Abstract
Skeletal myoblast fusion in vitro requires the expression of connexin43 (Cx43) gap junction channels. However, gap junctions are rapidly downregulated after the initiation of myoblast fusion in vitro and in vivo. In this study we show that this downregulation is accomplished by two related microRNAs, miR-206 and miR-1, that inhibit the expression of Cx43 protein during myoblast differentiation without altering Cx43 mRNA levels. Cx43 mRNA contains two binding sites for miR-206/miR-1 in its 3′-untranslated region, both of which are required for efficient downregulation. While it has been demonstrated before that miR-1 is involved in myogenesis, in this work we show that miR-206 is also upregulated during perinatal skeletal muscle development in mice in vivo and that both miR-1 and miR-206 downregulate Cx43 expression during myoblast fusion in vitro. Proper development of singly innervated muscle fibers requires muscle contraction and NMJ terminal selection and it is hypothesized that prolonged electrical coupling via gap junctions may be detrimental to this process. This work details the mechanism by which initial downregulation of Cx43 occurs during myogenesis and highlights the tight control mechanisms that are utilized for the regulation of gap junctions during differentiation and development.
INTRODUCTION
Skeletal muscle formation in vertebrates involves an interplay between the maturation of the contractile and the neural apparatus which is intricate and interdependent (1–3). Coordinated events in myogenesis and neuromuscular junction formation are facilitated by intercellular communication between myoprogenitors and neurons via signaling molecules on membrane surfaces as well as between neighboring myoprogenitors and myotubes via gap junctions. This communication has been repeatedly demonstrated to be essential for the formation of mature muscle and the development of a functional neural control mechanism (4–6).
Communication between developing muscle fibers is extensive during prenatal embryonic development. In fact, coupling is so extensive prenatally that excitation spreading laterally between myotubes gives rise to waves of excitation that propagate across the entire muscle (1). In developing intercostals muscles, for instance, electrical current applied to one muscle fiber can cause the contraction of the entire muscle; this depolarization can even spread to neighboring muscles in the next intercostal space through gap junctional communication (1). Immediately after birth, gap junctional communication in skeletal muscle sharply declines. This downregulation corresponds to the period of retraction of redundant nerve terminals from muscle fibers. During myoneural maturation, each stage requires the presence and contractile activity of functional muscle. Secondary myotube formation requires innervation, and contractile activity occurs very early after myotube formation. In utero, contraction of antagonistic muscles occurs in rhythmic alternation via reflex pathways in the spinal cord (7). Unified contraction of embryonic muscle fibers is facilitated by low-resistance electrical coupling via gap junctions composed primarily of connexin43 (Cx43). Gap junctions are hypothesized to be required in embryonic skeletal muscle to allow passage of signaling molecules and metabolites and for the coordinated maturation of contractile capabilities (1,8,9). Gap junctional coupling in hindlimb muscles disappears around birth. At this time, the only remaining mononuclear cells are the resident stem cells of skeletal muscle, the satellite cells. These cells remain quiescent until induced to proliferate and differentiate in response to injury. Upon injury, Cx43 is rapidly upregulated and functional gap junction channels couple proliferating satellite cells in vivo (10). High levels of Cx43 protein remain until after fusion of myoblasts.
In vivo observations have largely agreed with studies done on isolated primary myoblast cultures and with myoblast cell lines. Cx43 is expressed in established myoblast cell lines, and in isolated primary myoblast cultures prior to fusion (11,12). The fusion of myoblasts in vitro has been shown to require the presence of functional gap junctions (4,6,13). In the presence of heptanol or octanol, known gap junction channel blockers, myoblast differentiation is prevented (14). Under these conditions, the myogenic regulatory factors, myogenin and MRF4, also fail to be upregulated in response to induction (11). These results of chemical blockade suggest that functional channels are required for myogenesis in vitro. Elsewhere, it has been demonstrated that targeted gene deletion of Cx43, using a Cre-Lox system, inhibits the regenerative capability of myoblasts in vitro (15) and in vivo.
