Abstract
Dlx2, Lymphoid Enhancer Factor (Lef-1) and Msx2 transcription factors are required for several developmental processes. To understand the control of gene expression by these factors, chromatin immunoprecipitation (ChIP) assays identified Msx2 as a downstream target of Dlx2 and Lef-1. Dlx2 activates the Msx2 promoter in several cell lines and binds DNA as a monomer and dimer. A Lef-1 β-catenin-dependent isoform minimally activates the Msx2 promoter and a Lef-1 β-catenin-independent isoform is inactive, however co-expression of Dlx2 and both Lef-1 isoforms synergistically activate the Msx2 promoter. Co-immunoprecipitation and protein pull-down experiments demonstrate Lef-1 physically interacts with Dlx2. Deletion analyses of the Lef-1 protein reveal specific regions required for synergism with Dlx2. The Lef-1 β-catenin binding domain (βDB) is not required for its interaction with Dlx2. Msx2 can auto-regulate its promoter and repress Dlx2 activation. Msx2 repression of Dlx2 activation is dose-specific and both bind a common DNA-binding element. These transcriptional mechanisms correlate with the temporal and spatial expression of these factors and may provide a mechanism for the control of several developmental processes. We demonstrate new transcriptional activities for Dlx2, Msx2 and Lef-1 through protein interactions and identification of downstream targets.
INTRODUCTION
The vertebrate Msx genes originally cloned from mice, are homologous to the Drosophila muscle segment homeobox gene (msh) (1,2). There are three unlinked mammalian Msx gene family members consisting of Msx1, Msx2 and Msx3 (3,4). Msx3 has a restricted expression pattern and is seen only in the dorsal neural tube (5,6). However, Msx1 and Msx2 are expressed in many organs and strongly expressed in developing craniofacial regions (7–9). Msx2 is expressed in epithelial-mesenchymal tissue interactions of several organs including teeth (8,10). Unlike Msx1, which is confined to the mesenchyme throughout tooth development, Msx2 expression is observed in the epithelial and mesenchymal regions of the developing tooth germs. Msx2 mutations are associated with tooth defects and Msx2 deficient mice demonstrate developmental defects in ectodermal organs (11). Consistent with Msx2 expression occurring after the early expression of Pitx2, Lef-1 and Dlx2 in the dental epithelium, Msx2 mutant mice teeth develop normally through the early stages of development but have defects associated with late tooth morphogenesis and amelogenesis (8,12–16).
Msx2 homeobox gene expression can be observed in the apical ectodermal ridge (AER) and other regions of the developing limb (8,10). The expression of the chick Msx2 gene in the AER and other regions of the developing limb is dependent on multiple closely spaced regulatory elements (17). Interestingly, Msx2 and Dlx genes have highly localized expression patterns in the AER (18,19).
Dlx2, a member of the distal-less gene family, has been established as a regulator of branchial arch development (20,21). Homozygous mutants of Dlx2 have abnormal development of forebrain cells and craniofacial abnormalities in developing neural tissue (22). Dlx2 can regulate Dlx5 and/or Dlx6 expression and ectopic expression of Dlx2 induced Dlx5 expression in slice cultures of the mouse embryonic cerebral cortex (23,24). Furthermore, ectopic expression of Dlx2 can induce the expression of glutamic acid decarboxylase in brain slices (24). Analyses of the Dlx1/2 mutant mice demonstrate a decrease in expression of Aristaless, which may control the development of GABAergic neurons (25).
Dlx2 is also expressed in the first branchial arch and is involved in tooth development (21,26). Dlx genes are believed to play a role in tooth morphogenesis because homozygous Dlx1/Dlx2 mutants are missing maxillary molars (27). The overlapping expression patterns of Msx and Dlx genes in the branchial arches, AER, teeth and brain suggest an interactive role for these transcription factors (18,19,22).
Lymphoid enhancer-binding factor 1 (Lef-1) is a cell type-specific transcription factor expressed in lymphocytes of the adult mouse and in the neural crest, mesencephalon, tooth germs, whisker follicles and other sites during embryogenesis (13,28–32). Lef-1 is a member of the high mobility group (HMG) family of proteins and activates transcription only in collaboration with other DNA-binding proteins and may promote the assembly of a higher-order nucleoprotein complex by juxtaposing non-adjacent factor binding sites (33–35). Lef-1 expression overlaps that of Dlx2 and Msx2 in the dental epithelium, branchial arches and limb buds (13,31,36).
Msx2 expression is detected in tissues specific for Dlx2 and Lef-1 expression, which suggests that they functionally interact or regulate the expression of each other. The coordinated expression of genes during development is not regulated by one molecule or transcription factor but by several factors acting together through direct physical contact or through independent DNA-binding to their respective DNA elements. Thus, individual genes are regulated by complexes of factors interacting with regulatory elements consisting of multiple binding sites. This regulatory network provides a mechanism for the temporal and spatial control of gene expression that is absolutely required for embryo development.
We use chromatin immunoprecipitation (ChIP) assays to identify downstream targets of transcription factors involved in embryogenesis and tooth development. The ChIP assay identified Dlx2 binding to the Msx2 promoter in two different cell lines expressing Dlx2 and we confirmed this binding by electrophoretic mobility shift assays (EMSAs) and transient transfection of Msx2 promoter constructs and expression constructs. New transcriptional mechanisms are demonstrated through direct protein interactions between Dlx2 and Lef-1 in the activation of Msx2 promoter activity. ChIP assays also revealed Lef-1 binding to the Msx2 promoter. Structure and function analyses demonstrate specific interactions between Dlx2 and Lef-1. Deletion analyses of the Lef-1 protein demonstrate that the β-catenin binding region is not required for its interaction with Dlx2 or synergistic activation of the Msx2 promoter. Thus, Dlx2 is a new partner for Lef-1 and appears to be independent of β-catenin. However, two regions of Lef-1 have been identified that interact with Dlx2. Furthermore, Msx2 can auto-regulate its promoter and Msx2 can attenuate Dlx2 activation. Our research demonstrates a functional interaction between Dlx2 and Lef-1 in regulating gene expression and provides a new role for Lef-1.
MATERIALS AND METHODS
Electrophoretic mobility shift assay
Complementary oligonucleotides containing a Dlx2 binding site within the Msx2 promoter with flanking partial BamHI ends were annealed and filled with Klenow polymerase to generate 32P-labeled probes for EMSAs, as described (37). The sense primer was 5′-GATCCGGGTAGAGATTAGTTGAATATCCCTTGGG-3′ and the anti-sense primer was 5′-GATCCCCAAGGGATATTCAACTAATCTCTACCCG-3′ with the Dlx2 binding site underlined. The Dlx2 binding site selected for the EMSA is located at −260 to −265 (5′-TAGTTG-3′) of the Msx2 promoter. Standard binding assays were performed as previously described (38). A titration of the bacteria expressed and purified Dlx2 and Msx2 proteins were used in the assays. The samples were electrophoresed, visualized and quantitated as described previously, except quantitation of dried gels was performed on the Molecular Dynamics STORM PhosphoImager (37).
Chromatin immunoprecipitation analysis
The ChIP analysis was performed as described using the ChIP Assay Kit (Upstate) with the following modifications. CHO and LS-8 cells were fed for 24 h, harvested and plated in 60 mm dishes. Cells were cross-linked with 1% formaldehyde for 10 m at 37°C the next day. The cells were washed with cold phosphate-buffered saline with mammalian protease inhibitor cocktail (SIGMA) and lysed in 200 μl SDS lysis buffer for 10 m on ice and sonicated three times for 10 s. The average DNA fragments ranged between ∼200 and 2000 bp. The lysates were then clarified by centrifugation at 13 000 r.p.m. for 10 m at 4°C and diluted 10-fold in ChIP dilution buffer. The sonicated samples were precleared using Salmon Sperm DNA/Protein A Agarose-50% Slurry for 1 h at 4°C and 1% cell lysate was used for input control. Samples were incubated with Dlx2 goat polyclonal IgG (Santa Cruz Biotechnology) or Lef-1 mouse monoclonal IgG (Upstate), overnight at 4°C. Immune complexes were washed consecutively for 5 m with each of the following solutions: Low salt immune complex wash buffer, High salt immune complex wash buffer, LiCl immune complex wash buffer and TE buffer twice. Complexes were then eluted twice at room temperature for 15 m in elution buffer (1% SDS, 0.1M NaHCO3). Immunoprecipitated DNA was reverse cross-linked at 65°C for 4 h in 200 mM NaCl and phenol/chloroform purified. An aliquot of the immunoprecipitated DNA (5 μl) from non-transfected cells was used for PCR (40 cycles). All reactions were done under an annealing temperature of 57°C. Two primers for amplifying the Dlx2 and Lef-1 binding sites in the Msx2 promoter are as follows: sense- 5′-AAGGGAGAAAGGGTAGAG-3′ and antisense, 5′-CCCGCCTGAGAATGTTGG-3′. All the PCR products were evaluated on a 1% agarose gel in 1× TBE for appropriate size (272 bp) and confirmed by sequencing. As controls the Msx2 primers were used without chromatin, normal rabbit IgG was used replacing the Dlx2 or Lef-1 antibody to reveal non-specific immunoprecipitation of the chromatin and primers to an unrelated gene were used to demonstrate the specificity of the immunoprecipitated chromatin.
Expression and promoter constructs
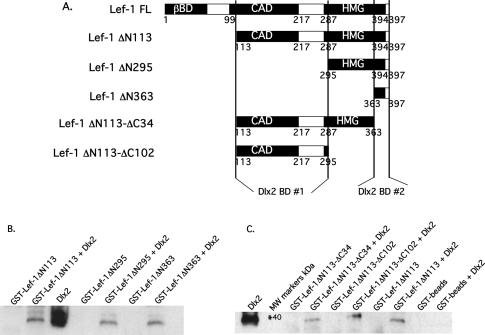
Expression plasmids containing the cytomegalovirus (CMV) promoter linked to the Msx2 and Lef-1 cDNA were constructed in pcDNA 3.1 MycHisC (Invitrogen) (37–39). The Lef-1 deletion constructs were prepared by PCR amplification of the full-length Lef-1 cDNA and cloned into the pcDNA3.1 MycHisC vector. The full-length Lef-1 cDNA was PCR amplified using a sense primer containing the Kozak sequence, initiation codon and an XbaI site and an anti-sense primer without the terminator codon containing a KpnI site. This clone is 1193 bp, encodes a protein of 397 residues and includes the β-catenin binding domain (βBD) in the N-terminus. The Lef-1 ΔN113 construct represents a smaller alternative Lef-1 transcript found in tissues and was cloned using a sense primer containing a kozak sequence, initiation codon and a NotI site. The anti-sense primer contained a HindIII site without the terminator codon is 854 bp and encodes a protein of 284 amino acids. This clone does not include the βBD in the N-terminus. The Lef-1 ΔN295 construct was PCR amplified using a NotI sense primer and a HindIII anti-sense primer; the product was 306 bp and 102 amino acids. It contains the HMG domain. The Lef-1 ΔN363 construct was made using a sense primer containing a BamHI site and HindIII anti-sense primer, is 102 bp and encodes a C-terminal peptide of 34 residues. Lef-1 ΔN113-ΔC34 was made using a NotI sense primer and HindIII anti-sense primer and contains the CAD and HMG domains but not the βBD. It is 752 bp and encodes a protein of 250 residues. Lef-1 ΔN113-ΔC102 was made using a BamHI sense primer and HindIII anti-sense primer and includes only the CAD domain. It is 546 bp and encodes a 182 amino acid protein. All PCR products were digested with the appropriate restriction enzymes and inserted into the digested vector.
