Abstract
Endonuclease IV encoded by denB of bacteriophage T4 is implicated in restriction of deoxycytidine (dC)-containing DNA in the host Escherichia coli. The enzyme was synthesized with the use of a wheat germ cell-free protein synthesis system, given a lethal effect of its expression in E.coli cells, and was purified to homogeneity. The purified enzyme showed high activity with single-stranded (ss) DNA and denatured dC-substituted T4 genomic double-stranded (ds) DNA but exhibited no activity with dsDNA, ssRNA or denatured T4 genomic dsDNA containing glucosylated deoxyhydroxymethylcytidine. Characterization of Endo IV activity revealed that the enzyme catalyzed specific endonucleolytic cleavage of the 5′ phosphodiester bond of dC in ssDNA with an efficiency markedly dependent on the surrounding nucleotide sequence. The enzyme preferentially targeted 5′-dTdCdA-3′ but tolerated various combinations of individual nucleotides flanking this trinucleotide sequence. These results suggest that Endo IV preferentially recognizes short nucleotide sequences containing 5′-dTdCdA-3′, which likely accounts for the limited digestion of ssDNA by the enzyme and may be responsible in part for the indispensability of a deficiency in denB for stable synthesis of dC-substituted T4 genomic DNA.
INTRODUCTION
Infection of Escherichia coli by T4 phage is followed by degradation of genomic DNA of the host cell to provide nucleotides for the synthesis of T4 genomic DNA, in which all deoxycytidine (dC) residues are replaced by glucosylated deoxyhydroxymethylcytidine (gluc-dHMC) (1,2). At least two T4 endonucleases [Endo II and Endo IV, encoded by denA (3) and denB (4), respectively] are thought to participate in the degradation of host DNA, with T4 genomic DNA being normally protected from cleavage by the presence of gluc-dHMC (1,2,5,6). The Endo II protein is essential for this degradation of host DNA whereas the Endo IV protein is not (3,7,8).
The Endo II and Endo IV proteins are composed of 136 and 185 amino acid residues, respectively, and their biochemical properties have been examined with partially purified enzyme preparations from E.coli cells infected with T4 (9–11). Both enzymes require Mg2+ for activity and do not cleave T4 genomic DNA containing gluc-dHMC. Although Endo II acts on double-stranded (ds) DNA and generates 5′ termini containing predominantly dG or dC, Endo IV acts on single-stranded (ss) DNA and generates 5′ termini containing exclusively dC. Specific roles for Endo II and Endo IV in the degradation of host genomic DNA have been proposed on the basis of their biochemical properties (9,10). Endo II is thought to introduce a nick in host genomic dsDNA, and the 46/47 exonuclease, encoded by gene 46 and gene 47, subsequently removes mononucleotides from the internal 3′-hydroxyl terminus and thereby exposes a region of ssDNA on the opposite strand. This exposed ssDNA region is cleaved by Endo IV and yields dsDNA fragments.
T4 phage with mutations in gene 42 (which encodes dCMP hydroxymethylase), gene 56 (which encodes dCTPase) and denB synthesize a completely dC-substituted T4 (T4dC) genomic DNA (12). An additional mutation in alc (unf), whose product shuts off transcription of T4dC DNA (13), results in the generation of a plaque-forming T4 phage containing dC-substituted DNA (T4dC phage). T4dC phage is fully susceptible to restriction endonucleases in vivo and in vitro (12,14,15). A deficiency in denB, but not in denA, is required for synthesis of stable T4dC genomic DNA in a gene 56− (6,16,17) or gene 42−, gene 46−, gene 56− (6) background, even though the denA product (Endo II) plays a major role in the degradation of dC-containing host DNA (5). These observations suggest the possibility that the denB product (Endo IV) plays a crucial role in inhibition of the replication of dC-containing DNA rather than in its degradation.
The mechanism of substrate recognition by Endo II has been studied extensively in vivo (18–20) and in vitro (9,21,22), given the major role of the enzyme in degradation of dC-containing T4 (5) and host (3,7,8) DNA. In contrast, the substrate recognition mechanism of Endo IV has been less well characterized (10,11,23,24). Moreover, the small number of Endo IV-related proteins in the genome sequence databases has limited the amount of insight provided by such proteins into the mechanism of Endo IV action.
We have now shown that a low level of expression of denB is highly toxic to E.coli cells. We therefore synthesized Endo IV with a wheat germ cell-free protein synthesis system (25) and purified it to homogeneity without the need for cloning of denB into an expression vector (26,27). Analysis of the substrate specificity and sequence preference of the highly purified enzyme indicated that it specifically cleaves the 5′ phosphodiester bond of dC in ssDNA with an efficiency that depends markedly on the surrounding nucleotides. A preference of the enzyme for a 5′-dTdCdA-3′ trinucleotide sequence was revealed.
MATERIALS AND METHODS
Materials
Restriction and other enzymes for recombinant DNA technology were obtained from Takara Shuzo. T4 and T4dC genomic DNA were prepared as described previously (28).
Cells and plasmids
The plasmid pEUGFP was constructed as described previously (27) and pGEX-6P-1 was obtained from Amersham Biosciences. E.coli strain KH5402-1 [ilv, thyA, thr (amber), trpE9829 (amber), metE, deo, supF6 (Ts)] (29) was provided by the National Institute of Genetics (Japan). T4dC phage [gene 42 (amC87), gene 56 (amE51), denB (s19), alc (unf39)] was constructed as described previously (15).
The plasmid pBRdenBam was constructed for the present study. The nucleotide sequence of denB has been deposited in EMBL/GenBank/DDBJ under the accession number NC_000866.4 (167103–167660) and in Entrez Gene with the GeneID 1258726. The denBam gene, in which the TAC codon for Tyr38 is replaced with a TAG nonsense codon, was constructed by the PCR with a 5′ primer containing a BamHI site (5′-CTACAGGATCCGAAGGAGATATACATATGCAGAAAACGAATCCTGG-3′, with the BamHI site underlined, the initiation codon in boldface and the Shine–Dalgarno sequence in italics) and a 3′ primer containing a SalI site (5′-AAAACCATGGGACGTCGACTTAAATGGAAAGATACCATCCGTTG-3′, with the complementary sequence of the termination codon in boldface and the SalI site underlined) as well as 5′ and 3′ mutagenic primers (5′-TGCAAGATAGTCTTTAAAACAAAACC-3′ and 5′-TTTTAAAGACTATCTTGCAGTATCAGC-3′, with the sequence and complementary sequence, respectively, of the TAG nonsense codon that replaces the TAC codon for Tyr38 shown in boldface). The upstream and downstream regions of denB in T4 genomic DNA were amplified by PCR with the 5′ primer and the 3′ mutagenic primer and with the 5′ mutagenic primer and the 3′ primer, respectively. The amplified DNA fragments were purified by gel electrophoresis, mixed (each at a final concentration of 5 nM) and subjected to a second round of PCR with the 5′ and 3′ primers. The DNA fragment generated by the second-round PCR was introduced into the BamHI–SalI sites of pBR322 (Takara Shuzo) to yield pBRdenBam and was cloned in non-amber-suppressing host cells. The nucleotide sequence of the denBam gene was determined by the dideoxy chain termination method. All PCR primers were synthesized by Sigma-Aldrich. PCR was performed for 25 cycles of incubation at 96°C for 30 s, at 55°C for 30 s and at 72°C for 60 s with Ex Taq DNA polymerase (Takara Shuzo) in a Takara PCR Thermal Cycler MP.
