Abstract
RNA interference and the microRNA (miRNA) pathway can induce sequence-specific mRNA degradation and/or translational repression. The human genome encodes hundreds of miRNAs that can post-transcriptionally repress thousands of genes. Using reporter constructs, we observed that degradation of mRNAs bearing sites imperfectly complementary to the endogenous let-7 miRNA is considerably stronger in human HEK293 than HeLa cells. The degradation did not result from the Ago2-mediated endonucleolytic cleavage but it was Dicer- and Ago2-dependent. We used this feature of HEK293 to address the size of a pool of transcripts regulated by RNA silencing in a single cell type. We generated HEK293 cell lines depleted of Dicer or individual Ago proteins. The cell lines were used for microarray analyses to obtain a comprehensive picture of RNA silencing. The 3′-untranslated region sequences of a few hundred transcripts that were commonly up-regulated upon Ago2 and Dicer knock-downs showed a significant enrichment of putative miRNA-binding sites. The up-regulation upon Ago2 and Dicer knock-downs was moderate and we found no evidence, at the mRNA level, for activation of silenced genes. Taken together, our data suggest that, independent of the effect on translation, miRNAs affect levels of a few hundred mRNAs in HEK293 cells.
INTRODUCTION
Post-transcriptional RNA silencing pathways, the RNA interference (RNAi) and the microRNA (miRNA) pathway, regulate gene expression by inducing degradation and/or translational repression of target mRNAs. These pathways are generally initiated by various forms of double-stranded RNA (dsRNA), which are processed by Dicer, an RNase III family endonuclease, to 21–22 nt long RNA molecules that serve as sequence-specific guides for silencing [reviewed in (1,2)]. RNAi operates in mammalian cells but its role is not well defined. RNAi effects induced by long dsRNA are generally masked by a sequence-independent response that is mediated by the interferon (IFN) and other defense pathways and results in a general translational block and RNA degradation (3,4). The vast majority of mammalian short RNAs known to date are represented by miRNAs [for recent data see (5,6)]. MiRNAs are transcribed as long primary transcripts (pri-miRNAs), which are processed by a nuclear RNase III Drosha-containing complex into short hairpin intermediates (pre-miRNAs). Pre-miRNAs are transported to the cytoplasm where they are further processed by the Dicer-containing complex [reviewed in (7)]. Mammals have only one Dicer protein, which produces both siRNAs and miRNAs (8,9).
Both siRNAs and miRNAs are loaded onto an Argonaute-containing effector ribonucleoprotein (RNP) complex, referred to as miRNP or RISC (RNA-induced silencing complex), which is capable of recognizing cognate mRNAs and inhibiting protein expression. The mammalian Argonaute protein family consists of eight members, four of which are ubiquitously expressed (Ago subfamily) while the remaining four (Piwi subfamily) are expressed in germ cells (10). All four mammalian Ago proteins, Ago1 through Ago4, associate with miRNAs and are implicated in translational repression (11–13). However, only one, Ago2, can mediate specific endonucleolytic cleavage of a target mRNA in the middle of the sequence that base pairs with a short RNA (11,12,14). Whether a short RNA will cause endonucleolytic mRNA degradation via the RNAi mechanism or will act as an miRNA inducing the translational repression depends on the degree of its complementarity with the mRNA target, rather than on the origin of the short RNA. The Ago2-mediated endonucleolytic cleavage requires formation of a perfect or nearly perfect siRNA–mRNA duplex, while imperfect base pairing generally results in translational repression (15,16). The predicted hybrids between animal miRNAs and their cognate mRNAs typically contain bulges and mismatches and result in translational repression. On the other hand, the extensive pairing of miR-196 with HoxB8 mRNA results in the endonucleolytic mRNA cleavage by the RNAi mechanism (17). Importantly, recent findings indicate that miRNAs can induce substantial mRNA degradation even in the absence of extensive base paring to their targets (18,19), and shortening of the poly(A) tail was proposed to be the initial step leading to the miRNA-mediated mRNA destabilization (20,21). Repressed mRNAs, miRNAs and Ago proteins localize to discrete cytoplasmic foci known as P-bodies, likely as a consequence of translational repression (22,23). P-bodies contain mRNA degrading enzymes such as a decapping complex, a deadenylase and the 5′–3′ exonuclease XRN1 [reviewed in (24)], and it is conceivable that the observed degradation of some miRNA targets is a consequence of their relocation to these structures [reviewed in (25,26)].
Numerous miRNAs have been identified in different species. The miRNA database (27) currently contains 462 human miRNAs (release 8.2) but some computational studies estimate that the number of miRNAs operating in humans is as much as 2- to 4-fold higher (28). MiRNAs are implicated in the regulation of many cellular processes and changes in their expression are observed in various diseases [reviewed in (29–31)]. However, the function of most of the human miRNAs remains unknown. Similarly, it is not known how many genes are regulated by miRNAs in humans. Profiling of mRNAs in Drosophila S2 cells depleted of Drosha or AGO1, revealed up-regulation (>1.5-fold) of 8.75 and 4.05% of transcripts, respectively (32). Transcripts up-regulated (>1.5-fold) in either Drosha- or AGO1-depleted cells represented 2.3% of detectable mRNAs (32). The estimates of miRNA targets in mammalian cells are based on computational target predictions (33–36), and on the effects of either ectopic expression of miRNAs (18) or inhibition of individual miRNAs by antisense molecules (37). Productive interaction of miRNAs with their mRNA targets appears to involve little beyond the ‘seed’ region between nt 1 and 8 of the miRNA (38,39). Given the limited information about what makes a functional miRNA target site, prediction algorithms generally estimate that hundreds of mRNAs are targeted by individual miRNAs (33–36). However, it is very likely that the number of mRNAs that have functional (detectable and/or biologically significant) interaction with miRNAs is much smaller. For instance, the abundance of miRNAs and their cognate transcripts will certainly affect the extent of the miRNA regulation (40). In addition, the efficiency of miRNA-mediated silencing depends on a number of miRNA-binding sites in the 3′-untranslated region (3′-UTR) (15,41). When taking such aspects into consideration, the number of genes controlled by miRNAs in a single cell type will be much smaller than one predicts simply by searching for individual miRNA ‘seed’ matches in the entire transcriptome.
The purpose of the experiments presented here was to identify transcripts that are sensitive to inhibition of RNA silencing and to correlate changes in mRNA levels with the presence of miRNAs in the same cell type. We used HEK293 cells (for simplicity referred to as 293) to generate cell lines allowing for an inducible knock-down of Dicer and individual Ago proteins, Ago1 through Ago4. These lines were used to analyze the effects of down-regulation of Dicer and Ago proteins on the efficiency of RNA silencing and on the profile of cellular mRNAs. Our results indicate that Ago2 is the most important Argonaute acting in the miRNA pathway in 293 cells, and that its knock-down effect is comparable to that of Dicer. The miRNA-mediated mRNA degradation in mammalian cells does not depend on the direct Ago2-mediated endonucleolytic cleavage, but it is probably a consequence of decapping and exonucleolytic processing of repressed mRNAs. Profiling of mRNA populations in the knock-down cell lines produced sets of Dicer- and Ago-dependent transcripts that show a significant enrichment of miRNA seeding regions.
MATERIALS AND METHODS
Plasmids
Constructs expressing anti-Argonaute, anti-Dicer and control short hairpins (sh)
Plasmids pTER-Ago1_sh, pTER-Ago2_sh2, pTER-Ago3_sh, pTER-Ago4_sh, pTER-Dcr_sh2 and pTER-control_sh were generated by cloning annealed synthetic 63mer oligonucleotides (Supplementary Table 1) into BglII and HindIII sites within pTER [a gift of M. van de Wetering (42)]. The Argonaute shRNAs were designed according to siRNA sequences described by Meister et al. (12).
Renilla (RL) and Firefly luciferase (FL) reporter constructs
pFL-Con, pRL-Con, pRL-Perf and pRL-3xBulgeB (formerly referred to as pRL-3xBulge) were described previously (23). pRL-Perf and pRL-3xBulgeB harbor in their 3′-UTR one perfect or three bulged binding sites complementary to let-7a miRNA, respectively. pRL-3xBulgeA carries in its 3′-UTR three bulged let-7a-binding sites different from those in pRL-3xBulgeB (Figure 1A); it was cloned as described for pRL-3xBulgeB (23). Similarly, an additional set of reporters under control of the thymidine kinase (TK) promoter was made using phRL-TK (Promega). TK promoter containing constructs were used for an experiment shown in Figure 3C.
