Abstract
The RNA-dependent protein kinase (PKR) is activated by binding to double-stranded RNA (dsRNA). Activation of PKR by short-interfering RNAs (siRNAs) and stimulation of the innate immune response has been suggested to explain certain off-target effects in some RNA interference experiments. Here we show that PKR's kinase activity is stimulated in vitro 3- to 5-fold by siRNA duplexes with 19 bp and 2 nt 3′-overhangs, whereas the maximum activation observed for poly(I)•poly(C) was 17-fold over background under the same conditions. Directed hydroxyl radical cleavage experiments indicated that siRNA duplexes have at least four different binding sites for PKR's dsRNA binding motifs (dsRBMs). The location of these binding sites suggested specific nucleotide positions in the siRNA sense strand that could be modified with a corresponding loss of PKR binding. Modification at these sites with N2-benzyl-2′-deoxyguanosine (BndG) blocked interaction with PKR's dsRBMs and inhibited activation of PKR by the siRNA. Importantly, modification of an siRNA duplex that greatly reduced PKR activation did not prevent the duplex from lowering mRNA levels of a targeted message by RNA interference in HeLa cells. Thus, these studies demonstrate that specific positions in an siRNA can be rationally modified to prevent interaction with components of cellular dsRNA-regulated pathways.
INTRODUCTION
The RNA-dependent protein kinase (PKR) is a component of the interferon antiviral response (1). PKR inhibits translation initiation through the phosphorylation of the alpha subunit of the initiation factor eIF2 (eIF2α) (2). However, PKR is synthesized in a latent, inactive form that requires association with double-stranded RNA (dsRNA) or other activator molecules (3). This activation manifests itself initially by autophosphorylation followed by the phosphorylation of protein substrates (4). In addition to its antiviral role, PKR has effects on the normal maintenance of cell growth, differentiation in myogenic cells, apoptosis and signal transduction pathways that control transcription (5–11). Some of these activities may arise from PKR phosphorylation of proteins other than eIF2α, since a number of alternate in vitro substrates have been reported including IκB, p53, B56α, TAT and histone (9,12–15).
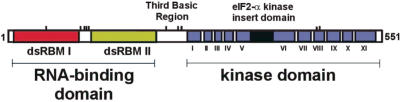
Human PKR is 62 kDa, consisting of a 20 kDa N-terminal RNA-binding domain and a C-terminal kinase domain (6,16). The RNA-binding domain is composed of two dsRNA-binding motifs (dsRBMs) linked by a stretch of 20 amino acids (Figure 1). dsRBMs are ∼70 amino acid segments commonly found in dsRNA-binding proteins that do not require specific sequences of dsRNA for binding (17–20). These motifs bind 16 bp of dsRNA via interaction with two different minor groove sites and phosphodiester contacts across the intervening major groove (21–24). In addition to RNA binding, activation of PKR also involves dimerization (25,26). The binding of an activating RNA ligand is thought to facilitate PKR self-association. Importantly, although only ∼16 bp of dsRNA is required for binding to PKR, activation of the enzyme requires a longer duplex region (27,28). It has been suggested that the length effect arises from the requirement of an activating RNA ligand to support binding to more than one PKR dsRBM (29). However, precisely how PKR assembles on an activating RNA is poorly defined at this time.
Figure 1.
Domain map for the dsRNA-dependent protein kinase, PKR.
Interestingly, the failure to induce a specific RNAi effect in certain cells with long dsRNA molecules can be attributed, at least in part, to the activation of PKR and the resulting nonspecific inhibition in translation (30). To overcome this obstacle, short-interfering RNAs (siRNAs) are used that mimic the Dicer products of the natural RNAi pathway (31). These 19 bp duplex RNAs with 2 nt 3′ overhangs were thought previously to be too short to activate PKR, since the minimum length of an RNA duplex shown to activate the enzyme in vitro was 33 bp (27). However, recent studies have indicated that siRNAs can activate PKR both in vitro and in cultured cells (32,33). Sledz et al. (32) showed that this activation occurred with a corresponding stimulation of JaK-STAT signaling and changes in the expression patterns of a number of interferon-regulated genes. Furthermore, these effects were diminished in a PKR null cell line. However, there are conflicting reports on the extent to which activation of PKR specifically, or other components of the dsRNA-induced antiviral pathways in general, is the cause of off-target effects observed in RNAi experiments with mammalian cells (32,34–36). Furthermore, the activation of PKR in vitro by RNA duplexes of the length found in siRNAs (19–21 bp) itself has been called into question recently (37).
In this paper, we describe the PKR activation properties of both unmodified and chemically modified siRNAs. The unmodified siRNAs studied in this work are PKR activators, stimulating autophosphorylation to a level 3- to 5-fold above background. Directed hydroxyl radical cleavage experiments were used to define the binding sites on siRNAs for PKR's dsRBMs. Site-specific modification at these locations with a nucleoside analog that projects steric bulk into the duplex RNA minor groove prevents activation of PKR. Greater than 80% inhibition of the activation observed with an unmodified siRNA can occur with as few as two purine N2-benzyl modifications located at specific positions in the siRNA sense strand. Furthermore, enhanced phosphorylation of PKR observed upon transfection of siRNA into human tissue culture cells is reduced by N2-benzyl modifications shown to block PKR activation in vitro. We present a model for siRNA activation of PKR that is consistent with these observations. Importantly, modifications that inhibit PKR activation do not prevent the siRNA from causing RNA interference. These observations indicate that chemical modification of the nucleobases found in siRNAs can be used to prevent the interaction with intracellular dsRNA-binding proteins without blocking RNA interference.
MATERIALS AND METHODS
Oligonucleotide synthesis
All oligoribonucleotides were synthesized in the DNA/Peptide core facility of University of Utah using an Applied Biosystems Model 394 DNA/RNA synthesizer. 2-FdI phosphoramidite was purchased from Glen Research Inc, VA. Substitution with benzylamine on resin-bound 2-FdI oligonucleotide was carried out as described by Allerson et al. (38). The crude deprotected oligonucleotides were purified using denaturing PAGE. The samples were analyzed by negative-ion electrospray mass spectrometry (ESI/MS) using a quadrupole mass spectrometer (Quattro-II; Micromass, Inc.). The resulting multiple-charge mass spectrum was then processed into a neutral molecular mass spectrum using MaxEnt software (Micromass, Inc.). For the molecular masses of individual BndG-modified RNA see Supplementary Table 1.
siRNA duplex formation and purification
Hybridization to form siRNA duplexes was accomplished by combining 3–6 nmol of sense and antisense strands in 80–100 μl of 10 mM Tris, pH 7.4 and 100 mM NaCl. Samples were heated to 95°C for 5 min in a dry bath incubator following which the dry bath block was removed from the heating unit and allowed to cool to room temperature. The samples were then electophoresed on a 16% native polyacrylamide gel. Samples were visualized by UV shadowing and hybridization confirmed by comparison with single strand standards. Duplex RNAs were extracted from the gel via crushing and then soaking overnight at room temperature in 200 mM NaCl and 0.1 mM EDTA. The resulting solution was phenol: chloroform extracted, ethanol precipitated, suspended in 10 mM Tris–HCl, pH 7.5 and 50 mM KCl, and quantified by UV-visible spectroscopy.
