Abstract
Progesterone receptor (PR) plays a critical role in cell proliferation and differentiation, and its transcriptional activity is known to be modulated by cofactor proteins. In the present study, we demonstrated that in the presence of progesterone, protein inhibitor of activated STAT-3 (PIAS3) significantly inhibited the PR transcriptional activity and the expression of progesterone-responsive genes. Reduction of endogenous PIAS3 by PIAS3 small-interfering RNA enhanced PR transactivation in a ligand-dependent manner. PIAS3 interacted with PR both in vitro and in vivo and the interaction was enhanced by progesterone. Furthermore, our findings suggested that PIAS3 strongly induced PRB sumoylation at three sites, Lys-7, Lys-388 and Lys-531. In addition, novel roles in PRB nuclear retention and transactivation were identified for these sites. Our data also suggested that PIAS3 was recruited in a largely hormone-dependent manner in response to a progesterone-responsive promoter. Finally, we demonstrated that PIAS3 inhibited the DNA-binding activity of PR and influenced its nuclear export as well as PR transactivation. Taken together, these data strongly suggested that PIAS3 played an important physiological role in PR function.
INTRODUCTION
Human progesterone receptor (PR) is a member of the nuclear receptor superfamily of ligand-dependent transcription factors and plays an important role in tissue development, reproduction and homeostasis (1). Similar to other steroid receptors, PR also contains a highly conserved DNA-binding domain (DBD) located in the center and a C-terminal hormone-binding domain (HBD). In addition, PR contains a ligand-independent activation function (AF) domain, AF1, located in the N-terminus upstream of the DBD and a hormone-dependent domain, AF2, in the C-terminal HBD. The inhibitory function (IF) domain is flanked at AF1 N-terminus, which auto-inhibits the function of PR (2,3). PRs are expressed in two isoforms, PRA and PRB. The two isoforms of PR are identical in sequence except that PRA is missing the far 164 residues N-terminal B-upstream segment (BUS) region. This segment is the third AF domain, AF3, which contributes to the different transcription activities of PRA and PRB (4–7). Consequently, PRB is in general a stronger transcription activator than PRA (8–12).
Similar to other steroid hormone receptors, PR is transcriptionally inactive and remains sequestered in a complex of heat-shock proteins in the absence of a ligand. Progestin binding to PR causes a conformational change and dimerization, resulting in the association of PR dimer with specific co-activators and general transcription factors. Ligand receptors then bind to DNA via specific progesterone response elements (PREs) located within the regulatory regions of target genes (13–17).
Apart from specific ligand regulation, different types of post-translational modifications, such as phosphorylation, acetylation and ubiquitination, also regulate the steroid hormone receptors transcriptional activation and/or stability (18–23). Recently, a new covalent modification of PR has been described: SUMO (small ubiquitin-like modifier) modification. SUMO modification is accomplished by the reversible attachment of SUMO family members to the acceptor lysine residues located in the target proteins, similar to ubiquitination, with the help of a set of enzymes. Even if mechanistically similar to ubiquitination, the two processes involve distinct enzymes and sumoylation does not promote protein degradation. Sumoylation appears to regulate diverse cellular processes, including specification of the subcellular localization of proteins, formation of subnuclear structures, interactions between proteins, stability of proteins and modulation of transcription factors (24–26).
Several studies have reported that SUMO-1 regulates the hormone-induced transactivation of PR, since its overexpression promotes sumoylation of PRB at the site of Lys-388, and very strongly enhances PR-mediated gene transcription. However, the mutation of sumoylation site in the K388R mutant also increased PR transactivation. Although it has been speculated that the enhancement of PR transactivation by SUMO-1 overexpression may be realized by the sumoylation of the coactivator SRC-1 (7,27), the molecular basis by which SUMO-1 regulates PR transcriptional capacity remains unknown and further studies are necessary to clarify the process.
Recently, a family of PIAS proteins was described as SUMO-E3 ligases for critical target proteins such as p53 (28,29), c-jun (29,30), LEF1 (31), androgen receptor (32–34) and estrogen receptor (35). However, it remains to be established whether PIAS3 was responsible for PR sumoylation. In the present study, we showed for the first time the pattern of PRB sumoylation at three sites was strongly induced by PIAS3. Overexpression of PIAS3 strongly inhibited the progesterone-triggered transactivation of PRB with different promoter or cells, and knockdown of endogenous PIAS3 with small-interfering RNA (siRNA) increased the PR transactivation. However, PIAS3 inhibited gene activation by ligand-stimulated PRB in a manner that was independent of PRB SUMO modification. Our results indicated that PIAS3 was recruited in a hormone-dependent manner to a progesterone-responsive promoter. Finally, we demonstrated that PIAS3 inhibited the DNA-binding activity of PR and influenced its nuclear export, which mechanistically resulted in PR transactivation.
MATERIALS AND METHODS
Plasmid constructions
PRA and PRB cDNA kindly provided by Professor O'Malley (Baylor College of Medicine) were cloned into pXJ40-Myc and pcDNA3.0-GAL4-DBD vectors. SUMO-1 and PIAS3 cDNA fragments generated by PCR were transferred into pXJ40-HA and pcDNA3.0-Flag vectors. HA-tagged PIAS3 C334S mutant was generated by PCR-mediated site-directed mutagenesis, where cysteine 334 was converted to serine. Myc-tagged PRB mutants (K388R, K7/531R and K7/388/531R) were generated by PCR-mediated site-directed mutagenesis, in them lysine 7, 388 and 531 were converted to arginine. For subcellular localization assays, the PIAS3 cDNAs were amplified and transferred into the pEGFP-N1 vector (Clontech Laboratories, Inc.). For Yeast two-hybrid assay, PRB and its mutants Δ456–556 (ΔAF1), Δ556–642 (ΔDBD), Δ456–642 (ΔAF1-DBD), 456–642 (AF1-DBD) were generated by PCR and cloned into pGADT7 vector, and PIAS3 cDNA was inserted into pGBKT7 vector (Clontech Laboratories, Inc.). To generate bacterial expression vector for GST–PIAS3, the corresponding PIAS3 cDNA was cloned in frame into pGEX-KG vector (Amersham Pharmacia Biotech). The Renilla vector (pRL-TK) was purchased from Promega and the luciferase reporter plasmid pMMTV-Luc was kindly provided by Professor Palvimo Jorma (University of Helsinki).
Immunofluorescence and fluorescence microscopy
For endogenous PIAS3 and PR location, T47D cells were seeded in 60 mm plates on to glass coverslips and cultured in medium containing phenol red-free DMEM (Hyclone) supplemented with 5% charcoal dextran-treated fetal bovine serum (BiochROM AG, Germany). The cells were treated, or not, with progesterone (100 nM) (Sigma) for 2 h before harvesting. Examining the effect of PIAS3 on PRB location, HeLa cells were treated as described above. Twenty-four hours after transfection using Lipofectamine 2000 (Invitrogen) with GFP–PIAS3 and Myc-PRB, the cells were treated with progesterone (100 nM) for various time period.
For immunofluorescence staining, the cells were fixed for 20 min at room temperature in 4% paraformaldehyde in PBS, permeabilized in 0.1% Triton X-100 in PBS for 15 min and probed with primary and secondary antibodies. Primary mouse monoclonal antibodies were used at the following concentrations: 1:100 anti-Myc antibody (Santa Cruz Biotechnology), 1:100 anti-PR antibody (Cell Signaling). Primary rabbit polyclonal antibodies were used at the following concentrations: 1:100 anti-PIAS3 antibody (Santa Cruz Biotechnology). Secondary antibodies were FITC-labeled anti-mouse or anti-rabbit antibodies and TRITC-labeled anti-mouse antibody (Santa Cruz Biotechnology) and were used at the following concentrations: 1:100 secondary antibodies.
Confocal imaging was performed using a Bio-Rad Radiance 2100TM confocal laser system connected to a Nikon TE300 microscope. The green fluorescence was excited with an argon laser (488 nm excitation line with 515 nm long pass barrier filter) and red fluorescence was simultaneously excited with an He–Ne laser (543 nm excitation line with 570 nm long pass barrier filter).
Transfection and luciferase assay
293T, COS7, T47D and HeLa cells were routinely cultured in DMEM containing 10% newborn calf serum at 37°C in a humidified atmosphere of 5% CO2 in air. For transfection, the cells were seeded in 12-well plates containing phenol red-free DMEM supplemented with 5% charcoal dextran-treated fetal bovine serum. The cells were transfected with 0.2 μg of pMMTV-Luc reporter or DBD-Luc reporter plasmids, 0.05 μg of pXJ40-Myc-PRB (wild-type or mutants) or pXJ40-Myc-PRA or pcDNA3.0-GAL4-DBD-PRB expression vectors, 0.02 μg of Renilla reporter pRL-TK (Promega), and 0.05–1.0 μg of the pXJ40-HA-PIAS3 expression vectors and the respective empty vector was used to adjust the total amount of DNA. The transfected cells were harvested 24 h after treatment with 10 nM progesterone, and Firefly and Renilla luciferase activities were determined using a Dual-Luciferase Reporter Assay System (Promega) and a TD-20/20 luminometer (Turner Designs). Renilla luciferase activity was used as an internal control for transfection efficiency. All experiments were repeated at least three times.
Yeast two-hybrid assay
To map the region of PRB interacting with PIAS3, the yeast two-hybrid assay was performed with a full-length PRB and its mutants ΔAF1, ΔDBD, ΔAF1-DBD, AF1-DBD as prey and pGBKT7-PIAS3 as the bait according to the manufacturer's instructions (Clontech Laboratories, Inc.). AH109 yeast strain was co-transformed with pGBKT7-PIAS3 as the GAL4–DBD and PRB fused to the pGADT7 plasmid. All the transformants were plated on selective medium lacking tryptophan, leucine, histidine and adenine, followed by β-galactosidase assays.
