Abstract
To support efforts to develop a ‘synthetic biology’ based on an artificially expanded genetic information system (AEGIS), we have developed a route to two components of a non-standard nucleobase pair, the pyrimidine analog 6-amino-5-nitro-3-(1′-β-D-2′-deoxyribofuranosyl)-2(1H)-pyridone (dZ) and its Watson–Crick complement, the purine analog 2-amino-8-(1′-β-D-2′-deoxyribofuranosyl)-imidazo[1,2-a]-1,3,5-triazin-4(8H)-one (dP). These implement the pyDDA:puAAD hydrogen bonding pattern (where ‘py’ indicates a pyrimidine analog and ‘pu’ indicates a purine analog, while A and D indicate the hydrogen bonding patterns of acceptor and donor groups presented to the complementary nucleobases, from the major to the minor groove). Also described is the synthesis of the triphosphates and protected phosphoramidites of these two nucleosides. We also describe the use of the protected phosphoramidites to synthesize DNA oligonucleotides containing these AEGIS components, verify the absence of epimerization of dZ in those oligonucleotides, and report some hybridization properties of the dZ:dP nucleobase pair, which is rather strong, and the ability of each to effectively discriminate against mismatches in short duplex DNA.
INTRODUCTION
As it was formulated by Watson and Crick over 50 years ago, the standard nucleobase pair follows two rules of complementarity: size complementarity (large purines pair with small pyrimidines) and hydrogen bonding complementarity (hydrogen bond donors pair with hydrogen bond acceptors) (1,2). Many groups have worked to deviate from this formula (3). For example, Rappaport (4), Ishikawa et al. (5),Fujiwara et al. (6), Hirao et al. (7) and Sismour et al. (8) have sought to change hydrogen-bonding patterns and to control the outcome with sulfur and/or steric interactions. Kool and his group have sought to dispense with hydrogen bonding entirely (9,10), as have Romesberg, Schultz and their coworkers (11,12) using hydrophobic interactions. In the opposite direction, Minakawa et al. have sought to increase the number of interpair hydrogen bonds (13).
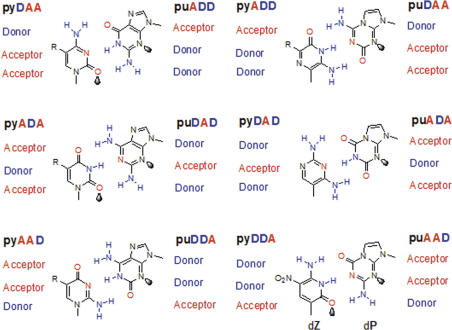
Despite the success of this work, alternative nucleic acid-like systems that deviate less drastically from the standard design have had, to date, the technological success and supported best the emerging field of synthetic biology (14). If nitrogen and oxygen are used as the only heteroatoms, six different hydrogen-bonding patterns can be directly placed within the standard Watson–Crick geometry. Here, they support mutually exclusive nucleobase pairing in an ‘artificially expanded genetic information system’ (AEGIS) (15–17) (Figure 1).
Figure 1.
The AEGIS that follows closely the geometry and hydrogen bonding architecture of natural nucleotides (1,2). The various hydrogen bonding patterns are named pu or py, depending on whether they are presented on a large (purine-like) or small (pyrimidine-like) heterocyclic ring system, with hydrogen bond donor (D) and acceptor (A) groups listed starting in the major groove and ending in the minor groove. Unshared pairs of electrons (or ‘electron density’) presented to the minor groove are shown by the shaded lobes. The pyDDA:puAAD hydrogen bonding pattern, the subject of this paper, is at the bottom right.
Two AEGIS components built according to this design, 2′-deoxyisoguanosine and 2′-deoxyisocytosine, are now incorporated into Bayer's branched DNA diagnostics tools that quantitate the levels of human immunodeficiency, hepatitis B and hepatitis C viruses in blood. These help improve the health care of over 400 000 patients annually (18,19). This pair also supports assays in development to detect certain genetic defects that cause cystic fibrosis (20). Further, using mutant polymerases that accept the non-standard nucleobases, and special strategies to manage undesired tautomerism of isoguanosine, AEGIS supports now two versions of the polymerase chain reaction that incorporate 6 nt ‘letters’ (8,21). Thus, these versions of DNA can support a primitive form of evolution.
This and other work has uncovered certain structural features of the nucleobases that make different AEGIS components more or less likely to be accepted by natural DNA polymerases. Most important is the ability of the component to present an unshared pair of electrons (or, more formally, ‘electron density’) to the minor groove (22). In the standard purines, adenosine and guanosine, this density is presented by nitrogen 3 (Figure 1). In the standard pyrimidines, thymidine and cytidine, this density is presented by the exocyclic carbonyl oxygen at the 2-position (23).
Only one AEGIS hydrogen bonding pair (other than the standard A:T and G:C pairs) can present unshared electron pairs to the minor groove from both components. This is the pyDDA and puAAD pair, implemented on heterocycles where the purine analog has a nitrogen at the position analogous to N3 on standard purines, and the pyDDA component has an exocyclic carbonyl oxygen at the position analogous to the 2-position on standard pyrimidines.
