Abstract
SLX5 and SLX8 encode RING-finger proteins that were previously identified based on their requirement for viability in yeast cells lacking Sgs1 DNA helicase. Slx5 and Slx8 proteins are known to be required for genome stability and to physically interact in yeast extracts; however, their biochemical functions are unknown. To address this question we purified and characterized recombinant Slx5 and Slx8 proteins. Here we show that Slx5 and Slx8 form a heterodimeric complex with double-stranded DNA (dsDNA)-binding activity. Individually, only the Slx8 subunit displays this activity. Structure–function studies indicate that the DNA-binding activity requires only the N-terminal 160 amino acids of Slx8, but not its C-terminal RING-finger domain. Alleles of SLX8 that express the RING-finger domain alone show almost complete complementation in yeast indicating that this DNA-binding domain is not essential for this in vivo function. Consistent with these findings we show that Slx5 immunolocalizes to the nucleus and that a portion of the Slx8 protein co-fractionates with chromatin. These results suggest that Slx5–Slx8 may act directly on DNA to promote genome stability.
INTRODUCTION
The Sgs1 protein of budding yeast has been used as a model system to understand how eukaryotic RecQ DNA helicases function to maintain genome integrity. Loss of SGS1 has been shown to result in increased rates of recombination, chromosome loss and missegregation, and a decrease in sporulation efficiency (1–3). These strains also display hypersensitivity to a variety of DNA damaging agents such as methyl methanesulfonate (MMS) and UV light, and they are hypersensitive to the DNA synthesis inhibitor hydroxyurea (HU) (4,5). Genetic and biochemical evidence suggests that the RecQ DNA helicases, such as Sgs1 and human BLM, cooperate with DNA topoisomerase III (Top3) (3,6–8) and the Rmi1/BLAP75 subunit (9–11) to resolve recombination intermediates in a pathway leading to non-crossovers. Enzymatically, this could be accomplished using the RecQ DNA helicase activity to branch-migrate double Holliday junctions into a hemi-catenane structure that is decatenated by Top3 (12–16).
The yeast system has been exploited to identify mutations that are synthetically lethal with Sgs1 (17–19). SLX5 and SLX8 encode proteins with a single RING-finger motif and no obvious biochemical function. On their own, null mutations in SLX5 or SLX8 produce nearly identical phenotypes. Both slx5 and slx8 mutants display a heterogeneous colony morphology consisting of a mixture of large and small colonies with nibbled edges (19). Interestingly, this phenotype may be related to a recently reported role of these genes in regulating the SUMO pathway (20), since SUMO mutants were originally characterized as having the nibbled phenotype (21–24). The slx5 or slx8 mutants also share phenotypes with sgs1 and top3 mutants such as reduced sporulation efficiency and hypersensitivity to prolonged exposure to HU (19). Moreover, SLX5 and SLX8 act in the same pathway to suppress gross chromosomal rearrangements (25). These phenotypes suggest that Slx5 and Slx8 may be required for DNA repair and/or recombination like Sgs1–Top3–Rmi1. However, the synthetic lethality of sgs1 slx5 or sgs1 slx8 cells cannot be suppressed by eliminating homologous recombination as is observed in sgs1 mus81 or sgs1 srs2 strains (26–29). Thus, at least one function of either Sgs1–Top3 or Slx5–Slx8 must be upstream of, or independent of, homologous recombination. Biochemically, the Slx5 and Slx8 proteins were shown to co-immunoprecipitate from cell extracts when overproduced in yeast, suggesting that the two proteins may work as a complex (19). Such an idea would explain their shared phenotypes.
Proteins homologous to Slx5 and Slx8 have been identified in multiple species, suggesting that they are conserved throughout eukaryotes [(19) and data not shown]. Both proteins contain a single RING-finger motif of the C3HC4 type at their C-termini. RING fingers are found in proteins of diverse function and it has been suggested that this zinc-binding domain may help to mediate DNA binding or protein–protein interactions. Such may be the case in the human PML, Cbl, TRAF2 or RAG1 proteins. By far, the largest class of RING-finger proteins is composed of ubiquitin E3 ligases such as the well-known BRCA1, Mdm2 and SCF proteins (30). More recently, variant RING-finger domains (SP-RING) have been found in SUMO E3 ligases, including the human PIAS1 and the yeast Siz1 and Siz2 proteins (31–34). The presence of the RING-finger motif suggests that Slx5 and Slx8 may interact with other proteins, bind DNA, or function as ubiquitin or SUMO E3 ligases.
Based on the presumed role of Slx5 and Slx8 in DNA metabolism, we tested the possibility that these proteins interact with DNA. Slx5 and Slx8 were purified as recombinant proteins and shown to form a stable complex when co-expressed in Escherichia coli. An electrophoretic mobility shift assay (EMSA) revealed that the Slx5–Slx8 complex (Slx5–Slx8) binds double-stranded DNA (dsDNA), but not single-stranded DNA (ssDNA), and that Slx8 mediates this activity. Structure–function studies indicated that DNA binding by Slx8 was independent of its RING domain whereas in vivo analysis confirmed the essential function of the RING domain. These studies represent the first biochemical characterization of Slx5 and Slx8 and show that these proteins directly interact and bind DNA.
MATERIALS AND METHODS
Yeast strains and plasmids
The yeast strains used in this study are isogenic derivatives of W303-1a (MATa ade2-1 ura3-52 his3-11, trp1-1 leu2-3,112 can1-100) (35). W303-1a, LYY2022 (SLX5-V5) and SIY778(slx8Δ + pJM6864 GAL1p::slx8-4) were maintained in 1% yeast extract, 2% peptone, 2% dextrose (YPD) or selective medium as appropriate. Yeast strain construction, growth and transformation was carried out using standard procedures (36,37). Mutant alleles of SLX8 were tested by functional complementation of the synthetic-lethal phenotype of strain VCY1525 [MATa ade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 leu2-3,112 lys2 sgs1-20::hphMX4 slx8-10::KAN pJM500 (URA3/ADE3/SGS1)]. Recombinant proteins were synthesized in E.coli using the T7 expression system (38) and the following plasmids: Plasmid pJM6813, which expresses N-terminally tagged His6-Slx8 protein (Slx8), was constructed by inserting the SLX8 ORF into plasmid pET28a on an NdeI–BamHI fragment. Plasmid pJM6818, which expresses full-length untagged Slx8 protein, was constructed by inserting the same SLX8 fragment into the NdeI and BamHI sites of plasmid pET11a. Plasmid pJM6511, which expresses an N-terminally tagged His6-Slx5 protein (Slx5), was constructed by inserting the SLX5 ORF into pET28a on an NdeI–BamHI fragment. The bi-cistonic plasmid pJM6819, which expresses untagged Slx8 and His6-Slx5 protein (Slx5–Slx8 complex), was constructed by moving the promoter and SLX5 ORF of pNJ6511 on a BstEII–BamHI fragment into the BstEII and BglII sites of pNJ6818.
