Abstract
Pre-mRNA splicing is performed by the spliceosome. SR proteins in this macromolecular complex are essential for both constitutive and alternative splicing. By using the SR-related protein ZNF265 as bait in a yeast two-hybrid screen, we pulled out the uncharacterized human protein XE7, which is encoded by a pseudoautosomal gene. XE7 had been identified in a large-scale proteomic analysis of the human spliceosome. It consists of two different isoforms produced by alternative splicing. The arginine/serine (RS)-rich region in the larger of these suggests a role in mRNA processing. Herein we show for the first time that XE7 is an alternative splicing regulator. XE7 interacts with ZNF265, as well as with the essential SR protein ASF/SF2. The RS-rich region of XE7 dictates both interactions. We show that XE7 localizes in the nucleus of human cells, where it colocalizes with both ZNF265 and ASF/SF2, as well as with other SR proteins, in speckles. We also demonstrate that XE7 influences alternative splice site selection of pre-mRNAs from CD44, Tra2-β1 and SRp20 minigenes. We have thus shown that the spliceosomal component XE7 resembles an SR-related splicing protein, and can influence alternative splicing.
INTRODUCTION
Removal of introns from pre-mRNAs is performed by the spliceosome, a large complex consisting of five small nuclear ribonucleoprotein particles (snRNPs; U1, U2, U4, U5 and U6) and various other proteins. Purification and analysis of the multiple proteins in the spliceosome (1–4) shows that these can be divided into different groups: SR proteins, non-snRNP spliceosome assembly proteins, U1, U2, U5 and U4/U6 snRNP specific proteins, and Sm/LSm core proteins. Members of the SR family include ASF/SF2, SC35 and SRp20. Each is required for constitutive splicing and can regulate alternative splicing. SR proteins are essential splicing factors since they can complement splicing-deficient S100 extracts. They contain one or more RNA recognition motifs (RRMs) and a C-terminal arginine/serine (RS)-rich domain in which the serine residues are phosphorylated. As well as binding to other proteins in the spliceosome via their RS domains, certain SR proteins can bind to the pre-mRNA directly (5). There is also a class of splicing factors known as SR-related proteins, which contain RS-domains of varying length. Unlike SR-proteins these may or may not contain an RRM, and instead may contain other domains such as a DEXD/H Box or a zinc finger. U2AF35, U1-70K and SRm160 are all examples of SR-related proteins (6) and recently many other proteins have been shown to belong to this family (7–14). SR proteins are distinguished from SR-related proteins by their specific domain structure (RRM domain/s followed by RS domain) and activities in splicing-deficient S100 extracts.
We have previously identified the zinc finger- and RS domain-containing protein ZNF265 as being a novel component of the mRNA processing machinery (14). This SR-related family member immunoprecipitates with mRNA from splicing extracts, interacts with U1-70K and U2AF35 and regulates splicing of Tra2β1 transcripts in a dose-dependent manner. ZNF265 exists in at least two alternatively-spliced forms, with different C-termini. The short form, in this paper referred to as ZNF265SF (GenBank Accession number NP_005446), differs by 10 amino acids from the long form, here referred to as ZNF265LF (GenBank Accession number NP_976225), and includes an alternative exon 11, containing a stop codon.
In a yeast library screen using ZNF265SF as bait we pulled out the novel protein XE7. Its gene, XE7, is pseudoautosomal (15), and generates two alternatively spliced isoforms, one with 695 amino acid residues and the other with 385 (16). The latter includes an exon that introduces an in-frame stop codon, generating a truncated protein. Expression of the gene is ubiquitous. The long isoform of XE7 is RS rich with 30% of the last 311 amino acids being either arginine or serine (16). This suggests that XE7 might be involved in RNA processing. XE7 was identified in a large-scale proteomic analysis of the human spliceosome, but under the alternative name ‘B-lymphocyte antigen precursor’ (4). To date, no functional studies on XE7 have been carried out.
Here we describe, for the first time, the characterization of XE7 and show that it is a regulator of alternative splicing.
MATERIALS AND METHODS
Plasmid constructs and cloning
Plasmids were made using cDNA generated from Calu-6 cells with Superscript II (Invitrogen). PCR products were inserted into pGEM to generate pGEM-XE7, pGEM-ASF/SF2, pGEM-ZNF265SF and pGEM-ZNF265LF. Full-length XE7 and XE7ΔRS (generated using primers 5′-aaaaaagaattctaatggcagcggctaccatcgtg-3′ and 5′-aaaaaagagctcgcttgtgccggtcagcaag-3′) were ligated into pGBK, pGAD and pCMV-HA. Full-length ASF/SF2 was ligated into pEGFP-C2 to generate EGFP (enhanced green fluorescence protein)-ASF/SF2. pACT-ASF/SF2 (ASF/SF2 in DNA-activation domain plasmid) was kindly provided by N. Hastie and R. Davies. Togenerate pGBK-ZNF265SF the primers 5′-catatgatgtcgaccaagaatttccga-3′ and 5′-gaattcgtagtggtgttccgta-3′ were used and to generate pGBK-ZNF265LF we used 5′- catatgatgtcgaccaagaatttccga-3′ and 5′-gaattcgcttcatgcactgtact-3′. pEGFP-ZNF265SF was generated using primers 5′-ctcgagtatgtcgaccaagaattt-3′ and 5′-gaattcgtagtggtgttccgta-3′ and pEGFP-ZNF265LF by using 5′-ctcgagtatgtcgaccaagaattt-3′ and 5′-gaattcgcttcatgcactgtact-3′. All ZNF265LF fragments were cloned by gel-purifying the desired fragment, after PCR, to distinguish the LF from the SF. pCMV-Myc-ZNF265SF was generated using primers 5′-aaaaaagaattcatatgtcgaccaagaattttccgagt-3′ and 5′-aaaaaactcgaggcttgactcacaggctttatttaca-3′.
