Abstract
In the use of non-antibody proteins as affinity reagents, diversity has generally been derived from oligonucleotide-encoded random amino acids. Although specific binders of high-affinity have been selected from such libraries, random oligonucleotides often encode stop codons and amino acid combinations that affect protein folding. Recently it has been shown that specific antibody binding loops grafted into heterologous proteins can confer the specific antibody binding activity to the created chimeric protein. In this paper, we examine the use of such antibody binding loops as diversity elements. We first show that we are able to graft a lysozyme-binding antibody loop into green fluorescent protein (GFP), creating a fluorescent protein with lysozyme-binding activity. Subsequently we have developed a PCR method to harvest random binding loops from antibodies and insert them at predefined sites in any protein, using GFP as an example. The majority of such GFP chimeras remain fluorescent, indicating that binding loops do not disrupt folding. This method can be adapted to the creation of other nucleic acid libraries where diversity is flanked by regions of relative sequence conservation, and its availability sets the stage for the use of antibody loop libraries as diversity elements for selection experiments.
INTRODUCTION
It is believed that a new suite of technologies, generically termed the ‘display’ technologies will overcome many of the disadvantages associated with the generation of antibodies by immunization. In particular, they avoid animals, provide monoclonal reagents and since genes are cloned simultaneously with selection, can be easily manipulated to provide novel downstream reagents with additional properties.
Although antibody fragments were originally most commonly used as scaffolds, many other proteins have also been used successfully (1,2), with the most widely pursued being single domains based on the immunoglobulin fold: e.g. single VH (3) or VL (4) chains, camel VHH domains (5), CTLA4 (6) or fibronectin (7) domains. In general these tend to be relatively well expressed (1–10 mg/l) with affinities in the nanomolar range, although expression in intracellular compartments can be difficult due to the presence of disulfide bonds. Beyond immunoglobulin domains, nanomolar binders have also been selected from libraries based on a three helix bundle domain from protein A [Affibodies (8,9)], lipocalins [termed anticalins (10,11)], cysteine rich domains (12) and ankyrins [termed DARPINS (13,14)], with X-ray crystallography (13,15) of anticalins and ankyrins showing that the mutated residues undergo structural changes, when compared to the parent molecule, to accomodate binding.
Transformation of a protein into a binding scaffold requires the introduction of diversity at the site targeted to become the binding site. This has generally been either replacement diversity (3–6,8–11,13)—where amino acids present in the scaffold of interest, within the chosen loops or surfaces, are randomized—or insertional diversity, where a specific insertional site is chosen and stretches of random amino acids are inserted. The latter has been carried out both in antibody binding loops (16–19) and other proteins (20–24), with diversity derived from random peptides encoded by degenerate oligonucleotides or in rare cases by trinucleotide codons (25). Recently, antibodies with high affinities have also been selected from libraries where the introduced complementarity determining region (CDR) diversity is limited to only four (tyrosine, alanine, aspartate and serine) (26) or two (tyrosine and serine) (27) different amino acids at specific sites in multiple CDRs.
Nature provides a potential source of functional and well folding binding elements in the form of the binding loops which make up the antibody binding site. Antibodies contain six such binding loops, termed CDRs, which are involved in forming the antibody binding site. The first and second CDRs in both light and heavy chains are encoded by the germline V genes and subsequent mutation, while CDR3 is created as a result of recombination between V and J genes in the case of the light chain, and V, D and J genes for the heavy chain (28,29). Further diversity is created by the addition and loss of nucleotides at the junctions between the recombined gene segments (30,31) and somatic hypermutation (32). Structurally, each class of CDRs is similar in size and structure, with each adopting one or a few distinct or ‘canonical’ conformations (33–35). HCDR3 is an exception, showing wide variations in length, structure, shape and sequence (36,37), as well as intrinsic conformational diversity (38–40), reflecting the importance of HCDR3s in antibody binding specificity (41,42). Given this data, and the fact that HCDR3s also contain very few stop codons, they appear to represent a very effective form of diversity. This conclusion is bolstered by the structural conservation found at the ends of HCDR3s, revealed by the finding that the four N-terminal and six C-terminal residues from different HCDR3 regions demonstrate <2.75 Å r.m.s.d for >99.7% of all pair-wise comparisons examined (37). As a result, HCDR3s would be expected to be less disruptive to protein structure than random peptides of the same length. Furthermore, if a scaffold is able to accept a single HCDR3 at a specific site, it is likely that many different HCDR3s can also be accommodated at that same site.
Although libraries of HCDR3s have never been assessed for their effects on protein structure, a number of examples of the use of specific antibody CDRs as diversity elements able to transplant binding activity to a heterologous protein have been described. An HCDR3 from an integrin binding antibody has been inserted into an exposed loop in tissue-type plasminogen activator, conferring integrin binding activity to the plasminogen activator, without eliminating its normal enzymatic function (43). Similarly, a CDR3 from a camelid VHH recognizing lysozyme has been transplanted to neocarzinostatin, a bacterial chromoprotein with a beta sheet structure, allowing the chimeric molecule to recognize lysozyme (44). As camelid VHH CDR3s are very similar to traditional antibody HCDR3s, these two examples indicate the potential for using libraries of HCDR3s as diversity/binding elements, if means for harvesting that diversity can be developed. More recently, an HCDR1 loop from a CD4 binding antibody was inserted into three exposed loops of the protein inhibitor of neuronal nitric oxide synthase and each construct was shown to exhibit CD4 binding (45). This work was based on earlier work showing that peptides derived from five of the six CDRs of the anti-CD4 antibody, and not other regions of the variable region were able to bind CD4 as soluble, circularized peptides (46).
While these experiments show that in these specific examples, HCDR3s can be inserted into heterologous proteins without disruption of protein function, it does not demonstrate that this can be carried out generally.
In this paper we explore the possibility of using HCDR3s as a source of insertional diversity. Using the green fluorescent protein (GFP), which is not fluorescent unless correctly folded (47), as a reporter protein, we first show that the VHH CDR3 described above is also functional when inserted into two sites in GFP. Subsequently we describe a novel PCR method to harvest HCDR3 diversity, based on the fact that the N- and C-terminal HCDR3 amino acids (CXX … XXWG) are extremely well conserved at the DNA, protein and structural levels. We examine the effects of inserting antibody binding loops amplified using this method into GFP, and show that for most sites, and most HCDR3s, there is relatively little disruption to GFP function, validating HCDR3s as a potential source of diversity. These experiments set the stage for further exploration of the use of HCDR3s as diversity elements in a variety of different scaffold proteins.
MATERIALS AND METHODS
pET-CK3 expression vector construction
Four BpmI sites, one SphI and BssHII site were eliminated from pET-C6His (48), a pET-28 derivative. The SphI site was eliminated by digesting pETC6-His with SphI. The linear DNA fragment was treated with T4 DNA polymerase and re-ligated with T4 DNA ligase. The ligation was digested with SphI and transformed into DH5αFT cells. The BpmI sites and the BssHII site were mutated using the Stratagene mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's recommendation. Briefly, 100 ng of pETC-His6-ΔSphI template DNA was amplified in a 25 μl reaction using 1 μM of the primers indicated in Table 1. A total of 1 μl of dNTP mix, 0.75 μl of QuikSolution and 1 μl of QuikChange® Multi enzyme blend with the following temperature cycle: 95°C for 1 min followed by 30 cycles of 95°C for 1 min, 55°C for 1 min, 65°C for 10 min. The PCR product was digest with DpnI, BpmI and BssHII for 1 h and the mixture was transformed into XL-10 Gold® ultracompetent cells. The resulting pET-CK3 vector was checked by restriction mapping and sequencing.
Table 1.
The sequences of the primers used to create the different recipient GFP vectors
The portion in green corresponds to GFP and the portion in red, the conserved portion of the HCDR3. Underlined bases represent restriction sites (BpmI and SmaI).
