Abstract
DNA gyrase is the only topoisomerase able to introduce negative supercoils into DNA. Absent in humans, gyrase is a successful target for antibacterial drugs. However, increasing drug resistance is a serious problem and new agents are urgently needed. The naturally-produced Escherichia coli toxin CcdB has been shown to target gyrase by what is predicted to be a novel mechanism. CcdB has been previously shown to stabilize the gyrase ‘cleavage complex’, but it has not been shown to inhibit the catalytic reactions of gyrase. We present data showing that CcdB does indeed inhibit the catalytic reactions of gyrase by stabilization of the cleavage complex and that the GyrA C-terminal DNA-wrapping domain and the GyrB N-terminal ATPase domain are dispensable for CcdB's action. We further investigate the role of specific GyrA residues in the action of CcdB by site-directed mutagenesis; these data corroborate a model for CcdB action based on a recent crystal structure of a CcdB–GyrA fragment complex. From this work, we are now able to present a model for CcdB action that explains all previous observations relating to CcdB–gyrase interaction. CcdB action requires a conformation of gyrase that is only revealed when DNA strand passage is taking place.
INTRODUCTION
The development and spread of antibiotic resistance among bacteria has become a major health and economic burden [http://www.who.int/topics/drug_resistance/en/ (1)]. The majority of antimicrobials are directed at a small group of well-validated targets, suggesting that these are the most effective means to kill cells. As a result, research into these targets, either in the development of novel inhibitors or modification of existing agents, is crucial for future drug development.
The DNA topoisomerases are well-validated drug targets (2–5). Topoisomerases resolve the topological problems created by the double-helical structure of DNA in essential cellular processes. They act by forming a transient single- or double-stranded break in DNA, and catalysing passage of a second strand or DNA duplex through the break prior to resealing the DNA backbone (6). One of the best-characterized topoisomerases is bacterial DNA gyrase, which is a type IIA topoisomerase (i.e. it makes double-strand breaks in DNA) and is unique in its ability to introduce negative supercoils into DNA. Gyrase is found almost exclusively in bacteria, where it is essential to survival, and has been shown to be a very good target for antimicrobial drugs (2,5).
Escherichia coli gyrase has two subunits, GyrA (97 kDa) and GyrB (90 kDa), and functions as a heterotetramer (A2B2). Each of the subunits has two distinct domains, the N- and C-terminal domains (GyrA-NTD, GyrA-CTD, GyrB-NTD and GyrB-CTD). The GyrA-NTD is the DNA cleavage-religation domain and houses the active-site tyrosines that form covalent linkages to DNA in the cleavage reaction. The GyrA-CTD is a DNA-wrapping domain that confers gyrase's unique ability to introduce negative supercoils into DNA. The GyrB-NTD is an ATPase domain, which is required for DNA supercoiling. The GyrB-CTD mediates the GyrA–GyrB interaction, provides residues for the cleavage/religation reaction and is postulated to aid in the DNA-wrapping process (7). The mechanism of DNA supercoiling catalysed by gyrase involves a series of coordinated steps. The enzyme binds a DNA duplex (the ‘G’ or ‘gate’ segment) across the head-dimer interface (DNA gate) formed by the GyrA-NTDs. The region of DNA adjacent to this is wrapped around the GyrA-CTD, and forms a crossover (positive node) in which the ‘transported’ or ‘T’ segment lies across the G segment. ATP binds to the GyrB monomers and causes dimerization, trapping the T segment. The G segment is cleaved in both strands and is covalently attached to the protein at its 5′ ends. Conformational changes lead to opening of the DNA gate. The T segment passes into the GyrA cavity, the G segment is religated and the T segment released through the bottom gate (primary-dimer interface). ATP hydrolysis is thought to stimulate strand passage and is required for resetting of the enzyme for another cycle (6,8).
A number of classes of inhibitors have been shown to inhibit DNA gyrase. The aminocoumarins are naturally-occurring antibiotics that act by competitively inhibiting the ATPase reaction but have received limited clinical use (5). The quinolones are synthetic compounds that stabilize the ‘cleavage complex’ of gyrase. The newer fluoroquinolones e.g. ciprofloxacin (CFX), have been widely applied clinically in the treatment of bacterial infections (9). However a variety of quinolone-resistant strains have been isolated that exhibit multiple mechanisms of resistance (10).
Microcin B17 and CcdB are two naturally-produced proteinaceous inhibitors of gyrase, which are plasmid-encoded toxins and, like quinolones, stabilize the cleavage complex of gyrase (2). Microcin B17 (MccB17) is a post-translationally modified peptide that is thought to act during the strand passage step of gyrase-catalysed supercoiling, although its precise mode of action is not known (11,12,13)
CcdB is the toxin component of the ccd toxin-antitoxin (TA) module. TA modules are best-characterized by their roles in the process of post-segregational killing of plasmid-cured cells but are also believed to be involved in stress responses (14,15). In the process of post-segregational killing, the modules are encoded on a plasmid and their efficacy relies on the differential stability of the toxin (stable) and antitoxin (unstable). The antitoxin is rapidly degraded, with loss of constitutive expression of the operon as a result of loss of the plasmid, and the toxin is released to act on its target (14). The ccd module is found on the F plasmid with CcdB being the toxin and CcdA the antitoxin (13). Like the quinolones, CcdB has been shown to poison gyrase by stabilizing the cleavage complex (16–18), causing a ‘road-block’ to cellular polymerases (18), leading to double-strand DNA breaks, and induction of the SOS response (19) and ultimately cell death. The resemblance ends there, as there is no cross-resistance between mutants (16,18) and there are differences in the ATP requirement for cleavage complex stabilization (17,18,20), suggesting that CcdB acts by a novel mechanism. A structure was published for CcdB in 1999 (21) and, using the existing structure for the GyrA-NTD (22), a model for CcdB action was proposed (21). The model suggested that a dimeric CcdB binds within the cavity formed by the GyrA dimer, but only when the DNA gate is open, as steric conflicts would prevent the interaction with the gate closed (21). This places the three C-terminal residues of CcdB, which are critical to its toxicity (23), in close proximity to the GyrA Arg462 residue, which, when mutated, has been shown to confer resistance to CcdB (16,17). Resistance is also conferred by mutation of Gly214 of GyrA to Glu (24), which does not lie in the proposed CcdB binding site (21). The observation that CcdB and GyrA inefficiently form their complex in vitro, with a denaturation/renaturation treatment of the proteins being required to form the complex (25), supported the model that CcdB only bound to a transiently-revealed conformation of GyrA. A structure of CcdB with a fragment of GyrA (GyrA14; a fragment containing the bottom gate of GyrA-NTD) was published recently showing a globally symmetric complex, with an asymmetric centre where the GyrA Arg462 residue stacks between the Trp99 residues of the CcdB monomers (26). This supports the previous model (21), which suggests that a conformational change involving DNA gate opening is required for CcdB binding. However, other work has indicated that CcdB and gyrase can bind efficiently without a conformational change (27). Although alluded to, it has never been shown whether CcdB can inhibit the catalytic reactions of gyrase. To accommodate these observations two complexes between CcdB and GyrA have been proposed (27).
In this paper, we have investigated the action of CcdB against gyrase using biochemical assays. We show that CcdB action requires a conformation of gyrase that allows strand passage and that stabilization of the gyrase cleavage complex by CcdB leads to inhibition of the catalytic reactions of gyrase. The role of the GyrA Arg462 residue in CcdB binding has been further investigated by site-directed mutagenesis. This work supports the structural data and a model for a single complex in CcdB action, and provides further information for the rational design of novel antimicrobials to target DNA gyrase.