Yeast two-hybrid and library screen
Saccharomyces cerevisiae strain AH109 was grown in YPDA medium (20 g/l peptone, 10 g/l yeast extract, 20 g/l glucose, 0.03 g/l adenine) and cells were transformed with binding and activation domain plasmids using the lithium acetate method according to the MATCHMAKER two-hybrid system (Clontech). The cells were subsequently plated on synthetic defined medium deficient in adenine, histidine, leucine and tryptophan [SD (−A/−H/−L/−W)] and allowed to grow for 3–5 days at 30°C. After re-streaking, true positive colonies were identified by checking for β-galactosidase production from the lacZ reporter gene using the colony-lift filter assay. Results were confirmed by a liquid culture assay using Galacton-Star reaction mixture (Clontech). A library screen was performed by mating a culture of S.cerevisiae AH109 pretransformed with pGBK-XE7 (bait) with a pretransformed foetal brain cDNA library (Clontech). After incubating the mixture with gentle swirling for 24 h at 30°C, the cells were plated on SD (−A/−H/−L/−W) and incubated at 30°C for 18 days. To identify true positives, a β-galactosidase filter assay was employed as described above and positive clones were restreaked several times. Positive clones were rescued, plasmid was extracted and transformation was conducted by electroporation into bacterial JM109 cells. After plasmid extraction, clones were sent for sequencing to the Australian Genomics Research Facility using the primer GAL4AD (5′-taccactacaatggatg-3′).
Cell culture
Cell lines were obtained from the American Type Culture Collection and cultured at 37°C under 5% CO2. Calu-6 cells were cultured in Minimum Essential Medium (Invitrogen), supplemented with 0.1 mM non-essential amino acids. HeLa cells and HEK293 cells were cultured in DMEM (Invitrogen), supplemented with 0.1 mM non-essential amino acids and 1.0 mM sodium pyruvate. 10% fetal calf serum (FCS; Invitrogen) and penicillin/streptomycin (5000 U/ml; Invitrogen) were added to all media. Constructs were transfected by Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For the transcription inhibition experiment, cells were treated with α-amanitin (50 μg/ml) for 8 h.
Antibodies
Polyclonal antibodies to XE7 were generated by Alpha Diagnostics by inoculating New Zealand white rabbits with a KLH-tagged peptide (TGDGLADRHKRERS) corresponding to amino acids 591–604 of XE7 (GenBank Accession Number Q02040). Antiserum was affinity purified by immobilizing peptide antigen on Sepharose-4B beads followed by column chromatography. Mouse monoclonal anti-SR (1H4; sc-13509) was from Santa Cruz and rabbit polyclonal anti-haemagglutinin (HA) was from Clontech. Mouse monoclonal anti-cMyc (clone 9E10) was from Sigma. Mouse monoclonal anti-GFP was from Clontech and Roche Diagnostics. Secondary antibodies were: Alexa Fluor 488 goat anti-mouse IgG and Alexa Fluor 594 goat anti-rabbit or anti-mouse IgG (all from Molecular Probes), horseradish peroxidase conjugate of goat anti-mouse or anti-rabbit IgG (Amersham Pharmacia Biotech Inc).
Immunofluorescence
For visualization of fluorescence, cells were cultured on Lab-Tek II chamber slides (Nalge Nunc International) and fixed with 100% ice-cold methanol for 1 min. After blocking with either 5% BSA or 10% goat serum in phosphate-buffered saline (PBS) for 1 h, the primary antibodies were applied, followed by the secondary antibodies for 1 h each. Cells were then stained with 300 nM 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Molecular Probes) before being mounted with gelatine/glycerol in PBS, 50:50 vol/vol (Sigma). Two- or three-channel fluorescent images were acquired on a Zeiss Axioplan 2 microscope.
Coimmunoprecipitation and western blotting
Cell extracts from 107 HeLa or Calu-6 cells were prepared using the mammalian cell lysis kit (Sigma). Approximately 500 μg of the extract was precleared with 20 μl Protein A agarose, then 3 μg antibody was added. After 4 h of incubation at 4°C, 50 μl protein A agarose was added. The immunocomplexes were incubated overnight at 4°C with frequent mixing, washed five times with PBS and resolved by 12% SDS–PAGE. Proteins were electroblotted on to a PVDF membrane (Perkin Elmer). Membranes were blocked for 1 h in 5% blocking agent (Amersham Pharmacia), followed by incubation with primary antibody for 1 h and subsequent incubation with secondary antibody for 1 h, before detection by enhanced chemiluminescence (ECL) reagents (Amersham Pharmacia) according to the manufacturer's instructions. All washes were with PBS-Tween.
Splicing assays
In vivo splicing assays were performed essentially as described (17). Briefly, a splicing reporter minigene was cotransfected with an increasing amount of XE7 expression construct (pCMV-HA-XE7) in HeLa or HEK293 cells. Empty pCMV-HA plasmids were added to ensure that the same amount of DNA was transfected. Additional EGFP-ASF/SF2 plasmid (1 μg) was added where indicated. Twenty hours after transfection RNA was isolated using QIAGEN RNAeasy columns. Reverse transcriptase (RT)-PCR for the CD44 minigene was performed using primers N3INS (5′-ctcccgggccacctccagtgcc-3′) and N5INS (5′'-gagggatccgcttcctgcccc-3′) and a program involving 30 cycles of 94°C for 20 s, 68°C for 30 s and 72°C for 40 s. For Tra2β1 we used primer pCR3.1-RT-Rev (5′-gccctctagactcgagctcga-3′) for the RT and primers MGTra-Bam (5′-gggccagttgggcgaccggcgcgtcgtgcgcgggg-3′) and MGTra-R-Xho (5′-gggctcgagtacccgattcccaacatgacg-3′) for the PCR consisting of 35 cycles of 94° for 20 s, 65° for 20 s and 72° for 40 s. For SRp20 we used primers T7 (5′-taatacgactcactataggg-3′) and X16R (5′-cctggtcgacactctagatttcctttcatttgacc-3′) for 30 cycles of 94° for 50 s, 55° for 50 s and 72° for 1 min. The PCR products were analysed with either a 2% agarose gel or a 6% polyacrylamide gel and the splicing pattern was quantified using the Image J program (Java). To confirm results some samples from the Tra2β1 and SRp20 splicing assays were also quantified with the DNA 1000 chip from the 2100 Bioanalyzer (Agilent Technologies).