In myoblast cell lines, Cx43 is present prior to and during fusion but is rapidly downregulated after induction of differentiation despite only a slight reduction in mRNA levels (6). These studies suggested that the initial step in downregulation of Cx43 occurs at the translational level. In this study we present evidence that indicates a role for two microRNAs in the regulation of Cx43 during myogenesis in mouse tissues and from cultured myoblasts. This regulation occurs by inhibiting translation without targeting the message for degradation. MicroRNAs have been shown to regulate many other developmental processes. Despite the requirement for Cx43 during the initial phase of myogenesis, the subsequent downregulation of Cx43-dependent gap junctional communication is important in generating insulated muscle fibers that are singly innervated for fine motor control.
MATERIALS AND METHODS
Bioinformatics
Alignment of DNA sequence pairs was performed using Blast 2 Sequences available at the NCBI website (http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi) (16). Multiple DNA sequence alignments were performed using ClustalX v1.8 (17). All genomic, expressed sequence tags (ESTs), and other nucleotide sequences were obtained from the NCBI online database via Genomic Blast, Blastn and Unigene, respectively.
Plasmid construction
Plasmids designed to express full length Cx43 mRNA sequence were subcloned from cDNA clone B0C01C08 from ATCC into the Xba I and Kpn I sites within pcDNA3.1 (Invitrogen). Reporter plasmids were constructed by PCR-amplifying the 3′-untranslated region (3′-UTR) from the cDNA clone and inserting the product into pcDNA3.1 downstream of the firefly luciferase coding region using the following primer sequences: 5′-GATCTCTAGAACAGGCTTGAACATCAAG and 5′-GATCGAATTCATTATACTAAATTAAAATTTTATTG. Mutations of miR-206 binding sites were introduced by site-directed mutagenesis using the following primers: for BS-1 5′-CCTACATCCCCGCTAAAAAACTAAGCAGTGTTTAAAAACT and 5′-AGTTTTTAAACACTGCTTAGTTTTTTAGCGGGGATGTAGG; for BS-2 5′-AACTAATTTGTTTGACTAAGCATGTTAAACTACTGTCA and 5′-TGACAGTAGTTTAACATGCTTAGTCAAACAAATTAGTT. The RNA pol III based expression plasmid, pSUPER, was constructed as described (18). pSUPER 206pri was constructed by PCR amplifying a ∼180 nt region of genomic sequence surrounding pre-miR-206 and inserting it into pSUPER using the following primers: 5′-CCTACATCCCCGCTAAAAAACTAAGCAGTGTTTAAAAACT and 5′-AGTTTTTAAACACTGCTTAGTTTTTTAGCGGGGATGTAGG. pSUPER miR-1 was constructed similarly based on the genomic sequence of the primary transcript of miR-1-2 with the following primers: 5′-CACCAAGCTTTTTGAATTCAGAGTATGGAAGTCATC and 5′-TGATGGATCCCCAAGTAATCCAAATGTCCTAC.
Cell culture
C2C12 cells were a generous gift from Dr Charles Murry at the University of Washington were cultured as described (12). HeLa cells were cultured and transfected as described (19). Luciferase assays were performed as described (19). Antisense inhibitors of miR-1 and miR-206 as well as negative control oligonucleotides were purchased from Ambion. For antisense inhibition of microRNAs, C2C12 cells were cultured to 50% confluence and transfected with either 100 nM negative control antisense RNA, 100 nM anti-miR-206, 100 nM anti-miR-1, or 50 nM anti-miR-206 and 50 nM anti-miR-1 for 6 h. Transfections were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Subsequently, the cells were rescued in differentiation media [DMEM + 5% horse serum (HS) and ITS] and incubated for 48 h to induce differentiation (12).
RNase protection assay (RPA)
Total RNA was isolated from tissues using Trizol (Invitrogen) and a Tekmar homogenizer. Total RNA was isolated from cell cultures using Trizol. Total RNA from developmental stages of neonatal mice were purchased from Zyagen. The radiolabeled antisense RNA probe targeting miR-206 was prepared by using the pSUPER 206pri plasmid linearized with EcoRV as a template with the MaxiScript-T7 in vitro Transcription Kit (Ambion). RNase protection assays (RPAs) for miR-206 were performed by using the mirVana miRNA Detection Kit (Ambion). The antisense RNA probes against β-actin and Cx43 were performed using the MaxiScript in vitro Transcription Kit (Ambion) (19). RPAs targeting β-actin and Cx43 were done by using the RPA III Kit (Ambion).