SacB insertion into the GFP loops
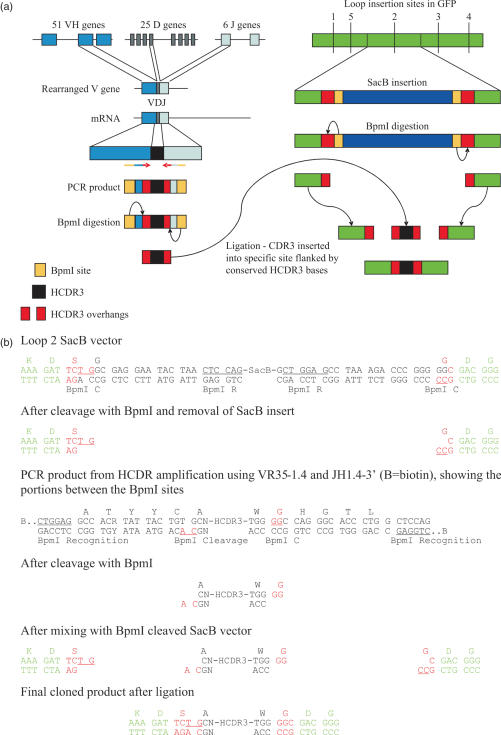
SacB is a negative selectable marker, which can be used to kill bacteria bearing by growth on sucrose (49). The SacB gene was inserted into superfolder GFP (50) at each of the different identified loop sites (Table 2) in such a way that it was flanked by two type BpmI restriction sites. These allowed the removal of the sacB gene and the creation of an appropriate cloning site for CDR3 sequences, which were also flanked with compatible BpmI sites. After BpmI cleavage, the N and C portions of a generic CDR3 were exposed, allowing the reassembly of a full CDR3 after ligation of amplified CDR3 inserts (see Figure 1). Since the sacB gene disrupts the GFP coding sequence, clones are not fluorescent unless permissive CDR3s have been inserted. These vectors were created by amplifying the full pET-CK3-sfGFP plasmid using pairs of primers flanking each insertion site. This created the following structure (illustrated for the insertion at loop 2), with the portion in green corresponding to GFP, the portion in red representing the primer encoded sequences which complement the cloned HCDR3, and the underlined bases the cleavage site for the indicated BpmI sites:
Table 2.
GFP insertion sites
| Loop | Amino acids | Loop | Amino acids | ||
|---|---|---|---|---|---|
| 1 | 23/24 | VN/GH | 1a | 22/24 | DV/N/GH |
| 5 | 51/52 | TG/KL | |||
| 2 | 102/103 | KD/DG | 2a | 101/102 | FK/DD |
| 3 | 173/174 | ED/GS | 3a | 172/173 | IE/DG |
| 4 | 213/214 | NE/KR |
The sites at which HCDR3 libraries were inserted into GFP are indicated. For loops 1, 2 and 3, two different insertion points were used as shown in the table. In loop 1a the underlined asparagine was deleted and the HCDR3s inserted between the valine and the glycine.
Figure 1.
(a) The genetic rearrangement which creates human VH genes, and the PCR strategy used to amplify, digest and purify HCDR3s is shown on the left, while on the right is shown the general scheme used to create a template for simple insertion of HCDR3s exploiting type IIs restriction sites and a negative selectable marker, such as SacB. (b) The detailed sequence of a recipient vector containing SacB inserted into loop 2, and the cloning strategy used to insert HCDR3s. The letters depicted in green represent GFP sequences, black are HCDR3 sequences, and red are junctional sequences which come together during the ligation procedure. As a result of the cloning procedure, the conserved cysteine present in the HCDR3 is converted into a serine. BpmI R (underlined) identifies the BpmI recognition site, while BpmI C identifies the cleavage site.
For each of the different loops, the primers in Table 1 were used. These PCR products were cleaved with BpmI and ligated to SacB amplified with sacB.2-5′ and sacB-3′ also cleaved with BpmI (Table 1). These SacB primers placed BpmI sites at equivalent positions, allowing the SacB gene to be removed by cleavage with BpmI in the ligated clone. After cloning, bacteria were tested for their inability to grow on both liquid and agar media containing 2–5% sucrose, as well as by restriction digestion.
HCDR3 amplification
Total RNA was prepared from 40 different samples of human peripheral blood lymphocytes purified by Ficoll Hypaque (Amersham Pharmacia Biotech, UK). Pathogens were deemed to be inactivated by the use of Trizol to purify RNA. This work was carried out under the auspices of the LANL IRB. cDNA was synthesized using random hexamers and reverse transcriptase following standard protocols. HCDR3s were amplified by nested PCR using the IgM for forward primer and a mixture of VH primers (4–6,10,12,14,22,51) with the following temperature cycle: 94°C, 60 s followed by 30 cycles of 94°C, 30 s, 55°C, 30 s, 72°C, 1 min followed by 72°C for 7 min. One microliter of the first PCR after gel purification was used as template in the second PCR. Biotinylated primers in Tables 2–5 were used to amplify the CDR3 sequences with the following temperature cycle: 94°C, 60 s followed by 30 cycles of 94°C, 30 s, 50°C, 30 s, 72°C, 1 min followed by 72°C for 7 min. The PCR product was phenol/chloroform extracted and ethanol precipitated. It was dissolved in 90 μl of water and digested with 50 U of BpmI for 2 h. The enzyme was heat inactivated at 65°C for 20 min. A total of 100 μl of M-280 Streptavidin Dynabeads (Dynal, Norway) was washed three times with TE and the beads were resuspended in the BpmI digested PCR products. The beads were mixed at room temperature for 30 min and collected with a magnet. The supernatant, which contains digested HCDR3s was used directly in the ligation reactions.
Library construction
pET-CK3-sfGFP-SacB in eight different loops were digested with BpmI, treated with Antarctic phosphatase and gel purified using the Qiagen gel purification kit. The concentration of the vector was measured by spectrofluorometer and ligations were set up with CDR3 fragment with a vector:insert ratio of 1:3 overnight at 4°C in 20 μl volume using 800 U of T4 DNA ligase (NEB). tRNA (1 μg) was added to the reactions and the total ethanol precipitated and redissolved in 50 μl of water. A total of 2 μl of each of the ligation reactions were electroporated into BL21 (DE3) Gold (Novagen) cells and plated on nitrocellulose filters on Luria–Bertani (LB) plates containing 50 μg/ml kanamycin/2% glucose/2% sucrose. Cells were grown overnight at 37°C. The filters were transferred onto kanamycin LB plates containing 1 μg/ml isopropyl-β-d-thiogalactopyranoside (IPTG) and induced for 4 h at 30°C.
Determination of c-lys affinity by flow cytometry
Streptavidin coated beads (50 μl) (Spherotec) were incubated with either 1 ng of biotinylated (Pierce Biotechnology) lysozyme (Sigma) or 15 ng of biotinylated 9E10 (anti-myc, Upstate, New York) antibody in a final volume of 100 μl phosphate-buffered saline (PBS), for 1 h at room temperature. A total of 100 μl of 5% BSA was added to block the bead surface and incubated for a further hour at room temperature. Beads were washed once in PBS and resuspended in 150 μl of PBS. GFP containing the anti-lysozyme CDR3 with an initial concentration of 0.6 mg/ml was 2-fold serially diluted and 5 μl of antigen coated beads added to 50 μl diluted protein per well. After 1 h incubation at room temperature the supernatant was removed by washing once in PBS and the beads were analyzed by flow cytometry using a FACSAria instrument (BD Biosciences). For the determination of affinity at each dilution, the mean fluorescence intensity of the non-specific binding of c-lys to the beads coated with an irrelevant target was subtracted from the specific fluorescence of the lysozyme coated ones. The resulting fluorescence values at each dilution were fitted to a logistic function using Origin (Microcal Software, Inc., Northampton, MA) and the affinity determined as the concentration at which half maximal fluorescence was obtained (52).
Flow cytometric analysis of bacterial fluorescence
Bacterial libraries expressing GFP clones were inoculated in minimal medium (53) and grown overnight at 37°C. The following day 1 ml of autoinduction medium (53) was inoculated with 10 μl of each library and grown at 30°C overnight. The cells were diluted in PBS and analyzed using a BD LSR II flow cytometer (BD Biosciences), using the 488 nm laser to excite GFP).
Protein expression, purification and characterization
All plasmids were transformed into Escherichia coli BL 21 Gold, plated on 2XTY/Kan/10% glucose and grown overnight at 37°C. Individual colonies were picked and grown overnight in liquid 2XTY/Kan/Glucose at 37°C. Confluent culture (1 ml) was used to inoculate 50 ml 2XTY/Kan/IPTG in 250 ml shaker flasks for expression at 30°C overnight.
Proteins were purified by low-salt immobilized metal affinity chromatography. Cultures were harvested by centrifugation, sonicated and resuspended in 10 mM Tris.HCl (pH 8.0) and recentrifuged at 3000 g for 30 min at 4°C. The supernatant was applied to IMAC columns pre-equilibrated with Tris for initial adhesion. The flow-through was reapplied three additional times and washed with 20 bed volumes Tris. An additional wash of 20 bed volumes of 10 mM Tris/300 mM NaCl/10% glycerol was performed preceding a final Tris wash before elution in 600 mM Imidazole. The buffers were exchanged from the eluted proteins using three passes of spin filtration with 10 000 MWCO Amicon Ultra centrifugal filtration devices at 4°C. The desalted proteins were diluted in prechilled Tris and stored at 4°C preceding further evaluation. Protein samples for SDS–PAGE comparison were diluted for equivalent fluorescence utilizing a Tecan Spectrafluor Plus plate fluorometer equipped with 485 nm excitation and 535 nm emission filters prior to standard denaturation and gel loading.
