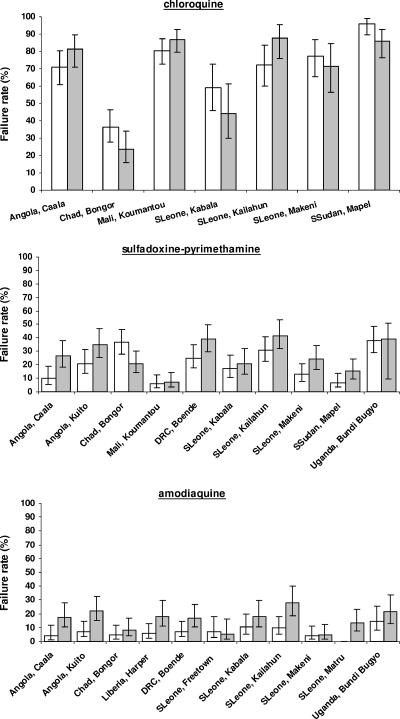
Because method 1 minimizes the number of patients lost in the analysis, we used it to compare risks of failure assessed at days 28 and 14 (Fig. 2); the median differences were −5.9% (range, −15.0 to +15.0) for CQ, +9.8% (range, −15.6 to +16.8) for SP, and +9.8% (range, −1.6 to +18.1) for AQ. The reason for the risk of failure being lower in some instances on day 28 than on day 14 is that we compared outcomes for the analyses of two different data sets (Table 1). Considering all studies, the coefficient of correlation was 0.94 between day 14 and day 28 results but was 0.89 for CQ, 0.67 for SP, and 0.50 for AQ.
FIG. 2.
Failure rates (95% CIs) on day 14 (crude) (white bars) and day 28 (PCR corrected, method 1) (gray bars) for chloroquine, sulfadoxine-pyrimethamine, and amodiaquine.
DISCUSSION
There has been extensive discussion as to how efficacy of malaria treatment should be assessed, particularly in areas of intense transmission. The “ideal”—follow-up sufficiently long for all true treatment failures to emerge and be identified, combined with the ability to discriminate correctly between recrudescence and new infection in all cases—is currently unattainable. Getting as close as possible to the ideal presents practical problems, especially concerning duration of posttreatment follow-up (feasibility and risk of new infection proportional to intensity of transmission), outcome measures (clinical or parasitological criteria), and discrimination of recrudescence from new infection. The revised WHO protocol with a 28-day follow-up and PCR genotyping of recurrent infections addresses some shortcomings of earlier protocols. However, questions are raised as to its feasibility in field conditions as well as optimal methods for analysis of PCR-corrected outcomes.
We attempted to address a series of methodological issues related to the evaluation of treatment outcomes in pediatric uncomplicated malaria in Africa by analyzing 28 treatment arms enrolling a total of 2,576 children on antimalarial mono-therapy.
We found that, with suitable resources, the currently recommended 28-day follow-up is feasible even in relatively problematical field conditions (dropout rate, <5%; total exclusions by day 28, 11%). Extending the period of observation from 14 to 28 days resulted in higher estimated risks of failure for SP and AQ but not for CQ, for which the failure rates by day 14 were already extremely high. For AQ, the drug with the lowest failure rates at day 14, the estimated failure rate was commonly increased twofold or more when the follow-up was increased to 28 days. The gain in information varied with the drug and the site (as a function of underlying levels of drug resistance and transmission intensity, etc.).
This longer follow-up requires, however, that recurrent isolates be genotyped. This too proved feasible and adequate in these studies. Without PCR genotyping, 36% of the recurrent parasitemias after day 14 (250/696 recurrences) would have been wrongly classified as failures. This would have led to 1,048 cases being considered failures by day 28 (352 by or before day 14 plus 696 between days 14 and 28), thus overestimating the risk of failure overall by about one-third. While not all recurrences could be ascribed to either a new infection or a recrudescence (for 6%, a paired sample was not available, and for an additional 10% of cases, genotypes could not be resolved due to DNA extraction problems or because the result was not interpretable), in these studies, PCR genotyping performed very well (overall use effectiveness, 84%; test performance, 90%). Samples were genotyped in different laboratories by using nonstandardized methods. While this may be seen as a methodological deficiency, it makes these results representative of the variety of conditions applied in real life. In order to optimize the use of PCR genotyping, it is important to calculate the pretest probability of the same genotype occurring in the same individual pre- and posttreatment.
