Abstract
We wish to understand the role of electrostatics in DNA stiffness and bending. The DNA charge collapse model suggests that mutual electrostatic repulsions between neighboring phosphates significantly contribute to DNA stiffness. According to this model, placement of fixed charges near the negatively charged DNA surface should induce bending through asymmetric reduction or enhancement of these inter-phosphate repulsive forces. We have reported previously that charged variants of the elongated basic-leucine zipper (bZIP) domain of Gcn4p bend DNA in a manner consistent with this charge collapse model. To extend this result to a more globular protein, we present an investigation of the dimeric basic-helix–loop–helix (bHLH) domain of Pho4p. The 62 amino acid bHLH domain has been modified to position charged amino acid residues near one face of the DNA double helix. As observed for bZIP charge variants, DNA bending toward appended cations (away from the protein:DNA interface) is observed. However, unlike bZIP proteins, DNA is not bent away from bHLH anionic charges. This finding can be explained by the structure of the more globular bHLH domain which, in contrast to bZIP proteins, makes extensive DNA contacts along the binding face.
INTRODUCTION
The DNA double helix is a relatively stiff biopolymer with a persistence length of ∼150 bp. Over this length the average deflection of the helix axis is one radian under physiological salt conditions and in the absence of proteins (1–3). In contrast, very long DNA molecules spontaneously collapse to conformations whose end-to-end distances are proportional to the square root of the contour length (4). These characteristics imply that DNA is locally stiff yet globally flexible. Fundamental cellular processes such as nucleosome-mediated compaction (5), assembly and regulation of transcriptional machinery (6–8), and genetic recombination (9,10) require energetically unfavorable DNA deformation. Protein binding must provide the energetic driving force required for sharp DNA bending. We are interested in how asymmetric electrostatic interactions between proteins and DNA contribute to the generation of DNA-bending forces (3,11–16). Protein-mediated DNA bending is a crucial element in many essential biological processes, yet the contributions of electrostatic effects remain poorly understood.
Previous results from our laboratory support the idea that simple DNA-binding proteins can be engineered to induce DNA bending in opposite directions by positioning positive or negative charges asymmetrically on one face of the DNA duplex (13). Considerable efforts are still focused on understanding how neighboring cations and the phosphate diester backbone affect the stiffness of DNA (17–23). These findings support the proposals of Rich, Manning and others who theorized that charge–charge repulsions between neighboring phosphate diesters are at least partially responsible for the intrinsic rigidity of DNA (24–26).
The basic-leucine zipper (bZIP) domain of the Saccharomyces cerevisiae transcription factor Gcn4p (Figure 1A) has been studied extensively as a biophysical model because of its simple secondary structure and orthology to the mammalian oncogenes Fos and Jun (27–33). The bZIP domain of Gcn4p is a 57 amino acid peptide sufficient for homo-dimerization and sequence-specific DNA binding (34). In the absence of DNA, the basic region of free bZIP peptides remains relatively disordered (35,36), whereas at high-protein concentration the leucine zipper region is natively folded into an alpha-helical coiled-coil. DNA binding potentiates folding, and crystal structures suggest that the basic region adopts a continuous alpha-helical structure when bound to DNA (34,37). Previous efforts in our laboratory have shown that the bZIP domain is adaptable as a scaffold for charge variations, while preserving the ability of the modified bZIP peptides to bind DNA (13,15,16).
Figure 1.
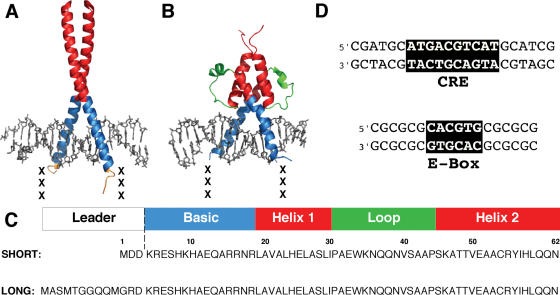
Comparison of the Gcn4p bZIP and Pho4p bHLH domains. (A) Crystal structure of the Gcn4p bZIP homodimer bound to the CRE DNA sequence (37). Leucine zipper (red) and basic (blue) domains are shown. Previous studies involved charge variants of bZIP peptides (XXX) introduced at the N-terminus of the basic region (gold) by mutating the wild-type sequence (PAA: neutral) to anionic (EEE) or a cationic (KKK) amino acids. (B) Crystal structure of the Pho4p bHLH homodimer bound to the E-box DNA sequence (58). Color-coding indicates the helical (red), basic (blue) and loop (green) domains. Three additional residues (XXX) were added at the N-terminus of the basic region to approximate the position of the XXX segment in bZIP peptides. Neutral (SAA), anionic (EEE) and cationic (KKK) bHLH charge variants were created at site XXX. (C) bHLH-XXX peptide sequence insertion site and domains. bHLHSHORT and bHLHLONG peptide variants are color-coded as in (B). The bHLHLONG peptides include an amino terminal segment previously shown to stabilize folding of bZIP peptides. (D) Comparison of CRE (Gcn4p bZIP) and E-box (Pho4p bHLH) binding sites sequences.
Electrophoretic assays have been widely used to detect DNA bending because mobility in native gels is related to DNA shape (38–40). Our own electrophoretic studies using DNA phasing probes were influenced by the work of Schepartz (30,41) and Kerppola (29,33,42) and have suggested a role for electrostatics in DNA bending. For example, the bZIP domain of transcription factor Gcn4p was used as a scaffold to position charged amino acids near one face of the DNA duplex to explore if local charges induce DNA bending (Figure 1A). When bound to DNA, charged residues on Gcn4p have the potential to create laterally asymmetric charge distributions that appear to modulate inter-phosphate repulsions in a manner consistent with predictions (3,11,24,25,43,44). Specifically, DNA bends toward cationic charges that asymmetrically reduce phosphate repulsions, and away from anionic charges that enhance these repulsions.
DNA bending interpretations in electrophoretic phasing assays involving bZIP proteins can be controversial due to concerns that observed mobility changes could reflect changes in the shape, flexibility and/or gel-matrix interactions of the elongated peptide structure (28,33,45,46). In particular, DNA cyclization kinetics experiments have suggested that certain bZIP proteins do not bend DNA (46,47). Our previous work on the effect of appending globular domains onto bZIP peptides showed that apparent charge-dependent DNA bending was preserved in these more symmetrical complexes but was not detected by minicircle binding experiments (15,16). We reconciled the results by proposing that the bending potential for bZIP–DNA complexes is quite flat, so that different average geometries are observed with different methods (15). In the present work, we study a relatively globular protein, the basic helix–loop–helix (bHLH) domain of yeast Pho4p (Figure 1B and C), as an alternative scaffold for positioning charges asymmetrically near DNA. Charge variants of Pho4p provide an independent opportunity for exploration of DNA bending by laterally asymmetric charge interactions and of protein–DNA complex flexibility.