The Dlx2 expression plasmid has been previously described (kindly provided by Dr John Rubenstein, University of California San Francisco, San Francisco, CA). The Msx2 promoter construct (kindly provided by Dr Y. Chen, Tulane University, New Orleans, LA) was digested with XhoI and NotI to isolate a 5.8 kB fragment, which was ligated into the luciferase vector [previously described (38)] using BamHI/XhoI and NotI/HindIII linkers. This created the Msx2-5820 luciferase reporter construct. The Msx2-872 construct was made by digesting Msx2-5820 with BamHI and HindIII and the resulting 872 bp fragment was ligated into the luciferase vector. The Msx2-238 construct was made by PCR using primers containing BamHI and HindIII sites. PCR products were TAE gel purified if necessary. All PCR products were ligated into the vector using the Rapid DNA ligation kit (Boehringer Mannheim, Indianapolis, IN, USA) and then transformed into DH5α competent cells (Invitrogen, Carlsbad, CA, USA). All constructs were confirmed by DNA sequencing. All plasmids were purified by CsCl gradient centrifugation for use in the transfection assays. A CMV or SV-40 β-galactosidase reporter plasmid (Clontech) was also purified for co-transfection in all experiments as a control for transfection efficiency.
Cell culture, transient transfections, luciferase and β-galactosidase assays
CHO, C3H10T1/2 and LS-8 cells were cultured in DMEM supplemented with 5% or 10% fetal bovine serum (FBS) and penicillin/streptomycin and transfected by electroporation. Cultures were fed 24 h prior to transfection, resuspended in PBS and mixed with 2.5 μg of expression plasmids, 5 μg of reporter plasmid and 0.5 μg of CMV or SV-40 β-galactosidase plasmid. Electroporation of CHO cells was performed at 360 V and 950 microfarads (μF) (Gene Pulser XL, Bio-Rad) and C3H10T1/2 cells at 340 V and 950 μF. LS-8 cells were transfected by electroporation as previously described (37). Transfected cells were incubated for 24 h in 60 mm culture dishes and fed with 5% FBS and DMEM and then lysed and assayed for reporter activities and protein content by Bradford assay (Bio-Rad). Luciferase was measured using reagents from Promega. β-Galactosidase was measured using the Galacto-Light Plus reagents (Tropix Inc.). All luciferase activities were normalized to β-galactosidase activity.
Expression and purification of GST fusion proteins
Lef-1 and Msx2 were PCR amplified from cDNA clones as described and ligated into the pGex6P-2 GST vector (Amersham Pharmacia Biotech, Piscataway, NJ) (37–39). Dlx2 was PCR amplified and cloned into the pGex6P-2 GST vector using BamHI and NotI restriction enzyme sites engineered into the primers. Lef-1 and Lef-1 deletion fragments were PCR amplified using primers with specific restriction endonuclease sites, digested and cloned into the GST digested vector. All pGex6P-2 GST plasmids were confirmed by DNA sequencing. The plasmids were transformed into BL21 cells. Proteins were isolated as described (40,41). Proteins were cleaved from the GST moiety using 80 U of PreScission Protease (Pharmacia Biotech) per ml of glutathione Sepharose. Cleaved proteins were stored in 10% glycerol. Protein concentration was quantitated with Bradford Reagent (Bio-Rad Laboratories, Hercules, CA). Proteins were examined by electrophoresis on denaturing SDS–polyacrylamide gels, followed by Coomassie Blue staining (50% methanol, 10% acetic acid and 0.5% coomassie brilliant blue stain).
GST pull-down assays
Immobilized GST-Dlx2, GST-PITX2 and GST-Lef-1 fusion proteins were prepared as described above and suspended in binding buffer (20 mM HEPES pH 7.5, 5% glycerol, 50 mM NaCl, 1 mM EDTA, 1 mM DTT, with or without 1% milk and 400 μg/ml of ethidium bromide). Purified bacteria expressed Lef-1 proteins (50–200 ng's) were added to 5 μg's immobilized GST–Dlx2 fusion proteins or GST in a total volume of 100 μl and incubated for 30 m at 4°C. The beads were pelleted and washed 4 times with 200 μl binding buffer. The bound proteins were eluted by boiling in SDS-sample buffer and separated on a 10% SDS–polyacrylamide gel. Approximately, 80 ng of purified Lef-1 proteins were analyzed in separate western blots. Following SDS gel electrophoresis, the proteins were transferred to PVDF filters (Millipore), immunoblotted and detected using appropriate antibodies, Lef-1, (Santa Cruz Biotechnology or Upstate), Dlx2 goat polyclonal IgG (Santa Cruz Biotechnology) and ECL reagents from Amersham.
Immunoprecipitation assay
Approximately 24 h after cell transfection with Dlx2 and Lef-1 ΔN113, CHO cells were rinsed with 1 ml of PBS, then incubated with 1 ml ice cold RIPA buffer for 15 m at 4°C. Cells were harvested and disrupted by repeated aspiration through a 25-gauge needle attached to a 1 ml syringe. The lysates were then incubated on ice for 30 m. Cellular debris was pelleted by centrifugation at 10 000× g for 10 m at 4°C. An aliquot of lysate was saved for analysis as input control. Supernatant was transferred to a fresh 1.5 ml microfuge tube on ice and precleared using the Preclearing Matrix B-goat (ExactaCruz B, Santa Cruz Biotechnology) for 30 m at 4°C. Matrix was removed by brief centrifugation and supernatant transferred to a new tube. An IP antibody-IP matrix complex was prepared as per manufacturer's instructions using primary anti-Dlx2 (25 μl) antibody (Santa Cruz Biotechnology). The IP antibody-IP matrix complex was incubated with the precleared cell lysate at 4°C for 12 h. After incubation the lysate was centrifuged to pellet the IP matrix. The matrix was washed three times with PBS and resuspended in 15 μl of ddH2O and 3 μl 6× SDS loading dye. Samples were boiled for 5 m and resolved on a 10% polyacylamide gel. A western blot assay was used with anti-Lef-1 antibody and HRP conjugated ExactaCruz reagent to detect immunoprecipitated proteins.
RESULTS
Dlx2 binds to the Msx2 promoter and is co-expressed with Msx2 in several cell lines
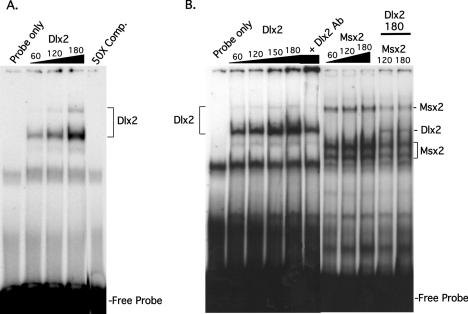
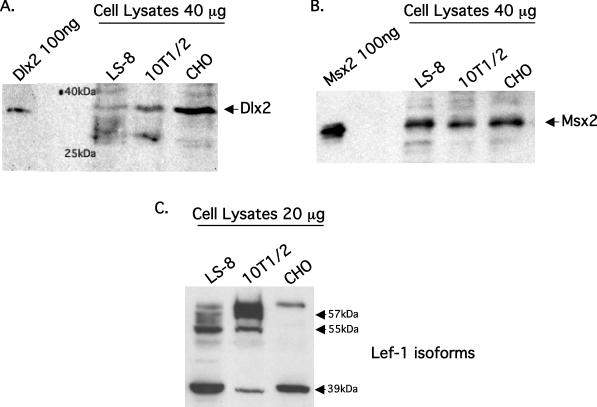
The 5.8 Kb Msx2 promoter contains multiple Dlx2 binding sites. To demonstrate Dlx2 binding to these sites, an EMSA was performed using purified Dlx2 protein and a probe derived from the Msx2 promoter containing a Dlx2 binding site, −260 to −265 bp (TAGTTG). Dlx2 protein was added to the probe at 60, 120 and 180 ng and two bands were detected that were not in the probe alone lane (Figure 1A). These bands increased in intensity with increasing Dlx2 protein and the slower migrating band appears to be a homodimer of Dlx2. Specificity of the complexes was demonstrated using 50-fold excess cold competitor probe (Figure 1A). Specificity of Dlx2 binding was further demonstrated using Dlx2 antisera and 180 ng of Dlx2 protein. The antisera reduced Dlx2 binding to the probe and both band intensities were decreased (Figure 1B). To determine if Msx2 and Dlx2 compete for binding to the probe, 180 ng of Dlx2 was incubated with 120 and 180 ng of Msx2. Msx2 bound as three bands, which increase in intensity with increasing amounts of protein (Figure 1B). Interestingly, the binding of both Dlx2 and Msx2 decreased when both proteins were present in the binding reaction, demonstrating a competition for the DNA-binding site (Figure 1B). The pure Msx2 protein used in the binding studies is shown in Figure 2B. The pure Dlx2 protein used in the EMSA experiment is shown in Figure 2A identified with the Dlx2 Ab on a western blot. Endogenous Dlx2 was observed in the tooth epithelial cell line LS-8, the pluripotent C3H10T1/2 cell line and the CHO cell line (Figure 2A). Dlx2 appears to be highly expressed in CHO cells but is also seen in the other two cell lines. We next determined if Msx2 was expressed in these cell lines and found similar levels of endogenous Msx2 expression in the three cell lines (Figure 2B). Lef-1 expression was observed in all three cell lysates, however only LS-8 and 10T1/2 cells express the larger molecular weight isoforms (Figure 2C).