We followed today's prevailing practice of genotyping only post-day 14 recurrences. However, we have (yet unpublished) evidence from Uganda and Burkina-Faso that new infections can be documented also pre-day 14. More research is needed to further clarify the role and conditions under which PCR genotyping should be done and analyzed, including the discrimination power of PCR to correctly classify recurrent parasites.
A number of approaches are available for analysis of PCR-corrected data. We compared current and alternative methods of analyzing outcomes; our data set was large and heterogeneous. Given the high rates of follow-up and the high use effectiveness of PCR achieved, the results obtained from the different approaches were very similar in most instances, but when the WHO protocol is applied in routine conditions, variable dropout rates can be expected. Based on the principle that one should discard as little information as possible, we favor the use of Kaplan-Meier analysis in conjunction with treating new infections as treatment successes (method 1). In our data, this resulted in only 6% of patient data being lost for the entire 28-day study; by contrast, the current method (with patients evaluable at target day and excluding new infections) results in a substantial reduction in sample size (25%). This has consequences for sample size calculations in studies, particularly when dropout rates during follow-up are high (particularly if follow-up is further extended for drugs with long half-lives). A further extension to this approach, to avoid losing all the information provided by patients for whom PCR fails, would be to use multiple imputation, and we believe that this is worthy of further investigation. Indeed, there are pending issues with the criteria used to ascribe a recurrent isolate to a new or recrudescent infection.
Our results are in line with the WHO recommendation for use of life tables in analyzing in vivo study results (19).
When comparing outcomes on days 14 and 28, the correlation coefficient calculated for all studies appears reasonable but is misleading (influenced by the spread of data and the high failure rates for CQ at both day 14 and day 28). There was poor correlation with AQ. It should be noted that these analyses pertain to single-agent treatments which are no longer recommended. The WHO is now encouraging a review of drug policy when PCR-corrected failure rates at day 28 exceed 10% and the use of effective treatments, notably artemisinin-based combinations, including some with drugs with long residence times (20). The new policies will exacerbate the need for extended follow-up and for accurate assessment of true failure rate.
In conclusion, our results support the implementation of the latest WHO protocol (28-day follow-up with PCR genotyping), which proved feasible in field conditions. While this protocol is more demanding for staff and carries extra costs, it is clear that it provides a clearer picture of the level of resistance than a follow-up of only 14 days, notably in the initial phase of the establishment of resistance. To make maximum use of the data, we favor analyzing PCR-corrected outcomes with the Kaplan-Meier method and retaining new infections as treatment successes.
Acknowledgments
The original studies were conducted in collaboration with the National Ministries of Health. We are grateful to MSF personnel at headquarters and field staff who actively contributed to the studies. Special thanks to the Epicentre and MSF researchers responsible for each individual study (Catherine Bachy, Maryline Bonnet, Francesco Grandesso, Xavier de Radiguès, Julia Sonia Ampuero, Valérie Gaboulaud, Paul Roddy, Guy Morineau, Martin de Smet, and Christa Hook). We also thank the staffs of the different institutions who performed the PCR analyses: Faculté de Médecine-Pharmacie, Rouen, Hôpital Bichat, Paris, and Hôpital Ambroise Paré, Boulogne, France; Prince Leopold Institute of Tropical Medicine, Antwerp, Belgium; Malaria Research and Training Centre, Bamako, Mali; University of Glasgow, Glasgow, United Kingdom; and Shoklo Malaria Research Unit, Mae Sod, Thailand. We thank Pascal Ringwald (WHO/RBM) for useful discussion and exchange of information. Thank you to Nick White and Kasia Stepniewska (Wellcome Trust, Mahidol University and Oxford University) for critically reviewing the manuscript. Special thanks to Emmanuel Baron, Head of the Medical Department of MSF-France, for the support provided.