Western blots
Total protein extracts from developmental stages of neonatal mice were purchased from Zyagen. Protein extracts were prepared in RIPA buffer [1% NP-40, 1% Deoxycholate, 0.1% SDS, 500 mM Tris, 150 mM NaCl, 1 mM PMSF, 1× Protease Inhibitor Cocktail (Roche)]. 10 μg of total protein was separated on precast 12% Tris–HCl PAGE gels (BioRad) and electrotransferred to PVDF membranes. Primary antibodies binding to Cx43 (Sigma, rabbit polyclonal, 1:1000 dilution), myogenin (Sigma, rabbit polyclonal, 1:1000), α-tubulin (Sigma, mouse monoclonal, 1:1000) were detected with HRP-conjugated anti-rabbit (Sigma) or HRP-conjugated anti-mouse antibodies (Sigma) and the ECLplus Western Blotting Detection Kit (Amersham).
Northern blotting
Northern blots designed to detect Cx43, Luciferase and β-actin mRNA were performed on 20 μg of total RNA from transfected cells using the NorthernMax Gly Kit (Ambion). Northern blots designed to detect miR-206 and U6 RNA from transfected HeLa cells were performed on 20 μg of total RNA by separating RNA on precast 15% TBE 8M urea gels (BioRad). Northern blots designed to detect U6 RNA from tissue samples utilized 1 μg of total RNA similarly. Fractionated RNA was transferred to positively charged nylon membrane by electroblotting. The membranes were blocked in a 1:1 mixture of Ultrahyb:2× SSC, 7% SDS and probed with 5′-biotinylated DNA oligonucleotides [anti-miR-206 probe (Biotin-CCACACACTTCCTTACATTCCA) and/or an anti-U6 probe (Biotin-GCTAATCTTCTCTGTATCGTTCCAA)] at a concentration of 10 pM in the same solution. Biotinylated probe was detected with the BrightStar Detection Kit (Ambion).
Immunocytochemistry
C2C12 cells cultured in 35 mm dishes (Nunc) were induced to differentiate by switching to differentiation medium at ∼70% confluency. Cells were fixed at various time points by incubation for 5 min in −10°C methanol and air dryed. Cells were washed with PBS, further permeabilized for 5 min in 0.1% Triton in PBS and blocked with 5% HS in PBS for 1 h at room temperature. Cells were incubated with 1:100 dilution of anti-Cx43 rabbit polyclonal antibody (Sigma) and 1 μg of anti-myogenin mouse monoclonal antibody (Sigma) overnight at 4°C in 5% HS in PBS. Dishes were washed three times in PBS and incubated with 1:100 dilution of goat anti-rabbit-TRITC conjugated secondary antibody and a 1:100 dilution of goat anti-mouse-FITC conjugated secondary antibody in PBS with 5% HS. Cells were washed three times in PBS and incubated for 5 min with PBS-DAPI. Finally cells were washed three times in PBS and examined by fluorescence microscopy.