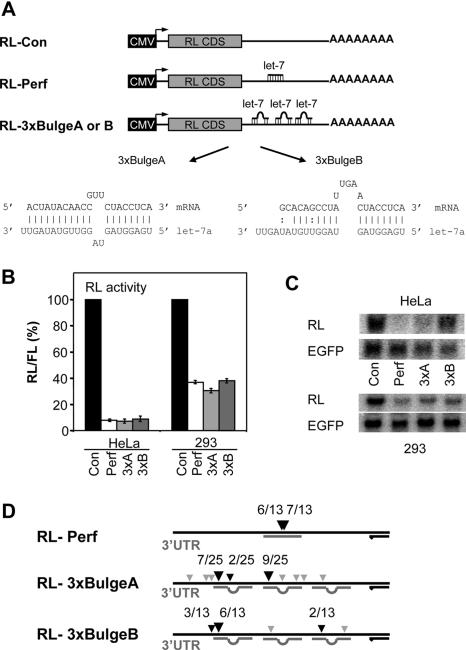
Figure 1.
Endogenous let-7 miRNA represses translation of RL reporters transcripts and leads to their partial degradation in different human cell lines. (A) Schematic representation of RL reporter constructs. The 3′-UTR of the RL reporter contains either one perfect or three bulged binding sites for let-7a miRNA. Binding sites are indicated as straight or bulged lines. Differences in target sequence complementarity between the RL-3xBulgeA and B reporters are indicated. Not drawn to scale. CMV, cytomegalovirus promoter; RL CDS, RL coding region. (B) Activity of different RL reporters in transfected HeLa and 293 cells. RL activity values are expressed relative to the activity of firefly luciferase (FL) encoded by the co-transfected pFL-Con. RL/FL value in cells transfected with the pRL-Con was set to 100%. Histogram shows normalized mean values (±SEM) of RL/FL activity from a minimum of three experiments performed in duplicates. (C) Northern blot analysis of expression of RL reporters in 293 and HeLa cells. The EGFP mRNA expression from the co-transfected plasmid (pGFP) served as normalization control. For quantification see Figure 3B and Supplementary Figure S1A. (D) 5′RACE analysis of mRNA expressed from RL reporters in 293 cells. Small gray arrowheads indicate single mapped cleavages, whereas larger black arrowheads are diagnostic of cleavages cloned repeatedly. For exact cleavage positions see Supplementary Figure S2. Positions of regions complementary to let-7a miRNA and the reverse PCR primer (black arrow) are indicated. Abbreviations of RL reporters: Con, pRL-Con; Perf, pRL-Perf; 3xA, pRL-3xBulgeA; and 3xB, pRL-3xBulgeB.
HA-tagged Argonaute and Renilla luciferase expression plasmids
Except of HA-Ago1, plasmids expressing HA-tagged proteins were described previously: HA-Ago2 (11–13), HA-Ago3 and HA-RL (23), and FLAG-HA-Ago4 (12). Plasmid expressing HA-Ago1 protein was cloned as described for HA-Ago2. Further details for all plasmids are available on request.
Cell culture, preparation of stable cell lines and transfections
Cell culture
HEK 293T-REx cells (HEK293 line expressing Tet repressor; referred to hereafter as 293; obtained from Invitrogen) were grown according to the manufacturer's protocol in DMEM (Gibco-BRL) supplemented with 2 mM l-glutamine and 10% heat-inactivated fetal calf serum (FCS), and 10 µg/ml blasticidin (Invitrogen). HeLa and MCF-7 cells were grown under the same conditions but in the absence of blasticidin. A-549 cells were grown in Roswell Park Memorial Institute (RPMI) medium supplemented with 10% heat-inactivated FCS.
Ago1-kd, Ago2-kd, Ago3-kd, Ago4-kd and 293-control_sh cell lines
To generate stably transfected tetracycline (Tet) inducible cell lines, 293 cells were co-transfected with pTER-Ago1_sh, pTER-Ago2_sh2, pTER-Ago3_sh, pTER-Ago4_sh or pTER-control_sh, and pBABE-puro (Clontech), encoding a puromycin resistance marker (43). Cells were grown in the presence of 2 µg/ml puromycin and 10 µg/ml blasticidin according to the manufacturer's protocol to select stably transfected clones. Single clones were selected to generate monoclonal cell lines, which were tested in the presence of Tet (10 µg/ml) for a specific knock-down effect. Cell line 293-control_sh, carrying a stably integrated pTER-control_sh plasmid, was used as a control in subsequent Ago knock-down experiments.
Dicer-kd/2-2, Dicer-kd/2b2 and 293-control cell lines
The 293 cells were co-transfected with pTER-Dcr_sh2 or pTER. Cells were grown in the presence of 50 µg/ml zeocin and 10 µg/ml blasticidin according to the manufacturer's protocol to select stably transfected clones. Dicer knock-down effect in stable transformed lines was tested upon induction with 1 µg/ml of doxycyclin (Dox) (Sigma). Cell line 293-control, carrying a stably integrated pTER plasmid, was used as a control in subsequent Dicer knock-down experiments.
Transfection of RL reporter constructs
Typically, at least three independent transfection experiments in duplicates were performed. Stable Argonaute knock-down cell lines were induced with 10 µg/ml of Tet 12 h before transfection. For luciferase assays and northern blot analyses, cells were transfected in 12- or 6-well plates with 5 or 10 ng pRL reporter constructs, 35 or 75 ng pGFP and 50 or 100 ng pFL-Con using Lipofectamine PLUS reagent (Invitrogen). pFL-Con, encoding FL was a control reporter. Dicer stable knock-down cell lines were induced with 1 µg/ml Dox for 1 or 5 days and transiently transfected with 50 ng of RL reporter constructs and 50 ng of pFL-Con in 12-well plates using Lipofectamine PLUS reagent (Invitrogen). All luciferase assays were performed 48 h post-transfection.
Western blot analysis
We verified the knock-down of Argonaute proteins in stable cell lines using plasmids expressing tagged Argonaute proteins. Knock-down cell lines were induced 12 h before transfection with 100 ng of a plasmid expressing an Argonaute protein tagged at the N-terminus with the hemagglutinin (HA)-tag (13). In addition, 100 ng of HA-RL expressing plasmid was co-transfected to serve as transfection and normalization control (13). Cells were harvested 48 h after transfection and knock-down was compared with uninduced cells and parental 293 cells. Whole cell extracts were separated on SDS–PAGE and amounts of HA-Ago and HA-RL proteins were analyzed by western blotting. HA-tagged proteins were detected with monoclonal anti-HA antibody (HA-3F10 Roche) used at 1:1000 dilution followed by secondary rat antibodies conjugated to horseradish peroxidase and ECL (Amersham Bioscience). The signal was detected using X-OMAT LS or BioMax Light films (Kodak). Dicer levels were analyzed using rabbit polyclonal anti-Dicer antibody D349 (44). Tubulin was visualized using anti-β-tubulin antibody (Sigma).
Luciferase assays
Luciferase assays were performed using the Dual-Luciferase Reporter Assay kit (Promega) as per the manufacturer's instructions. RL activity was normalized according to FL activity expressed from pFL-Con as described previously (13). Normalized RL activity in cells transfected with pRL-Con was always set to 100%.
RNA isolation and northern blotting
Total RNA from induced knock-down cells 48 h post-transfection was isolated using Absolutely RNA Miniprep kit (Stratagene). Purified total RNA (15 µg) was run on a 1% agarose gel under denaturing conditions, transferred to Hybond N+ membranes (Amersham) and subjected to hybridization using [α-32P]dATP-labeled random-primed probes specific for RL or EGFP (normalization control) coding regions. Quantification was done using Storm 860 PhosphorImager and ImageQuant software (Molecular Dynamics).
5′RACE
The 293 cells were transfected for 48 h with pRL-Perf, pRL-3xBulgeA or pRL-3xBulgeB. RNA was extracted as described above. Total RNA (4 µg) was ligated to an RNA adaptor (400 ng) using 20 U of T4 RNA Ligase (New England BioLabs) in the presence of 25% v/v PEG 8000 in the final volume of 10 µl. An aliquot of the ligation mixture (3 µl) was taken for reverse transcription using 200 U of Superscript III Reverse Transcriptase (Invitrogen) and RL-gene specific RT-primer in the final volume of 20 µl. Subsequently, one-tenth of the RT reaction mixture was used as template for the first round of nested PCR, using Outer Primer, RL-nested1 primer and High Fidelity Polymerase (Roche) in the final volume of 50 µl. PCR amplification conditions were as follows: initial denaturation at 94°C for 2 min was followed by 20 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 1 min, followed by final extension at 72°C for 5 min. An aliquot (2 µl) of the first round PCR was used as a template for a second nested PCR using Inner Primer and RL-nested2 primer. All primer sequences are listed in Supplementary Table 1. The final concentration of all primers was 250 nM. PCR products were analyzed on an agarose gel and were then cloned into TOPO TA vector (Invitrogen) according to manufacturer's instructions. Individual clones were sequenced using a T7 primer.