MATERIALS AND METHODS
Proteins
DNA gyrase subunits GyrA and GyrB were produced as described previously (28) with some material being a kind gift from Mrs A. J. Howells (John Innes Enterprises Limited, Norwich). Site-directed mutagenesis was performed using the QuickChange kit (Stratagene) and plasmid pPH3 (29) as a template. The mutations were confirmed by sequencing (JIC Genome Laboratory). Topoisomerase IV (topo IV) was produced as described previously (30) and was supplied by John Innes Enterprises. Topoisomerase II (topo II) was produced as described previously (31) and was a kind gift from Dr Melisa Wall (JIC). CcdB was purified from strain B462 (pULB2250), which has the ccdB gene under the tight control of the tac promoter, using the published protocol as a guide (32), with some modifications. Overnight cultures were diluted 30-fold into Terrific Broth plus ampicillin (100 μg/ml) and grown at 30°C. After 4 h, over-expression was induced by addition of isopropylthio-β-d-galactoside to 0.5 mM. After a further 3 h at 30°C, cells were harvested by centrifugation. Cell pellets were resuspended in 5 ml of 50 mM Tris–HCl (pH 7.8), 1 mM EDTA, 1 mM DTT, 150 mM NaCl and 10% glycerol. The cells were disrupted using a French press and the cell debris was pelleted by centrifugation. The supernatant was dialysed against 50 mM Tris–HCl (pH 8.0) and 150 mM NaCl. The protein sample was applied to a HiPrep 26/60 Sephacryl S-200 High Resolution gel filtration column (Pharmacia) at a flow rate of 1 ml/min with the same buffer. Fractions containing CcdB were pooled and dialysed against 20 mM Tris–HCl (pH 8.0). The dialysed sample was then applied to a MonoQ High Resolution 5/5 anion exchange column (Pharmacia) at a flow rate of 1 ml/min with the same buffer. Proteins were eluted with a 0–0.5 M NaCl linear gradient; CcdB eluted at ∼0.2 M NaCl. Fractions containing CcdB were pooled and dialysed against 25 mM MOPS, at pH 7.0. The sample was applied to an XK 16/20 column packed with CM Sepharose Fast Flow cation exchange media (Pharmacia), at a flow rate of 2 ml/min with the same buffer and eluted with a 0.1–0.3 M NaCl linear gradient. CcdB was eluted at ∼0.16 M NaCl. Fractions containing CcdB were pooled and dialysed against Enzyme Buffer [50 mM Tris–HCl (pH 7.5), 100 mM KCl, 2 mM DTT, 1 mM EDTA and 10% glycerol]. CcdB was judged to be >95% pure by SDS–PAGE with a typical yield of 5 mg/l of culture. CcdA was purified from strain SG22623 (pKK223CcdA), which has the ccdA gene under the tight control of the tac promoter, using the published protocol as a guide (33). CcdA was judged to be >95% pure by SDS–PAGE with a typical yield of 2 mg/l of culture. The CcdB and CcdA over-producing strains and plasmids were gifts from Dr L. Van Melderen (Brussels, Belgium).
DNA and drugs
Negatively supercoiled and relaxed pBR322 plasmid DNA were kind gifts from Mrs A. J. Howells (John Innes Enterprises). Linear pBR322 was generated by digesting negatively supercoiled DNA with EcoRI (New England BioLabs), followed by phenol extraction and ethanol precipitation. Positively supercoiled pBR322 was generated by incubating relaxed pBR322 with a large excess of DNA gyrase under relaxation conditions (no ATP): 35 mM Tris–HCl (pH 7.5), 24 mM KCl, 4 mM MgCl2, 5 mM DTT, 1.8 mM spermidine, 6.5% (v/v) glycerol, 0.36 mg/ml acetylated BSA for 3 h at 37°C, followed by phenol extraction and ethanol precipitation. CFX (Bayer) was dissolved in water.
Enzyme assays
DNA gyrase supercoiling assays were carried out as described previously (34), except that reactions were incubated at 25 or 37°C for various times up to 4 h and contained 3.5 nM relaxed pBR322, 1.5–20 nM DNA gyrase and 0–12.5 μM CFX or CcdB, as indicated. DNA gyrase relaxation assays were carried as described previously (35), except that reactions were incubated at 25 or 37°C for various times up to 4 h and contained 3.5 nM negatively supercoiled pBR322, 20–200 nM DNA gyrase and 0–12.5 μM CFX or CcdB, as indicated. A novel high-throughput microtitre plate-based fluorescence assay was carried out as described previously (36). Briefly, a triplex-forming oligonucleotide was immobilized onto the wells of a streptavidin-coated microplate. Supercoiling assays (as described above) were carried out in the wells using plasmid pNO1 as substrate (37). Subsequent incubation at pH 5.0 enabled triplex formation between the oligonucleotide and supercoiled pNO1. Unbound plasmid DNA was removed by washing and the wells stained with SYBR Gold (Invitrogen) and the fluorescence read using a SPECTRAmax Gemini fluorimeter. Topo IV and topo II relaxation assays were carried as described previously (37,38). Drug-induced DNA cleavage assays with gyrase were carried out as detailed above, except for post treatment with SDS (0.2%) and proteinase K (1 mg/ml) at 37°C for 30 min. Reversibility of CcdB-stabilized cleavage complex assays were carried out as per gyrase supercoiling assays, except that CFX and CcdB were at 2 μM and there was post treatment with either water, EDTA (10, 50 and 100 mM), NaCl (0.1, 0.5 and 1 M) or CcdA (1, 2 and 5 μM), with duplicates of each incubated at 37°C and 80°C for 30 min, followed by a further treatment with SDS (0.2%) and proteinase K (1 mg/ml) at 37°C for 30 min. With the exception of the fluorescence assay, the DNA products of all assays were analysed by electrophoresis in 1% agarose gels. Ethidium bromide (1 μg/ml) was sometimes added to the gel and running buffer, as indicated. Where required, the fraction of DNA distributed between the various topological forms was quantified using GeneTools software (Syngene). Where appropriate, data were fitted assuming simple 1:1 binding between ligand (CFX or CcdB) and gyrase, i.e. we did not presume a particular mechanism of binding. In one instance (Figure 5B) data were fitted to sigmoidal curves, i.e. indicating >1 binding site and cooperativity.
RESULTS
Nucleotide-dependence of CcdB-stabilized cleavage complexes of DNA gyrase
Previous work had reported a nucleotide-dependence for CcdB stabilization of gyrase cleavage complexes (17,18). With linear DNA as substrate, CcdB stabilization of cleavage complexes is ATP-dependent; with negatively supercoiled DNA, the presence of at least the non-hydrolysable analogue of ATP, ADPNP (5′-adenylyl β,γ-imidodiphosphate), is required (18). The nucleotide requirement for stabilization of gyrase cleavage complexes with two other substrates, relaxed DNA and positively supercoiled DNA, had not been investigated.