RESULTS
Alignment of the RS-rich region of XE7 with the RS domain of SR/SR-related proteins
Comparison of the very RS-rich C-terminal of XE7 with the RS domain of various splicing proteins, including ZNF265 and ASF/SF2, showed that XE7 is most similar to SC35 and SRp55, and least similar to ASF/SF2 (Figure 1). The comparison also showed the strong dipeptide conservation amongst RS domain containing/RS-rich splicing proteins. By having a large RS-rich region, XE7 is similar in structure to other SR-related splicing factors, such as SRm160 (18) and SRrp53 (7) (data not shown).
Figure 1.
Strong dipeptide conservation between SR and SR-related proteins. Alignment of part of the RS-rich region of XE7 with that of SR/SR-related proteins. Black boxes show regions with >53% homology.
Subcellular localization of XE7
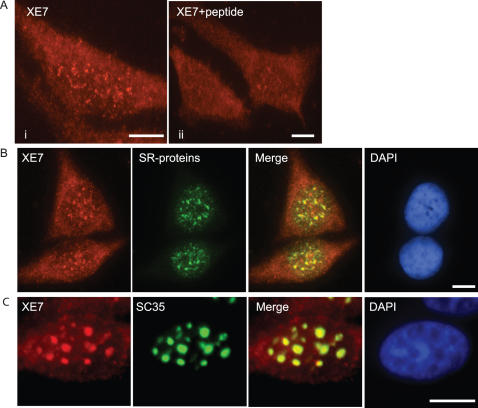
For intracellular localization studies we produced a polyclonal anti-XE7 antibody directed towards the C-terminus of the long isoform of XE7 (just 5′ of the RS-rich region) and found that endogenous XE7 was expressed in the nucleus of HeLa cells (Figure 2A, i). The staining from the XE7 antibody suggested that XE7 was also weakly expressed in the cytoplasm. However, competition with the XE7 oligopeptide antigen did reduce staining in the nucleus, showing the specificity of the antibody and suggested to us that the staining in the cytoplasm was only background (Figure 2A, ii). When performing immunolocalization using only the secondary antibody as a control we also noted staining in the cytoplasm, confirming that the cytoplasmic staining is only background (data not shown). XE7 also colocalized with SR proteins in speckles in the nucleus of Calu-6 cells (Figure 2B). XE7 localization was modified by inhibition of transcription using α-amanitin, and resulted in colocalization of XE7 with SC35 in well-defined round compartments (Figure 2C). This tighter association with round speckles upon inhibition of transcription is typical for other SR-proteins.
Figure 2.
Localization of XE7 in the nucleus (A) Endogenous localization of XE7 in HeLa cells. XE7 was detected with anti-XE7 rabbit polyclonal antibody (i). XE7 oligopeptide antigen (5 μg/ml) competes out XE7 (ii). (B) Colocalization of endogenous XE7 and SR proteins in Calu-6 cells using anti-XE7 and anti-SR. Yellow in the merged image indicates colocalization of XE7 with SR-proteins. (C) Effect of blocking transcription. HeLa cells transfected with pCMV-HA-XE7 were treated with α-amanitin (50 μg/ml) for 8 h, followed by addition of anti-HA and anti-SC35 antibodies. Yellow in the merged image indicates colocalization of XE7 with SC35 in transcriptionally inactive cells. The nucleus was stained with DAPI. Scale bars: 5 μm.
Subcellular localization of mutant XE7
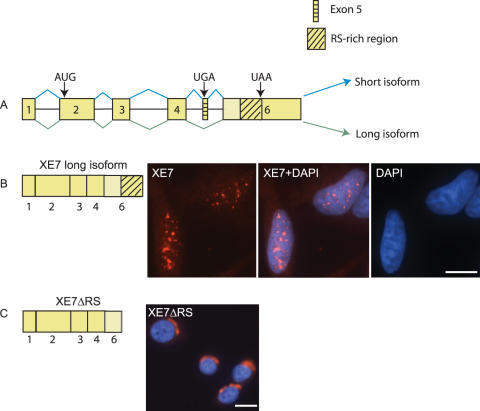
To determine which part of XE7 is responsible for its nuclear localization we used pCMV-HA constructs of XE7 and of XE7 with the RS-rich region deleted (XE7ΔRS). Figure 3A shows a schematic representation of XE7 and the constructs used are depicted in Figure 3B and C (left panels). XE7 was again found exclusively in the nucleus and exhibited a speckle-like pattern (Figure 3B). In contrast XE7ΔRS was found in the perinuclear region of the cytoplasm (Figure 3C), suggesting that the RS-rich region is responsible for XE7's nuclear localization. Localization of the short form of XE7 was also determined with a pCMV-HA construct and we found that the cellular localization was the same as for XE7ΔRS (data not shown).
Figure 3.
Cellular localization of mutant XE7. (A) XE7 pre-mRNA. The exons are shown as yellow boxes and introns as horizontal lines. The blue and green lines connecting the exons indicate the splicing patterns, either including or omitting exon 5. The blue lines depict inclusion of the alternative exon (exon 5), resulting in a premature stop codon and leading to production of the short isoform of XE7. The green lines show production of the long isoform. The start and stop codons are marked with arrows. (B) Full-length XE7 is localized to nuclear speckles in HeLa cells. A schematic representation of the full-length construct used is shown on the left. (C) Deletion of the RS-rich region causes XE7 to localize to the perinuclear region of the cytoplasm. XE7ΔRS indicates the XE7 form with the RS-rich region deleted, shown on the left. Scale bars: 10 μm.
XE7 interacts with ZNF265 isoforms and ASF/SF2
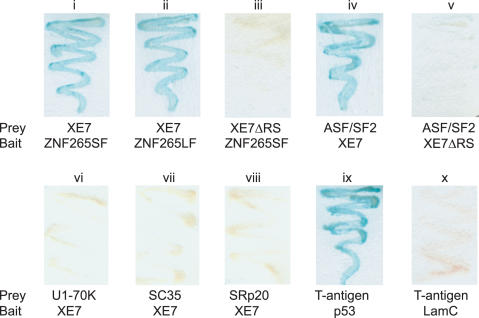
By yeast two-hybrid assay XE7 displayed a positive interaction with both forms of ZNF265, as determined by growth on SD (−L/−W/−A/−H) plates and the production of blue precipitate on a β-galactosidase filter assay (Figure 4, i and ii). Deletion of the RS-rich region from XE7 abolished the interaction (Figure 4, iii). To identify other proteins that interact with XE7 a library screen with a human brain cDNA library was performed using XE7 as bait. At least 3.5 × 106 colonies were screened and 30 positive clones were rescued and sequenced. We found that XE7 also interacts with the well-characterized SR protein ASF/SF2 (Figure 4, iv). This interaction was abolished when an XE7 construct without the RS-rich region was transformed together with ASF/SF2 (Figure 4, v). We also tested the ability of XE7 to interact with a number of other splicing proteins in a head to head yeast-two hybrid assay and found that XE7 does not interact with U1-70K, SC35, SRp20 (Figure 4, vi,vii and viii), U2AF35 or U2AF65 (data not shown). T-antigen/p53 and T-antigen/LamC were used as positive and negative controls (Figure 4, ix and x).