Pho4p, a 312 amino acid transcription factor in S.cerevisae, belongs to the PHO gene family, which encodes >30 proteins responsible for regulation of exogenous inorganic phosphate uptake (48,49). At high exogenous phosphate, Pho4p is rendered inactive through phosphorylation by CDK complex orthologs, Pho80p–Pho85p (50). In phosphate-poor media Pho4p is hypophosphorylated and imported into the nucleus, where it acts together with Pho2p to activate genes required for phosphate scavenging (51) by specific recognition of the E-box sequence, 5′-CACGTG (Figure 1D) (52,53). The C-terminal bHLH domain of Pho4p, the domain responsible for both selective DNA binding and homodimerization, includes a basic residue-rich region followed by a HLH fold (54–57). The X-ray co-crystal structure of the 62 amino acid Pho4p bHLH bound to DNA has been solved at 2.8 Å (58). The resulting model (Figure 1B) shows that the loops of the bHLH dimers play a critical role in conferring stability and specificity to Pho4p DNA binding through extensive protein-phosphodiester backbone contacts.
Here we show that, as seen for the bZIP domain of Gcn4p, charge variants of the bHLH domain of Pho4p appear to induce DNA bending toward appended cations. Unlike bZIP peptides, anionic charge variants of the bHLH peptide do not cause DNA bending in the opposite direction. We propose that DNA bending toward the bound protein is prevented by DNA contacts made by the loop region of the bHLH dimer.
MATERIALS AND METHODS
Plasmids
Pho4p expression plasmid pJ1080, derived from a pET21d (Novagen), encodes the 62 amino acid bHLH domain of Pho4p and was the gift of D. Wemmer (59). Site-directed mutagenesis using standard methods was used to generate plasmids encoding the bHLHSHORT Pho4p charge variants pJ1350, pJ1351 and pJ1352 (encoding residues SAA, EEE and KKK appended near the N-terminus). Longer charge variant peptides, the bHLHLONG (SAA, EEE, KKK) set, were generated from bHLHSHORT peptides by replacement of the first 3 N-terminal residues (MDD) with 12 bZIP N-terminal residues (MASMTGGQQMGRD). Plasmids pDP-AP-1-21, pDP-AP-1-23, pDP-AP-1-25, pDP-AP-1-26, pDP-AP-1-28 and pDP-AP-1-30 were modified to include an E-box site for bHLH binding in order to generate probes for electrophoretic phasing analyses (30).
Oligonucleotides
Unmodified oligonucleotides were synthesized using standard phosphoramidite chemistry. Oligonucleotides were purified by denaturing PAGE (20% acrylamide; 29:1 acrylamide/bisacrylamide, 8 M urea), extracted from gel fragments (10 mM Tris–HCl, 1 mM EDTA and 300 mM NaOAc, pH 5.2), and desalted using C18 reverse phase cartridges (Sep-Pak; Waters Corporation). Oligonucleotide concentrations were determined at 260 nm using nearest neighbor molar extinction coefficients as described previously (11). Oligonucleotides were annealed at ∼50 μM in 250 mM NaCl, by heating to 95°C and cooling to room temperature overnight.
Protein expression and purification
bHLH peptides were expressed in BL21(DE3) cells in Luria–Bertani media (59). Cultures were grown at 37°C to an A600 of 0.6–0.7 and induced with 5 mM isopropyl-β-d-thiogalactopyranoside. Induced cultures were then incubated 8 h at 37°C before centrifugation (5000 × g, 20 min). Cell pellets were frozen at −80°C, thawed and resuspended (20 ml/l culture) in lysis buffer (50 mM Tris–HCl, pH 6.0, 1 mM EDTA and 20 mM DTT). Initial lysis of the cells by sonication (15 s each, 10 × 4°C; Fisher Sonic Dismembrator 60) was followed by pneumatic sheering using an Avestin Emulsiflex-C5 disruptor driven by pressurized N2 (140 psi, 4°C). The resulting crude lysates were clarified by centrifugation (20 000 × g, 45 min, 4°C) and further purified on a gravity flow SP sepharose column (Pharmacia Biotech, Sweden). After loading, the cationic exchange column was washed with 40 ml steps of lysis buffer supplemented with increasing salt concentrations (0.5, 1.0, 1.5 and 2.0 M NaCl). Eluted fractions containing the bHLH peptides (1.0 M NaCl for bHLHSHORT and 1.5 M NaCl for bHLHLONG) were concentrated using 5000 molecular weight cutoff centrifugal concentrators (VivaSpin 20; VivaScience) and then purified by HPLC [C18 preparative reverse phase column (250 × 21.2 mm), Beckman 127P System Gold, buffer A: 0.1% trifluoroacetic acid (TFA), buffer B: 80% CH3CN, 0.1% TFA, gradient 10–70% buffer B over 50 min]. Peptide purity was assessed by ESI GC-MS to be >95%. Purified peptides were lyophilized for storage.
bHLH:DNA-binding affinity studies
DNA probes for binding affinity assays containing either an E-box (5′-CGCGCGCACGTGCCG; E-box underlined) or CRE (5′-CGATGCATGACGTCATGC; CRE underlined) sequence were prepared by annealing a self-complementary oligonucleotide to generate duplexes with 3′ recessed termini. The annealed probe (400 pmole) was end-labeled using 20 U of the Klenow fragment of E.coli DNA polymerase I and 10 μCi each of [α-32P]dGTP and dCTP (3000 Ci/mmol) in 70 mM Tris–HCl, pH 7.6, 10 mM MgCl2 and 5 mM DTT for 45 min at 37°C, followed by incubation with 5 mM each of dGTP and dCTP for 15 min to polymerize all termini. Radiolabeled probes were purified from unincorporated nucleotides using spin columns (Chromaspin TE-10; Clontech) and diluted for binding affinity assays. bHLH samples were dissolved in water and concentrations were determined by the method of Pace et al. (60) followed by dilution into binding buffer [1× phosphate-buffered saline containing 5% glycerol and 0.025% NP-40]. Binding reactions were performed by incubation of peptide dilutions (10 pM to 1 μM) with annealed radiolabeled DNA probes (100 pM) in a total volume of 20 μl of binding buffer supplemented with 100 μg/ml BSA. Binding reactions were incubated on ice for 30 min before analysis on native polyacrylamide gels (12% acrylamide; 29:1 acrylamide/bisacrylamide) in 0.5× TBE buffer run for 750 V-h at 22°C. Dried gels were analyzed using a STORM 840 scanner (Amersham). Equilibrium dissociation constant (Kd) values were estimated by measuring the fractional occupancy (θ) of the radiolabeled duplex [D] as a function of the bHLH concentration [P]. The Kd was estimated with non-linear least squares fitting to Equation 1, using Kaleidagraph™ software (Synergy Software).
| 1 |
Circular dichroism spectroscopy
CD spectra were recorded using a Jasco 810 spectropolarimeter. Far UV-CD spectra were obtained in the continuous mode, collecting measurements every 1 nm with an averaging time of 5 s at 295 K between 200–260 nm. bHLH samples were 10 μM in 0.2 cm path length cells. DNA probes containing the E-box sequence (underlined) were generated by annealing the single self-complementary oligonucleotide (5′-CGCGCGCACGTGCCGCGCG) at 500 μM. Spectra were acquired in binding buffer with either 10 μM free bHLH peptides or bHLH complexes with E-box DNA at a molar ratio of 1:1.1. All baseline buffer and nucleic acid contributions to ellipticity were subtracted after spectral collection. Mean residual ellipticity (ΘMRE, deg cm2/dmol) was calculated using the relation:
| 2 |
where Θmdeg is the ellipticity in mdeg, l is the path length in cm, c is the molar peptide concentration and n is the number of residues in the peptide. Alpha-helical estimates of bHLH peptides were based upon the previously published equation (61):
| 3 |
where ΘMRE is the mean residual ellipticity at 222 nm. For Θ0 and Θ100 representing 0 and 100% helicity, values of −2340 and −30 300 (deg cm2/dmol) were used.
