Abstract
Glutamate racemase (MurI) catalyses the conversion of l-glutamate to d-glutamate, an important component of the bacterial cell wall. MurI from Escherichia coli inhibits DNA gyrase in presence of the peptidoglycan precursor. Amongst the two-glutamate racemases found in Bacillus subtilis, only one inhibits gyrase, in absence of the precursor. Mycobacterium tuberculosis has a single gene encoding glutamate racemase. Action of M.tuberculosis MurI on DNA gyrase activity has been examined and its mode of action elucidated. We demonstrate that mycobacterial MurI inhibits DNA gyrase activity, in addition to its precursor independent racemization function. The inhibition is not species-specific as E.coli gyrase is also inhibited but is enzyme-specific as topoisomerase I activity remains unaltered. The mechanism of inhibition is different from other well-known gyrase inhibitors. MurI binds to GyrA subunit of the enzyme leading to a decrease in DNA-binding of the holoenzyme. The sequestration of the gyrase by MurI results in inhibition of all reactions catalysed by DNA gyrase. MurI is thus not a typical potent inhibitor of DNA gyrase and instead its role could be in modulation of the gyrase activity.
INTRODUCTION
Topoisomerases are essential enzymes, responsible for maintenance of the level of supercoiling of DNA in cells. DNA gyrase is unique amongst all topoisomerases in its ability to catalyze negative supercoiling of DNA in an ATP dependent fashion (1–3). Besides supercoiling, the enzyme catalyzes catenation/decatenation and knotting/unknotting reactions in vitro (4,5). It is also known to relax negatively supercoiled DNA in absence of ATP (6). The functional holoenzyme is a heterotetramer (A2B2) comprising of two GyrA and GyrB subunits (7,8).
DNA gyrase is an indispensable enzyme in prokaryotes and is a proven target for diverse classes of antibacterial agents. Mechanistically, gyrase inhibitors have been classified mainly into two broad categories. The first category includes coumarins and cyclothialidines that inhibit ATP hydrolysis catalysed by DNA gyrase. These antibiotics bind to GyrB at a region overlapping to ATP binding site, thus preventing ATP binding. As a result, they inhibit only the supercoiling activity of the enzyme with no effect on the relaxation activity (9,10). The second class includes the synthetic quinolones, which function as gyrase poisons, by stabilizing enzyme–DNA covalent intermediates (9,10). The protein–DNA adducts hinder the progress of replication and transcription complexes (11,12). They also lead to widespread chromosome fragmentation due to the release of DNA ends from the ternary complexes, resulting in rapid quinolone-mediated cell death (13). In addition, proteinaceous toxins such as ribosomally synthesized peptide antibiotic, microcin B17 (14), ParE from RK2 plasmid (15) and CcdB encoded by F plasmid (16) inhibit gyrase by arresting the enzyme–DNA covalent intermediates, leading to the accumulation of double-strand breaks, upon removal of the protein constraints. New inhibitors of DNA gyrase have been reported recently. These proteins appear to inhibit DNA gyrase in a manner distinct from the other two classes of inhibitors. For example, GyrI from Escherichia coli (17,18), MfpA from Mycobacterium sp. (19,20) inhibit DNA gyrase by interfering with gyrase–DNA interaction.
Glutamate racemase (MurI) catalyses the conversion of l-glutamate to d-glutamate, an essential component of the peptidoglycan. Besides racemization activity, E.coli MurI possesses an additional DNA gyrase inhibitory function. The inhibition of DNA gyrase requires the presence of peptidoglycan precursor (21). Studies with Bacillus subtilis revealed that it possesses two genes encoding glutamate racemases, the poly-gamma-glutamate synthesis-linking Glr enzyme and YrpC (22,23). Only the YrpC (MurI) isozyme, but not the Glr, negatively influences the activity of DNA gyrase, that too in the absence of the precursor (24).
Mycobacterium tuberculosis genome sequence revealed the presence of a single gene for glutamate racemase. Further, mycobacterial DNA gyrase shows distinctive features with respect to quinolone susceptibility and resistance to the action of toxins like CcdB and microcin B17 (25–27). Therefore, in this study, we have examined the effect of glutamate racemase from M.tuberculosis on DNA gyrase activity and elucidate its mechanism of action.
MATERIALS AND METHODS
Bacterial strains and plasmids
E.coli strains DH5α and BL26(DE3) were used for cloning and overexpression of mycobacterial MurI respectively. Genomic DNA from M.tuberculosis H37Ra was isolated by the method described earlier (28). The murI gene was cloned in pET11d vector. pBR322, pUC18 plasmids were used for the biochemical assays.
Enzyme and substrate preparation
E.coli DNA gyrase subunits, GyrA and GyrB were purified as described previously (29). Mycobacterium smegmatis DNA gyrase was purified as described previously (30). Specific activity of purified DNA gyrase was defined to be 1 U as the amount of the enzyme required to supercoil 300 ng of relaxed pUC18 DNA at 37°C in 30 min. Supercoiled pUC18 and pBR322 were prepared by standard DNA purification protocols (31). M.smegmatis topoisomerase I and relaxed pUC18 were prepared as described in (32). E.coli topoisomerase I was purified as described previously (33).
Cloning of murI
The murI gene was PCR amplified using M.tuberculosis H37Ra genomic DNA as a template and primers (forward primer 5′-GAAGTCATGAATTCGCCGTTG) and (reverse primer 5′-AAGATCTCTTCCATGGCCTAATG) containing RcaI and BglII sites, respectively. The amplification reaction was carried out with Pfu DNA polymerase, RcaI and BglII digested PCR product was ligated to NcoI-BamHI cut pET11d vector.
Expression and purification of MurI
The recombinant mycobacterial MurI was overexpressed from pET11d construct in E.coli BL26 (DE3) strain. Cells were harvested and resuspended in a buffer containing 50 mM Tris–HCl pH 8.0, 1 mM dl-glutamic acid, 0.1 mM phenylmethyl-sulfonyl fluoride, 0.5 mM MgCl2 and disrupted by sonication. The extract was centrifuged at 20 000 g for 30 min at 4°C (S20 fraction). The S20 pellet fraction containing MurI was subjected to 12% denaturing PAGE (SDS–PAGE). The band corresponding to the overexpressed protein was excised and eluted from the gel using a Bio-Rad electroelutor. The eluted protein was then subjected to acetone-precipitation to remove SDS, denatured in buffer containing 50 mM Tris–HCl pH 8.0, 0.2% 2-mercaptoethanol, 0.1 mM phenyl methane sulfonyl fluoride and 6 M urea. Subsequently the protein was renatured by stepwise dialysis against buffer containing 4, 2 and 1 M urea, respectively. Finally, the purified protein was dialyzed against the same buffer lacking urea.