The effect of denB expression on E.coli cell growth was examined by comparison of the plating efficiencies of E.coli KH5402-1 cells harboring pBRdenBam and grown at 30 or 42°C. The E.coli KH5402-1 cells were transformed with pBRdenBam and the transformants were spread on a Luria–Bertani (LB) agar plate containing thymine and ampicillin at 2 and 100 μg/ml, respectively (LB-Thy-Amp), and were incubated at 42°C. A single transformant was selected and grown overnight at 42°C in LB-Thy-Amp liquid broth. The resulting cells were spread on two LB-Thy-Amp agar plates, one of which was incubated at 42°C and the other at 30°C. Given that E.coli KH5402-1 cells harbor a temperature-sensitive (Ts) allele of the supF suppressor gene (29), the cells transformed with pBRdenBam would be expected to produce the active Endo IV protein when grown at 30°C but not at 42°C.
Construction of a DNA fragment encoding a GST-endo IV fusion protein
To construct a DNA fragment encoding a glutathione S-transferase (GST) fusion protein of Endo IV, we amplified DNA fragments corresponding to the 5′-untranslated region (5′-UTR), GST, Endo IV and 3′-UTR by PCR and assembled them with a second-round PCR. For construction of the DNA fragment corresponding to the 5′-UTR, the region of pEUGFP containing the promoter sequence for SP6 RNA polymerase and the omega sequence was amplified by PCR with a 5′ primer (5′-ATACATAAGCTTATGTATCATACACATACG-3′, primer 1) and a 3′ primer (5′-CATATGACTAGTGGCTGTAG-3′, with the complementary sequence of the initiation codon for the GST-Endo IV fusion protein shown in boldface).
For construction of the open reading frame for the GST portion of the GST-Endo IV fusion protein, the GST gene in pGEX-6P-1 was amplified by PCR with a 5′ primer (5′-CTACAGCCACTAGTCATATGGAATCCCCTATACTAGG-3′, with the sequence of the initiation codon for the fusion protein shown in boldface and the first 20 bases of the GST gene in italics) and a 3′ primer (5′-TCCCAGGGGCCCCTGGAACAGAACTTCCAGATCCGATTTTGGAGG-3′, with the complementary sequence for the PreScission protease recognition site underlined and the complementary sequence of the last 15 bases of the GST gene italicized).
For construction of the open reading frame for the Endo IV portion of the fusion protein, the denB gene was amplified from T4 genomic DNA by PCR with a 5′ primer (5′-TTCCAGGGGCCCCTGGGATCCATGCAGAAAACGAATCCTGG-3′, with a partial sequence for the PreScission protease recognition site underlined and the sequence of the first 20 bases of denB in italics) and a 3′ primer (5′-AAAACCATGGGACGTCGACTTAAATGGAAAGATACCATCCGTTG-3′, with the complementary sequence of the termination codon for the fusion protein shown in boldface and the last 22 bases of denB italicized).
For construction of the DNA fragment corresponding to the 3′-UTR, the region of pEUGFP containing the 3′-UTR of tobacco mosaic virus was amplified by PCR with a 5′ primer (5′-TAAGTCGACGTCCCATGGTTTTG-3′, with the termination codon of the fusion protein shown in boldface) and a 3′ primer (5′-GGAGAAAGGCGGACAGGTAT-3′, designated primer 2, which hybridizes 1175 bases downstream of the termination codon of the GFP gene in pEUGFP).
The four DNA fragments corresponding to the 5′-UTR, coding region and 3′-UTR of the GST-Endo IV fusion protein were purified by gel electrophoresis, mixed (each at a final concentration of 5 nM) and subjected to a second round of PCR without primers. The resulting DNA fragment was further amplified by a third round of PCR with primer 1 and a 3′ primer (5′-CCGGTAAGCGGCAGGGTCGG-3′, designated primer 3, which hybridizes 1 base upstream of the sequence corresponding to primer 2). Given that the yield of the third-round PCR was low, the amplicon was further amplified by a fourth round of PCR with a 5′ primer (5′-ACATACGATTTAGGTGACACTA-3′, with a partial sequence of the promoter element for SP6 RNA polymerase underlined) and primer 3. The DNA fragment for the GST-Endo IV fusion protein generated by the fourth-round PCR was transcribed as described previously (26), and the resulting mRNA was translated in a wheat germ cell-free protein synthesis system.
Cell-free protein synthesis
Conditions for the wheat germ cell-free protein synthesis system with a dialysis cup (molecular size cutoff of 12 000 Da; Daiichi Pure Chemicals) were as described previously (27), with slight modifications. The internal reaction mixture (100 μl) was as described previously (27) with the exception that 20% wheat germ extract (Zoegene) was included and GTP was omitted. The external solution (1 ml) contained the same components as the internal reaction mixture with the exception that creatine kinase, RNasin (RNase inhibitor, Promega) and wheat germ extract were omitted. The dialysis unit containing the reaction mixture was incubated at 26°C for 96 h, with the original amount of substrate mRNA being supplemented and the external solution changed every 24 h.
Protein purification
For purification of the GST-Endo IV fusion protein synthesized in the wheat germ cell-free protein synthesis system, the translation mixture was centrifuged at 20 000 g for 15 min at 4°C and the resulting supernatant was diluted 6-fold with phosphate-buffered saline and applied to a glutathione–Sepharose 4B MicroSpin column (Amersham Biosciences). The GST-Endo IV fusion protein was then cleaved in the column by incubation with PreScission protease (10 U/ml; Amersham Biosciences) for 4 h at 4°C. The flow-through fraction obtained after protease treatment contained a protein of the predicted size for Endo IV (21.1 kDa) as revealed by SDS–PAGE on a 12% gel and staining with Coomassie brilliant blue. The protein concentration of this fraction was estimated by densitometric analysis with NIH Image software, with the use of trypsin inhibitor (20.1 kDa) as a standard.