Expression and purification of recombinant proteins
Recombinant proteins were produced using the T7 expression system of Studier (38). Plasmids pJM6819 (His6-Slx5 + Slx8), pJM6813 (His6-SLX8) and pJM6511 (His6-SLX5) were transformed into E.coli BL21-RIL cells (Stratagene). Cells with plasmid pJM6819 were grown in Luria–Bertani (LB) media containing 100 μg/ml ampicillin, whereas cells with plasmids pJM6813 and pJM6511 were grown in LB media containing 35 μg/ml kanamycin. Cells were shaken at 37°C to an OD600 of 0.4. To induce the expression of the recombinant protein, cultures were treated with isopropyl-1-thio-d-galactopyranoside at a final concentration of 0.4 mM overnight at 15°C. Induced cells were pelleted and resuspended in Buffer A [25 mM Tris–HCl (pH 7.5), 1 mM EDTA, 0.01% (v/v) NP-40, 10% glycerol, 0.1 mM phenylmethlysulfonyl fluoride (PMSF) and 1 mM DTT] containing 150 mM NaCl and the following protease inhibitors: 10 μg/ml pepstatin, 5 μg/ml leupeptin, 10 mM benzamidine and 100 μg/ml bacitracin. Extractions and chromatography were performed at 4°C, except where noted. Cell suspensions were subjected to three freeze-thaw cycles and then sonicated three times for 1 min using a Branson sonifier 450 microtip at setting 4, 60% duty cycle. Lysed cells were clarified by centrifugation at 12 000 g for 20 min and the supernatant collected as extract. The supernatant was bound to 2 ml of Ni Probond resin (Invitrogen) in the presence of 10 mM imidazole for 2 h, after which the resin was poured into a column. The column was washed with 10 column volumes (CVs) of Buffer N [25 mM Tris–HCl (7.5), 500 mM NaCl, 0.01% NP-40, 10% glycerol and 0.1 mM PMSF] plus 10 mM imidazole. The column was developed with 7 CVs of Buffer N plus 50 mM imidazole, 7 CVs of Buffer N plus 100 mM imidazole, 7 CVs of Buffer N containing 200 mM imidazole and 5 CVs of Buffer N containing 500 mM imidazole. Peak fractions were identified by SDS–PAGE and Coomassie blue staining. The peak fractions were pooled and dialyzed in Buffer A plus 150 mM NaCl. The Slx5–Slx8 complex was further purified by gel-filtration chromatography using a 24 ml Superose 6 HR10/30 column in Buffer A lacking glycerol but containing 150 mM NaCl. Peak Superose 6 fractions were pooled and subjected to 15–35% glycerol gradient sedimentation by centrifugation for 24 h at 45 000 r.p.m. in a Beckman SW55 Ti rotor. Complex stability was tested by subjecting a portion of the Ni-purified material to Superose 6 fractionation in the presence of RIPA buffer [150 mM NaCl, 50 mM Tris–HCl (pH 7.5), 1% (v/v) NP-40, 0.5% (w/v) deoxycholate and 0.1% (w/v) SDS]. Protein concentrations were determined by the Bradford assay using BSA as standard.
EMSA assay and DNA substrates
Unless otherwise indicated, Slx5–Slx8 protein was incubated with 30 000 c.p.m. 32P-labeled DNA substrate (5 fmol) in a final volume of 20 μl containing 25 mM HEPES (pH 7.5), 50 mM NaCl, 10 μM ZnSO4, 0.5 mM DTT, 0.1% NP-40, 0.1 mg/ml BSA and 5% glycerol at room temperature (RT) for 30 min. The reaction products were electrophoresed through 2.5% polyacrylamide gel (20:1 acrylamide/bis) containing 0.3% agarose and 0.5× Tris–borate–EDTA at 4°C. The gel was then dried and visualized by a phosphorimager. All DNA substrates were prepared from labeled oligonucleotides (nt) that were annealed and purified essentially as described (22). The oligonucleotides used for this study were as follows: 1313 (30 nt), TGGCGTTAGGAGATACCGATAAGCTTCGGC; 1314 (30 nt), GCCGAAGCTTATCGGTATCTCCTAACGCCA; 1315 (45 nt), GGGTCAACGTGGGCAAAGATGTCCTAGCAAGCCAGAATTCGGCAG; 1316 (45 nt), CTGCCGAATTCTGGCTTGCTAGGACATCTTTGCCCACGTTGACCC; 1317 (90 nt), GATCCGAATTCTGGCTTGCTAGGACATCTTTGCCCACGTTGACCCGGGTTGGCGTTAGGAGATAGTCAGTTATAGCTGCGGCTGCTAAGG; 1319 (90 nt), GATCCCTTAGCAGCCGCAGCTATAACTGACTATCTCCTAACGCCAACCCGGGTCAACGTGGGCAAAGATGT CCTAGCAAGCCAGAATTCG;1312 (15 nt), TATCTCCTAACGCCA; 1320 (15 nt) TGGCGTTAGGAGATA; 891 (49 nt), ATCGATGTCTCTAGACAGCACGAGCCCTAACGCCAGAATTCGGCAGCGT994 (25 nt), GCTCGTGCTGTCTAGAGACATCGAT; 888 (49 nt), GACGCTGCCGAATTCTGGCGTTAGGAGATACCGATAAGCTTCGGCTTAA; 992 (24 nt), TTAAGCCGAAGCTTATCGGTATCT; 1254 (50 nt), TGCCGAATTCTACCAGTGCCAGTGATGGACATCTTTGCCCACGTTGACCC; 1258 (25 nt), ATCACTGGCACTGGTAGAATTCGGC; 1311 (25 nt), TGGGTCAACGTGGGCAAAGATGTCC; 1253 (50 nt), TGGGTCAACGTGGGCAAAGATGTCCTAGCAATGTAATCGTCTATGACGTT; 1256 (50 nt), CAACGTCATAGACGATTACATTGCTACATGGAGCTGTCTAGAGGATCCGA; 1302 (25 nt), GTCGGATCCTCTAGACAGCTCCATG; 892 (49 nt), GACGCTGCCGAATTCTGGCTTGCTAGGACATCTTTGCCCACGTTGACCC; 893 (50 nt), TGGGTCAACGTGGGCAAAGATGTCCTAGCAATGTAATCGTCTATGACGTT; 894 (51 nt), CAACGTCATAGACGATTACATTGCTAGGACATGCTGTCTAGAGACTATCGA; and 895 (50 nt), ATCGATAGTCTCTAGACAGCATGTCCTAGCAAGCCAGAATTCGGCAGCGT.