Racemization activity
The racemization activity of the purified glutamate racemase was assessed as described previously (34). Purified MurI samples were incubated in presence of d-glutamate and then rapidly heated to inactivate the enzyme and assayed for l-glutamate using NAD+/l-glutamate dehydrogenase (GDH). Varying concentrations (1 and 2 μM) of MurI were incubated with 10 mM d-glutamate in a buffer containing 100 mM Tris–HCl pH 8.0, 2 mM DTT at 37°C for 30 min. Samples were then heated at 95°C for 15 min. Denatured protein was removed by centrifugation for 10 min at 14 000 r.p.m. The l-glutamate formed was then measured by adding 5 mM of NAD+ and 10 U of GDH. Increase in absorbance at 340 nm was monitored for 6 min at 25°C using Beckman DU640 UV/vis spectrophotometer. One aliquot of purified MurI sample (2 μM) was treated with 400 μM methyl methanethiosulfonate (MMTS, cysteine modifying agent) at 37°C for 30 min, dialyzed and then assessed for its racemization activity.
DNA supercoiling, relaxation and decatenation reactions
Supercoiling assays were carried out at 37°C with 300 ng of relaxed pUC18 and 10 nM DNA gyrase from either E.coli or M.smegmatis, in supercoiling buffer [35 mM Tris–HCl pH 7.5, 5 mM MgCl2, 25 mM potassium glutamate, 2 mM spermidine, 2 mM ATP, 50 mg/l BSA and 90 mg/l yeast tRNA in 5% (v/v) glycerol]. Relaxation assays were carried out with 75 and 150 nM of E.coli enzyme using supercoiled pBR322 as the substrate in the supercoiling buffer devoid of ATP. The reactions were carried out either in presence of MurI or BSA (control) for 60 min at 37°C and terminated with 0.6% SDS. Decatenation reactions were carried out with 300 ng kinetoplast DNA and 100 nM enzyme in supercoiling buffer containing 10 mM MgCl2, at 37°C for 60 min. Relaxation assays with topoisomerase I were carried out in buffer containing 20 mM Tris–HCl pH 7.4, 40 mM NaCl, and 5 mM MgCl2. 20 nM of either E.coli or M.smegmatis topoisomerase I were incubated with supercoiled pUC18 DNA either in presence of MurI or BSA. The reactions were carried out at 37°C for 60 min and stopped with 0.6% SDS. The assay mixtures were resolved on 1% agarose gel in 40 mM Tris–actetate buffer containing 1 mM EDTA.
Electrophoretic mobility shift assay
Assays were carried out using a 240 bp DNA fragment encompassing the strong gyrase site (SGS) from pBR322 (35). The DNA fragment was PCR amplified with the end labeled forward primer. End-labeling was performed with [γ-32P]ATP (3000 Ci/mmol) and T4 polynucleotide kinase. In order to assess the effect of MurI on gyrase–DNA non-covalent complex, labeled DNA (1 × 10−9 M) was incubated with 100 nM of DNA gyrase either in absence or presence of different concentrations of MurI for 30 min at 4°C followed by electrophoresis on 4% native polyacrylamide gel in 45 mM Tris–borate buffer containing 5 mM MgCl2 at 4°C. For assessing gyrase–DNA covalent complex, the reactions were carried out with 40 nM DNA gyrase and in presence of 30 μg/ml ciprofloxacin at 30°C. One of these reaction mix was treated with 0.2% SDS followed by proteinase K (90 μg/ml) digestion for 30 min. The samples were electrophoresed on 4% native polyacrylamide gel in 45 mM Tris–borate buffer at 25°C. The free DNA and bound complexes were then quantitated using a phosphorimager.
Cleavage reactions
DNA cleavage assays were carried out in the supercoiling buffer with supercoiled pBR322 substrate for 30 min at 30°C and the gyrase–DNA complex was trapped by adding 0.2% SDS followed by proteinase K (90 μg/ml) digestion for 30 min. In case of drug-induced cleavage reactions, ciprofloxacin (30 μg/ml) was included in the assay. The reaction mixtures were then resolved on 1% agarose gel. Cleavage reactions using radiolabelled DNA substrate (240 bp SGS) were performed in a similar fashion. The reaction buffer contained 5 mM Ca2+ ions instead of Mg2+ ions, while monitoring the calcium-induced cleavage. The reaction products were resolved on 8% denaturing polyacrylamide gel (urea-PAGE). The substrate and cleaved DNA products were then quantitated using phosphorimager.
ATPase activity measurements
The ATPase reactions were carried out in supercoiling buffer containing 2 mM ATP, 10 μg ml−1 DNA (240 bp from pBR322) and 0.02 μCi of [γ-32P]ATP (3000 Ci/mmol). The DNA-stimulated ATPase activity was monitored with 40 nM DNA gyrase either in absence or presence of varying amounts of MurI. To assess the intrinsic ATPase activity, 1.4 μM GyrB was used and GyrA as well as DNA were omitted from the assay mixture. The assays were terminated by adding equal volume of chloroform. The aqueous layer (1.0 μl) was resolved on polyethyleneimine-cellulose thin layer chromatography with 1.2 M LiCl, and 0.1 mM EDTA. The spots corresponding to ATP and Pi were quantitated using a phosphorimager.
Far-western analysis
Thirty-four picomoles each of MurI and KpnI Restriction endonuclease (used as a negative control) were resolved in 12% SDS–polyacrylamide gel and then transferred onto nitrocellulose membrane. The proteins were then allowed to refold on the membrane followed by blocking with buffer containing 10 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 2% (w/v) BSA. The membrane was then incubated with 200 pmol of either mycobacterial GyrA or GyrB subunits individually, washed three times with phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4) containing 0.1% Tween-20 [PBST]. The membrane was then incubated with either anti-GyrA rabbit IgG in case of GyrA or anti-GyrB rabbit IgG at 1:10 000 dilutions followed by three washes with PBST. The membrane was incubated with secondary peroxidase-conjugated anti-rabbit antibody at 1:20 000 dilution. ECLplus (Amersham) was used to detect the bound secondary antibody.