Microarray analysis
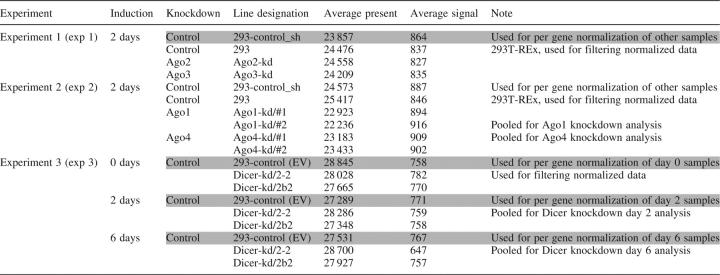
All cell lines (Table 1) were analyzed in duplicates, which were independently induced and harvested for RNA isolation. Two Dicer-kd lines were analyzed at day 0, 2 and 6 after induction. Identically induced parental 293 line carrying an empty pTER vector (293-control) was used as a reference. Ago knock-down lines (six in total, Table 1) were analyzed in two experiments 2 days after induction to minimize the accumulation of secondary effects of the knock-down. For reference, we used a 293-derived stable cell line (293-control_sh) expressing a short hairpin with a random (scrambled) sequence (pTER-control hairpin) and the parental 293-control cell line (Table 1). All original microarray data were deposited in the NCBI GEO database (series record GSE4246).
Table 1.
Overview of microarray samples
Each sample was analyzed in a duplicate. Control samples used for per gene normalization are highlighted in gray. EV, empty vector. This designation is used in the MIAME file.
Total RNA (5 µg) from each replicate was reverse transcribed with the Affymetrix cDNA synthesis kit and cRNA was produced by in vitro transcription (IVT) by T7 RNA polymerase using the Affymetrix IVT kit as per manufacturer's instructions. Biotinylated cRNA (20 µg) was fragmented by heating with magnesium (as per Affymetrix's instructions) and 15 µg of fragmented cRNA was hybridized to Human U133 plus 2.0 GeneChips™. Selected parameters of original hybridization data are shown in Figure 4A and Supplementary Data. Quality control and background normalization were performed using Refiner from Genedata AG (Basel, Switzerland). In order to compare data from three independent experiments within one environment, all arrays were condensed together using the GC-RMA algorithm with quantile normalization composed of 100 bins using Genedata's Refiner application. Per chip normalization was performed by scaling the median of the genes called present (detection P-value < 0.04) to a value of 500. The per chip normalized data are referred to as ‘raw’ expression values in this paper. In addition, a per gene normalization was performed using a point-wise division of gene (probe set) in each experimental sample with the median expression profile of the same gene from the matched control samples processed on the same day. The results of this point-wise division are referred to here as ‘normalized’ data. Argonaute knock-down lines were normalized to 293-control_sh and Dicer lines were normalized to the 293-control line from the same time point (Table 1). Unless noted, all subsequent analyses were performed using the normalized data.
Subsequent data analysis was performed using Analyst from Genedata AG. Genes were required to pass a t-test (1-way ANOVA) with a P < 0.05 and/or have a median fold change of 1.5 or greater between one or more pairs of conditions. Additional evaluation of similarity of Analyst-generated gene lists was performed using Genespring 7 (Agilent Technologies, USA). Similar gene lists in the Gene List Inspector window of the Genespring are gene lists that contain a significant number of overlapping genes with the one selected. The P-value is calculated using the hypergeometric distribution as the probability of k or more genes overlapping when one randomly samples a gene list of n genes and a gene list of m genes from a universe of u genes:
Motif analysis for 3′-UTRs
To assign a 3′-UTR sequence to each probe of the microarray (U133 plus 2.0) we took the latest annotation table provided on the Affymetrix website (http://www.affymetrix.com). For most of the probes, at least one identifier from the RefSeq or UniGene sequence databases was available. Whenever possible, we extracted the 3′-UTR sequence from the RefSeq file distributed via the NCBI website (http://www.ncbi.nih.gov). When a RefSeq identifier was not available, or the RefSeq sequence did not contain a 3′-UTR, we extracted the 3′-UTR sequence from the representative sequence of the corresponding UniGene cluster, also available via the NCBI interface. In the rare cases where several distinct sequences were compatible with the provided probe annotation, we chose the longest 3′-UTR for our subsequent analysis.
The currently available annotation of the microarray used in this work allowed finding one 3′-UTR sequence for ∼85% (45337/54675) of the probes, 85% (39134/45337) of which are based on the RefSeq database, which is highly reliable because it is manually curated. We obtained the following 3′-UTR sets for significantly (P < 0.05) up-regulated probes: Ago1-kd, 1482 probes/1243 3′-UTR sequences; Ago2-kd, 2782/2113; Ago3-kd, 1805/1403; Ago4-kd, 926/833; Dcr-kd day 2, 3104/2540; Dcr-kd day 6, 3298/2605.
Expression values could be directly attributed to 3′-UTRs since most of them had only one corresponding probe on the microarray. Redundancies only occurred in the form of multiple probes corresponding to a single mRNA. In this case the expression values were taken simply as the sum of the raw contributions of all related probes. In order to analyze motif over-representation in 3′-UTR sequences we needed a set of ‘control’ probes, the expression of which was not affected by the various knock-down experiments that have been carried out. We constructed this set by extracting 1144 probes for which the expression level varied by <10% over all experiments. They corresponded to a set of 925 non-redundant 3′-UTR sequences.
The evaluation of the statistical significance of the frequency of occurrence of different sequence motifs in the various sets of up-regulated 3′-UTR sequences always refers to a comparison with the control set described above. We always compared two lists: one describing the occurrences of the considered motif in each of the 3′-UTR sequences from the set of up-regulated transcripts and the other in the 3′-UTR sequences of the control set. The statistical significance of their dissimilarity is quantified by a P-value, which gives the probability of observing the data by chance if the two sets of motif occurrences were drawn from the same population. This quantity is calculated using the non-parametric Wilcoxon test implemented in the R statistical software (www.r-project.org).
RESULTS AND DISCUSSION
Activity of let-7 reporters in 293 cells
To evaluate the effects of Dicer and Ago protein deletion on the function of miRNAs in 293 cells, we characterized a set of reporters whose activity is controlled by the let-7 miRNAs endogenously expressed in these cells. Reporter constructs harbor one perfect (RL-Perf) or three bulged (RL-3xBulge) binding sites for let-7 in the 3′-UTR positioned downstream of the Renilla luciferase (RL) coding region (Figure 1A) (23).Two versions of the RL-3xBulge reporters, RL-3xBulgeA and RL-3xBulgeB were examined; they differ in the structure of the central bulge and base pairing of the 3′ portion of let-7 miRNAs in the predicted miRNA-mRNA duplex [RL-3xBulgeB was referred previously to as RL-3xBulge (23)]. The RL expression from RL-Perf and from both RL-3xBulge constructs in 293 cells was 60–70% lower than that from RL-Con that lacks the let-7 binding sites (Figure 1B). A stronger, ∼90% repression was observed for RL-Perf and RL-3xBulgeA and B in HeLa cells (Figure 1B) (23). The difference may be explained by different levels of let-7 miRNAs in 293 and HeLa cells [(12) and data not shown)].
We examined the levels of reporter mRNAs accumulating in transfected HeLa and 293 cells (Figure 1C). As reported previously (23), mRNA containing a perfectly complementary site, RL-Perf, accumulated in HeLa cells to a very low level, whereas the level of RL-3xBulgeB was comparable to that of the control mRNA (RL-Con). The level of RL-3xBulgeA mRNA was 44% lower (for quantification see Supplementary Figure S1A) from that of RL-Con. Examination of reporter mRNAs in 293 cells revealed a strong decrease in RL-Perf and RL-3xBulgeA transcript levels. Interestingly, RL-3xBulgeB mRNA also showed a strong decrease in 293 cells (Figures 1C and 3B). Such reduction of RL-3xBulgeB mRNA level was not observed in HeLa cells (Figure 1C and Supplementary Figure S1A). Activities of different reporters and their mRNA levels were also compared in MCF-7 (human breast carcinoma) and A-549 (human lung carcinoma) cells (Supplementary Figure S1B). In both cell lines, insertion of one perfect or three bulged (either 3xBulgeA or 3xBulgeB) sites inhibited RL expression to similar extent. Notably, like in HeLa cells, in both cell lines the level of RL-3xBulgeB mRNA was similar to that of RL-Con while the level RL-3xBulgeA mRNA was decreased by 40% (MCF-7 cells) or 60% (A-549 cells) (for quantification see legend to Supplementary Figure S1C). Taken together, these data indicate that relative steady-state levels of reporters bearing bulged let-7 sites may differ between various mammalian cell lines, and that the accumulation of reporter mRNAs may also be affected by the nature of bulged duplexes formed between let-7 miRNA and its target. The let-7 RNA hybrids formed with BulgeB sites are less stable than those with BulgeA sites (Figure 1A). It is possible that let-7 interaction with the RL-3xBulgeB reporter is sufficient for its translational repression but lower stability of the interaction negatively affects the final steady-state transcript level, at either the stage of relocalization to P-bodies or subsequent nucleolytic processing. Regarding the differences in mRNA degradation between different cell lines, it is possible that the cells differ in the concentration of active RISC complexes in the cytoplasm, rate of aggregation of repressed mRNAs into P-bodies and/or dynamics of mRNA degradation in P-bodies.