GST pull-down assays
GST and GST fusion proteins were expressed and purified according to the manufacturer's instructions (Pharmacia). Myc-PRB protein, obtained from the whole cell lysis of 293T cells, which were transfected with pXJ40-Myc-PRB plasmid, was mixed with GST and GST–PIAS3 fusion protein bound to Sepharose beads in 1 ml of binding buffer (20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 10% glycerol and 0.1% NP-40). The binding reaction was carried out for 4 h at 4°C. Beads were washed for three times in 1 ml washing buffer (the same as the binding buffer), eluted in 10 μl of 2× SDS–PAGE sample buffer and detected by immunoblotting.
Immunoprecipitation and immunoblotting
For immunoprecipitation experiments, 293T cells were transfected with the indicated plasmids. At 24 h post-transfection, cells were washed twice with phosphate-buffered saline (PBS) and lysed in IP lysis buffer (20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.1% NP-40, 10% glycerol, 1 mM DTT and 1× cocktail). After brief sonication, the lysates were cleared by centrifugation at 4°C. Supernatants were incubated with mouse monoclonal anti-HA or anti-Myc antibody for 4 h at 4°C and protein A/G–Sepharose beads for 2 h at 4°C. The immunocomplexes were washed three times with the same IP lysis buffer. The immunoprecipitated proteins were removed from the protein A/G beads by boiling for 10 min in SDS-sample buffer and immunoblotted with a monoclonal anti-Myc antibody, anti-HA antibody and anti-Flag antibody (Santa Cruz Biotechnology). For sumoylation experiments, cells were lysed in 0.5 ml of RIPA buffer [50 mM Tris–HCl (pH 8.0), 150 mM NaCl, 5 mM EDTA, 15 mM DTT, 20 mM NEM and 1× cocktail] followed by immunoprecipitation and immunoblotting.
Chromatin immunoprecipitation analysis
ChIP analysis was carried out using the ChIP assay kit from Upstate Biotechnologies with minor modifications of the protocol. HEK-293 cells were transfected with pMMTV-Luc vector using Lipofectamine 2000 according to the manufacturer's protocol. Transfected cells were selected in 500 μg/ml G418 (Invitrogen) for 2–3 weeks. Pooled clones were cultured and then used for ChIP analysis. HEK-293 cells (1 × 107) stably expressing the MMTV promoter were plated onto 60 mm dishes and were transfected with 1.0 μg of pXJ40-Myc-PRB or together with 2.0 μg of pXJ40-HA-PIAS3. HEK-293 cells were treated for 1 h with vehicle or 100 nM progesterone, cross-linked with 1% formaldehyde, and lysed in buffer containing SDS. Lysates were sonicated to shear chromatin and cleared by centrifugation. Clear lysates were incubated with a control unrelated antibody (IgG) or antibodies to Myc or HA (Santa Cruz Biotechnologies), with protein A–Sepharose as an adsorbent. Resins were washed in multiple buffers containing various salts and detergents, followed by elution, reversal of cross-links and isolation of DNA fragments. Following purification, the DNA was subjected to PCR amplification using primers MMTV-187 5′-TGGTTACAAACTGTTCTTAAAACGAGGATG-3′ and MMTV + 1 5′-GCAAGTTTACTCAAAAAATCAGCACTCTTT-3′ under the following conditions: 30 cycles, 55°C, 1.5 mM MgCl2, 0.2 mM dNTPs, 5 U Taq polymerase and 25 pmol of each primer. Amplified products were normalized to similarly amplified input DNA of the corresponding treatment groups (vehicle or progesterone). An aliquot of 10 μl of each reaction was analyzed on 1.5% 1× Tris–borate/EDTA–agarose gels.
Real-time RT–PCR analysis
T47D cells were seeded in six-well plates containing phenol red-free DMEM supplemented with 5% charcoal dextran-treated fetal bovine serum and transfected using Lipofectamine 2000 with 3.0 μg of the pXJ40-HA-PIAS3 expression vectors, and the respective empty vector was used to adjust the total amount of DNA. Eighteen hours after transfection, the cells were treated with 10 nM progesterone for another 6 h.
Cell pellets were collected and RNA was extracted using Trizol reagent (Invitrogen). The diluted RNA (1:100 for cyclin D1 and GAPDH) was analyzed using real-time PCR (Bio-Rad iCycler iQ real-time PCR system) with SYBR Green Jumpstart™ Taq ReadyMix™ (Sigma) and probe for the cyclin D1 gene (Forward primer, 5′-CCAGAGTGATCAAGTGTGAC-3′; Reverse primer, 5′-GATGTCCACGTCCCGCAC-3′) or the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene (Forward primer, 5′-GGCTGAGAACGGGAAGCTTGTCTAT-3′; Reverse primer, 5′-CAGCCTTCTCCATGGTGGTGAAGA-3′). The program used was 40 cycles at 95°C for 2 min, 94°C for 15 s, 58°C for 30 s and 72°C for 30 s.
RNA interference
From siRNA to target human PIAS3 was chemosynthesis and from the PIAS3 target sequence was 5′-TTGGTCATCTGAGTTCGGA-3′, the scrambled sequence was 5′-GCTTGTCCCGACAGAGTAG-3′. The siRNA was transfected into 293T cells using Lipofectamine 2000 according to the manufacturer's protocol. After 48 h, the cells were transfected with PRB (0.05 μg), PRB-luciferase reporter (0.2 μg) and siRNA again. Treated with progesterone for another 24 h, cells were lysed for PRB-luciferase reporter assay. The non-specific RNA duplex was used in control experiments. The relative expression of endogenous PIAS3 was monitored by Western blotting of cell extracts isolated from control or siRNA-treated cells, using a polyclonal antibody against PIAS3.
Gel shift assay
DNA-binding activity of PR was analyzed by electrophoresis gel mobility shift assay. Briefly, in vitro translated protein PR or the same amount of unprogrammed lysate (Promega) was combined with the 32P-labeled PRE (5′-GTTACAAACTGTTCT-3′) double-stranded oligonucleotide in binding buffer [10 mM Tris, pH 7.5, 50 mM NaCl, 1 mM MgCl2, 4% glycerol, 0.5 mM EDTA, 0.5 mM dithiothreitol and 0.1 μg/μl poly(dT–dC)] in the presence or absence of progesterone (10 nM). The binding reaction (20 μl total volume) was incubated at room temperature for 20 min and increasing amounts of in vitro purified GST–PIAS3 fusion protein or the same amount of GST were added to the binding reaction. Reactions in antibody supershift analyses were incubated with 1 μg of anti-PIAS3 antibody for 15 min at 4°C. Protein–DNA complexes were resolved on a 5% native polyacrylamide gel and visualized by autoradiography.
RESULTS
PIAS3 inhibited hormone-dependent PR-mediated gene transactivation
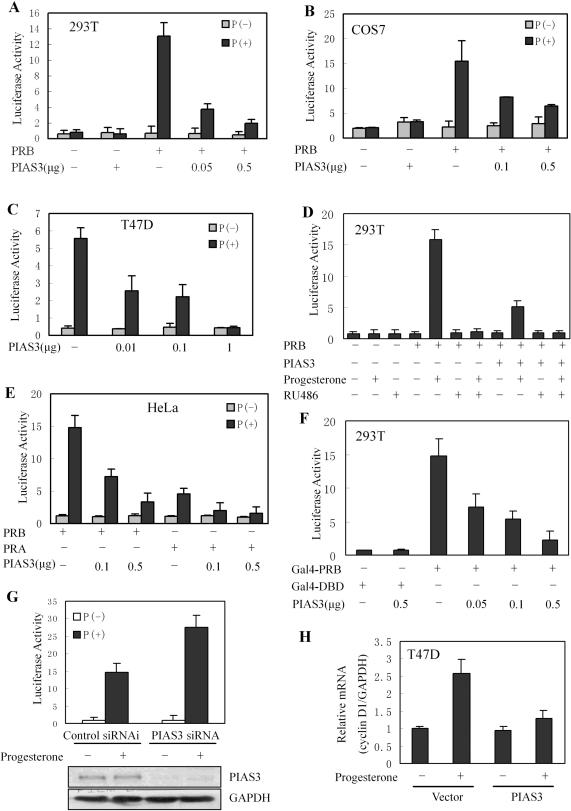
Previous studies in our laboratory suggested that, in the presence of progesterone, the localization of PRB was altered by PIAS3. To further elucidate the influence of PIAS3 on PR-mediated gene transactivation, PR-negative 293T cells were transfected with PRB, a MMTV luciferase reporter containing PRE in the promoter and PIAS3. As shown in Figure 1A, PRB exhibited a 25-fold induction of reporter gene activation in response to progesterone. In addition, the ligand-dependent transactivation of PRB was strongly repressed, in a dose-dependent manner, by PIAS3 expression. The effects of PIAS3 on PRB transactivation were not restricted to a single cell type, as similar results were noted in COS7 cells (Figure 1B). No differences were observed in PRB protein levels between 293T and COS7 cells containing transiently expressed PIAS3 and non-transfected controls (data not shown), suggesting that the inhibition of PRB transactivation by PIAS3 was not due to a modulation of PRB protein levels. Further examination of the transactivation of endogenous PR in PR-positive T47D breast cancer cells demonstrated that PIAS3 repressed progesterone-triggered reporter gene transcriptional activity by 90% (Figure 1C). In order to elucidate whether PIAS3 inhibition of PR transactivation was dependent on ligand stimulation, 293T cells were co-transfected with PRB, PIAS3 and MMTV-Luc reporter then treated with antiprogestin, RU486 (Figure 1D). The data suggested that, in the presence of progesterone, RU486 completely blocked the effects of PIAS3 on PRB transcriptional activity. The effect of PIAS3 on transactivation of PRA was also examined in HeLa cells, and PIAS3 had the similar inhibitory effect on PRA transactivation (Figure 1E).