Surface plasmon resonance
E.coli GyrA was immobilized on a CM5 sensor surface via amine-coupling in acetate buffer (pH 3.0). The surface was blocked with ethanolamine hydrochloride. The interaction was assessed in a buffer containing 35 mM Tris–HCl pH8.0, 1 mM EDTA, 0.05% Tween-20, 100 mM NaCl. Varying amounts of MurI were passed over the immobilized GyrA and the subsequent changes in the resonance units were recorded in a BIAcore 2000 system (Pharmacia). All the proteins were dialyzed against the buffer (35 mM Tris–HCl pH 8.0, 1 mM EDTA) prior to the experiment. 10 mM NaOH was used for surface regeneration.
RESULTS
Purification and racemization activity of purified MurI
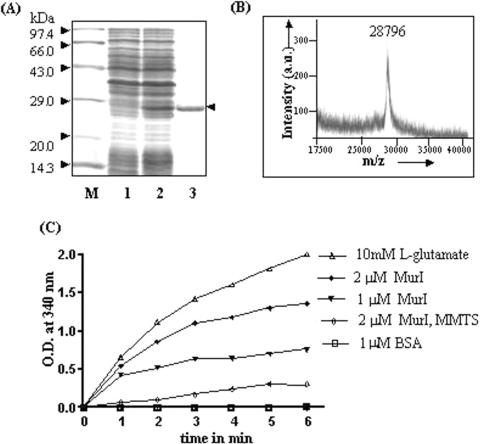
M.tuberculosis MurI was overexpressed in E.coli and purified from the inclusion bodies as described in the Materials and Methods section (Figure 1A). The authenticity of the protein was verified by tryptic mass fingerprinting analysis (data not shown). MALDI-TOF analysis revealed the molecular mass of the purified protein to be 28 796 Da (Figure 1B). The racemization activity was assessed by monitoring the absorbance of NADH at 340 nm. For this, MurI was initially incubated in presence of d-glutamate. l-Glutamate formed as a result of MurI racemase activity was measured by adding NAD+ and GDH. GDH-mediated conversion from l-glutamate to α-ketoglutarate led to the reduction of NAD+ to NADH. As shown in Figure 1C, the samples incubated with MurI, showed a significant increase in OD at 340 nm with time in a dose-dependent manner. The reaction with l-glutamate as a substrate served as a positive control while the reaction in presence of BSA and d-glutamate served as a negative control. Glutamate racemases are known to employ two active-site cysteine residues as acid/base catalysts during the interconversion of glutamate enantiomers (36). MMTS was used to modify the cysteine residues of M.tuberculosis MurI. MMTS-treated MurI was compromised in its racemization function revealing the importance of cysteine residues in catalysis (Figure 1C). The racemase activity of the enzyme was also monitored by circular dichroism spectra at 204 nm (data not shown). The results showed that M.tuberculosis MurI racemization activity does not require any peptidoglycan precursor, similar to B.subtilis glutamate racemases (22,23). As with other glutamate racemases, the activity is also independent of cofactor requirement.
Figure 1.
(A) Expression profile of MurI, M: protein molecular weight marker, lane 1: BL26 cell extract harbouring vector pET11d, lane 2: BL26 cell extract harbouring pET11d-murI construct, induced with 0.3 mM IPTG, lane 3: purified MurI; (B) molecular weight of M.tuberculosis MurI determined by MALDI-TOF analysis, (C): Racemization activity of MurI. The assay was carried out as described in Materials and Methods section. Reduction of NAD to NADH during the course of the reaction was monitored by a measuring absorbance at 340 nm. □: reaction with 1 μM BSA, ▾: reaction with 1 μM MurI and •: reaction with 2 μM MurI, ⋄: reaction with MMTS treated 2 μM MurI, ▵: in presence of 10 mM l-glutamate substrate.
MurI inhibits DNA gyrase activity
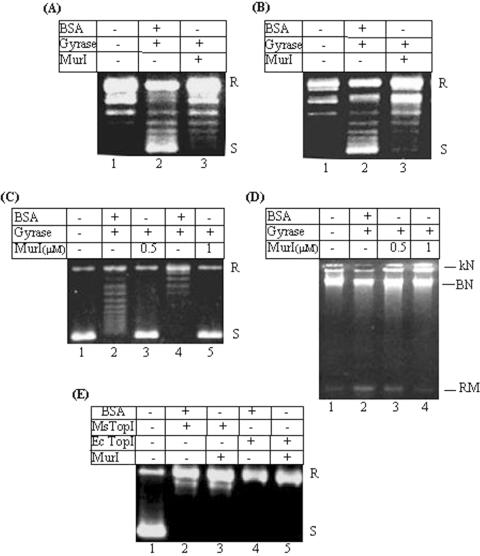
In order to test the effect of MurI on supercoiling activity, M.smegmatis DNA gyrase was preincubated with MurI for 15 min on ice prior to the addition of DNA substrate (relaxed pUC18) as described in the Materials and Methods section. The results presented in Figure 2A show that MurI inhibits supercoiling activity of mycobacterial DNA gyrase. When its effect on E.coli DNA gyrase was tested, mycobacterial MurI was observed to inhibit even the E.coli enzyme (Figure 2B). The inhibition of DNA gyrase by MurI is thus not species-specific in contrast to other proteinaceous inhibitors such as CcdB, microcin B17 (25).
Figure 2.