The pyDDA hydrogen-bonding pattern has proven to be especially difficult to implement in a form that can support synthetic biology, however. Our first effort, implementing the pyDDA hydrogen bonding pattern on a pyridine heterocycle gave easily oxidized compounds (24). The pyrazine implementation of the pyDDA hydrogen bonding pattern was stable to oxidation, but suffered epimerization (25,26). The pyDDA pattern implemented on a pyrimidine heterocycle would give rise to tautomeric ambiguity.
One promising solution to these problems exploits the nitro group to moderate the reactivity of the aminopyridone heterocycle. The electron withdrawing properties of the nitro group, attached to an aminopyridone 2′-deoxyriboside, were indeed found to diminish the oxidizability of the heterocycle, and decrease the rate of specific-acid catalyzed epimerization (27). This was shown in model studies using 6-amino-5-nitro-3-(1′-β-D-2′-deoxyribofuranosyl)-2(1H)-pyridone (here called dZ) (27).
The hydrogen bonding partner for dZ must implement the puAAD hydrogen-bonding pattern. The 2-amino-8-(2′-deoxy-β-D-erythro-pentofuranosyl)-imidazo[1,2-a]-1,3,5-triazin-4(8H)-one, referred to here as dP, has previously been proposed for this purpose (28). This N-glycoside is stable to epimerization, is rather stable to ‘depurination’ under acidic conditions, and has previously been studied as an antiviral agent (29,30).
Here we present an improved synthesis of dZ and dP nucleosides. The dZ nucleoside is prepared by gram scale Heck-coupling of an oxygen-protected iodinated heterocycle and a suitable glycal (31), followed by reduction. We also describe the preparation of the protected phosphoramidites and triphosphates of dZ and dP. Both phosphoramidites were used to synthesize DNA oligonucleotides with good coupling efficiency. We verified that dZ remains stable against epimerization during the synthesis of the oligonucleotides. Finally, we report the hybridization properties of DNA oligonucleotides containing the dZ:dP nucleobase pair, and some data that demonstrate the ability of each nucleobase to discriminate against mismatches in short duplex DNA. These experiments suggest that the dZ:dP nucleobase pair contributes more to duplex stability than any of the standard nucleobase pairs.
MATERIALS AND METHODS
Nucleotide and oligonucleotide synthesis
Full experimental procedures describing the synthesis of the compounds used in this work are available in Supplementary Data.
RESULTS
Synthesis of nucleoside phosphoramidite of dZ
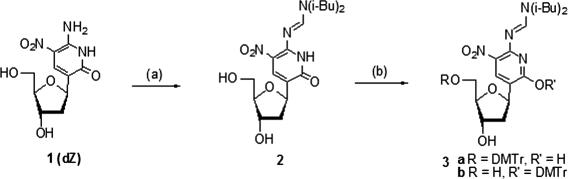
The synthesis of the C-nucleoside 1 (dZ) to implement the non-standard pyDDA hydrogen-bonding pattern was recently reported (27). This nucleoside was stable with respect to epimerization for hours, even at low pH (pH 3). When we attempted to synthesize the phosphoramidite starting with that fully deprotected nucleoside, however, we observed that the strong nucleophilicity of the heterocyclic keto group prevented a selective reaction during dimethoxytritylation; the DMT group went to the 2-position of the heterocycle as well as the 5′-position of the sugar (Figure 2). This nucleophilicity also caused problems during the preparation of the phosphoramidite (32).
Figure 2.
Conditions: (a) N, N-Diisobutylformamide dimethyl acetal, MeOH, rt; (b) (MeO)2TrCl, Py, Et3N, DMAP, 0°C to rt.
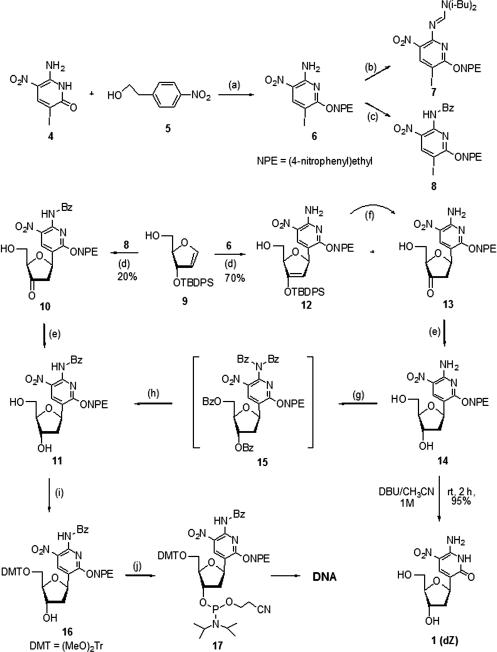
To circumvent these problems, the published procedure was modified to protect the oxygen on the heterocycle prior to Heck coupling (Figure 3) using the p-nitrophenethyl (NPE) protecting group of Pfleiderer and co-workers (33). These p-NPE ethers are stable under acidic and mild hydrolytic conditions (e.g. ammonia and triethylamine), but are cleaved by DBU in aprotic solvents.
Figure 3.
Conditions: (a) DEAD, Ph3P, dioxane/DMF (4/1, v/v), 0°C to rt, 92%; (b) N, N-Diisobutyl formamide dimethyl acetal, MeOH, rt; (c) i) BzCl, DMAP, Py, Et3N, 0°C to rt; ii) H2O, NH4OH (29%), 0°C, 73% (over two steps); (d) Pd(OAc)2, Ph3As, Et3N, DMF, 55°C; (e) NaBH(OAc)3, AcOH, CH3CN, 0°C, 98%; (f) Bu4NF, AcOH, THF, 0°C, 90%; (g) BzCl, DMAP, Py, Et3N, rt, 85%; (h) 1 M NaOH / H2O, 1,4-dioxane, rt, 90%; (i) (MeO)2TrCl, Py, Et3N, rt, 80%; (j) iPr2N(OC2H4CN)PCl, DIPEA, CH2Cl2, 0°C to rt, 71%.