Figure 1.
PIAS3 strongly inhibited the transactivation of PR. (A) Inhibition of PRB transcriptional activity by PIAS3 in 293T cells. All treatments were as follows, except when noted: cells were transiently transfected with pMMTV luciferase reporter construct together with an empty expression vector, Myc-PRB, or varying amounts of HA-PIAS3 vectors, alone or in combination, as indicated. After transfection, cultures were treated with progesterone (10 nM) for 24 h (P+) or not treated (P−), and cell extracts were prepared and measured for luciferase activity. (B) Inhibition of PRB transcriptional activity by PIAS3 in COS7 cells. (C) Inhibition of PR transcriptional activity by PIAS3 in T47D cells with endogenous PR expression, PR was replaced by endogenous expression. (D) 293T cells were transiently transfected with Myc-PRB expression vector together with HA-PIAS3 expression construct; antiprogestin RU486 (100 nM) was added as indicated. (E) Inhibition of PRA transcriptional activity by PIAS3 in HeLa cells. (F) 293T cells were co-transfected with Gal4DBD–PRB chimeric constructs, together with the DBD-luc reporter, or varying amounts of HA-PIAS3 vectors. (G) PIAS3 RNAi increased ligand-induced PRB transactivation. 293T cells were transfected with either siRNA control or siRNA directed against PIAS3 and were co-transfected, 48 h later, with pMMTV-luc reporter plasmid and PRB plasmid were then treated with progesterone (10 nM) or ethanol for 24 h, and luciferase activity was determined and normalized for transfection efficiency. Western-blot analysis using PIAS3 antibody was performed to determine PIAS3 protein levels following siRNA transfection. (H) PIAS3 decreased cyclin D1 mRNA levels in the presence of progesterone. T47D cells were transfected with PIAS3 expression construct as indicated. The cells were treated with control (0.1% ethanol) vehicle or progesterone (10 nM, 6 h) before harvest, and total RNA was extracted. A real-time RT–PCR was performed for the cyclin D1 gene and normalized to total GAPDH RNA, as described in Materials and Methods. The value for vehicle-treated cells was set at 1 (column 1). Mean ± SD (n = 3).
We further investigated whether the influence of PIAS3 on PR function was independent of the promoter construct, utilizing a Gal4–PRB fusion protein and a reporter plasmid containing a Gal4-driven promoter. Similar to the MMTV promoter, PIAS3 repressed Gal4–PRB transcriptional activity, but not Gal4–DBD (Figure 1F). These results indicated that PIAS3 strongly repressed PR-dependent transactivation, regardless of the promoter or cell type.
In order to investigate the role of endogenous PIAS3 in PR-mediated transcriptional activation, 293T cells were transfected with siRNA directed against PIAS3, or a control siRNA, then co-transfected, 48 h later, with a MMTV luciferase reporter construct together with the PRB expression vector. Cells were then stimulated with progesterone (24 h). The results demonstrated that there was an obvious increase in ligand-mediated PRB activity in PIAS3 siRNA-transfected cells, compared to control siRNA (Figure 1G), suggesting that the reduction of PIAS3 leads to increased ligand-mediated PRB activity. Western-blot analysis demonstrated that PIAS3 siRNA, but not control, specifically reduced the expression of endogenous PIAS3 (Figure 1G), further suggesting that PIAS3 inhibited PR transcriptional activity in the presence of progesterone.
Previous research has suggested that cyclin D1 played an important role in normal mammary gland development (36,37). Cell cycle analyses of biphasic cell growth patterns indicated that, following a single dose of progesterone, cyclin D1 and cyclin E levels increased initially, as cells entered S-phase (38). In order to corroborate luciferase reporter assay data, the effects of PIAS3 on the expression of endogenous PR target genes were examined. T47D cells were transfected with PIAS3 in the presence and absence of progesterone (10 nM, 6 h) before harvesting; RNA was extracted, transcription of cyclin D1 was measured by real-time RT–PCR and expression was normalized to GAPDH. Although cyclin D1 mRNA levels were upregulated in response to progesterone, with no changes noted in the absence of progesterone, PIAS3 overexpression reduced the ligand-induced increase in cyclin D1 mRNA (Figure 1H). These data demonstrated PIAS3 efficiently decreased the expression of endogenous PR-responsive genes, suggesting that PIAS3 functioned as an inhibitor of PR-mediated transactivation.
PIAS3 physically interacted with PRB
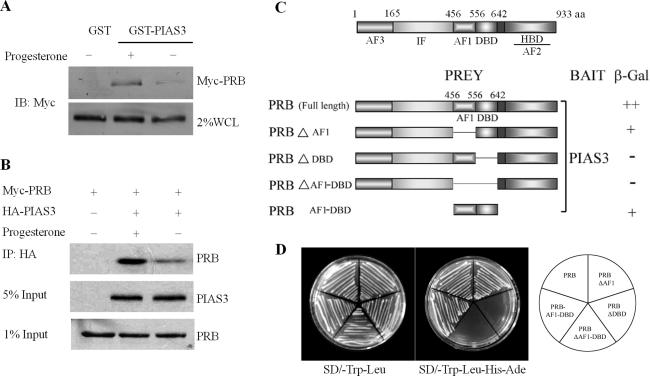
To further investigate the mechanism by which PIAS3 inhibited PRB transactivation, we examined whether there was physical association between PRB and PIAS3. Although previous research suggested that steroid nuclear receptors physically interacted with PIAS family proteins (39), detailed data of PR interaction with PIAS3 was not presented. We utilized a GST pull-down assay to further elucidate the physical association between PRB and PIAS3 in vitro. Our results demonstrated that the recombinant fusion protein glutathione S-transferase with PIAS3 (GST–PIAS3), but not GST alone, was able to bind to PRB (Figure 2A). In addition, we further verified the interaction between PRB and PIAS3 in intact cells (Figure 2B). A Myc-tagged PRB (Myc-PRB) was co-expressed with HA-tagged PIAS3 in 293T cells. Antibodies against the HA epitope precipitated a complex of PRB and PIAS3, suggesting that the two proteins physically interacted in living cells. Progesterone treatment significantly enhanced the association between PRB and PIAS3, as demonstrated by both GST pull-down and co-immunoprecipitation assays, which was consistent with previous results that the inhibition of PRB transactivation by PIAS3 was only observed in cells stimulated with ligands.
Figure 2.
PIAS3 interacts with PRB in vitro and in vivo. (A) Interaction of PRB with PIAS3 in vitro. GST–PIAS3 fusion proteins, immobilized on beads, were mixed with cell lysates with Myc-PRB expression in the absence or presence of progesterone (100 nM). Bound proteins were subjected to SDS–PAGE followed by western-blot analysis. (B) Interaction of PRB with PIAS3 in vivo. 293T cells were co-transfected with expression vectors for the Myc-tagged PRB and HA-tagged PIAS3 as indicated. Lysates from transfected cells were immunoprecipitated using an anti-HA antibody, and the immunoprecipitates were probed with an anti-Myc antibody. (C) Confirmation of the interaction of PRB with PIAS3 in yeast. The AH109 strain was co-transformed with GAL4-DBD–PIAS3 plus different GAL4AD–PRB constructs as indicated in the schematic diagram, then scored for β-galactosidase (β-gal) activity. (++, positive strongly interaction; +, positive interaction; -, no interaction). (D) PRB specifically interacted with PIAS3 in yeast. The AH109 strain was co-transformed as (C), then co-transformants were isolated on the SD medium deficient in leucine and tryptophan (SD/−leu/−trp, left plate) or on the same medium lacking histidine and adenine (selective medium, right plate).
In order to further identify the region of the PRB protein that binds to PIAS3, PRB and its deletions were cloned into a pGADT7 fusion protein expression vector then transformed into S.cerevisiae strain AH109, together with pGBKT7 plasmids encoding GAL4-BD fused to PIAS3. As shown in Figure 2C and D, the physical association between PRB and PIAS3 was observed in yeast two-hybrid assays, with amino acids 556–642 determined to be essential for the association of PRB with PIAS3 in yeast (Figure 2C).
PIAS3-induced sumoylation of PRB in vivo
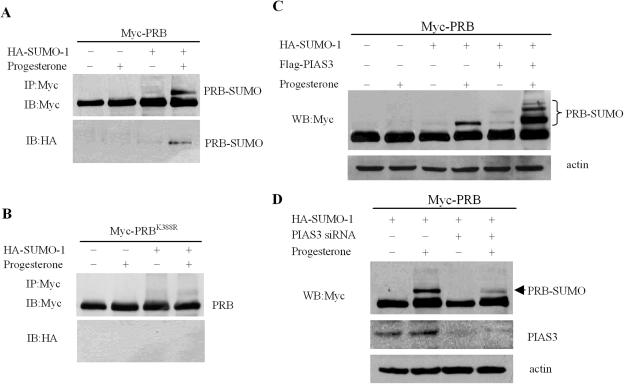
Recently, the PIAS protein family was described as SUMO-E3 ligases for critical target proteins such as p53 (28,29), c-jun (29,30), LEF1 (31), as well as the androgen (32–34) and estrogen receptors (35). Although PIAS proteins were shown to act as regulators of PR (39), it has not been reported whether PIAS family proteins mediate the sumoylation of PR. In order to clarify whether the effects of PIAS3 on PR transactivation were associated with the SUMO modification of PR, 293T cells were transfected with a Myc-PRB expression vector in the absence or presence of a vector encoding HA-tagged SUMO-1. Blotting with an anti-Myc monoclonal antibody detected a single protein band of ∼115 kDa in cells expressing only PRB (Figure 3A). In contrast, in the presence of progesterone, cells co-expressing both PRB and SUMO-1 displayed an additional slower-migrating band representing the sumoylated PRB (Figure 3A). An HA-SUMO-conjugated PRB band (Figure 3A) was detected, with an anti-HA antibody, at the same position on the gel (Figure 3A). These data suggested that the band represented sumoylated PRB. Additional studies demonstrated that, compared with the wild-type control, the PRB K388R mutant was not sumoylated (Figure 3B), which suggests that PRB underwent sumoylation at Lys-388, located at its N-terminal domain.