Immunological techniques
Purified recombinant Slx5 and Slx8 proteins were used as antigens to raise rabbit antisera. To supershift DNA–protein complexes, EMSAs were performed as above and following the 30 min incubation, the indicated anti-Slx5, anti-Slx8 and anti-Slx1 sera were added and incubated on ice for 20 min. The products were then resolved by native PAGE as described above. Yeast immunofluorescence was performed as described previously (39).
Protein chromatin-binding assay and western blotting
Wild-type cells were grown in 50 ml YPD until OD600 = 1.0. SIY778 cells containing pJM6864 were grown in SD medium lacking leucine with 2% raffinose to OD 0.2 before induction with 2% galactose for 6 h. Cells were harvested and sodium azide was added to 0.1%. Cells were resuspended in 3 ml of prespheroplasting buffer [100 mM PIPES (pH 9.4) and 10 mM DTT] and incubated at RT for 10 min. This was followed by an incubation in 2 ml of spheroplasting buffer [50 mM KPO4 (pH 7.5), 0.6 M sorbitol and 10 mM DTT], containing 100 μl of 1 mg/ml of zymolyase (100T) (ICN Biomedicals, Aurora, OH) in 50 mM KPO4 (pH 7.5) for 20 min at 30°C with occasional mixing. Spheroplasts were washed, lysed, digested by micrococcal nuclease (MNase) and fractionated by centrifugation at low or high speeds as described previously (40). For immunoblotting, proteins were transferred on to nitrocellulose and blocked with 5% dry milk in Tris-buffered saline containing 0.1% Tween-20 (TBST). Monoclonal antibodies anti-V5 (Invitrogen) and anti-Orc5 (a gift from Dr Steve Bell) were used at dilutions of 1:5000 and 1:1000, respectively. The antiserum against Slx8 was used at a 1:2000 dilution. Horseradish peroxidase-coupled anti-mouse secondary antibody (Orc5) and anti-rabbit secondary antibody (Slx8) was diluted at 1:10 000 in TBST containing 0.5% dry milk. Reaction products were visualized with a SuperSignal chemiluminescent reagent (Pierce).
RESULTS
Purification of Slx5 and Slx8
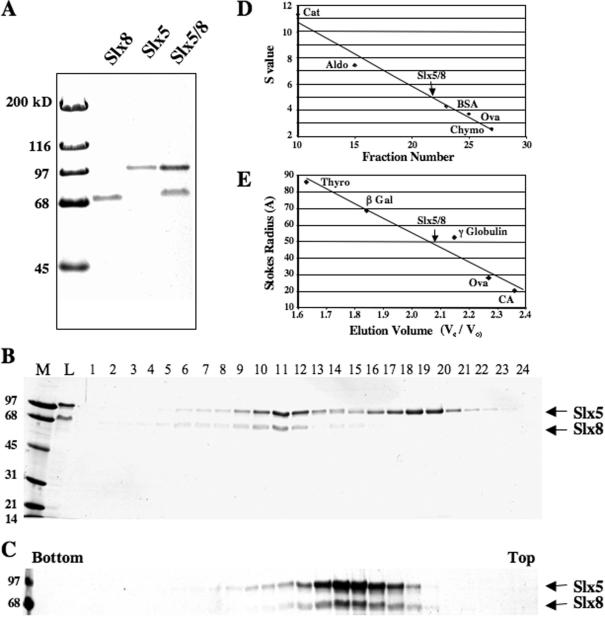
His6-tagged versions of Slx5 and Slx8 were expressed in E.coli to produce either Slx5, or Slx8 or the Slx5–Slx8 complex. These proteins were well-expressed and are soluble allowing for purification by Ni–agarose chromatography (Figure 1A). Slx5 and Slx8 were previously observed to migrate anomalously in SDS–PAGE when immunoblotted from yeast extracts (19). As shown in Figure 1A, the recombinant proteins showed the same effect where (His6)Slx5 (639 amino acids; pI 7.0; MWcalc = 73 kDa) and (His6)Slx8 (294 amino acids; pI 5.0; MWcalc = 33 kDa) migrated at MWr = 100 and 69 kDa, respectively. When co-expressed, the Slx5 and Slx8 proteins appeared to purify as a complex (Figure 1A). To test this idea, this material was subjected to gel-filtration chromatography in the presence of 0.15 M NaCl (Figure 1B and E). This treatment revealed a complex of Slx5–Slx8 protein and a peak of free Slx5 that had bound alone to the Ni affinity resin (Figure 1B). In separate gel-filtration experiments, the Slx5 and Slx8 subunits co-fractionated even in the presence of 0.1% SDS or 0.5 M NaCl (data not shown). Thus, the stability of the complex resembles that observed in yeast cell extracts (19). The Slx5 and Slx8 proteins also co-sedimented during glycerol gradient centrifugation (Figure 1C and D). These hydrodynamic data (s = 5.0S; Rs = 48A) yield a native molecular weight of 106 kDa using the method of Siegel and Monty (41). This value closely approximates that calculated for a simple 1:1 complex of (His6)Slx5 and Slx8 (104 kDa). We conclude that these proteins form a stable heterodimeric complex.
Figure 1.