PKR activity assay
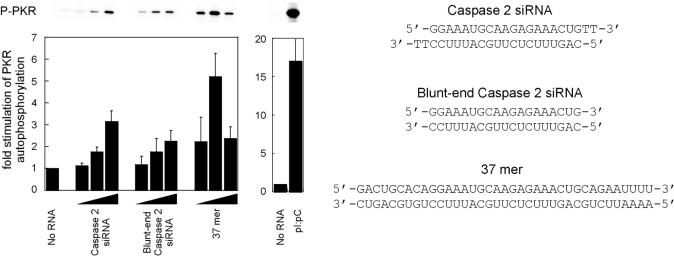
To compare difference in activities between siRNA, blunt-ended siRNA, a 37mer duplex and poly(I)•poly(C), a PKR activity assay was performed under identical condition as described by Gunnery and Mathews (39) in the presence of 1 μM eIF2α substrate (Figure 2). Vector pET-PKR/λPPase was obtained by cloning wild-type PKR into pET-λPPase (16). The resulting expression vector was transformed into Rosetta 2(DE3) cells (Novagen). Protein co-expression was induced by 1 mM isopropyl-β-d-thiogalactopyranoside in Luria–Bertani medium at 37°C for 4 h. PKR purification was performed as described previously (40) replacing the size exclusion column with a heparin column for the second purification step as follows. Following purification with the Ni-NTA column, the buffer was exchanged with phosphate-buffered saline. The resulting protein solution was applied to a heparin HP column (Amersham) pre-equilibrated in 10 mM Na2HPO4, pH 7.0. PKR was eluted using a NaCl gradient. To compare the activity of PKR by different modified siRNAs (Figures 6 and 7), we used assay conditions that gave slightly higher activation above background (200 nM PKR, 20 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 50 mM KCl, 2 mM DTT, 100 μM ATP, and 0.34 mCi/ml and 250 μg/ml histone type II-A) (41). The reactions were performed by mixing the selected siRNA duplex with a cocktail consisting of purified, dephosphorylated PKR, substrate [either histone type II-A (Figures 6 and 7) or eIF2α (Figure 2)], and reaction buffer. 32P-ATP was then added and the reactions were incubated at 30°C for 5 min and quenched with pre-heated (95°C) SDS–PAGE loading buffer. Samples were heated at 95°C for 5 min and electrophoresed on a 13.5% SDS–PAGE gel followed by exposure to a storage phosphor autoradiography screen. Labeled proteins were visualized using a Typhoon PhosphorImager (Pharmacia) and bands were quantified using ImageQuant software.
Figure 2.
siRNA activates PKR. (Upper panel) PKR autophosphorylation as a function of dsRNA concentration. (Lower panel) Quantification of the stimulation of PKR autophosphorylation as a function of dsRNA concentration. The extent of phosphorylation of PKR was measured by storage phosphor autoradiography and plotted as the ratio to an unstimulated (no RNA) sample. The concentration of each dsRNA tested from left to right is 20 nM, 200 nM and 2 μM except poly(I)•poly(C), which was tested in the absence (−) or presence (+) of 10 μg/ml. This concentration of poly(I)•poly(C) resulted in the highest stimulation of PKR autophosphorylation under these conditions (data not shown).
Preparation of dsRBD and directed hydroxyl radical cleavage experiments
PKR dsRBD E29C-EDTA•Fe and Q120C-EDTA•Fe were prepared and used in directed hydroxyl radical cleavage experiments as described previously (42,43). All cleavage experiments were performed with 5′-end-labeled RNA. Quantification of the cleavage data was performed using Image Quant software (Molecular Dynamics). To quantify the cleavage efficiency at each nucleotide, the intensity of the corresponding band in the lane with modified protein and RNA without hydroxyl radical generating reagents (hydrogen peroxide and sodium ascorbate) was subtracted from the lane with RNA treated with modified protein and these reagents. The length of the line adjacent to the nucleotides represents the resulting cleavage efficiency. The pixel count from the most efficient cleavage site was quantified and normalized to one. All the lines depicting the cleavage efficiency were drawn relative to the most efficient cleavage site. Since all the lanes in each figure were run on the same gel, the cleavage efficiencies can be directly compared for the different siRNAs.
siRNA-mediated gene knockdown
HeLa cells (8000 cells per well) were reverse-transfected in 96-well plates with 30–100 nM siRNA using 0.4 μl siPORT NeoFX (Ambion, Austin, TX) per well. The medium was replaced with fresh culture medium (DMEM supplemented with fetal bovine serum and penicillin/streptomycin) 24 h after transfection. The cultures were harvested for mRNA analysis 48 h after transfection. Total RNA from siRNA-transfected cells was isolated using the RNAqueous® MagMAX-96 Total RNA Isolation kit (Ambion). The purified, DNase-treated RNA was reverse transcribed with random decamers using the RETROscript® Kit (Ambion). Gene expression levels were determined by real-time PCR using SuperTaq™ reagents (Ambion) on the ABI Prism 7900 SDS (Applied Biosystems, Foster City, CA). The data were collected using primers and a dual-labeled probe specific for caspase 2 (Assays-on-Demand; Applied Biosystems). 18S rRNA was amplified (forward, 5′-ttgactcaacacgggaaacct-3′; reverse, 5′-agaaagagctatcaatctgtcaatcct-3′; probe, 5′-VIC-acccggcccggacacgga-NFQ-3′) as an internal reference to adjust for well-to-well variances in amount of starting template. The corrected values were normalized to a sample transfected with the Silencer™ Negative Control #1 siRNA (Ambion).
PKR activation in cultured cells by transfected siRNAs
Human A172 cells (3.3 × 105 cells per well) were transfected with 50 nM siRNAs using oligofectamine (Invitrogen) according to the manufacturer's protocol in a 6-well plate. After 2.5 h, cells were lysed in buffer containing 50 mM Tris, pH 7.4, 150 mM NaCl, 50 mM NaF, 0.1 mM EDTA, 0.1% Triton X-100, 10% glycerol, 2 mM orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 10 mM β-glycerophosphate and Roche's complete protease inhibitor cocktail. The lysate was clarified by centrifugation at 12 000 g for 10 min at 4°C. Identical amounts of protein (50 μg) were resolved by 12% SDS–PAGE and protein bands were transferred on to a PVDF membrane. The membrane was subjected to western blotting with antibodies specific for phospho-PKR (pThr451) (Cell Signaling Technology), PKR (Santa Cruz Biotechnology) and GAPDH (Santa Cruz Biotechnology).
RESULTS
PKR binding to siRNA: kinase activation and directed hydroxyl radical cleavage
We prepared siRNA duplexes and tested their ability to activate PKR in in vitro kinase assays. This was carried out using human PKR expressed in Escherichia coli and dephosphorylated with protein phosphatase I, as described previously (41) or by co-expressing PKR with lambda phosphatase (16,40). Dephosphorylation of activated PKR isolated from bacterial expression systems generates a form of the enzyme that is responsive to RNA stimulation of its in vitro kinase activity (16,37,41,44,45). siRNA duplexes were prepared by chemical synthesis and purification of the sense and antisense strands followed by hybridization and gel purification of the resulting duplex RNAs. PKR autophosphorylation activity was then measured as a function of siRNA concentration in the presence of substrate eIF2α (Figure 2) or histone type II-A (Figures 6 and 7). A 19 bp siRNA with 2 nt 3′ overhangstargeting human caspase 2 showed a 3-fold increase in autophosphorylation of PKR compared to a control sample with no RNA added (Figure 2). PKR substrate phosphorylation was also enhanced by added siRNA (data not shown). Maximum activation of PKR was observed at an siRNA concentration near 1 μM under these in vitro conditions. A 37 bp duplex RNA activated PKR 5-fold over background levels under these same conditions. This duplex exceeds the previously reported minimum length for PKR activation (33 bp) and was included as a positive control. In addition, the well-known PKR activator poly(I)•poly(C) was capable of 17-fold stimulation over background at its concentration of maximum activation (10 μg/ml). Thus, an siRNA duplex is capable of activating PKR in vitro at levels near that of a 37 bp duplex, but substantially less effectively than the duplex RNA polymer poly(I)•poly(C). When the 3′ overhangs of the siRNA were removed and a 19 bp blunt-ended duplex was evaluated, stimulation of PKR autophosphorylation was reduced slightly to 2-fold over background (Figure 2). Stimulation of PKR activity by siRNAs is not highly dependent on sequence, as similar activation levels were observed for two additional, unrelated siRNA duplexes targeting either human papilloma virus E7 protein (HPV-E7) or green fluorescent protein (GFP) (Figures 6 and 7).