Figure 3.
Sumoylation of PRB and effects of PIAS3 on PRB. (A and B) 293T cells were transfected with expression plasmids encoding HA-SUMO-1 and Myc-PRB or Myc-PRBK388R, incubated with or without progesterone (100 nM, 2 h), then lysed with RIPA buffer and immunoprecipitated (IP) with anti-Myc antibody. The immunoprecipitates were probed with anti-Myc and anti-HA antibody, respectively. (C) 293T cells were transiently transfected with Myc-PRB, HA-SUMO-1 and Flag-PIAS3 alone or in combination as indicated. After 24 h, the cells were treated with or without progesterone (100 nM, 2 h), then lysed and immunoblotted with anti-Myc antibody. (D) 293T cells were transfected with either siRNA control or siRNA directed against PIAS3 and were co-transfected, 48 h later, with Myc-PRB and HA-SUMO-1 plasmids. Before being harvested, cells were treated with progesterone (100 nM) or ethanol for 2 h, and then lysed. Anti-Myc and PIAS3 antibodies were used in western-blot analysis of cell lysates.
To further investigate the role of PRB sumoylation, we performed transient transfection assays in 293T cells and found that SUMO-1 expression significantly increased PRB transactivation in the presence of progesterone (Supplementary Figure 1A). Furthermore, mutation of sumoylation site in the K388R mutant also increased the transactivation ability of PRB (Supplementary Figure 1B). The result that the non-sumoylated mutant was more active than the wild-type receptor was similar to that reported previously (7,27).
In addition, we investigated whether PIAS3 protein was capable of enhancing SUMO-1 attachment to PRB in intact cells by co-expressing Myc-PRB, HA-SUMO-1 and Flag-PIAS3 proteins in 293T cells, in the absence or presence of progesterone. As shown in Figure 3C, in cells without co-expression of the PIAS3 protein, a single band representing SUMO-modified PRB was obviously enhanced by the addition of progesterone (Figure 3C). However, three slower-migrating bands, representing sumoylated PRB, appeared with PIAS3 co-expression, and the band representing sumoylation of PRB in Lys-388 site was strongly enhanced with progesterone treatment (Figure 3C). Given that this result was based on overexpression of PIAS3, we examined whether endogenous PIAS3 was involved in the sumoylation of PRB. Knockdown of PIAS3 by RNAi decreased the level of PRB sumoylation in the presence of progesterone, as shown in Figure 3D. PIASy, an alternative PIAS family member protein, only marginally enhanced PRB sumoylation in the presence of progesterone (Supplementary Figure 3), suggesting that PIAS3-promoted PRB sumoylation was specific.
This is, to our knowledge, the first report that, in the presence of progesterone, PIAS3 expression strongly induced a pattern of PRB sumoylation at three sites, including Lys-388 and two novel sites.
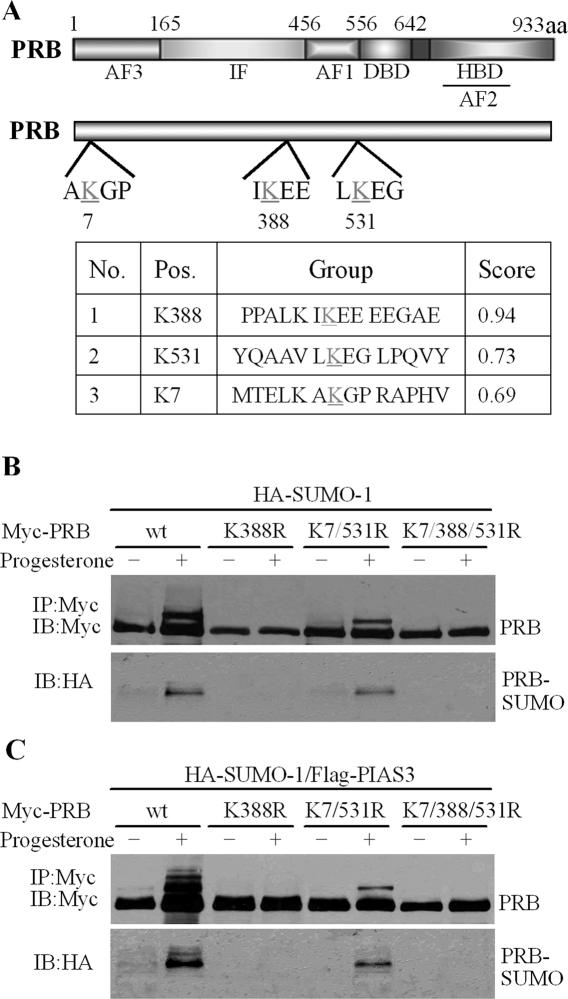
Determination of PIAS3-induced PRB sumoylation sites
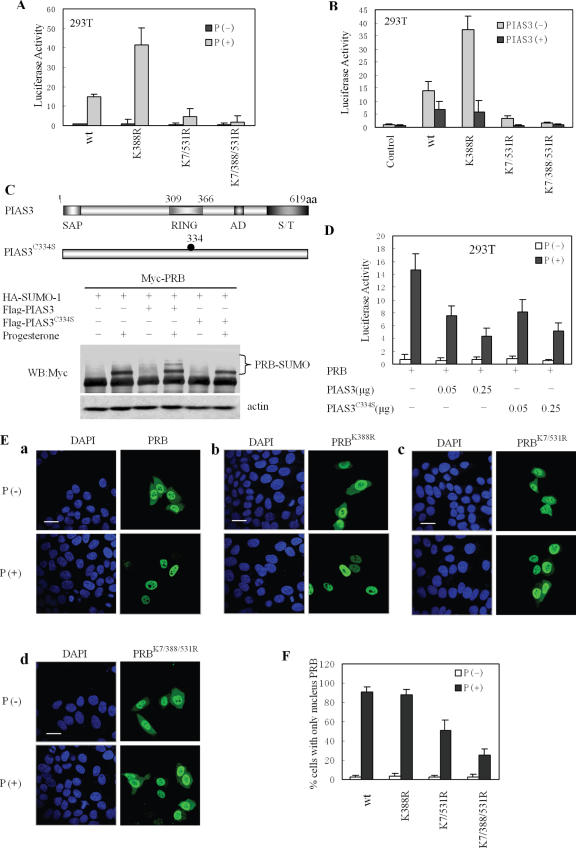
Mutagenesis studies were undertaken to further investigate the sumoylation sites of PRB mediated by PIAS3 in vivo. Examination of the PRB sequence (www.abgent.com/doc/sumoplot) showed the possible presence of three consensus motifs for SUMO-1 conjugation (ψKXE) (24), including Lys-388, as well as Lys-7 and Lys-531, which partially fit the consensus sumoylation sites (Figure 4A). 293T cells were co-transfected with expression vectors encoding various mutants of Myc-PRB (K388R or K7/531R or K7/388/531R) and HA-SUMO-1 with or without Flag-PIAS3 (Figure 4B and C). As shown in Figure 4B, substitution on Lys-388, including the K388R and K7/388/531/R mutants, abolished the ability of SUMO-1 conjugated to PRB, even when treated with progesterone. In contrast, the double substitution on Lys-7 and Lys-531 (K7/531R) did not abolish sumoylation at Lys-388, although it was slightly lessened, suggesting that mutations of Lys-7 and Lys-531 influenced Lys-388 sumoylation.
Figure 4.
Determination of PIAS3-induced sumoylation sites in PRB. (A) A schematic representation of PRB shows the sites corresponding to consensus sequences for SUMO-1 conjugation. (B and C) 293T cells were transfected with HA-SUMO-1 and Myc-PRB, Myc-PRB K388R, Myc-PRB K7/531R, or Myc-PRB K7/388/531R in the presence or absence of PIAS3. Cells were treated, or not, with progesterone (100 nM, 2 h) before harvesting. Cell extracts were lysed with RIPA buffer and immunoprecipitated with anti-Myc antibody. The immunoprecipitates were probed with anti-Myc and anti-HA antibody, respectively.
Ectopical expression of PIAS3 resulted in the strong enhancement of PRB sumoylation in the presence of progesterone (Figure 4C). However, the effects of PIAS3 on sumoylation were still abolished in the PRB K388R mutant (Figure 4C). Interestingly, sumoylation levels in the K7/531R mutant were strongly diminished compared to wild-type PRB, with only one band noted indicating Lys-388 sumoylation. The single band in the K7/531R mutant was similar to the band in the absence of PIAS3, which was relatively weaker than that in wild-type PRB and which was further abolished in the K7/388/531R. These results indicated that Lys-7 and Lys-531 were also involved in the PIAS3-mediated sumoylation of PRB. Notably, the ability of PIAS3 to stimulate PRB sumoylation was dependent on Lys-388 sumoylation.