Purification and characterization of recombinant Slx5–Slx8 complex. Slx5 and Slx8 were expressed either individually or together in E.coli and purified by chromatography on Ni–agarose as described in Materials and Methods. (A) Approximately 1 μg (His6)Slx8, 1 μg (His6)Slx5 and 2 μg (His6)Slx5–Slx8 complex were resolved by 12.5% SDS–PAGE and subjected to Coomassie blue staining. (B) An aliquot of 150 μg of the Ni-purified complex was subjected to Superose 6 size exclusion chromatography and analyzed as in (A). The positions of (His6)Slx5 (100 kDa) and Slx8 (69 kDa) subunits are indicated. Lanes: M, MW marker; L, Column Load. (C) The Slx5–Slx8 complex (Superose 6 pool) was subjected to 15–35% glycerol gradient sedimentation and fractions analyzed as above. Also shown are the sedimentation (D) and gel-filtration (E) profiles of the Slx5–Slx8 complex relative to the indicated standards.
Slx5–Slx8 binds dsDNA
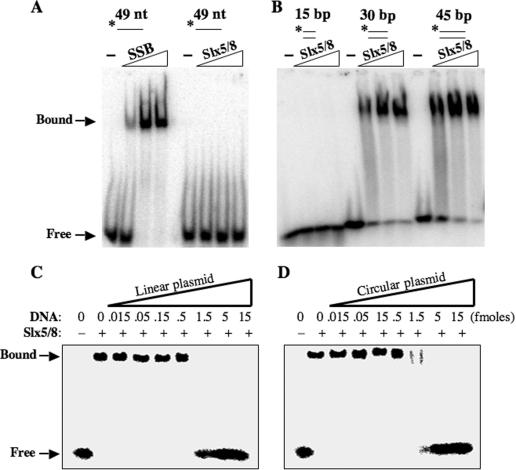
Slx5–Slx8 complex was tested for DNA-binding activity using an EMSA. Increasing amounts of protein were incubated with 32P-labeled ssDNA and the free and bound DNA were resolved by PAGE. As shown in Figure 2A, no interaction with ssDNA was apparent. However, binding of Slx5–Slx8 to dsDNA probes was observed (Figure 2B). Slx5–Slx8 failed to bind a 15 bp probe, but it bound to probes of 30 and 45 bp. Thus, the DNA-binding activity of Slx5–Slx8 requires a substrate of between 15 and 30 bp. A competition experiment was performed to test whether binding was specific to the ends of the DNA molecule or to internal sites. Binding to the 45 bp dsDNA probe could be competed with either linear (Figure 2C) or circular (Figure 2D) plasmid DNA. We conclude that Slx5–Slx8 is a dsDNA-binding protein.
Figure 2.
Slx5–Slx8 binds dsDNA by gel-mobility shift assay. (A) An aliquot of 5 fmol of a 49 nt radiolabeled ssDNA oligonucleotide was incubated with either E.coli SSB (0, 0.014, 0.7 or 1.4 pmol) or Slx5–Slx8 (0, 0.01, 0.6 or 1.2 pmol) prior to EMSA. Protein–DNA complexes were resolved by 2.5% polyacrylamide/0.3% agarose gel electrophoresis and analyzed by phosphorimaging. The positions of Slx5–Slx8/DNA complexes (Bound) or unbound substrate (Free) are indicated. The asterisk indicates 5′-32P-labeling of the probes. (B) An aliquot of 5 fmol of the indicated dsDNA probe was incubated with Slx5–Slx8 (0, 1.2, 2.45 or 4.9 pmol) before EMSA. (C) Gel-mobility shift assays were performed using 4 pmol Slx5–Slx8, 5 fmol of a 45 bp duplex probe and increasing amounts of linearized unlabeled competitor plasmid DNA (3.2 kb). (D) Gel-mobility shift assays were performed as in (C) but the competitor plasmid was undigested. The substrates were assembled from the following oligonucleotides: *888 (49 nt); *1312/1320 (15 bp); *1313/1314 (30 bp); and *1315/1316 (45 bp).
Subunit specificity of Slx5–Slx8
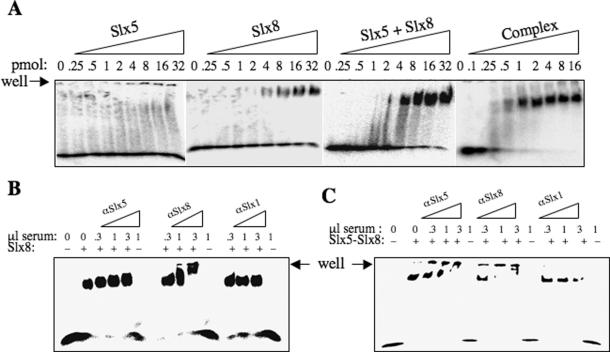
To determine the relative contribution of the two proteins to DNA-binding activity, we tested the individual subunits of Slx5–Slx8. As shown in Figure 3A, Slx5 alone showed no DNA-binding activity by EMSA. In contrast, the isolated Slx8 subunit was capable of binding the probe on its own. This result suggests that Slx8 is the main DNA-binding subunit. When the two subunits were incubated together with probe, Slx5 appeared to have little or no effect on the DNA-binding activity of Slx8. In contrast to this result, purified Slx5–Slx8 complex was capable of binding similar amounts of probe at ∼10-fold lower protein concentrations. In this experiment, ∼8 pmol of Slx8 was required for half-maximal DNA binding, whereas only 0.5 pmol of Slx5–Slx8 bound most of the probe (Figure 3A). The fact that Slx5 and Slx8 are more active when purified as a complex suggests that reconstitution of the complex does not occur efficiently in vitro. Taken together, these results suggest that Slx5–Slx8 has a higher affinity for DNA than Slx8 alone; however, we cannot rule out the possibility that these samples differ in the fraction of active molecules present in the two preparations.
Figure 3.
Slx8 is responsible for the DNA-binding activity of the Slx5–Slx8 complex. (A) An aliquot of 5 fmol of 32P-labeled dsDNA (45 bp) was incubated with indicated amounts of Slx5, Slx8, Slx5 and Slx8 mixed together (Slx5 + Slx8), or Slx5–Slx8 (Complex) followed by EMSA analysis. (B and C) Supershift analysis was performed to identify proteins present in the protein–DNA complex. A total of 32 pmol of Slx8 (B) or 8 pmol of Slx5–Slx8 complex (C) were incubated with 5 fmol of 32P-labeled dsDNA (45 bp). The reaction products were then incubated with anti-Slx5, anti-Slx8 or anti-Slx1 serum as indicated, followed by EMSA analysis.