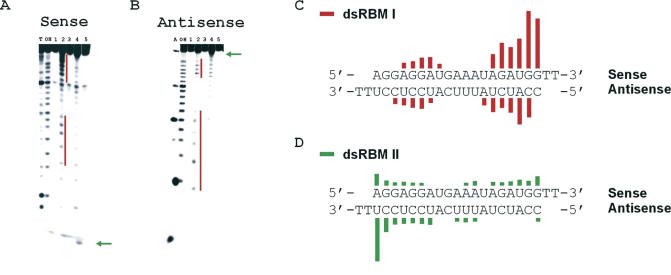
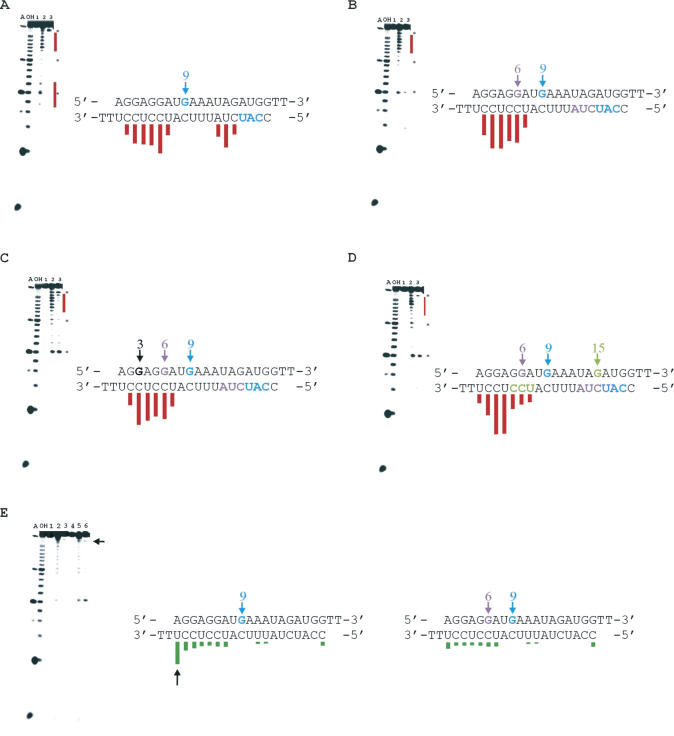
Since PKR binds RNA through its dsRBM-containing RNA-binding domain, we endeavored to define the dsRBM-binding sites present on the siRNA using directed hydroxyl radical cleavage with EDTA•Fe tethered to PKR's dsRBMs. Both ends of the HPV-E7 siRNA duplex were extensively cleaved with PKR RBD E29C-EDTA•Fe, (46) which comprises both dsRBMs including the intervening linker and bears the tethered EDTA•Fe at amino acid position 29 in dsRBM I (Figure 3A–C). The cleavage patterns observed arise from EDTA•Fe attached to bound PKR RBD, since these bands are not observed in control reactions lacking the reagents necessary to induce hydroxyl radical formation (Figure 3A and B). Multiple cleavage sites at both ends indicate little orientation preference and more than one binding register in each orientation. This is consistent with PKR's low sequence preference for binding perfectly matched RNA duplexes. The cleavage pattern was suggestive of four different binding sites on the siRNA for PKR's dsRBM I. A similar pattern was observed with an siRNA targeting GFP (data not shown).
Figure 3.
Directed hydroxyl radical cleavage to probe the binding of an siRNA with the PKR RBD. (A) Storage phosphor autoradiogram of a 19% denaturing polyacrylamide gel separating the RNA cleavage products from the HPV-E7 siRNA duplex with the sense strand 5′-end-labeled. Major cleavage sites are identified with lines and arrows. The lanes had the following reaction conditions as labeled: T, T1 RNAse (G-lane); OH, alkaline hydrolysis; lane 1, RNA with no added reagents (protein, hydrogen peroxide or sodium ascorbate); lane 2, 8 μM PKR RBD E29C-EDTA•Fe (46) in the presence of 0.001% hydrogen peroxide and 5 mM sodium ascorbate (cleavage reagents); lane 3, 8 μM PKR RBD E29C-EDTA•Fe in the absence of 0.001% hydrogen peroxide and 5 mM sodium ascorbate; lane 4, 10 μM PKR RBD Q120C-EDTA•Fe (42) in the presence of 0.001% hydrogen peroxide and 5 mM sodium ascorbate; lane 5, 10 μM PKR RBD Q120C-EDTA•Fe in the absence of 0.001% hydrogen peroxide and 5 mM sodium ascorbate. (B) Conditions are the same as in (A) with antisense strand 5′ end-labeled with the exception that A refers to RNAse A cleavage products (A>C lane) (C) Sites of cleavage on the HPV-E7 siRNA induced by PKR RBD modified with EDTA•Fe at position 29 of dsRBM I. Length of each line is proportional to the extent of cleavage at that nucleotide. (D) Sites of cleavage on the HPV-E7 siRNA induced by PKR RBD modified with EDTA•Fe at position 120 of dsRBM II.
When the HPV-E7 siRNA was cleaved with PKR RBD Q120C-EDTA•Fe (42), which bears the EDTA•Fe at amino acid position 120 in dsRBM II, the 3′ end of the antisense strand (5′ end of the sense strand) was the most efficiently cleaved site (Figure 3A, B and D). This cleavage site for dsRBM II is closer to the end of the duplex than those identified for dsRBM I. Other minor cleavage sites are also apparent, including some that appear to overlap with those observed for dsRBM I.
Directed hydroxyl radical cleavage of modified siRNA
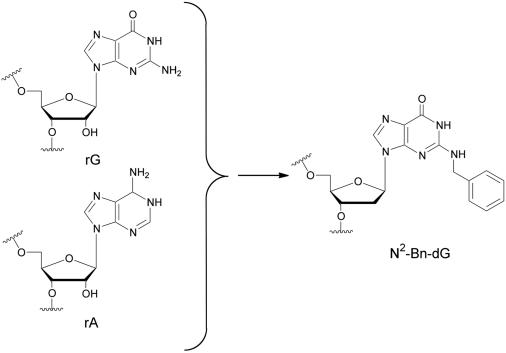
Our laboratory has shown previously that individual dsRBM-binding sites can be selectively disrupted by site-specific steric occlusion of minor groove sites by replacing specific purines in the duplex RNA with N2-benzylguanine (Figure 4) (47,48). Introduction of this bulky modification is advantageous, since single mutations that cause only a change in sequence typically have minimal effect on dsRBM binding. Furthermore, replacing purines with N2-benzylguanine maintains base pairing (G•C or G•U) and, thus, does not alter the overall helical structure of the duplex.
Figure 4.
Introduction of N2-benzyl-2′-deoxyguanosine into duplex RNA occludes the minor groove and disrupts dsRBM-RNA binding.