Figure 4.
Interaction of XE7 with ZNF265 and ASF/SF2 in yeast two-hybrid assays. Yeast were cotransformed with DNA-binding-plasmids (vector pGBK = bait) and activation-plasmids (vector pGAD or pACT =prey) encoding the proteins indicated, and colonies were grown on SD (−A/−H/−L/−W) dropout plates prior to β-galactosidase assay. p53 and T-antigen interaction was used as a positive control, and T-antigen and LamC as negative control. ZNF265SF = ZNF265 short form, ZNF265LF = ZNF265 long form. XE7ΔRS = XE7 with the RS-rich region deleted.
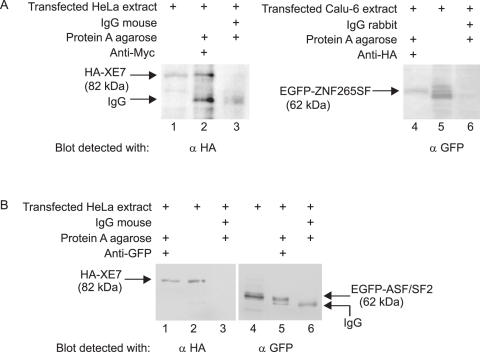
Next, we performed coimmunoprecipitation (co-IP) studies to confirm the interaction between XE7 and ZNF265 in HeLa and Calu-6 cells and between XE7 and ASF/SF2 in HeLa cells. A cell extract was prepared from cells transfected with pCMV-HA-XE7 and pCMV-Myc-ZNF265SF. Using an antibody towards the Myc epitope tag and Protein A agarose in the immunoprecipitation, we were able to detect HA-XE7 in the western blot (Figure 5A, lane 2). The XE7 and ZNF265SF association was specific since XE7 was absent in the control immunoprecipitate obtained using mouse IgG (Sigma) (Figure 5A, lane 3). A Calu-6 cell extract transfected with pCMV-HA-XE7 and pEGFP-ZNF265SF was used in another co-IP. With an HA antibody and Protein A agarose we were able to pull down EGFP-ZNF265SF in the immunoprecipitate (Figure 5A, lane 4). ZNF265SF was absent in the control immunoprecipitate obtained using rabbit IgG, again showing the specificity of the XE7 and ZNF265SF association (Figure 5A, lane 6). For the XE7–ASF/SF2 interaction, cells were transfected with pCMV-HA-XE7 and pEGFP-ASF/SF2 and incubated with GFP antibody and Protein A agarose. In a western blot of the immunoprecipitate HA-XE7 was detected using an HA antibody (Figure 5B, lane 1). The presence of ASF/SF2 in the immunoprecipitate was shown by incubating the extract with a GFP antibody together with Protein A agarose, and performing the western blot with a GFP antibody (Figure 5B, lane 5). Association of XE7 and ASF/SF2 was specific, since XE7 as well as ASF/SF2 were absent in the control immunoprecipitate using IgG mouse (Figure 5B, lanes 3 and 6). These results confirm the interaction of XE7 with ASF/SF2 and the short form of ZNF265.
Figure 5.
Interaction of XE7 with ZNF265SF and with ASF/SF2 by coimmunoprecipitation. (A) Coimmunoprecipitation of XE7 and ZNF265SF. HeLa cells were transfected with pCMV-HA-XE7 and pCMV-Myc-ZNF265SF, and Calu-6 cells with pCMV-HA-XE7 and pEGFP-ZNF265SF and cell extracts (500 μg) immunoprecipitated with 5 μg antibodies or control IgG (negative control) (lanes 3 and 6) using Protein A agarose. Ten microlitres of the complexes were resolved on SDS–PAGE gel, blotted and the presence of XE7 was confirmed with anti-HA antibody (lane 2). The presence of ZNF265SF was confirmed with a GFP antibody (lane 4). Positive control was cell extract only (lanes 1 and 5). (B) Coimmunoprecipitation of XE7 and ASF/SF2. HeLa cells were transfected with pCMV-HA-XE7 and EGFP-ASF/SF2 and cell extracts (500 μg) were immunoprecipitated with 5 μg antibodies or control IgG (lanes 3 and 6) using Protein A agarose. Ten microlitres of the complexes were resolved on SDS–PAGE gel, blotted, and the presence of HA-XE7 was confirmed with anti-HA (lane 1). Positive control was cell extract alone (lane 2). The presence of ASF/SF2 was shown when incubating the cell extract with anti-GFP in the immunoprecipitation, followed by detection of the blot with the same antibody (lane 5, upper band). Positive control for ASF/SF2 was cell extract alone (lane 4).
XE7 colocalizes with ZNF265 isoforms and ASF/SF2
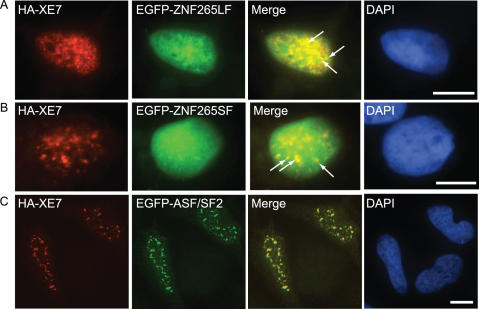
ZNF265 has a highly diffuse localization in the nucleus, thus obscuring a speckled pattern somewhat. However, we were able to detect a degree of specific colocalization of ZNF265 (both isoforms) with XE7 in HeLa cells, using pCMV-HA-XE7 and EGFP-ZNF265 constructs, which we have highlighted with arrows (Figure 6A and B). Using HA-XE7 and EGFP-ASF/SF2 constructs we found extensive colocalization of XE7 and ASF/SF2 in nuclear speckles of HeLa cells (Figure 6C) and HEK293 cells (data not shown).