We next tested whether or not both subunits were present in the DNA–protein complexes formed with Slx5–Slx8. In this experiment we first incubated the 45 bp probe with Slx8 and then added increasing amount of antiserum directed against Slx5 or Slx8. As shown in Figure 3B, the Slx8 protein shifted the 45 bp probe as before and was unaffected by anti-Slx5 serum. In contrast, the anti-Slx8 serum produced a progressive retardation, or supershift, of Slx8–DNA complexes. A control serum against Slx1 protein was inactive in supershifting this complex. Gel-shifted complexes with Slx5–Slx8 were treated similarly. When incubated with increasing amounts of anti-Slx5 serum, the DNA–protein complex was gradually supershifted until it remained in the well (Figure 3C). Similarly, increasing amounts of anti-Slx8 caused a supershifting of these protein–DNA complexes (Figure 3C) whereas the control serum against Slx1 had no effect. We conclude that Slx5–Slx8 binds DNA as a complex with both subunits present in the protein–DNA complex.
Substrate specificity of Slx5–Slx8
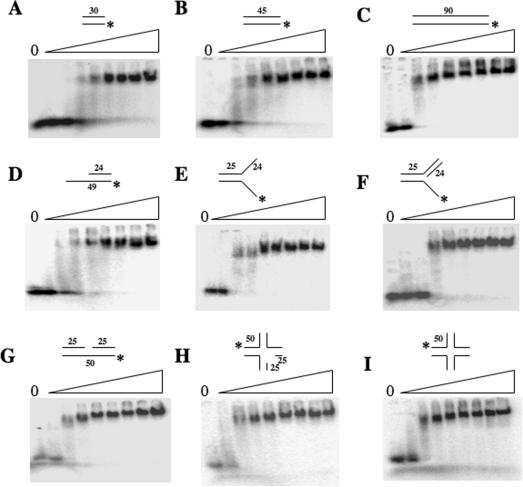
Other proteins identified in SGS1-synthetic-lethal screens have been shown to have structure-specific DNA endonuclease activity (42,43). To see if the Slx5–Slx8 complex preferred to bind branched substrates, we examined the substrate specificity of Slx5–Slx8 using a variety of radiolabeled substrates. As shown in Figure 4, Slx5–Slx8 bound to all dsDNA substrates tested including branched and nicked forms with no significant difference in affinity. As expected for a simple DNA-binding activity, larger dsDNA substrates (e.g. 45 and 90 bp) were bound at lower protein concentrations than the smaller 30 bp substrate (Figure 4A–C). This preference is presumably due to a greater availability of binding sites. Consistent with this idea, the use of a 90 bp dsDNA probe revealed a second lower-mobility band at higher protein concentrations. This second band-shift suggests that the larger 90 bp dsDNA is capable of two binding events that are not possible using smaller DNAs. Incubation of Slx5–Slx8 with these substrates in the presence of Mg2+ failed to reveal endonuclease activity (data not shown).
Figure 4.
Structure-independent DNA binding of Slx5–Slx8. Increasing amounts of purified Slx5–Slx8 (0, 0.1, 0.25, 0.5, 1, 2, 4, 8 or 16 pmol) were incubated with 5 fmol of the indicated 5′-32P-labeled substrates and analyzed as in Figure 2: (A) 30 bp blunt-ended dsDNA, (B) 45 bp dsDNA, (C) 90 bp dsDNA, (D) dsDNA with 3′-extension, (E) Y structure, (F) 5′-flap, (G) nicked duplex, (H) nicked Holliday junction and (I) Holliday junction. The substrates were assembled from the following oligonucleotides: (A) *1313/1314, (B) *1315/1316, (C) *1317/1319, (D) *891/994, (E) *891/888, (F) *891/888/992, (G) *1254/1258/1311, (H) *1254/1253/1256/1258/1302 and (I) *892/893/894/895.
We tested the role of Zn in DNA binding by modifying the proteins' cysteine residues with the reversible reagent p-Hydroxymercuriphenylsulfonate (PMPS) (44). Covalent modification of cysteine residues by PMPS results in the release of Zn in a reaction that is often reversible by treatment with the reducing agent DTT in the presence of Zn (44). Although the DNA-binding activity of Slx5–Slx8 was inactivated by PMPS, we were unable to restore activity by subsequent treatment with DTT and increasing amounts of Zn (data not shown). Because PMPS treatment was not informative, we performed a structure–function analysis to map the DNA-binding domain.
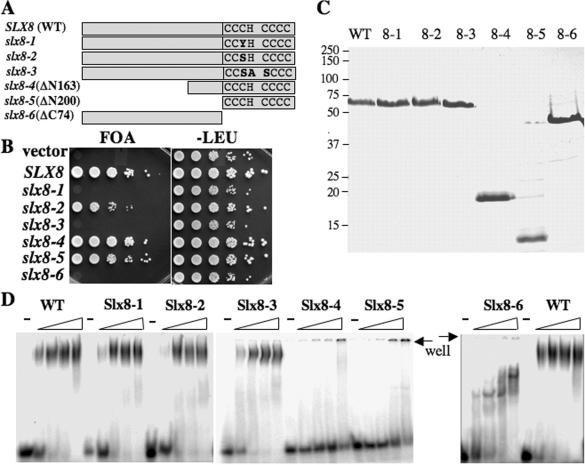
The role of the Zn-binding motif in Slx8 function was confirmed by functional analysis in yeast. DNA sequencing of the original slx8-1 allele revealed a single missense mutation, C221Y, that targets the third conserved cysteine residue in the RING-finger sequence C3HC4 (Figure 5A). Thus, the RING-finger motif of Slx8 appears to be critical for function in vivo. To confirm this idea, we created additional RING-finger mutations, as well as truncations of the N- and C-termini of Slx8. One hypomorphic allele, slx8-2 (C221S), was identified that conferred a slow-growth phenotype in the sgs1 mutant background, while an allele that altered three conserved RING domain residues (slx8-3) appeared null in vivo (Figure 5B). In contrast, truncation of the N-terminal 163 residues of Slx8 conferred a slight growth defect. Truncation of all residues N-terminal to the RING domain generated an allele that was also viable (slx8-5), but even more slow-growing in the sgs1 background (Figure 5B). The slx8-4 and slx8-5 alleles lacked the HU-sensitivity of slx8 null mutants (data not shown). These results indicate that the RING domain is essential for in vivo function and that the N-terminus of Slx8 is largely dispensible for viability in this assay. Interestingly, Slx8 lacking the RING domain fails to bind Slx5 in a recombinant expression system (data not shown). This suggests that dimer formation may be essential for in vivo function.