We used the benzyl modification strategy to disrupt PKR dsRBM-binding sites present on the siRNA inferred from directed hydroxyl radical cleavage patterns (Figure 5). From our experience, a benzyl group incorporated ∼8 nt from the center of a cleavage pattern generated by the PKR RBD E29C-EDTA•Fe protein blocks binding to that site most efficiently (47). Thus, a benzyl group was introduced at position 9 of the siRNA sense strand to disrupt the complex leading to cleavage at the extreme 3′ end of this strand (5′ end of the antisense strand). This was accomplished via incorporation of 2-fluoro-2′-deoxyinosine into the RNA strand using a commercially available phosphoramidite and subsequent substitution with benzylamine to generate an oligonucleotide with the requisite purine N2-benzyl modification (Materials and Methods) (38). Control experiments indicated that deoxy substitutions alone at the sites chosen for modification have no effect on PKR activation (data not shown). In addition, to avoid complications from analyzing cleavage data on the strand containing benzyl modifications, we modified only the sense strand and present cleavage results only for the unmodified antisense strand. The introduction of N2-benzyl-2′-deoxyguanosine (BndG) at position 9 of the sense strand led to disruption of cleavage at the nucleotides corresponding to the extreme 5′ end of the antisense strand (nt 2–4), with the cleavage at nt 5–7 unchanged (Figure 5A). This confirmed that the cleavage observed at this location of the duplex (nt 2–7 of the antisense strand) arises from at least two different dsRBM–RNA complexes. This was further supported by introduction of BndG at position 6 of the sense strand along with the position 9 modification (Figure 5B). The two BndG modifications completely absolved the 5′ end of the antisense strand from efficient cleavage by the PKR RBD E29C-EDTA•Fe protein. A BndG modification at position 3, in addition to positions 6 and 9, did not cause an appreciable change in the cleavage pattern (Figure 5C). However, the cleavage was altered when BndG was introduced at position 15 in addition to positions 6 and 9. A reduction of cleavage efficiency at nt 13–15 of the antisense strand was observed, while cleavage of nt 16–19 increased (Figure 5D). Thus, like at the 5′ end, it appeared the cleavage patterns observed at the 3′ end of the antisense strand arise from binding at two different sites.
Figure 5.
Directed hydroxyl radical cleavage to probe PKR RBD binding of benzyl-modified siRNAs. (A–D) Directed hydroxyl radical cleavage by PKR RBD E29C-EDTA•Fe to probe PKR dsRBM I binding of benzyl-modified siRNAs. Conditions for A, OH, lane 1, lane 2 and lane 3 for (A–D) were identical to the conditions of A, OH, lane 1, lane 2 and lane 3 for Figure 3B. (A) Cleavage pattern on HPV-E7 siRNA benzylated at position 9 on the sense strand. (B) Cleavage pattern on HPV-E7 siRNA benzylated at positions 6 and 9 on the sense strand. (C) Cleavage pattern on HPV-E7 siRNA benzylated at positions 3, 6 and 9 on the sense strand. (D) Cleavage pattern on HPV-E7 siRNA benzylated at positions 6, 9 and 15 on the sense strand. (E) Directed hydroxyl radical cleavage by PKR RBD Q120C-EDTA•Fe to probe PKR dsRBM II binding of benzyl-modified siRNAs. A and OH were RNAse A and alkaline hydrolysis, respectively. Conditions for lane 1, HPV-E7 siRNA benzylated at position 9 with no added cleavage reagents (protein, hydrogen peroxide or sodium ascorbate); lane 2, same as lane 4 of Figure 3; lane 3, same as lane 5 for Figure 3; lane 4, HPV-E7 siRNA benzylated at position 6 and 9 with no added cleavage reagents; lane 5, same as lane 4 of Figure 3; lane 6, same as lane 5 for Figure 3. Asterisk indicates a hyper-reactive nucleotide showing cleavage independent of directed hydroxyl radical cleavage. The cleavage sites disrupted on the antisense strand are shown in the same color as the benzyl-modified nucleotide on the sense strand that causes the change.
The ability of the PKR RBD E29C-EDTA•Fe protein to cleave duplexes bearing BndG at multiple positions indicated that the modifications do not disrupt the duplex structure, but only limit the ways in which PKR can bind. To address directly the effect these modifications have on duplex stability, we measured the thermal melting temperatures (Tm's) of unmodified and certain BndG modified HPV-E7 siRNAs. The Tm of the unmodified HPV-E7 siRNA was measured to be 79.6°C, whereas the duplex bearing BndG at positions 6, 9 and 15 had a Tm = 70.6°C. The duplex with BndG modifications at positions 3, 6 and 9 had a Tm = 69.7°C. These observations are consistent with other results suggesting that a single BndG modification leads to an ∼3°C decrease in the Tm of an siRNA duplex (Supplementary Table 2).
When PKR RBD Q120C-EDTA•Fe was used to cleave the modified siRNAs, we found that BndG modification at position 9 caused a decrease in cleavage efficiency at the 3′ end of the antisense strand (Figure 5E). The effect modification at position 9 has on the dsRBM I-directed and dsRBM II-directed cleavage suggests a binding site on the siRNA that can be occupied by either motif with one orientation favored for dsRBM I (cleavage at 5′ end of the antisense strand) and the other favored for dsRBM II (cleavage at 3′ end of the antisense strand). Cleavage observed on the duplex bearing BndG at position 9 was nearly completely inhibited by an additional modification at position 6 (Figure 5E).
The effect of siRNA modification on PKR activation
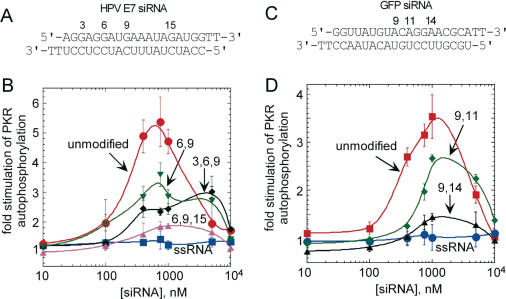
To determine the effect of BndG modification of the siRNA on PKR activation, we evaluated the ability of the modified siRNAs to stimulate PKR's kinase activity in vitro (Figure 6A–D). For the HPV E7 siRNA, activation of PKR was reduced from a maximum stimulation of over 5-fold to ∼3-fold when BndG was introduced at positions 6 and 9 of the sense strand (Figure 6A and B). When the same siRNA was additionally modified at position 3, little further reduction in PKR activation was observed, although a shift in the concentration of maximal activation was apparent (Figure 6A and B). Importantly, when BndG was introduced at positions 6, 9 and 15, an ∼85% reduction in maximum stimulation of PKR autophosphorylation was observed compared to unmodified siRNA.
Figure 6.
Introduction of BndG in the siRNA sense strand inhibits PKR activation. (A) The positions selected for modification based on the affinity cleavage data are shown on HPV-E7 siRNA. (B) Activation of PKR by (closed circle) unmodified HPV-E7 siRNA, (inverted closed triangle—6,9) (closed diamond—3,6,9) (closed triangle—6,9,15) modified HPV-E7 siRNA and (closed square) HPV-E7 sense single strand. (C) Positions of nucleotides replaced by BndG are shown for GFP siRNA. (D) Activation of PKR by (closed square) unmodified GFP siRNA and (closed diamond—9,11) (closed triangle—9,14) modified GFP siRNA and (closed circle) GFP sense single strand.
An siRNA targeting GFP was also shown to activate PKR in vitro (Figure 6C and D). We modified this siRNA with BndG to determine if the effects on PKR activation observed for the HPV E7 siRNA would be reproduced for an unrelated sequence. From the effect of modifications described above, the cleavage patterns observed on the various siRNAs and molecular models for dsRBM binding based on these cleavage results (see below), it appeared that modification of nucleotides near positions 6, 9, 11 and 14 of the sense strand would have the greatest effect on PKR binding. Thus, we also wished to determine if modification at subsets of these critical nucleotide positions would vary in effect. The BndG modification was introduced at two positions in the sense strand in two different duplexes (9, 11 and 9, 14) (Figure 6C). Interestingly, while double modification at positions 9 and 11 reduced PKR activation only by ∼30%, modification at positions 9 and 14 caused an 85% reduction in activation (Figure 6D).