Absorption spectra were collected on a ThermoSpectronic Genesys2 and exported to Microsoft Excel for comparison. Excitation and emission spectra were generated by either a QuantumMaster 6SE (Photon Technologies Incorporated; Edison, NJ) or a SPEX Fluorolog spectrofluorometer utilizing 1 cm2 cuvettes. Excitation scans were evaluated with 509 nm emission wavelength. Emission scans were generated with 488 nm excitation. All emission scans were normalized to the maximum value obtained at the main emission peak for each sample.
Surface plasmon resonance analysis of anti-lysozyme HCDR3 clones
SPR analysis was performed on a BIAcore 2000, using a Streptavidin chip (purchased from BIAcore). Our in-house biotinylated lysozyme was used as ligand on the Streptavidin chip (flow cells 1,2,3), and our biotinylated myoglobin was used as the negative control (non-specific binding control) ligand on the same chip (flow cell 4). Approximately 4000 RUs of both ligands were bound to the chip, under which conditions specific binding could be demonstrated.
RESULTS
Inserting a defined CDR3 into GFP to confer binding activity
The HCDR3 from a VHH recognizing lysozyme has been transplanted to neocarzinostatin, a bacterial chromoprotein with a beta sheet structure, with the chimeric molecule recognizing lysozyme with an affinity of 500 nM (44). We attempted to replicate this finding, by transferring the same HCDR3 to two surface exposed loops in ‘Superfolder’ GFP (sfGFP) (50), a GFP mutant selected to be resistant to the destabilizing effects of poorly folding proteins fused to its N-terminus, and hence more stable than other forms of GFP. In order to effectively use HCDR3s as diversity elements, both structural and sequence conservation must exist at the N- and C-terminal ends of the isolated HCDR3. Structural conservation is required to ensure that once a permissive site has been chosen, different HCDR3s can be inserted equally effectively at the same site, while sequence conservation is required to allow effective cloning of the isolated HCDR3s. Within the four N-terminal and six C-terminal amino acids from different HCDR3 regions found to be structurally similar by Morea et al. (37), the DNA sequences encoding the N-terminal cysteine and C-terminal tryptophan and glycine are extremely conserved. As the cysteine 104 [IMGT numbering (54)] usually forms a double hydrogen bond with the glycine 119 (37), these two amino acids were chosen to be the limits of the cloned HCDR3. However, to avoid the presence of an unpaired cysteine (the HCDR3 N-terminal cysteine normally disulfide bonds with another cysteine in framework one), this codon was mutated to a serine. This is identical to cysteine, except for the replacement of sulfur by oxygen, and so is able to form the same hydrogen bonds. In order to create recipient GFPs which could be used for cloning HCDR3 libraries, as well the specific anti-lysozyme HCDR3, we inserted (see Figure 1 and below) a SacB gene at each targeted insertion site flanked by BpmI sites. The SacB gene is a negative selector able to reduce vector background by 105-fold by plating bacteria on sucrose after transformation (49,55). BpmI is a type IIs restriction site which cleaves 14/16 bp away from its recognition site. The cleavage sites were designed to include conserved 5′- and 3′-HCDR3 sequences, which were exposed after digestion by BpmI, allowing the reconstruction of full-length HCDR3s within the GFP from either annealed oligonucleotides or amplified PCR fragments.
The anti-lysozyme HCDR3 described above was synthesized as a pair of overlapping phosphorylated oligonucleotides. These were annealed and ligated directly into two of the BpmI cut vectors (Figure 1). Loops 1 and 3 (see Table 2 for nomenclature) were both independently targeted. Both clones, named c-lys1 and c-lys3, depending upon the loop insertion site, yielded fluorescent proteins. These were expressed in BL21 using either 100 μM IPTG or autoinduction media (53), and subsequently purified by immobilized metal affinity chromatography using the C-terminal His6 tag.
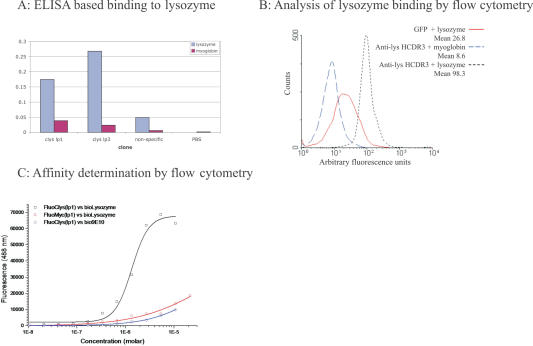
Binding between GFP containing this HCDR3 was demonstrated in an enzyme-linked immunosorbent assay (ELISA) format in which the lysozyme was biotinylated and interacted with the modified GFP prior to capture on a neutravidin coated plate (Figure 2A). Specific binding could also be demonstrated using a flow cytometric bead based method, in which detection was carried out by measuring the fluorescence of streptavidin coated beads to which biotinylated lysozyme and GFP containing the anti-lys HCDR3 had bound (Figure 2B). Unlike ELISA, this method relies on the intrinsic fluorescence of the binder, demonstrating that binding activity and fluorescence reside in the same protein, and that, at least in this case, HCDR3 insertion has not disrupted GFP function. This technique can also be used to determine affinity (52) by incubating microspheres coated with antigen with increasing concentrations of fluorescent binder. As concentration increases, the bead bound fluorescence reaches a plateau, as all target sites on the microspheres are bound. By subtracting background binding and determining the concentration of fluorescent binder at which half maximum fluorescence is obtained, we were able to estimate the affinity of this interaction to be 1.34 μM for c-lys1 (Figure 2C), similar to the estimate, obtained by isothermal calorimetry, for neocarzinostatin containing the same HCDR3 (44). Binding was also examined using surface plasmon resonance. Although, specific binding could be demonstrated when chips were densely coated with lysozyme, similar to the neocarzinostatin results (44), affinity was not high enough to show binding when the lower levels of coating required to determine affinity were used (data not shown).
Figure 2.
(A) ELISA was carried out by first interacting biotinylated antigen with the GFP clone of interest and then capturing on a neutravidin coated plate. GFP binding was revealed using SV5, a monoclonal antibody (73), which recognizes a tag appended to the C-terminus. The non-specific clone was GFP containing the myc epitope (74) at the loop 3 position, and indicates the level of binding due to a similarly disrupted GFP molecule. clyslp1 and clyslp3 have the lysozyme binding HCDR3 inserted into loop 1 and loop 3, respectively. (B) Streptavidin coated beads were incubated with either biotinylated lysozyme or myoglobin, (which serves as the negative control for non-specific binding to coupled beads) and subsequently with GFP or GFP containing the anti-lysozyme HCDR3 inserted at loop 1 (c-lys1). Analysis was carried out using a FACSCalibur. (C) As in (B), except that different concentrations of c-lys1 were used and the mean fluorescence of the non-specific (myoglobin) beads was subtracted from the fluorescence of the lysozyme coated beads and plotted. The affinity is calculated from the concentration of c-lys1, which gives half maximal fluorescence.
These results indicate that the orientation and structure of the HCDR3 is maintained at both insertion sites and is similar to that in the original VHH, suggesting that HCDR3s are a valid potential source of diversity, and may alone provide sufficient binding energy to yield micromolar binders.
Analyzing human HCDR3 flanking sequences
In order to determine the best way to clone HCDR3s, 5669 human heavy chain variable genes were downloaded from the IMGT web site (56), using ‘human heavy chain variable genes of any specificity’ as search criteria. These were pared down to 5646 full-length VH genes representing a wide spectrum of different V genes, and encompass the full range of mutations found at all different sites within the V genes, including potential primer sites flanking the CDR3. Of these 5646 VH genes, 4842 can be accounted for by searching for the following motifs (all based on the 10 bases finishing with the extremely conserved cysteine and the base which follows it—TGT G) found at the 3′ end of framework region 3 just before the CDR3 (number of times found):
TATTACTGTG (4061)
TATTATTGTG (462)
TATTTCTGTG (319)
As amplification usually requires more than 10 bases of homology, the 4842 sequences described above were extracted from the database and analyzed for homology upstream of these 10 bp sequences. Based on the homology found, the sequences described in Table 3 were designed. A similar procedure was carried out for the 3′ end of the HCDR3 in the JH gene sequences. However, in this case, the sequences were centered around the highly conserved CTGGGGCC sequence found in all JH genes. The alignment of these sequences with germline V and J genes is given in Table 4.
Table 3.