PIAS3 inhibition of PRB transactivation was independent of PRB sumoylation
In order to further examine the role of PIAS3-enhanced sumoylation on PR transcription regulation, 293T cells were transfected with the same amount of PRB or its mutants (K388R or K7/531R or K7/388/531R) and treated, or not, with progesterone (10 nM, 24 h). Similar to previous results, mutation of the sumoylation site in the K388R mutant increased the transactivation ability of PRB (7,27). Intriguingly, with the double substitution on both Lys-7 and Lys-531, the activation of the reporter gene mediated by K7/531R mutant was significantly downregulated (Figure 5A). Moreover, the K7/388/531/R mutant almost failed to be transcriptionally activated by progesterone (Figure 5A) whereas the protein levels of PR and its mutants in 293T cells were quite similar (data not shown). We then examined the ability of PIAS3 to inhibit gene activation mediated by mutants unable to be modified by SUMO-1. To this end, 293T cells were co-transfected with PRB or its mutants (K388R, K7/531R or K7/388/531R), the pMMTV-Luc reporter construct, and equal amounts of PIAS3 expression vector. Cell lysates were prepared for luciferase assays 24 h after progesterone treatment. Surprisingly, the assays revealed that the transactivation of these mutants, without sumoylation, was still inhibited by PIAS3 (Figure 5B). Most significantly, the inhibition levels were comparable with those observed in the wild-type PRB. In addition, all PRB mutants still interacted with PIAS3 in the presence of progesterone (Supplementary Figure 4).
Figure 5.
PIAS3 inhibited PRB transactivation, independent of its sumoylation. (A) 293T cells were transiently transfected with pMMTV luciferase reporter construct, Myc-PRB or various Myc-PRB mutant vectors. After transfection, cultures were untreated (P−) or treated with progesterone (10 nM, 24 h) (P+), and cell extracts were prepared and measured for luciferase activity. Methods and treatment similar for all figure, except when noted. (B) Inhibition of PRB mutant transactivation by PIAS3; HA-PIAS3 was co-transfected in 293T cells. (C) The RING finger domain of PIAS3 was required for increasing SUMO-1 modification of PRB. 293T cells were transiently transfected with expression vectors containing Myc-PRB, HA-SUMO-1 and Flag-tagged wild-type or C334S mutant of PIAS3, as indicated. Cell lysate was subjected to immunoblotting. (D) Inhibition of PRB transactivation by PIAS3 or its mutant; similar to Figure 1A, except PIAS3 C334S mutant was added as indicated. (E and F) The subcellular localization of PRB and its mutants. Cultured HeLa cells were placed in a medium containing charcoal-stripped serum then transiently transfected with Myc-PRB or its mutant expression vector. Cells were untreated (P−) or treated with progesterone (100 nM, 2 h) (P+), 24 h after transfection, then subjected to immunofluorescence and confocal microscopy using a Myc-specific monoclonal antibody. Scale bar: 20 μm. At least 300 transfected cells were scored for PRB and mutant subcellular distribution (E).
We further investigated whether the PIAS3 C334S mutant lacking E3 ligase activity (28,40) was able to inhibit PRB transactivation as well as the wild-type. Similar to previous assays, 293T cells were co-transfected with PRB, SUMO-1, PIAS3 or PIAS3 C334S mutant then treated with or without progesterone. As shown in Figure 5C, the PIAS3 C334S mutant lost the ability to enhance PRB sumoylation in the presence of progesterone, whereas PIAS3 inhibited the transcriptional activity of PRB in a dose-dependent manner (Figure 5D), with comparable capacity displayed by the C334S mutant. These results indicated that the PIAS3 protein was able to inhibit ligand-induced gene activation by PRB in a manner that was independent of PRB SUMO modifications.
The nuclear localization of the transcription factor PR is important for its activity (41–43); consequently, we further investigated the intracellular localization of PRB and its mutants. Confocal microscopy data indicated that both wild-type PRB and its mutants predominantly resided in the nucleus, with some cytoplasmic staining noted in the absence of progesterone (Figure 5E, a–d). However, 2 h after progesterone treatment, the wild-type PRB was completely translocated into the nucleus, but excluded from nucleoli (Figure 5E, a and F). Although a similar distribution pattern was observed with the PRB K388R mutant after treatment with progesterone (Figure 5E, b and F), >60% of the PRB K7/388/531R mutant was still localized to the cytoplasm in the ligand-treated HeLa cells (Figure 5E, d and F), and ∼40% of the PRB K7/531R mutant staining was also cytoplasmic, in the presence of progesterone (Figure 5E, c and F). These results suggested that the localization of PRB K7/531R and K7/388/531R differed from that of wild-type PRB, and were consistent with the ligand-stimulated transactivation of PRB and its mutants (Figure 5A). These data also suggested that the three arginine substitutions on Lys-7, Lys-388 and Lys-531 might have caused a conformational change in PR, resulting in the inability of the mutant to translocate to the nucleus in response to ligand stimulation.
PIAS3 destabilized PRB retention in the nucleus
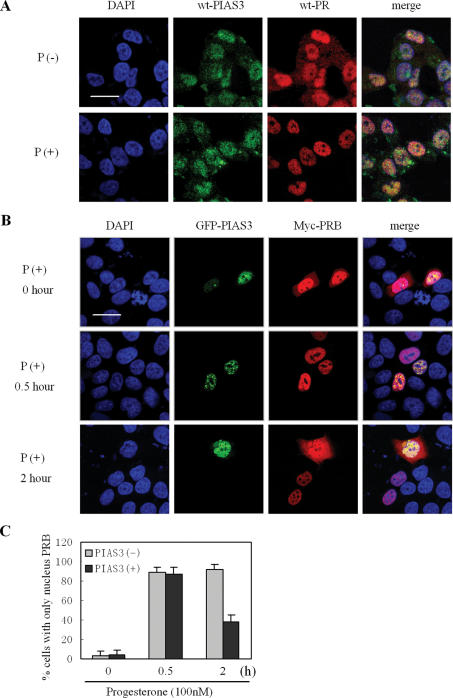
Immunofluorescence microscopy was used to visualize the subcellular localization of endogenous PR and PIAS3 in T47D cells treated, or not, with progesterone (2 h, 100 nM). As shown in Figure 6A, endogenous PR was localized mainly in the nucleus and cytosolic PR staining was visible in the absence of progesterone. Progesterone treatment resulted in the complete translocation of liganded-PRB from the cytoplasm to the nucleus. Detection of Myc-tagged PRB with an anti-Myc antibody yielded the same localization as endogenous PR (Figure 5E). PIAS3 resided mainly in the nucleus, as dotted structures, with no changes induced by progesterone (Figure 6A). Overlay of PR and PIAS3 clearly showed co-localization of both proteins as yellow-stained areas (Figure 6A). These data indicated that the two endogenous proteins co-localized in nuclear dot structures in T47D cells, both in ligand-treated and in control untreated cells.
Figure 6.
PIAS3 destabilized PRB retention in the nucleus. (A) The co-localization of wild-type PR and PIAS3 in T47D cells. T47D cells were cultured on coverslips, placed in medium containing charcoal-stripped serum, and untreated (P−) or treated (P+) with progesterone (100 nM, 2 h) then subjected to immunofluorescence using specific primary antibodies to PR and PIAS3. (B and C) PIAS3 destabilized PRB retention in the nucleus. HeLa cells were cultured in medium containing charcoal-stripped serum, and co-transfected with Myc-PRB and GFP–PIAS3, treated with progesterone (100 nM) for 0, 0.5 or 2 h, then subjected to immunofluorescence staining with Myc-specific monoclonal antibody. Scale bar: 20 μm. At least 300 transfected cells were scored for PRB subcellular distribution (C).
The effects of PIAS3 overexpression on PRB localization at the subcellular level were then examined. HeLa cells were co-transfected with GFP–PIAS3 and Myc-PRB, then treated with progesterone (100 nM) and examined by immunofluorescence (Figure 6B). GFP-tagged PIAS3 expression was also observed as nuclear dots. Although Myc-PRB mainly resided in the nucleus, some cytoplasmic staining was noted, which did not change as a result of ectopical expression of PIAS3 in the absence of progesterone (Figure 6B, 0 h). After 30 min of progesterone treatment, >85% of PRB (Figure 6C) translocated to the nucleus, and co-localized with PIAS3 as nuclear dot structures (Figure 6B). Interestingly, after 60 min of progesterone treatment, some PRB staining was noted in the cytoplasm of cells co-transfected with PIAS3, compared with cells, which only expressed PRB (Figure 6B and C). Similar results were also found in the PIAS3 C334S mutant, which lost the SUMO-E3-ligase ability (Supplementary Figure 5). These data suggested that, in the presence of progesterone, PIAS3 influenced the localization of PRB and that one of the mechanisms by which PIAS3 inhibited PR transcriptional activity was by promoting the nuclear export of PR.