To obtain predominantly O-alkylation, heterocycle 4 was treated with 2-(4-nitrophenyl)-ethyl iodide and silver carbonate in benzene (34,35). After optimization of the conditions, the yield was only about 40%. Since the pKa of heterocycle 4 (≈7.8) meets the requirement of a pKa < 13 for a Mitsunobu reaction, we investigated this reaction as a way of achieving O-alkylation (36–38). NPE protection at O2 was achieved in over 90% yield by treatment of 4 with 4-NPE alcohol (5) under Mitsunobu conditions (39). Protection of the exocyclic amino group of 6 as the diisobutylformamidine to give 7 occurred again in low yield, perhaps due to instability of the product during flash column purification (40,41). The amino group was therefore benzoylated to give monobenzoate 8 in over 70% yield after mild hydrolysis of the dibenzoate that was formed initially (42,43).
Heck couplings of iodinated heterocycles to the glycal 9 and subsequent deprotection and reduction, have been reported previously by several groups for several C-glycosides (44–46), and have been used in our previous synthesis of dZ (27). The use of the tert-butyldiphenylsilyl protecting group at the 3′-position blocks one side of the sugar glycal so that the Heck coupling occurs exclusively from the ‘top’ to give the β-C-nucleoside (Figure 3). The free 5′-hydroxyl of the sugar then leads to stereospecific reduction of the intermediate ketone by complexation with the boron.
Following literature procedures, palladium acetate and triphenylarsine were used as the catalyst system, with anhydrous DMF as the solvent. The Heck reaction was run at 55°C for three days. The desilylated 2′-deoxy-3′-keto-β-C-nucleoside 10 was isolated as the only product, in 20% yield, and the major byproduct was the deiodination of the heterocycle. This was presumably generated via the reaction of iodide to desilylate the silyl enol ether derivative during the coupling reaction (47,48). Reduction of the ketone with triacetoxyborohydride yielded the 2′-deoxy-β-C-nucleoside 11 in quantitative yield.
The low yield of the Heck coupling was ascribed to the electron withdrawing benzoyl group, which renders the heterocycle electron-poor. Therefore, the Heck coupling between non-benzoylated heterocycle 6 and glycal 9 was explored. After 3 days at 55°C, β-C-nucleosides 12 and 13 were obtained in 31 and 39% yield, respectively. As with 10, the 2′-deoxy-3′-keto-β-C-nucleoside 13 was generated in situ from desilylation of intermediate 12. Rapid removal of the silyl protecting group of 12 with TBAF at 0°C gave ketone 13. Reduction of the ketone with triacetoxyborohydride gave the 2′-deoxy-β-C-nucleoside 14. Perbenzoylation of 14 with benzoyl chloride gave 15, which was selectively deprotected with 1 M aqueous NaOH to give monobenzoate 11 in 80% overall yield. Two more steps led to the 5′-DMT protected phosphoramidite derivative 17, ready for solid-phase DNA synthesis.
To test the suitability of NPE as a protecting group in oligonucleotide synthesis, the NPE of 14 was removed by DBU (1 M in CH3CN) to give 2′-deoxy-β-C-nucleoside 1 (dZ) in 95% yield after flash chromatography (Figure 3). The 1H-NMR, 13C-NMR and high-performance liquid chromatography (HPLC) trace were the same as reported in literature for 1 (27).
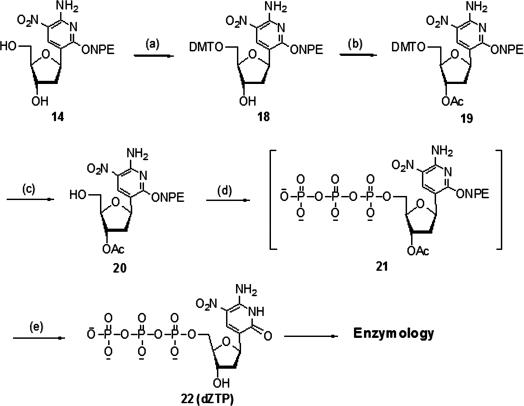
Synthesis of the nucleoside 5′-triphosphate of dZ
According to the classical procedure developed by Ludwig and Eckstein (49), the protection of functional groups on the standard nucleobase heterocycles is not required during the synthesis of the corresponding 5′-triphosphates. Thus, we began the synthesis of the dZ triphosphate without protecting the heterocycle by selectively protecting the 5′-OH of nucleoside 14 with DMT-Cl to give 18 in 80% yield. Selective acetylation to give the 3′-acetate 19 with excess Ac2O (10 equivalent) in pyridine at room temperature was accomplished in nearly quantitative yield after column chromatography. Surprisingly, the exocyclic amino group did not react with either DMT-Cl or Ac2O under these particular reaction conditions.