Primer analysis
| Primer | Sequence | Degeneracy | Rearranged V | Germline V |
|---|---|---|---|---|
| 5′ set | ||||
| VR35-1.4 | Gccacrtattactgtg | 2 | 123 (72, 51) | 3 |
| VR35-2.4 | Gccatntattactgtg | 4 | 434 (107, 30, 258, 39) | 3 |
| VR35-3.4 | Gccgthtattactgtg | 3 | 577 (356, 148, 73) | 1 |
| VR35-4.4 | Gccttgtattactgtg | 1 | 66 | 2 |
| VR35-5.4 | Gctgthtattactgtg | 3 | 391 (120, 162, 109) | 0 |
| VR35-6.4 | Gcygtgtattactgtg | 2 | 2118 (974, 1144) | 35 |
| VR35-7.4 | GCYGTVTATTATTGTG | 6 | 303 (33, 36, 95, 23, 49, 67) | 0 |
| VR35-8.4 | GCYGTNTATTTCTGTG | 8 | 170 (17, 23, 37, 17, 8, 13, 47, 9) | 0 |
| Total | 4182/5646 74% | 44/49 90% | ||
| 3′ set | ||||
| JH1.4-3′ | CCAGGGTGCCCTGGCCCCA | 1 | 170 | JH4, JH5 |
| JH2.4-3′ | CCAGGGTGCCACGGCCCCA | 1 | 94 | JH6 |
| JH3.4-3′ | CCATTGTCCCTTGGCCCCA | 1 | 487 | JH3 |
| JH4.4-3′ | CCAGGGTTCCCTGGCCCCA | 1 | 1958 | JH1 |
| JH6.4-3′ | CCGTGGTCCCTTGGCCCCA | 1 | 802 | JH2 |
| Total | 3511/5646 62% | 6/6 100% | ||
Sequences of those portions of the 5′ and 3′ primers corresponding to the VH or JH genes suitable for the amplification of HCDR3s are shown. These were analyzed by using only the portion of the primer which recognizes the V or J gene and searching against the database of 5646 rearranged V genes or the 49 germline V genes or 6 JH genes. These analyses are stringent (100%), so it is likely that in real experimental situations, more sequences are likely to be amplified, as in each case the 3′ primer sequences are extremely well conserved. Under ‘Rearranged V’, the total numbers of the 5646 rearranged VH genes downloaded from IMGT with absolute homology to each of the primers are given. In brackets are given the number of VH genes recognized by each of the individual primers making up the degenerate pool. Under ‘Germline V’, the number of germline VH genes with 100% homology to the primers is given.
Table 4.
Primer localization
The sequences shown represent the full diversity found in the germline V genes centered around the conserved cysteine (TGT). All six JH sequences are indicated. For both VH and JH, the region of primer recognition is given, and the cut site when BpmI is used is underlined. The IMGT numbering system is used.
Seven of the thirteen primer sequences were degenerate, with each component sequence recognizing V genes. Table 3 shows the numbers of the 5646 rearranged and germline VH genes recognized by each of these sequences, assuming 100% homology. In experimental use, it is likely that many more genes will be recognized by each individual primer, since single mismatches do not usually prevent PCR amplification, especially when found upstream of nine homologous bases.
Cloning design
The strategy used to clone HCDR3s amplified from B cells into GFP is described in Figure 1a and detailed in Figure 1b. The GFP recipient vectors contain the SacB gene flanked by the highly conserved 5′ and 3′ portion of the HCDR3. GFP containing SacB at the different insert sites is non-fluorescent, and the only way fluorescence can be restored is by removal and replacement of the sacB negative selector with a sequence that encodes a peptide permissive for GFP folding at the targeted insertion site. In order to isolate these sequences, without flanking framework sequences, and to recreate full-length HCDR3s, a type IIs restriction site, BpmI, was used. This cuts 16/14 bases away from its recognition site, allowing it to be placed upstream of an amplifying oligonucleotide in such a way that the majority of the oligonucleotide sequence can be removed after PCR, leaving a 2 bp 3′ extension for ligation (see Figure 1b). Based on the sequences described in Tables 4 and 5, the BpmI site in the 5′ HCDR3 primer was placed to cleave across the conserved cysteine and adjacent codon (TGTG) (Tables 4 and 5), while at the 3′ end it was placed to cleave within the conserved tryptophan codon (TGGGGC). Overhanging bases are underlined in both cases. Altogether, eight vectors containing SacB insertions at eight different sites, comprising five different loops, were created (Table 2 and below).
Table 5.
HCDR3 amplification primer sequences
The 5′ and 3′ primer sequences used are shown. Each primer is biotinylated at the 5′ end. The biotin is followed by four bases to assist in recognition and cleavage by the type IIs enzyme, BpmI (recognition sequence CTGGAG16/14). The cut site is underlined.
In order to eliminate the primer ends removed by BpmI cleavage (corresponding to the framework sequences), the oligonucleotides were biotinylated at their 5′ ends (Table 5), allowing their removal with streptavidin Dynabeads.
PCR amplification and cloning
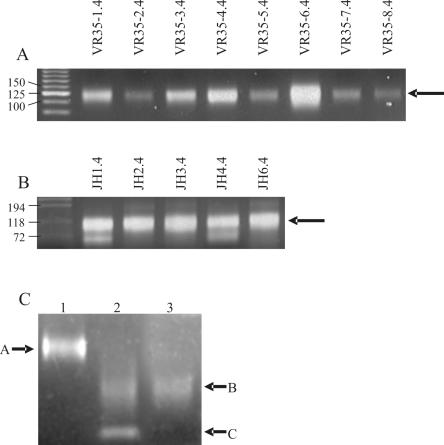
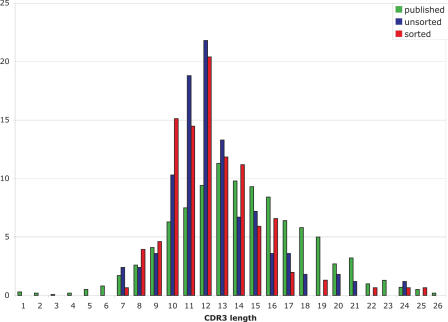
Non-biotinylated primers were tested by amplifying HCDR3s from the peripheral blood lymphoctye cDNA of 40 donors using non-biotinylated primers. Using each individual primer with a pool of the complementary primers yielded a 75–150 bp smear for all primers (Figure 3A and B), which is more visible for the primers recognizing more rearranged V genes (e.g. VR-35–36.4). When the length of the primers is taken into account, this corresponds to HCDR3s ranging from ∼20–95 bp, similar to the previously published range of HCDR3 lengths (36) (24–90 bp and Figure 5).
Figure 3.
PCR amplification of lymphocyte cDNA using a mixture of the 3′ J region primers and individual 5′ V gene primers (a), or a mixture of the 5′ V gene primers and the individual J region primers (b). In (c), the pooled PCR product prior to digestion is shown in lane 1. In lane 2 the PCR product after digestion with BpmI, and in lane 3 the product after the biotinylated primers are removed. In each case the arrows show the amplified HCDR3s except C in 3c.
Biotinylated versions of the primers were created and pooled for amplification before purification and cloning. Unfortunately, primer JH4.4-3 had to be omitted, due to the sub-optimal quality of the biotinylated primer. Lane 1 in Figure 3C shows the HCDR3 smear prior to digestion with BpmI, ranging from 75 to 150 bp (arrow A). After digestion (lane 2), the smear is reduced in size by 55 bp (range 25–95 bp, arrow B), corresponding to the primer portions removed by BpmI, which can be seen as an additional sharp lower band (arrow C). Lane 3 shows the final purified HCDR3 preparation after removal of the biotinylated primers using magnetic streptavidin beads (Dynabeads). The extent and intensity of the smear is essentially identical to that in lane 2, except that the lower band is eliminated, indicating the efficiency of the use of streptavidin to remove the biotinylated external primer portions.
HCDR3 libraries
In order to assess the effects of the insertion of many different HCDR3s on protein folding in general, and GFP folding and function in particular, five different loops in GFP were targeted for the insertion of HCDR3 libraries (Table 2). The insertion sites were identified by an examination of the structure of GFP (57), with the goal of placing the HCDR3s at the tips of the loops, and so hopefully continue the GFP beta strand structure into the first part of the HCDR3. In addition, one alternative site, differing by a single amino acid, and consequently slightly off the tip center, was also targeted for three of the loops (1, 2 and 3 in Table 2), to see whether insertion within loops had to be precisely localized, or whether it was sufficient to target a loop, without concern for the exact insertion sites.