Endo IV assay
Assay of Endo IV activity was performed by incubation of enzyme and substrate for 30 min at 37°C in a reaction mixture (20 μl) containing 10 mM Tris–HCl (pH 8.0), 10 mM MgCl2, 1 mM DTT and BSA (0.1 mg/ml). Purified Endo IV was diluted in a solution containing 10 mM Tris–HCl (pH 8.0), 1 mM DTT and BSA (0.1 mg/ml) and was included in the reaction mixture in a volume of 1 μl. phiX174 replicative form I (RFI) circular dsDNA (New England BioLabs), phiX174 RFI linear dsDNA prepared by PstI digestion, phiX174 virion circular ssDNA (New England BioLabs), linear ssRNA used for synthesis of the GST-Endo IV fusion protein, and heat-denatured T4 or T4dC genomic dsDNA were used as substrates at a final concentration of 5 μg/ml. The final concentration of the enzyme was varied from 0.05 to 1 μg/ml. The reaction was stopped by the addition of 20 μl of 25 mM EDTA (pH 8.0), and the reaction products were separated by electrophoresis on a 1.0% agarose gel and stained with SYBR Gold (Invitrogen).
Cleavage of a phiX174-based oligonucleotide by Endo IV
A 45 base oligonucleotide (5′-TTGGATGAGGAGAAGTGGCTTAATATGCTTGGCACGTTCGTCAAG-3′) based on the phiX174 virion ssDNA sequence and labeled at the 5′ end with Cy5 was used as a substrate. Hydrolysis of the substrate (10 μM) was performed as described above for the Endo IV assay at an enzyme concentration of 0.33–6.6 μg/ml. The reaction products were fractionated by electrophoresis on a 10% polyacrylamide gel containing 7 M urea and were visualized with a Variable Image Analyzer Typhoon 8600 (Amersham Biosciences).
Kinetic analysis of Endo IV activity
Hydrolysis of unlabeled oligonucleotide substrates (10 μM) was performed as described above for the Endo IV assay. The reaction was stopped by the addition of 30 μl of 25 mM EDTA (pH 8.0) and 50 μl of 10% trichloroacetic acid. The resulting mixture (100 μl) was maintained on ice for 15 min and then centrifuged at 5000 g for 15 min at 4°C. The amount of acid-soluble nucleotides in the supernatant fraction was quantified by measurement of absorbance at 260 nm, with molar absorption coefficients of 15 200, 7050, 12 010 and 8400 M−1cm−1 for dA, dC, dG and dT, respectively. One unit was defined as the amount of enzyme producing 1 μmol of acid-soluble nucleotides per minute, and specific activity was defined as the enzymatic activity per milligram of enzyme. The amount of enzyme was varied such that the amount of product increased in proportion to that of the enzyme. For determination of kinetic parameters, unlabeled oligonucleotides with a single cleavage site were used as the substrate. The concentration of the substrate was also varied from 2.5 to 25 μM such that it spanned the Km. All oligonucleotide substrates were synthesized by Sigma-Aldrich.
RESULTS
Detrimental effect of denB expression on E.coli growth
Attempts to clone the wild-type denB gene in several different expression vectors, including pET21a (under strict control of the T7 promoter), were unsuccessful, presumably because of a highly toxic effect of the denB product (Endo IV) on the host E.coli cells. We therefore examined the effect of Endo IV on E.coli by transformation of KH5402-1 (supF6Ts) cells, which harbor a temperature-sensitive allele of the supF suppressor gene (29), with a plasmid containing an amber mutant allele of denB, in which the TAC codon for Tyr38 is mutated to a TAG nonsense codon. Cells cultured at 30°C would thus be expected to produce the intact Endo IV protein whereas those cultured at 42°C would not.
Culture at 30°C of the cells transformed with pBRdenBam reduced colony-forming ability by a factor of ∼104 compared with that apparent at 42°C (Table 1). However, cells of most (99%) of the original colonies that formed at 30°C did not give rise to colonies in the presence of the selection drug (ampicillin) at 42°C, indicating that most of the colonies that formed at 30°C consisted of cells that did not harbor pBRdenBam. These results thus suggested that no E.coli cells that expressed the active Endo IV protein were viable and that Endo IV targets an activity, such as DNA replication, that is indispensable for host cell growth.
Table 1.
Effect of denB expression on E.coli growth
| Plasmid | No. of colonies formed at 30°C | No. of colonies formed at 42°C | 30°/42°C ratio |
|---|---|---|---|
| pBRdenBam | 795a | 281 × 104 | 2.8 × 10−4 |
| pBR322 | 309 × 104 | 445 × 104 | 0.69 |
The effect of denB expression on E.coli cell growth was examined as described in Materials and Methods. Data are means from three independent experiments.
aA total of 156 colonies formed at 30°C by cells transformed with pBRdenBam was tested for colony-forming ability on LB-Thy or LB-Thy-Amp agar plates at 42°C. Cells of all the original 156 colonies formed colonies on the LB-Thy agar plate at 42°C, but cells of 154 of these 156 original colonies (∼99%) did not form colonies on the LB-Thy-Amp agar plate at 42°C, indicating that most of the original colonies formed at 30°C consisted of cells lacking pBRdenBam.
Cell-free synthesis and purification of Endo IV
Given that Endo IV was found to be highly toxic to E.coli cells, we synthesized the enzyme as a GST fusion protein by in vitro translation with a wheat germ cell-free protein synthesis system. The yield of the fusion protein was ∼400 μg per milliliter of the translation mixture, as estimated by SDS–PAGE and densitometric analysis (Figure 1). The synthesized fusion protein was recovered in soluble form, purified to homogeneity by glutathione affinity chromatography, and cleaved to yield the Endo IV moiety. The amount of Endo IV purified from 1 ml of the translation mixture was ∼100 μg.
Figure 1.
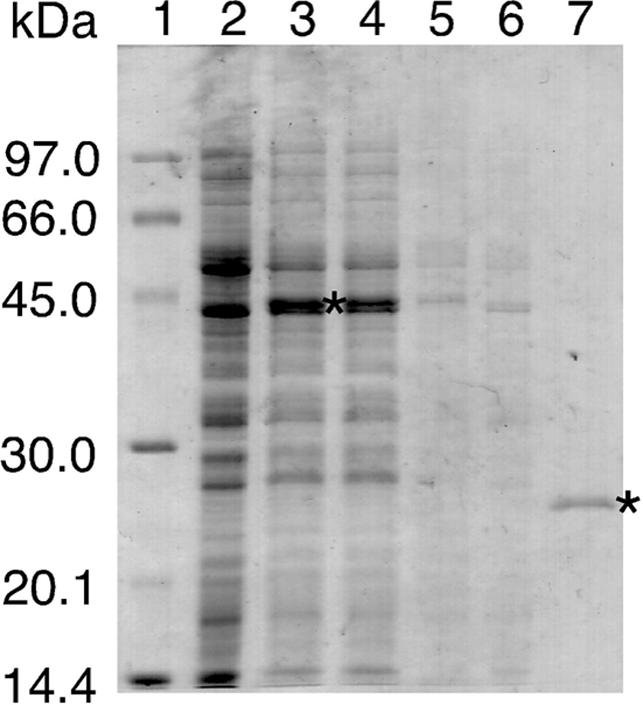
SDS–PAGE analysis of the cell-free synthesis and purification of Endo IV. Endo IV was synthesized as a GST fusion protein by in vitro translation and purified as described in Materials and Methods. Samples at various stages of the purification procedure were subjected to SDS–PAGE on a 12% gel and stained with Coomassie brilliant blue. Lane 1, molecular size standards; lane 2, a translation mixture incubated in the absence of mRNA; lane 3, a translation mixture incubated in the presence of mRNA; lanes 4 and 5, the soluble and insoluble fractions, respectively, of the translation mixture incubated in the presence of mRNA; lanes 6 and 7, the flow-through fractions of a glutathione–Sepharose 4B MicroSpin column for the sample in lane 4 before and after, respectively, treatment of column-bound proteins with PreScission protease. The bands corresponding to the GST-Endo IV fusion protein in lane 3 and the purified Endo IV protein in lane 7 are indicated by asterisks.