Figure 1.
Dlx2 binds to an element in the Msx2 Promoter. (A) Dlx2 proteins (60, 120 and 180 ngs) were incubated with the Msx2 promoter sequence containing a Dlx2 binding element (TAGTTG) as the radioactive probe. The 50× comp. lane was performed with 180 ngs of Dlx2 protein and 50-fold excess of cold competitor probe. (B) Dlx2 protein (180 ng) was incubated with 2 μl of Dlx2 Ab, which decreased Dlx2 binding demonstrating the specificity of Dlx2. Msx2 protein at 60, 120 and 180 ng bound the Msx2 probe as three distinct bands. Msx2 titration (120 and 180 ng) with 180 ng Dlx2 revealed competitive binding for the same element by these proteins. The EMSA experiments were analyzed in 8% native polyacrylamide gels. The free and bound forms of DNA were quantitated using the Molecular Dynamics STORM PhosphoImager. The free probe, bound and dimer complexes are indicated.
Figure 2.
Endogenous expression of Dlx2, Msx2 and Lef-1 in three cell lines. (A) Western blot of endogenous Dlx2 protein in the LS-8 tooth epithelial cell line, C3H10T1/2 pluripotent cell line, CHO cell line using the Dlx2 antibody. Whole cell lysates from each cell line were prepared and 40 μg of protein run on a 10% SDS–polyacrylamide gel. The proteins were visualized using ECL reagents from Amersham. Pure Dlx2 protein was used as a control at 100 ng and two molecular weight markers are noted (40 and 25 kDa). (B) Western blot of endogenous Msx2 protein from the same cell lines and experimental procedure as in (A). Pure Msx2 protein was used as a control at 100 ng. (C) Western blot of endogenous Lef-1 protein from the same cell lines and experimental procedures as in (A). The approximate molecular weights of the Lef-1 isoforms are noted.
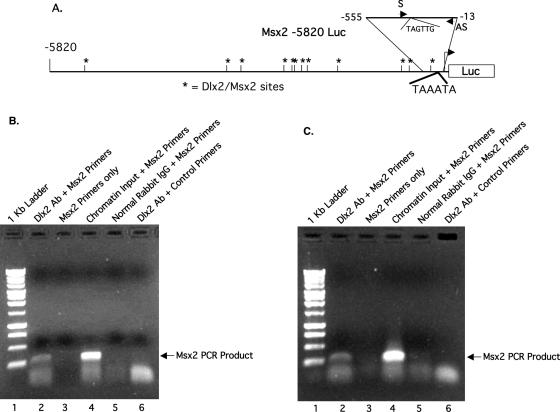
ChIP analyses were done to demonstrate Dlx2 binding to the Msx2 promoter in the cell chromatin because these three cell lines endogenously express Dlx2 and Msx2. A sense primer and an anti-sense primer, which flank the Dlx2 binding site located at −260 to −265 in the Msx2 promoter, were designed and produce a 272 bp product (Figure 3A). The sense primer anneals to the sequence located at −268 to −285 and the anitsense primer at −13 to −29 of the Msx2 promoter (Figure 3A). The Dlx2 binding site selected for the ChIP assay is identical to the EMSA probe sequence (Figure 1A). Chromatin isolated from cells prior to immunoprecipitation served as a control; a sample without treatment of Dlx2 antibody was used as input control template and the primers were used to demonstrate the presence of chromatin in the sample. The primer set amplified the Msx2 promoter from chromatin input derived from the CHO cell line (lane 4, Figure 3B). A PCR was performed without chromatin and primers only as a negative control (lane 3, Figure 3B). Endogenous Dlx2 is bound to the Msx2 promoter in vivo as shown using Dlx2 antibody and Msx2 primers (lane 2, Figure 3B). Normal rabbit IgG was used as a control and did not immunoprecipitate the Msx2 promoter (lane 5, Figure 3B). Furthermore, using primers to an unrelated gene did not amplify a product from the Dlx2 antibody immunoprecipitated chromatin (lane 6, Figure 3B). Lane 1 contains 1 Kb ladder (Promega) to confirm sizes of the PCR products (Figure 3B). All PCR products were sequenced to confirm their identity.
Figure 3.
Dlx2 binds to the Msx2 promoter in vivo. (A) Schematic of the 5.8 Kb Msx2 promoter with the Dlx2 binding sites noted by asterisks. The location of the sense primer (S) and the anti-sense primer (AS) are shown in the blowup of the −555 to −13 bp region of the proximal promoter used to amplify the immunoprecipiated chromatin. (B) ChIP assays were performed using CHO cells. Lane 1 contains the 1 Kb ladder and lane 2 is the Dlx2 immunoprecipitated chromatin amplified using the specific Msx2 promoter primers and produced the correct size product of 272 bp. Lane 3 is Msx2 primers only; lane 4 is the chromatin input using the Msx2 promoter primers. Lane 5 is the immunoprecipitation using normal rabbit IgG and Msx2 primers and lane 6 is the Dlx2 immunoprecipitated chromatin amplified with primers to an unrelated gene. (C) ChIP assays were performed using LS-8 cells and the same experimental procedures as in panel B. Experiments were repeated three separate times.
The tooth epithelial LS-8 cell ChIP assay was also performed to demonstrate endogenous Dlx2 binding to the Msx2 promoter in these cells. The ChIP results with LS-8 and CHO cells were identical. The results reveal in vivo binding of Dlx2 to the Msx2 promoter in CHO cells (lane 2, Figure 3C). The appropriate controls were performed as in Figure 3B.
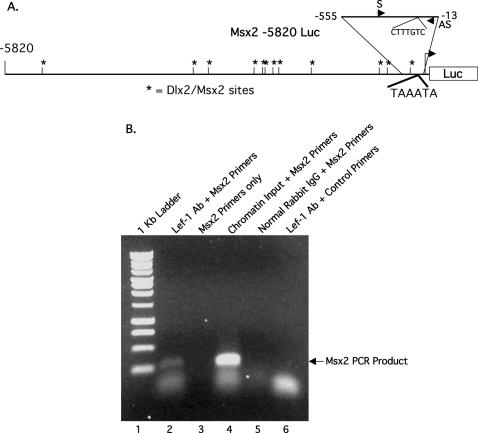
Chromatin immunoprecipitation assays reveal Lef-1 binding to the Msx2 promoter
Sequence analysis of the Msx2 promoter revealed a Lef-1 binding site (CTTTGTC) at −220 to −226 (Figure 4A). Because Lef-1 is endogenously expressed in many cell lines, the ChIP assay was used to determine if Lef-1 bound to the Msx2 promoter. Lef-1 is endogenously expressed in the LS-8 cell line and the Lef-1 Ab immunoprecipitates the Lef-1/chromatin complex containing the Msx2 promoter (lane 2, Figure 4B). The control lanes are identical to Figure 3.
Figure 4.
Lef-1 binds to the Msx2 promoter in vivo. (A) Schematic of the 5.8 Kb Msx2 promoter with the Lef-1 binding site (CTTTGTC) shown in the proximal promoter. The location of the sense primer (S) and the anti-sense primer (AS) are shown in the blowup of the −555 to −13 bp region of the proximal promoter used to amplify the immunoprecipitated chromatin. (B) ChIP assays were performed using LS-8 cells. Lane 1 contains the 1 Kb ladder and lane 2 is the Lef-1 immunoprecipitated chromatin amplified using the specific Msx2 promoter primers and produced the correct size product of 272 bp. Lane 3 is Msx2 primers only; lane 4 is the chromatin input using the Msx2 promoter primers. Lane 5 is the immunoprecipitation using normal rabbit IgG and Msx2 primers and lane 6 is the Lef-1 immunoprecipitated chromatin amplified with primers to an unrelated gene.
Dlx2 and Lef-1 synergistically activate the Msx2 promoter
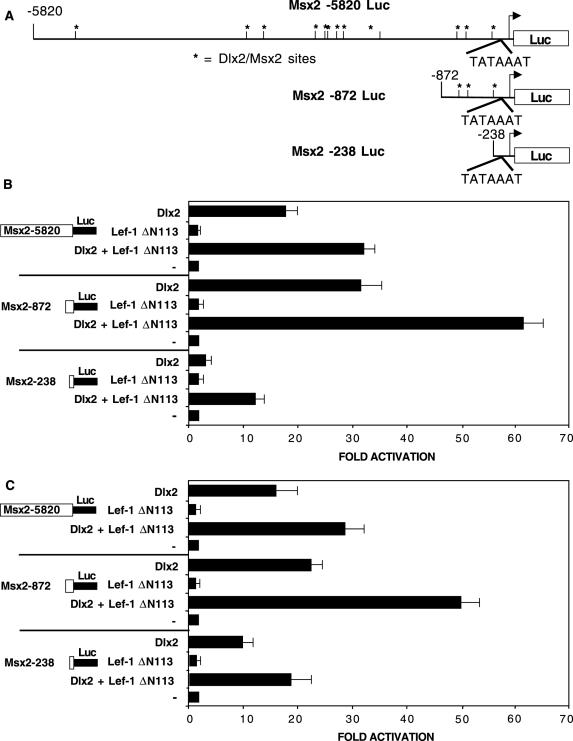
Dlx2 activation of Msx2 was demonstrated using three promoter constructs linked to the luciferase gene (Figure 5A). The Msx2-5820 Luc promoter contains 5.8 Kb of the Msx2 promoter and has ∼12 consensus (TAATTA) and non-consensus Dlx2 binding sites (TAGTTG, TATTTG). Two truncated Msx2 promoters were constructed; Msx2-872 Luc contains 872 bp of the Msx2 promoter and three Dlx2 binding sites and Msx2-238 Luc, which does not contain Dlx2 binding sites (Figure 5A). Dlx2 activated the full-length Msx2 promoter at ∼20-fold in CHO cells (Figure 5B). Interestingly, Dlx2 activated the truncated Msx2-872 promoter at higher levels (∼30-fold) compared to the full-length promoter (Figure 5B). These results may indicate the presence of other factors binding to upstream Msx2 promoter sequences that attenuate Dlx2 activation. However, Dlx2 did not activate the minimal Msx2-238 promoter (Figure 5B). Because Lef-1 is co-expressed with Dlx2 and Msx2 in several tissues, we asked if Lef-1 could activate the Msx2 promoter. In these experiments we used the alternatively spliced Lef-1 ΔN113 construct, which does not contain the βBD in its N-terminus. This naturally occurring Lef-1 transcript was used in order to eliminate the effect of β-catenin activating Lef-1. Lef-1 ΔN113 alone did not activate the Msx2 promoters however, co-transfection with Dlx2 resulted in a 33-fold synergistic activation of the Msx2-5820 promoter and 62-fold activation of the Msx2-872 promoter (Figure 5B). The Msx2-238 minimal promoter was activated at ∼5-fold by Dlx2 and Lef-1 ΔN113, due to the presence of the Lef-1 binding site in this promoter construct. There is no difference in the synergistic activation of the Msx2 promoter between the Lef-1 ΔN113 or Lef-1 FL (full length) constructs (see Figure 8).