The 5′-DMT protecting group was removed by saturated methanolic HCl to give 20 in 90% yields. The nucleoside 5′-triphosphate 21 was prepared by the ‘Ludwig–Eckstein procedure’ (49). The NPE group was then removed by treatment with DBU (0.8 M in acetonitrile) at room temperature for 4 h. Subsequent treatment with concentrated ammonium hydroxide at room temperature furnished the fully deprotected 5′-triphosphate 22 (dZTP) in 10% overall yield after HPLC purification on DEAE-Sephadex and C-18 (Figure 4).
Figure 4.
Conditions: (a) (MeO)2TrCl, Py, Et3N, rt, 80%; (b) Ac2O, Py, rt, 94%; (c) HCl, MeOH, CH2Cl2, 0°C, 90%; (d) i) 2-Chloro-4H-1,3,2-benzodioxaphosphorin-4-one, Py, dioxane, rt; ii) tributylammonium pyrophosphate, n-Bu3N, DMF, rt; iii) I2, H2O, Py, rt; (e) i) DBU, CH3CN, rt; ii) H2O, NH4OH, rt, 10% (over two steps).
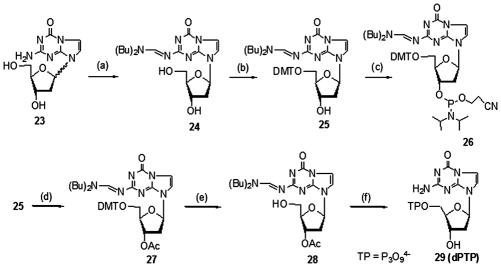
Synthesis of the phosphoramidite and 5′-triphosphate of dP
The synthesis of 26, the phosphoramidite of dP, followed the procedure developed by Seela and co-workers (28) with minor modifications (Figure 5). Seela's procedure calls for a delicate chromatographic resolution of the epimeric mixture obtained after attaching the heterocycle to the sugar, separating two compounds that have only slight differences in their Rf values. We found that adding the formamidine protecting group before the resolution of the epimers greatly improved the isolation of the pure β-epimer 24.
Figure 5.
Conditions: (a) Dibutylformamide dimethyl acetal, MeOH, 40°C, β isomer 53%, α isomer 37%; (b) (MeO)2TrCl, Py, Et3N, rt, 75%; (c) iPr2N(OC2H4CN)PCl, DIPEA, CH2Cl2, 0°C to rt, 76% ; (d) Ac2O, Py, rt, 93%; (e) HCl, MeOH, CH2Cl2, 0°C, 99%; (f) i) 2-Chloro-4H-1,3,2-benzodioxaphosphorin-4-one, Py, dioxane, rt; ii) tributylammonium pyrophosphate, n-Bu3N, DMF, rt; iii) I2, H2O, Py, rt; iv) H2O, NH4OH, 45°C, 20% (over two steps).
The corresponding 5′-triphosphate 29 (dPTP) was synthesized as described in Figure 5, applying the Ludwig–Eckstein procedure. Although an enzymatic study using this triphosphate has been reported in literature (50), a full experimental procedure yielding the triphosphate has never, to our knowledge, been published.
Oligonucleotide synthesis
Oligonucleotides containing the AEGIS components were prepared by using the standard conditions, except that an extended coupling time (6 min) was used for the nonstandard nucleoside phosphoramidites. Based on the amount of DMT released, the coupling efficiency of the dZ and dP phosphoramidites was found to be similar to that obtained with commercial phosphoramidites of the standard nucleosides. The oligonucleotides containing dP were deprotected in concentrated ammonium hydroxide (60°C, 12 h). The products containing dZ were first treated with DBU (1 M in acetonitrile, 10 h, rt) prior to ammonium hydroxide deprotection (51). The crude oligonucleotides were purified by ion-exchange HPLC (Dionex DNAPac™ PA-100 column, 9 × 250 mm) and desalted over SepPak C18-cartridges. The homogeneity of the purified chimeras was verified by analytical ion-exchange HPLC. Their composition was determined by tandem digestion with snake-venom phosphodiesterase (SVPDE) I and alkaline phosphatase, followed by reverse-phase HPLC (Nova-Pak C18 column 3.9 × 150 mm) (52). The analyses in all cases verified the incorporation of the correct number of nonstandard nucleosides (Table 1).
Table 1.
Analysis of the oligonucleotides by enzymatic digestiona
| Calculated | Found | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sequence | C | G | T | A | Z or P | C | G | T | A | Z or P |
| 3′-TGTGZTGAAAGAGG-5′ | 0 | 6 | 3 | 4 | 1 | 0.0 | 6.1 | 3.0 | 4.0 | 0.9 |
| 5′-CACPACTTTCTCCT-3′ | 6 | 0 | 5 | 2 | 1 | 6.0 | 0.0 | 5.0 | 2.0 | 1.1 |
| 3′-CAGTTCACZGATTGC-5′ | 4 | 3 | 4 | 3 | 1 | 4.0 | 3.0 | 3.9 | 2.9 | 0.9 |
| 5′-GTCAAGTGPCTAACG-3′ | 3 | 4 | 3 | 4 | 1 | 3.0 | 4.0 | 3.0 | 3.9 | 1.1 |
| 3′-CAGTTCACZZATTGC-5′ | 4 | 2 | 4 | 3 | 2 | 4.0 | 2.0 | 4.0 | 3.0 | 1.8 |
| 5′-GTCAAGTGPPTAACG-3′ | 2 | 4 | 3 | 4 | 2 | 2.0 | 4.0 | 3.0 | 3.9 | 2.2 |
aConditions: oligonucleotide 1.5 nmol, SVPDE 6 × 10−3 U, alkaline phosphatase 1.5 U, Digestion buffer [0.1 M Tris–HCl (pH 8.3) and 20 mM MgCl2], followed by reversed phase HPLC.