Effect of MurI on activities of topoisomerases: Inhibition of DNA gyrase activities by MurI (A–D). Effect on supercoiling activities of (A) mycobacterial DNA gyrase and (B) E.coli DNA gyrase respectively. 10 nM DNA gyrase used for the supercoiling reaction; lane 1: relaxed pUC18, lane 2: gyrase in presence of 1 μM BSA, lane 3: gyrase in presence of 1 μM MurI; (C) Relaxation activity. Lane 1: supercoiled pBR322, lanes 2 and 4: 75 and 150 nM E.coli DNA gyrase in presence of 1 μM BSA respectively; lanes 3 and 5: gyrase in presence of 0.5 and 1 μM MurI respectively; (D) Decatenation activity. 100 nM E.coli DNA gyrase used for the reactions, lane 1: kinetoplast DNA, lane 2: DNA gyrase and 1 μM BSA, lanes 3 and 4: DNA gyrase in presence of 0.5 and 1 μM MurI respectively. (E) Effect on topoisomerase I activity. 20 nM of M.smegmatis and E.coli topoisomerase I were used for the relaxation assays, lane 1: supercoiled pUC18; lane 2: M.smegmatis topoisomerase I and 1 μM BSA; lane 3: M.smegmatis topoisomerase I with 1 μM MurI; lane 4: E.coli topoisomerase I and 1 μM BSA; lane 5: E.coli topoisomerase I and 1 μM MurI. S and R represent supercoiled and relaxed plasmid DNA, kN: kinetoplast, kDNA network, BN: broken network, RM: released minicircles. All the assays were repeated at least thrice. The representative figures have been presented.
Apart from the supercoiling reaction, DNA gyrase is known to catalyze other reactions in vitro, namely ATP-independent relaxation and decatenation reactions. To analyze the effect of MurI on gyrase-catalyzed relaxation, DNA gyrase was preincubated with MurI prior to the addition of supercoiled pBR322 DNA substrate. As shown in Figure 2C, MurI inhibited the relaxation activity as well. To monitor its effect on the decatenation activity of DNA gyrase, the reactions were carried out using the catenated kinetoplast DNA as substrate. From the data presented in Figure 2D, it is evident that MurI inhibited decatenation activity of DNA gyrase as well. Since all the three catalytic activities of DNA gyrase are inhibited by M.tuberculosis MurI, a step common to all these processes is likely to be the target for MurI action.
MurI has no effect on the topoisomerase I activity
Bacterial topoisomerase I relaxes negatively supercoiled DNA in an ATP-independent manner. In order to assess the effect of MurI on the relaxation activity of topoisomerase I, both M.smegmatis as well as E.coli topoisomerase I were preincubated with MurI and then supercoiled pUC18 DNA substrate was added for the relaxation assays as described in the Materials and Methods section. As shown in Figure 2E, the topoisomerase I-mediated relaxation reactions were not inhibited in presence of MurI. Therefore, MurI appears to be a specific inhibitor of DNA gyrase.
Probing the mechanism of inhibition
The reaction cycle of DNA gyrase involves a series of coordinated steps (2,3). After initial wrapping of DNA around the A2B2 complex, cleavage of the G segment DNA in both the strands results in the formation of a DNA–protein covalent complex. Cleavage reaction is followed by the passage of an intact duplex T segment DNA through the double-stranded break. Religation of the broken ends of DNA after the strand passage results in the introduction of two negative supercoils. Hydrolysis of ATP is required to reset the reaction cycle for catalytic enzyme turnover (3). Inhibitors for DNA gyrase, thus, could potentially interfere at any one of the steps in the gyrase reaction cycle to arrest the chain of events.
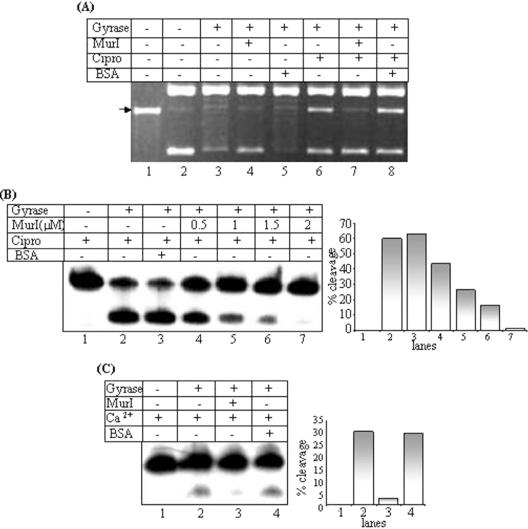
Effect of MurI on cleavage reaction
The quinolone class of DNA gyrase inhibitors (e.g. nalidixic acid and ciprofloxacin) inhibit both the supercoiling and relaxation reactions of DNA gyrase. They act by interfering with the rejoining of double-stranded breaks in DNA and promote the rate of double-stranded DNA cleavage by DNA gyrase (10). In order to test the effect of mycobacterial MurI at the cleavage step, DNA gyrase was preincubated with MurI and the cleavage assays were performed both with the supercoiled plasmid DNA as well as the radiolabelled linear DNA substrates. In Figure 3, effect of MurI on DNA gyrase-mediated cleavage has been summarized. Unlike ciprofloxacin, MurI did not stimulate DNA gyrase-mediated cleavage on its own (Figure 3A, compare lanes 4 and 6). Ciprofloxacin is known to arrest DNA gyrase at the cleavage step. SDS and proteinase K treatment removes the covalently attached protein and cleaved DNA fragment can be visualized on the gel. The extent of ciprofloxacin-induced cleavage on a supercoiled plasmid DNA substrate was monitored either in absence or presence of MurI. The drug-induced DNA cleavage was reduced in presence of MurI (Figure 3A, lanes 6–8). MurI also abrogated the cleavage even on a linear DNA substrate, in a dose-dependent manner (Figure 3B). Calcium (Ca2+) ions inhibit the religation and known to stimulate DNA cleavage activity of the enzyme (37). MurI exhibited a similar inhibitory effect on calcium-induced cleavage reaction (Figure 3C). From these observations, we conclude that MurI inhibits DNA gyrase by interfering with the DNA cleavage reaction or a step preceding it.
Figure 3.
Effect of MurI on DNA gyrase mediated cleavage reaction: (A) cleavage reactions with supercoiled pBR322 substrate, 50 nM E.coli DNA gyrase used; lane 1: linear substrate, lane 2: supercoiled substrate, lanes 3 and 6: DNA gyrase, lanes 4 and 7: gyrase in presence of 1 μM MurI, lanes 5 and 8: gyrase in presence of 1 μM BSA. Ciprofloxacin (30 μg/ml) added in the lanes 6–8; (B) cleavage with linear radiolabelled DNA (SGS). 100 nM E.coli DNA gyrase used; lane 1: 240 bp SGS, lane 2: DNA gyrase, lane 3: gyrase in presence of 2 μM BSA, lanes 4–7: gyrase in presence of increasing concentrations of MurI. Ciprofloxacin (30 μg/ml) added in all the lanes. The bar diagrams show the quantitative representations of the % cleavage observed in each lane; (C) cleavage reactions in presence of calcium ions; lane 1: 240 bp SGS lane 2: DNA gyrase, lane 3: gyrase in presence of 1 μM MurI, lane 4: gyrase in presence of 1 μM BSA. Mg2+ ions omitted and 5 mM Ca2+ ions added in the reactions. A quantitative representation is shown in the form of bar diagram adjacent to the figure. A representative set of data is presented based on several sets of experiments.