Electrophoretic phasing assays of DNA bending
DNA probes for phasing analyses were generated and radiolabeled by PCR from derivatives of plasmids pDP-AP-1-21, pDP-AP-1-23, pDP-AP-1-25, pDP-AP-1-26, pDP-AP-1-28 and pDP-AP-1-30 (30) that had been modified through site-directed mutagenesis of the original AP-1 binding site to generate E-box-binding sites suitable for bHLH binding. DNA probes were incubated with sufficient peptide for ∼50% mobility shift in 20 μl reactions containing binding buffer and 100 μg/ml BSA. Reactions were incubated on ice for 30 min before native PAGE (6% acrylamide; 29:1 acrylamide/bisacrylamide) in 0.5× TBE buffer for 2000 V-h at 22°C. Dried gels were analyzed as above.
Phasing analysis calculations
Phasing analysis was performed essentially as described previously (28,62,63). Migration distances for free and bound DNA probes were measured and normalized to the average mobility of each set of DNA probes according to:
| 4 |
where μrel is the normalized mobility, μ is the measured mobility of each probe and μavg is the average mobility of a group of phasing probes sharing a common bound peptide.
Values of μrel were plotted against the spacer distance (bp spacing between the center of the E-box site and the center of the proximal A5-tract) for fitting to the phasing function:
| 5 |
where μrel is the normalized mobility of the species, S is the spacer length (bp), PPH is the phasing period (10.5 bp/turn), ST is the trans spacer length (predicted length in bp where protein-induced and A5-tract bends are in trans), and APH, the amplitude of the phasing function. The value of APH was approximated by least squares curve fitting using KaleidaGraph. The bend angle, αB, was then estimated from the empirical relation:
| 6 |
where αC is the reference bend angle of the phased array of A5 tracts (54°), and k is an empirically determined constant (∼1.09 in this case).
DNA bending energy predictions
The ΔΔG° contributions to the overall process of protein–DNA complex formation, defined as ΔG°(variant) − ΔG°(SAA), were estimated from experimental data or else were proposed as reasonable guesses that rationalize experimental observations. The individual ΔΔG° contributions that come from the experiment were estimated as follows:
For protein folding, the folding reaction is considered to be two-state, with the equilibrium constant calculated as follows:
| 7 |
where fnative is the fraction of folded protein. The relative free energy for folding is calculated from the ratio of equilibrium constants:
| 8 |
The free energy cost of DNA bending is calculated as described previously (64). The equation assumes a persistence length of 150 bp:
| 9 |
where the bend angle Θ, in degrees, is evenly distributed over L base pairs. For example, the free energy cost of 600 cal/mol shown in Figure 5 corresponds to a bend of 24.5° over 14 bp.
The total of all the ΔΔG° contributions associated with protein–DNA complex formation is given by the observed dissociation constants.
| 10 |
RESULTS AND DISCUSSION
Experimental design
The native bHLH domain of Pho4p (Figure 1B and C), representing residues 248–312 from the wild-type sequence (59), was modified to laterally position asymmetric charges near one face of the DNA. bHLHSHORT peptides were generated by insertion of three additional residues (XXX, Figure 1B and C) N-terminal to the basic domain to position charge modifications near the phosphodiester backbone of DNA. This strategy placed charged residues at positions similar to those studied previously using the bZIP domain of Gcn4p (Figure 1A and B) (13). Comparison of the crystal structures shows that the bHLH peptide, despite its longer primary sequence, folds into a more globular structure (Figure 1A and B). The bHLHLONG peptide group, while positioning charges in a manner identical to bHLHSHORT, was predicted from studies of bZIP peptides (unpublished data) to stabilize folding by addition of 12 N-terminal residues (Figure 1C). The sequences of the CRE and E-box binding sites for Gcn4p and Pho4p, respectively, are shown in Figure 1D.
Binding affinities of bHLH charge variants
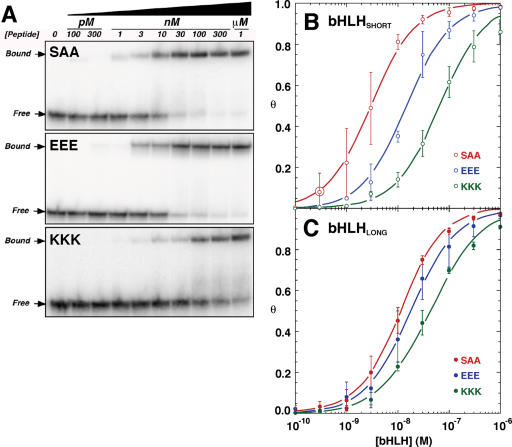
Before assessing their abilities to bend DNA, the DNA-binding affinities of Pho4p bHLH charge variants were determined by protein titrations against a constant concentration of an 18 bp DNA duplex containing the E-box-binding site (Figure 1D), and were assayed by electrophoretic mobility shift (Figure 2A). The fits shown in Figure 2B (bHLHSHORT) and 2C (bHLHLONG) were used to estimate apparent dissociation constants (Kd) for each of three charge variants for both peptide lengths (Table 1).
Figure 2.
Determination of bHLH charge variant binding affinities for E-box DNA. (A) Examples of electrophoretic gel mobility shift assays with increasing concentrations of bHLHSHORT (PAA, EEE and KKK) charge variants binding to 100 pM radiolabeled E-box DNA duplex. bHLH:DNA complexes were resolved from free E-box DNA by electrophoresis through 8% native polyacrylamide gels. (B and C) Estimation of DNA binding affinities of bHLHSHORT (C) and bHLHLONG (D) peptides for E-box DNA. Fractional DNA occupancy (θ) was plotted against the concentration of bHLH (XXX) peptide. Average Kd values ±1 SD derived from five datasets were estimated through curve fitting to Equation 1 as described in Materials and Methods.
Table 1.
Binding affinities of bHLH charge variants for E-box duplex DNA
| Kd (nM)a | |||
|---|---|---|---|
| SAA | EEE | KKK | |
| bHLHSHORT | 3 ± 2 | 15 ± 2 | 67 ± 7 |
| bHLHLONG | 10 ± 2 | 19 ± 2 | 55 ± 6 |
aKd values were estimated through curve fitting to Equation 1 as described in Materials and Methods. Mean and standard deviations were obtained from at least three independent measurements.
The data for both short and long bHLH peptides show that the neutral charge variant has the highest affinity for the E-box DNA, followed by the anionic form (Table 1). For both peptides, the cationic bHLH displayed 3- to 4-fold lower affinities for DNA (Table 1). The weaker binding affinities of cationic (KKK) charge variants were initially puzzling, as positive charges near the DNA surface might be expected to promote protein–DNA interactions. It is noteworthy that the appendage of the 12 N-terminal residues in bHLHLONG peptides reduces the differences in DNA binding affinities among the charge variants (Figure 2B and C and Table 1). Control experiments verified that all peptides retained proper DNA sequence specificity in binding assays (data not shown).
