Abstract
The genomic stability of the rDNA tandem array is tightly controlled to allow sequence homogenization and to prevent deleterious rearrangements. In this report, we show that the absence of the yeast CTD kinase I (CTDK-I) complex in null mutant strains leads to a decrease in the number of tandem rDNA repeats. Reintroduction of the missing gene induces an increase of rDNA repeats to reach a copy number similar to that of the original strain. Interestingly, while expansion is dependent on Fob1, a protein required for replication fork blocking activity in rDNA, contraction occurs in the absence of Fob1. Furthermore, silencing of class II genes at the rDNA, a process connected to rDNA stability, is not affected. Ctk1, the kinase subunit of the CTDK-I complex is involved in various steps of mRNA synthesis. In addition, we have recently shown that Ctk1 is also implicated in rRNA synthesis. The results suggest that the RNA polymerase I transcription defect occurring in a ctk1 mutant strain causes rDNA contraction.
INTRODUCTION
In the yeast Saccharomyces cerevisiae, the genes encoding the ribosomal RNAs are organized as a 9.1 kb unit repeated tandemly about 150 times on chromosome XII (1). Each unit contains a gene coding the 35S rRNA precursor transcribed by RNA polymerase I (Pol I), and a 5S gene transcribed by RNA polymerase III. These two transcription units are separated by non-transcribed spacers: NTS1 and NTS2. NTS1 contains replication fork blocking (RFB) sequences (2) and an RNA polymerase II (Pol II) promoter (3), whereas NTS2 contains an origin of replication (4) and a cohesin associating region (CAR) (5).
The number of ribosomal units is highly regulated in order to maintain the integrity of the rDNA locus. Whereas extensive recombination may be harmful to cells, a limited and regulated level of recombination is important for cellular adaptation, repeat number maintenance and also to keep sequence homogeneity among rRNA genes. If repeats are either inserted or deleted, a wild-type number is quickly restored (6). Fob1 and Sir2, two non-essential nucleolar factors (7,8), together with DNA topoisomerase I and proteins required for general recombination, are engaged in rDNA recombination. Fob1 (Fork barrier binding protein) can bind RFB sites (9), thus displaying a RFB activity required to prevent collision between DNA replication and rDNA transcription (10). It is postulated that such a block allows the formation of double strand breaks (DSBs) that can initiate recombination (11). Recombination through unequal, as opposed to equal, gene conversion, will lead to changes in rDNA copy number. Kobayashi et al. (6) have shown that in the absence of Pol I, rDNA copy number decreases to approximately one-half of the normal value. Restoration of Pol I activity leads to rDNA expansion until repeat number returns to the normal level. Fob1 has been shown to be required for both rDNA contraction and expansion processes (6).
Sir2, a NAD+-dependent histone deacetylase, was first identified as a silent information regulator (SIR) gene required for silencing Pol II transcription of genes inserted into the rDNA (12). Sir2 plays an essential role in forming higher-order repressive chromatin structure that prevents general access to Pol II machinery, thus causing silencing in the chromosomal rDNA repeats. Sir2 decreases the rate of recombination within rDNA repeats (13), regulates the initiation of DNA replication (14), and suppresses recombination at replication forks stalled at the RFB (15). It has been shown recently that rDNA amplification is dependent on transcription from a non-coding bidirectional promoter (E-pro) that is located in the rDNA spacer NTS1 (3). The authors suggested that transcription of E-pro by Pol II stimulates unequal recombination by disrupting cohesin association with the rDNA, thus allowing a change in rDNA copy number. In normal situations, Sir2 is a negative regulator of E-pro transcription, its activity thus allowing cohesin to associate with rDNA, thereby preventing recombination.
An unanswered question is why cells keep such a high content of rDNA units. In growing yeast cells, only a fraction of the 35S genes are actively transcribed by Pol I. Indeed, regulation of 35S synthesis can basically occur at two main levels: the ratio of active genes and the transcription rate of each gene. The percentage of active genes decreases dramatically as cells enter stationary phase (16), whereas in exponential cells, rRNA transcription rate is determined by the overall initiation rate, rather than the number of active genes (17). The major step controlling the rate of 35S synthesis is transcription initiation. Specific initiation requires Pol I-Rrn3, the upstream activator complex (UAF), the TATA binding protein and the core factor (18). Pol I-specific subunits of UAF are not absolutely required for cell growth, and UAF is not required in vitro for basal transcription (19,20). In the absence of functional UAF, efficient growth can be achieved by transcribing endogenous rDNA by Pol II, a process known as polymerase switched state (PSW) (21). It has been further shown that the alteration responsible for the PSW phenotype resides in expansion of chromosomal rDNA repeats (22).
CTD kinase I (CTDK-I) is a cyclin-dependent kinase complex composed of three subunits, Ctk1 encoding the catalytic subunit (23), Ctk2 and Ctk3, a cyclin and a co-cyclin, respectively, each required for kinase activity (24). Although CTDK-I is not essential for cell viability, deletion of each gene leads to cold sensitivity and severe growth defects. Ctk1 is one of the four kinases involved in phosphorylation of the C-terminal domain (CTD) of Pol II. It has been implicated in various aspects of mRNA synthesis. Ctk1 is the major kinase involved in CTD phosphorylation during transcription elongation (25), and it has also been connected to splicing (26), 3′ end processing (27,28), DNA damage-induced transcription (29), histone methylation (30) and nuclear export of mRNA (31).
We have recently shown that, similar to other factors previously described as specific components of the Pol II transcriptional machinery (32–36), Ctk1 is involved in rRNA synthesis: (i) Ctk1 is present in the nucleolus and interacts directly with Pol I, (ii) Δctk cells exhibit defects in nucleolar structure and (iii) in the absence of Ctk1, Pol I transcription is affected both in vivo and in vitro (37). In this report, we show that Ctk1 is required for the integrity of the rDNA locus. Null ctk mutant strains display rDNA contraction phenotypes that are fully reversed when the null mutation is reversed. Surprisingly, contraction does not require Fob1, suggesting a mechanism distinct from that leading to contraction observed in the absence of rDNA transcription by Pol I. The possible connection between a Pol I transcriptional defect and rDNA contraction is discussed.
MATERIALS AND METHODS
Strains and cultures
The yeast strains used in this study were derived from FY1679-18B (MATα his3Δ200 leu2Δ1 trp1Δ63 ura3-52) and W303−1B (MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1). Cells were grown in either YPD or SD medium supplemented with appropriate nutrients. All yeast constructs were obtained by one-step gene replacement (38). The MCD1-HA and SIR2-HA tagged-strains were constructed by fusion of sequences encoding three HA-tags at the 3′ end of each gene using the plasmid pFA6a-3HA-His3MX6 (39). Δctk1, Δctk2 and Δctk3 null mutants are described in (40). fob1 and ctk1 null mutations were created by replacing each open reading frame by the Schizosaccharomyces pombe HIS5 [pFA6a-His3MX6, (39)] and the TRP1 genes, respectively. Complementation of ctk1 and ctk2 null mutations were performed by replacing the TRP1 marker by either CTK1 or CTK2 open reading frame carried on a PCR-fragment. The resulting strains were grown for ∼60 generations before chromosome analysis. Complementation of Δctk1-Δfob1 cells was obtained by transformation with the CTK1 gene born on a LEU2 CEN plasmid (40). For psoralen cross-linking and primer extension analyses, we used a clone of Δfob1-Δctk1 cells that had not undergone rDNA contraction. Cells were grown for 100 generations by serial culturing (dilution when cultures reached OD 1.6): dilutions were performed every day for Δctk strains (doubling time of 3 h) and twice a day for CTK strains (doubling time of 1.5 h). OD measurements were performed with a BioPhotometer (Eppendorf) at a wavelength of 600 nm.