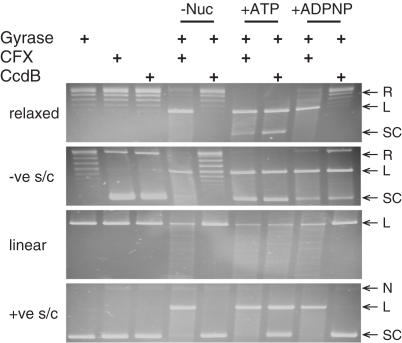
CcdB's nucleotide-dependence for stabilizing gyrase cleavage complexes was investigated by adding CcdB to a fixed concentration of the enzyme with linear, negatively supercoiled, positively supercoiled or relaxed DNA as substrates, each in either the absence or presence of ATP or ADPNP; CFX was used as a control. As reported previously (17,18), CcdB required ATP with a linear DNA substrate and at least ADPNP with a negatively supercoiled DNA substrate (Figure 1). Relaxed and positively supercoiled DNA substrates also showed ATP-dependence for stabilization of the cleavage complex (Figure 1). CcdB would appear to require nucleotide in order to stabilize the DNA gyrase cleavage complex, under any conditions (but see below).
Figure 1.
Nucleotide-dependence of CcdB-stabilized, DNA gyrase-mediated cleavage of DNA. Relaxed, negatively supercoiled (−ve s/c), linear or positively supercoiled (+ve s/c) pBR322 (3.5 nM) was incubated with gyrase (30 nM), either with no nucleotide, ATP (1.4 mM) or ADPNP (1.4 mM) and either CFX (13.5 μM) or CcdB (3.6 μM), as indicated, for 1 h at 37°C. Cleavage complexes were revealed by the addition of SDS and proteinase K and incubation at 37°C for 30 min. DNA was subjected to phenol extraction and analysed on 1% agarose gels. L, linear; N, nicked; R, relaxed; SC, supercoiled.
Estimation of IC50 value for CcdB in comparison to CFX
Previous work, and that detailed in the previous section, has highlighted significant differences in the action of CcdB in comparison to quinolones (17,18). We therefore assessed the relative potency of the inhibitors by estimating a value for the IC50 (inhibitor concentration required to see a 50% reduction in activity) for cleavage complex stabilization.
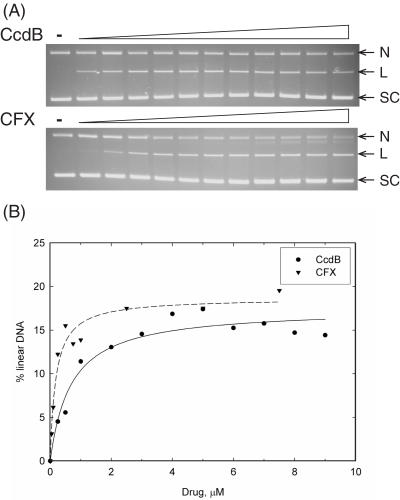
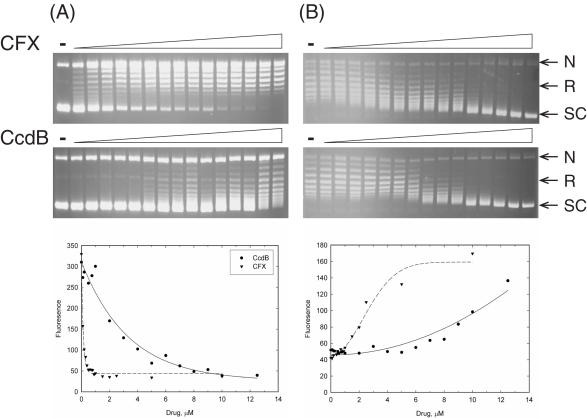
CcdB's ability to stabilize the gyrase cleavage complex was investigated by titrating CcdB into a fixed concentration of gyrase and DNA. The linear DNA band was estimated as a % of total DNA and the data plotted and fitted to a 1:1 ligand-binding curve (Figure 2). IC50s were estimated to be ∼190 nM and ∼680 nM for CFX and CcdB, respectively. A further point to note is that the maximum level of linear product for CcdB was lower than for CFX (Figure 2). Thus CcdB is not as potent an inhibitor of gyrase as CFX and stabilizes a lower proportion of the enzyme in the cleavage complex (see Discussion).
Figure 2.
Estimation of IC50s for CcdB stabilization of the gyrase cleavage complex. Relaxed pBR322 (3.5 nM) was incubated with gyrase (30 nM), ATP (1.4 mM) and various concentrations of CFX (0, 0.01, 0.05, 0.1, 0.25, 0.5, 0.75, 1, 2.5, 5, 7.5 and 10 μM) or CcdB (0, 0.25, 0.5, 1, 2, 3, 4, 5, 6, 7, 8 and 9 μM) as indicated, for 1 h at 37°C. Cleavage complexes were revealed by the addition of SDS and proteinase K and incubation at 37°C for 30 min. DNA was subjected to phenol extraction and analysed on (A) 1% agarose gels run in the presence of ethidium bromide (1 μg/ml). (B) The % of linear DNA was quantified, data plotted and data fitted to single rectangular hyperbolae.
Inhibition of DNA gyrase-catalysed supercoiling and relaxation of DNA by CcdB
Previous work has shown that CcdB acts to poison gyrase by stabilizing the cleavage complex (17,18). Although it has been alluded to (27), data have never been published to show whether CcdB can inhibit the catalytic reactions of gyrase. The model for CcdB action involves it binding in the GyrA cavity when the DNA gate is open and acting as a wedge (21,26). If this is the case, how a T segment can bypass this block is unclear; a model where there are two distinct CcdB–GyrA complexes has also been proposed (27). To further examine this question, the supercoiling and relaxation reactions of gyrase were challenged with CcdB.
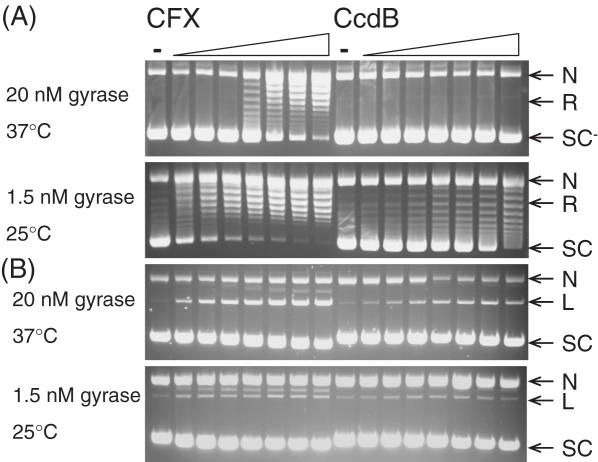
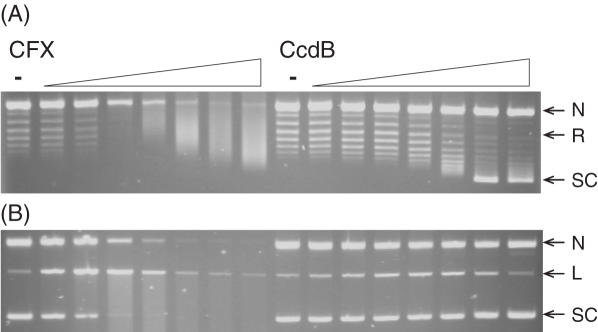
CcdB's ability to inhibit the supercoiling and relaxation reactions of gyrase was investigated by titrating CcdB into various concentrations of gyrase, with incubation at different temperatures. In standard supercoiling assays at 37°C with 20 nM gyrase, a relaxed DNA substrate is completely negatively supercoiled in 1 h; CFX inhibited this reaction (Figure 3A). CcdB did not apparently inhibit this reaction (Figure 3A) but did stabilize cleavage complex formation (Figure 3B). The concentration of gyrase was reduced to 1.5 nM and the reactions incubated at 25°C, in order to slow down the reaction. Under these conditions the relaxed DNA substrate was fully negatively supercoiled in 4 h, and both CFX and CcdB showed inhibition of the catalytic supercoiling reaction of gyrase (Figure 3A). Control experiments showed that this effect was not a consequence of the reaction temperature but depended on the rate of the reaction (data not shown). At these concentrations of gyrase, there are so few CcdB-stabilized cleavage complexes that it is difficult to visualize them by ethidium staining (Figure 3B). Subsequent detection using Southern blotting showed that CcdB increased the intensity of the cleaved linear band (data not shown). A fluorescence-based microplate assay (36) was used to better estimate the IC50s for supercoiling inhibition by the two inhibitors (see Figure 5): ∼135 nM and ∼2.98 μM, for CFX and CcdB, respectively.