Mapping of cleavage sites in RL-3xBulge RNAs
To gain insight into the mechanism of the decreased accumulation of RL-3xBulge reporter RNAs in 293 cells, we mapped potential RNA cleavage sites using the 5′RACE approach. The 5′ RACE yielded a single fragment in cells transfected with the RL-Perf construct and several bands or a smear in cells expressing RL-3xBulgeA or B (data not shown). All sequenced RL-Perf-specific clones corresponded to mRNAs cleaved at position complementary to nucleotides either 10 or 11 (counting from the 5′ end) of let-7 miRNA (Figure 1D and Supplementary Figure S2). These cleavage positions are consistent with the endonucleolytic processing catalyzed by Ago2 (45). In contrast, sequencing of clones originating from the analysis of RL-3xBulgeA and RL-3xBulgeB revealed clusters of degradation products mainly centered close to the 5′-proximal end of the first or second let-7-binding site in the 3′-UTR; more rare cleavages were mapped to positions scattered along all three let-7 sites (Figure 1D and Supplementary Figure S2).
Our results indicate that degradation of mRNAs containing bulged let-7 sites does not involve the endonucleolytic Ago2-mediated cleavage characteristic of RNAi. Similarly, down-regulation of Lim28 mRNA, a putative target of miR-125, also does not involve a specific Ago2-mediated endonucleolytic cleavage (20). Accumulation of the mRNA degradation products terminating at upstream regions of let-7-binding sites (Figure 1D) is consistent with the degradation being mediated by the 5′→3′ exonuclease, which is prevented from further progression by miRNPs bound to the 3′-UTR. Similar pattern of degradation, consistent with the decapping of the mRNA followed by its 5′→3′ exonucleolytic processing, was observed for some miRNA targets investigated in Caenorhabditis elegans (19). In mammalian cells, mRNA targets repressed by miRNAs were shown to be localized in P-bodies (22,23), structures containing the decapping complex and the 5′→3′ exonuclease XRN1 (24,46). It is likely that the observed degradation of RL-3xBulge reporters in 293 cells is a consequence of the relocation of the translationally repressed mRNAs to P-bodies.
Biochemical analysis of miRNA effects in Ago and Dicer knock-down cell lines
To examine the importance of individual Ago proteins and Dicer in the miRNA-mediated regulation in 293 cells, we generated cell lines in which these proteins can be knocked down by the tetracycline(Tet)-inducible expression of shRNAs targeting their mRNAs. Plasmids expressing the shRNAs were stably integrated into the genome of 293 cells that express the Tet repressor. In these cells, addition of Tet or doxycycline (Dox) induces expression of shRNAs that are subsequently processed to siRNA-like molecules (42). Although the knock-down approach has limitations due to the incomplete repression of targeted genes, it represents a practical and rapid strategy to analyze importance of different RNA silencing components in cultures cells. Since antibodies capable of distinguishing between the individual Ago family members are not available, we used HA-epitope tagged Ago proteins, expressed from transfected plasmids (12,23), to assess the effectiveness of individual knock-downs. Western analyses of Ago knock-down (kd) cell lines indicated that levels of targeted proteins were strongly reduced (Figure 2A). In addition, western analysis with an Ago2-specific antibody indicated a strong down-regulation of the endogenous Ago2 protein in Ago2-kd cells but not in Ago3-kd cells (Supplementary Figure S3). Specific Ago targeting was later also confirmed by microarray analysis (Figure 4B). It should be noted that a partial loss of some Ago proteins already occurred in non-induced knock-down lines, likely due to a limited leakiness of shRNA expression (Figure 2A).
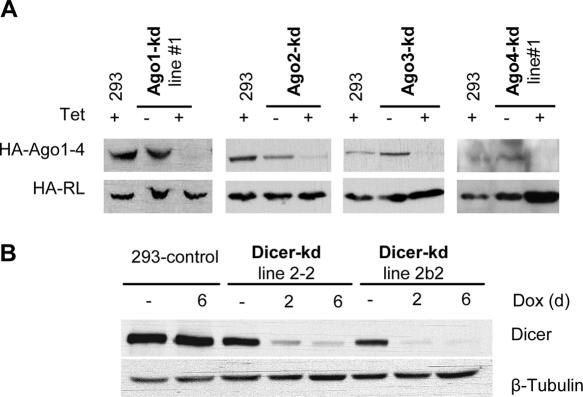
Figure 2.
Ago and Dicer protein reduction in inducible knock-down cell lines. (A) Western blot analysis of Ago proteins in knock-down cell lines. The cell lines were co-transfected with plasmids expressing corresponding HA-epitope tagged versions of Ago1 through Ago4. Levels of individual HA-Ago proteins were assayed 48 h post-transfection (60 h post-induction with Tet). Co-transfected plamsid encoding HA-RL served as control. Expression of transiently transfected HA-Ago4 was consistently very low and difficult to detect. 293 is the parental 293T-Rex line. (B) Western blot analysis of endogenous Dicer in knock-down cell lines. 293-control, a cell line carrying an empty pTER vector. Experiments shown in (A and B) were reproduced several times; shown are representative results.
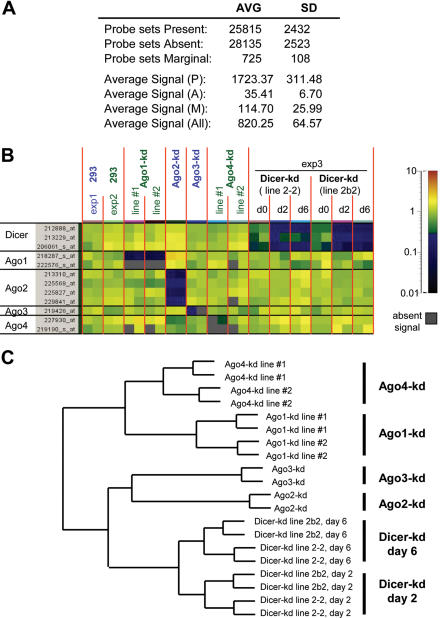
Figure 4.
Microarray analysis of transcriptomes of Dicer and Ago knock-down lines. (A) Selected characteristics of original microarray hybridization signal used for analysis (before per chip normalization) calculated from values of all arrays. AVG, average. Present (P), Absent (A) and Marginal (M) calls for Affymetrix probe sets are generated based on statistical analysis of hybridization signal of oligonucleotides from one probe set where Present, P < 0.04; Marginal, 0.06 > P > 0.04; Absent P > 0.06. Hybridization signal units are arbitrary, derived from the ‘per chip’ scaling to the median. (B) Specificity of knock-down of individual studied components of RNA silencing pathways. A tile plot depicts values normalized ‘per gene’ for probe sets for Dicer and Ago transcripts. Partial decrease of Dicer mRNA is detectable in Dicer-kd lines at day 0. Note that Ago4 hybridization data are inconclusive due to a low expression level of Ago4 mRNA. Although probe 227930_at is annotated as Ago4 and shows reduction in one of the Ago4-kd line, it does not match the human Ago4 sequence (NM_017629). The second Ago4 probe (matching NM_017629) yields a low signal, which does not pass the detection T-test. Individual microarray experiments (exp) are distinguished by colored letters: exp1, blue; exp2, green; and exp3, black. (C) Hierarchical clustering analysis based on a pooled list of genes differentially expressed between controls and knock-down samples. Probe lists for the analysis included all probes in the lane ‘changed (t-test, P < 0.05)’ in Table 2.