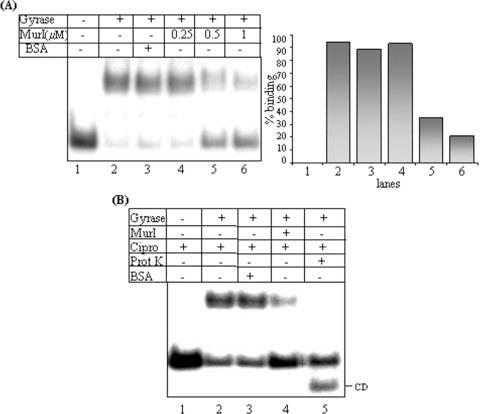
Effect of MurI on DNA-binding
Electrophoretic mobility shift assays (EMSAs) were employed to assess the effect of MurI on gyrase–DNA interaction. To monitor the effect of MurI on both the non-covalent and covalent gyrase–DNA complex formation, reactions were performed as described, either in absence or presence of ciprofloxacin and DNA gyrase was preincubated with MurI prior to the addition of DNA. As shown in Figure 4A, the amount of retarded gyrase–DNA non-covalent complex on the polyacrylamide gel was significantly reduced in presence of MurI, with concomitant increase in the free DNA species. MurI inhibited the formation of non-covalent enzyme–DNA complex in a dose-dependent manner. Similarly, the covalent gyrase–DNA complex formation was hindered by MurI (Figure 4B). From these results, it appears that by preventing gyrase–DNA interactions, M.tuberculosis MurI inhibits all the catalytic reactions of DNA gyrase.
Figure 4.
Effect of MurI on gyrase–DNA interaction: (A) Effect of MurI on non-covalent complex formation, EMSAs carried out with 100 nM E.coli DNA gyrase and radiolabelled 240 bp SGS at 4°C; lane1: free SGS, lane 2: DNA gyrase, lane 3: gyrase in presence of 1 μM BSA, lane 4–6: gyrase in presence of increasing concentrations of MurI. (B) Effect on gyrase–DNA covalent complex, 40 nM DNA gyrase incubated with radiolabelled SGS in presence of ciprofloxacin (30 μg/ml) at 30°C to arrest at the cleavage step and covalent enzyme–DNA complexes resolved on 4% native polyacrylamide gel; lane 1: free SGS, lane 2: DNA gyrase, lane 3: gyrase in presence of 1 μM BSA, lane 4: DNA gyrase in presence of 1 μM MurI, lane 5: gyrase–DNA covalent complex after proteinase K treatment; CD: cleaved DNA released after proteinase K treatment. All the assays were repeated at least thrice. The representative figures are shown.
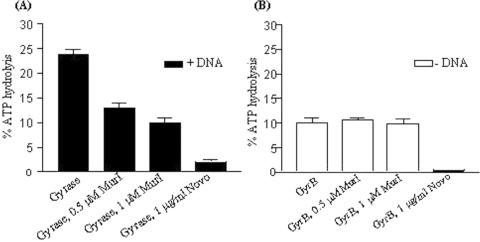
Effect of MurI on ATPase activity
GyrB subunit of DNA gyrase exhibits intrinsic ATPase activity, which is further enhanced in presence of GyrA and DNA (38,39). Coumarins and cyclothialidines inhibit supercoiling activity of the enzyme by interfering with ATPase activity (intrinsic as well as DNA-stimulated). If DNA-binding by gyrase is indeed the target of MurI action, the DNA-stimulated ATPase activity of the enzyme would be affected while its intrinsic activity would be unaltered. Reactions were carried out with DNA gyrase holoenzyme in presence of linear DNA substrate as described in the Materials and Methods section. As shown in Figure 5A, MurI inhibited the DNA-stimulated ATPase activity of DNA gyrase. To test the effect of MurI on intrinsic ATPase activity, reactions were carried out with prior incubation of MurI and only GyrB subunit. MurI had no effect on intrinsic ATPase activity of the enzyme, in contrast to the pattern observed with novobiocin (Figure 5B). Together, these data demonstrate that the MurI mode of inhibition is distinct from that of the ATPase inhibitors. These results also confirm that the inhibition of gyrase by MurI is at the step of DNA-binding.
Figure 5.
Effect of MurI on ATPase activity: (A) DNA-stimulated ATPase activity. Reactions were performed with 40 nM M.smegmatis DNA gyrase and 10 μg ml−1 DNA (240 bp SGS from pBR322), lane 1: gyrase, lanes 2 and 3:gyrase in presence of 0.5 and 1 μM MurI respectively, lane 4: gyrase in presence of 1 μg/ml novobiocin, (B) Intrinsic ATPase activity. Reactions were performed with 1.4 μM E.coli GyrB subunit. DNA and GyrA were omitted; lane 1: GyrB, lanes 2 and 3: GyrB in presence of 0.5 and 1 μM MurI respectively, lane 4: GyrB in presence of 1 μg/ml novobiocin. 2 mM ATP present in all the reactions. The average of three independent experiments is depicted graphically.
