Abstract
Understanding transcriptional regulation in early developmental stages is fundamental to understanding mammalian development and embryonic stem (ES) cell properties. Expression surveys suggest that the putative SCAN-Zinc finger transcription factor Zfp206 is expressed specifically in ES cells [Zhang,W., Morris,Q.D., Chang,R., Shai,O., Bakowski,M.A., Mitsakakis,N., Mohammad,N., Robinson,M.D., Zirngibl,R., Somogyi,E. et al., (2004) J. Biol., 3, 21; Brandenberger,R., Wei,H., Zhang,S., Lei,S., Murage,J., Fisk,G.J., Li,Y., Xu,C., Fang,R., Guegler,K. et al., (2004) Nat. Biotechnol., 22, 707–716]. Here, we confirm this observation, and we show that ZFP206 expression decreases rapidly upon differentiation of cultured mouse ES cells, and during development of mouse embryos. We find that there are at least six isoforms of the ZFP206 transcript, the longest being predominant. Overexpression and depletion experiments show that Zfp206 promotes formation of undifferentiated ES cell clones, and positively regulates abundance of a very small set of transcripts whose expression is also specific to ES cells and the two- to four-cell stages of preimplantation embryos. This set includes members of the Zscan4, Thoc4, Tcstv1 and eIF-1A gene families, none of which have been functionally characterized in vivo but whose members include apparent transcription factors, RNA-binding proteins and translation factors. Together, these data indicate that Zfp206 is a regulator of ES cell differentiation that controls a set of genes expressed very early in development, most of which themselves appear to be regulators.
INTRODUCTION
Preimplantation development comprises the period from fertilization to implantation, including the zygote, 2-cell, morula and blastocyst stages. Cells from embryos before the 8-cell stage are totipotent, giving rise to all cell types (1). Preimplantation development is characterized by three major developmental transitions that occur after fertilization: zygotic genome activation (ZGA) (also known as maternal to zygotic transition), compaction during the 8-cell stage, and differentiation of the morula into the blastocyst (2). The transition from the totipotent morula to the blastocyst marks the first cell differentiation event, specifying the pluripotent cells of the inner cell mass (ICM) that will give rise to the embryo proper, and the trophectoderm, which will give rise to extra-embryonic tissue including the placenta (3). Typically, embryonic stem (ES) cells are derived from the ICM and are capable of differentiating into all fetal and adult cell lineages (4). ES cells can also be derived from 8-cell and morula stages (5,6). The developmental potentiality of ES cells provides a powerful system to study early embryonic development, to generate experimental models with specific gene alternations, and potentially to obtain cells for transplantation therapies.
Since maintenance of ES cell self-renewal and pluripotency involves transcriptional programming (7,8), and each of these developmental transitions is accompanied by major changes in the pattern of gene expression (9,10), dissecting the full regulatory circuitry of ES cells and preimplantation embryos will be a key step in understanding cellular pluripotency and early embryonic development. Transcription factors that are important for cellular pluripotency and preimplantation development, such as Oct4 (Pou5f1), Nanog and Sox2, are predominantly expressed in preimplantation embryos and the ICM (11), and appear to regulate large groups of genes, many of which are developmentally important homeodomain proteins (12). However, these transcription factors are unlikely to account for all observed transcriptional regulation in early development. Moreover, post-transcriptional regulatory pathways are also likely to be mediators of fertilization and early development (13). For example, consistent with the increase in both transcription and translation in ZGA, large groups of genes, whose functions are enriched in post-transcriptional regulatory activities, such as ‘RNA binding’ and ‘translation initiation factor’, are transiently induced in ZGA (9). Among these genes, the translation initiation factor eIF-1A (also known as eIF-4C) is a well-established example (9,10,14). In vitro, eIF-1A increases translational efficiency by binding 40S ribosomes and recruiting mRNA (15).
Here, we have re-examined our previous microarray-based survey of expression of known and predicted mouse genes in diverse tissues and cell types (16), and identified ZFP206, encoding an apparent SCAN-zinc finger transcription factor, as being expressed almost exclusively in ES cells in these data. Upon further investigation and experimentation, we have found that ZFP206 strongly regulates levels of a handful of transcripts whose expression is also specific to ES cells and other stages of very early development, and which themselves encode apparent regulators. These include genes encoding eIF-1A and Tcstv1, which have previously been reported to be expressed in preimplantation embryos (10,14), as well as a Zscan4 (encoding another apparent SCAN-zinc finger transcription factor) and an RNA-binding protein similar to Thoc4. Consistent with a role in very early development and/or stem cell function, we found that ZFP206 expression levels also impact the differentiation state of cultured ES cells.
MATERIALS AND METHODS
ES cell culture and colony forming assays
ES cells were cultured as described (17). For colony forming assays, single-cell suspensions were prepared using trypsin–EDTA solutions, gently suspended, and seeded at 200 cells/well in 12-well plates. After 6 days, culture plates were stained for alkaline phosphatase (Chemicon) (18) and individual colonies scored for being uniformly undifferentiated.