For silencing studies, strains were constructed as in (41). The mURA3-HIS3 cassette was inserted in FY1679-18B cells either 50 nt before the left side of the rDNA array, or replacing the trp1Δ63 gene to generate WT-50L and WT-C, respectively. Transformants were selected by histidine prototrophy. CTK1 open reading frame was then replaced by the TRP1 marker to yield Δctk1-50L and Δctk1-C, respectively. Further SIR2 disruption was performed by replacing SIR2 open reading frame by the KanMX6 marker (39). To assay mURA3 expression, 5 μl of 10-fold serial dilutions of cells grown to early exponential phase (OD 0.4) were spotted onto SD medium supplemented with tryptophane and leucine, plus either histidine (SD-URA) or uracil (SD+URA).
For homologous recombination studies, CTK1 was deleted in RAD5+ W303-1B derived D444-1D cells (arg4ΔBglII URA3::arg4EcoRV::ura3-1). Wild-type and two independent Δctk1 strains were plated on YPD prior to replica on medium without arginine when colonies were 1.5 mm diameter. The number of papillae was scored after 2 day and 4 day incubation for wild-type and Δctk1 strains, respectively.
Contour-clamped homogeneous electric field electrophoresis
7 × 107 cells grown in YPD were washed with 50 mM EDTA (pH 8) and suspended in 100 μl agarose mixture (1% agarose SeaPlaque GTG, 1 M sorbitol and 20 mM EDTA, pH 8) at 45°C, supplemented with lyticase (Sigma) and β-mercaptoethanol at final concentration of 0.2 mg/ml and 0.1%, respectively. Plugs were incubated for 2 h at 37°C with gentle agitation in buffer A (1 M sorbitol, 20 mM EDTA, pH 8, and 10 mM Tris, pH 7.5) supplemented with lyticase (0.2 mg/ml) and β-mercaptoethanol (0.1%). Plugs were further incubated in lysis buffer (1% lithium lauryl sulfate, 100 mM EDTA, pH 8, and 10 mM Tris, pH 8) for 1 h at 37°C. After overnight incubation at 37°C in fresh lysis buffer, plugs were washed for 30 min with TE buffer pH 8 (three times at 50°C and three times at room temperature). DNA was subjected to gel electrophoresis in a 0.8% agarose gel (SeaKem GTG agarose), 0.5× TBE, using a contour-clamped homogeneous electric field (CHEF) Mapper (Bio-Rad) at a linear pulse of 5–15 min, 3 V/cm, an angle of 120°, at 12°C for 68 h. The gel was then stained with 1 μg/ml ethidium bromide for 20 min at room temperature and washed twice for 10 min before being photographed. The gel was eventually further analysed by Southern hybridization.
Isolation of DNA and Southern blot analysis
Yeast cells were washed in cold water, suspended at a concentration of 2.5 × 108 cells per ml in Yeast Wash Buffer (1 M sorbitol, 40 mM KPO4, pH 7.5 and 20 mM β-mercaptoethanol). Cells were then incubated with 0.1 mg/ml zymolyase 100T (ICN) for 30 min at 37°C. The spheroplasts were washed in Yeast Wash Buffer and lysed with 500 μl TE buffer (pH 7.5) containing 0.5% SDS. Suspension was further treated with 50 μg/ml RNase (Eurogentec) for 30 min at room temperature and then with 1 mg/ml proteinase K (Roche) for 3 h at 50°C. DNA was extracted twice with phenol:chloroform:isoamyl alcohol and precipitated with propanol. For telomere analysis, 2 μg DNA was digested with 30 U XhoI, separated on 1% agarose gel, transferred on to Hybond N+ membrane (Amersham Biosciences), cross-linked by UV and hybridized with telomere Y' probe. For Extrachromosomal Ribosomal Circles (ERCs) analyses, DNA extraction from 5 × 107 W303-1B derived cells, and one-dimensional gel electrophoresis, were performed as in (42), except that phenol/chloroform and chloroform extraction steps were omitted during DNA preparation. Hybridization of PCR fragments labelled with [α-32P]dCTP by random priming (Prime-it II; Stratagene), was performed using standard methods. ERC quantification from two independent experiments was performed with a PhosphorImager.
rDNA quantification
For rDNA quantification, 10 μg DNA was incubated overnight with 30 U EcoRI at 37°C and analysed by Southern blot with PCR fragments specific for rDNA, and genes located on chromosome XII (VPS13 and SIR3). For real-time PCR analysis, three serial dilutions of genomic DNA were amplified four times with two sets of primers specific for rDNA, and two sets specific for chromosome XII (VPS13 and SIR3). Oligonucleotides' sequences are available on request.
Psoralen cross-linking
Psoralen cross-linking procedure was adapted from (16). Briefly, 5 × 108 early exponential growth-phase cells were collected by centrifugation, washed twice with ice-cold water and suspended in 1.4 ml ice-cold TE buffer, pH 7.5. Cell suspensions were then placed in a six-well culture plate and irradiated 5 min on ice with an UV-lamp (125 watts, 365 nm) at a distance of 6 cm after addition of 70 μl of a 200 μg/ml stock solution in ethanol of 4,5′,8-trimethylpsoralen (Aldrich). This irradiation was performed five times. Genomic DNA was then extracted and digested with EcoRI prior to Southern blotting analysis using either a 35S or an intergenic rDNA-specific probe (16).
Primer extension
Total RNA was extracted from 1 × 108 cells grown to OD 1 by hot acid-phenol treatment as described previously (43). After suspension, 3 μg wild-type total RNA, and the corresponding volume for each sample, were analysed by primer extension as in (20) with 0.2 pmol of end-labelled 35S (ACACGCTGTATAGAGACTAGGC) and U6 (AAACGAAATCTCTTTG) oligonucleotides, plus 5 pmol of ice-cold oligonucleotide. Extension products were separated by electrophoresis on an 8% acrylamide gel containing 7 M urea in TBE. The gel was dried, submitted to autoradiography and analysed with a PhosphorImager (Molecular Dynamics).
Western immunoblotting
Proteins were extracted in TCA by mechanical lysis with glass beads and analysed by western immunoblotting as described previously (40) with either HA-11 monoclonal antibodies (Babco), or polyclonal antibodies directed against the A43 subunit of Pol I (44). After washing, membranes were incubated with horseradish peroxidase-conjugated goat anti-mouse (HA) or anti-rabbit (A43) immunoglobulin G at 1/10 000 (Promega). The blots were visualized by chemoluminescence (Amersham), and signals were quantified with a FluorChem FC (Alpha Innotech Corporation).
Chromatin immunoprecipitation
Wild type-MCD1-HA and ctk1-MCD1-HA strains were grown in YPD medium to OD 2. Cross-link, cell lysis, chromatin preparation, cross-link reversal and immunoprecipitation were performed as previously described (45,46). Immunoprecipitation was realised with protein Dynabeads Pan mouse IgG (Dynal Biotech) coupled with HA-12CA5 antibodies. Immuno-precipitated and total DNA were analysed by real-time PCR with the Platinium SYBR Green kit (Invitrogen) and the 7300 Real-Time PCR System (Applied Biosystems), using 3 pairs of primers amplifying a 122 bp fragment specific of the RFB region (GTGTGAGGAAAAGTAGTTGGGAGGTA, GACGAGGCCATTTACAAAAACATAAC), a 83 bp fragment specific of the CAR region located in NTS2 (CGGAAACGCAGGTGATATGAG, AAAATTGTCCTCCACCCATAACAC), and a 101 bp fragment specific of the CUP1 gene (GGTCATGAGTGCCAATGCCAATGT, GGGCATTTGTCGTCGCTGTTACAC). Two independent experiments were performed and triplicates were analysed 3 times for each experiment. The relative occupancy level is calculated as the ratio between the IP signal and the respective total DNA signal.