Figure 5.
Dlx2 and LEF-1 synergistically activate the Msx2 promoter. (A) Schematic of the Msx2 promoter constructs used in transient transfection assays showing the location of Dlx2/Msx2 shared binding sites by asterisks. Note that the Msx2-238 Luc minimal promoter does not contain a Dlx2/Msx2 binding element. (B) CHO cells were transfected with the Msx2-5820, Msx2-872 or Msx2-238 luciferase reporter gene (5 μg). The cells were co-transfected with the CMV-Dlx2 and/or CMV-Lef-1 ΔN113 short isoform expression plasmids or the CMV plasmid without Dlx2 or Lef-1 (−) (2.5 μg). (C) C3H10T1/2 cells were transfected as in (B) to determine if the activation was cell dependent. To control for transfection efficiency, all transfections included the SV-40 β-galactosidase reporter (0.5 μg). Cells were incubated for 24 h and then assayed for luciferase and β-galactosidase activities. The activities are shown as mean fold activation compared to the Msx2 promoter plasmids without Dlx2 or Lef-1 expression and normalized to β-galactosidase activity (+/− SEM from eight independent experiments for (B) and from five experiments in (C). The mean Msx2 promoter luciferase activity with Dlx2 expression was ∼150 000 light units per 15 μg protein and the β-galactosidase activity was ∼75 000 light units per 15 μg protein.
These experiments were repeated in the pluripotent C3H10T1/2 embryonic cell line. Transfection of these cells with the same constructs reveal similar results as with the CHO cells indicating activity in CHO cells was not cell specific (Figure 5C). Dlx2 activation and synergism with Lef-1 ΔN113 was similar to that observed in CHO cells. The combination of this data and the ChIP assay demonstrate specific regulation of the Msx2 promoter by Dlx2 and a synergy between Dlx2 and Lef-1. Furthermore, these experiments were performed in the LS-8 tooth epithelial cell line. The activation levels were decreased however, the relative activation of the Msx2 promoter by Dlx2 and Lef-1 were similar (Figure 11B).
Dlx2 and Lef-1 physically interact
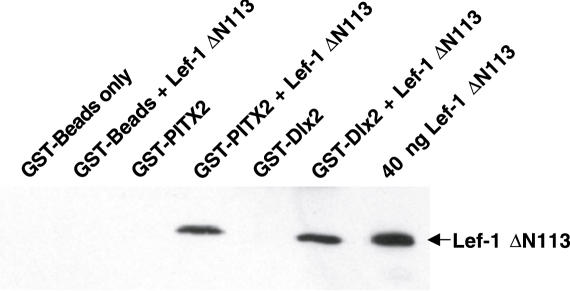
The finding that Lef-1 ΔN113 alone did not activate the Msx2 promoter but synergized with Dlx2 suggested a physical interaction between Dlx2 and Lef-1 independent of β-catenin. A GST-pull-down assay was performed using immobilized GST-Dlx2 on Sepharose beads and incubated with pure Lef-1 ΔN113 protein under stringent binding conditions. As a control, GST-beads alone did not bind Lef-1 ΔN113, however, Lef-1 ΔN113 bound to GST-Dlx2 demonstrating a direct protein interaction between these two proteins (Figure 6). Previously shown to interact with Lef-1, immobilized GST-PITX2 was used as a positive control (39) (Figure 6). Lef-1 binds to both proteins at similar levels. The binding of Lef-1 ΔN113, a smaller native transcript, indicates that its interaction with Dlx2 does not require the βDB or the N-terminus.
Figure 6.
Lef-1 physically interacts with Dlx2. (A) GST-Dlx2 pull-down assay with bacterial expressed and purified Lef-1 ΔN113 protein (40 ng). As a control GST-beads were incubated with purified Lef-1 to demonstrate the specificity of Lef-1 binding to the fusion proteins. As a positive control Lef-1 was incubated with GST-PITX2, which has previously been shown to bind Lef-1 (39). Lef-1 binds to immobilized GST-Dlx2 demonstrating that Lef-1 can physically interact with Dlx2. The bound protein was detected by western blot using the Lef-1 antibody. Lef-1 binds equally to both proteins. Three independent experiments were performed.
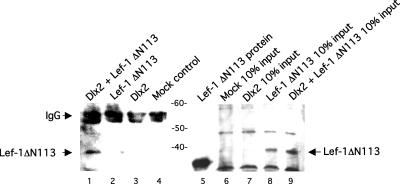
Immunoprecipitation experiments demonstrate Dlx2 and Lef-1 protein interactions. CHO cells co-transfected with Dlx2 and Lef-1 ΔN113 were assayed using the Dlx2 antibody to immunoprecipitate a Dlx2/Lef-1 complex. The immunoprecipitation experiments reveal a Dlx2 interaction with Lef-1 ΔN113 (lane 1, Figure 7). Because CHO cells do not express the larger molecular weight Lef-1 proteins but do express Dlx2 (Figure 2A), a small amount of Lef-1 ΔN113 is immunoprecipitated in the Lef-1 only transfected cells (Figure 7, lane 2). As controls, empty vector and Dlx2 transfection did not immunoprecipitate Lef-1. Lef-1 ΔN113 expression is shown in transfected CHO cell lysates (10% input) and also seen when co-transfected with Dlx2 (Figure 7, lanes 8 and 9). Mock and Dlx2 input lanes were used as controls with the Lef-1 antibody.
Figure 7.
Lef-1 and Dlx2 physically interact in CHO cells. Lef-1 and/or Dlx2 expression plasmids (2.5 μg) were transfected into CHO cells and incubated for 24 h. Cells were harvested and lysed and the Lef-1/Dlx2 protein complex immunoprecipitated (IP) using the Dlx2 antibody. The immunoprecipitated complexes were resolved on a 10% polyacrylamide gel and transferred to a PVDF filter and western blotting was done using the Lef-1 antibody. Lane 1 is the Lef-1 ΔN113 and Dlx2 co-transfected IP, lane 2 is the Lef-1 ΔN113 transfected IP, lane 3 is the Dlx2 transfected IP, lane 4 is the mock-transfected cell lysate IP. Input controls are shown in lanes 6–9. As a molecular weight size control, 150 ng of purified Lef-1 protein was immunoblotted (lane 5). The transfected Lef-1 migrates slightly slower due to the presence of a C-terminal Myc/His tag on the mammalian expression plasmids. Transfected Dlx2 is expressed in CHO cells and a representative lysate is shown in Figure 9C. Molecular weight markers are shown between lanes 4 and 5. Experiments were repeated more than three times.
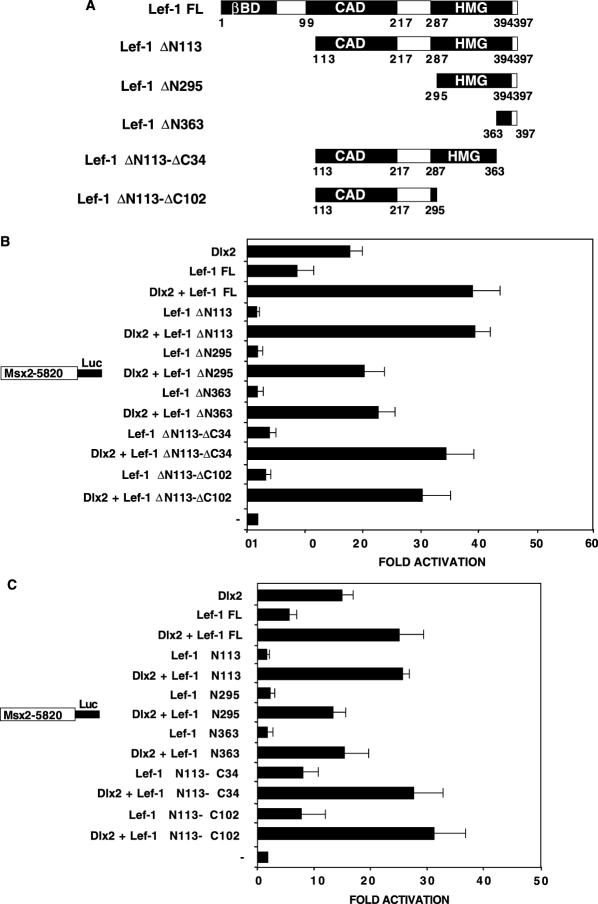
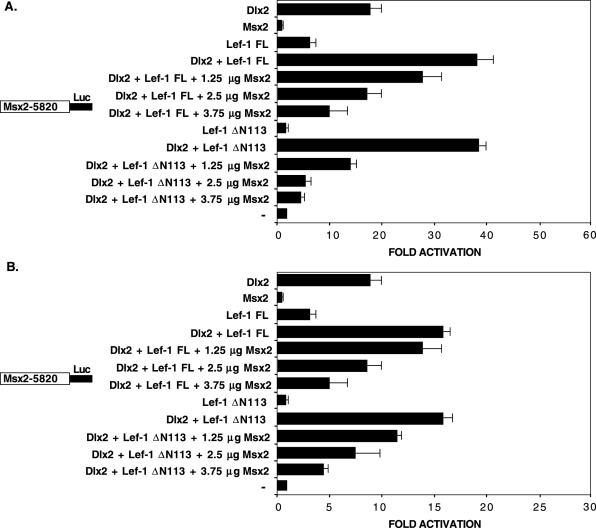
The Lef-1 CAD domain and 3′ flanking residues are required for synergism with Dlx2
We have shown that an alternative endogenous Lef-1 transcript (Lef-1 ΔN113) containing the context-dependent activation domain (CAD) and HMG domain while lacking the βBD interacts with Dlx2 and synergistically activates the Msx2 promoter (Figure 5B and C and Figure 8B and C). In CHO cells, Lef-1 ΔN113 co-transfected with Dlx2 synergistically activates the Msx2-5820 full-length promoter at ∼40-fold (Figure 8B). The full-length Lef-1 cDNA containing the βBD activated the Msx2 promoter at ∼6-fold and co-expression of Dlx2 synergistically activated the Msx2 promoter at ∼40-fold in CHO cells (Figure 8B). Similar activation was observed in C3H10T1/2 cells (Figure 8C). Thus, the βBD is not required for the Lef-1 synergistic activation with Dlx2. Deletion of 295 residues from the Lef-1 N-terminus (Lef-1 ΔN295), which removes the CAD domain and leaves most of the HMG domain intact, results in a loss of synergism in CHO cells (Figure 8B) and C3H10T1/2 cells (Figure 8C). These results suggest that the HMG domain alone is not sufficient for synergistic activation of the Msx2 promoter with Dlx2. Further truncation of the Lef-1 N-terminus deleting most of the HMG domain (Lef-1 ΔN363) resulted in a loss of synergism (Figure 8B and C). These results demonstrate that the CAD domain is required for Lef-1 activation and/or synergism with Dlx2. In contrast, deleting part or most of the HMG domain, Lef-1 ΔC34 or Lef-1 ΔC102 results in low activity alone (∼3-fold) and synergism with Dlx2 in both cell types (Figure 8B and C). The Lef-1 ΔC34 and Lef-1 ΔC102 constructs contain the CAD domain including 70 residues between the CAD and HMG domains (Figure 8A) that are necessary for synergism with Dlx2.