MurI interacts with GyrA subunit of DNA gyrase
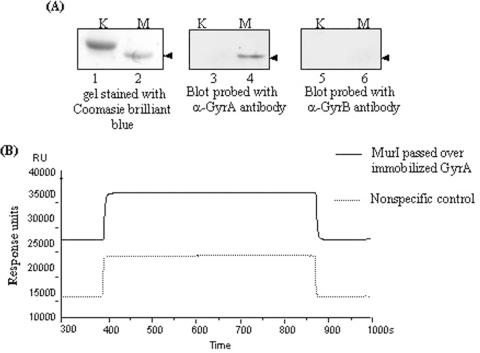
To investigate whether MurI-mediated inhibition is mediated by direct interaction between the two proteins, two experimental approaches were employed. In the far-western analysis presented in Figure 6A, upon co-incubation of immobilized MurI on the membrane with GyrA subunit, we detected a signal for GyrA at the position of MurI on the membrane. Similar analysis with GyrB subunit did not give any positive signal. In order to verify further the direct interaction, surface plasmon resonance reftractrometric (SPR) studies were performed. SPR experiments also revealed a direct interaction between MurI and GyrA subunit of DNA gyrase, with an increase of 60–70 RU upon binding of MurI to immobilized GyrA (Figure 6B). The physical interaction between MurI and DNA gyrase is therefore independent of the presence of GyrB subunit, DNA and ATP.
Figure 6.
Interaction between MurI and DNA gyrase: (A) Far-western analysis. 34 picomoles of MurI and KpnI restriction endonuclease immobilized on nitrocellulose membrane and then incubated with 3 μM of either mycobacterial GyrA or GyrB subunits and then probed with polyclonal antibodies raised against the mycobacterial DNA gyrase subunits. (lanes 1 and 2): gel stained with coomasie brilliant blue dye, showing the presence of KpnI (K) and MurI (M); (lanes 3 and 4): the nitrocellulose membrane with immobilized KpnI and MurI probed with anti-GyrA (α-GyrA) antibodies after incubation with GyrA, (lanes 5 and 6): the nitrocellulose membrane with immobilized KpnI and MurI probed with anti-GyrB (α-GyrB) antibodies after incubation with GyrB, (B) Surface plasmon resonance refractometry. Interaction was assessed in a buffer containing 35 mM Tris–HCl pH 8.0, 1 mM EDTA, 0.05% Tween-20, and 100 mM NaCl. (—) MurI passed over E.coli GyrA immobilized on CM5 sensor surface, (…….) non-specific control.
DISCUSSION
Mycobacterial glutamate racemase exhibits a dual role like its E.coli homologue. However, unlike the E.coli enzyme, mycobacterial MurI inhibits DNA gyrase in a precursor independent manner. In this respect, it is similar to the B.subtilis glutamate racemase, which affects DNA gyrase activity in absence of any precursor. Previous studies with E.coli and B.subtilis enzymes have not addressed the mechanism of inhibition of gyrase by glutamate racemase (21,24). Here, we demonstrate that the inhibition of gyrase by MurI is not species-specific. We have then investigated the mechanism of MurI-mediated inhibition of DNA gyrase. It binds to GyrA subunit and this interaction prevents gyrase from accessing the DNA substrate, thereby inhibiting all its catalytic reactions.
DNA gyrase is responsible for the maintenance of steady-state levels of negative supercoiling that is essential for chromosome condensation, transcription initiation and enzyme complex movement during replication and transcription. By virtue of being an essential enzyme, it is an ideal target for different classes of inhibitors. Amongst a large repertoire of the gyrase inhibitors, coumarins and quinolones have been studied extensively with respect to their mechanism of action (9,10). A noteworthy point is that new inhibitors with different mechanism of action are often discovered for DNA gyrase. Recently, a new class of antibiotics with an aminocoumarin moiety in its structure has been reported to inhibit gyrase in a mode distinct from the other known coumarins. Simocyclinone D8 inhibits an early step in gyrase catalytic cycle by preventing DNA-binding by the enzyme (40). Several proteinaceous inhibitors of DNA gyrase, discovered so far, could be categorized into two major groups. The first category includes the toxins like CcdB, microcin B17, ParE, which are encoded by the selfish plasmids to ensure their stable maintenance inside the cell (14–16). These toxins act as gyrase poisons and their mode of inhibition is akin to that of quinolones but not identical in all the details. The second group of inhibitors includes the chromosomally encoded inhibitors such as GyrI, MfpA and plasmid encoded Qnr. The quinolone resistance protein, Qnr, discovered from a clinical isolate of Klebsiella pnemoniae, binds gyrase holoenzyme thereby altering its DNA-binding property (41–43). Based on the crystal structure of MfpA, it is considered to be a DNA mimic, sequestering gyrase away from DNA (20). GyrI, an endogenous DNA gyrase inhibitor from E.coli binds to the holoenzyme and hinders the DNA-binding (18). None of these proteins are cytotoxic as their mode of inhibition is to prevent DNA gyrase from binding to DNA. Cytotoxicity usually arises due to accumulation of double-strand breaks in the genome. Bacterial cells tolerate some reduction of gyrase activity, whereas only few double-strand breaks in the genome could be lethal (44). In this context, amongst all the inhibitors of DNA gyrase studied so far, quinolones, which form toxic lesions, are the only commercially successful chemical entities.
From the present studies, it is clear that MurI mode of action resembles the other chromosomally encoded gyrase inhibitors. These inhibitors essentially influence the enzyme activity by sequestering the enzyme away from DNA. A comparative analysis of these proteinaceous inhibitors does not reveal a common motif or structural fold, which might be involved in their ability to inhibit DNA gyrase. Thus, at present, it appears that they belong to a group of very disparate proteins having a similar function as far as gyrase inhibition is concerned.
Apparently, presence of such endogenous inhibitors of an essential enzyme appears to be a severe burden for the cell. The intracellular optimal activity of DNA gyrase is very crucial for cell survival. These endogenous gyrase inhibitory proteins might be playing a role in regulating DNA gyrase activity. For example, in case of thioredoxins (TrxA and TrxC) from Rhodobacter sp., a change in oxygen tension influences their redox state, resulting in altered gyrase activity which in turn infuences the expression of puf and puc operon (45). GyrI in E.coli acts as an antidote to the plasmid encoded toxin microcin B17 and also involved in reducing DNA damages from diverse agents (18,46).
Glutamate racemase is the new member of chromosomally encoded gyrase inhibitors. Like others, it appears to sequester away DNA gyrase from its site of action. The physiological basis for MurI-mediated inhibition is not known. Since, mycobacterial MurI inhibits gyrase in absence of any peptidoglycan precursor; its expression would have to be under strict control to avoid any abnormality resulting from uncontrolled inhibition of gyrase. E.coli MurI is active and capable of inhibiting DNA gyrase, only upon accumulation of the peptidoglycan precursor (21). The gyrase inhibitory YrpC from B.subtilis is also poorly expressed in comparison to Glr, which is abundantly expressed but has no influence on DNA gyrase activity (24). When cell wall synthesis and chromosome segregation go hand in hand during cell division, gyrase activity may need to be controlled. The observed bifunctionality of the glutamate racemase might be a measure employed by the cell to avoid excess gyrase activity, rather than inhibiting gyrase in true sense. MurI-mediated modulation might be occurring transiently at the time of cell division to coordinate the process of cell wall biosynthesis and DNA replication.