Figure 5.
Structure–function analysis of Slx5–Slx8. (A) Schematic diagram of the wild-type and mutant Slx8 proteins indicating the conserved RING-finger residues. (B) Strain VCY1525 (sgs1Δ slx8Δ), which contains the complementing plasmid pJM500 (SGS1/URA3), was first transformed with centromere-based vector (pRS415) or vector containing the indicated SLX8 allele. Complementation of slx8Δ phenotype was then tested by serial dilution and spotting onto media containing 5-FOA to select against pJM500 or onto media lacking leucine as control. (C) An aliquot of 2 μg of the indicated Slx8 protein was resolved by SDS–PAGE and stained with Coomassie blue. (D) The indicated Slx8 protein (0, 0.4, 1.2 and 4 pmol) was incubated with 5 fmol of the 45 bp 5′-32P-labeled substrate and analyzed using EMSA as in Figure 2.
To localize the DNA-binding domain of Slx8, we expressed and purified these six mutant proteins (Figure 5C). As shown in Figure 5D, mutations in RING domain had no effect on the ability of Slx8 to bind DNA, although deletion of the N-terminus (Slx8-4 and Slx8-5 proteins) eliminated this activity. The N-terminal domain was sufficient for binding as the Slx8-6 protein bound the probe with an affinity similar to wild-type Slx8.
In vivo localization of Slx5–Slx8
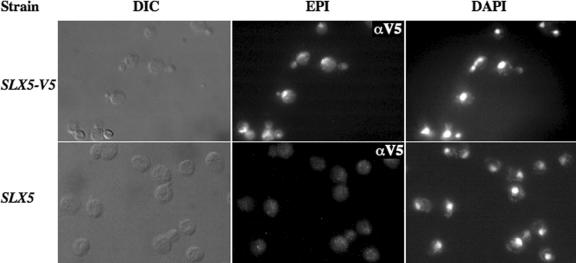
The DNA-binding activity of Slx5–Slx8 implies a role in the yeast nucleus. A nuclear function is also suggested by the genetic redundancy between Slx5–Slx8 and the Sgs1–Top3–Rmi1 complex. We tested this idea by immunolocalizing Slx5 within yeast cells using a strain that expresses epitope-tagged Slx5 under its own promoter. Treatment of these cells with anti-V5 monoclonal antibody resulted in specific nuclear staining that was present in unbudded and budded cells (Figure 6). Often the Slx5-V5 staining appeared punctate. This result indicates that a significant proportion of Slx5 protein resides in the nucleus and that localization is independent of cell-cycle position.
Figure 6.
Slx5 is a nuclear protein. Spheroplasted yeast cells of the indicated genotype were incubated with anti-V5 antibodies and processed for microscopy using differential interference contrast (DIC) and epifluorescence (EPI). The following strains were used: LYY2022 (SLX5-V5) and W303-1a (SLX5).
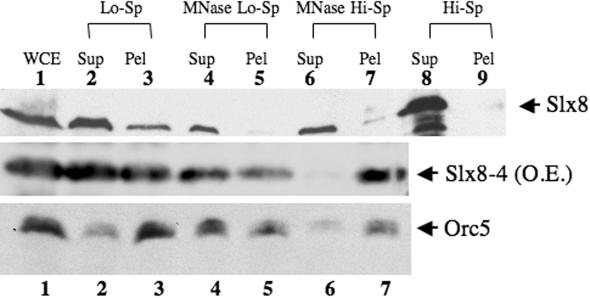
We next tested the prediction that Slx8 bound chromatin in vivo by immunoblotting chromatin fractions that had beenisolated by differential centrifugation (40). Upon lysis of cell spheroplasts, the majority of Slx8 protein was observed to fractionate with the soluble pool; however, a portion was also found in the pellet (Figure 7, upper panel). This fractionation differs from that of Orc5, which is found mostly in the pellet fraction (Figure 7, lower panel) as described originally in (40). If Slx8 was bound to chromatin it should be solubilized by treatment of the chromatin with MNase. As shown in Figure 7, lane 4, MNase treatment solubilized all the Slx8 material and a majority of the Orc5 material. These solubilized fractions were then subjected to high-speed centrifugation to test whether the liberated proteins were bound to polynucleosomes. As shown in lane 7, a small fraction of the Slx8 and all of the Orc5 pelleted under these conditions. The pelleted Slx8 material was not simply due to precipitation of the soluble protein since high-speed centrifugation of the initial soluble pool (fraction 2) was unable to pellet any Slx8 protein (Figure 7, lanes 8 and 9). Chromatin fractionation was also carried out on cells expressing the Slx8-4 protein that lacks DNA-binding activity. In order to overcome the difficulty of detecting this small protein on immunoblots we overexpressed Slx8-4 using the GAL1 promoter. Although Slx8-4 protein was again mostly soluble (fraction 2), we were surprised to find a strong signal in the polynucleosome fraction (Figure 7, middle panel). These results suggest that Slx5–Slx8 binds to chromatin even in the absence of the Slx8 DNA-binding domain. Such behavior is consistent with the ability of slx8-4 to largely complement the slx8 mutant phenotype and suggests that Slx8 may interact with other DNA-bound proteins. Such interactions are known to exists in ORC, for example, where subunits such as Orc3 fractionate with chromatin but do not contact DNA directly (40,45). Taken together, these results indicate that a portion of the Slx8 protein fractionates with chromatin and that this fractionation is independent of its DNA-binding domain.
Figure 7.