Figure 3.
CcdB can inhibit catalytic supercoiling of DNA by gyrase. Relaxed pBR322 (3.5 nM) was incubated with gyrase (1.5 or 20 nM), ATP (1.4 mM) and various concentrations of CFX and CcdB (0, 0.1, 0.2, 0.5, 1, 2, 5 and 10 μM) as indicated, for 1 h at 37°C (20 nM gyrase) or 4 h at 25°C (1.5 nM gyrase). Assays were either (A) stopped or (B) cleavage complexes were revealed by the addition of SDS and proteinase K and incubation at 37°C for 30 min. DNA was subjected to phenol extraction and analysed on 1% agarose gels run in the (A) absence or (B) presence of ethidium bromide (1 μg/ml).
In standard relaxation assays at 37°C with 20 nM gyrase, a negatively supercoiled DNA substrate is completely relaxed in 1 h. The concentration of gyrase was maintained but the incubation temperature was reduced to 25°C, which meant full relaxation took ∼4 h (Figure 4A). Both CFX and CcdB completely inhibited this reaction (Figure 4A) by the stabilization of cleavage complexes (Figure 4B). The fluorescence-based plate assay was used to obtain IC50s for inhibition of relaxation under these conditions (Figure 5B): ∼2.7 μM and ∼10.8 μM for CFX and CcdB, respectively. Note that data for inhibition of supercoiling were fitted to single exponentials (Figure 5A) and data for inhibition of relaxation to sigmoidal curves (Figure 5B). It is known that two molecules of CFX are likely to bind to the gyrase–DNA complex (39) and that this binding is cooperative; this cooperativity is apparent in Figure 5B but may be masked in Figure 5A due to the steepness of the curve. CcdB is thought to bind as a dimer to gyrase (26), although no cooperativity has been previously reported (27); it is not clear why the CcdB curve in Figure 5B shows a greater than first-order dependence on CcdB concentration.
Figure 4.
CcdB can inhibit catalytic relaxation of DNA by gyrase. Negatively supercoiled pBR322 (3.5 nM) was incubated with gyrase (20 nM) and various concentrations of CFX and CcdB (0, 0.1, 0.2, 0.5, 1, 2, 5 and 10 μM) as indicated, for 4 h at 25°C. Assays were either (A) stopped or (B) cleavage complexes were revealed by the addition of SDS and proteinase K and incubation at 37°C for 30 min. DNA was subjected to phenol extraction and analysed on 1% agarose gels run in the (A) absence or (B) presence of ethidium bromide (1 μg/ml).
Figure 5.
Estimation of IC50s for CcdB inhibition of the catalytic reactions of DNA gyrase. Relaxed (A) or negatively supercoiled (B) pN01 (3.5 nM) was incubated with gyrase [1.5 nM (A); 20 nM (B)], ATP [1.4 mM (A); 0 mM (B)] and various concentrations of CFX (0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1, 1.5, 2, 2.5, 5 and 10 μM) or CcdB (0, 0.1, 0.2, 0.5, 0.75, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 and 12.5 μM) as indicated, for 4 h at 25°C. DNA was subjected to phenol extraction and analysed on 1% agarose gels, or triplex formation was quantitatively analysed by SYBR fluorescence and data plotted (36). Data were fitted with (A) single exponential decay curves or (B) sigmoidal curves.
For the first time CcdB has been shown to inhibit both of the catalytic reactions of DNA gyrase via stabilization of the cleavage complex. Relaxation of negatively supercoiled DNA by DNA gyrase is an ATP-independent reaction and thus also indicates, in contrast to previous observations, that CcdB does not have an absolute requirement for ATP to exert its action.
The role of the GyrA-CTD in CcdB's action
Previous work had reported that the 35 kDa GyrA-CTD, the DNA-wrapping domain, was required for the action of CcdB (18). In this work, linear DNA was used as a substrate and, as such, the wrapping domain is essential to provide a node that will facilitate opening of the DNA gate. Gyrase with a truncated GyrA (A592B2) can still perform relaxation of negatively supercoiled DNA in both an ATP-independent (40) and ATP-dependent manner (41) manner. In both these situations, a node can be formed by the DNA itself, without the active help of the DNA-wrapping domain, thus allowing strand passage. To explore the role of the GyrA-CTD, the effect of CcdB on the reactions of the A592B2 complex was investigated.
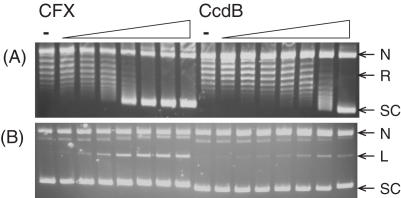
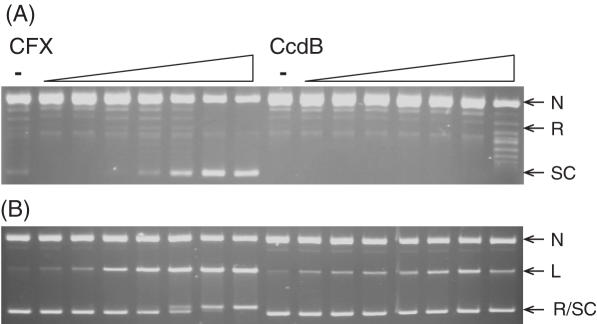
CcdB's ability to stabilize the A592B2 cleavage complex and inhibit the relaxation of negatively supercoiled DNA by the A592B2 enzyme was investigated by titrating CcdB into a fixed concentration of A592B2 and DNA. CcdB stabilized the A592B2 cleavage complex and inhibited the relaxation of negatively supercoiled DNA by the enzyme (Figure 6). This inhibition via cleavage complex stabilization by CcdB was observed both in the presence (Figure 6) and the absence of ATP (data not shown). (The streaking observed in panel A in the presence of CFX is a common feature of experiments involving GyrA59 and is likely to be associated with its DNA-binding properties.) It should be noted that the concentration of enzyme in these assays was 100 nM as the reactions are inefficient; hence, it is possible to see inhibition of the catalytic reaction under these conditions. Therefore CcdB does not require the GyrA-CTD for its action.
Figure 6.
CcdB can inhibit the ATP-dependent relaxation of DNA by an A592B2 gyrase complex. Negatively supercoiled pBR322 (3.5 nM) was incubated with A592B2 (100 nM), ATP (1.4 mM) and various concentrations of CFX and CcdB (0, 0.1, 0.2, 0.5, 1, 2, 5 and 10 μM) as indicated, for 1 h at 37°C. Assays were either (A) stopped or (B) cleavage complexes were revealed by the addition of SDS and proteinase K and incubation at 37°C for 30 min. DNA was subjected to phenol extraction and analysed on 1% agarose gels run in the (A) absence or (B) presence of ethidium bromide (1 μg/ml).