Figure 8.
Identification of Lef-1 domains required for synergism with Dlx2. (A) A schematic of the Lef-1 deletion constructs used in the transfection assays. βBD, β-catenin binding domain; CAD, context-dependent activation domain; HMG, high mobility group domain. The numbers under the constructs denote residues. (B) CHO cells were transfected with the Msx2-5820 luciferase reporter gene (5 μg). The cells were co-transfected with the CMV-Dlx2 and/or CMV-Lef-1 full-length (FL), Lef-1 short isoform (Lef-1 ΔN113), the Lef-1 deletion construct expression plasmids, the CMV plasmid without Dlx2 or Lef-1 (−) (2.5 μg). (C) C3H10T1/2 cells were transfected as in (B). To control for transfection efficiency, all transfections included the SV-40 β-galactosidase reporter (0.5 μg). Cells were incubated for 24 h and then assayed for luciferase and β-galactosidase activities. The activities are shown as mean fold activation compared to the Msx2 promoter plasmid without Dlx2 or Lef-1 expression and normalized to β-galactosidase activity (+/− SEM from five independent experiments).
Dlx2 interacts with the CAD and HMG domains of Lef-1
To determine specific regions of Lef-1 that interact with Dlx2, GST-pull-down experiments were performed using immobilized Lef-1 deletion constructs and purified Dlx2 protein. The Lef-1 constructs are identical to those used in the transfection experiments (Figure 8A). We have previously shown that the naturally occurring smaller Lef-1 protein, Lef-1 ΔN113 is sufficient for binding Dlx2 (Figure 6). Interestingly, Dlx2 can directly interact with the HMG domain and the C-terminal portion of the HMG domain, GST-Lef-1 ΔN295 and GST-Lef-1 ΔN363, respectively (Figure 9B). There is no reduction in binding compared to the GST-Lef-1 ΔN113 construct of these two N-terminal deletion mutants (Figure 9B). We were surprised to observe Dlx2 binding to Lef-1 ΔN363, as this is only a 34 amino acid peptide; however Dlx2 binding to this construct was observed in repeated experiments and at similar levels compared to wild type Lef-1. We term this C-terminal Dlx2 interaction region as Dlx2 binding domain (BD) #2. Deletion of the Lef-1 C-terminal region, which lacks the 34 amino acid sequence (GST-Lef-1 ΔC34) and GST-Lef-1 ΔC102, which lacks most of the HMG domain, bind Dlx2 at levels similar to wild type Lef-1 ΔN113 (Figure 9C). These data suggest that Dlx2 can bind to the CAD domain and/or the 70 residues that flank the CAD domain, termed Dlx2 BD #1. GST-beads do not bind Dlx2 as a control. Thus, Dlx2 may bind to several regions on Lef-1 or contact several residues of each region simultaneously. Further experiments will determine the exact binding sites and residues involved in the binding of each protein.
Figure 9.
Dlx2 binds to two regions of the Lef-1 protein. (A) Schematic of the Lef-1 deletion proteins used in the GST-pull-down assays. The location of the two Dlx2 interaction regions of Lef-1 are shown and bracketed by lines and designated Dlx2 binding domain (BD) #1 and 2. (B) GST-Lef-1 and truncated Lef-1 protein pull-down assay with bacterial expressed and purified Dlx2 protein (150 ng). To demonstrate Dlx2 binding to Lef-1, independent of the βBD, the Lef-1 short isoform (GST-Lef-1ΔN113) was incubated with pure Dlx2 protein. Dlx2 binds to the Lef-1 short isoform as well as the last 34 residues of the Lef-1 C-terminal tail (GST-Lef-1ΔN363). This region is termed Dlx2 BD #2. (C) Dlx2 binding to the Lef-1 C-terminal truncation mutants. Dlx2 binds to a region containing the CAD and 3′ flanking residues and termed Dlx2 BD #1. As a control GST-beads were incubated with purified Dlx2 to demonstrate the specificity of Dlx2 binding to the GST-Lef-1 fusion proteins.
Msx2 auto-regulation and attenuation of Dlx2 activation
Because the DNA-binding elements of Dlx2 and Msx2 are similar, we asked if Msx2 could regulate its promoter. Transfection of Msx2 in CHO cells resulted in a 3- to 4-fold repression of both the Msx2-5820 and Msx2-872 promoters (Figure 10A). These results demonstrate auto-regulation by this well-known repressor protein. Msx2 and Dlx2 directly interact to regulate their activities through protein interactions (18). However, they have not been shown to functionally regulate the Msx2 promoter. Because Msx2 and Dlx2 are co-expressed in similar tissues during development as well as in the CHO cells, can they antagonize each other? Equal expression of both factors revealed a decrease in Dlx2 activation of the full-length Msx2 promoter from 20- to 5-fold (Figure 10A). Repression of Dlx2 activation was also observed with the Msx2-872 promoter (Figure 10A). To determine if these activities were cell dependent, transfections were performed in C3H10T1/2 cells. No major differences were observed between these two different cell lines albeit a modest decrease in overall activation in the C3H10T1/2 cell line compared to the CHO cells (Figure 10B).
Figure 10.
Msx2 auto-regulation and repression of Dlx2 activation. (A) CHO cells were transfected with either the Msx2-5820 luciferase reporter gene or the Msx2-872 luciferase reporter gene. The cells were co-transfected with CMV-Dlx2 and/or CMV-Msx2 or the CMV empty vector. The concentrations of the reporter plasmids were 5 μg and 2.5 μg for the expression plasmids. (B) C3H10T1/2 cells were transfected as in (A) to determine if the activation was cell dependent. All transfection assays were performed as described in Figure 5. The activities are shown relative to Msx2 promoters without Dlx2 and Msx2 expression (+/− SEM from five independent experiments).
Msx2 repression of Dlx2 activation is dose-responsive and independent of Lef-1. Msx2 plasmid was titrated in CHO cells transfected with 2.5 μg of Dlx2 and Lef-1 FL or Lef-1 ΔN113 (Figure 11A). The synergistic activation of the Msx2 promoter by Dlx2 and Lef-1 FL was decreased from ∼38-fold without Msx2 to 28-fold (1.25 μg Msx2), 17-fold (2.5 μg Msx2) and 9-fold (3.75 μg Msx2). Interestingly, Msx2 repression of the synergistic activation by Dlx2 and the shorter Lef-1 isoform (Lef-1 ΔN113) was increased compared to Lef-1 FL (Figure 11A). Dlx2 and Lef-1 ΔN113 synergistically activate the Msx2 promoter at ∼38-fold; addition of 1.25 μg Msx2 repressed the activation to 13-fold, 2.5 μg Msx2 to 6-fold and 3.75 μg Msx2 to 5-fold in CHO cells (Figure 11A. The same experiment was repeated in the LS-8 tooth epithelial cell line. These cells demonstrate the synergistic activation of the Msx2 promoter by Dlx2 and Lef-1 FL or Lef-1 ΔN113 (Figure 11B). Msx2 represses the Msx2 promoter ∼3-fold as shown in CHO cells. Furthermore, a similar decrease in synergistic activation by Dlx2 and Lef-1 in LS-8 cells was observed by Msx2 titration as shown in CHO cells (Figure 11B). Together these data demonstrate that Msx2 can negatively auto-regulate its promoter and repress Dlx2 activation through direct binding to a common site or a physical interaction between the two proteins.
Figure 11.
Msx2 expression attenuates Dlx2 and Lef-1 synergistic activation of the Msx2 promoter. (A) CHO cells were co-transfected with the Msx2-5820 luciferase reporter gene, CMV Dlx2, CMV Lef-1 FL or CMV Lef-1 ΔN113. The CMV Msx2 expression plasmid was titrated from 1.25 to 3.75 μg in the indicated experiments. The concentration of the reporter plasmid was 5 μg and Dlx2 and Lef-1 expression plasmids were 2.5 μg. (B) LS-8 cells were transfected as in (A) to demonstrate the activation of the Msx2 promoter in these tooth epithelial cells. All transfection assays were performed as described in Figure 5. The activities are shown relative to Msx2 promoters without Dlx2, Lef-1 and Msx2 expression (+/− SEM from four independent experiments).
DISCUSSION
Several factors and molecules regulating Msx2 expression have been identified including signaling molecules, hormones and transcription factors. The FGF and BMP signaling molecules are involved in the differential regulation of Msx genes (42–45). BMP-dependent activation of Msx2 can be mediated through the cooperative binding of Smad4 and Lef-1; Lef-1 can synergize with Smad4 and Smad1 to activate the Msx2 promoter (46). Furthermore, Lef-1 synergizes with BMP2 to activate Msx2 expression. In contrast, YY1, a zinc finger transcription factor, can activate Msx2 independent of BMP signaling (47). Interestingly, glucocorticoid treatment can increase Msx2 transcripts and protein both in vitro and in vivo in embryonic mouse submandibular glands (48). Pax3 has been shown to bind to the Msx2 promoter and represses Msx2 expression in the murine cardiac neural crest (49). Genetic analysis suggests that Fox1 is required for BMP-dependent activation of Msx2 in calvarial tissues of mouse embryos (50). Recently, Dlx5 and Sox11 both expressed in the AER, have been identified as possible regulators of Msx2 gene expression (17). While BMP signaling plays a major role in Msx2 expression patterns during tooth development, little is known about the role and mechanisms of transcription factors regulating Msx2 expression in the developing tooth.