Epimerization of dZ
To estimate the extent of epimerization of dZ within an oligonucleotide during its synthesis and storage, a sample (4 nmol) of the oligonucleotide d(TGTGZTGAAAGAGG) containing a single dZ was dissolved in TEAA [25 mM (pH 7.0), 100 μl] and divided into two aliquots. One was stored at −20°C; the other was kept at room temperature for two months. Both samples were then digested with snake-venom phosphodiesterase (SVPDE) I and alkaline phosphatase (Supplementary Data) and analyzed by reversed phase HPLC. The HPLC trace showed only the β-nucleoside peak for both samples, indicating that the nucleotide dZ entered the DNA as only the beta epimer, and is stable with respect to epimerization at neutral pH for months once incorporated into DNA
Thermal denaturation studies
To determine the contribution of the dZ:dP nucleobase pair to the stability of duplex DNA, thermal denaturation conditions reported by Geyer et al. and Horn et al. were applied (17,53) (Supplementary Data). For comparison with their data, analogous oligonucleotides were used (Sequence pair 3). The variable position in the DNA duplex, denoted X:Y, was placed at least 4 nt from the end of the duplex, so as to eliminate ‘end effects’ (54). All experiments generated standard, single-transition melting, consistent with two-state melting behavior. This was expected, as the oligonucleotides are not able to form significant alternative duplex or hairpin structures.
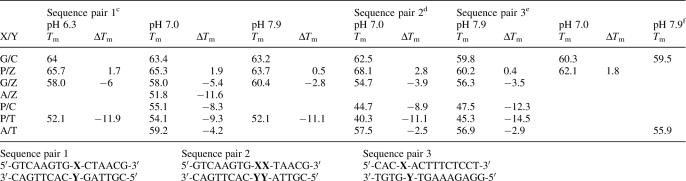
The melting temperatures for DNA duplexes were measured at pH 6.3, 7.0 and 7.9 (Table 2). These pH values were chosen both to enable a comparison with literature data, and to reflect the known pKa of the dZ heterocycle; the dP does not have an acidic hydrogen within this range, and the triazene skeleton is only very weakly basic. At pH 6.3, protonation of dZ is complete, while the standard nucleobases are only slightly protonated.
Table 2.
aConditions: 1.6 μM DNA strand concentration, 0.5 M NaCl at pH 6.3 and 7.0 (5 mM phosphate buffer), 0.05 mM EDTA; 0.45 M NaCl, 45 mM sodium citrate (pH 7.9).
bConditions: sequence pair 3, as reported by Geyer and Horn (17,53). 1.6 μM DNA strand concentration, 0.45 M NaCl, 45 mM sodium citrate, pH 7.9.
cData of columns 1, 2 and 3 are from Sequence pair 1.
dSequence pair 2, two consecutive Z/P base pairs in the same oligomer, ΔTm indicate the Tm difference of per base pair compare to C/G base pair.
eData of columns 5 and 6 are from Sequence pair 3.
The data in Table 2 indicate that a dZ:dP nucleobase pair is significantly stronger than a dC:dG base pair, increasing the Tm on the average by 1.8°C per pair at pH 6.3 and 7.0. The effect of two consecutive dZ:dP nucleobase pairs in the same oligomer is more than additive, with an increase in Tm of 2.8°C per pair at pH 7.0. Consistent with the acidity of dZ, the contribution of the dZ:dP nucleobase pair is less at pH 7.9.
DISCUSSION
These results establish that 6-amino-5-nitro-3-(1′-β-D-2′-deoxyribofuranosyl)-2(1H)-pyridone (dZ) and 2-amino-8-(1′-β-D-2′-deoxyribofuranosyl)-imidazo[1,2-a]-1,3,5-triazin-4(8H)-one (dP) implement the pyDDA and puAAD hydrogen bonding patterns as part of an AEGIS. The properties of both heterocycles allow them to be practically useful. Indeed, the dZ:dP nucleobase pair appears, in the studies done so far, to contribute more than the dG:dC nucleobase pair to duplex stability. In this respect, it is similar to the nucleobase pair between isoguanosine and isocytidine (puDDA and pyAAD, respectively). The ΔTm values of the dZ:dP and disoMeC:disoG nucleobase pairs are 1.8° and 2.0°C per nucleobase pair, respectively (relative to the C:G pair).
The dZ:dP nucleobase pair does not present the chemical problems that the disoMeC:disoG nucleobase pair presents, however, including tautomerization, depyrimidinylation (of isoC) and deamination (55). Further, both components of the dZ:dP nucleobase pair present unshared pairs of electrons to the minor groove. As noted in the Introduction, these are specificity elements for many polymerases at many points in the primer–template–triphosphate complex.
The stability of the dZ:dP nucleobase pair depends on pH, just as do the standard dA:dT and dG:dC nucleobase pairs. The heterocycle on the dZ nucleoside is an acid with a pKa value of ca. 7.8 (in the free nucleoside form). When deprotonated, the H-bonding pattern is changed. Further, Geyer et al. (17) showed that the introduction of a negative charge into a stack of nucleobases decreases the stability of the duplex. These are both consistent with the observed reduction in Tm for duplexes joined by the dZ:dP nucleobase pair at pH 7.9.