The role of the GyrB-NTD in CcdB's action
Previous work had reported that ATP was a crucial element in CcdBs action (17,18,20). With the observations detailed here showing that CcdB inhibits the ATP-independent relaxation reaction of gyrase, the requirement for ATP to allow CcdB action is called into question. Gyrase with the GyrB ATPase domain truncated (A2B472) can still relax negatively supercoiled DNA in an ATP-independent reaction (42,43). In this situation, a node can be formed by the DNA itself and opening of the DNA gate can occur without the ATPase domain. To investigate the importance of the GyrB-NTD in CcdB's action, reactions were carried out using A2B472.
CcdB's ability to stabilize the A2B472 cleavage complex and inhibit the relaxation of negatively supercoiled DNA was investigated by titrating CcdB into a fixed concentration of the A2B472 enzyme and DNA. CcdB-stabilized the A2B472 cleavage complex and inhibited the relaxation of negatively supercoiled DNA by A2B472 (Figure 7). Thus, CcdB does not require the GyrB-NTD, and thus ATP is not an absolute requirement for CcdB's action.
Figure 7.
CcdB can inhibit the ATP-independent relaxation of DNA by an A2B472 gyrase complex. Negatively supercoiled pBR322 (3.5 nM) was incubated with A2B472 complex (200 nM) and various concentrations of CFX and CcdB (0, 0.1, 0.2, 0.5, 1, 2, 5 and 10 μM) as indicated, for 2 h at 37°C. Assays were either (A) stopped or (B) cleavage complexes were revealed by the addition of SDS and proteinase K and incubation at 37°C for 30 min. DNA was subjected to phenol extraction and analysed on 1% agarose gels run in the (A) absence or (B) presence of ethidium bromide (1 μg/ml).
CcdB-stabilized cleavage complexes are irreversible without the presence of CcdA
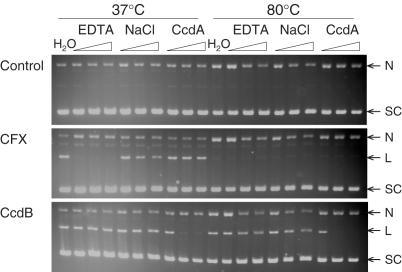
It is well known that CcdB-stabilized gyrase cleavage complexes are reversible by CcdA (17,18). However, the action of other agents, such as quinolones, can be reversed by heat and EDTA treatments (44,45). Therefore, the ability of various treatments to reverse CcdB-stabilized cleavage complexes has been assessed.
Following CcdB stabilization of the cleavage complex, assays where treated with various concentration of EDTA, NaCl or CcdA and incubated at either 37 or 80°C to investigate the requirements to reverse CcdB-stabilized cleavage complexes. CcdB-stabilized cleavage complexes were only reversed by the addition of equimolar, or greater, concentrations of CcdA (Figure 8); EDTA, NaCl and heat treatment had no effect (Figure 8). Cleavage stabilized with CFX was reversed by EDTA and heat treatment (Figure 8). Therefore CcdB-stabilized cleavage complexes are more stable than CFX-stabilized cleavage complexes and can only be reversed by CcdA treatment.
Figure 8.
CcdB-stabilized DNA gyrase cleavage complexes are only reversed by CcdA. Relaxed pBR322 (3.5 nM) was incubated with gyrase (20 nM), ATP (1.4 mM) and either no inhibitor (Control), CFX (2 μM) or CcdB (2 μM) for 1 h at 37°C. Assays were treated with either water, EDTA (10, 50 and 100 mM), NaCl (0.1, 0.5 and 1 M) or CcdA (1, 2 and 5 μM), with repeats of each incubated at 37°C or 80°C for a further 30 min. Cleavage complexes were revealed by the addition of SDS and proteinase K and incubation at 37°C for 30 min. DNA was subjected to phenol extraction and analysed on 1% agarose gels run in the presence of ethidium bromide (1 μg/ml).
Site-directed mutagenesis of GyrA Arg462 and Gly214 and biochemical analysis of their activity
Two mutations in GyrA have been isolated that confer resistance to CcdB: GyrA Arg462Cys (16,17) and Gly214Glu (24). The role of Arg462 is now well-established (26) and biochemical characterization of the protein has been performed indicating that CcdB is unable to bind to the mutant protein (17,27). Still the curiosity remains that the same mutation has been isolated a number of times, with the Arg always being substituted to Cys (16,17). No biochemical characterization of the GyrA Gly214Glu protein has been performed and the basis of conferring resistance is unknown. Therefore we have made GyrA proteins bearing mutations at these positions (Arg462 to Cys, Ser and Ala, and Gly214 to Glu and Ala) and analysed their properties.
The full-length gyrA gene, in plasmid pPH3 (29), was subjected to site-directed mutagenesis. Wild-type and mutant GyrA proteins were over-expressed and purified, with the exception of the Gly214Glu protein, which over-expressed poorly and did not bind to the purification columns. This protein was assumed to be mis-folded. We found two populations of GyrA: one (minor) fraction, which bound to the columns and behaved like the wild-type enzyme in assays; the other (major) fraction failed to bind to columns and could not be purified, suggesting that this was a mis-folded mutant protein.
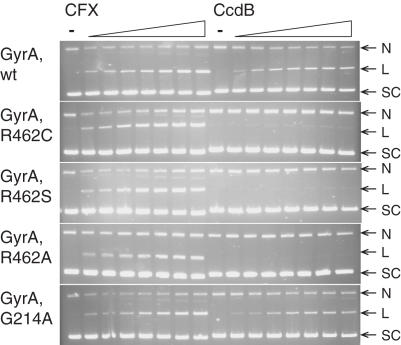
Reconstituted gyrase with the purified mutant GyrA proteins showed comparable supercoiling activity to wild-type GyrA (data not shown). CcdB was able to stabilize cleavage complexes with the wild-type and Gly214Ala enzymes (Figure 9); the Arg462Cys, Arg462Ser and Arg462Ala enzymes were refractory to CcdB (Figure 9). CcdB's effect on the supercoiling reaction mirrored that of its stabilization of the cleavage complex with only the wild-type and Gly214Ala enzymes being inhibited by CcdB (data not shown). These data therefore show that GyrA Arg462 is critical for stabilizing the CcdB–GyrA complex and that substitutions other than Cys at 462 (e.g. Ser and Ala) also result in CcdB-resistant GyrA. These data also suggest that Gly214 is not directly involved in CcdB–GyrA interaction.
Figure 9.
The effect of GyrA mutations on stabilization of the cleavage complex by CcdB. Relaxed pBR322 (3.5 nM) was incubated with various gyrase enzymes (wild-type or mutants, 20 nM), ATP (1.4 mM) and various concentrations of CFX and CcdB (0, 0.1, 0.2, 0.5, 1, 2, 5 and 10 μM), for 1 h at 37°C, as indicated. Cleavage complexes were revealed by the addition of SDS and proteinase K and incubation at 37°C for 30 min. DNA was subjected to phenol extraction and analysed on 1% agarose gels run in the presence of ethidium bromide (1 μg/ml).
DISCUSSION
DNA gyrase is a well-validated drug target in antimicrobial therapies. As with all drugs, the development of resistance is a problem that needs to be overcome and thus work to maintain the usefulness of such drugs and targets is of paramount importance. Bacteria themselves are excellent providers of antibiotics and, although clinically the most successful agents that target gyrase are synthetic (i.e. quinolones), there are many naturally-produced toxins that target gyrase. One of these, CcdB, is designed to target E.coli gyrase and kill the cell for the means of selfish maintenance of plasmid DNA, but its action is not yet fully understood. In this paper, we have investigated the mode of action of CcdB and have rationalized the behaviour of this inhibitor in terms of a model for its action on gyrase. Type IIA topoisomerases share high levels of sequence homology (46) and gyrase and topo IV are targeted by the same classes of drugs (4,47,48). We have therefore also challenged topo IV from E.coli and topo II from Saccharomyces cerevisiae with CcdB in relaxation and cleavage assays. With topo II we found no evidence of inhibitory activity; with topo IV we found inhibition of relaxation only at the highest concentration of CcdB tested (10 μM) with no evidence of cleavage complex formation (data not shown).