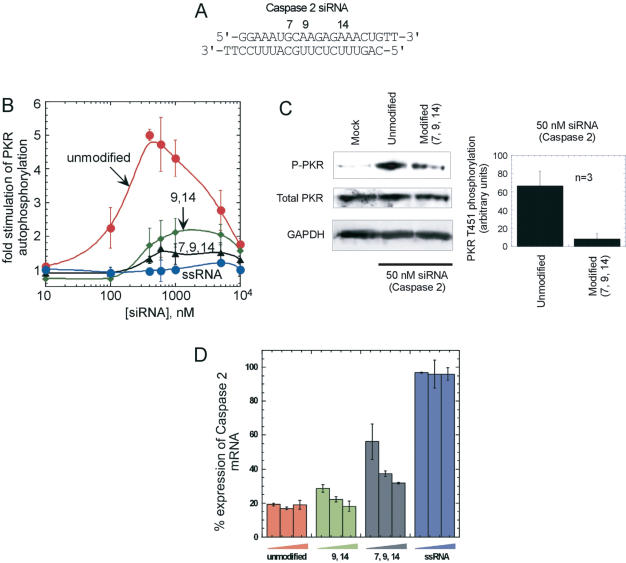
RNA interference with siRNAs modified to block PKR activation
To assess the effect on RNA interference activity caused by chemical modifications that decrease PKR activation, we analyzed the knockdown capability of modified siRNAs targeting the human caspase 2 message (Figure 7A). To decrease the ability of the siRNA to activate PKR, the sense strand was substituted at critical positions to generate two different modified duplexes bearing BndG at 2 (9, 14) or 3 (7, 9, 14) nt positions. Modification at positions 9 and 14 caused a large decrease in the maximum PKR activation (∼75%), as expected from results with the GFP siRNA (Figure 7B). Including the additional BndG at position 7 reduced the activation to near background levels (Figure 7B). We also compared the levels of phospho-PKR in human tissue culture cells (A172) transfected with unmodified caspase 2 siRNA and caspase 2 siRNA modified at positions 7, 9 and 14 (Figure 7C). The unmodified caspase 2 siRNA caused an increase in the phosphoryation of PKR at a critical activation loop threonine (T451) in comparison to a mock transfection with oligofectamine alone. Importantly, benzyl modification of the siRNA reduced the level of siRNA-stimulated PKR phosphorylation observed in these cells (Figure 7C).
Figure 7.
siRNAs modified to block PKR activation are capable of inducing RNA interference. (A) Nucleotides of the caspase 2 siRNA substituted by BndG. (B) Activation of PKR by (closed circle) unmodified caspase 2 siRNA, (closed diamond—9,14) (closed triangle—7,9,14) modified caspase 2 siRNA and (closed square) caspase 2 antisense single strand. (C) (Left) Western blot of cell lysate from A172 cells transfected with 50 nM caspase 2 siRNA. (Right) Quantification of western blot analysis of PKR phosphorylation in response to siRNA transfection. PKR T451 phosphorylation is plotted as an increase above background (mock transfection) in arbitrary units (mean ± SD for three independent experiments). (D) Caspase 2 message levels in HeLa cells transfected with varying concentrations of siRNAs. Concentration of each siRNA from left to right were 10, 30 and 100 nM.
The effect unmodified and BndG-modified siRNAs have on caspase 2 mRNA levels was monitored in HeLa cells 48h post transfection using real time RT–PCR (Figure 7D). The results were normalized to a scrambled sequence control siRNA. Unmodified caspase 2 siRNA reduced message levels by 80% at all three concentrations tested (10, 30 and 100 nM), whereas the antisense single strand alone had no effect. Importantly, the siRNA modified with BndG at positions 9 and 14 was nearly as effective as unmodified siRNA at these concentrations. Furthermore, the siRNA BndG-modified at positions 7, 9 and 14, which is an extremely poor activator of PKR, was capable of reducing caspase 2 message levels by 70% at 100 nM. However, some potency is lost for this modified siRNA, since only ∼45% knockdown is observed at 10 nM.
DISCUSSION
In this study, we set out to determine if two processes that involve the recognition of short dsRNAs (RNA activation of PKR and siRNA-mediated RNA interference) share similar structural requirements in the duplex. Our laboratory has developed methods for identifying binding sites for dsRBM proteins on duplex RNA and selectively blocking those sites by site-specific chemical modification (47,48). We applied this approach to control the manner in which PKR binds to siRNAs, which were shown to be PKR activators both in vitro and in cultured cells. Modified siRNAs with varying ability to activate PKR were then tested for their ability to induce an RNA interference effect. Results of these experiments suggest that the structural requirements for PKR activation and RNA interference are distinct, since chemical modification can be used to inhibit siRNA activation of PKR while maintaining the ability of the siRNA to knock down its RNAi target. These studies have also provided additional insight into the relationship between RNA structure and the efficacy of PKR activation.
SiRNAs activate PKR
Williams and co-workers (32) have presented data showing activation of PKR by siRNAs both in vitro and in cultured cells. Recently, Zhang et al. (33) also showed that PKR is activated in N9 cells when transfected with 50 nM siRNA. Our results are consistent with theirs and show activation of PKR by three unrelated siRNAs in vitro and by caspase 2 siRNA in cultured human cells. It is highly unlikely that the activation seen here comes from contaminating longer dsRNAs, as has been suggested in other studies (49), since the siRNA duplexes are purified on native gels before use and are prepared from gel purified, chemically synthesized strands. Furthermore, site-specific chemical modification that is shown to inhibit PKR binding also inhibits activation of PKR by these siRNA duplexes. To address the issue of 3′ overhangs playing a role in PKR activation, we showed that the blunt-ended siRNA still activates PKR, but to a lesser extent. This may be due to the stabilizing effect of 3′ overhangs or the 2 nt overhangs might be involved in the interactions with the PKR dsRBMs. The observation of activation of PKR by duplexes with only 19 bp appears to contradict other reports that indicated duplexes shorter than 33 bp cannot activate PKR (27,37). We would argue that this may simply be a result of our assay conditions that are optimized to observe activation by the short RNAs. For instance, our conditions include the use of a low concentration of PKR (200 nM) to reduce background activity from RNA-independent PKR dimerization at higher concentrations (16). Under these conditions, three different siRNAs with 19 bp and 2 nt 3′ overhangs were capable of activating PKR's kinase activity in a concentration dependent manner displaying the well-established bell-shaped curve for dependence on the concentration of the activator (Figures 2, 6 and 7). To compare the difference in activation between a 19 bp siRNA and a duplex that exceeds the previously reported minimal length required for activation, we generated a 37mer duplex and examined its ability to activate PKR under identical conditions. The 37mer proved to be <2-fold more effective in activating PKR compared to a 19 bp siRNA. However, the duplex RNA polymer poly(I)•poly(C) showed substantially greater activation of PKR under these conditions (17-fold versus 3-fold). We believe this is related to its ability to assemble multiple PKR dimers on the same RNA duplex molecule allowing both RNA-templated dimer formation and RNA-templated inter-dimer phosphorylation (see below).
Site-specific modifications inhibit formation of the kinase-activating PKR•RNA complex
Our directed hydroxyl radical cleavage data indicate that PKR's dsRBM I can bind an siRNA duplex in at least four different ways, with two overlapping cleavage patterns observed at each end of the duplex. By chemical modification at specific nucleotides within the putative dsRBM I-binding sites, the efficiency of cleavage was reduced in a predictable manner. Indeed, the benzyl modifications inhibited the cleavage reaction at specific nucleotides, consistent with inhibition of binding at distinct sites. If the modifications caused a global change in helical structure, one would expect to see a nonspecific decrease in cleavage efficiency throughout the duplex, which was not observed. Furthermore, although the BndG modifications do have an effect on the thermal stability of the siRNA, these effects cannot explain the differences in activation observed for duplexes of similar stability modified at different sites (Figure 6).