The data also show the high specificity of the dZ:dP nucleobase pair. In the limited studies that we have done so far, nucleobase combinations other than those having complementary pairing lead to reductions in Tm similar to those seen in mismatches between the standard nucleobases. This allows both of the non-standard nucleobases, dZ and dP, to discriminate against mismatches in short duplex DNA, at least in the contexts examined so far.
Last, the synthesis of the triphosphates of the nucleosides dZ and dP is reported. These syntheses support ongoing work investing the ability of DNA polymerases to copy DNA containing the dZ:dP nucleobase pair. Results from these investigations will be reported shortly.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
Support of the National Institutes of Health (GM-54048) and the NASA Exobiology program is gratefully acknowledged. Funding to pay the Open Access publication charges for this article was provided by Foundation for Applied Molecular Evolution.
Conflict of interest statement. None declared.
REFERENCES
- 1.Watson J.D., Crick F.H.C. Molecular structure of nucleic acids. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Watson J.D., Crick F.H.C. General implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 3.Hentry A.A., Romesberg F.E. Beyond A, C, G, and T: augmenting nature's alphabet. Curr. Opin. Chem. Biol. 2003;7:727–733. doi: 10.1016/j.cbpa.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Rappaport H.P. The 6-thioguanine/5-methyl-2-pyrimidinone base pair. Nucleic Acids Res. 1988;16:7253–7267. doi: 10.1093/nar/16.15.7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishikawa M., Hirao I., Yokoyama S. Synthesis of 3-(2-deoxy-β-Dribofuranosyl) pyridin-2-one and 2-amino-6-(N,Ndimethylamino)-9-(2-deoxy-β-D-ribofuranosyl)purine derivatives for an unnatural base pair. Tetrahedron Lett. 2000;41:3931–3934. [Google Scholar]
- 6.Fujiwara T., Kimoto M., Sugiyama H., Hirao I., Yokoyama S. Synthesis of 6-(2-thienyl)purine nucleoside derivatives that form unnatural base pairs with pyridin-2-one nucleosides. Bioorg. Med. Chem. Lett. 2001;11:2221–2223. doi: 10.1016/s0960-894x(01)00415-2. [DOI] [PubMed] [Google Scholar]
- 7.Hirao I., Harada Y., Kimoto M., Mitsui T., Fujiwara T., Yokoyama S. A two unnatural base pair system toward the expansion of the genetic code. J. Am. Chem. Soc. 2004;126:13298–13305. doi: 10.1021/ja047201d. [DOI] [PubMed] [Google Scholar]
- 8.Sismour A.M., Benner S.A. The use of thymidine analogs to improve the replication of an extra DNA base pair: a synthetic biological system. Nucleic Acids Res. 2005;33:5640–5646. doi: 10.1093/nar/gki873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moran S., Ren R.X.-F., Kool E.T. A thymidine triphosphate shape analog lacking Watson–Crick pairing ability is replicated with sequence selectivity. Proc. Natl Acad. Sci. USA. 1997;94:10506–10511. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kool E.T. Replacing the nucleobases in DNA with designer molecules. Acc. Chem. Res. 2002;35:936–943. doi: 10.1021/ar000183u. [DOI] [PubMed] [Google Scholar]
- 11.McMinn D.L., Ogawa A.K., Wu Y., Liu J., Schultz P.G., Romesberg F.E. Efforts toward expansion of the genetic alphabet: recognition of a highly stable, self-pairing hydrophobic base. J. Am. Chem. Soc. 1999;121:11585–11586. [Google Scholar]
- 12.Tae E.L., Wu Y.Q., Xia G., Schultz P.G., Romesberg F.E. Efforts toward expansion of the genetic alphabet: replication of DNA with three base pairs. J. Am. Chem. Soc. 2001;123:7439–7440. doi: 10.1021/ja010731e. [DOI] [PubMed] [Google Scholar]
- 13.Minakawa N., Kojima N., Hikishima S., Sasaki T., Kiyosue A., Atsumi N., Ueno Y., Matsuda A. New base pairing motifs. The synthesis and thermal stability of oligodeoxynucleotides containing imidazopyridopyrimidine nucleosides with the ability to form four hydrogen bonds. J. Am. Chem. Soc. 2003;125:9970–9982. doi: 10.1021/ja0347686. [DOI] [PubMed] [Google Scholar]
- 14.Benner S.A. Understanding nucleic acids using synthetic chemistry. Acc. Chem. Res. 2004;37:784–797. doi: 10.1021/ar040004z. [DOI] [PubMed] [Google Scholar]
- 15.Switzer C.Y., Moroney S.E., Benner S.A. Enzymatic incorporation of a new base pair into DNA and RNA. J. Am. Chem. Soc. 1989;111:8322–8323. [Google Scholar]
- 16.Piccirilli J.A., Krauch T., Moroney S.E., Benner S.A. Extending the genetic alphabet. Enzymatic incorporation of a new base pair into DNA and RNA. Nature. 1990;343:33–37. doi: 10.1038/343033a0. [DOI] [PubMed] [Google Scholar]