Quinolones and other gyrase-specific agents, such as the bacterial toxins CcdB and MccB17, inhibit gyrase by stabilizing the covalent complex between the enzyme and cleaved DNA. Previous work has suggested that CcdB shows significant differences in its action when compared with quinolones. For example, CcdB action was thought to be ATP- (or ADPNP-) dependent and require >160 bp of DNA (17,18). Also CcdB was not known to inhibit the catalytic reactions of gyrase. To account for these observations it was suggested that two modes of CcdB binding to gyrase occur (20,27): a complex with GyrA that is non-inhibitory, and a complex with gyrase (A2B2) that stabilizes the cleavage complex.
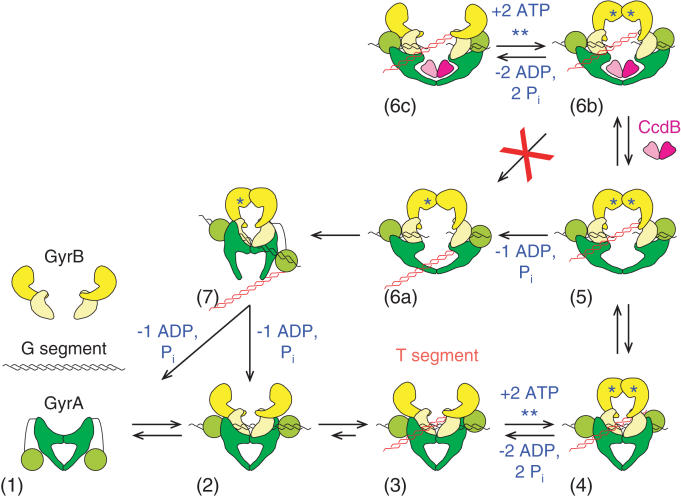
In this paper, we have shown that ATP (or ADPNP) is not required for CcdB action and, under appropriate conditions, inhibition of gyrase-catalysed supercoiling and relaxation can be detected. Furthermore, we found that neither the GyrA-CTD (DNA-wrapping domain) nor the GyrB-NTD (ATPase domain) is required. Taken together we propose that CcdB's action on gyrase can be explained by the model shown in Figure 10. Here the CcdB dimer binds within the GyrA cavity only when the DNA gate is open, thus forming a complex in which the DNA gate, and hence the broken G segment, is wedged open; several cycles of gyrase action may be required prior to CcdB binding. This complex (6b in Figure 10) can only form during strand passage (supercoiling or relaxation reactions) and is inefficiently formed, relying on a transiently-revealed conformation of the enzyme. The effect of CcdB binding is to stabilize the cleavage complex thus preventing strand passage (and hence supercoiling/relaxation). This complex can still undergo ATP hydrolysis cycles, albeit at a reduced rate (27). This complex is extremely stable (27) and we have found that it can only be reversed by the addition of CcdA (Figure 8). Such a mode of action is entirely consistent with the recently-published crystal structure of the complex between CcdB and GyrA14 (26). The apparent requirement for nucleotide in CcdB action reported from earlier work stems from the fact that the conformation of gyrase required for CcdB binding occurs much more efficiently in the presence of nucleotide (i.e. as part of the supercoiling cycle; Figure 10) than in its absence. However, this conformation can still be accessed in the absence of nucleotide when the enzyme is relaxing negatively supercoiled DNA.
Figure 10.
Model for CcdB action. The individual components (1) free in solution (7,50) come together (2) to form the holoenzyme with the G segment binding across the DNA gate (8). (3) The DNA is wrapped by the GyrA-CTD to present the T segment over the G segment and a positive node is formed. (4) Upon ATP binding, the GyrB monomers dimerise and (5) the G segment is cleaved and the DNA gate opens. Either (6a) ‘top-down’ passage of the T segment occurs upon hydrolysis of a single ATP or (6b) CcdB accesses its binding site to stabilize the cleavage complex. (7) The DNA gate closes, the G segment is religated and the T segment passes out through the bottom gate, with hydrolysis of the second ATP resetting the enzyme. (6c) A futile ATP hydrolysis/ATP binding cycle can occur as per Kampranis et al. (27). Several cycles of (5) to (6a) transition may occur prior to CcdB binding in a (5) to (6b) transition.
To test the proposed mode of interaction of CcdB with GyrA, we made mutations at residues in GyrA known to confer CcdB resistance (Arg462 and Gly214). We found that substitutions at GyrA Arg462 rendered the enzyme CcdB-resistant, consistent with its role in the interaction with Trp99 of CcdB (26). Substitution of Ala at Gly214 had no effect on GyrA; substitution of Glu produced an unstable protein that we could not purify. We therefore suggest that the Gly214Glu mutation affects GyrA folding and, in vivo, the protein is active but very inefficient at forming complexes with CcdB.
Complexes between CcdB and GyrA alone have been reported previously; this is surprising given that GyrA is a very stable dimer (49); however, these data can be rationalized in relation to the model in Figure 10. Maki et al. (50) found that cells over-producing CcdB had <1% of the supercoiling activity seen in normal cells, but that extracts from these cells could not inhibit purified gyrase. Further, Maki et al. (51) have shown that such supercoiling-deficient gyrase–CcdB complexes can be rescued in vitro by CcdA. These results can be explained by CcdB binding to GyrA as it is being translated and before it has a chance to dimerize; addition of CcdB to purified gyrase results in inefficient inhibition of supercoiling, as we have observed. Bahassi et al. (25) found that the inactive GyrA–CcdB complex could be reconstituted if GyrA was denatured and renatured: such treatment would dissociate the GyrA dimer and allow access of CcdB to its binding site upon refolding. Kampranis et al. (27) showed, using surface-plasmon resonance, that CcdB could bind to GyrA (or its N-terminal domain, GyrA59) immobilized on a Biacore chip (Kds∼10−10 M). Under these conditions, it is likely that at least some of the GyrA protein could be immobilized as a monomer or as an open dimer thus exposing the CcdB binding surface. Thus the reports of two complexes between CcdB and gyrase are likely to only reflect the possibility of CcdB binding to monomeric GyrA and to a conformation of gyrase revealed during the topoisomerase cycle; these are likely to be the same complex, i.e. that shown in Figure 10.
Two further features of CcdB–gyrase interaction deserve comment. It has been reported that CcdB interaction requires >160 bp of DNA to interact with gyrase (18). Referring to Figure 10, this can be simply explained by the need to have DNA long enough to form G and T segments and thus trigger the conformational changes required for strand passage that allow CcdB binding. It has also been shown that ADPNP can promote CcdB-stabilized cleavage only with negatively supercoiled DNA (18). However, in this paper we have shown that CcdB action is nucleotide-independent, providing that strand passage is able to occur (Figure 4). We suggest that the earlier observations are a consequence of the ‘bottom-up’ strand passage that occurs during relaxation of negatively supercoiled DNA by gyrase, generating intermediate 5 from 7 in Figure 10. The presence of ADPNP stabilizes this intermediate by closing the ATP-operated clamp. Under these conditions the T segment is in the upper part of the gyrase cavity, clear of the CcdB binding site permitting binding of the toxin. With other substrates (e.g. positively supercoiled DNA) ADPNP is more likely to trap intermediate 6a, where the T segment may occlude the CcdB binding site.