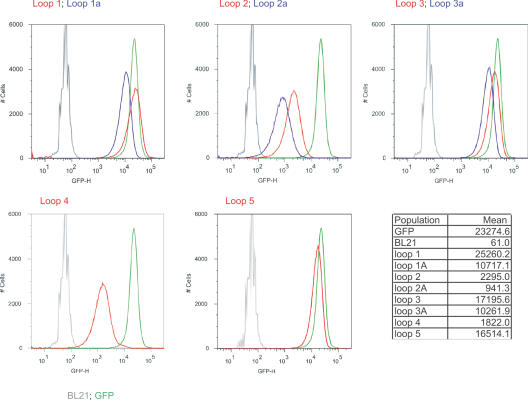
The HCDR3 fragments were gel purified and cloned into the eight recipient GFP vectors. The libraries were then induced and analyzed by flow cytometry. Each of the libraries showed significant numbers of fluorescent bacteria (Figure 4) when induced with IPTG, with loops 1, 1a, 3, 3a and 5 providing the greatest number, many of which overlapped with the fluorescence of bacteria expressing GFP. The mean fluorescence for these libraries ranged from 10 711–25 260, while GFP had a mean fluorescence of 23 274. The remaining loops (2, 2a, 4) were less fluorescent (941–2295), although still significantly more fluorescent than BL21 (mean 61). All the fluorescent profiles for bacteria containing GFP with inserts were broader than GFP with a slightly longer tail on the low fluorescence portion of the curve. The differences between libraries inserted at different loops sites differed by as much as 27-fold (loop 2a compared to loop 1), while the greatest difference between insertions in the same loop differing by a single amino acid were in no case greater than 2.5. This suggests that the primary determinant of fluorescence is the targeted loop, with the site within the loop playing a lesser role.
Figure 4.
Flow cytometric analysis of bacteria containing GFP with HCDR3 libraries at different insertion positions. In each panel the grey line represents BL21 bacteria, and the green line BL21 expressing GFP. Each panel represents libraries inserted at a different loop, with different positions indicated for loops 1, 2 and 3. The mean fluorescence for each of the populations, calculated from the plots, is given in the table.
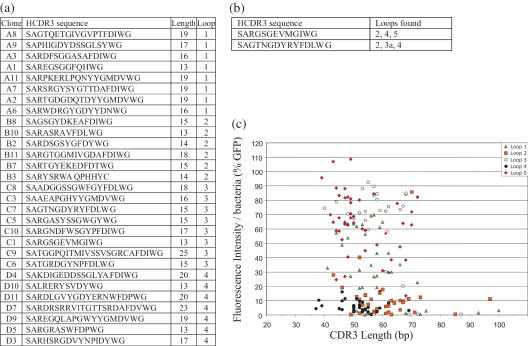
Sequence analysis of HCDR3s
302 random clones containing inserts were sequenced to further analyze the nature of the cloned diversity. The length distributions of the HCDR3s (Figure 5) showed a slight increase in shorter HCDR3s (14–16 amino acid), and a reduction in longer ones (>21 amino acid) compared to the HCDR3 length distributions reported in the literature (36). All inserts were in frame with GFP, and nucleotide blast searches showed homology to HCDR3s (Figure 6a shows representative sequences). Only 1.3% of the sequences contained stop codons, and 4.8% lacked the characteristic C(S) … . WG HCDR3 sequence. In addition there were 18 sequences repeated more than once, corresponding to 5.9% of all sequences. Interestingly, two of these duplicated HCDR3 sequences were each found in three different loops (Figure 6b) indicating duplication was present in the source cDNA, rather than a cloning artifact. This has been previously observed in sequencing human peripheral B cell V genes (58), and reflects different VH gene pools with different representations, such as those derived from recent infections, rather than the saturation of diversity.
Figure 5.
The length distribution of cloned HCDR3s derived from bacterial libraries, which were unsorted (blue), or sorted for fluorescence (red), with published distributions (36) in green.
Figure 6.
(a) Shows the sequence of random HCDR3s cloned into the different loops. In (b) are shown two HCDR3 sequences whose sequences were found in different loops. (c) Shows the correlation between HCDR3 length and bacterial fluorescence as expressed as a percentage of the fluorescence of bacteria expressing GFP.
A number of clones from each library were picked at random and analyzed for their fluorescence, and correlated with the length of the HCDR3 insert. Figure 6c shows the fluorescence of individual clones expressed as a percentage of GFP fluorescence, correlated with the length of the inserted HCDR3. As can be seen, the spread of HCDR3s ranges from 40 to 75 bp, with little correlation between length and fluorescence, except beyond 80 bp, where fluorescence is reduced. This indicates that providing the HCDR3 length is within the normal range (40–75 bp), larger HCDR3s do not reduce fluorescence more frequently than smaller HCDR3s.
In order to determine whether there were any differences between HCDR3s found in strongly fluorescent clones compared to weakly fluorescent ones, bacteria containing the libraries were flow sorted for fluorescence, and an additional 434 sequences analyzed. Although the length distributions of the HCDR3s in the unsorted and sorted libraries were extremely similar (see Figure 5), the percentages of repeated sequences (1.6%), non-characteristic HCDR3 sequences (2%) and sequences containing stop codons (0.7%) were all reduced.
Examination of protein properties
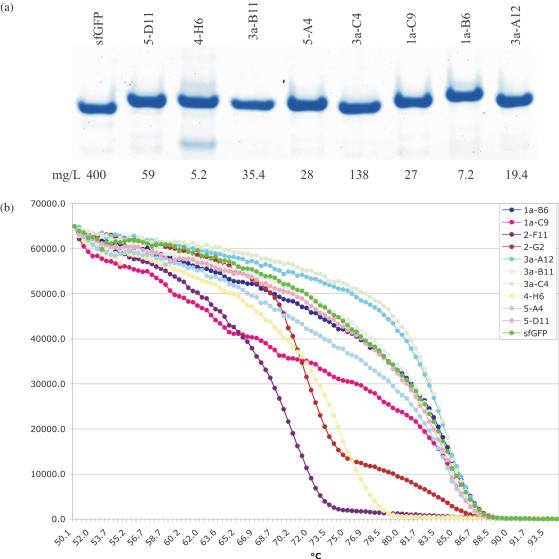
A number of fluorescent clones containing HCDR3 inserts were expressed and purified to study their properties compared to GFP. The expected size differences between GFP and clones containing inserts were apparent for all clones (Figure 7a). The expression levels of the different clones ranges from 5.2–138 mg/l, 1.3–30% the level of GFP. Normalized absorption/emission spectra were essentially identical to GFP (data not shown).
Figure 7.
(a) Shows a polyacrylamide gel of different purified clones containing HCDR3 inserts at different positions. (b) Shows the normalized fluorescent emission of different clones gradually heated up at the rate of 0.1°C/second. For each clone, the first figure indicates the loop insertion site.
The stability of some of these proteins was studied using a real time PCR machine in which protein fluorescence was monitored as temperature was gradually raised (0.1°C/s) (59). Figure 7d shows fluorescence levels for GFP and a number of different clones, scaled to start at the same fluorescent level. All proteins, with the exception of 2-G2, showed two phase melting curves with an initial slow phase for 15–30°C after 50°C, followed by a sharp transition to complete melting. The midpoint of the cooperative transition for GFP was 83.5°C, while clones containing HCDR3 inserts ranged from ∼71–83.5°C, with two clones (3a-A12 and 3a-C4) slightly more stable than GFP. By 88.5°C all clones had completely lost fluorescence. This melting pattern is similar to that shown by the engineered ankyrins, with a less steep first phase than that seen with the ankyrins (60). Although not examined in detail, the proteins with the lower thermal stabilities tended to come from the less fluorescent libraries (loops 2, 2a and 4).
DISCUSSION
Antibody CDRs, or hypervariable regions, are the portions of the variable regions, which constitute the antigen binding loops. Of the six CDRs, the HCDR3 is the most diverse, reflecting its complex genetic origin (Figure 1). It also plays the most important role in antigen recognition, as shown by the isolation of pairs of antibodies recognizing different antigens which differ only in their HCDR3s (41,42). This has not been shown for any of the other CDRs, indicating that HCDR3s alone, within the context of an antibody variable domain, are able to provide sufficient binding affinity to discriminate between different antigens. Furthermore, it has recently been shown that some antibodies are able to bind distinctly different antigens as a result of conformational flexibility, in which the HCDR3 undergoes large structural changes when binding (38), a property likely to be far more widespread than hitherto expected. Coupled with the known length (36), chemical (36) and structural (37) diversity of HCDR3s, as well as the relative lack of stop codons, these results indicate that HCDR3s may serve as useful sources of diversity if ways could be found to transplant them from antibodies to alternative scaffolds with different properties. In this regard, a tissue plasminogen activator (TPA) able to recognize integrins αvβ3 and αIIbβ3 was created by inserting the HCDR3 from a recombinant anti-integrin antibody (61) into a TPA loop flanked by a beta sheet structure (43), and a functional neocarzinostatin derivative able to recognize lysozyme was similarly created by grafting an HCDR3 from a camelid anti-lysozyme VHH to a surface loop (44). In the first experiments reported here, we were also able to transfer this VHH CDR3 to two different GFP loops and confer lysozyme-binding with an affinity comparable to that obtained when transferred into neocarzinostatin, indicating the generality of transferring HCDR3s with specific binding properties to alternative scaffolds.