Substrate specificity of Endo IV
Optimal pH, optimal concentrations of several divalent cations, and inhibition by several salts for Endo IV activity, which were reported previously in detail (10), were as follows. The enzyme exhibited the activity in the presence of several different divalent cations, Ca2+, Co2+, Mg2+ or Mn2+, and the optimal MgCl2 concentration was 10 mM. The enzyme exhibited a pH optimum between 8.4 and 9.2. The enzymatic activity was sensitive to several salts. The enzymatic activity in the presence of 50 mM NaCl or KCl was 15 or 20% of that in the absence of salts, respectively. Therefore, the Endo IV assay in the present study was performed under the conditions similar to the reported optimal conditions for Endo IV activity.
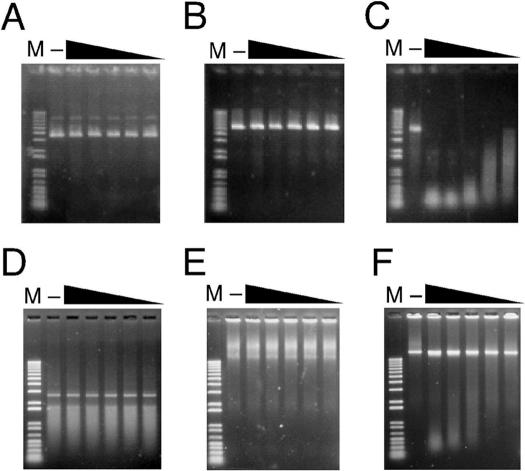
The substrate specificity of Endo IV was investigated previously with an enzyme preparation partially purified from T4 phage-infected E.coli B cells (10,11). Given the possibility that this preparation might have been contaminated with other nucleases, we examined the substrate specificity of the enzyme translated in vitro and purified to homogeneity. With phiX174 circular dsDNA (Figure 2A), phiX174 linear dsDNA (Figure 2B) or the linear ssRNA used for synthesis of the GST-Endo IV fusion protein (Figure 2D) as the substrate, no degradation was detected even in the presence of the enzyme at a concentration of 1 μg/ml. With phiX174 circular ssDNA as the substrate, however, a substantial amount of cleavage products was generated (Figure 2C). In addition, the enzyme mediated a marked extent of cleavage of heat-denatured T4dC genomic dsDNA (Figure 2F), but it had no effect on heat-denatured T4 genomic dsDNA containing gluc-dHMC (Figure 2E). These results indicated that Endo IV catalyzes endonucleolytic cleavage of dC-containing ssDNA.
Figure 2.
Substrate specificity of Endo IV. The activity of Endo IV (1, 0.5, 0.2, 0.1 or 0.05 μg/ml) was assayed with phiX174 RFI circular dsDNA (A), phiX174 RFI linear dsDNA (B), phiX174 virion circular ssDNA (C), linear ssRNA used for the in vitro synthesis of the GST-Endo IV fusion protein (D), heat-denatured T4 genomic dsDNA containing gluc-dHMC (E), or heat-denatured dC-substituted T4 (T4dC) genomic dsDNA (F) as the substrate (5 μg/ml). The reaction products were separated by electrophoresis on a 1.0% agarose gel and stained with SYBR Gold. Lanes M and (–) contain a 1 kb DNA ladder and a reaction mixture incubated in the absence of the enzyme, respectively.
We next analyzed the nucleotide specificity of Endo IV by quantifying the amount of acid-soluble nucleotides liberated by the enzyme from 45 base oligo(dA) [(dA)45], (dT)45, (dG)45 or (dC)45 substrates. All of the oligonucleotide substrates used in the present study were precipitable by trichloroacetic acid under the assay conditions before treatment with Endo IV. Of the four 45 base homo-oligomers tested, only (dC)45 was hydrolyzed by Endo IV (Table 2).
Table 2.
Enzymatic activity of Endo IV with various oligonucleotides
| Oligonucleotide | Relative activity (%) |
|---|---|
| (dA)45 | <0.3a |
| (dT)45 | <0.3a |
| (dG)45 | <0.3a |
| (dC)45 | 100 |
| (dC)25 | 100 |
| 5′-(dA)12dC(dA)12-3′ | 1.8 |
The activity of Endo IV was determined by measurement of the amount of acid-soluble nucleotides released from the substrate (10 μM). The specific activity of the enzyme for the (dC)45 substrate was ∼8.0 U/mg. Relative activity was calculated by dividing the enzymatic activity for each substrate by that for the (dC)45 substrate. Data are means from two independent experiments.
aActivity did not exceed the background level.
Together, our results indicated that Endo IV endonucleolytically and specifically cleaves ssDNA at dC, consistent with the previous results obtained with the partially purified enzyme (10,11), and that the enzyme does not possess ribonuclease activity. In addition, the enzyme processed both phiX174 circular ssDNA (Figure 2C) and heat-denatured T4dC genomic dsDNA (Figure 2F) substrates to oligonucleotides comprising several hundred bases, suggesting that digestion at dC was limited in each substrate and that the sequence surrounding dC or the size of dC-containing oligonucleotides might affect the cleavage efficiency.
Cleavage of a phiX174-derived oligonucleotide
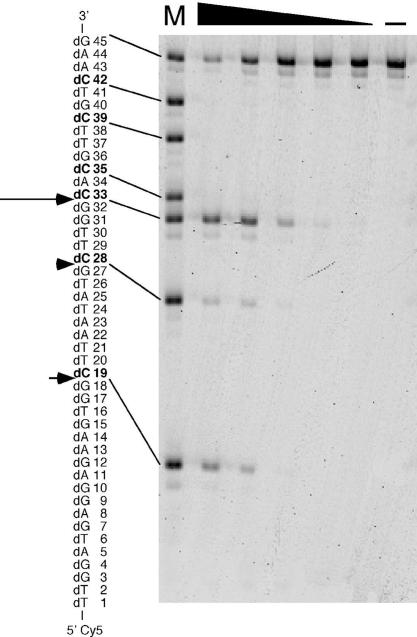
To confirm the dC-specific cleavage of ssDNA by Endo IV, we examined the cleavage of a 45 base oligonucleotide based on the phiX174 ssDNA sequence and labeled at the 5′ end with Cy5. The cleavage products were separated by electrophoresis on a polyacrylamide gel containing 7 M urea. The substrate contains six dC residues at positions 19, 28, 33, 35, 39 and 42. Cleavage at 5′ phosphodiester bonds of dA, dT or dG was not detected (Figure 3). The enzyme cleaved the substrate at three sites, dG18–dC19, dG27–dC28 and dG32–dC33, with an apparent preference for cleavage at the latter site. No cleavage at dA34–dC35, dT38–dC39 or dT41–dC42 was observed. At the maximal Endo IV concentration of 6.6 μg/ml, a proportion of the substrate molecules remained intact at the end of the 30 min incubation. If the enzyme efficiently cleaved the substrate at dA34–dC35, dT38–dC39 or dT41–dC42 under these conditions, the cleavage products would have been detected on the gel. It is therefore unlikely that the enzyme efficiently cleaves the substrate at these sites under these conditions. These results indicated that Endo IV specifically cleaves the 5′ phosphodiester bond of dC in ssDNA but that it does so with markedly different efficiencies depending on the sequence surrounding dC or the size of the dC-containing oligonucleotide.