Both dsRBM I- and dsRBM II-directed cleavage was reduced when BndG was substituted at position 6 of the sense strand (Figure 5). It is difficult to envision how modification at position 6 inhibits dsRBM II binding directly to this site. Rather we assert it is more likely that the dsRBM II binding is inhibited because benzylation at position 6 blocks dsRBM I binding in a complex that also involves the covalently linked dsRBM II contacting the RNA. Thus, it appears that BndG modification at positions 6 and 9 could block interaction of dsRBM I and dsRBM II, respectively, from a PKR monomer. We have observed this type of simultaneous binding by the two dsRBMs to another PKR ligand that activates the enzyme (42). These observations are consistent with a previous proposal that activation requires enough RNA to bind both of PKR's dsRBMs (29). Furthermore, given that PKR's dsRBMs show little sequence specificity, a similar complex can be envisioned with PKR's dsRBMs bound at the opposite end of the duplex. This complex should be disrupted by modification near the base pairs related by symmetry. These base pairs include positions 11 and 14 of the sense strand. Indeed, we found that combinations of BndG modifications near positions 6, 9, 11 or 14 in the sense strand prevent the duplex from activating PKR (Figures 6 and 7).
It has been shown recently that PKR undergoes dimerization at high concentrations in the absence of dsRNA leading to autophosphorylation and activation of the kinase (16). Others had demonstrated that RNA perturbs the PKR monomer/dimer equilibrium to favor formation of the dimer (50). These studies indicated that an important role for an activating RNA is to serve as a scaffold to assemble the PKR dimer at low concentrations. This explains the bell-shaped curve observed for the dependence of PKR activity on RNA concentration (i.e. at higher RNA concentrations the stoichiometry of the PKR:RNA complex changes from 2:1 to 1:1). Thus, full activation of the kinase may require enough RNA to bind both dsRBMs from both monomers of the dimer (51).
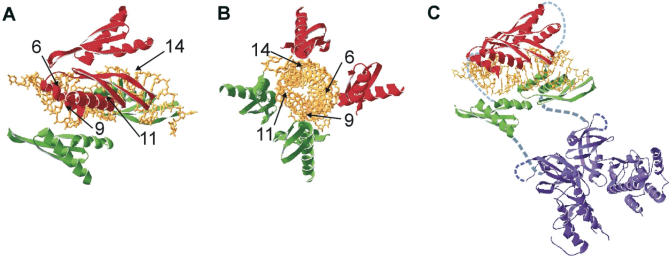
Given the previously described RNA duplex length requirements, the size and positioning of dsRBM-binding sites and the known requirement for dimerization, our model for activation of PKR by an siRNA is presented in Figure 8. We suggest that a PKR dimer assembles on the siRNA with the two dsRBM I components fully bound to the duplex. In contrast, the dsRBM II components of the dimer make partial contact to the duplex at opposite ends. This is consistent with the binding sites revealed by the hydroxyl radical cleavage experiments. Since a dsRBM binds 16 bp across one face of the duplex, four motifs can be accommodated on this relatively short RNA duplex, as shown in Figure 8A and B. Given the ‘back-to-back’ nature of the PKR catalytic domain dimer recently solved by X-ray crystallography, it appears PKR autophosphorylation occurs in a trans-interdimer fashion (52,53). The binding mode we suggest allows two monomers to form a dimer at low PKR concentrations (Figure 8C) that could phosphorylate another PKR dimer bound to a different siRNA molecule. The higher activation observed with the RNA duplex polymer poly(I)•poly(C) compared to the siRNAs most likely arises from its ability to bind more than one PKR dimer in an orientation that allows for templated inter-dimer phosphorylation.
Figure 8.
(A–C) Model for siRNA binding and activation of PKR (21,22,53). The model was generated using Swiss-PdbViewer (70) based on directed hydroxyl radical cleavage data of the PKR dsRBMs on unmodified HPV-E7 siRNA (Figure 3) and the observed effects of BndG modification on PKR binding and activation (Figures 5–7). Numbers indicate the positions on the siRNA sense strand where BndG modification disrupts PKR interactions.
Our model for the active complex involving an siRNA duplex does not include full contact to the RNA for dsRBM II from each of the two monomers. We estimate this would require an additional 4–5 bp at each end, resulting in a duplex RNA length of 27–29 bp, closer to that reported previously to be the minimal activation length (between 24 and 33 bp). This is consistent with our observation that a 37mer duplex is a slightly more effective PKR activator (Figure 2). The 37 bp would more easily accommodate all four dsRBMs of a PKR dimer. The model is also consistent with the observed effects of modification on kinase activation. Modification at positions 9 and 14 of the siRNA sense strand causes a 75–80% reduction in maximum activation (Figures 6D and 7B). Interestingly, this arrangement of modifications places the benzyl groups in dsRBM-binding sites on opposite faces of the helix. This is more effective at inhibition of PKR than positioning the modifications on adjacent faces (e.g. positions 9 and 11 or position 6 and 9). This could be because this arrangement prevents two dsRBMs from different PKR monomers from coming in proximity in the complex, since no pair of adjacent dsRBM-binding sites is unmodified.
The effect of PKR-inhibiting modifications on RNA interference
siRNAs must interact with multiple dsRNA-binding proteins to function in RNAi including proteins that are part of the RISC loading complex and RISC itself for the antisense strand (54). Part of the impetus for this study was to determine if base modifications could be made to an siRNA that inhibited PKR activation while maintaining RNA interference activity. A similar approach has been described recently where backbone modifications of an siRNA were shown to block RNAi ‘off-target’ effects arising from partial complementarity between the antisense strand and off-target messages (55). This requires detailed information on the RNA structure/activity relationships for both processes to determine the extent to which these relationships overlap. We (42) and others (27,28,37,56) have reported on the effect changes in duplex RNA structure have on PKR binding and kinase activation. In addition, there is substantial evidence in the literature that the RNA interference activity of an siRNA is maintained after extensive modification of both strands, but particularly for modifications on the sense strand (57–60). Modifications that stabilize the siRNA duplex have had the effect of increasing potency, probably by slowing nuclease degradation of the component strands during the experiment (59–61). Multiple BndG modifications on the siRNA sense strand are shown here to decrease RNAi potency (Figure 7D). Since these modifications also have a small destabilizing effect on thermal stability of the duplex, the decreased RNAi potency observed could be a result of this stability defect (Supplementary Table 2). This would likely be overcome with additional modifications designed to increase duplex stability.
An important outcome of this study is the generation of siRNA duplexes with varying ability to activate PKR. These new reagents should prove valuable in defining further the extent to which PKR is involved in pathways activated by siRNAs. For instance, it will be informative to evaluate the effects on both JaK-STAT signaling and the expression levels of interferon-stimulated genes in various cell lines transfected with the modified siRNAs described here (32,34). In addition, it will be important to define their effect on the activity of other cellular components regulated by dsRNA such as Toll-like receptors, 2′, 5′-oligoadenylate synthases, RNA-editing adenosine deaminases and dsRNA-activated transcription factors (62–68). Indeed, it has been shown recently that the RNA-editing adenosine deaminase ADAR1, which is interferon inducible, decreases the efficiency of RNAi by forming a stable cytoplasmic complex with siRNAs (Kd = 0.21 nM) (68). This effect may be enhanced in cell lines expressing higher levels of ADARs (68,69). The BndG modified siRNAs described here likely have reduced binding affinity for ADAR1, since, like PKR, ADAR1 binds dsRNA via dsRBMs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
This work was funded by a grant from the National Institutes of Health to PAB (GM57214). Funding to pay the Open Access publication charges for this article was provided by University of Utah.
Conflict of interest statement. None declared.