Dlx2 regulates the Msx2 promoter
ChIP assays identified Dlx2 binding to the Msx2 promoter in vivo and co-transfection of Dlx2 with the Msx2 promoter reveals a direct activation by Dlx2. This transcriptional mechanism coincides with the distinct expression pattern of these two transcription factors during tooth development. Dlx2 is expressed initially in both the dental mesenchyme and epithelium and high expression is observed in the epithelial tissue at later stages. Msx2 expression occurs ∼1 day after Dlx2 expression in the epithelium, which would correspond to its potential activation by Dlx2 in this tissue. There is also overlapping expression patterns of Msx2 and Dlx2 in the first two branchial arches and the AER where Msx2 and Dlx2 overlap in the ectoderm (18).
Dlx2 binds to the Msx2 promoter as a monomer and possible homodimer as shown by the EMSA and correlates with the homodimerization revealed in previous GST-pull-down experiments (18). Furthermore, previous studies have shown Dlx2 binding to the TAATTG sequence as multiple bands suggesting the presence of dimers (22). There are approximately 12 Dlx2 binding sites in the Msx2 promoter including the proximal site in which the ChIP assay revealed Dlx2 binding was a non-consensus 5′-TAGTTG-3′ compared to the consensus 5′-TAATTG-3′ element. A previous report using ChIP analyses has shown that the 5′-TAATTA-3′ sequence was required for Dlx2 DNA-binding and activation in the Dlx5/Dlx6 intergenic enhancer (23,51). A consensus site for Xenopus Dlx3 was identified as (A/C/G)TAATT(G/A)(C/G) (52). The core is a TAAT motif, which is observed in many homeodomain DNA-binding proteins (53). However, the 3′ dinucleotide confers specificity to the binding of these factors. Our data demonstrate that perturbations in the core TAAT can be recognized by the Dlx2 homeodomain protein and activate the Msx2 promoter. However, there is a TAATCA sequence overlapping the TAGTTG element in the anti-sense strand. It may be possible for Dlx2 to bind to this region. Similarly, we have previously reported on the promiscuity of the Msx2 protein binding to non-consensus elements and regulating promoter activity (37). Our results further demonstrate the ability of Msx2 to bind to the non-consensus TAGTTG element recognized by Dlx2 in the Msx2 promoter.
Dlx2 directly interacts with Lef-1 to regulate Msx2
The Lef-1 FL construct minimally activates the Msx2 promoter at ∼6-fold and the alternative Lef-1 transcript, Lef-1 ΔN113 cannot activate the Msx2 promoter, however both can complex with Dlx2 to synergistically activate Msx2.
We specifically utilized the smaller native Lef-1 transcript (Lef-1 ΔN113) to dissociate the effects of β-catenin on Lef-1 interactions with Dlx2. Lef-1 transcripts can arise through alternative splicing and different promoters (54–56). A recent report describes the use of an internal ribosome entry site that mediates the translation of a full-length Lef-1 transcript. The full-length Lef-1 (Lef-1 FL) transcript contains a large 5′UTR synthesized using a separate PI promoter and utilizes a cap-independent mechanism for translation of the full-length Lef-1, containing the βBD (57). This isoform has been termed growth promoting because it is expressed during cell growth in undifferentiated, mitotically active cells. This Lef-1 activity is countered by the expression of a truncated Lef-1 protein that lacks the βBD and can compete for binding to Wnt target genes. The shorter Lef-1 isoform is produced from a second P2 promoter located in the second intron of the LEF-1 locus (54,57). This shorter Lef-1 transcript (Lef-1 ΔN113) could act as an inhibitory isoform due to its ability to interact with co-factors interacting with Lef-1 FL as well as compete for DNA-binding sites and thus been termed growth suppressing. Interestingly, in colon cancer cells only the LEF-1 FL transcript is produced and the loss of balance between the long and short forms may effect cancer progression (57).
It is well known that Lef-1 requires other factors to become transcriptional active. In response to Wnt signals, β-catenin is stabilized and interacts with the N-terminus of Lef-1 to activate transcription of Wnt-responsive genes (58–60). A previous report has demonstrated that Lef-1 can synergize with Smad4 to activate the Msx2 promoter (46). This activation was independent of BMP signaling and was in response to β-catenin activation. Recently, we demonstrated a role for Lef-1, β-catenin and PITX2 in regulating the LEF-1 promoter suggesting a role for β-catenin in regulating Dlx2 and Lef-1 regulation of the Msx2 promoter. Without Wnt signaling, Lef-1 can interact with the co-repressor Groucho to repress Wnt-responsive genes (61,62). We have previously identified a Lef-1 interaction with the homeodomain transcription factor PITX2 to synergistically regulate the LEF-1 promoter (39). In this report we demonstrate that both the Lef-1 Fl and Lef-1 ΔN113 proteins can interact with Dlx2 and that the βBD has no effect on this interaction. Thus, Dlx2 interaction with Lef-1 and synergistic activation of the Msx2 promoter is not limited to a specific Lef-1 isoform during development.
Lef-1 interaction domains
Several domains of Lef-1 have been identified that interact with other factors. The Lef-1 amimo terminus contains the β-catenin interaction domain (βBD) involved in Wnt signaling (63). The CAD (33,34) and a region flanking the CAD are required for association with the Groucho corepressor and HDAC (64). The C-terminal HMG domain is involved in DNA-binding (65) and two regions within the HMG domain mediate the interaction with Smad factors (66). Two regions of Lef-1 were found to interact with Smad3 in the HMG domain. One site mapped to residues 324–334 while another site was localized to a lysine- and arginine-rich region between residues 370 and 383 (66). These two regions bind to the MH2 and MH1 domains of Smad3, respectively. Our data suggests two regions of Lef-1 interact with Dlx2; the C-terminal region of the HMG domain containing 30 amino acids (Lef-1 ΔN363), which corresponds to the Smad3 MH1 binding domain and a second region containing the CAD and flanking sequences (Lef-1 ΔN113-ΔC102). These regions may be involved in the interactions with other factors, such as Smad3. Thus Dlx2 binding to Lef-1 could have an impact on the overall activity of Lef-1 protein. Competition between factors for these sites, could possibly add to the complexity of Lef-1 transcriptional activity. We are currently investigating the role of β-catenin in regulating the Dlx2/Lef-1 transcription complex by fine mapping the interaction sites.
An Msx2 feedback regulatory mechanism
Msx2 is a homeodomain transcription factor required for the development of several tissues and processes (15). Msx2 is a transcriptional repressor and can interact with other factors to regulate transcription (18,67). These effects can occur through their interactions with other factors without binding to DNA (18,67–72).
Our data has shown that Msx2 can negatively regulate its promoter. We have previously demonstrated Msx2 binding to the TAATTG sequence with high affinity, which is shared by Dlx2. However, Msx2 binding appears to be somewhat promiscuous by binding to other homeodomain transcription factor elements (37). We have previously reported that Msx2 and PITX2 can bind to the Dlx2 promoter bicoid elements (consensus PITX2 binding elements) independent of one another and the levels of binding are based on the relative amount of each protein (37). In this report we demonstrate that Msx2 competes with Dlx2 for binding to the TAGTTG element in the Msx2 promoter. Msx2 bound to this element as three bands, which is similar to Msx2 binding to the consensus TAATTG element (37). Msx1 and Dlx2 demonstrate mutual exclusive DNA-binding and we show that Msx2 competes with Dlx2 for DNA-binding sites on the Msx2 promoter in a similar fashion (18). Furthermore, Msx2 physically interacts with Dlx2, which may inhibit Dlx2 transcriptional activity (18). Our data demonstrates a reduction in DNA-binding by both Dlx2 and Msx2 when incubated together in the EMSA experiment. This would be expected as both proteins can directly interact and inhibit the DNA-binding activity of the other protein (18). The activity of the Msx2 promoter would be differentially regulated as the level of these two proteins change during development. Such as during tooth development, the level of Msx2 expression is initially high immediately after Dlx2 and Lef-1 expression (cap stage), restricted during the tooth bell stage, expressed at later stages and during amelogenesis (8,16,73). Moreover, Msx and Dlx proteins have been shown to bind identical elements on the GnRH promoter resulting in functional antagonism (74); Msx2 and Dlx5 on the osteocalcin promoter (75); and Pax3 and Msx1 on the MyoD promoter (76).
Based on the functional interaction between Dlx2 and Lef-1, the identification that Dlx2 binds the Msx2 promoter in vivo and the transcriptional activation of the Msx2 promoter in three cell types, we propose a model for the regulation of Msx2 by these factors (Figure 12). Furthermore, we have demonstrated that Msx2 can negatively regulate its promoter and attenuate the activation by Dlx2. Thus, Dlx2 and Lef-1 interact to synergistically activate Msx2; Msx2 can attenuate Dlx2 activation or directly repress Msx2 promoter activity. These interactions would regulate the spatial and temporal expression pattern of Msx2 observed during development.
Figure 12.
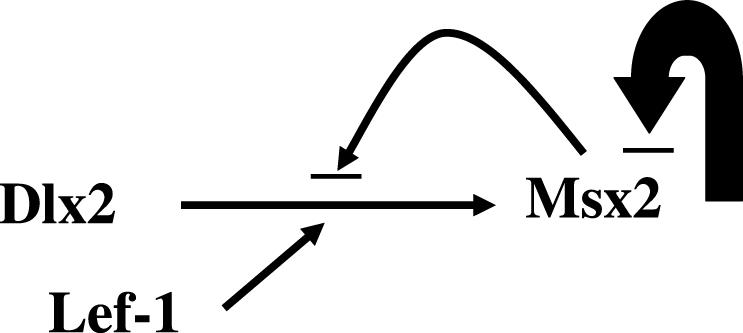
Model for the regulation of Msx2 expression by Dlx2, Lef-1 and Msx2. Dlx2 activates the Msx2 promoter and can synergistically activate Msx2 in concert with Lef-1. Msx2 can repress Dlx2 activation through competition for shared DNA-binding sites on the Msx2 promoter or direct protein interactions. Msx2 can negatively auto-regulate its promoter, shown by the large arrow.