A portion of Slx8 protein is bound to chromatin. Wild-type or GAL1p::slx8-4 yeast cells were lysed and the resulting extracts were fractionated by differential centrifugation and MNase treatment. Fractions were then immunoblotted for Slx8 (upper two panels) or Orc5 (lower panel) as follows: (1) whole cell extract; (2) first low-speed supernatant containing non-chromatin-binding proteins; (3) crude chromatin pellet; (4) polynucleosome-containing supernatant after limited MNase digestion of fraction 3; (5) chromatin pellet remaining after MNase digestion of fraction 3; (6) soluble proteins from polynucleosomes following ultracentrifugation of fraction 4; (7) pelleted polynucleosomes following ultracentrifugation of fraction 4; (8) soluble proteins following ultracentrifugation of fraction 2; and (9) insoluble proteins following ultracentrifugation of fraction 2. Immunoblotted material was normalized by loading cell equivalents in each lane.
DISCUSSION
The major conclusion from this work is that recombinant Slx5 and Slx8 form a stable complex that interacts with DNA. Although previous studies have observed physical interactions between Slx5 and Slx8, those experiments employed proteins that were overexpressed and did not address whether the Slx5–Slx8 interaction was direct (19,46). Here we have shown that the recombinant Slx5–Slx8 complex is stable to chromatography in the presence of 0.5 M NaCl or 0.1% SDS. Even though this interaction suggests that they act as a complex in vivo, it is still unknown whether Slx5 and Slx8 are bound together at all times in vivo, or if they have other binding partners. This will require chromatographic fractionation of soluble yeast extracts to quantitatively measure their co-fractionation and to determine the composition of Slx5–Slx8 complexes in vivo. At present, we have shown that Slx5 and Slx8 are predominantly nuclear.
The Slx8 subunit appears to be responsible for the DNA-binding activity of the complex, although we cannot rule outa role for Slx5. Supershift analysis indicates that both subunits are present in the DNA-bound complex. And although Slx5 did not improve the binding of Slx8 when the two proteins had been purified individually, the purified complex bound DNA ∼10-fold more efficiently than Slx8 alone. Thus, it is possible that Slx5 stimulates the DNA-binding activity of Slx8 in the complex. Such a cooperative interaction might also explain why the Slx8-4 protein, lacking its DNA-binding domain, still fractionated with chromatin. Our inability to reconstitute the Slx5–Slx8 complex in vitro from its individual subunits is reminiscent of other multi-subunit complexes (e.g. Mus81–Mms4, Slx1–Slx4 and RPA) that are assembled only when co-expressed in vivo (42,43,47).
The DNA-binding activity of Slx5–Slx8 is likely to be biologically relevant for several reasons. First, SLX5 and SLX8 were originally identified based on a genetic interaction with SGS1-TOP3, which encodes a protein complex that acts during recombination-mediated DNA repair. Second, both proteins have been shown to play a role in maintaining genome stability [(25) and Janet R. Mullen and Steven J. Brill, unpublished data] and both proteins immunolocalize to the nucleus. Finally, DNA binding is specific for dsDNA >15 bp with no preference for DNA structures that mimic recombinational intermediates. Although we cannot rule out the possibility that we did not test the optimal substrate, the data are most consistent with the idea that the complex binds dsDNA without any regard to structure or sequence. In chromatin fractionation experiments Slx8 behaved differently than a stable sequence-specific DNA-binding protein, Orc5, in that only a small fraction of it was bound to chromatin fragments. Thus, Slx8 appears to be a largely soluble protein that may bind DNA in a specific phase of the cell cycle, in response to DNA damage, or it may have additional activities that are independent of DNA.
The DNA-binding domain of Slx8 was localized to the N-terminal 200 amino acids, which does not include the RING finger. Amino acid sequence alignments of Slx8 homologs from a variety of yeast species have not revealed obvious conserved motifs in this region (19). As a result, more specific mutational analysis will be required to pinpoint the domain that directly interacts with DNA. Although the N-terminus is dispensible for yeast viability in the absence of SGS1, sgs1Δ cells containing N-terminal truncation alleles were slightly slow-growing. We suspect that this hypomorphic phenotype arises from the loss of DNA binding of this protein; however, it is possible that the N-terminus plays a role in maintaining the structure of the RING domain, in binding Slx5, or in some other Slx8 function. Genetic characterization of the alleles lacking the N-terminus should address these questions and provide clues as to the biological function of the DNA-binding domain.
The major unanswered question in this work concerns the mechanism by which Slx5–Slx8 controls genome stability. The fact that Slx5 and Slx8 both contain RING-finger motifs suggests the possibility that they have the same or related enzyme activity. And the strong physical interaction between these two proteins may explain why mutations in either SLX5 or SLX8 produce nearly identical phenotypes (19,20,25). One possibility that is being pursued relates to the fact that a large number of RING-finger proteins function as ubiquitin or SUMO E3 ligases. Although recent studies have revealed genetic interactions between SLX5-SLX8 and components of the sumoylation pathway (20), we have been unsuccessful in demonstrating a RING-dependent Ub or SUMO E3 ligase activity for either protein (Tatsuya Ii, Janet R. Mullen and Steven J. Brill, unpublished data). If Slx5–Slx8 does the function as a Ub or SUMO E3 ligase, it may be possible that Slx5 and Slx8 are mutually dependent or have some overlap in function in one of these pathways. Given this possibility, the DNA-binding activity of Slx8 may be involved in targeting Slx5 to the DNA where it could modify chromatin proteins or perform some other function. Clearly, it will be necessary to determine the enzymatic activity of Slx5 before we fully understand the role of this complex in controlling genome stability.