REFERENCES
- 1.Samuel C.E. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leroux A., London I.M. Regulation of protein synthesis by phosphorylation of eukaryotic initiation factor 2alpha in intact reticulocytes and reticulocyte lysates. Proc. Natl Acad. Sci. USA. 1982;79:2147–2151. doi: 10.1073/pnas.79.7.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaufman R. The double-stranded RNA-activated protein kinase PKR. In: Sonenberg N., Hershey J.W.B., Mathews M.B., editors. Translation Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2000. pp. 503–527. [Google Scholar]
- 4.Galabru J., Hovanessian A. Autophosphorylation of the protein kinase dependent on double-stranded RNA. J. Biol. Chem. 1987;262:15538–15544. [PubMed] [Google Scholar]
- 5.Koromilas A.E., Roy S., Barber G.N., Katze M.G., Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- 6.Meurs E.F., Galabru J., Barber G.N., Katz M.G., Hovanessian A.G. Tumor suppressor function of the interferon-induced double-stranded RNA-activated protein kinase. Proc. Natl Acad. Sci. USA. 1993;90:232–236. doi: 10.1073/pnas.90.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis S., Watson J.C. In vitro activation of the interferon-induced, double-stranded RNA-dependent protein kinase PKR by RNA from the 3′ untranslated regions of human alpha-tropomyosin. Proc. Natl Acad. Sci. USA. 1996;93:508–513. doi: 10.1073/pnas.93.1.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salzberg S., Vilchik S., Cohen S., Heller A., Kronfield-Kinar Y. Expression of a PKR dominant-negative mutant in myogenic cells interferes with the myogenic process. Exp. Cell Res. 2000;254:45–54. doi: 10.1006/excr.1999.4721. [DOI] [PubMed] [Google Scholar]
- 9.Kumar A., Haque J., Lacoste J., Hiscott J., Williams B.R.G. Double-stranded RNA-dependent protein kinase activates transcription factor NF-kappa B by phosphorylating I kappa B. Proc. Natl Acad. Sci. USA. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar A., Yang Y.L., Flati V., Der S., Kadereit S., Deb A., Haque J., Reis L., Weissmann C., Williams B.R.G. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-kappaB. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong A.H.T., Wai Ning Tam N., Yang Y.L., Cuddihy A.R. Physical association between STAT1 and the interferon-inducible protein kinase PKR and implications for interferon and double-stranded RNA signaling pathways. EMBO J. 1997;16:1291–1304. doi: 10.1093/emboj/16.6.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuddihy A.R., Wong A.H., Tam N.W., Li S., Koromilas A.E. The double-stranded RNA activated protein kinase PKR physically associates with the tumor suppressor p53 protein and phosphorylates human p53 on serine 392 in vitro. Oncogene. 1999;18:2690–2702. doi: 10.1038/sj.onc.1202620. [DOI] [PubMed] [Google Scholar]
- 13.Xu Z., Williams B.R.G. The B56 alpha regulatory subunit of protein phosphatase 2A is a target for regulation by double-stranded RNA-dependent protein kinase PKR. Mol. Cell. Biol. 2000;20:5285–5299. doi: 10.1128/mcb.20.14.5285-5299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brand S.R., Kobayashi R., Mathews M.B. The tat protein of human immunodeficiency virus type 1 is a substrate and inhibitor of the interferon-induced, virally activated protein kinase, PKR. J. Biol. Chem. 1997;272:8388–8395. doi: 10.1074/jbc.272.13.8388. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs B.L., Imani F. Histone proteins inhibit activation of the interferon-induced protein kinase by binding to double-stranded RNA. J. Interferon Res. 1988;8:821–830. doi: 10.1089/jir.1988.8.821. [DOI] [PubMed] [Google Scholar]
- 16.Leamaire P.A., Lary J., Cole J.L. Mechanism of PKR activation: dimerization and kinase activation in the absence of double-stranded RNA. J. Mol. Biol. 2005;345:81–90. doi: 10.1016/j.jmb.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 17.Fierro-Monti I., Mathews M.B. Proteins binding to duplexed RNA: one motif, multiple functions. Trends Biochem. Sci. 2000;25:241–246. doi: 10.1016/s0968-0004(00)01580-2. [DOI] [PubMed] [Google Scholar]
- 18.Stefl R., Skrisovska L., Allain F.H. RNA sequence- and shape-dependent recognition by proteins in the ribonucleoprotein particle. EMBO Rep. 2005;6:33–38. doi: 10.1038/sj.embor.7400325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saunders L.R., Barber G.N. The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J. 2003;17:961–983. doi: 10.1096/fj.02-0958rev. [DOI] [PubMed] [Google Scholar]
- 20.Tian B., Bevilacqua P.C., Diegelman-Parente A., Mathews M.B. The double-stranded-RNA-binding motif: interference and much more. Nature Rev. Mol. Cell Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- 21.Ryter J.M., Schultz S.C. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramos A., Grunert S., Adams J., Micklem D.R., Proctor M.R., Freund S., Bycroft M., St Johnston D., Varani G. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 2000;19:997–1009. doi: 10.1093/emboj/19.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu H., Henras A., Chanfreau G., Feigon J. Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III. Proc. Natl Acad. Sci. USA. 2004;101:8307–8312. doi: 10.1073/pnas.0402627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blaszczyk J., Gan J., Tropea J.E., Court D.L., Waugh D.S., Ji X. Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure. 2004;12:457–466. doi: 10.1016/j.str.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 25.Wu S., Kaufman R.J. A model for the double-stranded RNA (dsRNA)-dependent dimerization and activation of the dsRNA activated protein kinase PKR. J. Biol. Chem. 1997;272:1291–1296. doi: 10.1074/jbc.272.2.1291. [DOI] [PubMed] [Google Scholar]
- 26.Ung T.L., Cao C., Lu J., Ozato K., Dever T.E. Heterologous dimerization domains functionally substitute for the double-stranded RNA binding domains of the kinase PKR. EMBO J. 2001;20:3728–3737. doi: 10.1093/emboj/20.14.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manche L., Green S.R., Schmedt C., Mathews M.B. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maitra R.K., McMillan N.A., Desai S., McSwiggen J., Hovanessian A.G., Sen G., Williams B.R.G., Silverman R.H. HIV-1 TAR RNA has an intrinsic ability to activate interferon-inducible enzymes. Virology. 1994;204:823–827. doi: 10.1006/viro.1994.1601. [DOI] [PubMed] [Google Scholar]
- 29.Nanduri S., Rahman F., Williams B.R.G., Qin J. A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. EMBO J. 2000;19:5567–5574. doi: 10.1093/emboj/19.20.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Billy E., Brondani V., Zhang H., Ulrich M., Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl Acad. Sci. USA. 2001;98:14428–14433. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 32.Sledz C.A., Holko M., de Veer M.J., Silverman R.H., Williams B.R.G. Activation of the interferon system by short-interfering RNAs. Nature Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z., Weinschenk T., Guo K., Schluesener H.J. siRNA binding proteins of microglial cells: PKR is an unanticipated ligand. J. Cell. Biochem. 2006;97:1217–1229. doi: 10.1002/jcb.20716. [DOI] [PubMed] [Google Scholar]