Folding of free and DNA-bound bHLH charge variants
To test if the observed binding affinity changes were due to changes in the coupling between protein folding and DNA binding, we characterized the folding of bHLH charge variants using circular dichroism (CD) spectroscopy (65). Changes in protein conformation induced upon DNA binding can be detected as spectral changes in the far UV CD region between 210 and 230 nm, a region largely unaffected by DNA, but useful in observing protein secondary structure. Under conditions comparable to those used in binding studies, far-UV CD data were collected for both free peptide and DNA-peptide complexes to determine the structural effects of charge insertion and DNA binding.
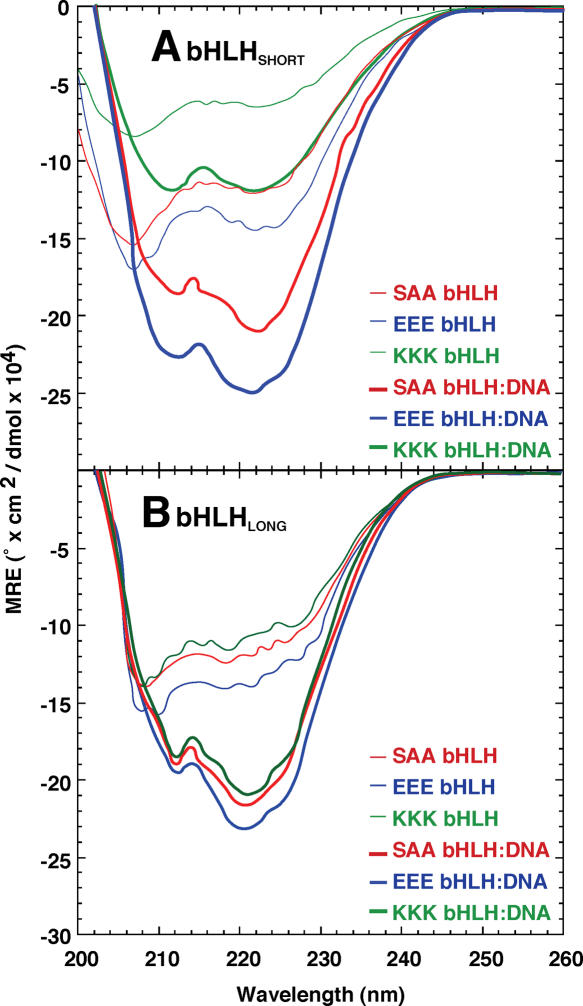
The far-UV CD spectra of the free bHLH peptides (10 μM) are shown in Figure 3. Based on the molar ellipticity values at 222 nm (Θ222) and published interpretations (66), the helical content of unbound bHLHSHORT charge variants was calculated to be 34, 40 and 16% for SAA, EEE and KKK, respectively. Helical content values of 32, 37 and 27% were observed for the bHLHLONG peptide charge variants SAA, EEE and KKK, respectively (Table 2). The results for the neutral charge variant (SAA) are consistent with the expected helical content of the peptides (free bHLHSHORT: 43%; free bHLHLONG: 37%) based upon the number of bHLH residues expected to be helical when not bound to DNA. For free bHLHSHORT peptides, insertion of cationic residues (KKK) destabilizes folding whereas addition of anionic residues (EEE) stabilizes folding (Figure 3A and Table 2). These effects are significantly reduced in bHLHLONG peptides, demonstrating the strong stabilizing effect the 12 amino acid leader segment. We have observed similar effects for corresponding charge variants of the bZIP domain of Gcn4p (data not shown). The ratio of molar ellipticity at 205 nm to the ellipticity at 222 nm, diagnostic for alpha helix content, confirms that unbound bHLH charge variants remain partially unfolded. Partial unfolding is consistent with a previous report that noted a stabilizing effect of buffer cation composition (66).
Figure 3.
Folding of bHLH charge variants. CD data (mean residual elipticity, ΘMRE) of free bHLH charge variants (thin lines) and corresponding bHLH:E-box (1:1.1) complexes (thick lines) are shown for both bHLHSHORT (A) and bHLHLONG (B) peptides.
Table 2.
CD and derived alpha helical content for free and DNA-bound bHLH peptides
| SAA | EEE | KKK | SAA:DNA | EEE:DNA | KKK:DNA | |
|---|---|---|---|---|---|---|
| bHLHSHORT MREa | −12 050 | −14 400 | −6350 | −21 100 | −25 200 | −11 950 |
| bHLHSHORT helicity (%)b | 34 | 40 | 16 | 62 | 76 | 33 |
| bHLHLONG MREa | −12 000 | −13 800 | −10 500 | −21 800 | −24 000 | −20 800 |
| bHLHLONG helicity (%)b | 32 | 37 | 27 | 64 | 71 | 61 |
aMean Residual Ellipticity (MRE) is calculated as described in Materials and Methods (Equation 2).
bHelical content, expressed as a fraction of the total number of residues, was determined as described in Materials and Methods (Equation 3). In the case of this bHLHSHORT peptide, the maximum helical content of the free peptide (Helix 1 + Helix 2 + loop helix) is predicted to be 28 of 65 residues (∼43%), in the case of bHLHLONG peptide, the maximum helical content of the free peptide (Helix 1 + Helix 2 + loop helix) is predicted to be 28 of 75 residues (∼37%). The maximum helical content of the bHLH:DNA peptide (Helix 1 + Helix 2 + loop helix + basic region) is predicted to be 43–47 of 65 residues (∼66–72%) for bHLHSHORT and 48–52 of 75 residues (∼64–69%).
Upon addition of 1.1 molar equivalents of cognate E-box DNA to the bHLH charge variant peptides, alpha-helical content was increased in all cases (Figure 3 and Table 2). For the bHLHSHORT peptides, comparison of the results for the cationic variant (KKK) to the neutral (SAA) and anionic (EEE) variants suggests that the cationic variant remains incompletely alpha-helical, despite a clear conformational change upon DNA binding (Figure 3A and Table 2). In contrast, bHLHLONG peptides displayed similar folding enhancements in the presence of DNA (Figure 3B and Table 2). In all cases, the random-coil minimum at 207 nm shifted (by 5 nm) to 212 nm and was less prominent relative to the alpha-helical signature with a Θ212:Θ222 ratio of ∼0.9 (66). This change is consistent with an increase in alpha-helical structure and a reduction in the random-coil folded state. The calculated helical content of the neutral bHLH:DNA complex is again consistent with predicted results in both long and short bHLH peptides. These findings confirm reports of far-UV CD spectral changes upon bHLH binding to DNA (66).
Our data highlight the importance of electrostatic effects in the folding of the bHLH domain. Placement of additional negative charges near the peptide N-terminus enhances the intrinsic helical content of the bHLH fold whereas additional positive charge reduces helical content. These results are consistent with the expected electrostatic interactions between charged residues near the terminus of an alpha helix and the intrinsic helix macrodipole (67,68), which are favorable for negatively charged residues near the N-terminus and unfavorable for positively charged residues. Our CD results show that cationic bHLH charge variant samples the unfolded state much more often than the other variants, explaining the ∼20-fold decrease in binding affinity for the E-box DNA in the case of the bHLHSHORT cationic variant. The reduction in the charge-dependent variation of folding stabilities of the bHLHLONG peptide group further supports this notion, as the inserted charges are placed further from the amino terminus, and so the electrostatic–macrodipole interaction is greatly reduced leading to more uniform folding stabilities and presumably binding affinities. The reduction in binding affinity seen for the SAA variant and the lack of the expected increase in affinity upon improved folding of the KKK variant may be due to steric interference between the extended helix and the DNA or to loss of contacts formed previously by less structured DNA tails.