Localization by immunodetection
We examined the subcellular localization of Zfp206 protein in cultured ES cells by tagging the cDNA at the 3′ end with three copies of the FLAG epitope and inserting it into the ES expression plasmid pPyCAGIP (19), which has a CAG [hybrid cytomegalovirus (CMV) enhancer/chicken-actin] promoter and a puromycin resistance marker. Transiently transfected R1 cells on glass coverslips were fixed with 4% paraformaldehyde, permeabilized with 0.25% Triton X-100, blocked with 10% BSA, and incubated with mouse monoclonal ANTI-FLAG® M2 (Sigma) for 2 h at 37°C. After washing, a fluorescein isothiocyanate (FITC) conjugated secondary anti-mouse antibody (Sigma) was used to detect the primary antibody. Samples were mounted in antifade solution and observed using a fluorescence microscopy. For western blotting, ZFP206-FLAG transfected R1 cells were washed with ice-cold phosphate-buffered saline (PBS) and harvested in 100 μl of RIPA lysis buffer. Lysates were resolved on 10–12% SDS–polyacrylamide gels. Protein was transferred to a nitrocellulose membrane (Bio-Rad) using an electroblotting procedure. The protein blot was blocked with 5% non-fat dry milk in TBS [50 mM Tris (pH 7.6) and 150 mM NaCl] overnight at 4°C, or in TBS-T (TBS and 0.1% Tween-20) for 1 h at room temperature. The blot was incubated with mouse monoclonal ANTI-FLAG® M2 (Sigma) 1 h at room temperature, thoroughly washed and incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies, and detected by enhanced chemiluminescence according to the manufacturer's instructions.
RT–PCR
Total RNA was isolated with Trizol (Invitrogen) and 2 μg of total RNA were reverse transcribed using Superscript™ II (Invitrogen). 1/20th of the reverse transcription product was amplified using SYBR Green PCR Master Mix (Applied Biosystems) and analyzed on 7900HT Sequence Detection System (Applied Biosystems). Primer sequences are posted in Supplementary Data at http://hugheslab.med.utoronto.ca/ZFP206 together with the sequences of RT–PCR products described in the Results.
Northern blotting
A total of 10 μg of total RNA was separated on 1.5% agarose–glyoxal gels, transferred to a Hybond N+ membrane (Amersham) by capillary transfer, ultraviolet (UV) cross-linked to the membrane, and probed with a 782 bp ZFP206 fragment that is common to all isoforms (sequence posted at http://hugheslab.med.utoronto.ca/ZFP206).
Whole-mount in situ hybridization
Whole-mount in situ hybridization was performed as described previously (20). Sense and antisense probes were in vitro transcribed from ZFP206 isoform 2 cDNA.
Overexpression and knockdown of ZFP206
We constructed a ZFP206 overexpression plasmid by cloning our sequenced ZFP206 isoform 1 cDNA into the plasmid pPyCAGIP (19). The shRNA expression plasmid was made by Kunath et al. (21) from pcDNA3.1 (Invitrogen) with neomycin selection marker. An shRNA targeting ZFP206 isoforms 1 and 2 (sequence TGAGTTACCTCCACCTCAG) was introduced into the Asp718 and XbaI sites and under the control of H1 RNA pol III promoter. Stable transfected overexpression clones (ZFP206 OX-1, OX-2 and OX-3) (derived from E14TG2a ES cells) and ZFP206 shRNA knockdown clone ZFP206KD-bF9 (derived from R1 ES cells) were selected by standard techniques.
Microarray analysis
Microarray analysis was as described (16) with two-color comparisons normalized using Lowess smoothing (22). Microarray data are posted in Supplementary Data at http://hugheslab.med.utoronto.ca/ZFP206. Microarray data have been submitted to NCBI GEO upon acceptance for publication.
Collection and analysis of preimplantation embryos
Mouse ICR embryos were isolated from superovulated outbred ICR (Harlan) mice as described previously (10). Unfertilized oocytes were collected 46–48 h after pregnant mare serum gonadotropin (PMSG) injection. The embryos were collected during the indicated time after human chorionic gonadotropin (hCG) treatment: 1-cell (21–22 h), 2-cell and 4-cell (50–51 h), 8-cell and morula (73–74 h) and blastocyst (96–97 h). The mixed 2-cell and 4-cell stage embryos, 8-cell and morula stage embryos were sorted according to their morphology under the microscope. A total of 20–50 embryos were collected at each stage and pooled. RNA was isolated from the different stage embryos using Absolutely RNA® Nanoprep Kit (Stratagene). Two rounds of linear amplification were performed for RNA isolated from each stage using the MessageAmp™ II aRNA Amplification Kit (Ambion).
RESULTS
ZFP206 expression and localization in mouse ES cells
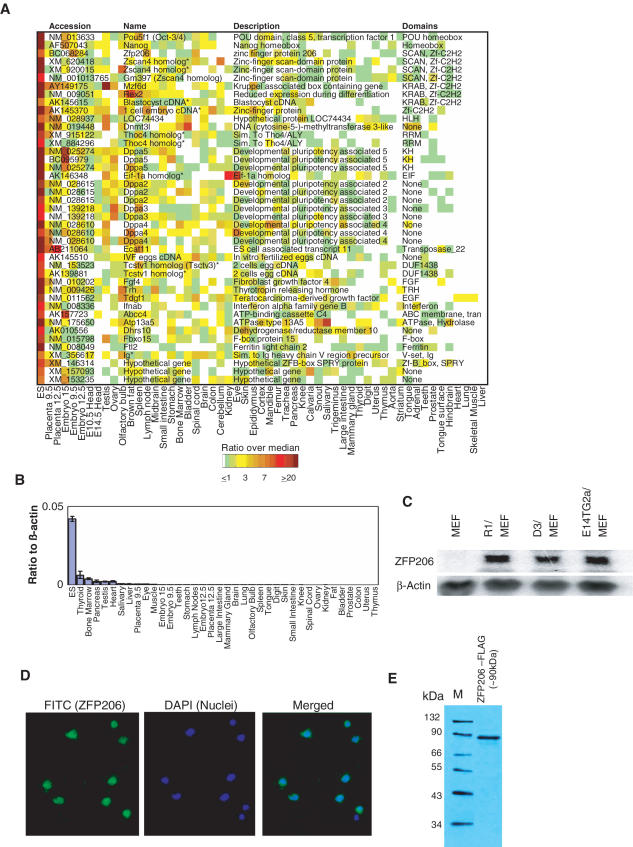
We previously described a microarray analysis of the expression of over 40 000 known and predicted genes across 55 mouse tissues and cell types, including cultured R1 ES cells (16). To identify new genes potentially involved in stem cell functions and/or very early development, we scored all of the genes on the array for highly specific expression in ES cells (Figure 1A). The 43 genes identified as most specific included many known ES cell related genes, including those encoding the transcription factors Oct4 and Nanog (7), as well as genes encoding a variety of putative novel regulatory proteins (Figure 1A). Among these was ZFP206 (NM_001033425). On the basis of its predicted domain structure, Zfp206 is likely to be a sequence-specific regulator of transcription, as it belongs to the SCAN-ZFP protein family (23): the predicted ORF of ZFP206 encodes a protein of 782 amino acids (88.4 kDa) containing a SCAN domain followed by fourteen C2H2 zinc fingers. The C2H2 zinc finger is one of the most versatile DNA-recognition elements (24), and it is currently impossible to predict the binding sequence in all but a few special cases [e.g. those with three C2H2 zinc fingers (25)]. The SCAN domain (26) (an acronym for the proteins in which it was first found—SRE-ZBP, CTfin-51, AW-1, Number 18 cDNA—and also known as LER, for Leucine Rich Repeat) is a highly conserved 84-residue motif that appears to be specific to vertebrates (27) and may play a role in the assembly and function of the SCAN-zinc finger transcription factors by mediating homo- and hetero-oligomerization (23,28). In support of a function for Zfp206 in transcription, we found that epitope-tagged full-length Zfp206 localized to the nucleus of cultured R1 ES cells (Figure 1D and E).