The individual studies were financed by Médecins sans Frontières and by Merlin in one of the sites in Sierra Leone. The present work is a desk-based analysis, supported by Médecins sans Frontières.
The opinions expressed in this paper are the authors' and do not necessarily reflect those of the employing organizations.
Footnotes
Published ahead of print on 5 September 2006.
REFERENCES
- 1.Bachy, C., F. Checchi, and P. Cavailler. 2003. Chloroquine and sulphadoxine-pyrimethamine drug sensitivity study for the treatment of uncomplicated malaria due to Plasmodium falciparum in Mapel, Sudan. Epicentre/MSF, Paris, France.
- 2.Basco, L. K., and P. Ringwald. 2000. Molecular epidemiology of malaria in Yaounde, Cameroon. VII. Analysis of recrudescence and reinfection in patients with uncomplicated falciparum malaria. Am. J. Trop. Med. Hyg. 63:215-221. [PubMed] [Google Scholar]
- 3.Bonnet, M. 2004. Efficacité de la sulfadoxine-pyriméthamine, amodiaquine, artesunate plus sulfadoxine-pyriméthamine et artesunate plus amodiaquine pour le traitement du paludisme non-compliqué à Plasmodium falciparum, dans le Centre de Santé de Boende, République Démocratique du Congo. Epicentre/MSF Report, Paris, France.
- 4.Checchi, F., S. Balkan, B. T. Vonhm, M. Massaquoi, P. Biberson, P. Eldin de Pecoulas, P. Brasseur, and J.-P. Guthmann. 2002. Efficacy of amodiaquine for uncomplicated Plasmodium falciparum malaria in Harper, Liberia. Trans. R. Soc. Trop. Med. Hyg. 96:670-673. [DOI] [PubMed] [Google Scholar]
- 5.Checchi, F., P. Piola, C. Kosack, E. Ardizzoni, D. Klarkowski, E. Kwezi, G. Priotto, S. Balkan, N. Bakyaita, A. Brockman, and J.-P. Guthmann. 2004. Antimalarial efficacy of sulfadoxine-pyrimethamine, amodiaquine and a combination of chloroquine plus sulfadoxine-pyrimethamine in Bundi Bugyo, western Uganda. Trop. Med. Int. Health 9:445-450. [DOI] [PubMed] [Google Scholar]
- 6.Checchi, F., P. Roddy, S. Kamara, A. Williams, G. Morineau, A. R. Wurie, B. Hora, N. Lamotte, T. Baerwaldt, A. Heinzelmann, A. Danks, L. Pinoges, A. Oloo, R. Durand, L. Ranford-Cartwright, and M. de Smet. 2005. Evidence basis for antimalarial policy change in Sierra Leone: five in vivo efficacy studies of chloroquine, sulphadoxine-pyrimethamine and amodiaquine. Trop. Med. Int. Health 10:146-153. [DOI] [PubMed] [Google Scholar]
- 7.de Radiguès, X., K. I. Diallo, P. A. Ngwakun, A. Djimdé, O. Doumbo, M. Diallo, M. van Herp, and J.-P. Guthmann. Submitted for publication.