DNA bending by Pho4p charge variants
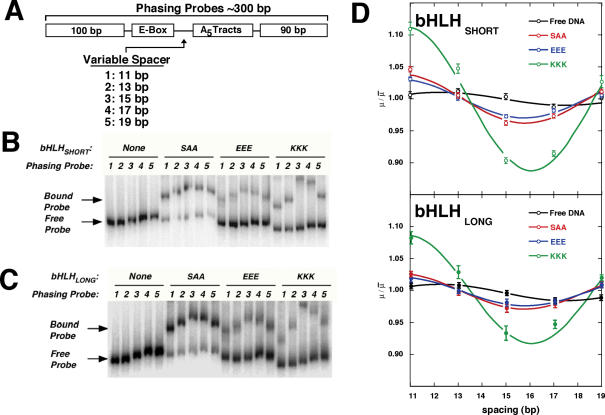
We employed a well-established electrophoretic DNA phasing assay that permits quantitative estimation of both the magnitude and direction of DNA bending. Our goal was to determine how asymmetric charge interactions at the Pho4p bHLH binding site (E-box) can lead to changes in observed DNA bending. Figure 4A illustrates the phasing probe design based on constructs originally developed by Schepartz (30) and Crothers (40). These ∼300 bp DNA duplexes contain a central binding site for Pho4p (E-box) variably spaced from an array of three phased A5-tracts that serves as a curved reference. This spacer distance is varied in the five phasing probes by a total of 10 bp (in 2–3 bp increments) in order to generate unique helical phasings between the phased A5 tract and the E-box. DNA probes in which the phased A5 tract and protein-induced bend occur on the same helical face (cis) will migrate more slowly due to a large composite helical deflection, whereas DNA probes in which the deformations lie on opposite helical faces (trans) will migrate more rapidly as the two deformations tend to cancel.
Figure 4.
Electrophoretic phasing assay of Pho4p bHLH charge variant homodimers. (A) Design of phasing probes 1–5. Phasing probes derived from previous constructs (Ref) were modified to contain the E-box sequence required for Pho4p binding. All probes also contained an array of three phased A5-tracts. The distance in bp between the Pho4p binding site and A5-tracts array varies as shown. (B) Typical autoradiogram obtained after 8% native polyacrylamide gel electrophoresis of phasing probes 1–5 incubated with the bHLHSHORT charge variants (neutral: SAA, anionic: EEE, cationic: KKK). (C) As in (B), except phasing probes were incubated with bHLHLONG charge variants. (D) Phasing analysis of electrophoretic data for probes 1–5 both free and bound to charge variants (SAA, EEE and KKK) of both bHLH peptide constructs. The relative mobilities of each of the five phasing probes in a given condition (free, bound to SAA, EEE, KKK) were plotted as a function of the spacing between the center of the curvature in the proximal A5 tract and the middle of the E-box binding site (Figure 1D). Data were fit to Equation 2 as described in Materials and Methods.
Results of electrophoretic phasing experiments for bHLH peptides are shown in Figure 4B and C. Probe migration rates were normalized, fit to a phasing function and plotted against spacer length (Figure 4D). The calculated bend angles (Table 3) show that the free E-box site has little intrinsic bending (estimated bend angle of 1°–2° toward the major groove). bHLH binding bends the E-box site slightly, with the neutral SAA:DNA complex showing a DNA bend toward the minor groove of 4°–5° (a 5°–7° difference from the free E-Box DNA; Table 3). These findings are consistent with previous crystallographic studies that show very little DNA bending in the bHLH:DNA complex (58). In contrast to the neutral variants, the cationic charge variants (KKK) showed a strong apparent bend, estimated at 26°–29°, toward the minor groove at the center of the E-box. This amounts to ∼24° of net bending relative to the SAA:DNA complex. These data are consistent with previous reports for cationic bZIP charge variants based on electrophoretic phasing assays (13,15,63). Both the anionic bHLH charge variants (EEE) caused little DNA bending, yielding minor-groove directed bends of 3°–4° (Table 3). This result contrasts with previous observations for bZIP charge variants, where approximately equal but opposite DNA bending (anionic: major groove; cationic: minor groove) was observed for opposite charges.
Table 3.
E-box bending using bHLH charge variants
| E-box | SAA:DNA | EEE:DNA | KKK:DNA | |
|---|---|---|---|---|
| bHLHSHORT bend angle1 | 1.2 ± 0.8° | 5.6 ± 1.1° | 4.1 ± 1.1° | 28.7 ± 3.7° |
| bHLHLONG bend anglea | 1.5 ± 0.6° | 4.2 ± 0.7° | 2.6 ± 1.5° | 25.5 ± 2.8° |
| Bend directionb | Major | Minor | Minor | Minor |
aBend angle estimates are generated from Equations 2–4 as described in Materials and Methods. Mean and standard deviations were obtained from three independent measurements.
bBend direction from Equations 4–6 refers to the DNA groove that is narrowed by bending, relative to a reference frame at the center of the E-box sequence.
Why does the bHLH scaffold permit DNA bending in only one direction? Since the DNA-binding affinities and specificities of the neutral (SAA) and anionic (EEE) bHLH variants are comparable, the lack of DNA bending by bHLH (EEE) is presumably not due to misfolding or loss of DNA-binding activity. Inspection of the crystal structures of both bZIP and bHLH suggests that the inability of the anionic charge variant of bHLH to bend DNA toward the major groove is explained by the bulky loop residues of the bHLH, which are known to be in contact with DNA and involved in enhancing DNA binding and specificity (58). The Pho4p bHLH loop directly contacts DNA on the major groove face of the E-box. Unlike the case for bZIP peptides, this presents a considerable steric obstacle to DNA bending in that direction.
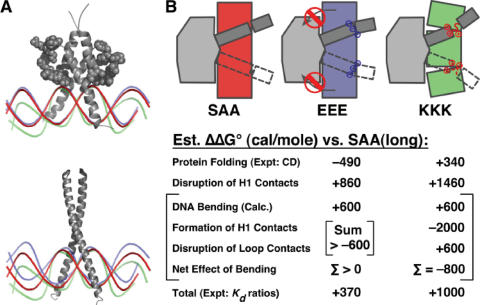
To explicate the linkages among charged bHLH peptide folding, DNA bending and changes in protein–DNA contacts, we sketch a plausible free energy balance model in Figure 5. Figure 5A compares DNA bending by bHLH charge variants (top) and bZIP charge variants (bottom), illustrating the steric hindrance that prevents DNA from binding toward the bHLH protein. Figure 5B rationalizes the bending, folding and binding affinity results coherently in terms of ΔΔG° contributions (relative free energies for protein–DNA complex formation) in the EEE and KKK complexes relative to the SAA complex. The total of all the ΔΔG° contributions relative to the SAA state must equal the observed ΔΔG° for binding, which is about +1.8 kcal/mol for bHLHSHORT and +1.0 kcal/mole for bHLHLONG. The description below uses measured or calculated ΔΔG° contributions where possible, as described in Materials and Methods. Otherwise, the values are chosen to be reasonable and to rationalize experimental observations. The point of the analysis is not to claim that these values are correct but simply to list some of the contributions that must be considered and to show that small free energy changes can rationalize DNA bending by the KKK variant, lack of bending by the EEE variant, and the experimentally determined ΔΔG° for bHLH:DNA complex formation.