The main problem with isolating libraries of HCDR3s, rather than specific ones, is the fact that their diversity is embedded within relatively conserved structured beta sheet framework regions that form extensive contacts with other VH region amino acids. In this paper we overcome this problem with a PCR based method that uses flanking type IIs restriction sites to remove the framework regions after amplification. This relies on the structural, DNA and amino acid sequence conservation found at either end of human HCDR3s: cysteine 104 and tryptophan 119 (IMGT numbering) are essentially 100% conserved at the amino acid and nucleotide levels. Structurally these amino acids are joined by two hydrogen bonds and so very close to one another, providing further justification for their use as diversity elements. This allowed us to design 13 primers annealing within the flanking framework regions which were able to amplify a large percentage of rearranged VH gene CDR3s. By adding BpmI sites at the ends of the primers, these framework regions could be removed, leaving two base pair overhangs residing within the conserved amino acids at either end of the HCDR3. These overhangs were ligated to sequences encoding the remaining portion of the HCDR3 (with the cysteine exchanged for serine), exposed when the negative selector (SacB) gene was removed from GFP by digestion with BpmI (Figure 1). This arrangement facilitated the cloning of a diverse set of HCDR3s, independently of knowledge of their sequences.
By inserting HCDR3 libraries into the tips of five loops on one end of GFP, we were able to assess the degree of disturbance these HCDR3s caused to folding, since GFP must fold correctly to become fluorescent (47). Of the eight sites examined, five were very permissive, giving mean bacterial fluorescence profiles within 2.3-fold of GFP. The remaining three sites, although up to 25-fold less fluorescent than GFP, were nevertheless significantly fluorescent. The mean fluorescence per cell is a combination of the number of fluorescent proteins per cell and the intrinsic fluorescence of those fluorescent proteins. In Figure 7a, the amount of each fluorescent protein loaded on the polyacrylamide gel was normalized for fluorescence. The similar intensity of the coomassie blue staining suggests that the greatest variability between different bacterial clones is in the amount of expressed fluorescent protein, rather than any difference in intrinsic fluorescence. This is supported by the differing levels of protein which can be purified from each clone.
For three of the sites (loops 1, 2 and 3) two insertion sites were tested. These differed by a single amino acid, and the rationale was to determine whether it was sufficient to place the HCDR3 within a permissive loop, or whether the exact site within that loop was critical. Although there was a small (<2.5-fold) difference in the fluorescence at the two sites for each loop, this was far less than the difference observed between different loops (up to 27-fold), suggesting, at least within this small set, that the loop targeted is more important than the precise position within the loop. We did not examine insertion at other sites, and it is possible that insertion of HCDR3s into secondary structures which are not loops, or perhaps at the loop extremities, may be significantly more disruptive, especially since loops tend to be less well conserved than more structured elements.
We sequenced over 300 cloned HCDR3s. Over 90% had the characteristic HCDR3 sequence: C(S)XX … XXWG. The sequence characteristics of HCDR3s more permissive for GFP fluorescence were identified by flow sorting bacteria expressing GFP containing HCDR3 libraries in different loops. We found that HCDR3s in the more fluorescent clones were less likely to contain stop codons, non-characteristic or repeated HCDR3 sequences. There was no bias in favor of either bulged or non-bulged HCDR3s (37), with the proportion remaining constant before and after sorting. As both non-characteristic and repeated HCDR3s are more likely to be derived from heavily mutated clones, this may explain their detrimental effect on GFP fluorescence.
GFP is known to be an extremely stable protein. The GFP we used, superfolder (50), was selected to be particularly resistant to the effects of poorly folding proteins fused to its N-terminus. This also confers improved stability to the protein generally (50). This was confirmed in our thermal stability studies, in which the cooperative transition midpoint was 83.5°C, not dissimilar to GFP containing loops which ranged from 71 to 83.5°C. This minimal disturbance likely reflects both the stability of GFP as well as the relatively non-perturbing nature of the HCDR3 inserts, which was also shown by the fact that the spectral properties of these proteins were essentially identical to GFP.
This study was carried out with human HCDR3s because of the great deal of sequencing information available. This made the design of appropriate framework primers relatively straightforward. However, with sufficient information on the sequences of appropriate flanking regions, this approach could be applied to the harvesting of diversity from the antibody genes of other species. This may be of more than academic interest, since the means by which antibody diversity has evolved in different species, although sharing some commonalities, has tended to be species-specific (62,63). As a result, different CDRs have different properties, lengths and amino acid distributions in different species, with tendencies to bind to different classes of antigens. Cow (64) and dromedary heavy chain genes (65,66), e.g. have far longer HCDR3s than humans. In camels, these have been shown to be important in the mediation of enzyme inhibition by direct insertion into active sites (67,68), as well the recognition of conserved cryptic epitopes of infectious agents, perhaps by penetration into conserved receptor binding sites (69). Although less dramatically different, murine HCDR3s tend to be shorter than human HCDR3s, with different amino acid compositions (36), resulting in more HCDR3s which have stabilized hydrogen bond ladder structures, as opposed to human HCDR3s which contain more prolines, preventing the formation of such ladders.
The method we describe here can also be adapted to other antibody CDRs, as well as to other immunological proteins, such as T cell receptors, which share similar primary structures: variable regions flanked by relatively conserved framework regions. γ and δ TCRs have CDR3s which resemble immunoglobulin heavy chain CDR3s in their length variability, while α and β TCRs have CDR3s, which are extremely homogenous in length (8–9 amino acids), in common with the other antibody CDRs (70). The common component of all such diversity elements is that they have evolved to bind, and so are likely to be more functional than random peptides, which generally contain more stop codons and are more likely to contain destabilizing inserts.
An alternative to the use of completely random amino acids, has been the use of restricted amino acid sets in the generation of antibody libraries (26,27,71,72). In these experiments it has been shown that different amino acid diversities at specific sites significantly affect the successful outcome of selection experiments, and in one of the most surprising results, specific high affinity antibodies can be selected from libraries in which heavy chain diversity is limited to only two amino acids (27). These careful studies of the roles of different amino acids in functional diversity, are similar to those which nature has been conducting over evolutionary time in the different molecules involved in immune recognition in many different species. The method described here enables the harvesting of such diversity for transplantation into heterologous proteins, setting the stage for the exploration of the use of libraries containing such sequences for selection experiments.