Figure 1.
Expression of ZFP206 in ES cells. (A) Forty-three known and predicted genes expressed with high specificity in ES cells were identified by sorting microarray profiles (16) to identify genes whose expression in ES cells is higher than in 54 other tissues examined, then clustering to remove those that are also appreciably expressed in embryos or other tissue sets. In some cases, multiple loci encode very similar proteins, which are represented here by a single cDNA Accession number. ‘*’ indicates genes that are not found in the MGI database and therefore have no standard name. (B) Real time PCR on 38 mouse tissues and ES cells confirming that ZFP206 is primarily expressed in R1 ES cells. (C) Northern blot analysis of ZFP206 expression in three pluripotent ES cell lines (R1, D3 and E14TG2a) co-cultured with MEF. (D) Subcellular localization of 3XFLAG tagged Zfp206 cDNA (isoform 1) in R1 ES cells. (E) Western blotting of 3XFLAG tagged Zfp206 cDNA (isoform 1) in R1 ES cells. The expected MW of the tagged protein is ∼90 kDa.
We verified expression of native ZFP206 in ES cells by RT–PCR (Figure 1B) and northern blotting (Figure 1C). ZFP206 mRNA was expressed in three different pluripotent ES cell/murine embryonic fibroblast (MEF) co-cultures, but not in MEF feeder cells alone (Figure 1C). ZFP206 was also recently reported to be expressed predominantly in the ICM of blastocysts, which is the source of ES cells (11), and large-scale cDNA analysis to characterize genes specific to human ES cells identified ZFP206 as a human ES cell-specific gene (29). Hence, although to our knowledge ZFP206 is functionally and biochemically uncharacterized in any organism, its expression pattern appears to be conserved between human and mouse, suggesting that its function is also conserved.
ZFP206 transcript levels are dramatically reduced in response to ES cell differentiation
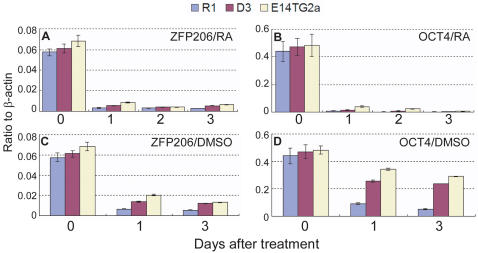
To ask whether ZFP206 expression responds to cues that trigger differentiation, we examined how its expression levels react when cultured ES cells are induced to differentiate by the addition of retinoic acid (RA) and dimethyl sulfoxide (DMSO) (Figure 2A and C). Relative to beta-actin control, ZFP206 expression levels are dramatically reduced, indicating that ZFP206 expression is specific for the pluripotent state of ES cells. The degree of change observed for ZFP206 was comparable to that for Oct4 (Figure 2B and D). Similar results were observed previously in human ES cells (29).
Figure 2.
ZFP206 expression decreases dramatically upon differentiation of ES cells. (A–D) ZFP206 and OCT4 expression levels measured using real time PCR in three different pluripotent ES cell lines (R1, D3 and E14TG2a) treated with 0.5 μM RA (All-trans RA) or 0.5% DMSO for up to three days, as indicated.
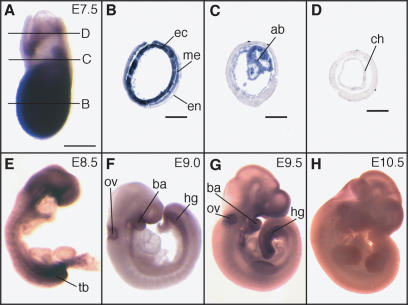
We also investigated the tissue distribution of ZFP206 mRNA during post-implantation mouse development using whole-mount in situ hybridization (Figure 3). At E7.5 ZFP206 is expressed throughout the embryonic ectoderm, mesoderm (Figure 3A and B), and allantois (Figure 3A and C). At E8.5 expression was reduced but widespread (Figure 3E). At E9.0 and E9.5, ZFP206 expression was further restricted to the otic vesicle, branchial arches and hindgut (Figure 3F and G). We detected no ZFP206 transcripts in 10.5 dpc embryos (Figure 3H). Therefore expression of ZFP206 in the embryo is restricted largely to the early stages of post-implantation development, in tissues that are presumably not highly differentiated, and its expression is quickly down-regulated at later stages of development. This is consistent with our previous microarray study and with analyses of ZFP206 expression in cultured ES cells upon differentiation.