RESULTS
The absence of CTDK-I leads to contraction of the rDNA locus
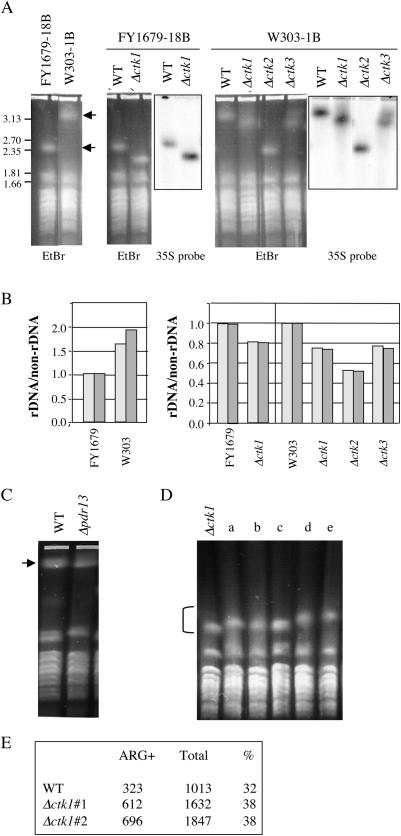
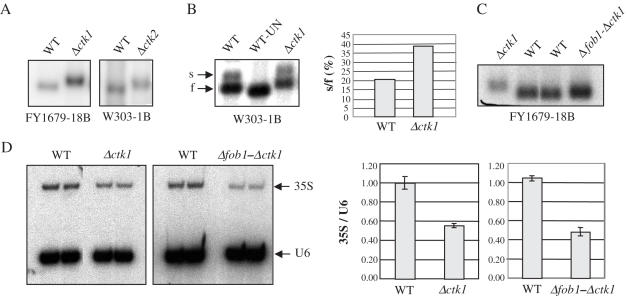
ctk1, ctk2 and ctk3 null mutant cells display identical phenotypes (47), as they all lack Ctk1 kinase activity (24). We have previously shown that the absence of Ctk1 kinase activity leads to defects in nucleolar structure (37). Because it is known that chromosomal rDNA deletion mutants contain a nucleolus that is not organized as a crescent-shaped structure (48), we sought to study the rDNA locus in Δctk1 cells. As the number of rDNA units varies from strain to strain (49), two strains with distinct genetic backgrounds were used: FY1679-18B (50) and W303-1B (51). The size of the XII chromosome, containing the rDNA locus, was analysed using CHEF electrophoresis. Chromosomes were visualized by ethidium bromide staining, and chromosome XII was further identified by Southern blot hybridization with an rDNA probe. To determine rDNA quantities relative to chromosome XII in each strain, genomic DNA was analysed by quantitative real-time PCR and by Southern blotting after enzymatic digestion. We used a probe specific for the 35S gene, and as references, we used probes specific for the left (VPS13) and the right (SIR3) arms of chromosome XII, respectively. We observed that FY1679-18B and W303-1B wild-type cells contained a different sized chromosome XII (Figure 1A), suggesting different contents of rDNA sequences (120 and 230 copies of rDNA genes, respectively). Experiments analysing genomic DNA further confirmed that FY1679-18B cells contained 50% less 35S genes than W303-1B cells (Figure 1B). Interestingly, chromosome XII from Δctk1, Δctk2 and Δctk3 cells, was systematically shorter than that from isogenic wild-type cells (Figure 1A, and data not shown). Specifically in FY1679-18B strains, containing a relatively small chromosome XII, wild-type and mutant chromosome XII each migrated as a sharp band, indicating a rather homogeneous chromosome population. Further analysis of genomic DNA by Southern blotting and quantitative PCR confirmed that the shortening of chromosome XII was owing to contraction of the rDNA tandem array (Figure 1B). For each null mutant strain, rDNA content varied from 50 to 80% of that found in isogenic wild-type cells. Analysis of a dozen independent additional null mutants gave similar results (data not shown). Examination of a catalytic mutant of the Ctk1 kinase (37) revealed also a chromosome XII shorter than wild type (data not shown), with an estimation of rDNA sequences being 40% of that of the isogenic wild type. Altogether, the results show that any ctk mutation leads to rDNA contraction.
Figure 1.
The rDNA tandem array is contracted in Δctk null mutant strains. (A–D) Chromosome XII analysis. FY1679-18B and W303-1B derived cells were grown in YPD. After cell digestion, agarose plugs were subjected to CHEF electrophoresis. (A) Gels were photographed after ethidium bromide straining (EtBr), and further analysed by Southern blotting with a 35S probe and autoradiographed (35S probe). Arrows point to chromosome XII. (B) rDNA was quantified using probes specific for the rDNA and for genes located on the extremities of chromosome XII (VPS13 and SIR3). Genomic DNA was analysed either by Southern blotting and signals were quantified with a PhosphorImager (light grey boxes), or by quantitative real-time PCR (dark grey boxes). Ratios of rDNA over non-rDNA values were reported. Units were set relative to the measurement for the FY1679 strain (left panel) or the wild-type isogenic strain (right panel), performed using Southern blotting. (C) Chromosomes from W303-1B (WT) and isogenic Δpdr13 cells were analysed by CHEF electrophoresis and ethidium bromide straining. The arrow points to chromosome XII species. (D) Five independent cultures of Δctk1 cells (FY1679-18B) were grown for 100 generations in YPD prior to chromosome analysis. Bracket points to chromosome XII. (E) Homologous recombination analysis. Wild-type and Δctk1 cells were spotted on YPD prior to replica-plating on SD-ARG. The number of papillae was scored and reported for each strain (ARG+).
Although the regulation of rRNA synthesis has not been connected to rDNA contraction and expansion (17), we wondered whether the contraction phenotype observed in the ctk mutant strains was the consequence of their reduced growth rate. To get an insight into this question, we analysed Δpdr13 cells. PDR13 encodes a member of the Hsp70 family that is implicated in pleiotropic drug resistance (52). PDR13 is required for normal growth, and Δpdr13 cells display a reduced growth rate similar to that of Δctk1 cells (WT: 1h30, Δpdr13: 2h35 and Δctk1: 2h50). We have previously shown that Δpdr13 cells do not display any defect in Pol I transcription (37). Chromosome analysis by CHEF electrophoresis showed that the size of chromosome XII was similar in Δpdr13 and isogenic wild-type cells (Figure 1C), indicating that Δpdr13 cells did not undergo rDNA contraction. This result suggests that rDNA contraction is not (at least not solely) the consequence of the reduced growth rate of the Δctk1 mutant strain.
Five independent cultures of Δctk1 cells (Figure 1A, FY1679-18B) were carried out for 100 generations, prior to chromosome analysis by CHEF electrophoresis. As previously observed in the FY1679-18B genetic background, each culture contained homogeneous chromosome XII populations migrating as sharp bands (Figure 1D). However, chromosomes from some of the clonal cultures exhibited significant size variations (clones d and e, Figure 1D). We conclude that rDNA contraction phenotypes are not completely stable as they can subsequently undergo slight clonal variations.
rDNA contraction may result either from unequal gene conversion occurring through homologous recombination, or repair of DSB by a single strand annealing mechanism, whereas in contrast, expansion has to result from unequal gene conversion (53). We thus asked whether the reduced size of the rDNA array observed in ctk1 mutant cells was linked to a putative incapacity to perform gene conversion, and thus to expand the rDNA array. To address this question, we tested homologous recombination within a non-rDNA gene duplication. CTK1 was deleted in a strain containing two distinct arg4 mutant alleles (D444-1D; S. Gangloff, unpublished data), so that only an event of homologous recombination can lead to a functional ARG4 gene. Generation of ARG4 cells was monitored by the apparition of papillae after replica-plating on medium lacking arginine. We observed that CTK1 deletion led to a number of papillae on medium without arginine similar to that observed for the wild type (Figure 1E), showing that homologous recombination is not affected in Δctk1 cells. Taken together with the fact that we observed clonal variation including limited expansion, following rDNA contraction in Δctk1 cells, these results suggest that mechanisms required for rDNA expansion are not defective in Δctk1 cells.