Figure 3.
Cleavage of a phiX174-based oligonucleotide by Endo IV. A 45 base oligonucleotide based on the sequence of phiX174 ssDNA and labeled at the 5′ end with Cy5 was used as the substrate at a concentration of 10 μM for assay of the activity of Endo IV (6.6, 3.3, 1.3, 0.66 or 0.33 μg/ml). The reaction products were separated by electrophoresis on a 10% polyacrylamide gel containing 7 M urea and visualized with an image analyzer. Lane (–) represents a reaction mixture incubated in the absence of enzyme. Lane M represents a mixture of oligonucleotides labeled at the 5′ end with Cy5 and with sequences identical to those of residues 1–18, 1–27, 1–32, 1–34, 1–38, 1–41 and 1–45 of the substrate. Cleavage sites of the substrate are indicated by arrows with a size proportional to the relative extent of cleavage at the corresponding position. Each dC in the substrate sequence is shown in boldface.
Design of an oligonucleotide with a single cleavage site
Given that oligo(dC) substrates are unsuitable for rigorous kinetic analysis of Endo IV activity because of the multiplicity of cleavage sites, we designed an oligonucleotide substrate with a single cleavage site. The activity of Endo IV with the 25 base oligonucleotide 5′-(dA)12dC(dA)12-3′ as substrate was 1.8% of that apparent with (dC)45, whereas the enzymatic activity with (dC)25 was identical to that for (dC)45 (Table 2). The decrease in the length of the oligo(dC) substrate from 45 to 25 bases thus did not affect cleavage efficiency. We confirmed that the 5′ and 3′ products of cleavage of 5′-(dA)12dC(dA)12-3′, presumably at the 5′ phosphodiester bond of the internal dC, were acid-soluble under the assay conditions (data not shown). The reduced activity of Endo IV with this substrate compared with that apparent for (dC)25 was likely due to the reduced number of potential cleavage sites. This notion was supported by the observation that the amount of enzyme required for cleavage of this substrate did not differ markedly from that required for cleavage at either dG18–dC19 or dG27–dC28 of the phiX174 oligonucleotide substrate (data not shown). We therefore used the basic configuration of the 5′-(dA)12 dC(dA)12-3′ substrate to examine the sequence preference and kinetic parameters of Endo IV.
Sequence preference of Endo IV
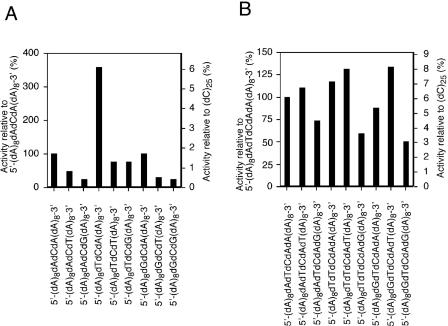
To examine whether combinations of individual nucleotides flanking each side of the dC residue in the substrate affect the cleavage efficiency of Endo IV, we determined enzymatic activity with a series of 19 base 5′-(dA)8dDdCdD(dA)8-3′oligonucleotides, where dD represents dA, dT or dG. The activity of Endo IV with 5′-(dA)8dAdCdA(dA)8-3′ as substrate was 1.8% of that apparent with (dC)25 (Figure 4A) and identical to that apparent with 5′-(dA)12dC(dA)12-3′ (Table 2). The decrease in the length of the latter substrate from 25 to 19 bases thus did not affect cleavage efficiency. Among all examined sequences, only 5′-dTdCdA-3′ was substantially (3.6 times) more susceptible to enzymatic cleavage than was 5′-dAdCdA-3′ (Figure 4A), indicating that Endo IV has a preference for the former trinucleotide sequence.
Figure 4.
Sequence preference of Endo IV. The activity of Endo IV was determined by measurement of the amount of acid-soluble nucleotides released from the substrate (10 μM). A series of 19 base 5′-(dA)8dDdCdD(dA)8-3′ oligonucleotides (A) and a series of 21 base 5′-(dA)8dDdTdCdAdD(dA)8-3′ oligonucleotides (B) were used as substrates, where dD represents dA, dT or dG. The relative activity was calculated by dividing the enzymatic activity for each oligonucleotide by that for 5′-(dA)8dAdCdA(dA)8-3′ or (dC)25 in (A) or by that for 5′-(dA)8dAdTdCdAdA(dA)8-3′ or (dC)25 in (B). Data are means from two independent experiments.
To examine further whether combinations of individual nucleotides flanking the 5′-dTdCdA-3′ sequence affect cleavage efficiency, we determined enzymatic activity with a series of 21 base 5′-(dA)8dDdTdCdAdD(dA)8-3′ oligonucleotides as substrates. Although both 5′-dTdTdCdAdT-3′ and 5′-dGdTdCdAdT-3′ sequences were slightly (1.3 times) more susceptible to enzymatic cleavage than was 5′-dAdTdCdAdA-3′, none of the sequences was substantially more susceptible (Figure 4B). On the other hand, for each set of sequences with a given nucleotide (dD) flanking the 5′ end of the 5′-dTdCdA-3′ sequence, the 5′-dDdTdCdAdG-3′ sequence was consistently less susceptible to cleavage than was the 5′-dDdTdCdAdA-3′ or 5′-dDdTdCdAdT-3′ sequence. These results indicate that, with the exception of the 5′-dDdTdCdAdG-3′ sequence, Endo IV tolerates a variety of combinations of individual nucleotides flanking the 5′-dTdCdA-3′ sequence and thus exhibits a broad preference for the 5′-dDdTdCdAdW-3′ pentanucleotide sequence, where dW represents dA or dT.