- 8.Grandesso, F., C. Bachy, I. Donam, J. Ntambi, J. Habimana, U. d'Alessandro, J. Maikere, V. Vanlerberghe, C. Hinzoumbe Kerah, and J.-P. Guthmann. Efficacy of chloroquine, sulfadoxine-pyrimethamine and amodiaquine for treatment of uncomplicated Plasmodium falciparum malaria in Bongor and Koumra, Chad. Trans. R. Soc. Trop. Med. Hyg., in press. [DOI] [PubMed]
- 9.Greenwood, B. M., K. Bojang, C. J. Whitty, and G. A. Targett. 2005. Malaria. Lancet 365:1487-1498. [DOI] [PubMed] [Google Scholar]
- 10.Guthmann, J.-P., J. Ampuero, F. Fortes, C. van Overmeir, V. Gaboulaud, S. Tobback, J. Dunand, N. Saraiva, P. Gillet, J. Franco, A. Denoncin, M. van Herp, S. Balkan, J.-C. Dujardin, U. D'Alessandro, and D. Legros. 2005. Antimalarial efficacy of chloroquine, amodiaquine, sulfadoxine-pyrimethamine, and the combinations of amodiaquine + artesunate and sulfadoxine-pyrimethamine + artesunate in Huambo and Bié provinces, central Angola. Trans. R. Soc. Trop. Med. Hyg. 99:485-492. [DOI] [PubMed] [Google Scholar]
- 11.Irion, A., I. Felger, S. Abdulla, T. Smith, R. Mull, M. Tanner, C. Hatz, and H. P. Beck. 1998. Distinction of recrudescences from new infection by PCR-RFLP analysis in a comparative trial of CGP 56 697 and chloroquine in Tanzanian children. Trop. Med. Int. Health 3:490-497. [DOI] [PubMed] [Google Scholar]
- 12.Ranford-Cartwright, L. C., J. Taylor, T. Umasunthar, L. H. Taylor, H. A. Babiker, B. Lell, J. R. Schmidt-Ott, L. G. Lehman, D. Walliker, and P. G. Kremsner. 1997. Molecular analysis of recrudescent parasites in a Plasmodium falciparum drug efficacy trial in Gabon. Trans. R. Soc. Trop. Med. Hyg. 91:719-724. [DOI] [PubMed] [Google Scholar]
- 13.Snounou, G. 2002. Genotyping of Plasmodium spp. Nested PCR. Methods Mol. Med. 72:103-116. [DOI] [PubMed] [Google Scholar]
- 14.Snow, R. W., C. A. Guerra, A. M. Noor, H. Y. Myint, and S. I. Hay. 2005. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature 434:214-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stepniewska, K., W. R. Taylor, M. Mayxay, R. Price, F. Smithuis, J.-P. Guthmann, K. Barnes, H. Y. Myint, M. Adjuik, P. Olliaro, S. Pukrittayakamee, S. Looareesuwan, T. T. Hien, J. Farrar, F. Nosten, N. P. Day, and N. J. White. 2004. In vivo assessment of drug efficacy against Plasmodium falciparum malaria: duration of follow-up. Antimicrob. Agents Chemother. 48:4271-4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Talisuna, A. O., P. Bloland, and U. D'Alessandro. 2004. History, dynamics, and public health importance of malaria parasite resistance. Clin. Microbiol. Rev. 17:235-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.WHO. 1973. Chemotherapy of malaria and resistance to antimalarials. Report of a WHO Scientific Group. WHO Tech. Rep. Ser. 529:1-21. [PubMed] [Google Scholar]
- 18.WHO. 1996. Assessment of therapeutic efficacy of antimalarial drugs for uncomplicated falciparum malaria in areas with intense transmission. Document WHO/MAL/96.1077. World Health Organization, Geneva, Switzerland.
- 19.WHO. 2003. Assessment and monitoring of antimalarial drug efficacy for the treatment of uncomplicated falciparum malaria. Document WHO/HTM/RBM/2003.50. World Health Organization, Geneva, Switzerland.
- 20.WHO. 2006. Guidelines for the treatment of malaria. Document WHO/HTM/MAL/2006.1108. World Health Organization, Geneva, Switzerland. [Online.] http://www.who.int/malaria/docs/TreatmentGuidelines2006.pdf.
- 21.Yeung, S., W. Pongtavornpinyo, I. M. Hastings, A. J. Mills, and N. J. White. 2004. Antimalarial drug resistance, artemisinin-based combination therapy, and the contribution of modeling to elucidating policy choices. Am. J. Trop. Med. Hyg. 71(Suppl. 2):179-186. [PubMed] [Google Scholar]