rDNA contraction is reversible and specific
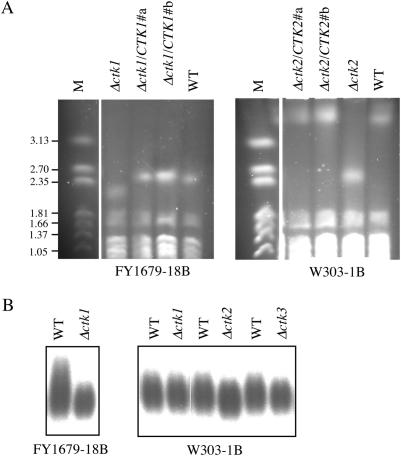
To study whether rDNA contraction observed in the absence of CTDK-I is a reversible process, we reintroduced the appropriate CTK allele at its native chromosomal locus, and subsequently analysed the size of chromosome XII in the resulting strains. For W303-1B genetic background, we chose to analyse complementation in Δctk2 cells that contained the largest rDNA deletion. Interestingly, introduction of the CTK1 and CTK2 genes into Δctk1 (FY1679-18B) and Δctk2 (W303-1B) cells, respectively, restored wild-type size for each strain's chromosome XII (Figure 2A). We conclude that the rDNA contraction induced by the absence of the CTDK-I kinase is fully reversed when the CTDK-I complex is restored, demonstrating that CTDK-I is required for the integrity of the rDNA locus.
Figure 2.
The rDNA contraction is reversible and specific. FY1679-18B and W303-1B derived cells were grown in YPD. (A) Complementation analysis. Chromosomes from two independent Δctk1/CTK1 (FY1679-18B) and Δctk2/CTK2 (W303-1B) clones, respectively, were analysed by CHEF electrophoresis and ethidium bromide staining. Size markers are Hansenula wingei chromosomes (M), and their sizes are indicated in megabase pairs. (B) Telomeres analysis. After digestion, genomic DNA was analysed by Southern blotting with a probe specific for telomeres. Bands are visualized by autoradiography.
We next wondered whether, among the different repeated sequences of the genome, the contraction phenotype in Δctk1 cells was specific of the rDNA locus. To address this question, we analysed telomeres that are also composed of sequence repetitions. We performed Southern blot experiments after restriction digestion of genomic DNA from Δctk1, Δctk2 and Δctk3 mutant strains. FY1679 wild-type cells exhibited a larger smear than any of the other strains (Figure 2B). However, we did not observe any substantial differences in the sizes of the telomeres from the different strains (Figure 2B). We note that this result is not in agreement with that described in a genome-wide analysis showing that Δctk1 deletion affects telomere length (54). Although this discrepancy could be owing to distinct genetic backgrounds, it is puzzling that neither Δctk2 nor Δctk3 deletion mutant strains were recovered in this systematic study. Taken together, these results suggest that CTDK-I is specifically required for the integrity of the rDNA locus.
Cohesin association at the rDNA locus
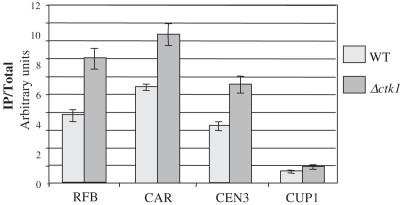
It has been shown recently that cohesin complexes dissociate from rDNA when this latter is undergoing amplification (3). Cohesin complexes are found to bind to specific regions of chromatin from S phase to mitosis in order to hold chromatids in place leading to equal (versus unequal) sister-chromatid recombination, and thereby preventing changes in copy number after the formation of DSB (11). We thus asked whether a defect of cohesin binding at rDNA in Δctk1 cells might be linked to rDNA contraction. To assess cohesin association at rDNA, we performed chromatin immunoprecipitation with strains containing a tagged version of Mcd1, one of the components of cohesin complexes (55). After immunoprecipitation, similar levels of Mcd1-HA proteins were recovered in wild-type and Δctk1 extracts (data not shown). Co-precipitation of 500 bp-long DNA fragments was analysed with pairs of oligonucleotides located at the RFB region and the CAR in NTS2, and as positive and negative controls we used oligonucleotides located in a centromeric region (CEN3), and in the CUP1 gene array, respectively. Fragments were quantified by real-time PCR and the relative occupancy of Mcd1-HA at a given locus was reported as the ratio of immunoprecipitated over total DNA (Figure 3). We observed a slight but significant increase of co-precipitation of the three specific loci analysed in Δctk1 compared to wild-type extract (1.5- to 1.8-fold), as well as for the CUP1 locus (1.4-fold). We do not know the biological relevance, if any, of such an increase. However, we conclude that cohesin binding at rDNA sequences is not decreased in Δctk1 compared to wild-type cells, indicating that rDNA contraction is not correlated to cohesin complex dissociation such as that taking place during expansion (3).
Figure 3.
Analysis of Mcd1 association to rDNA by chromatin immunoprecipitation. After cross-linking, cell lysis and sonication, wild-type MCD1-HA (light grey boxes) and Δctk1 MCD1-HA (dark grey boxes) extracts were immunoprecipitated with anti-HA antibodies. Total and immunoprecipitated DNA were analysed by real-time PCR using four sets of primers: RFB, CAR, CEN3 and CUP1, respectively. Ratio of immunoprecipitated over total DNA was reported.
Fob1 requirement
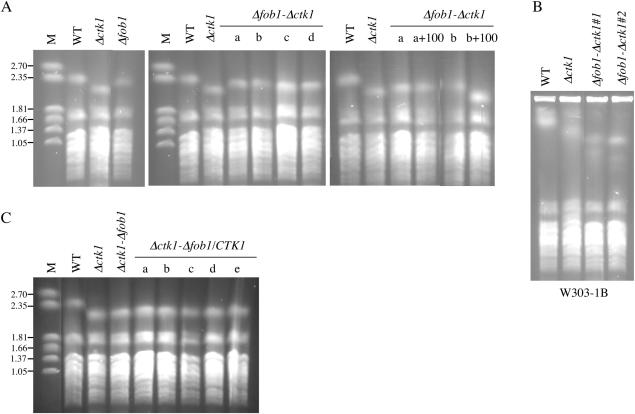
Two types of components are involved in rDNA contraction and expansion processes: (i) factors implicated in Pol I transcription and (ii) factors implicated in the recombination process. Although it is not systematically required for rDNA recombination (56), Fob1 has been shown to be required for the gradual decrease in rDNA repeat number that is observed in rpa135 deletion mutants (6). To analyse whether Fob1 is required for rDNA contraction in Δctk1 mutant cells, we deleted the FOB1 gene prior to deleting CTK1 in FY1679-18B cells. Unlike what has been reported recently (57), we observed that FOB1 deletion generated a moderate decrease in chromosome XII size (Figure 4A). Four independent Δfob1-Δctk1 double mutant strains were grown for 100 generations prior to chromosome XII analysis by CHEF electrophoresis. In all cases, the size of chromosome XII was smaller than that of the wild-type control, although slightly longer than that from Δctk1 cells (Figure 4A). Two of these strains were grown for 100 additional generations. CHEF analysis showed that in one case, chromosome XII size did not vary (Figure 4A, compare lane a+100 to lane a), whereas in the other case, the size decreased (Figure 4A, compare lane b+100 to lane b), and was significantly smaller than the Δfob1 control. Further analysis of genomic DNA by real-time PCR showed that Δfob1-Δctk1 a+100 and Δfob1-Δctk1 b+100 rDNA was contracted (75 and 50% rDNA compared to wild type, respectively). To confirm these results, we also analysed contraction in two independent Δfob1-Δctk1 clones derived from the W303-1B strain. Both strains showed a chromosome XII shorter than wild type (Figure 4B), indicating contraction of the rDNA array. Reduction of the number of 35S genes was confirmed by quantification of genomic DNA with real-time PCR experiments (63 and 68% rDNA compared to wild type, respectively). We conclude that contraction can still occur in the absence of Fob1, although it may be less efficient.