Figure 3.
Whole-mount in situ hybridization analysis of ZFP206 expression during post-implantation mouse development. (A) At E7.5 ZFP206 is expressed throughout the embryo proper, the allantoic bud and weakly in the chorion. Anterior is to the left. (B–D) Transverse sections of the embryo as shown in (A). (B) In the embryo, proper ZFP206 is expressed strongly in the ectoderm (ec), moderately in the mesoderm (me) and there is weak to no expression in the endoderm (en). (C) ZFP206 is expressed in the allantoic bud (ab). (D) There is weak to no expression of ZFP206 in the extra-embryonic ectoderm, however, some expression was detected in the chorion (ch). (E–H) Whole-mount in situ hybridization of embryos at the labeled stages. (E) Widespread expression of ZFP206 throughout out the embryo at E8.5 with strong expression in the tail bud (tb) region. (F–G) At E9.0 and E9.5 expression is reduced but with persistent expression in the otic vesicle (ov), branchial arch (ba) and hindgut (hg). (H) Weak to no expression was detected at E10.5. Scale bars: (A) 200 μm; (B–D) 100 μm. Embryonic age was determined by the number of days after the formation of the copulation plug. Whole-mount in situ hybridizations were performed following a standard procedure with Digoxygenin-labeled ZFP206 antisense RNA probes.
ZFP206 has at least six isoforms
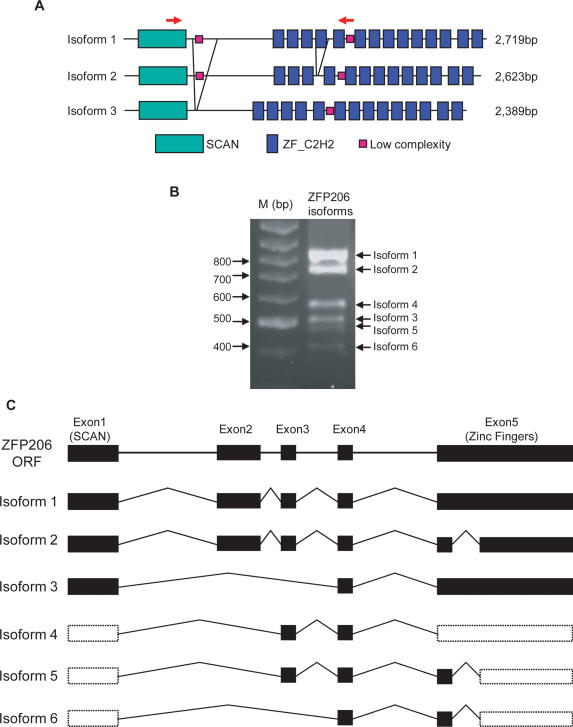
The ZFP206 gene has only recently been annotated and the GenBank sequence is based on two clones and gene prediction models. Our northern analysis (Figure 1C) appeared to show more than one band, suggesting that ZFP206 may have multiple isoforms. Based on the predicted ZFP206 sequence, we cloned and sequenced the ZFP206 open reading frame (ORF) by RT–PCR, and the 5′ and 3′ ends by RACE. Among our clones, we obtained three different isoforms generated by alternative splicing (Figure 4A) (GenBank entries DQ323929, DQ323929 and DQ323931). The full mouse ZFP206 transcript (isoform 1), whose coding sequence and exon structure is virtually identical to that of both the predicted gene and the FANTOM3 clone, is 2719 bases long, and encodes all 14 C2H2 zinc fingers and the SCAN domain (Figure 4A). ZFP206 isoform 2 is missing one of the zinc fingers, and isoform 3 is missing a low-complexity region between the SCAN domain and the zinc fingers.
Figure 4.
ZFP206 isoforms. (A) Domain organization of ZFP206 isoforms 1, 2 and 3. These three clones were sequenced in their entirety. Sequences have been deposited in GenBank (Accession nos DQ323929, DQ323930 and DQ323931). (B) RT–PCR products of the variable region of ZFP206, using primers as indicated in (A) and RNA from R1 ES cells. (C) Schematic diagram of the exon structure of ZFP206, and exon structures of isoforms determined by sequencing. The exons outside the amplified regions in isoforms 4, 5 and 6 (which were not examined) are in dashed lines.
In order to estimate the relative abundance of each isoform, we used semi-quantitative RT–PCR with a pair of primers that flank the entire alternative splicing regions (red arrows in Figure 4A), to amplify the three isoforms. Surprisingly, we obtained six isoforms instead of three (Figure 4B). All six bands in Figure 4B were sequenced and found to be ZFP206 transcripts generated by alternative splicing. The exon structures of each transcript are shown in Figure 4C. The top two bands and the fourth band from the top in Figure 4B are identical to the three we identified above. Since the efficiency of RT–PCR would normally be biased towards preferential amplification of shorter products, yet the longer products appear to be most predominant RT–PCR products, ZFP206 isoforms 1 and isoform 2 are apparently the most abundant.
Zfp206 expression levels influence ES cell differentiation
To ask whether Zfp206 influences properties of ES cells, we examined the differentiation state of single clones isolated from three independent ZFP206 (isoform 1) overexpression cell lines, as well as a ZFP206 shRNA knockdown cell line, using an AP (Alkaline Phosphatase) assay (undifferentiated ES cells produce high level of alkaline phosphatase) (18) (Figure 5A and B). The ZFP206 shRNA knockdown would affect isoforms 1 and 2, the two major isoforms. In comparison to empty vector and untransfected cells, we estimated by real time PCR (using primers that would detect all six ZFP206 isoforms described above) that the overexpression cultures used in this experiment had a ∼25-fold increase in ZFP206 transcript levels, whereas only 25–30% of the ZFP206 transcript remained in the ZFP206 shRNA knockdown (data not shown and see below).