Figure 6.
Colocalization of XE7 with ZNF265LF, ZNF265SF and ASF/SF2. (A) shows HeLa cells cotransfected with pCMV-HA-XE7 and EGFP-ZNF265LF (B) shows results for cotransfection of pCMV-HA-XE7 and EGFP-ZNF265SF and (C) shows pCMV-HA-XE7 and EGFP-ASF/SF2 cotransfection. The nucleus was stained with DAPI. Merged image (yellow) indicates colocalization in the nucleus of XE7 with, respectively, ZNF265LF, ZNF265SF, or ASF/SF2. Arrows denote specific colocalization of XE7 and ZNF265 isoforms in speckles. Scale bar: 10 μm.
XE7 can influence alternative splice site selection
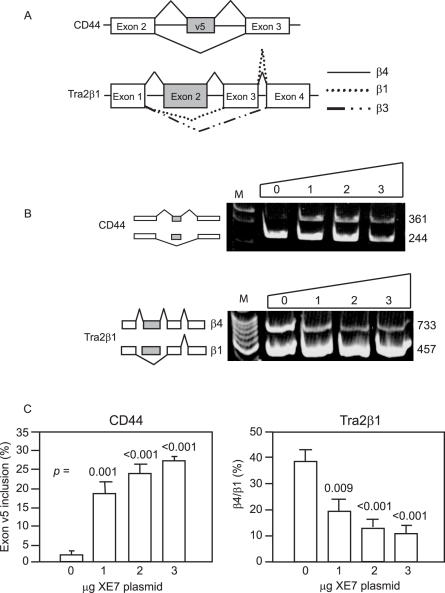
To further characterize XE7 we conducted in vivo splicing assays to test the ability of XE7 to influence alternative splicing. Several reporters containing alternative exons flanked by constitutive exons (minigenes) (Figure 7A) were transfected into HeLa cells with increasing amounts of pCMV-HA-XE7. After RNA extraction, RT–PCR was performed with primers directed at sequences located in the flanking constitutive exons. Figure 7B shows representative 6% polyacrylamide gels from the splicing assays. The most significant effect of XE7 was seen with the minigene CD44. When adding an increasing amount of XE7, inclusion of exon v5 in the pre-mRNA increased from 2% up to a maximum of 27% (Figure 7C). Overexpression of XE7 had the opposite effect on the Tra2β1 minigene, resulting in a reduction in exon 2 inclusion from 40 to 12%, so leading to a decrease in the production of the β4 alternatively spliced isoform (Figure 7C). The same effect on Tra2β1 was also seen with ZNF265 (14). Production of the β3 form represents <0.5% of total in HeLa cells and we could only detect this form using the more sensitive 2100 Bioanalyzer. Results showed that XE7 does not influence the abundance of the β3 form (data not shown). We also tested if ZNF265 could influence the effect XE7 has on splicing of Tra2β1 pre-mRNA by adding a constant amount (2 μg) of ZNF265 to the assay. However, this showed that ZNF265 influences Tra2β1 splicing independently of XE7 and does not affect the splicing performed by XE7 (data not shown). It is thus unlikely that XE7 and ZNF265 have a synergistic effect.
Figure 7.
XE7 influences alternative splice site selection of CD44 and Tra2β1 in HeLa cells in vivo. (A) Structure of the CD44 and Tra2β1 minigenes. Indicated are: alternative exon (grey) and introns (horizontal lines). Lines connecting the exons indicate the splicing patterns, either including or omitting alternative exons. (B) Promotion by XE7 of exon 5 inclusion on minigene CD44 and exclusion of exon 2 on Tra2β1 (decrease in β4 form). An increasing amount of pCMV-HA-XE7 (0–3 μg) was cotransfected with 1 μg of the minigene. Parental vector was added to ensure that similar amounts of cDNAs were transfected. The ethidium bromide stained polyacrylamide gels are from representative experiments. Sizes of the bands are shown in base pairs (bp) on the right side of the gels. M = 100 bp DNA marker. (C) Splicing results of three independent experiments using pCMV-HA-XE7 with minigenes CD44 and Tra2β1. Error bars indicate ± S.E. All P-values are from t-test vs 0 μg XE7 plasmid.
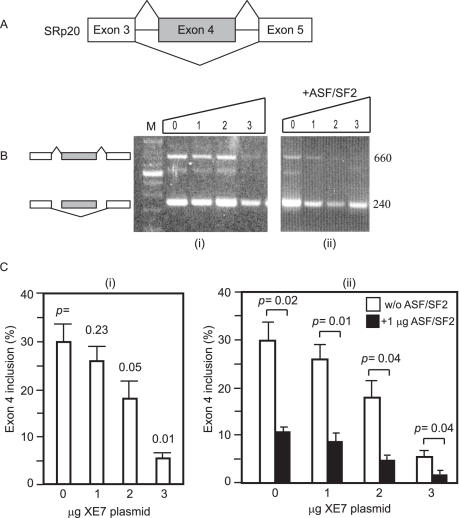
The ability of XE7 to influence the alternative splice pattern of SRp20 was tested in HEK293 cells. The SRp20 minigene construct is shown in Figure 8A, and 8B shows representative agarose gels. When adding an increasing amount of XE7, exon 4 inclusion for SRp20 decreased from 30 to 5% (Figure 8C, i). This effect on SRp20 was similar to that of ASF/SF2 on the same minigene (19). When the XE7-SRp20 splicing reaction was supplemented with a constant amount of ASF/SF2, exon 4 inclusion decreased even more (Figure 8C, ii), demonstrating that XE7 and ASF/SF2 have a synergistic effect. XE7 did not influence the splicing pattern of the minigene SMN2 (data not shown).
Figure 8.