Figure 4.
Fob1 is required for rDNA expansion, but not for contraction. Chromosomes from FY1679-18B (A and C) and W303-1B (B) derived cells grown in YPD were analysed by CHEF electrophoresis and ethidium bromide staining. (A and B) Contraction analysis. CTK1 was deleted in Δfob1 cells, and independent clones were analysed either after growth for 100 generations (A, a–d and B) or for 200 generations (A, a+100, b+100). (C) Expansion analysis. FOB1 was deleted in Δctk1 cells prior to re-introduction of the CTK1 gene born on a CEN, LEU2 plasmid. Five independent clones were analysed after growth for 100 generations. Size markers are Hansenula wingei chromosomes (M), and their sizes are indicated in megabase pairs.
To analyse rDNA expansion, we deleted FOB1 in Δctk1 cells, before reintroduction of a wild-type CTK1 gene borne on a centromeric plasmid. Five independent clones were analysed. We did not observe any rDNA expansion when Fob1 was absent (Figure 4C). Thus, rDNA expansion after re-introduction of the CTK1 gene is a process that strictly requires the Fob1 protein. Altogether, the results suggest that rDNA contraction generated by CTK1 deletion, and rDNA expansion following CTK1 re-insertion arise by two distinct mechanisms.
ERCs levels in Δctk1 cells
In a yeast DNA topoisomerase double mutant, over half of the rDNA is present as ERCs (58). ERCs are generated by a Fob1-dependent homologous recombination process initiated by DSB in the rDNA (8). To study whether rDNA contraction was related to ERC formation in Δctk1 cells, we analysed ERC abundance. After extraction from spheroplasts, DNA was analysed by Southern blotting with an rDNA-specific probe. As a negative control we used a Δfob1 (8) strain as fob1 mutations have been shown to reduce ERC formation (8,59), and as a positive control, we analysed an sgs1 mutation that stimulates rDNA recombination and the formation of ERCs (42). The levels of chromosomal rDNA were comparable in all four samples (Figure 5). As expected, the ratio of ERC monomers and dimers over chromosomal rDNA was significantly lower in Δfob1 cells compared to wild type (0.3-fold), whereas in contrast, it was higher in Δsgs1 cells (2.8-fold). Δctk1 cells contained ERCs at a level similar to isogenic wild-type cells (1.2-fold). Although this data slightly over-estimates the number of ERCs per cell (as this mutant strain contains 75% rDNA compared to wild type), the results show that ERC abundance is similar in wild-type and Δctk1 cells. We conclude that rDNA contraction in Δctk1 mutant strain might not be concomitant with ERC formation. This observation is consistent with the fact that ERC formation is a Fob1-dependent process, whereas contraction in Δctk1 cells is not, suggesting that contraction might occur by a mechanism distinct from the unequal chromatid exchange recombination pathway involved in ERC formation (60).
Figure 5.
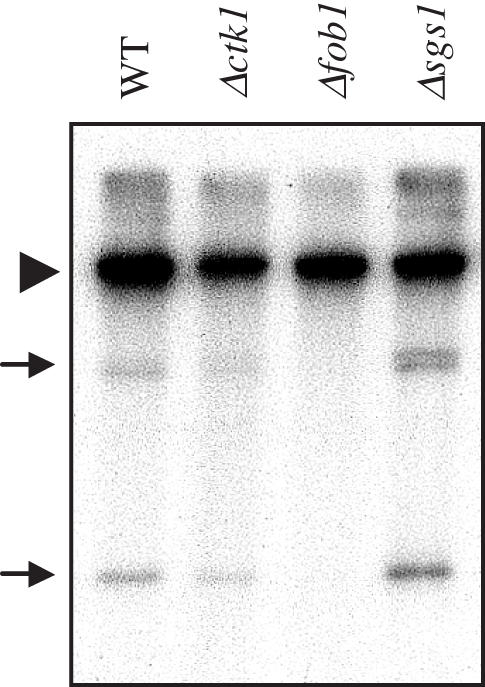
ERC analysis. DNA was extracted from W303-1B (WT), Δctk1, Δfob1 and Δsgs1 cells, respectively, prior to electrophoresis on an agarose gel and Southern blot analysis with a 35S probe. The chromosomal band is indicated by an arrowhead, and ERC species by arrows.
Pol I transcription in Δctk1 cells
In principle, changes in rDNA copy number neither affect the cell doubling time (6), nor rRNA synthesis (17). This is possible by modifying the proportion of active genes, thus maintaining a wild-type number of active genes. To study the possible connection between a defect in 35S synthesis and the rDNA contraction phenotype, we analysed the ratio of transcribed genes in Δctk1 cells. For this purpose, we performed in vivo cross-link of DNA to psoralen (16). Active and inactive genes are characterized by distinct chromatin structures (16). An increase of psoralen cross-linking at the rDNA reflects a more open chromatin structure, corresponding to transcribed genes, that is detected by a slower mobility of isolated rDNA fragments on native gels. Psoralen cross-linking of yeast cells and photo-reactions were carried out prior to genomic DNA isolation and restriction enzyme digestion. rDNA fragments were then analysed by Southern blotting with an intergenic rDNA-specific probe. Such a probe hybridizes to a 2.5 kb-EcoRI intergenic fragment whose migration is correlated with the extent of psoralen cross-linking (12,61). We observed that migration was slower for fragments derived from Δctk1 and Δctk2 cells compared to wild type (Figure 6A). Because ribosomal spacers between active genes have a different nucleosomal organization than spacers flanking inactive genes (16), these results suggest that the mutant cells contain a fraction of open genes higher than wild type. We next performed experiments with a 35S probe that leads to detection of a 2.8 kb EcoRI fragment as two distinct bands (16): a slower migrating band corresponding to heavily cross-linked DNA, and a faster migrating band corresponding to slightly cross-linked DNA molecules (Figure 6B). Quantification with a PosphorImager showed an increase in the ratio of slow/fast-migrating bands, indicating a higher proportion of transcribed genes in Δctk1 cells compared to the isogenic wild type (Figure 6B). Together with the estimation of the total number of 35S genes, the results suggest that the absolute number of open genes is similar in mutant and wild-type cells. We next analysed a Δfob1-Δctk1 strain deficient in Ctk1 kinase activity, but containing a wild-type rDNA copy number (see Materials and Methods). With the intergenic probe, we detected a labelled band displaying a migration similar to that resulting from the wild-type isogenic strain, suggesting similar levels of open genes (Figure 6C). Altogether, the results suggest that the total number of active genes is similar in mutant and wild-type cells, regardless of the number of rDNA genes.
Figure 6.
Pol I transcription analysis. (A–C) Psoralen cross-linking experiments. After psoralen treatment, genomic DNA from wild type (WT), Δctk1, Δctk2 and Δfob1−Δctk1 extracts were analysed by Southern blotting with either an intergenic rDNA probe (A and C) or a 35S rDNA probe (B). (B) As a control, non-crosslinked wild-type DNA was loaded (WT-UN). Arrows indicate the 2.8 kb-EcoRI slow (s) and fast (f) migrating fragments, respectively. Quantification from two independent experiments was performed with a PhosphorImager. Ratios of slow to fast fragments are reported. (D) Primer extension analysis. RNA was extracted from wild type (WT), Δctk1 and Δfob1−Δctk1 cells. Primer extensions were performed with oligonucleotides specific for the 35S and the U6 RNA, respectively. Two independent experiments were analysed by electrophoresis and autoradiography. Signals were quantified with a PhosphorImager and reported as the ratio of 35S over U6 signals.