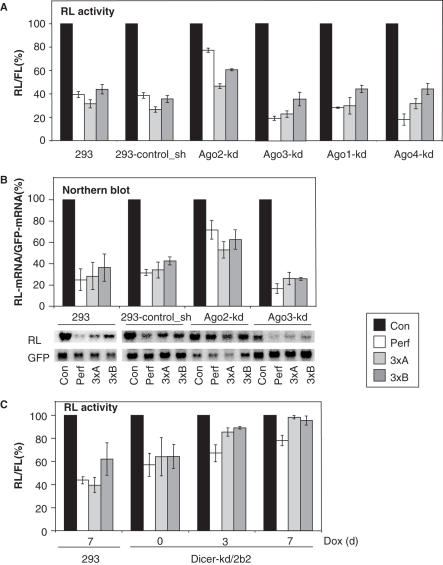
To functionally characterize the knock-down cell lines, we analyzed the effect of depletion of individual Ago proteins on repression of different RL reporters. As shown in Figure 3A, depletion of Ago1, Ago3 and Ago4 had no substantial effect on the strength of repression of RL reporters containing either perfect or bulged let-7 sites. Notably, the knock-down of Ago3 appeared to even slightly increase repression of the reporters. In contrast, the knock-down of Ago2 resulted in the substantial release of the inhibition of all tested let-7 reporters (Figure 3A). This effect was most pronounced for the RL-Perf, which was repressed by 60% in control cells and by only 20% in the Ago2-kd cells (Figure 3A). Importantly, northern analysis revealed that the Ago2 but not Ago3 knock-down resulted in a marked increase of RL-Perf and RL-3xBulge RNA reporter levels accumulating in transfected cell lines (Figure 3B). Generally, effects of Ago2 and Ago3 knock-downs on the level of reporter RNAs closely paralleled the effects on RL activity.
Figure 3.
Effects of Ago and Dicer protein knock-downs on expression of RL reporters. (A) Activity of RL reporter constructs in cells with knock-downs of individual Ago proteins (for Ago1-kd and Ago4-kd, lines #1 were used). Luciferase assays were performed 2 days after transfection (60 h post-induction with Tet). Histogram shows normalized mean values (±SEM) of RL/FL activity from three different experiments performed in duplicates (except for Ago3-kd done in two duplicate experiments). (B) Northern blot analysis of expression of RL reporters in Ago2-kd and Ago3-kd lines. mRNA was isolated from cells transfected and induced as described in (A). RL reporter mRNA expression was normalized to EGFP mRNA expressed from a co-transfected plasmid. Histogram shows normalized mean values of RL mRNAs relative to RL mRNA level in cells transfected with the control RL reporter (pRL-Con), which was set to 100%. Error bars (SEM) are derived from four northern blot experiments. Northern blot phosphorimager scans below the graph show results of one representative experiment. (C) Effect of Dicer knock-down on activity of RL reporters. Dicer-kd 2b2 cells were induced with Dox and harvested 3 and 7 days post-induction. Transfection with reporter constructs was performed 2 days before collection. Histogram shows normalized mean values (±SEM) of RL/FL activity from three different experiments performed in duplicates. Small repression of RL-Perf (but not of RL-3xBulgeA and RL-3xBulgeB reporters) still observed even at day 7 likely reflects activity of the residual RISC complex, which turns over when executing RNAi but is required in stochiometric amounts when mediating translational repression.
The generation and characterization of Dicer-kd cell lines followed procedures similar to those described for the Ago-kd lines. Two different clones (2-2 and 2b2) expressing the anti-Dicer shRNA were characterized by western analysis. Both clones showed a small knock-down effect even in the absence of induction, presumably due to leakiness of the shRNA transcription (Figures 3C and 4B). Induction of either of the Dicer-kd lines for 2 or 6 days resulted in a robust depletion of Dicer protein when compared to control cells (Figure 2B). Analysis of reporter activity in the Dicer-kd/2b2 line indicated a significant relief of the repression of both RL-Perf and RL-3xBulge constructs; partial relief was already observed in cells grown in the absence of Dox, consistent with the partial leakiness of shRNA expression (Figure 3C). The relief of the reporter repression in the Dicer-kd/2b2 line was comparable to that seen in the Ago2-kd line.
Dicer is required for biogenesis of miRNAs and possibly their assembly into the RISC (39). Hence, the suppressive effect of the Dicer knock-down on repression of reporters targeted by the endogenous let-7 miRNA was not surprising. However, the observation that depletion of Ago2 but not of the other Ago proteins has such a pronounced effect on the repression was rather unexpected. Although miRNAs are known to associate with all four mammalian Ago proteins (11,12), it is possible that Ago2 is a major component of functional miRNPs. It was revealed recently that Ago2 can effectively assemble into functional RISC/miRNP by cleaving the passenger strand of siRNA or miRNA duplex (47–49). In contrast to Ago2, other Ago proteins have to follow a less effective RISC assembly mechanism, involving the displacement of an uncut passenger strand, possibly requiring a helicase (48). In addition, raw microarray hybridization signal of Ago2 is the highest among Argonaute transcripts in 293 cells. Taken together, our results suggest that Ago2 is a principal Ago protein mediating the miRNA repression in 293 cells. However, it is possible that knock-downs of other Ago proteins have effects on miRNA-mediated silencing, which are not detected by the reporter system used in this work.
Microarray analysis of Ago and Dicer knock-down lines
To identify transcripts regulated by RNA silencing in 293 cells, we have analyzed individual knock-down lines in three microarray experiments using Affymetrix U133 Plus 2.0 microarrays (Table 1; additional information is provided in Supplementary Data). All three experiments yielded microarray hybridization results of comparable quality (Table 1, Figure 4A and Supplementary Table 1). First, all microarray data were condensed together and normalized by scaling the median. The per chip normalized data are referred to as ‘raw’ expression values. Subsequently, a per gene normalization of raw data was performed and results of this normalization are referred to as ‘normalized’ data. Unless indicated otherwise, all subsequent analyses were performed using the normalized data.
The analysis of probe sets for transcripts specifically targeted by shRNAs in Ago and Dicer knock-down lines revealed strong and specific mRNA down-regulation (Figure 4B). Ago4-kd line #2 appears to have a weak knock-down effect but it should be noted that Ago4 mRNA detection by microarrays is inconclusive due to its very low expression level (Figure 4B). As mentioned above, the Tet-inducible system suffers from a partial leakiness of shRNA expression in non-induced cells. This is apparent in both Dicer-kd lines, which showed a mild decrease of Dicer mRNA already at day 0 (Figure 4B).
To assess the similarity of array replicates and the relationships between knock-down lines, we performed condition clustering employing all the probe sets called present (i.e. detectable transcripts) by the Affymetrix software. This approach revealed a good concordance between replicates and also showed that Ago4-kd samples cluster closest to the controls. However, including all transcripts caused separate clustering of individual experiments (Supplementary Figure S3). To address the relationships between knock-down lines in a way less biased by variability between individual experiments, we have performed condition clustering based on a list of transcripts differentially expressed in any Ago or Dicer line (Figure 4C). Hierarchical clustering again revealed a good similarity between replicates. Interestingly, Ago1-kd and Ago4-kd lines clustered separately from Ago2-kd and Ago3-kd, which clustered with Dicer-kd lines (Figure 4C). Two Dicer-kd lines clustered together according to the time of induction. These data indicated that (i) replicates and different cell lines expressing the same shRNA behave consistently; (ii) knock-down of Ago4 causes the smallest effect (subsequent analysis has revealed that transcriptome changes upon the Ago4 knock-down are relatively small, comparable to the variability observed between replicates); and (iii) the effect of Ago3 knock-down is the closest to that of Ago2 while the effects of Ago2 and Ago3 knock-downs are more similar to the Dicer knock-down than are effects of Ago4 or Ago1 knock-downs.
Tables 2 and 3 summarize analysis transcripts changing expression levels upon knock-down of individual RNA silencing components (Table 2) and similarity of transcriptome changes between different knock-downs (Table 3). All results presented in Tables 2 and 3 were obtained with transcripts that were called present on at least 75% of arrays. However, we have also analyzed all raw data to ascertain that we do not miss silenced (absent) transcripts, which are reactivated (present) upon a knock-down. Figure 5A shows log scatter plots for knock-downs of Ago2 and Dicer (day 6), which we considered as most relevant since control experiments, discussed in the previous section, have demonstrated a partial relief of RNA silencing in these two knock-downs. The data show numerous small effects on gene expression, generally applying to genes expressed below the average raw signal (∼500). Differentially expressed transcripts (Supplementary Data) do not exhibit any apparent functional relationship. No silenced transcripts appeared to be reactivated upon Dicer or Ago2 knock-downs suggesting that a switch-off function of miRNAs may not be common in 293 cells. Strong induction of at least 5-fold was only observed for ∼10 transcripts in either knock-down experiment but none of them exhibited such up-regulation in both Dicer and Ago2-kd lines (for details see Supplementary Data). However, it should be kept in mind that the use of the knock-down approach makes it difficult to completely relieve the potential target mRNAs from the miRNA-mediated repression.