RESULTS
Bioinformatics
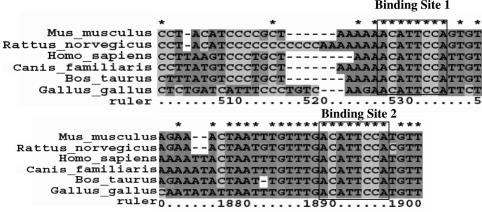
Based on evidence from previous work suggesting that downregulation of Cx43 during myogenesis occurs through a post-transcriptional mechanism, we looked for the presence of potential cis elements for translational regulation in the Cx43 mRNA sequence. Numerous investigators have published algorithms to detect possible interactions between microRNAs and target mRNA sequences. One such algorithm predicts the presence of two binding sites in the 3′-UTR of Cx43 for the microRNA, miR-206 (20). We aligned the 3′-UTR sequences of Cx43 mRNAs from the domesticated cow, (Bos taurus) (NM174068), the common dog (Canis familiaris) (NM_001002951 and genomic sequence from NW876269.1), mouse (Mus musculus) (NM010288), rat (Rattus norvegicus)(NM012567), chicken (Gallus gallus)(NM204586 and genomic sequence from NW060336.1) and human (Homo sapiens)(NM000165) from Genbank and/or the EST database. The chicken and dog sequences were did not contain polyadenylation signals as reported and were extended to conserved polyadenylation signals downstream (data not shown). These sequences were confirmed by the presence of overlapping EST sequences. The ‘seed’ sequence, corresponding to the most important region for miRNA:mRNA interactions, was completely conserved in all aligned sequences. The conserved binding regions are shown in Figure 1.
Figure 1.
Conserved predicted miR-206/miR-1 binding sites in Cx43 3′-UTR. Outlined boxes demark the region that is perfectly complimentary to miR-206/miR-1 and is most important for microRNA:mRNA interaction (28). This region is 2 nt larger than the ‘seed’ region used by Lewis et al. (25) to find potential miR-206/miR-1 targets.
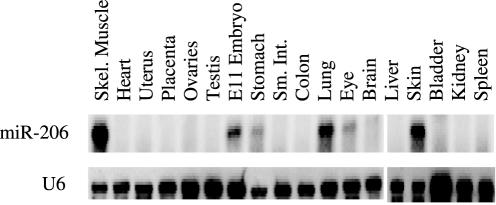
Tissue distribution of miR-206
To determine the expression pattern of miR-206, RPAs were performed on 5 μg of total RNA from various tissues. As shown in Figure 2, miR-206 is expressed predominantly in adult skeletal muscle; it could also be detected in skin, E11 whole embryo and lung, but not in heart muscle. The absence of miR-206 in heart is consistent with the high levels of Cx43 expression in heart tissue. Confirmation that miR-206 is indeed expressed in skeletal muscle, but is absent from cardiac muscle, was important because some published microarray data had suggested that miR-206 is expressed in heart, a tissue that expresses high levels of Cx43 (21,22). A northern blot directed against U6 snRNA demonstrates that small RNA molecules are represented in each sample.
Figure 2.
Tissue distribution of miR-206. RNase protection assay to detect miR-206 was performed on 5 μg of total RNA from various tissue samples. 5 μg of total RNA was subjected to RPA. 1 μg of total RNA was used to detect U6 snRNA by northern blotting.
Downregulation of Cx43 in vitro
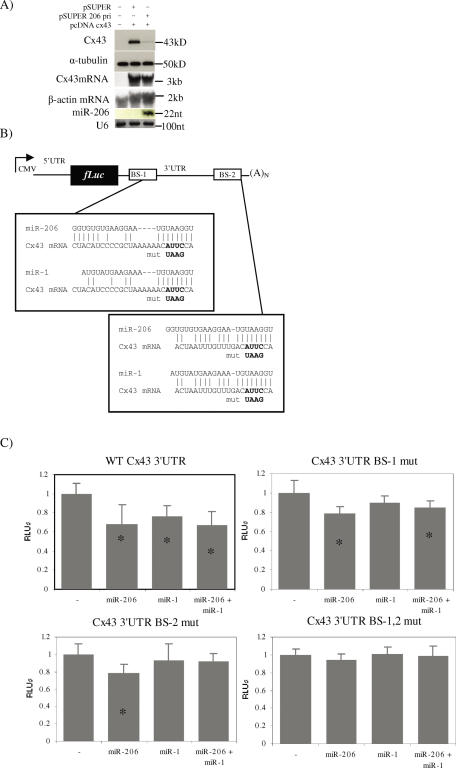
In order to test if miR-206 is indeed capable of regulating Cx43 protein expression in the context of its native mRNA sequence, we cloned the full-length cDNA corresponding to the entire Cx43 mRNA sequence from transcription start site to polyadenylation signal into pcDNA3.1 (pcDNA Cx43). Another plasmid designed to deliver miR-206 contained a ∼180 nt genomic insert under control of the H1 RNA promoter (pSUPER 206pri) (18). For expression studies, the human cervical cancer cell line HeLa was used because it expresses neither Cx43 nor miR-206. HeLa cells were grown in culture and transfected with pcDNA Cx43 and either pSUPER 206pri or pSUPER as a negative control. Forty-eight hours after transfection, protein extracts and total RNA were prepared. Western blot analysis against Cx43 shows a significant