Acknowledgments
This work was funded by a DOE GTL pilot grant, and LANL LDRD-DR funds, to A.R.M.B. The authors are grateful to the National Flow Cytometry Resource at LANL for help with sorting, and to Prof. C. Kado (UC Davis) for the SacB gene. Funding to pay the Open Access publication charges for this article was provided by the DOE GTL grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Skerra A. Engineered protein scaffolds for molecular recognition. J. Mol. Recognit. 2000;13:167–187. doi: 10.1002/1099-1352(200007/08)13:4<167::AID-JMR502>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 2.Nygren P.A., Skerra A. Binding proteins from alternative scaffolds. J. Immunol. Meth. 2004;290:3–28. doi: 10.1016/j.jim.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Davies J., Riechmann L. Antibody VH domains as small recognition units. Biotechnology (N Y) 1995;13:475–479. doi: 10.1038/nbt0595-475. [DOI] [PubMed] [Google Scholar]
- 4.van den Beucken T., van Neer N., Sablon E., Desmet J., Celis L., Hoogenboom H.R., Hufton S.E. Building novel binding ligands to B7.1 and B7.2 based on human antibody single variable light chain domains. J. Mol. Biol. 2001;310:591–601. doi: 10.1006/jmbi.2001.4703. [DOI] [PubMed] [Google Scholar]
- 5.Arbabi Ghahroudi M., Desmyter A., Wyns L., Hamers R., Muyldermans S. Selection and identification of single domain antibody fragments from camel heavy-chain antibodies. FEBS Lett. 1997;414:521–526. doi: 10.1016/s0014-5793(97)01062-4. [DOI] [PubMed] [Google Scholar]
- 6.Nuttall S.D., Rousch M.J., Irving R.A., Hufton S.E., Hoogenboom H.R., Hudson P.J. Design and expression of soluble CTLA-4 variable domain as a scaffold for the display of functional polypeptides. Proteins. 1999;36:217–227. [PubMed] [Google Scholar]
- 7.Xu L., Aha P., Gu K., Kuimelis R.G., Kurz M., Lam T., Lim A.C., Liu H., Lohse P.A., Sun L., et al. Directed evolution of high-affinity antibody mimics using mRNA display. Chem. Biol. 2002;9:933–942. doi: 10.1016/s1074-5521(02)00187-4. [DOI] [PubMed] [Google Scholar]
- 8.Nord K., Gunneriusson E., Ringdahl J., Stahl S., Uhlen M., Nygren P.A. Binding proteins selected from combinatorial libraries of an alpha- helical bacterial receptor domain. Nat. Biotechnol. 1997;15:772–777. doi: 10.1038/nbt0897-772. [DOI] [PubMed] [Google Scholar]
- 9.Wikman M., Steffen A.C., Gunneriusson E., Tolmachev V., Adams G.P., Carlsson J., Stahl S. Selection and characterization of HER2/neu-binding affibody ligands. Protein Eng. Des. Sel. 2004;17:455–462. doi: 10.1093/protein/gzh053. [DOI] [PubMed] [Google Scholar]
- 10.Beste G., Schmidt F.S., Stibora T., Skerra A. Small antibody-like proteins with prescribed ligand specificities derived from the lipocalin fold. Proc. Natl Acad. Sci. USA. 1999;96:1898–1903. doi: 10.1073/pnas.96.5.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogt M., Skerra A. Construction of an artificial receptor protein (‘anticalin’) based on the human apolipoprotein D. Chembiochem. 2004;5:191–199. doi: 10.1002/cbic.200300703. [DOI] [PubMed] [Google Scholar]
- 12.Silverman J., Lu Q., Bakker A., To W., Duguay A., Alba B.M., Smith R., Rivas A., Li P., Le H., et al. Multivalent avimer proteins evolved by exon shuffling of a family of human receptor domains. Nat. Biotechnol. 2005;23:1556–1561. doi: 10.1038/nbt1166. [DOI] [PubMed] [Google Scholar]
- 13.Binz H.K., Amstutz P., Kohl A., Stumpp M.T., Briand C., Forrer P., Grutter M.G., Pluckthun A. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat. Biotechnol. 2004;22:575–582. doi: 10.1038/nbt962. [DOI] [PubMed] [Google Scholar]
- 14.Kohl A., Binz H.K., Forrer P., Stumpp M.T., Pluckthun A., Grutter M.G. Designed to be stable: crystal structure of a consensus ankyrin repeat protein. Proc. Natl Acad. Sci. USA. 2003;100:1700–1705. doi: 10.1073/pnas.0337680100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korndorfer I.P., Beste G., Skerra A. Crystallographic analysis of an ‘anticalin’ with tailored specificity for fluorescein reveals high structural plasticity of the lipocalin loop region. Proteins. 2003;53:121–129. doi: 10.1002/prot.10497. [DOI] [PubMed] [Google Scholar]
- 16.Parhami-Seren B., Viswanathan M., Margolies M.N. Selection of high affinity p-azophenyarsonate Fabs from heavy-chain CDR2 insertion libraries. J. Immunol. Meth. 2002;259:43–53. doi: 10.1016/s0022-1759(01)00488-4. [DOI] [PubMed] [Google Scholar]
- 17.Knappik A., Ge L., Honegger A., Pack P., Fischer M., Wellnhofer G., Hoess A., Wolle J., Pluckthun A., Virnekas B. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J. Mol. Biol. 2000;296:57–86. doi: 10.1006/jmbi.1999.3444. [DOI] [PubMed] [Google Scholar]
- 18.Lamminmaki U., Pauperio S., Westerlund-Karlsson A., Karvinen J., Virtanen P.L., Lovgren T., Saviranta P. Expanding the conformational diversity by random insertions to CDRH2 results in improved anti-estradiol antibodies. J. Mol. Biol. 1999;291:589–602. doi: 10.1006/jmbi.1999.2981. [DOI] [PubMed] [Google Scholar]
- 19.Moroncini G., Kanu N., Solforosi L., Abalos G., Telling G.C., Head M., Ironside J., Brockes J.P., Burton D.R., Williamson R.A. Motif-grafted antibodies containing the replicative interface of cellular PrP are specific for PrPSc. Proc. Natl Acad. Sci. USA. 2004;101:10404–10409. doi: 10.1073/pnas.0403522101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scalley-Kim M., Minard P., Baker D. Low free energy cost of very long loop insertions in proteins. Protein Sci. 2003;12:197–206. doi: 10.1110/ps.0232003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minard P., Scalley-Kim M., Watters A., Baker D. A ‘loop entropy reduction’ phage-display selection for folded amino acid sequences. Protein Sci. 2001;10:129–134. doi: 10.1110/ps.32401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bessette P.H., Rice J.J., Daugherty P.S. Rapid isolation of high-affinity protein binding peptides using bacterial display. Protein Eng. Des. Sel. 2004;17:731–739. doi: 10.1093/protein/gzh084. [DOI] [PubMed] [Google Scholar]
- 23.Camaj P., Hirsh A.E., Schmidt W., Meinke A., von Gabain A. Ligand-mediated protection against phage lysis as a positive selection strategy for the enrichment of epitopes displayed on the surface of E. coli cells. Biol. Chem. 2001;382:1669–1677. doi: 10.1515/BC.2001.202. [DOI] [PubMed] [Google Scholar]
- 24.Lu Z., Murray K.S., Van Cleave V., LaVallie E.R., Stahl M.L., McCoy J.M. Expression of thioredoxin random peptide libraries on the Escherichia coli cell surface as functional fusions to flagellin: a system designed for exploring protein protein interactions. Biotechnology. 1995;13:366–372. doi: 10.1038/nbt0495-366. [DOI] [PubMed] [Google Scholar]
- 25.Virnekas B., Ge L., Plückthun A., Schneider K.C., Wellnhofer G., Moroney S.E. Trinucleotide phophoramidites: ideal reagents for the synthesis of mixed oligonucleotides for random mutagenesis. Nucleic Acids Res. 1994;22:5600–5607. doi: 10.1093/nar/22.25.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fellouse F.A., Wiesmann C., Sidhu S.S. Synthetic antibodies from a four-amino-acid code: a dominant role for tyrosine in antigen recognition. Proc. Natl Acad. Sci. USA. 2004;101:12467–12472. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fellouse F.A., Li B., Compaan D.M., Peden A.A., Hymowitz S.G., Sidhu S.S. Molecular recognition by a binary code. J. Mol. Biol. 2005;348:1153–1162. doi: 10.1016/j.jmb.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 28.Early P., Huang H., Davis M., Calame K., Hood L. An immunoglobulin heavy chain variable region gene is generated from three segments of DNA: VH, D and JH. Cell. 1980;19:981–992. doi: 10.1016/0092-8674(80)90089-6. [DOI] [PubMed] [Google Scholar]
- 29.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 30.Lieber M.R. Site-specific recombination in the immune system. FASEB J. 1991;5:2934–2944. doi: 10.1096/fasebj.5.14.1752360. [DOI] [PubMed] [Google Scholar]
- 31.Schatz D.G., Oettinger M.A., Schlissel M.S. V(D)J recombination: molecular biology and regulation. Annu. Rev. Immunol. 1992;10:359–383. doi: 10.1146/annurev.iy.10.040192.002043. [DOI] [PubMed] [Google Scholar]
- 32.French D.L., Laskov R., Scharff M.D. The role of somatic hypermutation in the generation of antibody diversity. Science. 1989;244:1152–1157. doi: 10.1126/science.2658060. [DOI] [PubMed] [Google Scholar]
- 33.Chothia C., Lesk A.M. Canonical structures for the hypervariable regions of immunoglobulins. J. Mol. Biol. 1987;196:901–917. doi: 10.1016/0022-2836(87)90412-8. [DOI] [PubMed] [Google Scholar]
- 34.Chothia C., Lesk A.M., Gherardi E., Tomlinson I.M., Walter G., Marks J.D., Llewelyn M.B., Winter G. The structural repertoire of the human VH segments. J. Mol. Biol. 1992;227:799–817. doi: 10.1016/0022-2836(92)90224-8. [DOI] [PubMed] [Google Scholar]
- 35.Chothia C., Lesk A.M., Tramontano A., Levitt M., Smith G.S., Air G., Sheriff S., Padlan E.A., Davies D., Tulip W.R., et al. Conformations of immunoglobulin hypervariable regions [see comments] Nature. 1989;342:877–883. doi: 10.1038/342877a0. [DOI] [PubMed] [Google Scholar]