To analyze the effects of sequences flanking the monodeoxycytidine of the substrate on Endo IV activity in more detail, we determined the kinetic parameters for the hydrolysis of 5′-(dA)7dAdAdCdAdA(dA)7-3′, 5′-(dA)7dAdTdCdAdA(dA)7-3′ or 5′-(dA)8dGdTdCdAdG(dA)8-3′ oligonucleotides, which were also used as substrates for the experiments shown in Figure 4. We confirmed that the 5′ and 3′ products of cleavage of these oligonucleotides, presumably at the 5′ phosphodiester bond of the internal dC, were acid-soluble under the assay conditions (Supplementary Data). The kcat for the 5′-dAdTdCdAdA-3′ sequence was increased 2.9-fold whereas the Km was largely unchanged compared with the values for the 5′-dAdAdCdAdA-3′ sequence, resulting in a 3.7-fold increase in the kcat/Km ratio (Table 3). Compared with the 5′-dAdTdCdAdA-3′ sequence, the 5′-dGdTdCdAdG-3′ sequence yielded a kcat reduced by ∼50% but a similar Km. These results suggest that the identity of the nucleotides in the 5′-dDdDdCdDdD-3′ sequence affects the rate of hydrolysis by Endo IV without markedly affecting the affinity for the substrate.
Table 3.
Effects of dC-flanking sequences of the substrate on kinetic parameters of Endo IV
| Oligonucleotide | Km (μM) | Relative Km | kcat (min−1) | Relative kcat | kcat/Km (μM−1 min−1) | Relative kcat/Km |
|---|---|---|---|---|---|---|
| 5′-(dA)7dAdAdCdAdA(dA)7-3′ | 23 | 1.0 | 0.34 | 1.0 | 0.015 | 1.0 |
| 5′-(dA)7dAdTdCdAdA(dA)7-3′ | 18 | 0.78 | 1.0 | 2.9 | 0.056 | 3.7 |
| 5′-(dA)8dGdTdCdAdG(dA)8-3′ | 17 | 0.74 | 0.52 | 1.5 | 0.031 | 2.1 |
The activity of Endo IV was determined by measurement of the amount of acid-soluble nucleotides released from the substrate. Kinetic parameters were determined by a least-squares fit of the data in Lineweaver–Burk plots. The relative Km, relative kcat and relative kcat/Km were calculated by dividing the Km, kcat and kcat/Km values for each substrate by those for 5′-(dA)7dAdAdCdAdA(dA)7-3′. Data are means from two independent experiments.
DISCUSSION
Substrate specificity of Endo IV
Previous biochemical analyses of Endo IV have been restricted to partially purified preparations of the enzyme from T4 phage-infected E.coli B cells (10,11,23,24). These previous studies revealed that the activity of the enzyme with ssDNA was greater than that with dsDNA (11). The enzyme did not cleave alkaline-denatured T4 genomic dsDNA containing gluc-dHMC, but it did cleave phage fd ssDNA and alkaline-denatured phage lambda dsDNA (10), both of which contain dC. Cleavage of fd ssDNA by Endo IV yielded oligonucleotides with 5′-phosphate and 3′-hydroxyl termini and an average length of ∼50 bases (11), most of which were not solubilized by trichloroacetic acid (10,11). The cleavage of fd ssDNA into various discrete fragments was also detected by PAGE analysis (23). The cleavage products obtained with fd (10,11,24), M13 (24) or phiX174 (24) ssDNA molecules exclusively contained dC at their 5′ termini.
In the present study, we found that expression of Endo IV in E.coli cells prevented cell growth and we therefore synthesized the enzyme in vitro with the use of a wheat germ cell-free protein synthesis system and purified it to homogeneity. Analysis of the activity of the purified enzyme demonstrated that Endo IV catalyzes endonucleolytic cleavage of ssDNA specifically at the 5′ phosphodiester bond of dC, consistent with the previous observations, and that the efficiency of such cleavage is highly dependent on the identity of the nucleotides surrounding the dC residue. The purified enzyme did not cleave heat-denatured T4 genomic dsDNA containing gluc-dHMC but did cleave heat-denatured dC-substituted T4 (T4dC) genomic dsDNA, showing that Endo IV alone is capable of distinguishing between the absence or presence of gluc-dHMC in ssDNA without a requirement for additional factors.
We also observed that the digestion of phiX174 ssDNA, heat-denatured T4dC genomic dsDNA and a phiX174-based oligonucleotide by Endo IV was limited. Indeed, the cleavage products of phiX174 ssDNA generated by Endo IV were largely acid-insoluble under our assay conditions, whereas subsequent treatment of these products with E.coli exonuclease I rendered them acid-soluble (data not shown), as demonstrated previously for Endo IV-mediated cleavage of fd ssDNA (10,11). This latter observation also supports the conclusion that Endo IV generates oligonucleotides containing 5′-phosphate and 3′-hydroxyl termini (10), given that E.coli exonuclease I activity requires a 3′-hydroxyl terminus of ssDNA (30).
Sequence preference of Endo IV
A decrease in size of oligonucleotide substrates from 45 to 19 bases did not substantially affect the enzymatic activity of Endo IV. However, the enzyme exhibited a marked preference for the 5′-dTdCdA-3′ trinucleotide sequence, consistent with previous analyses of the cleavage products of fd, M13 and phiX174 ssDNA molecules (24). The 3′ terminal mononucleotide and 5′ terminal dinucleotides of these products thus consisted predominantly of dT and 5′-dCdA-3′, respectively. In addition, the 5′-dTdCdA-3′ sequence appears in fd ssDNA with an average frequency of 1 per 75 bases, which may account for the previously identified average length (∼50 bases) of the products generated by Endo IV from fd ssDNA (11). The average frequencies of this trinucleotide sequence in M13 (1 per 76 bases) and phiX174 (1 per 74 bases) ssDNA molecules are also similar to that for fd ssDNA. We further demonstrated that Endo IV tolerated various combinations of individual nucleotides flanking the 5′-dTdCdA-3′ sequence, exhibiting a broad preference for the 5′-dDdTdCdAdW-3′ sequences. These results suggest that the enzyme preferentially recognizes short nucleotide sequences that include 5′-dTdCdA-3′.
On the other hand, although Endo IV cleaved the phiX174 oligonucleotide substrate used in the present study at the three sites, dG18–dC19, dG27–dC28 and dG32–dC33, within the 5′-dGdCdA-3′ or 5′-dGdCdT-3′ sequence, an efficient cleavage at the dT41–dC42 site within the 5′-dTdCdA-3′ sequence was not detected. This indicates that Endo IV prefers the 5′-dGdCdA-3′ or 5′-dGdCdT-3′ sequence to the 5′-dTdCdA-3′ sequence in this substrate. However, in contrast to the 5′-dGdCdA-3′ or 5′-dGdCdT-3′ sequence located at the middle of this substrate, the 5′-dTdCdA-3′ sequence is quite proximal to the 3′ end of this substrate, suggesting that the distance from a terminus to a cleavage site in oligonucleotides may substantially affect cleavage efficiency at the site.