To analyse Pol I transcription, we performed primer extension using primers specific of the 35S RNA precursor and of the U6 RNA as a control. As previously described (37), primer extension gave rise only to products corresponding to the expected start site (62), showing that Δctk1 cells do not contain 35S rRNA synthesized by Pol II (21). Gel analysis by autoradiography and with a PhosphorImager showed that levels of primer extension products corresponding to 35S RNA were decreased 2-fold in Δfob1-Δctk1 and Δctk1 cells, compared to wild type (Figure 6D). In contrast, levels of products corresponding to the control U6 remained unchanged (Figure 6D). The results show that despite a similar number of open genes, 35S RNA synthesis is affected in the absence of Ctk1, consistent with previous observations indicating that Pol I transcription is affected at the level of transcription initiation (37). Moreover, we conclude that the decrease in 35S RNA synthesis is not caused by rDNA contraction.
Silencing at the rDNA locus in Δctk1 cells
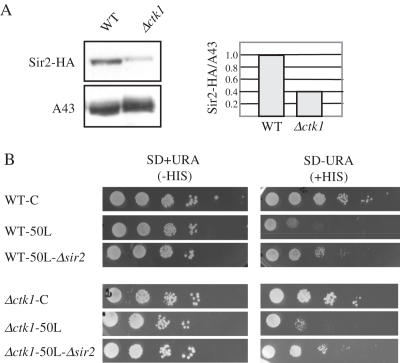
It was shown recently that mammalian Sir2 homolog is an activator of Pol I transcription (63). Furthermore, several studies in yeast have shown that Pol I transcription is required for Sir2-mediated rDNA silencing (41,64). To study silencing in Δctk1 cells, we first constructed strains containing a tagged-version of the Sir2 protein in order to analyse Sir2 expression by western blotting. Although a control protein was expressed at similar levels in wild-type and mutant strains, Sir2-HA levels were substantially lower in Δctk1 mutant cells (Figure 7A). Consistent with this observation, mRNA levels analysed by northern blot were decreased in Δctk1 cells (data not shown). We conclude that SIR2 expression is dramatically affected in Δctk1 mutant cells.
Figure 7.
Analysis of Pol II silencing at rDNA (A) Sir2 expression analysis. After extraction from wild-type SIR2-HA (WT) and Δctk1 SIR2-HA (Δctk1) cells, proteins were analysed by western blotting with anti-HA (upper panel) and polyclonal anti-A43 (lower panel) antibodies, respectively. Chemoluminescence was quantified with a Fluorchem FC and reported as the ratio of Sir2-HA over A43 signal. (B) mURA3 silencing analysis. 5 μl of 10-fold serial dilutions of the indicated strain grown in YPD were spotted on SD+URA (−HIS) and SD-URA (+HIS), respectively.
To get an insight into Pol II silencing at the rDNA locus, we took advantage of the fact that rDNA silencing spreads into the centromere-proximal unique sequence located downstream of Pol I transcription (41). We used a reporter cassette containing a modified URA3 gene (mURA3) to measure silencing, and the HIS3 gene for selecting transformants (41). The cassette was inserted into wild-type and Δctk1 cells, respectively, 50 bp distant from the left side of the rDNA array (WT-50L and Δctk1-50L). In non-silenced control strains, the cassette was inserted at the trp1Δ63 locus (WT-C and Δctk1-C). Growth on medium without uracil allowed monitoring of mURA3 expression. Comparison of growth of control WT-C and Δctk1-C cells on SD-URA (+HIS) and SD+URA (-HIS) media, showed that the mURA3 marker is properly expressed (Figure 7B). WT-50L cells on SD-URA (+HIS) displayed reduced growth compared to control, showing that when the mURA3 marker is inserted at the rDNA locus, it is subjected to silencing. We observed similar results for Δctk1 cells (Figure 7B), indicating that silencing is not affected in the absence of CTK1. As expected, further disruption of the SIR2 gene generated growth similar to isogenic control cells, showing that Pol II silencing was relieved (Figure 7B). The results show that despite a decrease in cellular level of Sir2, silencing at rDNA is not affected in Δctk1 cells. We thus conclude that the reduced rate of Pol I transcription in Δctk1 mutant cells is still able to spread silencing.
DISCUSSION
The absence of CTDK-I systematically induces a decrease in the number of rDNA copies. Interestingly, this specific effect is not dependent on Fob1. Complementation of the ctk null mutation leads to rDNA expansion until rDNA copy number reaches a value similar to that of the original strain. In contrast to the contraction, rDNA expansion requires Fob1. Despite reduced levels of Sir2 protein, silencing at the rDNA is not affected. The results suggest that a defect in Pol I transcription generates rDNA contraction.
Silencing at rDNA is not affected
SIR2 transcription and Sir2 protein levels are severely reduced in Δctk1 cells. Although it is possible that the Ctk1 kinase is specifically required for SIR2 transcription by Pol II, another hypothesis is that Sir2 reduced level is the direct consequence of the rDNA contraction. High levels of Sir2 are toxic to the cell, perhaps because it causes a profound decrease in chromosome stability (65), and its expression must be tightly regulated. Indeed, Michel et al. (66) have recently reported that SIR2 expression is down regulated by the Sir2 protein itself. Spontaneous rDNA contraction leads to a decrease in the fraction of nucleolar Sir2 and in the overall Sir2 protein levels, but resulting cells display normal rDNA silencing. The authors suggested that upon rDNA loss, a large nucleolar pool of Sir2 is released, and made available for down regulation of SIR2 expression. Altogether, our results suggest that the reduced levels of Sir2 are the consequence of rDNA contraction. Consistent with this hypothesis, silencing at rDNA is not affected in Δctk1 cells.
Fob1 is required for rDNA expansion but not for contraction
Gene conversion between non-aligned sister chromatids is the predominant mechanism responsible for the expansion/contraction of rDNA (53). Kobayashi et al. (6) proposed that Fob1 mediates DSB, the initial event of gene conversion, at the RBF sites of rDNA repeats, thus coupling expansion/contraction to replication. Although expansion following re-introduction of the missing CTK allele is dependent on Fob1, rDNA contraction after deletion of a CTK gene is not. Consistently, contraction of the rDNA tandem array is not correlated to an increase in ERC levels. Taken together, the results suggest that rDNA expansion may require Fob1 in order to increase DSB to initiate unequal gene conversion events that will progressively restore a wild-type number of rDNA genes. In contrast, rDNA contraction does not require Fob1 suggesting that it might arise as a single-step process, maybe by a mechanism such as single-strand annealing. In addition, a single event (or a limited number of events) leading to contraction is consistent with the observation of different degrees of rDNA contraction in mutant strains.
Interestingly, it has been described that spontaneous rDNA copy number variations arise as a single-step process at a very high frequency (1%) in W303 cells (66). These deletions remove up to 50% of the rDNA locus. W303 wild-type cells that have undergone rDNA contraction contain a rather homogeneous population of chromosome XII (66). Yet, W303 strains keep a constant number of rDNA genes. Thus, either cells with a large deletion exhibit a selective disadvantage, and in the long term, will not be maintained in the population, or these cells will recover at some time an rDNA locus of the initial size. The first hypothesis seems unlikely as it has been shown that yeast cellular growth rate is not correlated to the number of 35S genes: a strain carrying only 25% of the normal rDNA repeats displays a growth rate identical to that of its wild type (6). Δctk strains contain a reduced rDNA copy number that is in the order of 40–80% compared to wild type. This feature is strongly reminiscent of the spontaneous contraction described in W303 cells. It is possible that the contraction leads to a selective advantage in Δctk1 cells, thus generating a cell population with a shorter chromosome XII. Alternatively, although our results suggest that mechanisms leading to gene conversion are not affected in Δctk1 cells, it is possible that these mutant cells are unable to restore a wild-type number of rDNA copies after a spontaneous contraction.