- 36.Zemlin M., Klinger M., Link J., Zemlin C., Bauer K., Engler J.A., Schroeder H.W., Jr, Kirkham P.M. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J. Mol. Biol. 2003;334:733–749. doi: 10.1016/j.jmb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 37.Morea V., Tramontano A., Rustici M., Chothia C., Lesk A.M. Conformations of the third hypervariable region in the VH domain of immunoglobulins. J. Mol. Biol. 1998;275:269–294. doi: 10.1006/jmbi.1997.1442. [DOI] [PubMed] [Google Scholar]
- 38.James L.C., Roversi P., Tawfik D.S. Antibody multispecificity mediated by conformational diversity. Science. 2003;299:1362–1367. doi: 10.1126/science.1079731. [DOI] [PubMed] [Google Scholar]
- 39.James L.C., Tawfik D.S. The specificity of cross-reactivity: promiscuous antibody binding involves specific hydrogen bonds rather than nonspecific hydrophobic stickiness. Protein Sci. 2003;12:2183–2193. doi: 10.1110/ps.03172703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.James L.C., Tawfik D.S. Conformational diversity and protein evolution—a 60-year-old hypothesis revisited. Trends Biochem. Sci. 2003;28:361–368. doi: 10.1016/S0968-0004(03)00135-X. [DOI] [PubMed] [Google Scholar]
- 41.Davis M.M. The evolutionary and structural ‘logic’ of antigen receptor diversity. Semin. Immunol. 2004;16:239–243. doi: 10.1016/j.smim.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 42.Xu J.L., Davis M.M. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity. 2000;13:37–45. doi: 10.1016/s1074-7613(00)00006-6. [DOI] [PubMed] [Google Scholar]
- 43.Smith J.W., Tachias K., Madison E.L. Protein loop grafting to construct a variant of tissue-type plasminogen activator that binds platelet integrin alpha IIb beta 3. J. Biol. Chem. 1995;270:30486–30490. doi: 10.1074/jbc.270.51.30486. [DOI] [PubMed] [Google Scholar]
- 44.Nicaise M., Valerio-Lepiniec M., Minard P., Desmadril M. Affinity transfer by CDR grafting on a nonimmunoglobulin scaffold. Protein Sci. 2004;13:1882–1891. doi: 10.1110/ps.03540504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bes C., Troadec S., Chentouf M., Breton H., Dominique Lajoix A., Heitz F., Gross R., Pluckthun A., Chardes T. PIN-bodies: a new class of antibody-like proteins with CD4 specificity derived from the protein inhibitor of neuronal nitric oxide synthase. Biochem. Biophys. Res. Commun. 2006;343:334–344. doi: 10.1016/j.bbrc.2006.02.126. [DOI] [PubMed] [Google Scholar]
- 46.Bes C., Briant-Longuet L., Cerruti M., De Berardinis P., Devauchelle G., Devaux C., Granier C., Chardes T. Efficient CD4 binding and immunosuppressive properties of the 13B8.2 monoclonal antibody are displayed by its CDR-H1-derived peptide CB1. FEBS Lett. 2001;508:67–74. doi: 10.1016/s0014-5793(01)03036-8. [DOI] [PubMed] [Google Scholar]
- 47.Reid B.G., Flynn G.C. Chromophore formation in green fluorescent protein. Biochemistry. 1997;36:6786–6791. doi: 10.1021/bi970281w. [DOI] [PubMed] [Google Scholar]
- 48.Pedelacq J.D., Cabantous S., Tran T., Terwilliger T.C., Waldo G.S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 49.Gay P., Le Coq D., Steinmetz M., Berkelman T., Kado C.I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 1985;164:918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cabantous S., Terwilliger T.C., Waldo G.S. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 2005;23:102–107. doi: 10.1038/nbt1044. [DOI] [PubMed] [Google Scholar]
- 51.Sblattero D., Bradbury A. A definitive set of oligonucleotide primers for amplifying human V regions. Immunotechnology. 1998;3:271–278. doi: 10.1016/s1380-2933(97)10004-5. [DOI] [PubMed] [Google Scholar]
- 52.Sklar L.A., Edwards B.S., Graves S.W., Nolan J.P., Prossnitz E.R. Flow cytometric analysis of ligand-receptor interactions and molecular assemblies. Annu. Rev. Biophys. Biomol. Struct. 2002;31:97–119. doi: 10.1146/annurev.biophys.31.082901.134406. [DOI] [PubMed] [Google Scholar]
- 53.Studier F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 54.Lefranc M.P., Pommie C., Ruiz M., Giudicelli V., Foulquier E., Truong L., Thouvenin-Contet V., Lefranc G. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol. 2003;27:55–77. doi: 10.1016/s0145-305x(02)00039-3. [DOI] [PubMed] [Google Scholar]
- 55.Siegel R.W., Velappan N., Pavlik P., Chasteen L., Bradbury A. Recombinatorial cloning using heterologous lox sites. Genome Res. 2004;14:1119–1129. doi: 10.1101/gr.1821804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giudicelli V., Chaume D., Lefranc M.P. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2005;33:D256–D261. doi: 10.1093/nar/gki010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ormo M., Cubitt A.B., Kallio K., Gross L.A., Tsien R.Y., Remington S.J. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 58.Huang S.C., Jiang R., Glas A.M., Milner E.C. Non-stochastic utilization of Ig V region genes in unselected human peripheral B cells. Mol. Immunol. 1996;33:553–560. doi: 10.1016/0161-5890(95)00162-x. [DOI] [PubMed] [Google Scholar]
- 59.Utermark J., Karlovsky P. Quantification of green fluorescent protein fluorescence using real-time PCR thermal cycler. Biotechniques. 2006;41:150–154. doi: 10.2144/000112221. [DOI] [PubMed] [Google Scholar]
- 60.Binz H.K., Stumpp M.T., Forrer P., Amstutz P., Pluckthun A. Designing repeat proteins: well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J. Mol. Biol. 2003;332:489–503. doi: 10.1016/s0022-2836(03)00896-9. [DOI] [PubMed] [Google Scholar]
- 61.Barbas C.F., 3rd, Languino L.R., Smith J.W. High-affinity self-reactive human antibodies by design and selection: targeting the integrin ligand binding site. Proc. Natl Acad. Sci. USA. 1993;90:10003–10007. doi: 10.1073/pnas.90.21.10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cohn M. What are the commonalities governing the behavior of humoral immune recognitive repertoires? Dev. Comp. Immunol. 2006;30:19–42. doi: 10.1016/j.dci.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 63.Marchalonis J.J., Adelman M.K., Schluter S.F., Ramsland P.A. The antibody repertoire in evolution: chance, selection, and continuity. Dev. Comp. Immunol. 2006;30:223–247. doi: 10.1016/j.dci.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 64.Zhao Y., Jackson S.M., Aitken R. The bovine antibody repertoire. Dev. Comp. Immunol. 2006;30:175–186. doi: 10.1016/j.dci.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 65.De Genst E., Saerens D., Muyldermans S., Conrath K. Antibody repertoire development in camelids. Dev. Comp. Immunol. 2006;30:187–198. doi: 10.1016/j.dci.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 66.Conrath K.E., Wernery U., Muyldermans S., Nguyen V.K. Emergence and evolution of functional heavy-chain antibodies in Camelidae. Dev. Comp. Immunol. 2003;27:87–103. doi: 10.1016/s0145-305x(02)00071-x. [DOI] [PubMed] [Google Scholar]
- 67.Transue T.R., De Genst E., Ghahroudi M.A., Wyns L., Muyldermans S. Camel single-domain antibody inhibits enzyme by mimicking carbohydrate substrate. Proteins. 1998;32:515–522. doi: 10.1002/(sici)1097-0134(19980901)32:4<515::aid-prot9>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 68.Conrath K.E., Lauwereys M., Galleni M., Matagne A., Frere J.M., Kinne J., Wyns L., Muyldermans S. Beta-lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae. Antimicrob. Agents Chemother. 2001;45:2807–2812. doi: 10.1128/AAC.45.10.2807-2812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stijlemans B., Conrath K., Cortez-Retamozo V., Van Xong H., Wyns L., Senter P., Revets H., De Baetselier P., Muyldermans S., Magez S. Efficient targeting of conserved cryptic epitopes of infectious agents by single domain antibodies. African trypanosomes as paradigm. J. Biol. Chem. 2004;279:1256–1261. doi: 10.1074/jbc.M307341200. [DOI] [PubMed] [Google Scholar]
- 70.Rock E.P., Sibbald P.R., Davis M.M., Chien Y.H. CDR3 length in antigen-specific immune receptors. J. Exp. Med. 1994;179:323–328. doi: 10.1084/jem.179.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee C.V., Liang W.C., Dennis M.S., Eigenbrot C., Sidhu S.S., Fuh G. High-affinity human antibodies from phage-displayed synthetic Fab libraries with a single framework scaffold. J. Mol. Biol. 2004;340:1073–1093. doi: 10.1016/j.jmb.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 72.Sidhu S.S., Li B., Chen Y., Fellouse F.A., Eigenbrot C., Fuh G. Phage-displayed antibody libraries of synthetic heavy chain complementarity determining regions. J. Mol. Biol. 2004;338:299–310. doi: 10.1016/j.jmb.2004.02.050. [DOI] [PubMed] [Google Scholar]
- 73.Hanke T., Szawlowski P., Randall R.E. Construction of solid matrix-antibody-antigen complexes containing simian immunodeficiency virus p27 using tag-specific monoclonal antibody and tag-linked antigen. J. Gen. Virol. 1992;73:653–660. doi: 10.1099/0022-1317-73-3-653. [DOI] [PubMed] [Google Scholar]
- 74.Evan G.I., Lewis G.K., Ramsay G., Bishop J.M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]