Acknowledgments
The authors thank A. Maxwell for E.coli gyrase overexpression clones, H. K. Majumder for kDNA and acknowledge the Phosphorimager, Biacore and proteomics facilities supported by the Department of Biotechnology, Government of India. S.S. is the recipient of senior research fellowship from Council of Scientific and Industrial Research, Government of India. Funding to pay the Open Access publication charges for this article was provided by Funding for the work and to pay the Open Acess publication charges for this article was provided by grants from Department of Biotechnology and Indian Council of Medical Research, Government of India.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gellert M., Mizuuchi K., O'Dea M.H., Nash H.A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl Acad. Sci. USA. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang J.C. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 3.Champoux J.J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 4.Kreuzer K.N., Cozzarelli N.R. Formation and resolution of DNA catenanes by DNA gyrase. Cell. 1980;20:245–254. doi: 10.1016/0092-8674(80)90252-4. [DOI] [PubMed] [Google Scholar]
- 5.Liu L., Liu C.C., Alberts B.M. Type II DNA topoisomerases: enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell. 1980;19:697–707. doi: 10.1016/s0092-8674(80)80046-8. [DOI] [PubMed] [Google Scholar]
- 6.Gellert M., Mizuuchi K., O'Dea M.H., Itoh T., Tomizawa J. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc. Natl Acad. Sci. USA. 1977;74:4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klevan L., Wang J.C. Deoxyribonucleic acid gyrase-deoxyribonucleic acid complex containing 140 base pairs of deoxyribonucleic acid and an alpha 2 beta 2 protein core. Biochemistry. 1980;19:5229–5234. doi: 10.1021/bi00564a012. [DOI] [PubMed] [Google Scholar]
- 8.Krueger S., Zaccai G., Wlodawer A., Langowski J., O'Dea M.H., Maxwell A., Gellert M. Neutron and light-scattering studies of DNA gyrase and its complex with DNA. J. Mol. Biol. 1990;211:211–220. doi: 10.1016/0022-2836(90)90021-D. [DOI] [PubMed] [Google Scholar]
- 9.Maxwell A. DNA gyrase as a drug target. Biochem. Soc. Trans. 1999;27:48–53. doi: 10.1042/bst0270048. [DOI] [PubMed] [Google Scholar]
- 10.Lewis R.J., Tsai F.T., Wigley D.B. Molecular mechanisms of drug inhibition of DNA gyrase. Bioessays. 1996;18:661–671. doi: 10.1002/bies.950180810. [DOI] [PubMed] [Google Scholar]
- 11.Drilca K., Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pohlhaus J.R., Kreuzer K.N. Norfloxacin-induced DNA gyrase cleavage complexes block Escherichia coli replication forks, causing double-stranded breaks in vivo. Mol. Microbiol. 2005;56:1416–1429. doi: 10.1111/j.1365-2958.2005.04638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malik M., Zhao X., Drlica K. Lethal fragmentation of bacterial chromosomes mediated by DNA gyrase and quinolones. Mol. Microbiol. 2006;61:810–825. doi: 10.1111/j.1365-2958.2006.05275.x. [DOI] [PubMed] [Google Scholar]
- 14.Liu J. Microcin B17: posttranslational modifications and their biological implications. Proc. Natl Acad. Sci. USA. 1994;91:4618–4620. doi: 10.1073/pnas.91.11.4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang Y., Pogliano J., Helinski D.R., Konieczny I. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 2002;44:44971–44979. doi: 10.1046/j.1365-2958.2002.02921.x. [DOI] [PubMed] [Google Scholar]
- 16.Couturier M., Bahassi el-M., Van Melderen L. Bacterial death by DNA gyrase poisoning. Trends Microbiol. 1998;6:269–275. doi: 10.1016/s0966-842x(98)01311-0. [DOI] [PubMed] [Google Scholar]
- 17.Nakanishi A., Oshida T., Matsushita T., Imajoh-Ohmi S., Ohnuki T. Identification of DNA gyrase inhibitor (GyrI) in Escherichia coli. J. Biol. Chem. 1998;273:1933–1938. doi: 10.1074/jbc.273.4.1933. [DOI] [PubMed] [Google Scholar]
- 18.Chatterji M., Nagaraja V. GyrI: a counter-defensive strategy against proteinaceous inhibitors of DNA gyrase. EMBO Rep. 2001;3:261–267. doi: 10.1093/embo-reports/kvf038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montero C., Mateu G., Rodriguez R., Takiff H. Intrinsic resistance of Mycobacterium smegmatis to fluoroquinolones may be influenced by new pentapeptide protein MfpA. Antimicrob. Agents Chemother. 2001;45:3387–3392. doi: 10.1128/AAC.45.12.3387-3392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hegde S.S., Vetting M.W., Roderick S.L., Mitchenall L.A., Maxwell A., Takiff H.E., Blanchard J.S. A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science. 2005;308:1480–1483. doi: 10.1126/science.1110699. [DOI] [PubMed] [Google Scholar]
- 21.Ashiuchi M., Kuwana E., Yamamoto T., Komatsu K., Soda K., Misono H. Glutamate racemase is an endogenous DNA gyrase inhibitor. J. Biol. Chem. 2002;277:39070–39073. doi: 10.1074/jbc.C200253200. [DOI] [PubMed] [Google Scholar]
- 22.Ashiuchi M., Tani K., Soda K., Misono H. Properties of glutamate racemase from Bacillus subtilis IFO 3336 producing poly-gamma-glutamate. J. Biochem. (Tokyo) 1998;123:1156–1163. doi: 10.1093/oxfordjournals.jbchem.a022055. [DOI] [PubMed] [Google Scholar]
- 23.Ashiuchi M., Soda K., Misono H. Characterization of yrpC gene product of Bacillus subtilis IFO 3336 as glutamate racemase isozyme. Biosci. Biotechnol. Biochem. 1999;63:792–798. doi: 10.1271/bbb.63.792. [DOI] [PubMed] [Google Scholar]