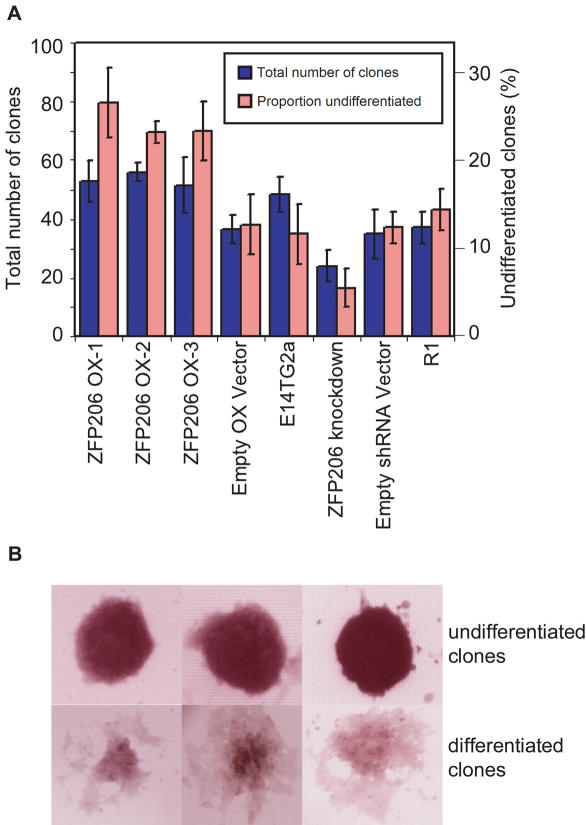
Figure 5.
Effect of ZFP206 expression on ES cell differentiation. (A) Parental ES cells (E14TG2a for overexpression clones and R1 for knockdown clones), three ZFP206 overexpression, one knockdown, and corresponding empty vector control cell lines were analyzed following plating at clonal density (see Materials and Methods) using normal ES cell medium with LIF. After 6 days of culture, the percentage of pure alkaline phosphatase positive colonies were calculated. Data are the mean and SDs of three repeats. (B) Examples of uniformly undifferentiated and differentiated clones.
While only ∼10% of the colonies produced by the overexpression empty vector control and its parental ES cells (E14TG2a) consisted of uniformly undifferentiated ES cells, ZFP206 overexpression transfectants contained ∼24% pure undifferentiated stem cell colonies. In contrast, the pure stem cell colonies were reduced to ∼5% in ZFP206 knockdown ES cells, in comparison to ∼12% of colonies produced by the knockdown empty vector control and its parental ES cells (R1), both comparable to the E14TG2a line. Zfp206 also positively influenced the overall number of clones obtained (Figure 5A), and to a lesser extent the number of differentiated clones, suggesting that it may affect survival or proliferation of individual cells in addition to influencing differentiation state. However, this effect is not as pronounced as the effect on proportion of undifferentiated versus differentiated clones (Figure 5A).
ZFP206 controls a small number of apparent regulators expressed in ES cells
We reasoned that if ZFP206 is a transcriptional regulator in ES cells, then perturbation of ZFP206 expression might affect expression of other genes in ES cells, which would represent likely downstream targets of ZFP206. We therefore used the same Agilent microarray design employed in our initial study (16) to analyze two overexpression clones (OX-1 and OX-2 from the AP assay above, and an independent knockdown clone using the same vector), under standard high-density culture conditions, in which these clones do not display the striking differentiation effects as above, presumably due to differences in local environment (data not shown). In each case, we used a two-color protocol, comparing to a corresponding empty vector control line. In these experiments, we confirmed by RT–PCR that ZFP206 overexpression cultures had a ∼25-fold increase in ZFP206 transcripts, whereas the shRNA line had a ∼5-fold reduction (i.e. ∼20% of total ZFP206 transcript remained). Differences in absolute levels of overexpression or knockdown from the cultures used for the AP assay may be due to slight variation in either experiments or measurements over time. To maximize the fidelity of measurements, unamplified cDNA from 2 μg of poly(A)-purified mRNA was hybridized to each channel of each array.
Figure 6A shows a scatter plot comparing the log2(ratio) values obtained for each gene in the overexpression experiments [with log2(ratios) averaged] to the log2(ratio) values obtained in the knockdown experiment. The blue points in the center of the plot resemble random scatter. However, there are a handful of clear outliers in the lower-right quadrant; these are microarray probes reporting transcript abundance that is increased by ZFP206 overexpression and decreased by ZFP206 knockdown. The separate data points for these ten array probes are shown in Figure 6B.
Figure 6.
ZFP206 regulates ES cell-specific genes. (A) Scatter plot of log2(expression ratios) comparing the effects of ZFP206 overexpression and knockdown. Array spots representing 19 220 XM sequences are shown, restricting to those expressed above negative controls with a P-value of 0.1. (B) Clustergram of all measurements for these ten microarray probes. (C) Real time PCR analysis of the indicated genes (or gene families) in the same samples shown in (A), compared to the same empty vector controls. Primer sequences are listed in the Supplementary Data.