Acknowledgments
The authors thank Val Carabetta for technical assistance and Steve Bell for providing antibodies. This work was supported by NIH grant GM071268. Funding to pay the Open Access publication charges for this article was provided by NIH grant GM071268.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cheok C.F., Bachrati C.Z., Chan K.L., Ralf C., Wu L., Hickson I.D. Roles of the Bloom's syndrome helicase in the maintenance of genome stability. Biochem. Soc. Trans. 2005;33:1456–1459. doi: 10.1042/BST0331456. [DOI] [PubMed] [Google Scholar]
- 2.Watt P.M., Hickson I.D., Borts R.H., Louis E.J. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gangloff S., McDonald J.P., Bendixen C., Arthur L., Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakraverty R.K., Hickson I.D. Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. Bioessays. 1999;21:286–294. doi: 10.1002/(SICI)1521-1878(199904)21:4<286::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 5.Mullen J.R., Kaliraman V., Brill S.J. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2000;154:1101–1114. doi: 10.1093/genetics/154.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu L., Davies S.L., North P.S., Goulaouic H., Riou J.F., Turley H., Gatter K.C., Hickson I.D. The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- 7.Bennett R.J., Noirot-Gros M.F., Wang J.C. Interaction between yeast Sgs1 helicase and DNA topoisomerase III. J. Biol. Chem. 2000;275:26898–26905. doi: 10.1074/jbc.M003137200. [DOI] [PubMed] [Google Scholar]
- 8.Fricke W.M., Kaliraman V., Brill S.J. Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J. Biol. Chem. 2001;276:8848–8855. doi: 10.1074/jbc.M009719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin J., Sobeck A., Xu C., Meetei A.R., Hoatlin M., Li L., Wang W. BLAP75, an essential component of Bloom's syndrome protein complexes that maintain genome integrity. EMBO J. 2005;24:1465–1476. doi: 10.1038/sj.emboj.7600622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang M., Bellaoui M., Zhang C., Desai R., Morozov P., Delgado-Cruzata L., Rothstein R., Freyer G.A., Boone C., Brown G.W. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J. 2005;24:2024–2033. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mullen J.R., Nallaseth F.S., Lan Y.Q., Slagle C.E., Brill S.J. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1–Top3 complex. Mol. Cell. Biol. 2005;25:4476–4487. doi: 10.1128/MCB.25.11.4476-4487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu L., Bachrati C.Z., Ou J., Xu C., Yin J., Chang M., Wang W., Li L., Brown G.W., Hickson I.D. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc. Natl Acad. Sci. USA. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harmon F.G., DiGate R.J., Kowalczykowski S.C. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol. Cell. 1999;3:611–620. doi: 10.1016/s1097-2765(00)80354-8. [DOI] [PubMed] [Google Scholar]
- 14.Harmon F.G., Kowalczykowski S.C. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 1998;12:1134–1144. doi: 10.1101/gad.12.8.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu L., Hickson I.D. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 16.Ira G., Malkova A., Liberi G., Foiani M., Haber J.E. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong A.H., Evangelista M., Parsons A.B., Xu H., Bader G.D., Page N., Robinson M., Raghibizadeh S., Hogue C.W., Bussey H., et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 18.Ooi S.L., Shoemaker D.D., Boeke J.D. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nature Genet. 2003;35:277–286. doi: 10.1038/ng1258. [DOI] [PubMed] [Google Scholar]
- 19.Mullen J.R., Kaliraman V., Ibrahim S.S., Brill S.J. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z., Jones G.M., Prelich G. Genetic analysis connects SLX5 and SLX8 to the SUMO pathway in Saccharomyces cerevisiae. Genetics. 2006;172:1499–1509. doi: 10.1534/genetics.105.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holm C. Sensitivity to the Yeast Plasmid 2u DNA is conferred by the nuclear allele nib1. Mol. Cell. Biol. 1982;2:985–992. doi: 10.1128/mcb.2.8.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X., Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl Acad. Sci. USA. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X.L., Reindle A., Johnson E.S. Misregulation of 2 micron circle copy number in a SUMO pathway mutant. Mol. Cell. Biol. 2005;25:4311–4320. doi: 10.1128/MCB.25.10.4311-4320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobson M.J., Pickett A.J., Velmurugan S., Pinder J.B., Barrett L.A., Jayaram M., Chew J.S. The 2 micron plasmid causes cell death in Saccharomyces cerevisiae with a mutation in Ulp1 protease. Mol. Cell. Biol. 2005;25:4299–4310. doi: 10.1128/MCB.25.10.4299-4310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang C., Roberts T.M., Yang J., Desai R., Brown G.W. Suppression of genomic instability by SLX5 and SLX8 in Saccharomyces cerevisiae. DNA Repair (Amst) 2006;5:336–346. doi: 10.1016/j.dnarep.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 26.Gangloff S., Soustelle C., Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nature Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- 27.Gangloff S., de Massy B., Arthur L., Rothstein R., Fabre F. The essential role of yeast topoisomerase III in meiosis depends on recombination. EMBO J. 1999;18:1701–1711. doi: 10.1093/emboj/18.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fabre F., Chan A., Heyer W.D., Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl Acad. Sci. USA. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bastin-Shanower S.A., Fricke W.M., Mullen J.R., Brill S.J. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol. Cell. Biol. 2003;23:3487–3496. doi: 10.1128/MCB.23.10.3487-3496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 31.Kahyo T., Nishida T., Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell. 2001;8:713–718. doi: 10.1016/s1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- 32.Johnson E.S., Gupta A.A. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi Y., Toh-e A., Kikuchi Y. A novel factor required for the SUMO1/Smt3 conjugation of yeast septins. Gene. 2001;275:223–231. doi: 10.1016/s0378-1119(01)00662-x. [DOI] [PubMed] [Google Scholar]
- 34.Hochstrasser M. SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell. 2001;107:5–8. doi: 10.1016/s0092-8674(01)00519-0. [DOI] [PubMed] [Google Scholar]
- 35.Thomas B.J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 36.Rose M.D., Winston F., Hieter P. Methods in Yeast Genetics. NY: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 1990. [Google Scholar]
- 37.Guldener U., Heck S., Fielder T., Beinhauer J., Hegemann J.H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Studier F.W., Rosenberg A.H., Dunn J.J., Dubendorff J.W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 39.Adams A., Gottschling D.E., Kaiser C., Stearns T. Methods in Yeast Genetics. NY: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 1977. [Google Scholar]
- 40.Liang C., Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siegel L.M., Monty K.J. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Biochem. Biophys. Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- 42.Kaliraman V., Mullen J.R., Fricke W.M., Bastin-Shanower S.A., Brill S.J. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 2001;15:2730–2740. doi: 10.1101/gad.932201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fricke W.M., Brill S.J. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 2003;17:1768–1778. doi: 10.1101/gad.1105203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diffley J.F., Stillman B. Similarity between the transcriptional silencer binding proteins ABF1 and RAP1. Science. 1989;246:1034–1038. doi: 10.1126/science.2511628. [DOI] [PubMed] [Google Scholar]
- 45.Lee D.G., Bell S.P. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol. Cell. Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 47.Henricksen L.A., Umbricht C.B., Wold M.S. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]