PIAS3 is recruited to endogenous PR-responsive promoters
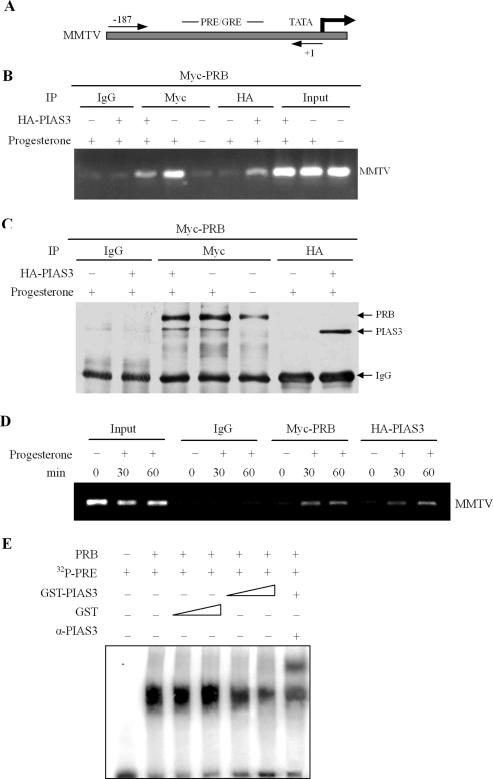
To further understand the mechanisms by which PIAS3 inhibited PRB transactivation, chromatin immunoprecipitation (ChIP) was used to determine whether PIAS3 and PRB were recruited to promoters of progesterone-regulated genes in vivo. HEK-293 cells, stably expressing the MMTV promoter, were transfected with Myc-PRB or HA-PIAS3, then treated, or not, with progesterone (100 nM, 2 h). Previous research demonstrated that the MMTV promoter was highly inducible by progesterone and contained multiple GRE/PREs, located between 190 and 80 bp from the transcription start site (44) (Figure 7A). Following formaldehyde cross-linking and chromatin precipitation with anti-Myc and anti-HA antibodies, the precipitated DNA was PCR amplified with specific primers flanking the PRE in the PR enhancer region. In the absence of hormone, ChIP assay detected minimal occupancy of the MMTV promoter by PRB, as well as non-specific IgG background, whereas progesterone treatment resulted in a substantial increase of PRB recruitment by the MMTV promoter (Figure 7B). At the same time, PIAS3 also associated with the MMTV promoter in the presence of hormone (Figure 7B). Both PIAS3 and PRB were specifically co-immunoprecipitated with HA- and Myc-specific antibodies, respectively (Figure 7C). In addition, HEK-293 cells stably expressing the MMTV promoter were co-transfected with Myc-PRB and HA-PIAS3, then treated, 24 h after transfection, with progesterone for various times points. As expected, PRB displayed a clear time-dependent recruitment to the MMTV promoter. Importantly, PIAS3 also revealed a distinct time-dependent recruitment to the MMTV promoter (Figure 7D). These results highlighted the finding that PIAS3 and PRB were recruited in a largely hormone-dependent manner to a progesterone-responsive promoter in vivo.
Figure 7.
PIAS3 and PRB physically interacted and were recruited to a progesterone-inducible promoter in vivo. (A and B) ChIP was performed using HEK-293 cells stably expressing the MMTV promoter in the presence or absence of progesterone hormones. Primers specific for the MMTV promoter were used to amplify the DNA associated with PR and PIAS3 in vivo. (C) Cleared ChIP lysis was first incubated with pre-immune serum, anti-Myc or anti-HA antibody as indicated, then with protein A–Sepharose as an adsorbent. Resins were washed and bound proteins were detected by immunoblotting with antibodies to Myc or HA. (D) Recruitment of PR and PIAS3 to progesterone-responsive promoters. HEK-293 cells stably expressing the MMTV promoter were cultured in the absence of progesterone, and were then treated without (time 0) and with 100 nM progesterone for 30 and 60 min followed by ChIP analysis. (E) PIAS3 decreased protein–DNA complex formation. Increasing amounts of in vitro purified GST–PIAS3 were mixed with in vitro translated PRB and 32P-labeled PRE in the presence of progesterone (10 nM). Supershift experiments utilized anti-PIAS3 antibodies in the binding reactions, as indicated.
To investigate whether PIAS3 inhibited the DNA-binding activity of PRB, in vitro translated PRB and 32P-labeled PRE were incubated with increasing amounts of in vitro purified GST–PIAS3 fusion protein. As shown in Figure 7E, PIAS3 inhibited PRB binding to the PRE sequence in a dose-dependent manner. Antibody supershift experiments showed that PIAS3 was present in the PR–PRE complexes (Figure 7E). These data suggested that one possible mechanism of PIAS3 action on PR transcription might be due to the inhibition of PR binding to PRE.
DISCUSSION
The PR transcription factor plays an important role in transcriptional regulation. In the current study, we have identified PIAS3 as a novel PR-interacting partner utilizing a number of in vitro and in vivo assays, including yeast two-hybrid, in vitro GST pull-down and in vivo co-immunoprecipitation. Overexpression of PIAS3 inhibited the transcriptional activity of PR and the expression of progesterone-responsive genes. In sharp contrast, knockdown of endogenous PIAS3 increased PR transactivation, suggesting that PIAS3 played an important role in PR signaling. Moreover, our results showed that, in the presence of progesterone, PIAS3 expression strongly induced a pattern of PRB sumoylation at three distinct sites. We further provided evidence that PIAS3 inhibited the DNA-binding activity of PRB and promoted its nuclear export.
Influence of PIAS3 on PRB transactivation and sumoylation
There have been diverse reports on the influence of the PIAS protein family on steroid receptor function. Kotaja et al. (39) reported that while all PIAS proteins were able to co-activate PR-dependent transcription, the degree of activation was dependent on the receptor, the promoter and the cell type. However, it was of note that with a more complex promoter (the mouse mammary tumor virus promoter, pMMTV), Tan et al. (45) found PIAS1 repressed PR-dependent transactivation. Controversial results of the effects of PIAS family proteins on steroid receptors might have resulted from the fact that these studies used different promoter constructs and cell types to measure transcriptional activity. Our findings, utilizing promoters of pMMTV-Luc and GAL4-Luc reporters, consistently indicated that PIAS3 strongly inhibited PR-mediated transactivation in 293T, HeLa, T47D and COS7 cells.
Sumoylation has emerged as an important mechanism of gene expression control. Studies on a number of SUMO targeted transcription factors revealed that steroid receptors underwent sumoylation in response to ligand stimuli, which involved the regulation of transcriptional activity (26,46), including AR (32–34), ER (35) and PR (7,27). On the other hand, PIAS family proteins were described as SUMO-E3 ligases for critical target proteins, such as p53 (28,29), c-Jun (29,30), LEF1 (31), androgen receptor (32–34) and estrogen receptor (35). However, it remains to be established whether PIAS3 was responsible for PR sumoylation while inhibiting its transactivation. This is, to our knowledge, the first demonstration that co-expression of PIAS3 strongly increased the sumoylation of PRB in the presence of progesterone.
Interestingly, SUMO-1 modification appears to have inconsistent effects on the transcriptional activity of PR (7,27). Abdel-Hafiz et al. (7) demonstrated that SUMO-1 modification repressed PR transcriptional activity, identifying the consensus SUMO-1 binding motif as 387IKEE, within the IF domain of PR. In contrast, SUMO-1 overexpression markedly enhanced PR-mediated gene transcription. The controversial results were partly due to the fact that SUMO-1 also exerted effects on other proteins involved in PR transcription regulation, such as SRC-1 (27). However, our findings clearly indicated that PIAS3-induced SUMO modification did not involve in the repression of PR transcriptional activity by PIAS3, as the PRB K388R and K7/388/531R mutants, lacking sumoylation ability, were still repressed by PIAS3. Moreover, the PIAS3 C334S mutant, lacking SUMO-E3-ligase activity, still repressed PR transcriptional activity. These findings clearly indicated that the PIAS3 protein inhibited PR transactivation through a mechanism that was independent of SUMO modification of PR. Similar mechanisms have also been reported with the regulation of other transcription factors by PIAS proteins, including p53 (47), STAT1 (48), ER (35), AR (32), LEF1 (31) and Elk-1 (49). Whereas SUMO-E3-ligase activity of PIAS1 and PIASx-α might be important for the regulation of androgen receptor activity (33), but in the current study, PIAS3-induced sumoylation had no effect on PR activity, suggesting that PIAS proteins regulated the activity of transcription factors through different mechanisms more than just SUMO-E3 ligases (50).
SUMO modification increased PRB stability
Although a relationship between SUMO modification of PR and PIAS3-mediated inhibition of its transcriptional activity was not demonstrated, our findings established a pattern of PRB sumoylation that was strongly induced by PIAS3, and identified the positions of SUMO modification at three distinct sites. PRB sumoylation is known to occur on Lys-388 (7,27). We found that PIAS3 also induced PRB sumoylation at other two sites, Lys-7 and Lys-531, which reside in a motif that partially fits the ΨKXE consensus sumoylation sites (24,26,46). Our data demonstrated that the double mutant (K7/531R) had only one slower-migrating band, corresponding to SUMO-modified PRB at Lys-388, which was much weaker than that in wild-type PRB, whereas the K7/388/531R completely eliminated PRB sumoylation in the presence of PIAS3 in vivo. Intriguingly, in the presence of PIAS3, the K388R mutant abrogated PRB sumoylation at the two additional sites. These results indicated that PRB sumoylation at these sites might occur in a sequential fashion as mutation of Lys-388 blocked the sumoylation of Lys-7 and Lys-531. Similar findings were reported with the NFAT1 protein (51).
Interestingly, our findings identified previously unexpected roles for Lys-7 and Lys-531 in the transcriptional activity and nuclear retention of PRB. Although the ligand-triggered translocation of PRB into nucleus (43,52) was still observed in the K388R mutant lacking sumoylation ability, the K7/388/531R mutant failed to translocate in response to progesterone stimulation. Moreover, it fitted well with the impaired transactivation capacity and the K7/388/531R mutant was no longer activated by ligand. Our results suggested that mutants were still able to dimerize in the presence of progesterone (Supplementary Figure 2), which contributed to the nuclear localization of PR. This may have resulted from the fact that the substitution of the three lysine sites with arginine influenced PR dimensional conformation. The results suggested that Lys-388, Lys-7 and Lys-531 were important for the ligand-triggered transactivation of PRB.
Our findings provided insights that allowed us to speculate on the role of SUMO modification of PRB. Overexpression of either SUMO-1 or PIAS3 significantly increased the levels of PRB protein (Figure 4B and C), suggesting that sumoylation increased the stability of PRB protein. Moreover, PRB mutants, with no sumoylation ability, were shown to be resistant to progesterone-induced degradation by ubiquitin-proteasome (data not shown), suggesting that SUMO-1 and ubiquitin compete for conjugation to the same lysines in PRB. Similar mechanisms have been observed for Mdm2 (53), the E3 ubiquitin ligase for p53 and IκBα (54).