Figure 5.
Proposed energetics of Pho4 variant binding and DNA bending. (A) Sketches based on the structures of Pho4 and Gcn4 bound to DNA. The red DNA resembles the DNA in the original structures, proposed to be the DNA shape for neutral appended residues. Steric interactions due to bulky loop residues of Pho4p allow appended cations (KKK charge variant) to cause DNA bending toward the minor groove (green), but do not allow substantial anion-mediated DNA bending (EEE charge variant) toward the major groove (blue). The bZIP charge variants have less steric hindrance and allow DNA bending in either direction. (B) Speculative energy balance for Pho4 EEE and KKK variants. ΔΔG° values relative to the SAA variant are shown. The values for protein folding, DNA-bending energy and the total ΔΔG° are calculated as described in Materials and Methods. The other ΔΔG° values, for the disruption of H1 contacts upon substitution, the formation of H1 contacts upon DNA bending and the disruption of loop contacts upon DNA bending, are reasonable guesses that rationalize bending only for the KKK variant and sum to the correct total ΔΔG°.
The elements of the model are as follows:
The KKK variant disrupts folding, which decreases the available binding free energy relative to SAA because more of the binding free energy must be coupled to folding and/or because contacts made by the N-terminal tail are lost. This free energy cost is larger for the bHLHSHORT variant because it does not fold as well. The free energy cost for folding can be estimated from the CD data by assuming that the EEE peptide is 95% folded and calculating the equilibrium constants for folding. The free energy cost shown for disrupting Helix 1 contacts is not known from experiment.
The free energy needed to bend DNA by ∼25° over ∼15 bp is small, only ∼0.6 kcal/mol [assuming a persistence length of 150 bp; (7)], and the bending is paid for by improved contacts with the N-terminal positive charges, again chosen arbitrarily to rationalize the existence of bending while being consistent with measured energetics.
The improved Helix 1 contacts in the KKK variants cause some distortion of contacts on the loop side, which costs binding free energy. The net effect is that as long as the sum of DNA bending energy and the loss of loop contacts is less than the free energy gained from electrostatic contacts, then the bent state will be a free energy minimum.
For the EEE variants, the small negative ΔΔG° expected from enhanced protein folding is opposed by unfavorable steric contacts made by the N-terminal helix and/or unfavorable EEE–DNA electrostatic interaction, leading to a slight decrease in binding affinity that is again smaller for the bHLHLONG variants. As discussed above, DNA bending is not observed because the free energy cost of compressing the protein–DNA interface is greater than the free energy gain from decreasing electrostatic repulsion by bending away from Helix 1.
Attempts to test some of these hypotheses directly using bHLH variants with either extensions of helix 1 or substituting residues with less bulky side-chains in the loop region were unsuccessful because these peptides failed to bind DNA (data not shown). These results are consistent with the model in Figure 5, although not conclusive. The essential result is that it is understandable how DNA bending can occur only with the KKK charge variant, even though the KKK peptide binds DNA less tightly than the SAA and EEE peptides: Contacts between the loop region and the DNA are disrupted by the apparent bending away from the KKK variants, and the disruption is reflected in the decreased binding affinity of the KKK variants. Electrostatic repulsion or structural changes destabilize the binding of EEE variants but cannot induce bending.
Summary
The work presented here supports the interpretations of previous experiments suggesting that electrostatic effects can cause DNA bending. Gel-based DNA bending assays have been compared previously to other DNA bending assays such as DNA cyclization and minicircle binding, with variable results (15,45,46). There has been concern that discrepancies might be explained by the elongated shape of bZIP peptides bound to DNA. The use of the more globular bHLH domain of Pho4p complements our previous elongated bZIP phasing assay results as it demonstrates that electrostatic effects in DNA bending are observed in the context of a less elongated, more globular peptide complex. We also observed electrostatic effects on DNA bending when we previously appended globular domains to the N-termini of bZIP peptides (15). Taken together, these results argue against the concern that electrophoretic phasing assay results are dominated by peptide shape. The difference in bending behavior between bHLH and bZIP charge variants likely results from a difference in how peptides pack against DNA, as the simple helical structure and lack of bulky domains on the bZIP peptide permits DNA bending in both directions, whereas the larger bHLH domain only permits bending away from the protein.
Our findings support the electrostatic collapse model of DNA bending and are consistent with (but do not prove) the somewhat controversial notion that DNA phosphate diester charges contribute significantly to DNA stiffness (3,11,25,26,44,69,70). Complementary approaches to these gel-based assays are currently under investigation.
Acknowledgments
We thank D. Wemmer for the Pho4p bHLH expression plasmid, J. Cave for the BL21 expression strain, A. Schepartz for the original pDP-AP1 phasing plasmids, M. Ramirez-Alvarado for instrument access and useful discussion, and G. Mer for technical discussion. The technical support of J. Zimmerman is acknowledged. We thank N. Becker, K. Riley and E. Smith for comments on the manuscript. This work was supported by the Mayo Foundation and NIH grants GM53620 (J.D.K.) and GM054411 (L.J.M). Funding to pay the Open Access publication charges for this article was provided by the Mayo Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hagerman P.J. Flexibility of DNA. Annu. Rev. Biophys. Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- 2.Maher L.J., III Mechanisms of DNA bending. Curr. Opin. Chem. Biol. 1998;2:688–694. doi: 10.1016/s1367-5931(98)80104-x. [DOI] [PubMed] [Google Scholar]
- 3.Williams L.D., Maher L.J., III Electrostatic mechanisms of DNA deformation. Annu. Rev. Biophys. Biomol. Struct. 2000;29:497–521. doi: 10.1146/annurev.biophys.29.1.497. [DOI] [PubMed] [Google Scholar]
- 4.Cantor C., Schimmel P. Biophysical Chemistry Part III: The Behavior of Biological Molecules. NY: W.H. Freeman and Company; 1980. [Google Scholar]
- 5.Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 6.Schultz S.C., Shields G.C., Steitz T.A. Crystal structure of a CAP–DNA complex: the DNA is bent by 90 degrees. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 7.Kahn J.D., Crothers D.M. DNA bending in transcription initiation. Cold Spring Harb. Symp. Quant. Biol. 1993;58:115–122. doi: 10.1101/sqb.1993.058.01.015. [DOI] [PubMed] [Google Scholar]