Table 2.
Distinct filters identify various numbers of differentially expressed (up-regulated and down-regulated) up-regulated probe sets in individual knock-down samples (pooled for each gene knock-down and/or timepoint)
| Ago1 | Ago2 | Ago3 | Ago4 | Dicer day 2 | Dicer day 6 | |
|---|---|---|---|---|---|---|
| All probes changed (up-regulated and down-regulated probes) | ||||||
| Changed (t-test, (P < 0.05) | 3414 | 5395 | 3868 | 1517 | 4936 | 5502 |
| Changed 1.5-fold | 3499 | 4163 | 2838 | 1406 | 3947 | 4903 |
| Changed 2.0-fold | 799 | 1084 | 621 | 151 | 886 | 1161 |
| Changed 1.5-fold (t-test, (P < 0.05) | 1687 | 2241 | 1304 | 285 | 2335 | 3006 |
| Changed 2.0-fold (t-test, (P < 0.05) | 530 | 751 | 384 | 56 | 716 | 939 |
| Up-regulated probes | ||||||
| Up-reguated (t-test, (P < 0.05) | 1482 | 2782 | 1805 | 926 | 3104 | 3298 |
| Up-regulated 1.5-fold | 1011 | 2124 | 1351 | 318 | 2400 | 3157 |
| Up-regulated 2.0-fold | 180 | 495 | 342 | 44 | 560 | 859 |
| Up-regulated 1.5-fold (t-test, (P < 0.05) | 317 | 1092 | 670 | 97 | 1437 | 1951 |
| Up-regulated 2.0-fold (t-test, (P < 0.05) | 104 | 325 | 214 | 28 | 463 | 705 |
Transcripts found in lists ‘up-regulated (t-test, P < 0.05)’ and ‘up-regulated 1.5 fold’ are included in Supplementary Data.
Table 3.
Analysis of similarity between lists of transcritps up-regulated in different knock-down cell lines (pooled for each gene knock-down and/or timepoint)
| Ago1 | Ago2 | Ago3 | Ago4 | Dicer day 2 | Dicer day 6 | |
|---|---|---|---|---|---|---|
| Ago1 1 | 1.6E-54 | 2.8E-100 | 2.4E-102 | 9.1E-05 | 2.2E-15 | |
| Ago2 | 1.6E-54 | 1.0E-267 | 1.2E-19 | 6.9E-129 | 1.1E-200 | |
| Ago3 | 2.8E-100 | 1.0E-267 | 3.0E-11 | 9.6E-71 | 3.2E-62 | |
| Ago4 | 2.4E-102 | 1.2E-19 | 3.0E-11 | 5.0E-07 | 2.1E-26 | |
| Dicer day 2 | 9.1E-05 | 6.9E-129 | 9.6E-71 | 5.0E-07 | 0.0E+00 | |
| Dicer day 6 | 2.2E-15 | 1.1E-200 | 3.2E-62 | 2.1E-26 | 0.0E+00 |
P-values expressing similarity of given gene lists were calculated in Genespring7 as described in Materials and Methods. The most significant overlaps (the lowest (P-value) are shown in boldface.
Figure 5.
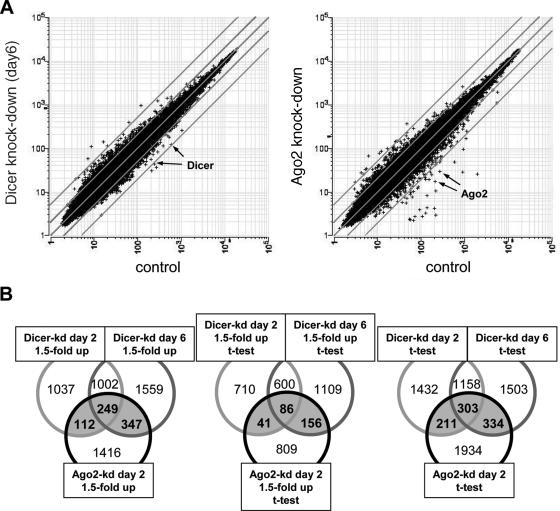
Transcriptome changes upon Dicer and Ago2 knock-down. (A) Raw data scatter plots of array signals after Dicer (left panel) and Ago2 (right panel) knock-down in 293 cells. Shown are all 54675 probes from Human U133 plus 2.0 GeneChips™. Each probe is represented by a cross depicting the mean raw expression in control and knock-down samples. Array samples: Dicer knock-down, a pool of Dicer-kd lines 2-2 and 2b2 at day 6; Ago2 knock-down, a duplicate of Ago2-kd induced for 2 days; control (Dicer), a pool of lines 2-2 and 2b2 at day 0 and the 293-control line induced for 0, 2 and 6 days; control (Ago2), a pool of 293 and 293-control_sh lines. Diagonal gray lines represent fold change borders (1-, 2- and 5-fold). Arrows indicate position of Dicer and Ago2 microarray probe sets in corresponding knock-downs. Expression units are arbitrary, the mean value of expressed transcripts is scaled to 500. (B) Venn diagrams showing overlaps of differently filtered gene lists. Filtering of up-regulated transcripts with 1.5-fold cut-off (left), t-test P < 0.05 (right) or their combination (middle). Gray area depicts transcripts up-regulated in Ago2 and at least one Dicer-kd line.
Global analysis of up-regulated transcripts
We sought evidence that the mRNAs that are up-regulated upon knock-downs of miRNA pathway components represent miRNA targets. We extracted and analyzed either mRNAs whose expression increases by some minimal factor (1.5 or 2) or mRNAs whose expression increases consistently and significantly in replicate arrays (P-value < 0.05 in t-test) upon a miRNA pathway component knock-down (Table 2). Either approach indicated that Dicer and Ago2 knock-downs affect larger sets of transcripts, while the number of transcripts affected by depletion of Ago3 and Ago1 is approximately one-half to one-third compared to Ago2 knock-down. The effect of Ago4 knock-down was very small, comparable to the variability observed between two control lines. Number of transcripts in Ago2-kd and Dicer-kd lines significantly up-regulated >1.5-fold was between 5 and 10% of present transcripts. Interestingly, this result is very similar to the recent study of mRNAs up-regulated upon Drosha and AGO1 knock-downs in Drosophila cells (32).
Since similar sets of transcripts are expected to be up-regulated upon targeting different components of the miRNA pathway, we used Genespring 7 to analyze the similarity between all lists of up-regulated probes (1.5-fold, P < 0.05) in knock-down cell lines (Table 3). We found that the Ago2-kd up-regulated transcript list shares a clear similarity with the lists from Ago3-kd and Dicer-kd at days 2 and 6. Similarity of both Dicer-kd lists to the Ago2-kd list is remarkably higher compared to other Ago knock-down lists (Table 3). Correspondingly, the most significant overlap between genes up-regulated in different knock-down lines is found between Dicer-kd and Ago2-kd lines. As expected, the size of this overlap in terms of the number of transcripts and fraction of each list depends on filtering applied for compared lists. Three examples of such overlaps in Ago2 and Dicer knock-downs are depicted in Figure 5B. Using either the 1.5-fold change (Figure 5B, left panel) or statistical significance P < 0.05 (Figure 5B, right panel), the fractions of Ago2 transcripts that were found also up-regulated in at least one Dicer knock-down time point reach 33% (708 transcripts) and 30% (848 transcripts), respectively. These numbers corresponds to ∼4% of transcripts detectable on at least three quarters of all arrays. Combination of the fold up-regulation and statistical significance data reveals 283 transcripts (1.4% of present transcripts) up-regulated >1.5-fold in Ago2-kd and Dicer-kd lines at day 2 or 6 (Figure 5B, middle Venn diagram). Interestingly, overlap of Ago2-kd with Dicer-kd at day 6 is always higher than with Dicer knock-down at day 2. This is consistent with an expected gradual relief of the miRNA-dependent RNA degradation after the induction of Dicer knock-down. These data suggest that hundreds of transcripts show common up-regulation upon Ago2 or Dicer knock-down in mammalian cells. For comparison, a similar study in Drosophila revealed that 2.3% of transcripts are commonly up-regulated (>1.5-fold) upon knock-down of either Drosha or AGO1 (32).