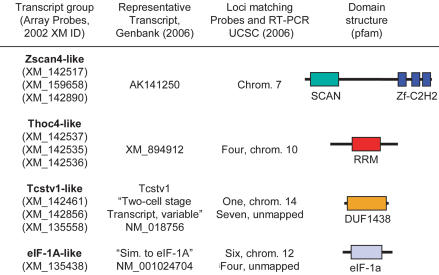
We mapped these ten array probes (which were based on mouse ‘XM’ genes and gene predictions) to current cDNA and genome annotations. Most of the probes were similar to several related sequences in the genome, i.e. each of the probes could potentially hybridize one or more members of a family of genes with related sequences. Likewise, in all but one case, each these gene families was represented by more than one probe among the ten. In this way, the ten probes correspond to what appear to be only four families of genes with very similar sequences that are distributed in multiple locations in the current build of the mouse genome (Figure 7). The group we refer to as ‘Zscan4-like’ corresponds to variations on a putative SCAN-C2H2 zinc finger protein sequence similar to the functionally uncharacterized human protein Zscan4. The ‘Thoc4-like’ group consists of a set of predicted transcripts that carry an RNA recognition motif (RRM) domain and bear sequence similarity to Thoc4 (also known as ALY), which has been identified as being involved in both transcriptional regulation (by increasing transcription factor binding) and RNA export from the nucleus (by interacting with the splicing exporting factor UAP56) (30–33). The ‘Tcstv1-like’ group has no informative domains; the DUF1438 motif is present only in this group of proteins. Tcstv is an acronym for 2-cell stage variable transcript; i.e. expression has been previously observed during the 2-cell/4-cell stage of the preimplantation mouse embryo (10). Likewise, the ‘eIF-1A-like’ group bears strong sequence similarity to translation factor eIF-1A, which has been previously observed to be transiently induced during the 2-cell stage of the preimplantation mouse embryo (14). Strikingly, expression of members of these families detected by these same microarray probes in our initial study (16) appears to be restricted to ES cells (Figure 1A), suggesting that they are bona fide physiological targets of ZFP206. Differential expression of members of these gene families in response to ZFP206 overexpression and depletion was confirmed by real time PCR (Figure 6C).
Figure 7.
Four putative gene families with at least one member positively regulated by ZFP206. The representative transcript was selected by searching GenBank (NR) for a known or predicted full-length cDNA using BLASTN. The mouse loci indicated (March 2005 assembly) were identified manually using the UCSC Genome Browser (35) as being nearly identical (95% or greater over their entire length) to the array probe sequences, or completely identical to the ∼100 base RT–PCR products, and having nearly complete known or predicted exon structures at the same locus that are similar to those of the representative transcript. Pfam domains were identified in the representative transcript using default settings.
Since our microarray probes would be unable to distinguish between genes in these apparent families, we sequenced the ∼100 base RT–PCR products (from primers used in Figure 6) from both normal ES cells and ZFP206 overexpressing ES cells, in an effort to distinguish the isoforms (sequences are posted in Supplementary Data). The Zscan4-like RT–PCR products with or without ZFP206 overexpression corresponded entirely to a single locus on mouse chromosome 7. A 1 kb RT–PCR product obtained using primers corresponding to predicted genes and partial cDNAs surrounding and including this locus amplified fragments of transcripts matching almost perfectly to AK141250, a partial cDNA derived from ES cells, and less perfectly to predicted Zscan4-like transcripts from the same region, all of which encode a protein with a SCAN domain and three C2H2 zinc fingers. However, differences were entirely nucleotide mismatches and may be due to errors resulting from PCR, sequencing, or polymorphisms between the sequenced strain background (C57/BL6) and that of the ES cells we analyzed (129/Ola).
The ∼100 bp RT–PCR products from the Thoc4-like proteins matched perfectly to four different locations on the bacterial artificial chromosome AC164629.14, each spaced almost exactly 9.5 kb apart, and (presumably as a consequence) to four such repeated loci on mouse chromosome 10. We have thus far been unable to distinguish between the predicted transcripts that would originate from these four loci even with expanded RT–PCR products; however, among the three predicted transcript variants from the four loci, the one expressed in ES cells appears to correspond to the XM_894912 variant.
Different ∼100 base RT–PCR products for the Tcstv1-like and the eIF-1A-like transcripts contained sequence differences indicating that they originated from several different loci in the genome, i.e. for each gene multiple isoforms originating from different copies in the genome are expressed simultaneously, as is already documented in GenBank entries for Tcstv1. We did not further pursue quantification of relative levels of isoforms for these two gene families.
Expression of ZFP206 and its putative targets in preimplantation embryos
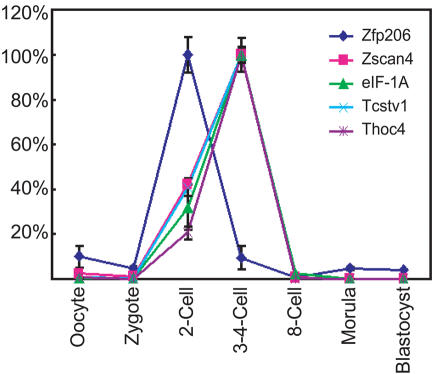
Two of these target genes (Tcstv1 and eIF-1A) have been reported to be expressed in mouse embryos at the 2-cell/4-cell stage (10,14) supporting a role in preimplantation development. We reasoned that if Zfp206 is responsible for induction of these genes, then it too may be expressed in early embryonic development, as may its other putative targets. We therefore examined the expression of Zfp206, Zscan4, Thoc4, Tcstv1 and eIF-1A in a time-course of early development, using RT–PCR on preimplantation embryos ex vivo (Figure 8). We observed a prominent spike in the expression of Zfp206 at the 2-cell stage, with its putative targets following rapidly, peaking at the 3–4-cell stage. This is consistent with regulation of these genes by Zfp206 in preimplantation development in vivo.
Figure 8.