- 24.Ashiuchi M., Kuwana E., Komatsu K., Soda K., Misono H. Differences in effects on DNA gyrase activity between two glutamate racemases of Bacillus subtilis, the poly-gamma-glutamate synthesis-linking Glr enzyme and the YrpC (MurI) isozyme. FEMS Microbiol. Lett. 2003;223:221–225. doi: 10.1016/S0378-1097(03)00381-1. [DOI] [PubMed] [Google Scholar]
- 25.Chatterji M., Unniraman S., Mahadevan S., Nagaraja V. Effect of different classes of inhibitors on DNA gyrase from Mycobacterium smegmatis. J. Antimicrob. Chemother. 2001;48:479–485. doi: 10.1093/jac/48.4.479. [DOI] [PubMed] [Google Scholar]
- 26.Manjunatha U.H., Mahadevan S., Visweswariah S.S., Nagaraja V. Monoclonal antibodies to mycobacterial DNA gyrase A inhibit DNA supercoiling activity. Eur. J. Biochem. 2001;268:2038–2046. doi: 10.1046/j.1432-1327.2001.02077.x. [DOI] [PubMed] [Google Scholar]
- 27.Manjunatha U.H., Maxwell A., Nagaraja V. A monoclonal antibody that inhibits mycobacterial DNA gyrase by a novel mechanism. Nucleic Acids Res. 2005;33:3085–3094. doi: 10.1093/nar/gki622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madhusudan K., Ramesh V., Nagaraja V. Cloning and sequence analysis of DNA gyrase genes from Mycobacterium tuberculosis. Curr. Sci. 1994;66:664–667. [Google Scholar]
- 29.Maxwell A., Howells A.J. Overexpression and purification of bacterial DNA gyrase. Methods Mol. Biol. 1999;94:135–144. doi: 10.1385/1-59259-259-7:135. [DOI] [PubMed] [Google Scholar]
- 30.Manjunatha U.H., Dalal M., Chatterji M., Radha D.R., Visweswariah S.S., Nagaraja V. Functional characterisation of mycobacterial DNA gyrase: an efficient decatenase. Nucleic Acids Res. 2002;30:2144–2153. doi: 10.1093/nar/30.10.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory manual. 2nd edn. NY: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 1989. [Google Scholar]
- 32.Jain P., Nagaraja V. Indispensable, functionally complementing N and C-terminal domains constitute site-specific topoisomerase I. J. Mol. Biol. 2006;357:1409–1421. doi: 10.1016/j.jmb.2006.01.079. [DOI] [PubMed] [Google Scholar]
- 33.Lynn R.M., Wang J.C. Peptide sequencing and site-directed mutagenesis identify tyrosine-319 as the active site tyrosine of Escherichia coli DNA topoisomerase I. Proteins. 1989;6:231–239. doi: 10.1002/prot.340060305. [DOI] [PubMed] [Google Scholar]
- 34.Gallo K.A., Knowles J.R. Purification, cloning, and cofactor independence of glutamate racemase from Lactobacillus. Biochemistry. 1993;32:3981–3990. doi: 10.1021/bi00066a019. [DOI] [PubMed] [Google Scholar]
- 35.Fisher L.M., Mizuuchi K., O'Dea M.H., Ohmori H., Gellert M. Site-specific interaction of DNA gyrase with DNA. Proc. Natl Acad. Sci. USA. 1981;78:4165–4169. doi: 10.1073/pnas.78.7.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glavas S., Tanner M.E. Active site residues of glutamate racemase. Biochemistry. 2001;40:6199–6204. doi: 10.1021/bi002703z. [DOI] [PubMed] [Google Scholar]
- 37.Gmunder H., Kuratli K., Keck W. Effect of pyrimido [1, 6-a] benzimidazoles, quinolones, and Ca2+ on the DNA gyrase-mediated cleavage reaction. Antimicrob. Agents Chemother. 1995;39:163–169. doi: 10.1128/aac.39.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staudenbauer W.L., Orr E. DNA gyrase: affinity chromatography on novobiocin-Sepharose and catalytic properties. Nucleic Acids Res. 1981;9:3589–3603. doi: 10.1093/nar/9.15.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maxwell A., Gellert M. The DNA dependence of the ATPase activity of DNA gyrase. J. Biol. Chem. 1984;259:14472–14480. [PubMed] [Google Scholar]
- 40.Flatman R.H., Howells A.J., Heide L., Fiedler H.P., Maxwell A. Simocyclinone D8, an inhibitor of DNA gyrase with a novel mode of action. Antimicrob. Agents Chemother. 2005;49:1093–1100. doi: 10.1128/AAC.49.3.1093-1100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez-Martinez L., Pascual A., Jacoby G.A. Quinolone resistance from a transferable plasmid. Lancet. 1998;351:797–799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 42.Martinez-Martinez L., Pascual A., Garcia I., Tran J., Jacoby G.A. Interaction of plasmid and host quinolone resistance. J. Antimicrob. Chemother. 2003;51:1037–1039. doi: 10.1093/jac/dkg157. [DOI] [PubMed] [Google Scholar]
- 43.Tran J.H., Jacoby G.A., Hooper D.C. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 2005;49:118–125. doi: 10.1128/AAC.49.1.118-125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartman P.S., Eisenstark A. Killing of Escherichia coli K-12 by near-ultraviolet radiation in the presence of hydrogen peroxide: role of double-strand DNA breaks in absence of recombinational repair. Mutat Res. 1980;72:31–42. doi: 10.1016/0027-5107(80)90217-1. [DOI] [PubMed] [Google Scholar]
- 45.Li K., Pasternak C., Hartig E., Haberzettl K., Maxwell A., Klug G. Thioredoxin can influence gene expression by affecting gyrase activity. Nucleic Acids Res. 2004;32:4563–4575. doi: 10.1093/nar/gkh794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chatterji M., Sengupta S., Nagaraja V. Chromosomally encoded gyrase inhibitor GyrI protects Escherichia coli against DNA-damaging agents. Arch. Microbiol. 2003;180:339–346. doi: 10.1007/s00203-003-0598-4. [DOI] [PubMed] [Google Scholar]