Effect of a proximal dC on Endo IV-mediated cleavage
The dG32–dC33 site within the 5′-dG31–dG32–dC33–dA34–dC35-3′ sequence of the phiX174 oligonucleotide substrate used in the present study was more susceptible to cleavage by Endo IV than was the 5′-(dA)8dGdCdA(dA)8-3′ substrate by a factor of ∼6, even though both cleavage sites are located within a 5′-dGdCdA-3′ sequence; this difference was estimated by comparison of the amounts of enzyme required for substrate cleavage (data not shown). In addition, Endo IV cleaved the 5′-dG31–dG32–dC33–dA34–dC35-3′ sequence efficiently at dG32–dC33 but inefficiently at dA34–dC35, suggesting that the presence of a dC residue proximal to a cleavage site may enhance cleavage efficiency at this site even when such a proximal dC is not cleaved efficiently. The enzyme may thus preferentially cleave the 5′-dTdCdA-3′ sequence within dC-rich regions.
How might such a proximal dC contribute to cleavage susceptibility? Kinetic analysis of Endo IV activity demonstrated that an increase in hydrolysis rate, not an increase in affinity for the substrate, was responsible for the preference of the enzyme for the 5′-dDdTdCdAdW-3′ sequence. We did not identify any sequence that contributed to enzyme-substrate affinity. To avoid a multiplicity of cleavage sites, we did not examine the cleavage of an oligonucleotide with multiple dC residues as a substrate for kinetic analysis of enzymatic activity. Determination of the kinetic parameters of Endo IV for hydrolysis of oligonucleotides containing the 5′-dGdGdCdAdC-3′ sequence of the phiX174 oligonucleotide and with a single cleavage site may provide insight into the effect of a proximal dC on cleavage efficiency.
Lack of stable T4dC DNA synthesis in the denA−, gene 56− background
A combination of detrimental mutations in denB and gene 56 allows synthesis of stable T4dC genomic DNA (6,16,17). T4dC genomic DNA synthesized in such a denB−, gene 56− background is highly resistant to cleavage by Endo II in vivo (6). Such a background reduces the incorporation of gluc-dHMC into T4 genomic DNA by 91–97% as a result of the deficiency in the gene 56 product (6). The presence of such a small amount of gluc-dHMC in T4 genomic DNA thus effectively inhibits Endo II activity, which was thought to preferentially target long nucleotide sequences consisting mostly of dC (6). The detection of only a few cleavage sites in T4dC genomic DNA synthesized in a denB−, gene 42−, gene 46−, gene 56− background (18) also supported this latter notion, given that deficiencies in both gene 42 and gene 56 result in a complete replacement of gluc-dHMC with dC and that deficiencies in denB and gene 46 prevent further degradation of cleavage products generated by Endo II. Indeed, Endo II was subsequently shown to prefer asymmetric dC-rich dsDNA sequences, typically 15 bp sequences that conform to 5′-dGdRdCdCdGdCdN/dTdTdGdGdCdNdGdC-3′, where dR and dN represent dA or dG and dA, dT, dG or dC, respectively, and the slash indicates the position of the preferred complementary-strand nick (21,22).
On the other hand, given that T4dC genomic DNA synthesis is not observed in the denA−, gene 56− background (6,16,17), it is likely that Endo IV effectively attacks T4dC genomic DNA even in the presence of a small amount of gluc-dHMC. Together, these observations suggest that Endo IV recognizes dC-containing sequences that are shorter and more frequent than are those targeted by Endo II. Indeed, we have now shown that Endo IV preferentially recognizes short nucleotide sequences that contain 5′-dTdCdA-3′.
Possible role of Endo IV in inhibition of replication of dC-containing DNA
A deficiency in denB suppresses the lack or arrest of T4dC genomic DNA synthesis in the gene 56− (6,16,17) or gene 42−, gene 46−, gene 56− (6) background, respectively. In addition, T4dC phage mutants with a denB+ background cease synthesizing their genomic DNA within 30 min after infection (6). Moreover, several point mutations in denB that allow stable T4dC genomic DNA synthesis in the gene 56− (4) or gene 42−, gene 56− (H. Ohshima et al., unpublished data) background result in a complete loss of Endo IV activity (confirmed in vitro). Together, these various observations suggest that a lack of Endo IV activity is required for stable T4dC genomic DNA synthesis.
Although the denB product, Endo IV, is dispensable for the normal shutoff of host genomic DNA synthesis after infection with T4 phage (8), our present study has shown that its expression in E.coli cells is lethal, indicating that the enzyme alone is able effectively to inhibit host DNA synthesis in the absence of other T4 phage-encoded proteins. Given that the presence of Endo IV has little effect on the degradation of T4dC (5) or host (3,7,8) genomic DNA, the enzyme appears to specialize in inhibition of the replication of dC-containing DNA.
Replication forks contain regions of ssDNA, most prominently in the lagging strand. Such ssDNA regions may become potential substrates for Endo IV. Given that cleavage of the lagging strand would result in a double-strand break, it is possible that the cleavage of several such adjacent ssDNA regions at dC by Endo IV results in the accumulation of damage to the replication fork and its eventual collapse. If so, the existence of a small number of such ssDNA regions in vivo will result in a limited number of sites cleaved by Endo IV that yield dsDNA fragments, which may account for the previous observations that the enzyme has little effect on the fragmentation (5) but effectively restricts the synthesis (6) of T4dC genomic DNA. In vivo studies of the effects of Endo IV on replication forks, such as by 2D gel analysis of the structure of replication bubbles in E.coli cells, should provide insight into the validity of this hypothesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank N. Arai and H. Mitsusawa for discussion and Zoegene Co. for providing the wheat germ extract for protein synthesis. This work was supported by the 21st Century COE Program of the Ministry of Education, Culture, Sports, Science and Technology of Japan. Funding to pay the Open Access publication charges for this article was provided by Nihon University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wiberg J.S. Mutants of bacteriophage T4 unable to cause breakdown of host DNA. Proc. Natl Acad. Sci. USA. 1966;55:614–621. doi: 10.1073/pnas.55.3.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kutter E.M., Wiberg J.S. Degradation of cytosine-containing bacterial and bacteriophage DNA after infection of Escherichia coli B with bacteriophage T4D wild type and with mutants defective in genes 46, 47 and 56. J. Mol. Biol. 1968;38:395–411. doi: 10.1016/0022-2836(68)90394-x. [DOI] [PubMed] [Google Scholar]