Acknowledgments
We thank Drs Tord A. Hjalt, YiPing Chen and John L.R. Rubenstein for reagents and helpful discussions. Support for this research was provided from grant DE13941 from the National Institute of Dental and Craniofacial Research and ES09106 from the National Institute of Environmental Health Sciences to Brad A. Amendt. Funding to pay the Open Access publication charges for this article was provided by grant DE13941 from NIDCR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hill R.E., Jones P.F., Rees A.R., Sime C.M., Justice M.J., Copeland N.G., Jenkins N.A., Graham E., Davidson D.R. A new family of mouse homeo box-containing genes: molecular structure, chromosomal location, and developmental expression of Hox-7.1. Genes Dev. 1989;3:26–37. doi: 10.1101/gad.3.1.26. [DOI] [PubMed] [Google Scholar]
- 2.Robert B., Sassoon D., Jacq B., Gehring W., Buckingham M. Hox-7, a mouse homeobox gene with a novel pattern of expression during embryogenesis. EMBO J. 1989;8:91–100. doi: 10.1002/j.1460-2075.1989.tb03352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davidson D.R., Hill R.E. Msh-like genes: a family of homeo-box genes with wide-ranging expression during vertebrate development. Sem Dev. Biol. 1991;2:405–412. [Google Scholar]
- 4.Grindley J.C., Davidson D.R., Hill R.E. The role of Pax-6 in eye and nasal development. Development. 1995;121:1433–1442. doi: 10.1242/dev.121.5.1433. [DOI] [PubMed] [Google Scholar]
- 5.Shimeld S.M., McKay I.J., Sharpe P.T. The murine homeobox gene Msx-3 shows highly restricted expression in the developing neural tube. Mech. Dev. 1996;55:201–210. doi: 10.1016/0925-4773(96)00505-9. [DOI] [PubMed] [Google Scholar]
- 6.Wang W., Chen X., Xu H., Lufkin T. Msx3: a novel murine homologue of the Drosophila msh homeobox gene restricted to the dorsal embryonic central nervous system. Mech. Dev. 1996;58:203–215. doi: 10.1016/s0925-4773(96)00562-x. [DOI] [PubMed] [Google Scholar]
- 7.MacKenzie A., Ferguson M.W., Sharpe P.T. Hox-7 expression during murine craniofacial development. Development. 1991;113:601–611. doi: 10.1242/dev.113.2.601. [DOI] [PubMed] [Google Scholar]
- 8.MacKenzie A., Ferguson M.J.W., Sharpe P.T. Expression patterns of the homeobox gene, Hox-8, in the mouse embryo suggest a role in specifying tooth initiation and shape. Development. 1992;115:403–420. doi: 10.1242/dev.115.2.403. [DOI] [PubMed] [Google Scholar]
- 9.Mina M., Gluhak J., Upholt W.B., Koller E.J., Rogers B. Experimental analysis of Msx-1 and Msx-2 gene expression during chick mandibular morphogenesis. Dev. Dyn. 1995;202:195–214. doi: 10.1002/aja.1002020211. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y.H., Ma L., Wu L.Y., Luo W., Kundu R., Sangiorgi F., Snead M.L., Maxson R. Regulation of the Msx2 homeobox gene during mouse embryogenesis: a transgene with 439 bp of 5′ flanking sequence is expressed exclusively in the apical ectodermal ridge of the developing limb. Mech. Dev. 1994;48:187–197. doi: 10.1016/0925-4773(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 11.Satokata I., Ma L., Ohshima H., Bei M., Woo I., Nishizawa K., Maeda T., Takano Y., Uchiyama M., Heaney S., et al. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat. Genet. 2000;24:391–395. doi: 10.1038/74231. [DOI] [PubMed] [Google Scholar]
- 12.Mucchielli M.-L., Mitsiadis T.A., Raffo S., Brunet J.-F., Proust J.-P., Goridis C. Mouse Otlx2/RIEG expression in the odontogenic epithelium precedes tooth initiation and requires mesenchyme-derived signals for its maintenance. Dev. Biol. 1997;189:275–284. doi: 10.1006/dbio.1997.8672. [DOI] [PubMed] [Google Scholar]
- 13.Kratochwil K., Dull M., Farinas I., Galceran J., Grosschedl R. Lef1 expression is activated by BMP-4 and regulates inductive tissue interactions in tooth and hair development. Genes Dev. 1996;10:1382–1394. doi: 10.1101/gad.10.11.1382. [DOI] [PubMed] [Google Scholar]
- 14.Thomas B.L., Porteus M.H., Rubenstein J.L., Sharpe P.T. The spatial localization of Dlx-2 during tooth development. Connect. Tissue Res. 1995;32:27–34. doi: 10.3109/03008209509013702. [DOI] [PubMed] [Google Scholar]
- 15.Bendall A.J., Abate-Shen C. Roles for Msx and Dlx homeo-proteins in vertebrate development. Gene. 2000;247:17–31. doi: 10.1016/s0378-1119(00)00081-0. [DOI] [PubMed] [Google Scholar]
- 16.Bei M., Stowell S., Maas R. Msx2 controls ameloblast terminal differentiation. Dev. Dyn. 2004;231:758–765. doi: 10.1002/dvdy.20182. [DOI] [PubMed] [Google Scholar]
- 17.Cheng H.-C., Wang C.-K.L., Upholt W.B. Transcriptional regulation of Msx2 in the AERs of developing limbs is dependent on multiple closely spaced regulatory elements. Dev. biol. 2004;270:513–524. doi: 10.1016/j.ydbio.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H., Hu G., Wang H., Sciavolino P., Iler N., Shen M.M., Abate-Shen C. Heterodimerization of Msx and Dlx homeoproteins results in functional antagonism. Mol. Cell. Biol. 1997;17:2920–2932. doi: 10.1128/mcb.17.5.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Capdevila J., Izpisula Belmonte J.C. Patterning mechanisms controlling vertebrate limb development. Annu. Rev. Cell Dev. Biol. 2001;17:87–132. doi: 10.1146/annurev.cellbio.17.1.87. [DOI] [PubMed] [Google Scholar]
- 20.Qiu M., Bulfone A., Martinez S., Meneses J.J., Shimamura K., Pedersen R.A., Rubenstein J.L.R. Null mutation of Dlx-2 results in abnormal morphogenesis of proximal first and second branchial arch derivatives and abnormal differentiation in the forebrain. Genes Dev. 1995;9:2523–2538. doi: 10.1101/gad.9.20.2523. [DOI] [PubMed] [Google Scholar]
- 21.Thomas B.L., Liu J.K., Rubenstein J.L.R., Sharpe P.T. Independent regulation of Dlx2 expression in the epithelium and mesenchyme of the first branchial arch. Development. 2000;127:217–224. doi: 10.1242/dev.127.2.217. [DOI] [PubMed] [Google Scholar]
- 22.Liu J.K., Ghattas I., Liu S., Chen S., Rubenstein J.L.R. Dlx genes encode DNA-binding proteins that are expressed in an overlapping and sequential pattern during basal ganglia differentiation. Dev. Dyn. 1997;210:498–512. doi: 10.1002/(SICI)1097-0177(199712)210:4<498::AID-AJA12>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Q.-P., Ngoc Le T., Qiu X., Spencer V., de Melo J., Du G., Plews M., Fonseca M., Sun J., Davie J.R., et al. Identification of a direct Dlx homeodomain target in the developing mouse forebrain and retina by optimization of chromatin immunoprecipitation. Nucleic Acids Res. 2004;32:884–892. doi: 10.1093/nar/gkh233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stuhmer T., Anderson S.A., Ekker M., Rubenstein J.L.R. Ectopic expression of the Dlx genes induces glutamic acid decarboxylase and Dlx expression. Development. 2002;129:245–252. doi: 10.1242/dev.129.1.245. [DOI] [PubMed] [Google Scholar]
- 25.Cobos I., Broccoli V., Rubenstein J.L.R. The vertebrate ortholog of aristaless is regulated by Dlx genes in the developing forebrain. J. Comp. Neuro. 2005;483:292–303. doi: 10.1002/cne.20405. [DOI] [PubMed] [Google Scholar]
- 26.Qiu M., Bulfone A., Ghattas I., Meneses J.J., Christensen L., Sharpe P.T., Presley R., Pedersen R.A., Rubenstein J.L.R. Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev. Biol. 1997;185:165–184. doi: 10.1006/dbio.1997.8556. [DOI] [PubMed] [Google Scholar]
- 27.Thomas B.L., Tucker A.S., Qiu M., Ferguson C.A., Hardcastle Z., Rubenstein J.L.R., Sharpe P.T. Role of Dlx-1 and Dlx-2 genes in patterning of the murine dentition. Development. 1997;124:4811–4818. doi: 10.1242/dev.124.23.4811. [DOI] [PubMed] [Google Scholar]
- 28.Travis A., Amsterdam A., Belanger C., Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor α enhancer function. Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 29.Waterman M.L., Fischer W.H., Jones K.A. A thymus-specific member of the HMG protein family regulates the human T-cell receptor Cα enhancer. Genes Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- 30.Oosterwegel M., van de Wetering M., Timmerman J., Kruisbeek A., Destree O., Meijlink F., Clevers H. Differential expression of the HMG boxfactors TCF-1 and LEF-1 during murine embryogenesis. Development. 1993;118:439–448. doi: 10.1242/dev.118.2.439. [DOI] [PubMed] [Google Scholar]
- 31.van Genderen C., Okamura R.M., Farinas I., Quo R.-G., Parslow T.G., Bruhn L., Grosschedl R. Development of several organs that require inductive epithelial-mesenchymal interactions is impared in LEF-1-deficient mice. Genes Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- 32.Zhou P., Byrne C., Jacobs J., Fuchs E. Lymphoid enhancer factor 1 directs hair follicle patterning and epithelial cell fate. Genes Dev. 1995;9:700–713. doi: 10.1101/gad.9.6.700. [DOI] [PubMed] [Google Scholar]
- 33.Carlsson P., Waterman M.L., Jones K.A. The hLEF/TCF-1α HMG protein contains a context-dependent transcriptional activation domain that induces the TCRα enhancer in T cells. Genes Dev. 1993;7:2418–2430. doi: 10.1101/gad.7.12a.2418. [DOI] [PubMed] [Google Scholar]