Before testing whether up-regulated transcripts correspond to miRNA targets, we have first converted partially redundant probe sets into non-redundant mRNA 3′-UTR sequences (Materials and Methods). On these, we then performed two computational motif analyses. The first one revealed that there were indeed n-mer motifs (n = 6, 7) that are over-represented in a statistically significant way in the sets of up-regulated transcripts in the Ago2-kd and both Dicer-kd lines. For the second analysis, we selected three motifs that are common to the three lines and correspond to ‘seeds’ of miRNAs expressed in 293 cells. Both analyses are described in the two following sections.
Analysis of hexamer and heptamer motifs in up-regulated transcripts
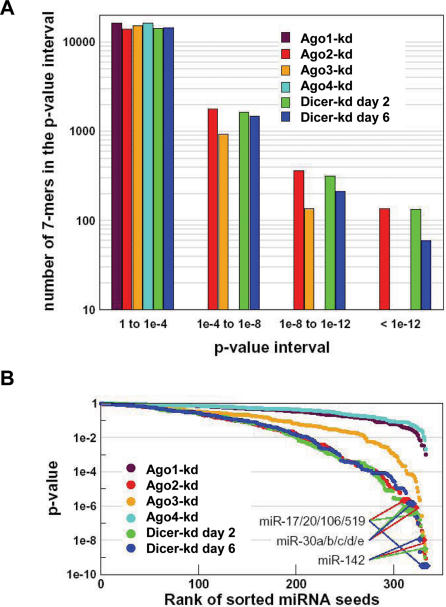
As a first test for establishing the involvement of the miRNA pathway in the mRNA up-regulation, we performed an analysis of over-representation for all possible n-mer sequences (n = 6, 7). For each of the 4n motifs, we generated a list of counts of its occurrences in each of the 3′-UTRs from the various sets of up-regulated mRNAs. We compared this list to the one obtained for a control set (unchanged transcripts) using the non-parametric Wilcoxon test. The P-value obtained in this test quantifies the statistical significance of the dissimilarity between the two sets (Materials and Methods). Figure 6A shows the results for the case of heptamers. Similar results were obtained when we analyzed hexamers and frequencies (number of motif occurrence divided by the corresponding 3′-UTR length) rather than ‘raw’ counts (data not shown). Moreover, similar results were also acquired when the over-representation analysis was restricted to miRNA seeds (nt 2–8) of known human miRNAs (Figure 6B). The 4n motif and the miRNA seeds P-value distributions in the six knock-down sets are consistent with conclusions stemming out from transcript analysis: Ago4 and Ago1 knock-downs have the smallest effect (no motifs with P-values < 1e-4). Ago2-kd line shows the strongest enriched motif signal, and Dcr-kd lines at days 2 and 6 are very close to it. The behavior of the Ago3-kd lies in between these two extremes. This order is consistent with condition clustering results (Figure 4C) and the analysis of overlaps of lists of up-regulated transcripts (Table 3). For the genes that are up-regulated in the Ago2 and Dicer knock-down experiments, it was further possible to make the connection with miRNA-mediated regulation by determining whether the most significantly enriched motifs indeed correspond to binding sites of miRNAs known to be expressed in 293 cells (www.ambion.com/miRNA). Remarkably, the sequences that are complementary to the seeds of several miRNAs expressed in 293 cells are found among the top 10% motifs from Ago2-kd, Dicer-kd day 2 and Dicer-kd day 6 3′-UTR lists: AAAGUGC for miR-17/20/106, GUAAACA for miR-30 and AUAAAGU for miR-142 (Figure 6B). We notice that they all share the AAA motif. These results are very similar to the ones obtained by Krutzfeldt et al. (37) when functionally knocking down miR-122 in liver cells. The miR-122 seed-matching motif was found to have one of the lowest P-values in mRNAs up-regulated upon inactivation of miR-122.
Figure 6.
Enrichment of specific heptanucleotides in 3′-UTRs of transcripts up-regulated upon Ago2 and Dicer knock-downs. (A) Distribution of enrichments of all heptamers (47 = 16384) in 3′-UTRs of mRNAs up-regulated in different knock-down lines. Enrichment of each heptamer is calculated as a P-value of the Wilcoxon tests that compares the number of occurrences of each heptamer in the sets of 3′-UTRs that are significantly up-regulated (t-test P-value < 0.05) in one of the six sample sets from knock-down lines relative to the control set of mRNAs showing <10% change in any of the knock-down lines. The horizontal axis shows four bins into which all heptamers were sorted according to their P-values. The vertical axis shows, on a logarithmic scale, a count of heptamers for a given P-value range and a sample set. (B) Distribution of enrichments of heptamers corresponding to known miRNA seed regions. P-values were calculated as in (A) but data are displayed as decreasing P-values of miRNA seeds. The vertical axis shows, on a logarithmic scale, P-values of miRNA seeds from the Wilcoxon test of heptamer enrichment in a given sample set. The horizontal axis corresponds to rank sorted (according to the descending P-value for each sample set) miRNA seeds (nt 2–8) from 328 mature miRNAs from Rfam 8.0 (27). Values for selected miRNAs known to be expressed in 293 cells are indicated.
Analysis of hexamer and heptamer motifs for miR-17/20/106, miR-30 and miR-142
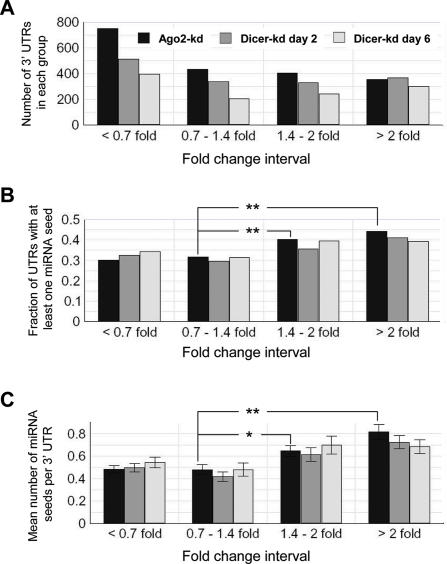
In a second test, we restricted the analysis to the set of miRNA seeds (miR-17/20/106, miR-30 and miR-142) that were significantly over-represented in Ago2-kd and Dicer-kd lines and determined whether these motifs are indeed predictive of the magnitude of up-regulation of the mRNAs. For each of the three experiments (Ago2-kd, Dicer-kd day 2 and Dicer-kd day 6) and their respective controls, we extracted the expression values based on the microarray probe data for all cases where mRNA 3′-UTR sequences were available (Materials and Methods). We then selected transcripts from the array that passed a signal detection t-test (P-value < 0.05) and binned them into four groups based on the values of the expression ratio r between knock-down and control lines: ‘down-regulated’ (r < 0.7), ‘unchanged’ (0.7 < r < 1.4), ‘weakly up-regulated’ (1.4 < r < 2) and ‘strongly up-regulated’ (r > 2). Each group contained at least 200 non-redundant transcripts (Figure 7A), making it possible to draw statistically solid conclusions.
Figure 7.
Analysis of the occurrence of motifs matching the seeds of miR-17/20/106, miR-30 and miR-142 in 3′-UTRs of transcripts that are up-regulated in Ago2-kd at day 2 and both Dicer knock-down cell lines at days 2 and 6. (A) Number of 3′-UTRs for transcripts in each of the analyzed expression categories in three studied knock-down datasets. Expression categories of transcripts were chosen as follows: ‘down-regulated’ (<0.7-fold), ‘unchanged’ (0.7- to 1.4-fold), ‘weakly up-regulated’ (1.4- to 2-fold) and ‘strongly up-regulated’ (>2-fold). (B) Fraction of the total number of 3′-UTRs [shown in (A)] containing at least one seed for the miR-17/20/106, miR-30 or miR-142. (C) Average number (±SEM) of the miR-17/20/106, miR-30 and miR-142 seeds per the 3′-UTR sequence in each of the analyzed expression categories in the three studied knock-down datasets. Asterisks indicate statistically significant difference (t-test, **P < 0.01, *P < 0.05) between ‘unchanged’ and ‘weakly up-regulated’ or ‘strongly up-regulated’ transcripts in Ago2-kd. The complete statistical analysis of categories in (B and C) is given in Supplementary Table 2.
We first considered the fraction of 3′-UTRs that contain at least one occurrence of one of the seed motifs. For both knock-downs, but more significantly for Ago2, the percentage increased with the degree of up-regulation of mRNA expression (Figure 7B and Supplementary Table 2). Similar results were obtained for the specific case of miR-122 (37). However, differences in the percentage of UTRs containing the seed motif are smaller in our analysis and, additionally, we find that fractions of ‘down-regulated’ and ‘unchanged’ mRNAs that contain at least one miRNA seed are statistically indistinguishable (Supplementary Table 2). This suggests that the observed down-regulation of transcripts in our experiments is unlikely to be directly linked to the miRNA-mediated regulation.