- 3.Hercules K., Munro J.L., Mendelsohn S., Wiberg J.S. Mutants in a nonessential gene of bacteriophage T4 which are defective in the degradation of Escherichia coli deoxyribonucleic acid. J. Virol. 1971;7:95–105. doi: 10.1128/jvi.7.1.95-105.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vetter D., Sadowski P.D. Point mutants in the D2a region of bacteriophage T4 fail to induce T4 endonuclease IV. J. Virol. 1974;14:207–213. doi: 10.1128/jvi.14.2.207-213.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson K., Wiberg J.S. In vivo cleavage of cytosine-containing bacteriophage T4 DNA to genetically distinct, discretely sized fragments. J. Virol. 1983;48:18–30. doi: 10.1128/jvi.48.1.18-30.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlson K., Overvatn A. Bacteriophage T4 endonucleases II and IV, oppositely affected by dCMP hydroxymethylase activity, have different roles in the degradation and in the RNA polymerase-dependent replication of T4 cytosine-containing DNA. Genetics. 1986;114:669–685. doi: 10.1093/genetics/114.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warner H.R., Snustad D.P., Jorgensen S.E., Koerner J.F. Isolation of bacteriophage T4 mutants defective in the ability to degrade host deoxyribonucleic acid. J. Virol. 1970;5:700–708. doi: 10.1128/jvi.5.6.700-708.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Souther A., Bruner R., Elliott J. Degradation of Escherichia coli chromosome after infection by bacteriophage T4: role of bacteriophage gene D2a. J. Virol. 1972;10:979–984. doi: 10.1128/jvi.10.5.979-984.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadowski P.D., Hurwitz J. Enzymatic breakage of deoxyribonucleic acid. Purification and properties of endonuclease II from T4 phage-infected Escherichia coli. J. Biol. Chem. 1969;244:6182–6191. [PubMed] [Google Scholar]
- 10.Sadowski P.D., Hurwitz J. Enzymatic breakage of deoxyribonucleic acid. II. Purification and properties of endonuclease IV from T4 phage-infected Escherichia coli. J. Biol. Chem. 1969;244:6192–6198. [PubMed] [Google Scholar]
- 11.Sadowski P.D., Bakyta I. T4 endonuclease IV. Improved purification procedure and resolution from T4 endonuclease III. J. Biol. Chem. 1972;247:405–412. [PubMed] [Google Scholar]
- 12.Wilson G.G., Tanyashin V.I., Murray N.E. Molecular cloning of fragments of bacteriophage T4 DNA. Mol. Gen. Genet. 1977;156:203–214. doi: 10.1007/BF00283493. [DOI] [PubMed] [Google Scholar]
- 13.Snyder L., Gold L., Kutter E. A gene of bacteriophage T4 whose product prevents true late transcription on cytosine-containing T4 DNA. Proc. Natl Acad. Sci. USA. 1976;73:3098–3102. doi: 10.1073/pnas.73.9.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Velten J., Fukada K., Abelson J. In vitro construction of bacteriophage and plasmid DNA molecules containing DNA fragments from bacteriophage T4. Gene. 1977;1:93–106. doi: 10.1016/0378-1119(76)90009-3. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi H., Saito H., Ikeda Y. Viable T4 bacteriophage containing cytosine substituted DNA (T4dC phage). I. Behavior towards the restriction-modification systems of Escherichia coli and derivation of a new T4 phage strain (T4dC) having the complete T4 genome. J. Gen. Appl. Microbiol. 1978;24:297–306. [Google Scholar]
- 16.Bruner R., Souther A., Suggs S. Stability of cytosine-containing deoxyribonucleic acid after infection by certain T4 rII-D deletion mutants. J. Virol. 1972;10:88–92. doi: 10.1128/jvi.10.1.88-92.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kutter E., Beug A., Sluss R., Jensen L., Bradley D. The production of undegraded cytosine-containing DNA by bacteriophage T4 in the absence of dCTPase and endonucleases II and IV, and its effects on T4-directed protein synthesis. J. Mol. Biol. 1975;99:591–607. doi: 10.1016/s0022-2836(75)80174-4. [DOI] [PubMed] [Google Scholar]
- 18.Krabbe M., Carlson K. Selectivity of in vivo restriction: sequence and structure of endonuclease II-dependent cleavage sites in bacteriophage T4 DNA. J. Biol. Chem. 1991;266:23407–23415. [PubMed] [Google Scholar]
- 19.Carlson K., Krabbe M., Nystrom A.C., Kosturko L.D. DNA determinants of restriction: bacteriophage T4 endonuclease II-dependent cleavage of plasmid DNA in vivo. J. Biol. Chem. 1993;268:8908–8918. [PubMed] [Google Scholar]
- 20.Carlson K., Kosturko L.D., Nystrom A.C. Short-range and long-range context effects on coliphage T4 endonuclease II-dependent restriction. J. Bacteriol. 1996;178:6419–6426. doi: 10.1128/jb.178.22.6419-6426.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlson K., Kosturko L.D., Nystrom A.C. Sequence-specific cleavage by bacteriophage T4 endonuclease II in vitro. Mol. Microbiol. 1999;31:1395–1406. doi: 10.1046/j.1365-2958.1999.01281.x. [DOI] [PubMed] [Google Scholar]
- 22.Carlson K., Lagerback P., Nystrom A.C. Bacteriophage T4 endonuclease II: concerted single-strand nicks yield double-strand cleavage. Mol. Microbiol. 2004;52:1403–1411. doi: 10.1111/j.1365-2958.2004.04062.x. [DOI] [PubMed] [Google Scholar]
- 23.Ling V. Partial digestion of 32P-fd DNA with T4 endonuclease IV. FEBS Lett. 1971;19:50–54. doi: 10.1016/0014-5793(71)80602-6. [DOI] [PubMed] [Google Scholar]
- 24.Bernardi A., Maat J., de Waard A., Bernardi G. Preparation and specificity of endonuclease IV induced by bacteriophage T4. Eur. J. Biochem. 1976;66:175–179. doi: 10.1111/j.1432-1033.1976.tb10437.x. [DOI] [PubMed] [Google Scholar]
- 25.Madin K., Sawasaki T., Ogasawara T., Endo Y. A highly efficient and robust cell-free protein synthesis system prepared from wheat embryos: plants apparently contain a suicide system directed at ribosomes. Proc. Natl Acad. Sci. USA. 2000;97:559–564. doi: 10.1073/pnas.97.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawasaki T., Ogasawara T., Morishita R., Endo Y. A cell-free protein synthesis system for high-throughput proteomics. Proc. Natl Acad. Sci. USA. 2002;99:14652–14657. doi: 10.1073/pnas.232580399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirano N., Sawasaki T., Tozawa Y., Endo Y., Takai K. Tolerance for random recombination of domains in prokaryotic and eukaryotic translation systems: limited interdomain misfolding in a eukaryotic translation system. Proteins. 2006;64:343–354. doi: 10.1002/prot.21008. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi H., Shimizu M., Saito H., Ikeda Y. Studies of viable T4 bacteriophage containing cytosine-substituted DNA (T4dC phage). II. Cleavage of T4dC DNA by endonuclease SalI and BamHI. Mol. Gen. Genet. 1979;168:49–53. doi: 10.1007/BF00267932. [DOI] [PubMed] [Google Scholar]
- 29.Kimura M., Yura T., Nagata T. Isolation and characterization of Escherichia coli dnaA amber mutants. J. Bacteriol. 1980;144:649–655. doi: 10.1128/jb.144.2.649-655.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehman I.R., Nussbaum A.L. The deoxyribonucleases of Escherichia coli. V. On the specificity of exonuclease I (phosphodiesterase) J. Biol. Chem. 1964;239:2628–2636. [PubMed] [Google Scholar]