- 17.Geyer C.R., Battersby T.R., Benner S.A. Nucleobase pairing in expanded Watson–Crick-like genetic information systems. Structure. 2003;11:1485–1498. doi: 10.1016/j.str.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Elbeik T., Surtihadi J., Destree M., Gorlin J., Holodniy M., Jortani S.A., Kuramoto K., Ng V., Valdes R., Valsamakis A., et al. Multicenter evaluation of the performance characteristics of the Bayer VERSANT HCV RNA 3.0 assay (bDNA) J. Clin. Microbiol. 2004;42:563–569. doi: 10.1128/JCM.42.2.563-569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gleaves C.A., Welle J., Campbell M., Elbeik T., Ng V., Taylor P.E., Kuramoto K., Aceituno S., Lewalski E., Joppa B., et al. Multicenter evaluation of the Bayer VERSANT (TM) HIV-1 RNA 3.0 assay: analytical and clinical performance. J. Clin. Virol. 2002;25:205–216. doi: 10.1016/s1386-6532(02)00011-2. [DOI] [PubMed] [Google Scholar]
- 20.Johnson S.C., Marshall D.J., Harms G., Miller C.M., Sherrill C.B., Beaty E.L., Lederer S.A., Roesch E.B., Madsen G., Hoffman G.L., et al. Multiplexed genetic analysis using an expanded genetic alphabet. Clin. Chem. 2004;50:2019–2027. doi: 10.1373/clinchem.2004.034330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sismour A.M., Lutz S., Park J.-H., Lutz M.J., Boyer P.L., Hughes S.H., Benner S.A. PCR amplification of DNA containing non-standard base pairs by variants of reverse transcriptase from Human Immunodeficiency Virus-1. Nucleic Acids Res. 2004;32:728–735. doi: 10.1093/nar/gkh241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steitz T. In: Biological Organization: Macromolecular Interactions at High Resolution. Burnett R.M., Vogel H.J., editors. NY: Academic Press; 1987. pp. 45–55. [Google Scholar]
- 23.Hendrickson C., Devine K., Benner S.A. Probing the necessity of minor groove interactions with three DNA polymerase families using 3-deaza-2′-deoxyadenosine 5′-triphosphate. Nucleic Acids Res. 2004;32:2241–2250. doi: 10.1093/nar/gkh542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piccirilli J.A., Krauch T., MacPherson L.J., Benner S.A. A direct route to 3-(ribofuranosyl)-pyridine nucleosides. Helv. Chim. Acta. 1991;74:397–406. [Google Scholar]
- 25.Voegel J.J., von Krosigk U., Benner S.A. Synthesis and tautomeric equilibrium of 6-amino-5-benzyl-3-methylpyrazin-2-one. An acceptor-donor-donor nucleoside base analog. J. Org. Chem. 1993;58:7542–7547. [Google Scholar]
- 26.Voegel J.J., Benner S.A. Synthesis and characterization of nonstandard nucleosides and nucleotides bearing the acceptor-donor-donor pyrimidine analog 6-amino-3-methylpyrazin-2-one. Helv. Chim. Acta. 1996;79:1863–1880. [Google Scholar]
- 27.Hutter D., Benner S.A. Expanding the genetic alphabet. Nonepimerizing nucleoside with the pyDDA hydrogen bonding pattern. J. Org. Chem. 2003;68:9839–9842. doi: 10.1021/jo034900k. [DOI] [PubMed] [Google Scholar]
- 28.Seela F., Amberg S., Melenewski A., Rosemeyer H. 5-Aza-7-deazaguanine DNA: recognition and strand orientation of oligonucleotides incorporating anomeric imidazo[1,2-a]-1,3,5-triazine nucleosides. Helv. Chim. Acta. 2001;84:1996–2014. [Google Scholar]
- 29.Kim S.H., Bartholomew D.G., Allen L.B., Robins R.K., Revankar G.R., Dea P. Imidazo[1,2-a]-s-triazine nucleosides. Synthesis and antiviral activity of the N-bridgehead guanine, guanosine, and guanosine monophosphate analogs of imidazo[1,2-a]-s-triazine. J. Med. Chem. 1978;21:883–889. doi: 10.1021/jm00207a009. [DOI] [PubMed] [Google Scholar]
- 30.Rosemeyer H., Seela F. 5-Aza-7-deaza-2′-deoxyguanosine: studies on the glycosylation of weakly nucleophilic imidazo[1,2-a]-s-triazinyl anions. J. Org. Chem. 1987;52:5136–5143. [Google Scholar]
- 31.Cameron M.A., Cush S.B., Hammer R.P. Facile preparation of protected furanoid glycals from thymidine. J. Org. Chem. 1997;62:9065–9069. [Google Scholar]
- 32.Jurczyk S.C., Horlacher J., Devine K.G., Benner S.A., Battersby T.R. Synthesis and characterization of oligonucleotides containing of 2′-deoxyxanthosine using phosphoramidite chemistry. Helv. Chim. Acta. 2000;83:1517–1524. [Google Scholar]
- 33.Himmelsbach F., Schulz B.S., Trichtinger T., Pfleiderer W. The p-nitrophenylethyl (NPE) group: a versatile new blocking group for phosphate and aglycone protection in nucleosides and nucleotides. Tetrahedron. 1984;40:59–72. [Google Scholar]