PIAS3 protein repression of PRB transactivation
The specific mechanism by which PIAS3 protein inhibited PRB-activated transcription in vivo remains intriguing. Our results showed that the interaction between PRB and PIAS3 protein destabilized PRB nuclear localization. It was noted that PRB translocation to the nucleus seemed to be markedly inhibited by the interaction with PIAS3 after progesterone treatment (Figure 6B). However, evaluation of PRB and PIAS3 using confocal microscopy showed that within a short time period (0.5 h) PRB completely entered the nucleus, while at a later time point (2 h) PRB was partly cytosolic. Interestingly, co-localization of PR and PIAS3 was observed predominantly in the nucleus (Figure 6A). These data suggested it was unlikely that PIAS3 inhibited PRB translocation to the nucleus, but rather destabilized PRB retention in the nucleus. The results might explain, at least in part, the strong inhibition of PR transcriptional activity by PIAS3 in the presence of progesterone.
The PIAS family is known to inhibit DNA-binding activity (55). Consequently, it was important to determine whether PIAS3 could form a complex with PRB on DNA, and whether the presence of PIAS3 could, in part, affect PR DNA-binding activity. Data from ChIP and gel shift assays demonstrated that PIAS3 was recruited to endogenous PR-responsive promoters and inhibited the DNA-binding activity of PR (Figure 7). We then hypothesized that PIAS3 inhibited the interaction of PR with its target DNA by binding to its DBD. It has been previously reported that the signal for nuclear export of PR is contained within its DBD domain, which functions as the export signal in multiple steroid receptor family members (56,57). Our data showed PIAS3 which influenced the nuclear export of PRB (Figure 6). One possible mechanism of action is that PIAS3 might contribute to the DBD structure, which augments recognition of the signal by the export machinery and by interacting with the DBD domain of PR.
Alternatively, it is still possible that the observed inhibition of PRB involved PIAS3-mediated SUMO modification of transcription regulators other than PRB, as PIAS3 overexpression might enhance the SUMO modification of many proteins. Although the precise identity of these SUMO conjugates is not known, they are likely to include numerous transcriptional regulators known to undergo SUMO modification (24,26,27,46). It is conceivable that the enhanced modification of some of these factors can indirectly regulate PRB activity. Consistent with this model, p53 (29) and LEF1 (31) are both inhibited by PIAS3 protein overexpression, independent of their ability to be directly modified by SUMO.
In summary, our findings demonstrated that PIAS3 protein significantly inhibited PR-mediated transactivation, which was accompanied by a strong induction of PR sumoylation in a progesterone-dependent manner. The pattern of PIAS3-triggered PRB SUMO modification was identified at three sites, Lys-7, Lys-388 and Lys-531. We further identified a novel role for Lys-7 and Lys-531 in the nuclear retention of PRB and the ligand-triggered transactivation of PRB. Our data demonstrated that PIAS3 inhibited gene activation by ligand-stimulated PRB in a manner that was independent of PRB SUMO modification. In addition, SUMO modifications of PRB significantly increased PRB stability, rendering it resistant to progesterone-induced degradation. Finally, we demonstrated that the mechanism by which PIAS3 protein strongly inhibited PRB-activated transcription was mediated by the interaction between PRB and PIAS3, which inhibited the DNA-binding activity of PRB and influenced its nuclear export.
SUPPLEMENTARY MATERIAL
Supplemenatry data are available at NAR Online.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science foundation of China (30321003) and granted from National 973 Project (2004CB518800). Funding to pay the Open Access publication charges for this article was provided by National 973 Project (2004CB518800).
Conflict of interest statement. None declared.
REFERENCES
- 1.Evans R. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sartorius C., Groshong S., Miller L., Powell R., Tung L., Takimoto G., Horwitz K. New T47D breast cancer cell lines for the independent study of progesterone B- and A-receptors: only antiprogestin-occupied B-receptors are switched to transcriptional agonists by cAMP. Cancer Res. 1994;54:3868–3877. [PubMed] [Google Scholar]
- 3.Tung L., Shen T., Abel M., Powell R., Takimoto G., Sartorius C., Horwitz K. Mapping the unique activation function 3 in the progesterone B-receptor upstream segment. Two LXXLL motifs and a tryptophan residue are required for activity. J. Biol. Chem. 2001;276:39843–39851. doi: 10.1074/jbc.M106843200. [DOI] [PubMed] [Google Scholar]
- 4.Horwitz K., Alexander P. In situ photolinked nuclear progesterone receptors of human breast cancer cells: subunit molecular weights after transformation and translocation. Endocrinology. 1983;113:2195–2201. doi: 10.1210/endo-113-6-2195. [DOI] [PubMed] [Google Scholar]
- 5.Krett N., Wei L., Francis M., Nordeen S., Gordon D., Wood W., Horwitz K. Human progesterone A-receptors can be synthesized intracellularly and are biologically functional. Biochem. Biophys. Res. Commun. 1988;157:278–285. doi: 10.1016/s0006-291x(88)80044-5. [DOI] [PubMed] [Google Scholar]
- 6.Kastner P., Krust A., Turcotte B., Stropp U., Tora L., Gronemeyer H., Chambon P. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J. 1990;9:1603–1614. doi: 10.1002/j.1460-2075.1990.tb08280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdel-Hafiz H., Takimoto G.S., Tung L., Horwitz K.B. The inhibitory function in human progesterone receptor N termini binds SUMO-1 protein to regulate autoinhibition and transrepression. J. Biol. Chem. 2002;277:33950–33956. doi: 10.1074/jbc.M204573200. [DOI] [PubMed] [Google Scholar]
- 8.Tung L., Mohamed M., Hoeffler J., Takimoto G., Horwitz K. Antagonist-occupied human progesterone B-receptors activate transcription without binding to progesterone response elements and are dominantly inhibited by A-receptors. Mol. Endocrinol. 1993;7:1256–1265. doi: 10.1210/mend.7.10.8123133. [DOI] [PubMed] [Google Scholar]
- 9.Sartorius C., Tung L., Takimoto G., Horwitz K. Antagonist-occupied human progesterone receptors bound to DNA are functionally switched to transcriptional agonists by camp. J. Biol. Chem. 1993;268:9262–9266. [PubMed] [Google Scholar]
- 10.Sartorius C., Melville M., Hovland A., Tung L., Takimoto G., Horwitz K. A third transactivation function (AF3) of human progesterone receptors located in the unique N-terminal segment of the B-isoform. Mol. Endocrinol. 1994;8:1347–1360. doi: 10.1210/mend.8.10.7854352. [DOI] [PubMed] [Google Scholar]
- 11.Richer J., Jacobsen B., Manning N., Abel M., Wolf D., Horwitz K. Differential gene regulation by the two progesterone receptor isoforms in human breast cancer cells. J. Biol. Chem. 2002;277:5209–5218. doi: 10.1074/jbc.M110090200. [DOI] [PubMed] [Google Scholar]
- 12.Arnett-Mansfield R.L., de Fazio A., Wain G., Jaworski R., Byth K., Mote P., Clarke C. Relative expression of progesterone receptors A and B in endometrioid cancers of the endometrium. Cancer Res. 2001;61:4576–4582. [PubMed] [Google Scholar]
- 13.Smith D.F., Faber L.E., Toft D.O. Purification of unactivated progesterone receptor and identification of novel receptor-associated proteins. J. Biol. Chem. 1990;265:3996–4003. [PubMed] [Google Scholar]
- 14.DeMarzo A.M., Beck C.A., Onate S.A., Edwards D.P. Dimerization of mammalian progesterone receptors occurs in the absence of DNA and is related to the release of the 90-kDa heat shock protein. Proc. Natl Acad. Sci. USA. 1991;88:72–76. doi: 10.1073/pnas.88.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKenna N.J., O'Malley B.W. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 16.Bagchi M.K., Elliston J.F., Tsai S.Y., Edwards D.P., Tsai M.J., O'Malley B.W. Steroid hormone-dependent interaction of human progesterone receptor with its target enhancer element. Mol. Endocrinol. 1988;2:1221–1229. doi: 10.1210/mend-2-12-1221. [DOI] [PubMed] [Google Scholar]
- 17.Tsai M.J., O'Malley B.W. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu. Rev. Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 18.Weigel N.L. Steroid hormone receptors and their regulation by phosphorylation. Biochem. J. 1996;319:657–667. doi: 10.1042/bj3190657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nirmala P.B., Thampan R.V. Ubiquitination of the rat uterine estrogen receptor: dependence on estradiol. Biochem. Biophys. Res. Commun. 1995;213:24–31. doi: 10.1006/bbrc.1995.2093. [DOI] [PubMed] [Google Scholar]
- 20.Syvala H., Vienonen A., Zhuang Y.H., Kivineva M., Ylikomi T., Tuohimaa P. Evidence for enhanced ubiquitin-mediated proteolysis of the chicken progesterone receptor by progesterone. Life Sci. 1998;63:1505–1512. doi: 10.1016/s0024-3205(98)00417-2. [DOI] [PubMed] [Google Scholar]