To further quantify the representation of seed motifs in the four groups of transcripts, we calculated the mean and standard error of the number of occurrences of the miRNA seeds in all 3′-UTR sequences present in each set. We again observed a trend compatible with the inhibition of the miRNA pathway, particularly clear for the Ago2 case: the more seed matches are present, the stronger is the up-regulation of the transcript (Figure 7C and Supplementary Table 2). No statistically significant similar correlation could be detected for ‘down-regulated’ and ‘unchanged’ 3′-UTRs (Supplementary Table 2). Similar conclusions were reached when we defined miRNA seeds as hexamers instead of heptamers and when we analyzed frequencies of motifs (number of ‘raw’ counts per kilobase) rather than ‘raw’ counts of motifs (data not shown). The correlation between the magnitude of up-regulation and the number of seed occurrences was also observed in a recent Dicer knock-out experiment in zebrafish embryos (21). Our data reproduce this observation, although the magnitude of the effect in our experiments is not as large.
The motif analysis presented above supports the hypothesis that the observed up-regulation of mRNAs is due to their release from the miRNA-mediated repression upon the knock-down of important components of the pathway. Although our results are consistent with previous analyses, the signals observed by us were generally weaker than those recorded from experiments in which individual miRNAs were either up- or down-regulated (21,31,33,37). This quantitative discrepancy could be due to the use of 293 cells, a cell type in which no miRNA is as predominant as miR-124 in brain (18), miR-122 in liver (37) or miR-430 in zebrafish embryo (21). The seed motifs uncovered in our analysis have a potential to base pair with several different human miRNAs (miR-17, miR-20a/b, miR-106, miR-30a/b/c/d/e and miR-142), but none of them displays a particularly strong and specific expression in 293 cells (www.ambion.com/miRNA). This situation may result in signals of lower magnitude, with the general features of the miRNA regulation being preserved.
CONCLUSIONS
Using reporter constructs, we observed that degradation of mRNAs bearing sites imperfectly complementary to the endogenous let-7 miRNA is considerably stronger in human 293 than HeLa cells. Mapping of the cleavage sites occurring in 293 cells revealed that the degradation does not result from the Ago2-mediated endonucleolytic cleavage but is rather a consequence of the relocation of the translationally repressed mRNAs to P-bodies combined with decapping and 5′→3′ exonucleolytic processing. Dicer and Ago2 knock-downs resulted in a significant relief of silencing of reporter RNAs as measured at both protein activity and mRNA levels. Knock-down of other Ago protein had much weaker effect, indicating that Ago2-contaning complexes may represent a major fraction of functional miRNPs in 293 cells. Similarly, microarray analysis identified most pronounced changes in cellular mRNA levels in Ago2 and Dicer knock-down lines. Analysis of the transcriptome of knock-down lines indicated that up to a few thousand genes may change their expression pattern upon inhibition of RNA silencing. Since inhibition of the miRNA pathway should correlate with increased abundance of mRNAs targeted by miRNAs, we have searched for putative miRNA targets among genes up-regulated in Ago2 and Dicer knock-downs. Over 250 transcripts were significantly up-regulated >1.5-fold in both Ago2 and Dicer knock-downs. Analysis of potential miRNA targets in lists of transcripts significantly up-regulated upon knock-down of Ago2 or Dicer revealed that the magnitude of the miRNA seed over-representation in the 3′-UTR sets correlates well with the magnitude of up-regulation of corresponding mRNAs. Taken together, our analysis suggests that miRNAs affect expression of several hundred genes in 293 cells. As expected for randomly evolved miRNA targets, up-regulated genes did not show any apparent functional relationship. Most of the statistically significant changes were small (<2-fold) and, at the mRNA level, we found no evidence for activation of silenced genes, suggesting that miRNAs generally have a tuning role in regulating gene expression in 293 cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Kaifu Tang for providing Dicer knock-down cell lines and other material, Azeddine Si-Ammour for assistance with the 5′RACE, Herbert Angliker for processing Affymetrix microarrays, Suvendra N. Bhattacharyya for providing an Ago2 antibody, Tabea Zoller for assistance and the FMI sequencing facility. Friedrich Miescher Institut is supported by the Novartis Research Foundation. P.S. was supported by an EMBO Long Term Fellowship. A.S. was supported by SNF grant #205321-105945 to M.Z. Support by the EC FP6 STREP Program LSHG-CT-2004 to W.F. is also acknowledged. Funding to pay the Open Access publication charges for this article was provided by Novartis Research Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sontheimer E.J., Carthew R.W. Silence from within: endogenous siRNAs and miRNAs. Cell. 2005;122:9–12. doi: 10.1016/j.cell.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 2.Zamore P.D., Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 3.Marques J.T., Devosse T., Wang D., Zamanian-Daryoush M., Serbinowski P., Hartmann R., Fujita T., Behlke M.A., Williams B.R. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat. Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 4.Wang Q., Carmichael G.G. Effects of length and location on the cellular response to double-stranded RNA. Microbiol. Mol. Biol. Rev. 2004;68:432–452. doi: 10.1128/MMBR.68.3.432-452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mineno J., Okamoto S., Ando T., Sato M., Chono H., Izu H., Takayama M., Asada K., Mirochnitchenko O., Inouye M., et al. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 2006;34:1765–1771. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cummins J.M., He Y., Leary R.J., Pagliarini R., Diaz L.A., Jr, Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., et al. The colorectal microRNAome. Proc. Natl Acad. Sci. USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim V.N. MicroRNA biogenesis: coordinated cropping and dicing. Nature Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 8.Hutvagner G., McLachlan J., Pasquinelli A.E., Balint E., Tuschl T., Zamore P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 9.Billy E., Brondani V., Zhang H., Muller U., Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl Acad. Sci. USA. 2001;98:14428–14433. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall T.M. Structure and function of argonaute proteins. Structure. 2005;13:1403–1408. doi: 10.1016/j.str.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 12.Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 13.Pillai R.S., Artus C.G., Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 15.Doench J.G., Petersen C.P., Sharp P.A. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutvagner G., Zamore P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 17.Yekta S., Shih I.H., Bartel D.P. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 18.Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 19.Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 20.Wu L., Fan J., Belasco J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl Acad. Sci. USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 22.Liu J., Valencia-Sanchez M.A., Hannon G.J., Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nature Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 24.Newbury S.F., Muhlemann O., Stoecklin G. Turnover in the Alps: an mRNA perspective. Workshops on mechanisms and regulation of mRNA turnover. EMBO Rep. 2006;7:143–148. doi: 10.1038/sj.embor.7400628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valencia-Sanchez M.A., Liu J., Hannon G.J., Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 26.Pillai R.S. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nature Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 29.Wienholds E., Plasterk R.H. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 30.Alvarez-Garcia I., Miska E.A. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 31.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 32.Rehwinkel J., Natalin P., Stark A., Brennecke J., Cohen S.M., Izaurralde E. Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster. Mol. Cell. Biol. 2006;26:2965–2975. doi: 10.1128/MCB.26.8.2965-2975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., et al. Combinatorial microRNA target predictions. Nature Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 34.Kiriakidou M., Nelson P.T., Kouranov A., Fitziev P., Bouyioukos C., Mourelatos Z., Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 37.Krutzfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 38.Brennecke J., Stark A., Russell R.B., Cohen S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sontheimer E.J. Assembly and function of RNA silencing complexes. Nature Rev. Mol. Cell Biol. 2005;6:127–138. doi: 10.1038/nrm1568. [DOI] [PubMed] [Google Scholar]
- 40.Doench J.G., Sharp P.A. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng Y., Wagner E.J., Cullen B.R. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 42.van de Wetering M., Oving I., Muncan V., Pon Fong M.T., Brantjes H., van Leenen D., Holstege F.C., Brummelkamp T.R., Agami R., Clevers H. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003;4:609–615. doi: 10.1038/sj.embor.embor865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haase A.D., Jaskiewicz L., Zhang H., Laine S., Sack R., Gatignol A., Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kotaja N., Bhattacharyya S.N., Jaskiewicz L., Kimmins S., Parvinen M., Filipowicz W., Sassone-Corsi P. The chromatoid body of male germ cells: Similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc. Natl Acad. Sci. USA. 2006;103:2647–2652. doi: 10.1073/pnas.0509333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elbashir S.M., Martinez J., Patkaniowska A., Lendeckel W., Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parker R., Song H. The enzymes and control of eukaryotic mRNA turnover. Nature Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 47.Rand T.A., Petersen S., Du F., Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 48.Leuschner P.J., Ameres S.L., Kueng S., Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matranga C., Tomari Y., Shin C., Bartel D.P., Zamore P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.