XE7 influences alternative splice site selection of SRp20 in synergy with ASF/SF2 in HEK293 cells in vivo. (A) Structure of the SRp20 minigene. Indicated are: alternative exon (grey) and introns (horizontal lines). Lines connecting the exons indicate the splicing patterns, either including or omitting the alternative exon. (B) XE7 promotes exon 4 exclusion (i) and addition of ASF/SF2 leads to a further decrease in the form with exon 4 included (ii). An increasing amount of pCMV-HA-XE7 (0–3 μg) was cotransfected with 1 μg of the minigene. Parental vector was added to ensure that similar amounts of cDNAs were transfected. One μg of EGFP-ASF/SF2 was added in (ii). The ethidium bromide stained agarose gels are from representative experiments. Sizes of the bands are shown in bp on the right side of the gels. M = 100 bp DNA marker. (C) Splicing results of three independent experiments using pCMV-HA-XE7 (i) and of 3 independent experiments using pCMV-HA-XE7 with or without (w/o) adding 1 μg ASF/SF2 to the splicing reaction (ii). Error bars indicate ± S.E.
DISCUSSION
We describe the initial characterization of the protein XE7, encoded by one of the 24 genes (cloned or putative) located in the pseudoautosomal region 1 (PAR1) near the terminal of the short arm of the X and Y chromosomes. The human PAR1 is essential for meiotic pairing and recombination, and its deletion causes male sterility. Genes in the pseudoautosomal region escape the process of X-inactivation and thus two active copies of each gene are required for normal function in both males and females. Over 15 putative orthologues for XE7 have been found, ranging from Drosophila melanogaster (34% homology), to Canis familiaris (88% homology) and Macaca mulatta (97% homology). Little is known about the function of individual proteins from the PAR1 genes. Ours represents the first report of a protein expressed from a pseudoautosomal gene that functions in alternative splicing.
XE7 was identified in 1992 (15,16) and later that year the predicted amino acid sequence of a protein termed B-lymphocyte antigen precursor/721P was reported (20). B-lymphocyte antigen precursor was shown to be a plasma-membrane-associated glycoprotein, expressed on activated lymphocytes, most vascular endothelia and syncytiotrophoblasts (20). The sequence of B-lymphocyte antigen precursor is the same as that of XE7 up to residue 460, where a frame-shift in B-lymphocyte antigen precursor results in a different C-terminus. While it is possible that this is a genetic variant, it is more likely that this published sequence contains a sequencing error.
In support of our findings, XE7 was found recently in the human spliceosome, and was listed under the name B-lymphocyte antigen precursor (SWISS_PROT: Q02040). Herein we show that XE7 is localized to speckles in the nucleus of human cells and that localization is most likely due to the RS-rich region, as has been found for many other RS-domain-containing proteins (11,21–23). Speckles (also termed interchromatin granule clusters) are compartments that store pre-mRNA processing factors [for a review see (24)]. Our observation that localization of XE7 was modified from nuclear speckles into more defined compartments in transcriptionally inactive cells has been shown for other splicing factors (24–27). It has also been shown that SC35 and the large subunit of RNA polymerase II accumulate simultaneously in these large speckled domains that lack interconnections (28), and in this study we show that XE7 colocalizes with SC35 in the same compartments.
The spliceosome assembles onto the pre-mRNA through a series of complexes [as depicted in a recent review on alternative splicing (29)]. Pre-mRNAs most likely become incorporated co-transcriptionally into an H complex together with hnRNPs. In the E complex all consensus elements and exon splicing enhancers (ESEs) are recognized. U1 snRNP binds the 5′ splice site, U2AF65/35 binds the polypyrimidine tract and the 3′ splice site, and SR proteins bind to ESEs and contact U2AF, U1 snRNP and the branch point. Exons are further recognized in the A complex where binding of U2snRNP takes place. U4/U5 and U6 accompany B complex formation. Finally, catalysis of the splicing reaction takes place in the C complex following conformational rearrangements of RNA.
We show that XE7 interacts and colocalizes with the SR protein ASF/SF2 and the SR-related protein ZNF265. ASF/SF2 is important for almost every step of the splicing process. The RNA-binding domains of ASF/SF2 specifically recognize the exonic enhancer element (30–32) and are capable of binding 5′ splice sites (33). ASF/SF2 has also been shown to interact with the intact U1 snRNP (33,34). It has been shown that ZNF265 and ASF/SF2 interact with the U1 snRNP-specific protein U1-70K (14,33). ZNF265 and ASF/SF2 also each interact with U2AF35 (14,35). Since U1 snRNP recognizes the 5′ splice site and U2AF the 3′ splice site, the binding of ASF/SF2 and ZNF265 to U1-70K and U2AF35 may help in defining splice sites and aid in recruitment of other factors to the pre-mRNA. In contrast to most factors that bind pre-mRNA directly, XE7 does not have an RRM domain and then may not bind RNA directly. However, it is an intriguing possibility that XE7 could stabilize or strengthen the interaction between the four proteins ASF/SF2, U1-70K, ZNF265 and U2AF35. There is of course the possibility that XE7 does not interact with ASF/SF2 and ZNF265 at the same time, although it has been shown that ASF/SF2 can simultaneously interact with U1-70K and U2AF35 (35). That the RS domain of XE7 is responsible for the interaction between XE7 and ZNF265, and ASF/SF2, is not unexpected. It has been shown previously that both ASF/SF2 and SC35 interact with U2AF35 in an RS-domain-dependent manner (35). Lately, there has been some controversy regarding the function of the RS domain as an exclusive protein–protein interaction domain. Shen and Green (36) showed that the RS domain of SR proteins and U2AF65 engage directly in protein–RNA interactions. Further experiments are needed to show if the RS-rich region of XE7 interacts with pre-mRNA directly or if it instead forms a kind of ‘glue’ between proteins as postulated above.
We have also shown that XE7 has a significant effect on the alternative splicing of the minigene constructs CD44, Tra2β1 and SRp20. It seems that XE7 affects splicing of Tra2β1 pre-mRNA independently of ZNF265 and it is possible that the two proteins only bind to each other when they are not functioning as splice factors. Increased levels of CD44v5 have been found in certain tumors (37–39) and siRNA-mediated depletion of CD44v5 decreases tumour cell invasion (40). Since addition of XE7 increases exon 5 inclusion it would be interesting to test if depletion of this protein results in a decrease of CD44v5 inclusion and possibly that of tumour cell invasion, as has been shown for the SR-related protein SRm160 (40). SRm160 often exists in a complex with SRm300, and both proteins are similar to XE7 in structure since they contain RS, but not RRM, domains. The SRm160/300 complex has been shown to promote splicing through interactions with SR proteins (18), and we propose that this is also the case for XE7.