- 34.Daniel L.C., Gao J. N- versus O-alkylation in the Mitsunobu reaction of 2- pyridone. Tetrahedron Lett. 1994;35:2819–2822. [Google Scholar]
- 35.Chung N.M., Tieckelmann H. Alkylations of heterocyclic ambident anions. IV. Alkylation of 5-carbethoxy- and 5-nitro-2-pyridone salts. J. Org. Chem. 1970;35:2517–2520. [Google Scholar]
- 36.Mitsunobu O. The use of diethyl azodicarboxylate and triphenylphosphine in synthesis and transformation of natural products. Synthesis. 1981:1–28. [Google Scholar]
- 37.Hughes D.L. Progress in the Mitsunobu reaction. Org. React. 1992;42:335–656. [Google Scholar]
- 38.Tsunoda T., Yamamiya Y., Ito S. 1,1′-(Azodicarbonyl)dipiperidine-tributylphosphine, a new reagent system for Mitsunobu reaction. Tetrahedron Lett. 1993;34:1639–1642. [Google Scholar]
- 39.Lan T., McLaughlin L.W. Synthesis of a dA-dT base pair analogue and its effects on DNA-ligand binding. Bioorg. Chem. 2001;29:198–210. doi: 10.1006/bioo.2001.1209. [DOI] [PubMed] [Google Scholar]
- 40.Froehler B.C., Matteucci M.D. Dialkylformamidines: depurination resistant N6-protecting group for deoxyadenosine. Nucleic Acids Res. 1983;11:8031–8036. doi: 10.1093/nar/11.22.8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Froehler B.C., Jones R.J., Cao X., Terhorst T.J. Oligonucleotides derived from 5-(1-propynyl)-2′-O-allyl-uridine and 5-(1-propynyl)-2′-O-allyl-cytidine: synthesis and RNA duplex formation. Tetrahedron Lett. 1993;34:1003–1006. [Google Scholar]
- 42.Koster H., Kulikowski K., Liese T., Heikens W., Kohli V. N-acyl protecting groups for deoxynucleosides: a quantitative and comparative study. Tetrahedron. 1981;37:363–369. [Google Scholar]
- 43.Krosigk U.V., Benner S.A. pH-Independent triple helix formation by an oligonucleotide containing a pyrazine donor-donor- acceptor base. J. Am. Chem. Soc. 1995;117:5361–5362. [Google Scholar]
- 44.Hsieh H.-P., McLaughlin L.W. Syntheses of two pyridine C-nucleosides as ‘deletion-modified’ analogues of dT and dC. J. Org. Chem. 1995;60:5356–5359. [Google Scholar]
- 45.Chen D.L., McLaughlin L.W. Use of pKa differences to enhance the formation of base triplets involving C-G and G-C base pairs. J. Org. Chem. 2000;65:7468–7474. doi: 10.1021/jo000754w. [DOI] [PubMed] [Google Scholar]
- 46.Coleman R.S., Madaras M.L. Synthesis of a novel coumarin C-Riboside as a photophysical probe of oligonucleotide dynamics. J. Org. Chem. 1998;63:5700–5703. [Google Scholar]
- 47.Farr R.N., Outten R.A., Cheng J.C.-Y., Daves G.D., Jr C-Glycoside synthesis by palladium-catalyzed iodoaglycon-glycal coupling. Organometallics. 1990;9:3151–3156. [Google Scholar]
- 48.Chen J.J., Walker J.A., Liu W., Wise D.S., Townsend L.B. An efficient and stereospecific synthesis of novel pyrazine cnucleosides. Tetrahedron Lett. 1995;36:8363–8366. [Google Scholar]
- 49.Ludwig J., Eckstein F. Rapid and efficient synthesis of nucleoside 5′-O (1-thiotriphosphates), 5′-triphosphates and 2′,3′-cyclophosphorothioates using 2-chloro-4H- 1,3,2-benzodioxaphosphorin-4-one. J. Org. Chem. 1989;54:631–635. [Google Scholar]
- 50.McDougall M.G., Sun L., Livshin I., Hosta L.P., McArdle B.F., Samols S.-B., Fuller C.W., Kumar S. Analogs of guanine nucleoside triphosphates for sequencing applications. Nucleosides Nucleotides. 2001;20:501–506. doi: 10.1081/NCN-100002325. [DOI] [PubMed] [Google Scholar]
- 51.Wagner T., Pfleiderer W. Synthesis of 2′-deoxyribonucleoside 5′-phosphoramidites: new building blocks for the inverse (5′–3′)-oligonucleotide approach. Helv. Chim. Acta. 2000;83:2023–2035. [Google Scholar]
- 52.Seela F., Lampe S. 3-Deazaguanine N7-and N9-(2′-Deoxy-β-D-ribofuranosides): building blocks for solid-phase synthesis and incorporation into oligodeoxyribonucleotides. Helv. Chim. Acta. 1991;74:1790–1800. [Google Scholar]
- 53.Horn T., Chang C.A., Collins M.L. Hybridization properties of the 5- methyl-isocytidine/isoguanosine base pair in synthetic oligodeoxynucleotides. Tetrahydron Lett. 1995;36:2033–2036. [Google Scholar]
- 54.Peyret N., Seneviratne P.A., Allawi H.T., SantaLucia J., Jr. Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A·A, C·C, G·G, and T·T mismatches. Biochemistry. 1999;38:3468–3477. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- 55.Jurczyk S.C., Kodra J.T., Rozzell J.D., Benner S.A., Battersby T.R. Synthesis of oligonucleotides containing 2′-deoxyisoguanosine and 2′-deoxy-5-methylisocytidine using phosphoramidite chemistry. Helv. Chim. Acta. 1998;81:793–811. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.