Reduced rate of Pol I transcription and rDNA contraction
In this report, we show that Pol I transcriptional impairment is not the result of rDNA contraction, consistent with previous studies showing that rDNA contraction does not affect 35S synthesis (17). This is also consistent with the fact that Ctk1 interacts directly with Pol I in the nucleolus, strongly suggesting that Ctk1 is involved directly in Pol I transcription (37). Thus, although we do not rule out the possibility that rDNA contraction might be connected to growth rate, and although we have not established a direct connection between Pol I transcription and rDNA contraction in Δctk1 cells, it is tempting to speculate that a Pol I transcription defect accounts for rDNA contraction in these cells. The absence of an essential subunit of Pol I triggers a reversible gradual decrease in rDNA repeat number to reach approximately one-half of the normal value (6). Similar phenotypes are observed in uaf N-PSW mutant strains in which rDNA is very weakly transcribed by Pol II (22). Both studies demonstrate that rDNA undergoes contraction when it is not transcribed by Pol I. Unlike uaf null mutations, ctk1 mutation does not lead to rDNA transcription by Pol II. It is thus the first description of rDNA contraction occuring in a Pol I mutant strain in which 35S RNA is transcribed by Pol I. Importantly, unlike rDNA contraction previously described in Pol I mutant strains, that induced by ctk1 mutation does not seem to be gradual and is not Fob1-dependent, suggesting distinct mechanisms. Interestingly, our results are consistent with the hypothesis that Pol I controls repeats number, perhaps by stabilizing rDNA with the normal repeat numbers as a stable nucleolar structure (6).
Acknowledgments
The authors thank N. Alic for critical reading of the manuscript, P-A. Defossez for technical advice and S. Gangloff for fruitful discussions and the gift of D444-1D strain. The authors also thank C. Carles and M. Riva for their constant interest in this work. C.B. was supported by the Fondation pour la Recherche Médicale and S.G. by a fellowship from the French Ministère de la Recherche et des Technologies. Funding to pay the Open Access publication charges for this article was provided by CEA/DBJC/SBGM.
Conflict of interest statement. None declared.
REFERENCES
- 1.Petes T.D. Yeast ribosomal DNA genes are located on chromosome XII. Proc. Natl Acad. Sci. USA. 1979;76:410–414. doi: 10.1073/pnas.76.1.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brewer B.J., Fangman W.L. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi T., Ganley A.R. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science. 2005;309:1581–1584. doi: 10.1126/science.1116102. [DOI] [PubMed] [Google Scholar]
- 4.Skryabin K.G., Eldarov M.A., Larionov V.L., Bayev A.A., Klootwijk J., de Regt V.C., Veldman G.M., Planta R.J., Georgiev O.I., Hadjiolov A.A. Structure and function of the nontranscribed spacer regions of yeast rDNA. Nucleic Acids Res. 1984;12:2955–2968. doi: 10.1093/nar/12.6.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laloraya S., Guacci V., Koshland D. Chromosomal addresses of the cohesin component Mcd1p. J. Cell. Biol. 2000;151:1047–1056. doi: 10.1083/jcb.151.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi T., Heck D.J., Nomura M., Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gotta M., Strahl-Bolsinger S., Renauld H., Laroche T., Kennedy B.K., Grunstein M., Gasser S.M. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 1997;16:3243–3255. doi: 10.1093/emboj/16.11.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Defossez P.A., Prusty R., Kaeberlein M., Lin S.J., Ferrigno P., Silver P.A., Keil R.L., Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi T. The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol. Cell. Biol. 2003;23:9178–9188. doi: 10.1128/MCB.23.24.9178-9188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeuchi Y., Horiuchi T., Kobayashi T. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 2003;17:1497–1506. doi: 10.1101/gad.1085403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi T., Horiuchi T., Tongaonkar P., Vu L., Nomura M. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell. 2004;117:441–453. doi: 10.1016/s0092-8674(04)00414-3. [DOI] [PubMed] [Google Scholar]
- 12.Smith J.S., Boeke J.D. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 13.Gottlieb S., Esposito R.E. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 14.Pappas D.L., Jr, Frisch R., Weinreich M. The NAD(+)-dependent Sir2p histone deacetylase is a negative regulator of chromosomal DNA replication. Genes Dev. 2004;18:769–781. doi: 10.1101/gad.1173204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benguria A., Hernandez P., Krimer D.B., Schvartzman J.B. Sir2p suppresses recombination of replication forks stalled at the replication fork barrier of ribosomal DNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:893–898. doi: 10.1093/nar/gkg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dammann R., Lucchini R., Koller T., Sogo J.M. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.French S.L., Osheim Y.N., Cioci F., Nomura M., Beyer A.L. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 2003;23:1558–1568. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nomura M. Transcription factors used by Saccharomyces cerevisiae RNA polymerase I and the mechanism of initiation. In: Paule M.R., editor. Transcription of Ribosomal RNA Genes by Eucaryotic RNA Polymerase I. Berlin, Germany: Springer Verlag; 1998. pp. 155–172. [Google Scholar]
- 19.Keys D.A., Lee B.S., Dodd J.A., Nguyen T.T., Vu L., Fantino E., Burson L.M., Nogi Y., Nomura M. Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 1996;10:887–903. doi: 10.1101/gad.10.7.887. [DOI] [PubMed] [Google Scholar]
- 20.Keener J., Josaitis C.A., Dodd J.A., Nomura M. Reconstitution of yeast RNA polymerase I transcription in vitro from purified components. TATA-binding protein is not required for basal transcription. J. Biol. Chem. 1998;273:33795–33802. doi: 10.1074/jbc.273.50.33795. [DOI] [PubMed] [Google Scholar]
- 21.Vu L., Siddiqi I., Lee B.S., Josaitis C.A., Nomura M. RNA polymerase switch in transcription of yeast rDNA: role of transcription factor UAF (upstream activation factor) in silencing rDNA transcription by RNA polymerase II. Proc. Natl Acad. Sci. USA. 1999;96:4390–4395. doi: 10.1073/pnas.96.8.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oakes M., Siddiqi I., Vu L., Aris J., Nomura M. Transcription factor UAF, expansion and contraction of ribosomal DNA (rDNA) repeats, and RNA polymerase switch in transcription of yeast rDNA. Mol. Cell. Biol. 1999;19:8559–8569. doi: 10.1128/mcb.19.12.8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J.M., Greenleaf A.L. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1991;1:149–167. [PMC free article] [PubMed] [Google Scholar]
- 24.Hautbergue G., Goguel V. Activation of the cyclin-dependent kinase CTDK-I requires the heterodimerization of two unstable subunits. J. Biol. Chem. 2001;15:8005–8013. doi: 10.1074/jbc.M010162200. [DOI] [PubMed] [Google Scholar]
- 25.Cho E.J., Kobor M.S., Kim M., Greenblatt J., Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 2001;15:3319–3329. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris D.P., Greenleaf A.L. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 2000;275:39935–39943. doi: 10.1074/jbc.M004118200. [DOI] [PubMed] [Google Scholar]
- 27.Skaar D.A., Greenleaf A.L. The RNA Polymerase II CTD Kinase CTDK-I Affects Pre-mRNA 3′ Cleavage/Polyadenylation through the Processing Component Pti1p. Mol. Cell. 2002;10:1429–1439. doi: 10.1016/s1097-2765(02)00731-1. [DOI] [PubMed] [Google Scholar]