The SR protein SRp20 autoregulates its own mRNA by activating production of transcripts containing exon 4, and ASF/SF2 suppresses the use of this exon (19). In the present study we have shown that XE7 has the same effect as ASF/SF2 on the SRp20 transcript. The RNA binding domain of ASF/SF2 is responsible for specificity of exon 4 splicing (41). Since XE7 does not have an RNA domain it is possible that XE7 exerts its function by interacting with ASF/SF2 and functions as a coactivator of splicing, just as the SRm160/300 coactivator subunits described above. XE7 and ASF/SF2 do indeed seem to act synergistically, since we observed an even more defined decrease in exon 4 inclusion when a constant amount of ASF/SF2 was added to the SRp20-XE7 splicing reaction. It is possible that the binding of XE7 to ASF/SF2 helps in defining the splice sites on SRp20 and/or recruits other factors to the pre-mRNA.
Our study has characterized the full-length isoform of XE7. The alternative (small) isoform has been predicted by Hillman et al. (42) to undergo nonsense-mediated mRNA decay (NMD). This eukaryotic mRNA surveillance mechanism detects and degrades mRNAs with premature termination codons (PTCs), a termination codon being recognized as premature if it is located more than ∼50 nt upstream of the final exon. This is the case for the alternative small transcript of XE7.
Some splicing factors have been shown to autoregulate their own mRNA by promoting alternative splicing of transcripts containing PTCs, which are then degraded by NMD (43,44). This is an effective way of ensuring that correct amounts of splicing proteins are made where needed and more importantly are not expressed where not needed (or where they could do harm). Cycloheximide, a chemical inhibitor of translation, has been found to inhibit NMD (45), since NMD depends on translation. Thus it would be interesting to treat cells with cycloheximide to confirm that the short form of XE7 is degraded by NMD. It has also been shown that the RS domain of various SR proteins, amongst them ASF/SF2, is involved in enhancing NMD (46). Further experiments may show whether overexpression of ASF/SF2 has the same effect on XE7.
In summary, we show that XE7 binds to, and colocalizes with, the SR protein ASF/SF2 and the SR-related protein ZNF265. Full-length XE7 localizes to the nucleus in all cells examined and localization is modified during inhibition of transcription. We also show that XE7 influences alternative splice site selection of each of several pre-mRNA transcripts. Together, these results show that XE7 is a novel spliceosomal component, resembling SR-related splicing factors.
Acknowledgments
This work was supported by a grant from the Australian Research Council (to B.J.M). We thank Prof. N. Hastie and Prof. R. Davies, MRC, Edinburgh, UK for providing us with pACT-plasmids and Prof. S. Stamm, Erlangen, Germany, for the gift of minigene plasmids. Funding to pay the Open Access publication charges for this article was provided by the Australian Research Council grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hartmuth K., Urlaub H., Vornlocher H.P., Will C.L., Gentzel M., Wilm M., Luhrmann R. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl Acad. Sci. USA. 2002;99:16719–16724. doi: 10.1073/pnas.262483899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou Z., Licklider L.J., Gygi S.P., Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 3.Jurica M.S., Licklider L.J., Gygi S.P., Grigorieff N., Moore M.J. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rappsilber J., Ryder U., Lamond A.I., Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen H., Kan J.L.C., Green M.R. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol. Cell. 2004;13:367–376. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 6.Blencowe B.J., Bowman J.A.L., McCracken S., Rosonina E. SR-related proteins and the processing of messenger RNA precursors. Biochem. Cell Biol. 1999;77:277–291. [PubMed] [Google Scholar]
- 7.Cazalla D., Newton K., Caceres J.F. A novel SR-related protein is required for the second step of pre-mRNA splicing. Mol. Cell. Biol. 2005;25:2969–2980. doi: 10.1128/MCB.25.8.2969-2980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maita H., Kitaura H., Ariga H., Iguchi-Ariga S.M.M. CIR, a corepressor of CBF1, binds to PAP-1 and effects alternative splicing. Exp. Cell Res. 2005;303:375–387. doi: 10.1016/j.yexcr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Youn H.G., Matsumoto J., Tanaka Y., Shimotohno K. SR-related protein TAXREB803/SRL300 is an important cellular factor for the transactivational function of human T-cell lymphotropic virus type 1 Tax. J. Virol. 2003;77:10015–10027. doi: 10.1128/JVI.77.18.10015-10027.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sampson N.D., Hewitt J.E. SF4 and SFRS14, two related putative splicing factors on human chromosome 19p13.11. Gene. 2003;305:91–100. doi: 10.1016/s0378-1119(02)01230-1. [DOI] [PubMed] [Google Scholar]
- 11.Umehara H., Nishii Y., Morishima M., Kakehi Y., Kioka N., Amachi T., Koizumi J., Hagiwara M., Ueda K. Effect of cisplatin treatment on speckled distribution of a serine/arginine-rich nuclear protein CROP/Luc7A. Biochem. Biophys. Res. Commun. 2003;301:324–329. doi: 10.1016/s0006-291x(02)03017-6. [DOI] [PubMed] [Google Scholar]
- 12.Katsu R., Onogi H., Wada K., Kawaguchi Y., Hagiwara M. Novel SR-rich-related protein clasp specifically interacts with inactivated Clk4 and induces the exon EB inclusion of Clk. J. Biol. Chem. 2002;277:44220–44228. doi: 10.1074/jbc.M206504200. [DOI] [PubMed] [Google Scholar]
- 13.Barnard D.C., Patton J.G. Identification and characterization of a novel serine-arginine-rich splicing regulatory protein. Mol. Cell. Biol. 2000;20:3049–3057. doi: 10.1128/mcb.20.9.3049-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams D.J., van der Weyden L., Mayeda A., Stamm S., Morris B.J., Rasko J.E. ZNF265--a novel spliceosomal protein able to induce alternative splicing. J. Cell Biol. 2001;154:25–32. doi: 10.1083/jcb.200010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellison J., Passage M., Yu L.C., Yen P., Mohandas T.K., Shapiro L. Directed isolation of human genes that escape X inactivation. Somat. Cell Mol. Genet. 1992;18:259–268. doi: 10.1007/BF01233862. [DOI] [PubMed] [Google Scholar]