inhibition of Cx43 expression as shown in Figure 3A. A control western blot against α-tubulin showed that this protein remained unchanged. Northern blot analysis against total RNA revealed that the presence of miR-206 did not affect the level of Cx43 mRNA. As expected, miR-206 was only present when the cells were transfected with pSUPER 206pri. As a control for the efficiency of small RNA purification, the blot was stripped and rehybridized with a probe against U6 snRNA. Data from three independent experiments yielded a reproducible and considerable inhibition of Cx43 expression by miR-206. This experiment unequivocally shows that Cx43 can be regulated at the post-transcriptional level by miR-206 and further supports the hypothesis that miR-206 is responsible for the initial downregulation of Cx43 during myogenesis.
Figure 3.
Regulation of Cx43 by miR-206/1 in vitro. (A) HeLa cells were mock transfected or transfected with cDNA for Cx43 in addition to empty pSUPER plasmid or pSUPER 206pri. After 48 h Western blots and RNA analysis was performed as described in the Materials and Methods section. (B) Diagram of luciferase reporter construct with putative miR-206 binding sites and mutations. Binding sites were mutated in pcDNA Luc 43-3′-UTR and the 5′-UTR was individually cloned into each mutant to make the constructs containing both the 5′- and 3′-UTRs. (C) Effect of binding site mutants on luciferase expression in the presence of miR-206, miR-1 or both miR-206 and miR-1. HeLa cells were transfected with pcDNA Luc constructs along with either pSUPER or pSUPER 206pri and analysed 24 h later. Data is presented as relative firefly luciferase units (RLUs). Relative firefly luciferase values were determined by a ratio of firefly to renilla luciferase with the negative control set at 1. Error bars—standard deviation; 0 ≥ 9. *A student's t-test performed between microRNA transfected samples and the negative control yielded a P-value < 0.05 (actual P-value < 0.01).
To test the specific regulation of Cx43 through the two predicted binding sites, we inserted the Cx43 3′-UTR sequence downstream of the firefly luciferase coding region. Mutants of the putative binding sites were then prepared (Figure 3B). Known sequence similarity between miR-206 and miR-1 caused us to also consider miR-1 as a potential regulator of Cx43 despite the suggestions that miR-1 and Cx43 coexist in cardiac tissue. Therefore, a plasmid designed to deliver miR-1 utilizing the pSUPER vector was constructed for comparison. As shown in this figure, there is less complimentarity between miR-1 and binding site 1 than between miR-206 and the same sequence. As expected, the wild-type 3′-UTR (pcDNA Luc 43-3′-UTR) construct was significantly inhibited by the presence of miR-206 and to a lesser degree by miR-1 (Figure 3C). Mutating either one of the binding sites alone had a partial effect on the ability of miR-206 to inhibit the expression of the luciferase reporter but completely abolished the ability of miR-1 to regulate luciferase expression. A double mutant completely abolished the inhibitory effect of miR-206 on the reporter expression. There was no statistically significant effect of combining miR-1 and miR-206 in this experimental model, suggesting that there is no synergistic or additive effect of the microRNAs when they are overexpressed together. In another series of experiments, two other non-related microRNAs, miR-183 and miR-137, were compared to miR-206 tested for their effect on luciferase expression. These microRNAs (also produced from pSUPER based plasmids) produced no significant difference in luciferase expression when compared to the empty vector (data not shown).