The mode of action of CcdB suggested in Figure 10 is quite unlike that of quinolones, which are thought to bind to a pocket involving GyrA, GyrB and DNA, prior to DNA cleavage and without the requirement of DNA-wrapping [Figure 10, step 2; (52)]. There is currently no structural information on the binding of MccB17 to gyrase, but it is thought that this toxin also binds during DNA strand passage by gyrase in a complex involving the GyrB-CTD (11,12). The action of CcdB on gyrase shares a number of similarities with that of MccB17: both toxins appear to bind to a transient intermediate that is revealed during strand passage and are rather inefficient inhibitors of the enzyme allowing several strand passage cycles before a substantial amount of cleavage complex is trapped. Whereas the MccB17 binding site involves GyrB, GyrA would appear to be the exclusive target for CcdB. Despite its comparative inefficiency of action, CcdB is an effective toxin that is essentially irreversible in its action; small molecule inhibitors based on CcdB would be promising candidates for novel drugs.
Acknowledgments
A.B.S. was funded by a BBSRC-CASE studentship, supported by the BBSRC and GlaxoSmithKline. The authors thank Lionel Costenaro, Remy Loris and Olivier Pierrat for comments on the manuscript. Funding to pay the Open Access publication charges for this article was provided by BBSRC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Howard D.H. Resistance-induced antibiotic substitution. Health Econ. 2004;13:585–595. doi: 10.1002/hec.856. [DOI] [PubMed] [Google Scholar]
- 2.Maxwell A. DNA gyrase as a drug target. Biochem. Soc. Trans. 1999;27:48–53. doi: 10.1042/bst0270048. [DOI] [PubMed] [Google Scholar]
- 3.Hande K.R. Clinical applications of anticancer drugs targeted to topoisomerase II. Biochim. Biophys. Acta. 1998;1400:173–184. doi: 10.1016/s0167-4781(98)00134-1. [DOI] [PubMed] [Google Scholar]
- 4.Pommier Y., Pourquier P., Fan Y., Strumberg D. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim. Biophys. Acta. 1998;1400:83–105. doi: 10.1016/s0167-4781(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 5.Maxwell A., Lawson D.M. The ATP-binding site of type II topoisomerases as a target for antibacterial drugs. Curr. Top. Med. Chem. 2003;3:283–303. doi: 10.2174/1568026033452500. [DOI] [PubMed] [Google Scholar]
- 6.Corbett K.D., Berger J.M. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 7.Chatterji M., Unniram S., Maxwell A., Nagaraja V. The additional 165 amino acids in the B protein of Escherichia coli DNA gyrase have an important role in DNA binding. J. Biol. Chem. 2000;275:22888–22894. doi: 10.1074/jbc.M001047200. [DOI] [PubMed] [Google Scholar]
- 8.Heddle J.G., Mitelheiser S., Maxwell A., Thomson N.H. Nucleotide binding to DNA gyrase causes loss of DNA wrap. J. Mol. Biol. 2004;337:597–610. doi: 10.1016/j.jmb.2004.01.049. [DOI] [PubMed] [Google Scholar]
- 9.Drlica K., Malik M. Fluoroquinolones: action and resistance. Curr. Top. Med. Chem. 2003;3:249–282. doi: 10.2174/1568026033452537. [DOI] [PubMed] [Google Scholar]
- 10.Ruiz J. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J. Antimicrob. Chemother. 2003;51:1109–1117. doi: 10.1093/jac/dkg222. [DOI] [PubMed] [Google Scholar]
- 11.Pierrat O.A., Maxwell A. The action of the bacterial toxin microcin B17: insight into the cleavage-religation reaction of DNA gyrase. J. Biol. Chem. 2003;278:35016–35023. doi: 10.1074/jbc.M304516200. [DOI] [PubMed] [Google Scholar]
- 12.Pierrat O.A., Maxwell A. Evidence for the role of DNA strand passage in the mechanism of action of microcin B17 on DNA gyrase. Biochemistry. 2005;44:4204–4215. doi: 10.1021/bi0478751. [DOI] [PubMed] [Google Scholar]
- 13.Ogura T., Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl Acad. Sci. USA. 1983;80:4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science. 2003;301:1496–1499. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- 15.Couturier M., Bahassi E.M., Van Melderen L. Bacterial death by DNA gyrase poisoning. Trends Microbiol. 1998;6:269–275. doi: 10.1016/s0966-842x(98)01311-0. [DOI] [PubMed] [Google Scholar]
- 16.Bernard P., Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of the DNA-topoisomerase II complex. J. Mol. Biol. 1992;226:735–745. doi: 10.1016/0022-2836(92)90629-x. [DOI] [PubMed] [Google Scholar]
- 17.Bernard P., Kézdy K.E., Van Melderen L., Steyaert J., Wyns L., Pato M.L., Higgins N.P., Couturier M. The F plasmid CcdB protein induces efficient ATP-dependent DNA cleavage by gyrase. J. Mol. Biol. 1993;234:534–541. doi: 10.1006/jmbi.1993.1609. [DOI] [PubMed] [Google Scholar]
- 18.Critchlow S.E., O'Dea M.H., Howells A.J., Couturier M., Gellert M., Maxwell A. The interaction of the F-plasmid killer protein, CcdB, with DNA gyrase: induction of DNA cleavage and blocking of transcription. J. Mol. Biol. 1997;273:826–839. doi: 10.1006/jmbi.1997.1357. [DOI] [PubMed] [Google Scholar]
- 19.Karoui H., Bex F., Drèze P., Couturier M. Ham22, a miniF mutation which is lethal to host cell and promotes recA-dependent induction of lambdoid prophage. EMBO J. 1983;2:1863–1868. doi: 10.1002/j.1460-2075.1983.tb01672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheirer K., Higgins N.P. The DNA cleavage reaction of DNA gyrase. Comparison of stable ternary complexes formed with enoxacin and CcdB protein. J. Biol. Chem. 1997;272:27202–27209. doi: 10.1074/jbc.272.43.27202. [DOI] [PubMed] [Google Scholar]
- 21.Loris R., Dao-Thi M.-H., Bahassi E.M., Van Melderen L.V., Poortmans F., Liddington R.C., Couturier M., Wyns L. Crystal structure of CcdB, a topoisomerase poison from E. coli. J. Mol. Biol. 1999;285:1667–1677. doi: 10.1006/jmbi.1998.2395. [DOI] [PubMed] [Google Scholar]
- 22.Morais Cabral J.H., Jackson A.P., Smith C.V., Shikotra N., Maxwell A., Liddington R.C. Structure of the DNA breakage-reunion domain of DNA gyrase. Nature. 1997;388:903–906. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 23.Bahassi E.M., Salmon M.A., Van Melderen L., Bernard P., Couturier M. F plasmid CcdB killer protein: ccdB gene mutants coding for non-cytotoxic proteins which retain their regulatory functions. Mol. Microbiol. 1995;15:1031–1037. doi: 10.1111/j.1365-2958.1995.tb02278.x. [DOI] [PubMed] [Google Scholar]
- 24.Miki T., Park J.A., Nagao K., Murayama N., Horiuchi T. Control of segregation of chromosomal DNA by sex factor F in Escherichia coli. J. Mol. Biol. 1992;225:39–52. doi: 10.1016/0022-2836(92)91024-j. [DOI] [PubMed] [Google Scholar]