- 21.Lange C.A., Shen T., Horwitz K.B. Phosphorylation of human progesterone receptors at serine-294 by mitogen-activated protein kinase signals their degradation by the 26S proteasome. Proc. Natl Acad. Sci. USA. 2000;97:1032–1037. doi: 10.1073/pnas.97.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X.Y., Boudjelal M., Xiao J.H., Peng Z.H., Asuru A., Kang S., Fisher G.J., Voorhees J.J. 1,25-Dihydroxyvitamin D3 increases nuclear vitamin D3 receptors by blocking ubiquitin/proteasome-mediated degradation in human skin. Mol. Endocrinol. 1999;13:1686–1694. doi: 10.1210/mend.13.10.0362. [DOI] [PubMed] [Google Scholar]
- 23.Boudjelal M., Wang Z., Voorhees J.J., Fisher G.J. Ubiquitin/proteasome pathway regulates levels of retinoic acid receptor gamma and retinoid X receptor alpha in human keratinocytes. Cancer Res. 2000;60:2247–2252. [PubMed] [Google Scholar]
- 24.Hilgarth R.S., Murphy L.A., Skaggs H.S., Wilkerson D.C., Xing H.Y., Sarge K.D. Regulation and function of SUMO modification. J. Biol. Chem. 2004;279:53899–53902. doi: 10.1074/jbc.R400021200. [DOI] [PubMed] [Google Scholar]
- 25.Yang S.H., Jaffray E., Senthinathan B., Hay R.T., Sharrocks A.D. SUMO and transcriptional repression: dynamic interactions between the MAP kinase and SUMO pathways. Cell Cycle. 2003;2:528–530. doi: 10.4161/cc.2.6.597. [DOI] [PubMed] [Google Scholar]
- 26.Verger A., Perdomo J., Crossley M. Modification with SUMO: a role in transcriptional regulation. EMBO J. 2003;4:137–142. doi: 10.1038/sj.embor.embor738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chauchereau A., Amazit L., Quesne M., Guiochon-Mantel A., Milgrom E. Sumoylation of the progesterone receptor and of the steroid receptor coactivator SRC-1. J. Biol. Chem. 2003;278:12335–12343. doi: 10.1074/jbc.M207148200. [DOI] [PubMed] [Google Scholar]
- 28.Kahyo T., Nishida T., Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell. 2001;8:713–718. doi: 10.1016/s1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt D., Muller S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl Acad. Sci. USA. 2002;99:2872–2877. doi: 10.1073/pnas.052559499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kotaja N., Karvonen U., Janne O.A., Palvimo J.J. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 2002;22:5222–5234. doi: 10.1128/MCB.22.14.5222-5234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sachdev S., Bruhn L., Sieber H., Pichler A., Melchior F., Grosschedl R. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 2001;15:3088–3103. doi: 10.1101/gad.944801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gross M., Yang R., Top I., Gasper C., Shuai K. PIASy-mediated repression of the androgen receptor is independent of sumoylation. Oncogene. 2004;23:3059–3066. doi: 10.1038/sj.onc.1207443. [DOI] [PubMed] [Google Scholar]
- 33.Nishida T., Yasuda H. PIAS1 and PIASxα function as SUMO-E3 ligases toward androgen receptor and repress androgen receptor-dependent transcription. J. Biol. Chem. 2002;277:41311–41317. doi: 10.1074/jbc.M206741200. [DOI] [PubMed] [Google Scholar]
- 34.Gross M., Liu B., Tan J.A., French F.S., Carey M., Shuai K. Distinct effects of PIAS proteins on androgen-mediated gene activation in prostate cancer cells. Oncogene. 2001;20:3880–3887. doi: 10.1038/sj.onc.1204489. [DOI] [PubMed] [Google Scholar]
- 35.Sentis S., Romancer M.L., Bianchin C., Rostan M.C., Corbo L. Sumoylation of the estrogen receptor α hinge region regulates its transcriptional activity. Mol. Endocrinol. 2005;19:2671–2684. doi: 10.1210/me.2005-0042. [DOI] [PubMed] [Google Scholar]
- 36.Sicinski P., Weinberg R.A. A specific role for cyclin D1 in mammary gland development. J. Mammary Gland Biol. Neoplasia. 1997;2:335–342. doi: 10.1023/a:1026391128117. [DOI] [PubMed] [Google Scholar]
- 37.Sutherland R.L., Musgrove E.A. Cyclin D1 and mammary carcinoma: new insights from transgenic mouse models. Breast Cancer Res. 2002;4:14–17. doi: 10.1186/bcr411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Groshong S.D., Owen G.I., Grimison B., Schauer I.E., Todd M.C., Langan T.A., Sclafani R.A., Lange C.A., Horwitz K.B. Biphasic regulation of breast cancer cell growth by progesterone: role of the cyclin-dependent kinase inhibitors, p21 and p27 (Kip1) Mol. Endocrinol. 1997;11:1593–1607. doi: 10.1210/mend.11.11.0006. [DOI] [PubMed] [Google Scholar]
- 39.Kotaja N., Aittomaki S., Silvennoinen O., Palvimo J.J., Janne O.A. ARIP3 (androgen receptor-interacting protein 3) and other PIAS (protein inhibitor of activated STAT) proteins differ in their ability to modulate steroid receptor- dependent transcriptional activation. Mol. Endocrinol. 2000;14:1986–2000. doi: 10.1210/mend.14.12.0569. [DOI] [PubMed] [Google Scholar]
- 40.Nakagawa K., Yokosawa H. PIAS3 induces SUMO-1 modification and transcriptional repression of IRF-1. FEBS Lett. 2002;530:204–208. doi: 10.1016/s0014-5793(02)03486-5. [DOI] [PubMed] [Google Scholar]
- 41.Hager G.L., Nagaich A.K., Johnson T.A., Walker D.A., John S. Dynamics of nuclear receptor movement and transcription. Biochim. Biophys. Acta. 2004;1677:46–51. doi: 10.1016/j.bbaexp.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 42.Qiu M., Olesn A., Faivre E., Horwitz K.B., Lange C.A. Mitogen-activated protein kinase regulates nuclear association of human progesterone receptors. Mol. Endocrinol. 2003;17:628–642. doi: 10.1210/me.2002-0378. [DOI] [PubMed] [Google Scholar]
- 43.Wan Y.H., Coxe K.K., Thackray V.G., Housley P.R., Nordeen S.K. Separable features of the ligand-binding domain determine the differential subcellular localization and ligand-binding specificity of glucocorticoid receptor and progesterone receptor. Mol. Endocrinol. 2001;15:17–31. doi: 10.1210/mend.15.1.0584. [DOI] [PubMed] [Google Scholar]
- 44.Wardell S.E., Boonyaratanakornkit V., Adelman J.S., Aronheim A., Edwards D.P. Jun dimerization protein 2 functions as a progesterone receptor N-terminal domain coactivator. Mol. Cell. Biol. 2002;22:5451–5466. doi: 10.1128/MCB.22.15.5451-5466.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan J., Hall S.H., Hamil K.G., Grossman G., Petrusz P., Liao J., Shuai K., French F.S. Protein inhibitor of activated STAT-1 (signal transducer and activator of transcription-1) is a nuclear receptor coregulator expressed in human testis. Mol. Endocrinol. 2000;14:14–26. doi: 10.1210/mend.14.1.0408. [DOI] [PubMed] [Google Scholar]
- 46.Johnson E.S. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 47.Megidish T., Xu J.H., Xu C.W. Activation of p53 by protein inhibitor of activated Stat1 (PIAS1) J. Biol. Chem. 2002;277:8255–8259. doi: 10.1074/jbc.C200001200. [DOI] [PubMed] [Google Scholar]
- 48.Rogers R.S., Horvath C.M., Matunis M.J. SUMO modification of STAT1 and its role in PIAS-mediated inhibition of gene activation. J. Biol. Chem. 2003;278:30091–30097. doi: 10.1074/jbc.M301344200. [DOI] [PubMed] [Google Scholar]
- 49.Yang S.H., Sharrocks A.D. PIASx acts as an Elk-1 coactivator by facilitating derepression. EMBO J. 2005;24:2161–2171. doi: 10.1038/sj.emboj.7600690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharrocks A.D. PIAS proteins and transcriptional regulation—more than just SUMO E3 ligases? Genes Dev. 2006;20:754–758. doi: 10.1101/gad.1421006. [DOI] [PubMed] [Google Scholar]
- 51.Terui Y., Saad N., Jia S.D., McKeon F., Yuan J.Y. Dual role of sumoylation in the nuclear localization and transcriptional activation of NFAT1. J. Biol. Chem. 2004;279:28257–28265. doi: 10.1074/jbc.M403153200. [DOI] [PubMed] [Google Scholar]
- 52.Carol S.L., Christopher T.B., Htun H., Xian W.J., Irie M., Catharine L.S., Gordon L.H. Differential localization and activity of the A- and B-forms of the human progesterone receptor using green fluorescent protein chimeras. Mol. Endocrinol. 1999;13:366–375. doi: 10.1210/mend.13.3.0247. [DOI] [PubMed] [Google Scholar]
- 53.Buschmann T., Fuchs S.Y., Lee C.G., Pan Z.Q., Ronai Z. SUMO-1 modification of Mdm2 prevents its self-ubiquitination and increases Mdm2 ability to ubiquitinate p53. Cell. 2000;101:753–762. doi: 10.1016/s0092-8674(00)80887-9. [DOI] [PubMed] [Google Scholar]
- 54.Desterro J.M., Rodriguez M.S., Hay R.T. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 55.Shuai K., Liu B. Regulation of gene activation pathways by PIAS proteins in the immune system. Nat. Immunol. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- 56.Black B.E., Holaska J.M., Rastinejad F., Paschal B.M. DNA binding domains in diverse nuclear receptors function as nuclear export signals. Curr. Biol. 2001;11:1749–1758. doi: 10.1016/s0960-9822(01)00537-1. [DOI] [PubMed] [Google Scholar]
- 57.Holaska J.M., Black B.E., Love D.C., Hanover J.A., Leszyk J., Paschal B.M. Calreticulin is a receptor for nuclear export. J. Cell Biol. 2001;152:127–140. doi: 10.1083/jcb.152.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.