- 34.Giese K., Grosschedl R. LEF-1 contains an activation domain that stimulates transcription only in a specific context of factor-binding sites. EMBO J. 1993;12:4667–4676. doi: 10.1002/j.1460-2075.1993.tb06155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giese K., Kingsley C., Kirshner J.R., Grosschedl R. Assembly and function of a TCRα enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 36.Galceran J., Farinas I, Depew MJ, Clevers H, R G. Wnt3a-/--like phenotype and limb deficiency in Lef1(−/−)Tcf1(−/−) mice. Genes Dev. 1999;13:709–717. doi: 10.1101/gad.13.6.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Green P.D., Hjalt T.A., Kirk D.E., Sutherland L.B., Thomas B.L., Sharpe P.T., Snead M.L., Murray J.C., Russo A.F., Amendt B.A. Antagonistic regulation of Dlx2 expression by PITX2 and Msx2: implications for tooth development. Gene Expr. 2001;9:265–281. doi: 10.3727/000000001783992515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amendt B.A., Sutherland L.B., Russo A.F. Multifunctional role of the Pitx2 homeodomain protein C-Terminal tail. Mol. Cell. Biol. 1999b;19:7001–7010. doi: 10.1128/mcb.19.10.7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vadlamudi U., Espinoza H.M., Ganga M., Martin D.M., Liu X., Engelhardt J.F., Amendt B.A. PITX2, β-catenin, and LEF-1 interact to synergistically regulate the LEF-1 promoter. J. Cell Sci. 2005;118:1129–1137. doi: 10.1242/jcs.01706. [DOI] [PubMed] [Google Scholar]
- 40.Amendt B.A., Sutherland L.B., Semina E., Russo A.F. The molecular basis of rieger syndrome: analysis of Pitx2 homeodomain protein activities. J. Biol. Chem. 1998;273:20066–20072. doi: 10.1074/jbc.273.32.20066. [DOI] [PubMed] [Google Scholar]
- 41.Cox C.J., Espinoza H.M., McWilliams B., Chappell K., Morton L., Hjalt T.A., Semina E.V., Amendt B.A. Differential regulation of gene expression by PITX2 isoforms. J. Biol. Chem. 2002;277:25001–25010. doi: 10.1074/jbc.M201737200. [DOI] [PubMed] [Google Scholar]
- 42.Vainio S., Karavanova I., Jowett A., Thesleff I. Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell. 1993a;75:45–58. [PubMed] [Google Scholar]
- 43.Jowett A.K., Vainio S., Ferguson M.W., Sharpe P.T., Thesleff I. Epithelial-mesenchyme interactions are required for msx1 and msx2 gene expression in the developing murine molar tooth. Development. 1993b;117:461–470. doi: 10.1242/dev.117.2.461. [DOI] [PubMed] [Google Scholar]
- 44.Tucker A.S., Al Khamis A., Sharpe P.T. Interactions between Bmp-4 and Msx-1 act to restrict gene expression to odontogenic mesenchyme. Dev. Dyn. 1998;212:533–539. doi: 10.1002/(SICI)1097-0177(199808)212:4<533::AID-AJA6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 45.Bei M., Maas R. FGFs and BMP4 induce Msx1-dependent and Msx1-independent signaling pathways in early tooth development. Development. 1998;125:4316–4327. doi: 10.1242/dev.125.21.4325. [DOI] [PubMed] [Google Scholar]
- 46.Hussein S.M., Duff E.K., Sirard C. Smad4 and β-catenin co-activators functionally interact with lymphoid-enhancing factor to regulate graded expression of Msx2. J. Biol. Chem. 2003;278:48805–48814. doi: 10.1074/jbc.M305472200. [DOI] [PubMed] [Google Scholar]
- 47.Tan D.P., Nonaka K., Nuckolls G.H., Liu Y.H., Maxson R.E., Slavkin H.C., Shum L. YY1 activates Msx2 gene independent of bone morphogenetic protein signaling. Nucleic Acids Res. 2002;30:1213–1223. doi: 10.1093/nar/30.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaskoll T., Luo W., Snead M.L. Msx-2 expression and glucocorticoid-induced overexpression in embryonic mouse submandibular glands. J. Craniofac. Genet. Dev. Biol. 1998;18:79–87. [PubMed] [Google Scholar]
- 49.Kwang S.J., Brugger S.M., Lazik A., Merrill A.E., Wu L.-Y., Liu Y-H., Ishil M., Sangiorgi F.O., Rauchman M., Sucov H.M., et al. Msx2 is an immediate downstream effector of Pax3 in the development of the murine cardiac neural crest. Development. 2002;129:527–538. doi: 10.1242/dev.129.2.527. [DOI] [PubMed] [Google Scholar]
- 50.Ishii M., Merrill A.E., Chan Y.-S., Gitelman I., Rice D.P.C., Sucov H.M., Maxson R.E., Jr Msx2 and Twist cooperatively control the development of the neural crest-derived skeletogenic mesenchyme of the murine skull vault. Development. 2003;130:6131–6142. doi: 10.1242/dev.00793. [DOI] [PubMed] [Google Scholar]
- 51.Zerucha T., Stuhmer T., Hatch G., Park B.K., Long Q., Yu G., Gambarotta A., Schultz J.R., Rubenstein J.L.R., Ekker M. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J. Neurosci. 2000;20:709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feledy J.A., Morasso M.I., Jang S.-I., Sargent T.D. Transcriptional activation by the homeodomain protein distal-less 3. Nucleic Acid Res. 1999;27:764–770. doi: 10.1093/nar/27.3.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duboule D. Guidebook to the Homeobox Genes. New York: Oxford University Press Inc.; 1994. [Google Scholar]
- 54.Hovanes K., Li T.W.H., Waterman M.L. The human LEF-1 gene contains a promoter preferentially active in lymphocytes and encodes multiple isoforms derived from alternative splicing. Nucleic Acids Res. 2000;28:1994–2003. doi: 10.1093/nar/28.9.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hovanes K., Li T.W., Munguia J.E., Truong T., Milovanovic T., Lawrence Marsh J., Holcombe R.F., Waterman M.L. Beta-catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat. Genet. 2001;28:53–57. doi: 10.1038/ng0501-53. [DOI] [PubMed] [Google Scholar]
- 56.Cordray P., Satterwhite D.J. TGF-b induces novel Lef-1 splice variants through a SMAD-independent signaling pathway. Dev. Dyn. 2005;232:969–978. doi: 10.1002/dvdy.20275. [DOI] [PubMed] [Google Scholar]
- 57.Jimenez J., Jang G.M., Semler B.L., Waterman M.L. An internal ribosome entry site mediates translation of lymphoid enhancer factor-1. RNA. 2005;11:1385–1399. doi: 10.1261/rna.7226105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cadigan K.M., Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 59.van de Wetering M., Cavallo R., Dooijes D., van Beest M., van Es J., Loureiro J., Ypma A., Hursh D., Jones T., Bejsovec A., et al. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 60.Hsu S., Galceran J., Grosschedl R. modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol. Cell. Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Levanon D., Goldstein R.E., Bernstein Y., Tang H., Goldenberg D., Stifani S., Paroush Z., Groner Y. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl Acad. Sci. USA. 1998;95:11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roose J., Molenaar M., Peterson J., Hurenkamp J., Brantjes H., Moerer P., Van de Wetering M., Destree O., Clevers H. The xenopus Wnt effector XTcf-3 interacts with groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 63.Behrens J., von Kries J.P., Kuhl M., Bruhn L., Wedlich D., Grosschedl R., Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 64.Billin A.N., Thirlwell H., Ayer D.E. Beta-catenin-histone deacetylase interactions regulate the transition of LEF-1 from a transcriptional repressor to an activator. Mol. Cell. Biol. 2000;20:6882–6890. doi: 10.1128/mcb.20.18.6882-6890.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Giese K., Cox J., Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992;69:185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- 66.Labbe E., Letamendia A., Attisano L. Association of Smads with lymphoid enhancer binding factor 1/T cell-specifc factor mediates cooperative signaling by the transforming growth factor-b and Wnt pathways. Proc. Natl Acad. Sci. 2000;97:8358–8363. doi: 10.1073/pnas.150152697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Newberry E.P., Latifi T., Battaile J.T., Towler D.A. Structure-function analysis of Msx2-mediated transcriptional suppression. Biochem. 1997;36:10451–10462. doi: 10.1021/bi971008x. [DOI] [PubMed] [Google Scholar]
- 68.Catron K.M., Iler N., Abate C. Nucleotides flanking a conserved TAAT core dictate the DNA binding specificity of three murine homeodomain proteins. Mol. Cell. Biol. 1993;13:2354–2365. doi: 10.1128/mcb.13.4.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Catron K.M., Zhang H., Marshall S.C., Inostroza J.A., Wilson J.M., Abate C. Transcriptional repression by Msx2 does not require homeodomain DNA-binding sites. Mol. Cell. Biol. 1995;15:861–871. doi: 10.1128/mcb.15.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Semenza G.L., Wang G.L., Kundu R. DNA binding and transcriptional properties of wild-type and mutant forms of the homeodomain protein MSX2. Biochem. Biophy. Res. Comm. 1995;209:257–262. doi: 10.1006/bbrc.1995.1497. [DOI] [PubMed] [Google Scholar]
- 71.Zhang H., Catron K.M., Abate-Shen C. A role for the Msx-1 homeodomain in transcriptional regulation: residues in the N-terminal arm mediate TATA binding protein interaction and transcriptional repression. Proc. Natl Acad. Sci. 1996;93:1764–1769. doi: 10.1073/pnas.93.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bendall A.J., Rincon-Limas D.E., Botas J., Abate-Shen C. Protein complex formation between Msx1 and Lhx2 homeoproteins is incompatible with DNA binding activity. Differentiation. 1998;63:151–157. doi: 10.1046/j.1432-0436.1998.6330151.x. [DOI] [PubMed] [Google Scholar]
- 73.Alappat S., Zhang Z.Y., Chen Y.-P. Msx homeobox gene family and craniofacial development. Cell Res. 2003;13:429–442. doi: 10.1038/sj.cr.7290185. [DOI] [PubMed] [Google Scholar]
- 74.Givens M.L., Rave-Harel N., Goonewardena V.D., Kurotani R., Berdy S.E., Swan C.H., Rubenstein J.L.R., Robert B., Mellon P.L. Developmental regulation of gonadotropin-releasing hormone gene expression by the MSX and DLX homeodomain protein families. J. Biol. Chem. 2005;280:19156–19165. doi: 10.1074/jbc.M502004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Newberry E.P., Latifi T., Towler D.A. reciprocal regulation of osteocalcin transcription by the homeodomain proteins Msx2 and Dlx5. Biochem. 1998;37:16360–16368. doi: 10.1021/bi981878u. [DOI] [PubMed] [Google Scholar]
- 76.Bendall A.J., Ding J., Hu G., Shen M.M., Abate-Shen C. Msx1 antagonizes the myogenic activity of Pax3 in migrating limb muscle precursors. Development. 1999;126:4965–4976. doi: 10.1242/dev.126.22.4965. [DOI] [PubMed] [Google Scholar]