- 28.Ahn S.H., Kim M., Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell. Biol. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 29.Ostapenko D., Solomon M.J. Budding yeast CTDK-I is required for DNA damage-induced transcription. Eukaryot. Cell. 2003;2:274–283. doi: 10.1128/EC.2.2.274-283.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao T., Hall H., Kizer K.O., Shibata Y., Hall M.C., Borchers C.H., Strahl B.D. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurt E., Luo M.J., Rother S., Reed R., Strasser K. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc. Natl Acad. Sci. USA. 2004;101:1858–1862. doi: 10.1073/pnas.0308663100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradsher J., Auriol J., Proietti de Santis L., Iben S., Vonesch J.L., Grummt I., Egly J.M. CSB is a component of RNA pol I transcription. Mol. Cell. 2002;10:819–829. doi: 10.1016/s1097-2765(02)00678-0. [DOI] [PubMed] [Google Scholar]
- 33.Hoogstraten D., Nigg A.L., Heath H., Mullenders L.H., van Driel R., Hoeijmakers J.H., Vermeulen W., Houtsmuller A.B. Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol. Cell. 2002;10:1163–1174. doi: 10.1016/s1097-2765(02)00709-8. [DOI] [PubMed] [Google Scholar]
- 34.Iben S., Tschochner H., Bier M., Hoogstraten D., Hozak P., Egly J.M., Grummt I. TFIIH plays an essential role in RNA polymerase I transcription. Cell. 2002;109:297–306. doi: 10.1016/s0092-8674(02)00729-8. [DOI] [PubMed] [Google Scholar]
- 35.Lin C.Y., Tuan J., Scalia P., Bui T., Comai L. The cell cycle regulatory factor TAF1 stimulates ribosomal DNA transcription by binding to the activator UBF. Curr. Biol. 2002;12:2142–2146. doi: 10.1016/s0960-9822(02)01389-1. [DOI] [PubMed] [Google Scholar]
- 36.Fath S., Kobor M.S., Philippi A., Greenblatt J., Tschochner H. Dephosphorylation of RNA polymerase I by Fcp1p is required for efficient rRNA synthesis. J. Biol. Chem. 2004;279:25251–25259. doi: 10.1074/jbc.M401867200. [DOI] [PubMed] [Google Scholar]
- 37.Bouchoux C., Hautbergue G., Grenetier S., Carles C., Riva M., Goguel V. CTD kinase I is involved in RNA polymerase I transcription. Nucleic Acids Res. 2004;32:5851–5860. doi: 10.1093/nar/gkh927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rothstein R.J. One step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 39.Longtine M.S., McKenzie A., Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 40.Hautbergue G., Goguel V. The yeast C-type cyclin Ctk2p is phosphorylated and rapidly degraded by the ubiquitin-proteasome pathway. Mol. Cell. Biol. 1999;19:2527–2534. doi: 10.1128/mcb.19.4.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buck S.W., Sandmeier J.J., Smith J.S. RNA polymerase I propagates unidirectional spreading of rDNA silent chromatin. Cell. 2002;111:1003–1014. doi: 10.1016/s0092-8674(02)01193-5. [DOI] [PubMed] [Google Scholar]
- 42.Sinclair D.A., Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 43.Schmitt M.E., Brown T.A., Trumpower B.L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buhler J.M., Huet J., Davies K.E., Sentenac A., Fromageot P. Immunological studies of yeast nuclear RNA polymerases at the subunit level. J. Biol. Chem. 1980;255:9949–9954. [PubMed] [Google Scholar]
- 45.Kuras L., Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 46.Kuras L., Borggrefe T., Kornberg R.D. Association of the Mediator complex with enhancers of active genes. Proc. Natl Acad. Sci. USA. 2003;100:13887–13891. doi: 10.1073/pnas.2036346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sterner D.E., Moon Lee J., Hardin S.E., Greenleaf A.L. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin–cyclin-dependent kinase complex. Mol. Cell. Biol. 1995;15:5716–5724. doi: 10.1128/mcb.15.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oakes M., Aris J.P., Brockenbrough J.S., Wai H., Vu L., Nomura M. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J. Cell. Biol. 1998;143:23–34. doi: 10.1083/jcb.143.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rustchenko E.P., Sherman F. Physical constitution of ribosomal genes in common strains of Saccharomyces cerevisiae. Yeast. 1994;10:1157–1171. doi: 10.1002/yea.320100904. [DOI] [PubMed] [Google Scholar]
- 50.Winston F., Dollard C., Ricupero-Hovasse S.L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 51.Thomas B.J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 52.Hallstrom T.C., Katzmann D.J., Torres R.J., Sharp W.J., Moye-Rowley W.S. Regulation of transcription factor Pdr1p function by an Hsp70 protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 1998;18:1147–1155. doi: 10.1128/mcb.18.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gangloff S., Zou H., Rothstein R. Gene conversion plays the major role in controlling the stability of large tandem repeats in yeast. EMBO J. 1996;15:1715–1725. [PMC free article] [PubMed] [Google Scholar]
- 54.Askree S.H., Yehuda T., Smolikov S., Gurevich R., Hawk J., Coker C., Krauskopf A., Kupiec M., McEachern M.J. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl Acad. Sci. USA. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guacci V., Koshland D., Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S.cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Merker R.J., Klein H.L. hpr1Delta affects ribosomal DNA recombination and cell life span in Saccharomyces cerevisiae. Mol. Cell. Biol. 2002;22:421–429. doi: 10.1128/MCB.22.2.421-429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johzuka K., Terasawa M., Ogawa H., Ogawa T., Horiuchi T. Condensin loaded onto the replication fork barrier site in the rRNA gene repeats during S phase in a FOB1-dependent fashion to prevent contraction of a long repetitive array in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:2226–2236. doi: 10.1128/MCB.26.6.2226-2236.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim R.A., Wang J.C. A subthreshold level of DNA topoisomerases leads to the excision of yeast rDNA as extrachromosomal rings. Cell. 1989;57:975–985. doi: 10.1016/0092-8674(89)90336-x. [DOI] [PubMed] [Google Scholar]
- 59.Johzuka K., Horiuchi T. Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S. cerevisiae. Genes Cells. 2002;7:99–113. doi: 10.1046/j.1356-9597.2001.00508.x. [DOI] [PubMed] [Google Scholar]
- 60.Park P.U., Defossez P.A., Guarente L. Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:3848–3856. doi: 10.1128/mcb.19.5.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith J.S., Caputo E., Boeke J.D. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 1999;19:3184–3197. doi: 10.1128/mcb.19.4.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klootwijk J., de Jonge P., Planta R.J. The primary transcript of the ribosomal repeating unit in yeast. Nucleic Acids Res. 1979;6:27–39. doi: 10.1093/nar/6.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ford E., Voit R., Liszt G., Magin C., Grummt I., Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20:1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cioci F., Vu L., Eliason K., Oakes M., Siddiqi I.N., Nomura M. Silencing in yeast rDNA chromatin: reciprocal relationship in gene expression between RNA polymerase I and II. Mol. Cell. 2003;12:135–145. doi: 10.1016/s1097-2765(03)00262-4. [DOI] [PubMed] [Google Scholar]
- 65.Holmes S.G., Rose A.B., Steuerle K., Saez E., Sayegh S., Lee Y.M., Broach J.R. Hyperactivation of the silencing proteins, Sir2p and Sir3p, causes chromosome loss. Genetics. 1997;145:605–614. doi: 10.1093/genetics/145.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Michel A.H., Kornmann B., Dubrana K., Shore D. Spontaneous rDNA copy number variation modulates Sir2 levels and epigenetic gene silencing. Genes Dev. 2005;19:1199–1210. doi: 10.1101/gad.340205. [DOI] [PMC free article] [PubMed] [Google Scholar]