Expression of ZFP206 and its putative targets in preimplantation embryos. The expression levels of Zfp206, Zscan4, Thoc4, Tcstv1 and eIF-1A were measured using real time PCR in a time-course of preimplantation development as indicated. Data were normalized within each time-point to β-actin then the values for each gene were scaled to percentage of maximum across the time-course as shown. Error bars represent the standard deviation over three assays.
DISCUSSION
Here, we have shown that the SCAN and Zinc finger domain containing gene ZFP206 has at least six isoforms, with the two longest being predominant, and is primarily expressed in undifferentiated ES cells and mouse embryos at early developmental stages. Overexpression of ZFP206 induces transcripts that encode proteins of the Zscan4, Thoc4, Tcstv1 and eIF-1A families, and knockdown of ZFP206 results in a decrease of exactly these same transcripts. Expression of these transcripts closely follows that of ZFP206 in preimplantation embryos. In addition, the proportion of undifferentiated clones obtained from plating ES cells at low density increases or decreases along with ZFP206 expression level, indicating that ZFP206 activity prevents differentiation of ES cells.
We propose, on the basis of these results, that ZFP206 functions as a positive regulator of pluripotency in ES cells and preimplantation embryos, and that the mechanism may involve a new regulatory cascade involving transcriptional and post-transcriptional mechanisms. Further studies will be required to fully elucidate the mechanism(s) by which Zfp206 regulates transcript levels of its putative targets and by which it influences ES cell differentiation, and to understand its relationship to other factors that function in ES cell pluripotency and early development. While a handful of key factors in ES cells have been identified (7), the molecular basis of the pluripotentiality, self-renewal and differentiation of ES cells is still poorly understood. Transcriptional regulation in extremely early embryogenesis is also largely uncharted. ZFP206 and other new factors in Figure 1 may play roles in these processes, or in previously unappreciated functions or activities of ES cells and early embryos.
On the basis of conservation scores in genomic alignments (34,35), the exon sequences of all four apparent Zfp206 target genes (including multiple members of the eIF-1A, Thoc4 and Tsctv1 families) appear to be under strong purifying selection in comparison to flanking introns, indicating that they are not pseudogenes. Some of the Thoc4-like and eIF-1A-like family members are conserved in at least some types of fish in genomic alignments. In contrast, Zscan4, like most other SCAN-ZNF domain transcription factors, are conserved along their entire coding length only in mammals, and the Tcstv1-like genes appear to be conserved only in rat, suggesting that they are rodent-specific. The full exon structure of ZFP206 itself is conserved only as far as opossum, suggesting that its role in regulation of Thoc4-like and eIF-1A-like family members arose subsequent to the genes themselves and may be specific to mammals.
To our knowledge, none of these putative Zfp206 targets is functionally characterized in vivo. However, general biochemical functions can be inferred on the basis of sequence features for all but the Tcstv1-like group. It is intriguing that three of these four families—Zscan4-like, Thoc4-like and Eif1a-like—encode potential transcription factors, RNA-binding proteins and translational regulators, respectively, on the basis of sequence features (Figure 7), thus representing three major levels of gene regulation. The observation that they are all transiently induced during ZGA raises the possibility that Zfp206 and its putative target genes might play a role in ZGA, which is marked by an increase in both transcription and translation. It is noteworthy that ZFP206 activity can evidently be controlled both positively and negatively by simply altering the level of its transcript. Overexpression and knockdown are now relatively easy to accomplish, and with a battery of such experiments applied uniformly to both known and potential regulators, it may be possible to begin deciphering the full regulatory structures in mammalian embryogenesis, as has been described in sea urchin (36). These data would form an invaluable complement to chromatin immunoprecipitation studies (37), and need not be restricted to analysis of transcription factors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
The authors thank Quaid Morris and Lourdes Pena Castillo for assistance with microarray data normalization, Martha Bulyk, Mike Berger, Anthony Philippakis, Gwenael Badis-Breard, Mariana Kekis, and Shaheynoor Talukder for helpful discussions, and Marina Gertsenstein for technical assistance with the early embryo time-course. This work was supported by grants from CIHR to T.R.H., W.L.S. and J.R. and a fellowship from CIHR to O.J.T. Funding to pay the Open Access publication charges for this article was provided by CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Carlson B. Human Embryology & Developmental Biology. Mosby: Elsevier; 1999. [Google Scholar]
- 2.Edwards R.G. Aspects of the molecular regulation of early mammalian development. Reprod. Biomed. Online. 2003;6:97–113. doi: 10.1016/s1472-6483(10)62061-5. [DOI] [PubMed] [Google Scholar]
- 3.Rossant J. Stem cells from the Mammalian blastocyst. Stem Cells. 2001;19:477–482. doi: 10.1634/stemcells.19-6-477. [DOI] [PubMed] [Google Scholar]
- 4.Bradley A., Evans M., Kaufman M.H., Robertson E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature. 1984;309:255–256. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- 5.Delhaise F., Bralion V., Schuurbiers N., Dessy F. Establishment of an embryonic stem cell line from 8-cell stage mouse embryos. Eur. J. Morphol. 1996;34:237–243. doi: 10.1076/ejom.34.4.237.13046. [DOI] [PubMed] [Google Scholar]