- 34.Persengiev S.P., Zhu X., Green M.R. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) RNA. 2004;10:12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scacheri P.C., Rozenblatt-Rosen O., Caplen N.J., Wolfsberg T.G., Umayam L., Lee J.C., Hughes C.M., Shanmugam K.S., Bhattacharjee A., Meyerson M., et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:1892–1897. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hornung V., Guenthner-Biller M., Bourquin C., Ablasser A., Schlee M., Uematsu S., Noronha A., Manoharan M., Akira S., de Fougerolles A., et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nature Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 37.Zheng X., Bevilacqua P.C. Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA. 2004;10:1934–1945. doi: 10.1261/rna.7150804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allerson C.R., Chen S.L., Verdine G.L. A chemical method for site-specific modification of RNA: the convertible nucleoside approach. J. Am. Chem. Soc. 1997;119:7423–7433. [Google Scholar]
- 39.Gunnery S., Mathews M.B. RNA binding and modulation of PKR activity. Methods. 1998;15:189–98. doi: 10.1006/meth.1998.0623. [DOI] [PubMed] [Google Scholar]
- 40.McKenna S.A., Kim I., Liu C.W., Puglisi J.D. Uncoupling of RNA binding and PKR kinase activation by viral inhibitor RNAs. J. Mol. Biol. 2006;358:1270–1285. doi: 10.1016/j.jmb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 41.Jammi N.V., Beal P.A. Phosphorylation of the RNA-dependent protein kinase regulates its RNA-binding activity. Nucleic Acids Res. 2001;29:3020–3029. doi: 10.1093/nar/29.14.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spanggord R.J., Vuyisich M., Beal P.A. Identification of binding sites for both dsRBMs of PKR on kinase-activating and kinase-inhibiting RNA ligands. Biochemistry. 2002;41:4511–4520. doi: 10.1021/bi0120594. [DOI] [PubMed] [Google Scholar]
- 43.Vuyisich M., Spanggord R.J., Beal P.A. The binding site of the RNA-dependent protein kinase (PKR) on EBER1 RNA from Epstein-Barr virus. EMBO rep. 2002;3:622–627. doi: 10.1093/embo-reports/kvf137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsui T., Tanihara K., Date T. Expression of unphosphorylated form of human double-stranded RNA-activated protein kinase in Escherichia coli. Biochem. Biophys. Res. Commun. 2001;284:798–807. doi: 10.1006/bbrc.2001.5039. [DOI] [PubMed] [Google Scholar]
- 45.Tan S.L., Tareen S.U., Melville M.W., Blakely C.M., Katze M.G. The direct binding of the catalytic subunit of protein phosphatase 1 to the PKR protein kinase is necessary but not sufficient for inactivation and disruption of enzyme dimer formation. J. Biol. Chem. 2002;277:36109–36117. doi: 10.1074/jbc.M205109200. [DOI] [PubMed] [Google Scholar]
- 46.Spanggord R.J., Beal P.A. Selective binding by the RNA binding domain of PKR revealed by affinity cleavage. Biochemistry. 2001;40:4272–4280. doi: 10.1021/bi002512w. [DOI] [PubMed] [Google Scholar]
- 47.Puthenveetil S., Véliz E.A., Beal P.A. Site-specific modification of Epstein–Barr virus-encoded RNA 1 with N2-benzylguanosine limits the binding sites occupied by PKR. ChemBioChem. 2004;5:383–386. doi: 10.1002/cbic.200300816. [DOI] [PubMed] [Google Scholar]
- 48.Stephens O.M., Haudenschild B.L., Beal P.A. The binding selectivity of ADAR2′s dsRBMs contributes to RNA-editing selectivity. Chem. Biol. 2004;11:1239–1250. doi: 10.1016/j.chembiol.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 49.Mellits K.H., Pe'ery T., Manche L., Robertson H.D., Mathews M.B. Removal of double-stranded contaminants from RNA transcripts: synthesis of adenovirus VA RNAI from a T7 vector. Nucleic Acids Res. 1990;18:5401–5406. doi: 10.1093/nar/18.18.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carpick B.W., Graziano V., Schneider D., Maitra R.K., Lee X., Williams B.R.G. Characterization of the solution complex between the interferon-induced, double-stranded RNA-activated protein kinase and HIV-I trans-activating region RNA. J. Biol. Chem. 1997;272:9510–9516. doi: 10.1074/jbc.272.14.9510. [DOI] [PubMed] [Google Scholar]
- 51.Robertson H.D., Mathews M.B. The regulation of the protein kinase PKR by RNA. Biochimie. 1996;78:909–914. doi: 10.1016/s0300-9084(97)86712-0. [DOI] [PubMed] [Google Scholar]
- 52.Dey M., Cao C., Dar A.C., Tamura T., Ozato K., Sicheri F., Dever T.E. Mechanistic link between PKR dimerization, autophosphorylation, and eIF2alpha substrate recognition. Cell. 2005;122:901–913. doi: 10.1016/j.cell.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 53.Dar A.C., Dever T.E., Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 54.Tang G. siRNA and miRNA: an insight into RISCs. Trends Biochem. Sci. 2005;30:106–114. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 55.Jackson A.L., Burchard J., Leake D., Reynolds A., Schelter J., Guo J., Johnson J.M., Lim L., Karpilow J., Nichols K., et al. Position-specific chemical modification of siRNAs reduces ‘off-target’ transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghadge G.D., Malhotra P., Furtado M.R., Dhar R., Thimmapaya B. In vitro analysis of virus-associated RNA I (VAI RNA): inhibition of the double-stranded RNA-activated protein kinase PKR by VAI RNA mutants correlates with the in vivo phenotype and the structural integrity of the central domain. J. Virol. 1994;68:4137–4151. doi: 10.1128/jvi.68.7.4137-4151.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prakash T.P., Allerson C.R., Dande P., Vickers T.A., Sioufi N., Jarres R., Baker B.F., Swayze E.E., Griffey R.H., Bhat B. Positional effect of chemical modifications on short interference RNA activity in mammalian cells. J. Med. Chem. 2005;48:4247–4253. doi: 10.1021/jm050044o. [DOI] [PubMed] [Google Scholar]
- 58.Allerson C.R., Sioufi N., Jarres R., Prakash T.P., Naik N., Berdeja A., Wanders L., Griffey R.H., Swayze E.E., Bhat B. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- 59.Chiu Y.L., Rana T.M. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Czauderna F., Fechtner M., Dames S., Aygun H., Klippel A., Pronk G.J., Giese K., Kaufmann J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Layzer J.M., McCaffrey A.P., Tanner A.K., Huang Z., Kay M.A., Sullenger B.A. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 63.Kariko K., Bhuyan P., Capodici J., Ni H., Lubinski J., Friedman H., Weissman D. Exogenous siRNA mediates sequence-independent gene suppression by signaling through toll-like receptor 3. Cells Tissues Organs. 2004;177:132–138. doi: 10.1159/000079987. [DOI] [PubMed] [Google Scholar]
- 64.Rebouillat D., Hovanessian A.G. The human 2′,5′-oligoadenylate synthetase family: interferon-induced proteins with unique enzymatic properties. J. Interferon Cytokine Res. 1999;19:295–308. doi: 10.1089/107999099313992. [DOI] [PubMed] [Google Scholar]
- 65.Knight S.W., Bass B.L. The role of RNA editing by ADARs in RNAi. Mol. Cell. 2002;10:809–817. doi: 10.1016/s1097-2765(02)00649-4. [DOI] [PubMed] [Google Scholar]
- 66.Tonkin L.A., Bass B.L. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science. 2003;302:1725. doi: 10.1126/science.1091340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Daly C., Reich N.C. Double-stranded RNA activates novel factors that bind to the interferon-stimulated response element. Mol. Cell. Biol. 1993;13:3756–3764. doi: 10.1128/mcb.13.6.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang W., Wang Q., Howell K.L., Lee J.T., Cho D.S., Murray J.M., Nishikura K. ADAR1 RNA deaminase limits siRNA efficacy in mammalian cells. J. Biol. Chem. 2004;280:3946–3953. doi: 10.1074/jbc.M407876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wagner R.W., Yoo C., Wrabetz L., Kamholz J., Buchhalter J., Hassan N.F., Khalili K., Kim S.U., Perussia B., McMorris F.A. Double-stranded RNA unwinding and modifying activity is detected ubiquitously in primary tissues and cell lines. Mol. Cell. Biol. 1990;10:5586–5590. doi: 10.1128/mcb.10.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.