- 25.Bahassi E.M., O'Dea M.H., Allali N., Messens J., Gellert M., Couturier M. Interaction of CcdB with DNA gyrase. Inactivation of GyrA, poisoning of the gyrase–DNA complex, and the antidote action of CcdA. J. Biol. Chem. 1999;274:10936–10944. doi: 10.1074/jbc.274.16.10936. [DOI] [PubMed] [Google Scholar]
- 26.Dao-Thi M.H., Van Melderen L., De Genst E., Afif H., Buts L., Wyns L., Loris R. Molecular basis of gyrase poisoning by the addiction toxin CcdB. J. Mol. Biol. 2005;348:1091–1102. doi: 10.1016/j.jmb.2005.03.049. [DOI] [PubMed] [Google Scholar]
- 27.Kampranis S.C., Howells A.J., Maxwell A. The interaction of DNA gyrase with the bacterial toxin CcdB: evidence for the existence of two gyrase–CcdB complexes. J. Mol. Biol. 1999;293:733–744. doi: 10.1006/jmbi.1999.3182. [DOI] [PubMed] [Google Scholar]
- 28.Maxwell A., Howells A.J. Overexpression and purification of bacterial DNA gyrase. In: Bjornsti M.-A., Osheroff N., editors. DNA Topoisomerase Protocols I. DNA Topology and Enzymes. Totowa, New Jersey: Humana Press; 1999. pp. 135–144. [Google Scholar]
- 29.Hallett P., Grimshaw A.J., Wigley D.B., Maxwell A. Cloning of the DNA gyrase genes under tac promoter control: overproduction of the gyrase A and B proteins. Gene. 1990;93:139–142. doi: 10.1016/0378-1119(90)90148-k. [DOI] [PubMed] [Google Scholar]
- 30.Peng H., Marians K.J. Escherichia coli topoisomerase IV. J. Biol. Chem. 1993;268:24481–24490. [PubMed] [Google Scholar]
- 31.Lindsley J.E. Overexpression and purification of Saccharomyces cerevisiae DNA topoisomerase II from yeast. Meth. Mol. Biol. 1999;94:187–197. doi: 10.1385/1-59259-259-7:187. [DOI] [PubMed] [Google Scholar]
- 32.Steyaert J., Van Melderen L., Bernard P., Thi M.H.D., Loris R., Wyns L., Couturier M. Purification, circular dichroism analysis, crystallization and preliminary X-ray diffraction analysis of the F plasmid CcdB killer protein. J. Mol. Biol. 1993;231:513–515. doi: 10.1006/jmbi.1993.1301. [DOI] [PubMed] [Google Scholar]
- 33.Van Melderen L., Dao Thi M.H., Lecchi P., Gottesman S., Couturier M., Maurizi M.R. ATP-dependent degradation of CcdA by Lon protease. J. Biol. Chem. 1996;271:27730–27738. doi: 10.1074/jbc.271.44.27730. [DOI] [PubMed] [Google Scholar]
- 34.Mizuuchi K., Mizuuchi M., O'Dea M.H., Gellert M. Cloning and simplified purification of Escherichia coli DNA gyrase A and B proteins. J. Biol. Chem. 1984;259:9199–9201. [PubMed] [Google Scholar]
- 35.Reece R.J., Maxwell A. Tryptic fragments of the Escherichia coli DNA gyrase A protein. J. Biol. Chem. 1989;264:19648–19653. [PubMed] [Google Scholar]
- 36.Maxwell A., Burton N.P., O'Hagan N. High-throughput assays for DNA gyrase and other topoisomerases. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl504. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hiasa H., Yousef D.O., Marians K.J. DNA strand cleavage is required for replication fork arrest by a frozen topoisomerase-quinolone-DNA ternary complex. J. Biol. Chem. 1996;271:26424–26429. doi: 10.1074/jbc.271.42.26424. [DOI] [PubMed] [Google Scholar]
- 38.Roca J., Berger J.M., Harrison S.C., Wang J.C. DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Proc. Natl Acad. Sci. USA. 1996;93:4057–4062. doi: 10.1073/pnas.93.9.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Critchlow S.E., Maxwell A. DNA cleavage is not required for the binding of quinolone drugs to the DNA gyrase–DNA complex. Biochemistry. 1996;35:7387–7393. doi: 10.1021/bi9603175. [DOI] [PubMed] [Google Scholar]
- 40.Reece R.J., Maxwell A. Probing the limits of the DNA breakage-reunion domain of the Escherichia coli DNA gyrase A protein. J. Biol. Chem. 1991;266:3540–3546. [PubMed] [Google Scholar]
- 41.Kampranis S.C., Maxwell A. Conversion of DNA gyrase into a conventional type II topoisomerase. Proc. Natl Acad. Sci. USA. 1996;93:14416–14421. doi: 10.1073/pnas.93.25.14416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown P.O., Peebles C.L., Cozzarelli N.R. A topoisomerase from Escherichia coli related to DNA gyrase. Proc. Natl Acad. Sci. USA. 1979;76:6110–6114. doi: 10.1073/pnas.76.12.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gellert M., Fisher L.M., O'Dea M.H. DNA gyrase: purification and catalytic properties of a fragment of gyrase B protein. Proc. Natl Acad. Sci. USA. 1979;76:6289–6293. doi: 10.1073/pnas.76.12.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gellert M., Mizuuchi K., O'Dea M.H., Itoh T., Tomizawa J. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc. Natl Acad. Sci. USA. 1977;74:4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugino A., Peebles C.L., Kruezer K.N., Cozzarelli N.R. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl Acad. Sci. USA. 1977;74:4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caron P.R. Appendix: compendium of DNA topoisomerase sequences. In: Bjornsti M.-A., Osheroff N., editors. DNA Topoisomerase Protocols. DNA Topology and Enzymes. Vol. 94. Towata, NJ: Humana Press; 1999. pp. 279–316. [Google Scholar]
- 47.Drlica K., Hooper D.C. Mechanisms of quinolone action. In: Hooper D.C., Rubinstein E., editors. Quinolone Antimicrobial Agents, 3rd edn. Washington DC: ASM Press; 2003. pp. 19–40. [Google Scholar]
- 48.Mitscher L.A. Bacterial topoisomerase inhibitors: quinolone and pyridone antibacterial agents. Chem. Rev. 2005;105:559–592. doi: 10.1021/cr030101q. [DOI] [PubMed] [Google Scholar]
- 49.Costenaro L., Grossmann J.G., Ebel C., Maxwell A. Small-angle X-ray scattering reveals the solution structure of the full-length DNA gyrase A subunit. Structure. 2005;13:287–296. doi: 10.1016/j.str.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 50.Maki S., Takiguchi S., Miki T., Horiuchi T. Modulation of DNA supercoiling activity of Escherichia coli DNA gyrase by F plasmid proteins. J. Biol. Chem. 1992;267:12244–12251. [PubMed] [Google Scholar]
- 51.Maki S., Takiguchi S., Horiuchi T., Sekimizu J., Miki T. Partner switching mechanisms in activation and rejuvenation of Escherichia coli DNA gyrase by F plasmid proteins, LetD (CcdB) and LetA (CcdA) J. Mol. Biol. 1996;256:473–482. doi: 10.1006/jmbi.1996.0102. [DOI] [PubMed] [Google Scholar]
- 52.Heddle J., Maxwell A. Quinolone-binding pocket of DNA gyrase: role of GyrB. Antimicrob. Agents Chemother. 2002;46:1805–1815. doi: 10.1128/AAC.46.6.1805-1815.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]