- 6.Tesar P.J. Derivation of germ-line-competent embryonic stem cell lines from preblastocyst mouse embryos. Proc. Natl Acad. Sci. USA. 2005;102:8239–8244. doi: 10.1073/pnas.0503231102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers I. The molecular basis of pluripotency in mouse embryonic stem cells. Cloning Stem Cells. 2004;6:386–391. doi: 10.1089/clo.2004.6.386. [DOI] [PubMed] [Google Scholar]
- 8.Niwa H. Molecular mechanism to maintain stem cell renewal of ES cells. Cell. Struct. Funct. 2001;26:137–148. doi: 10.1247/csf.26.137. [DOI] [PubMed] [Google Scholar]
- 9.Hamatani T., Carter M.G., Sharov A.A., Ko M.S. Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- 10.Zeng F., Baldwin D.A., Schultz R.M. Transcript profiling during preimplantation mouse development. Dev. Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 11.Yoshikawa T., Piao Y., Zhong J., Matoba R., Carter M.G., Wang Y., Goldberg I., Ko M.S. High-throughput screen for genes predominantly expressed in the ICM of mouse blastocysts by whole mount in situ hybridization. Gene Expr. Patterns. 2006;6:213–224. doi: 10.1016/j.modgep.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Latham K.E., Garrels J.I., Chang C., Solter D. Quantitative analysis of protein synthesis in mouse embryos. I. Extensive reprogramming at the one- and two-cell stages. Development. 1991;112:921–932. doi: 10.1242/dev.112.4.921. [DOI] [PubMed] [Google Scholar]
- 14.Davis W., Jr, De Sousa P.A., Schultz R.M. Transient expression of translation initiation factor eIF-4C during the 2-cell stage of the preimplantation mouse embryo: identification by mRNA differential display and the role of DNA replication in zygotic gene activation. Dev. Biol. 1996;174:190–201. doi: 10.1006/dbio.1996.0065. [DOI] [PubMed] [Google Scholar]
- 15.Hershey J.W. Protein phosphorylation controls translation rates. J. Biol. Chem. 1989;264:20823–20826. [PubMed] [Google Scholar]
- 16.Zhang W., Morris Q.D., Chang R., Shai O., Bakowski M.A., Mitsakakis N., Mohammad N., Robinson M.D., Zirngibl R., Somogyi E., et al. The functional landscape of mouse gene expression. J. Biol. 2004;3:21. doi: 10.1186/jbiol16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M., Pevny L., Lovell-Badge R., Smith A. Generation of purified neural precursors from embryonic stem cells by lineage selection. Curr. Biol. 1998;8:971–974. doi: 10.1016/s0960-9822(98)70399-9. [DOI] [PubMed] [Google Scholar]
- 18.Pease S., Braghetta P., Gearing D., Grail D., Williams R.L. Isolation of embryonic stem (ES) cells in media supplemented with recombinant leukemia inhibitory factor (LIF) Dev. Biol. 1990;141:344–352. doi: 10.1016/0012-1606(90)90390-5. [DOI] [PubMed] [Google Scholar]
- 19.Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 20.Lickert H., Kutsch S., Kanzler B., Tamai Y., Taketo M.M., Kemler R. Formation of multiple hearts in mice following deletion of beta-catenin in the embryonic endoderm. Dev. Cell. 2002;3:171–181. doi: 10.1016/s1534-5807(02)00206-x. [DOI] [PubMed] [Google Scholar]
- 21.Kunath T., Gish G., Lickert H., Jones N., Pawson T., Rossant J. Transgenic RNA interference in ES cell-derived embryos recapitulates a genetic null phenotype. Nat. Biotechnol. 2003;21:559–561. doi: 10.1038/nbt813. [DOI] [PubMed] [Google Scholar]
- 22.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams A.J., Blacklow S.C., Collins T. The zinc finger-associated SCAN box is a conserved oligomerization domain. Mol. Cell. Biol. 1999;19:8526–8535. doi: 10.1128/mcb.19.12.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolfe S.A., Nekludova L., Pabo C.O. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan T., Friedman N., Margalit H. Ab initio prediction of transcription factor targets using structural knowledge. PLoS Comput. Biol. 2005;1:e1. doi: 10.1371/journal.pcbi.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams A.J., Khachigian L.M., Shows T., Collins T. Isolation and characterization of a novel zinc-finger protein with transcription repressor activity. J. Biol. Chem. 1995;270:22143–22152. doi: 10.1074/jbc.270.38.22143. [DOI] [PubMed] [Google Scholar]
- 27.Letunic I., Copley R.R., Schmidt S., Ciccarelli F.D., Doerks T., Schultz J., Ponting C.P., Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sander T.L., Haas A.L., Peterson M.J., Morris J.F. Identification of a novel SCAN box-related protein that interacts with MZF1B. The leucine-rich SCAN box mediates hetero- and homoprotein associations. J. Biol. Chem. 2000;275:12857–12867. doi: 10.1074/jbc.275.17.12857. [DOI] [PubMed] [Google Scholar]
- 29.Brandenberger R., Wei H., Zhang S., Lei S., Murage J., Fisk G.J., Li Y., Xu C., Fang R., Guegler K., et al. Transcriptome characterization elucidates signaling networks that control human ES cell growth and differentiation. Nat. Biotechnol. 2004;22:707–716. doi: 10.1038/nbt971. [DOI] [PubMed] [Google Scholar]
- 30.Virbasius C.M., Wagner S., Green M.R. A human nuclear-localized chaperone that regulates dimerization, DNA binding, and transcriptional activity of bZIP proteins. Mol. Cell. 1999;4:219–228. doi: 10.1016/s1097-2765(00)80369-x. [DOI] [PubMed] [Google Scholar]
- 31.Bruhn L., Munnerlyn A., Grosschedl R. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRalpha enhancer function. Genes Dev. 1997;11:640–653. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- 32.Luo M.L., Zhou Z., Magni K., Christoforides C., Rappsilber J., Mann M., Reed R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- 33.Keys R.A., Green M.R. Gene expression. The odd coupling. Nature. 2001;413:583–585. doi: 10.1038/35098172. [DOI] [PubMed] [Google Scholar]
- 34.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., et al. The UCSC genome browser database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davidson E.H., Rast J.P., Oliveri P., Ransick A., Calestani C., Yuh C.H., Minokawa T., Amore G., Hinman V., Arenas-Mena C., et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 37.Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]