- 16.Ellison J.W., Ramos C., Yen P.H., Shapiro L.J. Structure and expression of the human pseudoautosomal gene XE7. Hum. Mol. Genet. 1992;1:691–696. [PubMed] [Google Scholar]
- 17.Stoss O., Stoilov P., Hartmann A.M., Nayler O., Stamm S. The in vivo minigene approach to analyze tissue-specific splicing. Brain Res. Brain Res. Protoc. 1999;4:383–394. doi: 10.1016/s1385-299x(99)00043-4. [DOI] [PubMed] [Google Scholar]
- 18.Blencowe B.J., Issner R., Nickerson J.A., Sharp P.A. A coactivator of pre-mRNA splicing. Genes Dev. 1998;12:996–1009. doi: 10.1101/gad.12.7.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jumaa H., Nielsen P.J. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997;16:5077–5085. doi: 10.1093/emboj/16.16.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voland J.R., Wyzykowski R.J., Huang M., Dutton R.W. Cloning and sequencing of a trophoblast-endothelial-activated lymphocyte surface protein: cDNA sequence and genomic structure. Proc. Natl Acad. Sci. USA. 1992;89:10425–10429. doi: 10.1073/pnas.89.21.10425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gama-Carvalho M., Carvalho M.P., Kehlenbach A., Valcarcel J., Carmo-Fonseca M. Nucleocytoplasmic shuttling of heterodimeric splicing factor U2AF. J. Biol. Chem. 2001;276:13104–13112. doi: 10.1074/jbc.M008759200. [DOI] [PubMed] [Google Scholar]
- 22.Cazalla D., Zhu J., Manche L., Huber E., Krainer A.R., Caceres J.F. Nuclear export and retention signals in the RS domain of SR proteins. Mol. Cell. Biol. 2002;22:6871–6882. doi: 10.1128/MCB.22.19.6871-6882.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caceres J.F., Misteli T., Screaton G.R., Spector D.L., Krainer A.R. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamond A.I., Spector D.L. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 25.Melcak I., Cermanova S., Jirsova K., Koberna K., Malinsky J., Raska I. Nuclear pre-mRNA compartmentalization: trafficking of released transcripts to splicing factor reservoirs. Mol. Biol. Cell. 2000;11:497–510. doi: 10.1091/mbc.11.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bubulya P.A., Prasanth K.V., Deerinck T.J., Gerlich D., Beaudouin J., Ellisman M.H., Ellenberg J., Spector D.L. Hypophosphorylated SR splicing factors transiently localize around active nucleolar organizing regions in telophase daughter nuclei. J. Cell Biol. 2004;167:51–63. doi: 10.1083/jcb.200404120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Keefe R.T., Mayeda A., Sadowski C.L., Krainer A.R., Spector D.L. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell Biol. 1994;124:249–260. doi: 10.1083/jcb.124.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bregman D.B., Du L., van der Zee S., Warren S.L. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matlin A.J., Clark F., Smith C.W.J. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 30.Sun Q., Mayeda A., Hampson R.K., Krainer A.R., Rottman F.M. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 31.Liu H.X., Zhang M., Krainer A.R. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kondo S., Yamamoto N., Murakami T., Okumura M., Mayeda A., Imaizumi K. Tra2 beta, SF2/ASF and SRp30c modulate the function of an exonic splicing enhancer in exon 10 of tau pre-mRNA. Genes Cells. 2004;9:121–130. doi: 10.1111/j.1356-9597.2004.00709.x. [DOI] [PubMed] [Google Scholar]
- 33.Kohtz J.D., Jamison S.F., Will C.L., Zuo P., Luhrmann R., Garcia-Blanco M.A., Manley J.L. Protein–protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 34.Xiao S.H., Manley J.L. Phosphorylation of the ASF/SF2 RS domain affects both protein–protein and protein–RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 35.Wu J.Y., Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 36.Shen H, Green M.R. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol. Cell. 2004;16:363–373. doi: 10.1016/j.molcel.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 37.Lucin K., Matusan K., Dordevic G., Stipic D. Prognostic significance of CD44 molecule in renal cell carcinoma. Croat. Med. J. 2004;45:703–708. [PubMed] [Google Scholar]
- 38.Kawano T., Yanoma S., Nakamura Y., Ozeki A., Kokatsu T., Kubota A., Furukawa M., Tsukuda M. Soluble CD44 standard, CD44 variant 5 and CD44 variant 6 and their relation to staging in head and neck cancer. Acta Otolaryngol. 2005;125:392–397. doi: 10.1080/00016480510026971. [DOI] [PubMed] [Google Scholar]
- 39.Lee S.C., Harn H.J., Lin T.S., Yeh K.T., Liu Y.C., Tsai C.S., Cheng Y.L. Prognostic significance of CD44v5 expression in human thymic epithelial neoplasms. Ann. Thorac. Surg. 2003;76:213–218. doi: 10.1016/s0003-4975(03)00319-9. [DOI] [PubMed] [Google Scholar]
- 40.Cheng C., Sharp P.A. Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol. Cell. Biol. 2006;26:362–370. doi: 10.1128/MCB.26.1.362-370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jumaa H., Nielsen P.J. Regulation of SRp20 exon 4 splicing. Biochim Biophys. Acta. 2000;1494:137–143. doi: 10.1016/s0167-4781(00)00233-5. [DOI] [PubMed] [Google Scholar]
- 42.Hillman R.T., Green R.E., Brenner S.E. An unappreciated role for RNA surveillance. Genome Biol. 2004;5:R8. doi: 10.1186/gb-2004-5-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sureau A., Gattoni R., Dooghe Y., Stevenin J., Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wollerton M.C., Gooding C., Wagner E.J., Garcia-Blanco M.A., Smith C.W.J. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- 45.Carter M.S., Doskow J., Morris P., Li S.L., Nhim R.P., Sandstedt S., Wilkinson M.F. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Z., Krainer A.R. Involvement of SR proteins in mRNA surveillance. Mol. Cell. 2004;16:597–607. doi: 10.1016/j.molcel.2004.10.031. [DOI] [PubMed] [Google Scholar]