- 8.Parkinson G., Wilson C., Gunasekera A., Ebright Y.W., Ebright R.E., Berman H.M. Structure of the CAP-DNA complex at 2.5 angstroms resolution: a complete picture of the protein–DNA interface. J. Mol. Biol. 1996;260:395–408. doi: 10.1006/jmbi.1996.0409. [DOI] [PubMed] [Google Scholar]
- 9.Goodman S.D., Nash H.A. Functional replacement of a protein-induced bend in a DNA recombination site. Nature. 1989;341:251–254. doi: 10.1038/341251a0. [DOI] [PubMed] [Google Scholar]
- 10.Paull T.T., Haykinson M.J., Johnson R.C. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993;7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- 11.Strauss J.K., Maher L.J., III DNA bending by asymmetric phosphate neutralization. Science. 1994;266:1829–1834. doi: 10.1126/science.7997878. [DOI] [PubMed] [Google Scholar]
- 12.Strauss J.K., Prakash T.P., Roberts C., Switzer C., Maher L.J. DNA bending by a phantom protein. Chem. Biol. 1996;3:671–678. doi: 10.1016/s1074-5521(96)90135-0. [DOI] [PubMed] [Google Scholar]
- 13.Strauss-Soukup J.K., Maher L.J., III Electrostatic effects in DNA bending by GCN4 mutants. Biochemistry. 1998;37:1060–1066. doi: 10.1021/bi972146p. [DOI] [PubMed] [Google Scholar]
- 14.Tomky L.A., Strauss-Soukup J.K., Maher L.J., III Effects of phosphate neutralization on the shape of the AP-1 transcription factor binding site in duplex DNA. Nucleic Acids Res. 1998;26:2298–2305. doi: 10.1093/nar/26.10.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardwidge P.R., Kahn J.D., Maher L.J., III Dominant effect of protein charge rather than protein shape in apparent DNA bending by engineered bZIP domains. Biochemistry. 2002;41:8277–8288. doi: 10.1021/bi020185h. [DOI] [PubMed] [Google Scholar]
- 16.Hardwidge P.R., Wu J., Williams S.L., Parkhurst K.M., Parkhurst L.J., Maher L.J., III DNA bending by bZIP charge variants: a unified study using electrophoretic phasing and fluorescence resonance energy transfer. Biochemistry. 2002;41:7732–7742. doi: 10.1021/bi020213w. [DOI] [PubMed] [Google Scholar]
- 17.Soto A.M., Kankia B.I., Dande P., Gold B., Marky L.A. Incorporation of a cationic aminopropyl chain in DNA hairpins: thermodynamics and hydration. Nucleic Acids Res. 2001;29:3638–3645. doi: 10.1093/nar/29.17.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gold B. Effect of cationic charge localization on DNA structure. Biopolymers. 2002;65:173–179. doi: 10.1002/bip.10215. [DOI] [PubMed] [Google Scholar]
- 19.Kosikov K.M., Gorin A.A., Lu X.J., Olson W.K., Manning G.S. Bending of DNA by asymmetric charge neutralization: all-atom energy simulations. J. Am. Chem. Soc. 2002;124:4838–4847. doi: 10.1021/ja0125218. [DOI] [PubMed] [Google Scholar]
- 20.Li Z., Huang L., Dande P., Gold B., Stone M.P. Structure of a tethered cationic 3-aminopropyl chain incorporated into an oligodeoxynucleotide: evidence for 3′-orientation in the major groove accompanied by DNA bending. J. Am. Chem. Soc. 2002;124:8553–8560. doi: 10.1021/ja0201707. [DOI] [PubMed] [Google Scholar]
- 21.Soto A.M., Kankia B.I., Dande P., Gold B., Marky L.A. Thermodynamic and hydration effects for the incorporation of a cationic 3-aminopropyl chain into DNA. Nucleic Acids Res. 2002;30:3171–3180. doi: 10.1093/nar/gkf430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Range K., Mayaan E., Maher L.J., III, York D.M. The contribution of phosphate-phosphate repulsions to the free energy of DNA bending. Nucleic Acids Res. 2005;33:1257–1268. doi: 10.1093/nar/gki272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams S.L., Parkhurst L.K., Parkhurst L.J. Changes in DNA bending and flexing due to tethered cations detected by fluorescence resonance energy transfer. Nucleic Acids Res. 2006;34:1028–1035. doi: 10.1093/nar/gkj498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mirzabekov A.D., Rich A. Asymmetric lateral distribution of unshielded phosphate groups in nucleosomal DNA and its role in DNA bending. Proc. Natl Acad. Sci. USA. 1979;76:1118–1121. doi: 10.1073/pnas.76.3.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manning G.S., Ebralidse K.K., Mirzabekov A.D., Rich A. An estimate of the extent of folding of nucleosomal DNA by laterally asymmetric neutralization of phosphate groups. J. Biomol. Struct. Dyn. 1989;6:877–889. doi: 10.1080/07391102.1989.10506519. [DOI] [PubMed] [Google Scholar]
- 26.Manning G.S. Is a small number of charge neutralizations sufficient to bend nucleosome core DNA onto its superhelical ramp? J. Am. Chem. Soc. 2003;125:15087–15092. doi: 10.1021/ja030320t. [DOI] [PubMed] [Google Scholar]
- 27.Oakley M.G., Dervan P.B. Structural motif of the GCN4 DNA binding domain characterized by affinity cleaving. Science. 1990;248:847–850. doi: 10.1126/science.2111578. [DOI] [PubMed] [Google Scholar]
- 28.Kerppola T.K., Curran T. DNA bending by Fos and Jun: the flexible hinge model. Science. 1991;254:1210–1214. doi: 10.1126/science.1957173. [DOI] [PubMed] [Google Scholar]
- 29.Kerppola T.K., Curran T. Selective DNA bending by a variety of bZIP proteins. Mol. Cell. Biol. 1993;13:5479–5489. doi: 10.1128/mcb.13.9.5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paolella D.N., Palmer C.R., Schepartz A. DNA targets for certain bZIP proteins distinguished by an intrinsic bend. Science. 1994;264:1130–1133. doi: 10.1126/science.8178171. [DOI] [PubMed] [Google Scholar]
- 31.Suckow M., Madan A., Kisters-Woike B., von Wilcken-Bergmann B., Muller-Hill B. Creating new DNA binding specificities in the yeast transcriptional activator GCN4 by combining selected amino acid substitutions. Nucleic Acids Res. 1994;22:2198–2208. doi: 10.1093/nar/22.12.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kerppola T., Curran T. Transcription. Zen and the art of Fos and Jun. Nature. 1995;373:199–200. doi: 10.1038/373199a0. [DOI] [PubMed] [Google Scholar]
- 33.Kerppola T.K. Fos and Jun bend the AP-1 site: effects of probe geometry on the detection of protein-induced DNA bending. Proc. Natl Acad. Sci. USA. 1996;93:10117–10122. doi: 10.1073/pnas.93.19.10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellenberger T.E., Brandl C.J., Struhl K., Harrison S.C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein–DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 35.Weiss M.A., Ellenberger T., Wobbe C.R., Lee J.P., Harrison S.C., Struhl K. Folding transition in the DNA-binding domain of GCN4 on specific binding to DNA. Nature. 1990;347:575–578. doi: 10.1038/347575a0. [DOI] [PubMed] [Google Scholar]
- 36.Metallo S.J., Paolella D.N., Schepartz A. The role of a basic amino acid cluster in target site selection and non-specific binding of bZIP peptides to DNA. Nucleic Acids Res. 1997;25:2967–2972. doi: 10.1093/nar/25.15.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konig P., Richmond T.J. The X-ray structure of the GCN4-bZIP bound to ATF/CREB site DNA shows the complex depends on DNA flexibility. J. Mol. Biol. 1993;233:139–154. doi: 10.1006/jmbi.1993.1490. [DOI] [PubMed] [Google Scholar]
- 38.Koo H.S., Crothers D.M. Calibration of DNA curvature and a unified description of sequence-directed bending. Proc. Natl Acad. Sci. USA. 1988;85:1763–1767. doi: 10.1073/pnas.85.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koo H.S., Drak J., Rice J.A., Crothers D.M. Determination of the extent of DNA bending by an adenine-thymine tract. Biochemistry. 1990;29:4227–4234. doi: 10.1021/bi00469a027. [DOI] [PubMed] [Google Scholar]
- 40.Crothers D., Drak J. Global features of DNA structure by comparative gel electrophoresis. Methods Enzymol. 1992;212:46–71. doi: 10.1016/0076-6879(92)12005-b. [DOI] [PubMed] [Google Scholar]
- 41.Paolella D.N., Liu Y., Fabian M.A., Schepartz A. Electrostatic mechanism for DNA bending by bZIP proteins. Biochemistry. 1997;36:10033–10038. doi: 10.1021/bi970515b. [DOI] [PubMed] [Google Scholar]
- 42.Kerppola T.K., Curran T. The transcription activation domains of Fos and Jun induce DNA bending through electrostatic interactions. EMBO J. 1997;16:2907–2916. doi: 10.1093/emboj/16.10.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rich A. Localized positive charges can bend double helical nucleic acid. FEBS Lett. 1978;51:71–81. [Google Scholar]
- 44.Podesta A., Indrieri M., Brogioli D., Manning G.S., Milani P., Guerra R., Finzi L., Dunlap D. Positively charged surfaces increase the flexibility of DNA. Biophys. J. 2005;89:2558–2563. doi: 10.1529/biophysj.105.064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hagerman P.J. Do basic region-leucine zipper proteins bend their DNA targets … does it matter? Proc. Natl Acad. Sci. USA. 1996;93:9993–9996. doi: 10.1073/pnas.93.19.9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sitlani A., Crothers D.M. Fos and jun do not bend the AP-1 recognition site. Proc. Natl Acad. Sci. USA. 1996;93:3248–3252. doi: 10.1073/pnas.93.8.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sitlani A., Crothers D. DNA-binding domains of Fos and Jun do not induce DNA curvature: an investigation with solution and gel methods. Proc. Natl Acad. Sci. USA. 1998;95:1404–1409. doi: 10.1073/pnas.95.4.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toh-e A., Inouye S., Oshima Y. Structure and function of the PHO82-pho4 locus controlling the synthesis of repressible acid phosphatase of Saccharomyces cerevisiae. J. Bacteriol. 1981;145:221–232. doi: 10.1128/jb.145.1.221-232.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ogawa N., Oshima Y. Functional domains of a positive regulatory protein, PHO4, for transcriptional control of the phosphatase regulon in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:2224–2236. doi: 10.1128/mcb.10.5.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaffman A., Herskowitz I., Tjian R., O'Shea E.K. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80–PHO85. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- 51.Magbanua J.P., Ogawa N., Harashima S., Oshima Y. The transcriptional activators of the PHO regulon, Pho4p and Pho2p, interact directly with each other and with components of the basal transcription machinery in Saccharomyces cerevisiae. J. Biochem. (Tokyo) 1997;121:1182–1189. doi: 10.1093/oxfordjournals.jbchem.a021713. [DOI] [PubMed] [Google Scholar]
- 52.Vogel K., Horz W., Hinnen A. The two positively acting regulatory proteins PHO2 and PHO4 physically interact with PHO5 upstream activation regions. Mol. Cell. Biol. 1989;9:2050–2057. doi: 10.1128/mcb.9.5.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fisher F., Jayaraman P.S., Goding C.R. C-myc and the yeast transcription factor PHO4 share a common CACGTG-binding motif. Oncogene. 1991;6:1099–1104. [PubMed] [Google Scholar]
- 54.Murre C., McCaw P.S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 55.Berben G., Legrain M., Gilliquet V., Hilger F. The yeast regulatory gene PHO4 encodes a helix–loop–helix motif. Yeast. 1990;6:451–454. doi: 10.1002/yea.320060510. [DOI] [PubMed] [Google Scholar]
- 56.Littlewood T.D., Evan G.I. Transcription factors 2: helix–loop–helix. Protein Profile. 1995;2:621–702. [PubMed] [Google Scholar]
- 57.Shao D., Creasy C.L., Bergman L.W. A cysteine residue in helixII of the bHLH domain is essential for homodimerization of the yeast transcription factor Pho4p. Nucleic Acids Res. 1998;26:710–714. doi: 10.1093/nar/26.3.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shimizu T., Toumoto A., Ihara K., Shimizu M., Kyogoku Y., Ogawa N., Oshima Y., Hakoshima T. Crystal structure of PHO4 bHLH domain–DNA complex: flanking base recognition. EMBO J. 1997;16:4689–4697. doi: 10.1093/emboj/16.15.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cave J.W., Kremer W., Wemmer D.E. Backbone dynamics of sequence specific recognition and binding by the yeast Pho4 bHLH domain probed by NMR. Protein Sci. 2000;9:2354–2365. doi: 10.1110/ps.9.12.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pace C.N., Vajdos F., Fee L., Grimsley G., Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Y.H., Yang J.T., Chau K.H. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 62.Kerppola T. DNA bending specificity among bZIP family protiens. In: Conaway R., Conaway J., editors. Transcription: Mechanisms and Regulation. NY: Raven Press, Ltd; 1994. pp. 387–424. [Google Scholar]
- 63.Strauss-Soukup J.K., Maher L.J., III DNA bending by GCN4 mutants bearing cationic residues. Biochemistry. 1997;36:10026–10032. doi: 10.1021/bi970215u. [DOI] [PubMed] [Google Scholar]
- 64.Liu-Johnson H.N., Gartenberg M.R., Crothers D.M. The DNA binding domain and bending angle of E.coli CAP protein. Cell. 1986;47:995–1005. doi: 10.1016/0092-8674(86)90814-7. [DOI] [PubMed] [Google Scholar]
- 65.Fasman G., editor. Circular Dichroism and the Conformational Analysis of Biomolecules. NY: Plenum Press; 1996. [Google Scholar]
- 66.Winston R.L., Millar D.P., Gottesfeld J.M., Kent S.B. Characterization of the DNA binding properties of the bHLH domain of Deadpan to single and tandem sites. Biochemistry. 1999;38:5138–5146. doi: 10.1021/bi982856a. [DOI] [PubMed] [Google Scholar]
- 67.Munoz V., Serrano L. Elucidating the folding problem of helical peptides using empirical parameters. II. Helix macrodipole effects and rational modification of the helical content of natural peptides. J. Mol. Biol. 1995;245:275–296. doi: 10.1006/jmbi.1994.0023. [DOI] [PubMed] [Google Scholar]
- 68.Lacroix E., Viguera A.R., Serrano L. Elucidating the folding problem of alpha-helices: local motifs, long-range electrostatics, ionic-strength dependence and prediction of NMR parameters. J. Mol. Biol. 1998;284:173–191. doi: 10.1006/jmbi.1998.2145. [DOI] [PubMed] [Google Scholar]
- 69.Hagerman P.J. Straightening out the bends in curved DNA. Biochim. Biophys. Acta. 1992;1131:125–132. doi: 10.1016/0167-4781(92)90066-9. [DOI] [PubMed] [Google Scholar]
- 70.Mills J.B., Hagerman P.J. Origin of the intrinsic rigidity of DNA. Nucleic Acids Res. 2004;32:4055–4059. doi: 10.1093/nar/gkh740. [DOI] [PMC free article] [PubMed] [Google Scholar]