Regulation of Cx43 during development
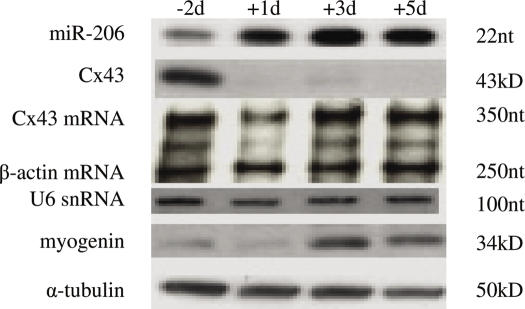
Since we determined that miR-206 and to a lesser extent miR-1 are capable of regulating Cx43 expression through direct binding to the 3′-UTR of its mRNA and that miR-206 is expressed in skeletal muscle, the next step was to look in vivo to see if miR-206 regulates Cx43 expression during myogenesis. We predicted that miR-206 RNA and Cx43 protein levels should be inversely related in a manner consistent with muscle development. To this end, we performed an RPA on total RNA isolated from skeletal muscle derived from 2 days prenatal mice (E16), and the first, third, and fifth days after birth. At 2 days prenatal, miR-206 became visible reaching a maximum level by day 3 postnatal (Figure 4). Western blots for Cx43 protein show that its levels decrease significantly after birth. In contrast, Cx43 mRNA levels remains unchanged during this period, as determined by RPA. The observed downregulation of Cx43 during this time period is in agreement with previous reports that have shown that myoblasts and developing myotubes express high levels of Cx43 gap junctions prior to and during fusion and growth (8,11). A high level of expression of miR-206 during myogenesis in vivo supports the notion that this miRNA is involved in the downregulation of connexin during perinatal development, the time when the bulk of skeletal muscle is being formed. It has been shown elsewhere that miR-1 shows a similar pattern of upregulation during in neonatal mouse muscle (23).
Figure 4.
miR-206 regulates Cx43 during myoblast fusion and skeletal muscle development. Total RNA and total lysates were isolated from skeletal muscle from Swiss-Webster mice at E16 (−2days), day 1, 3, or 5 after birth. RNase protection assay was performed against miR-206 (500 ng total RNA) or Cx43 mRNA and β-actin (10 μg total RNA). Western blots were performed on 10 μg total protein as described in the Materials and Methods section.
Downregulation of Cx43 in C2C12 cells
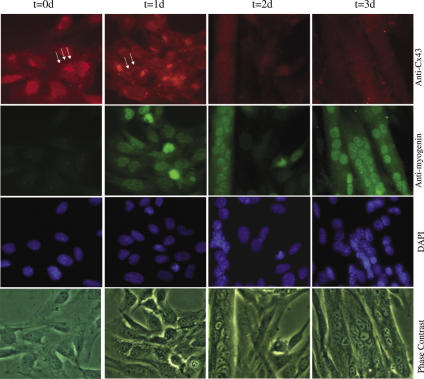
To confirm and extend our hypothesis that miR-206 plays an inportanat role in muscle cell development, we studied the expression of miR-206 in myoblasts in vitro, using a highly myogenic subclone of the mouse cell line C2C12. These cells are propagated as undifferentiated, mononucleated myoblasts under high serum conditions. When cultures are switched to low serum, they rapidly fuse into contractile myotubes. It has been reported that Cx43 is highly expressed in the mononucleated C2C12 myoblasts but is rapidly downregulated after induction of differentiation. C2C12 cells are often used to recapitulate the muscle differentiation pathway in vitro. To confirm the pattern of Cx43 expression in this in vitro model for muscle development, C2C12 cells were cultured to ∼70% confluency in growth media and then induced to differentiate by switching to differentiation medium. Samples were fixed at day 0 (day of induction) and every 24 h until day 3. Immunofluorescence studies were performed to detect Cx43 protein and myogenin, a marker for differentiation in skeletal muscle. As shown in Figure 5, Cx43 was highly expressed in proliferating myoblasts at time 0. As expected, Cx43 protein is localized at the nuclear periphery, where it is synthesized on the endoplasmic reticulum and processed in the Golgi apparatus. It can also be seen at punctuate loci at cellular borders. Myogenin was undetectable at this time. After 1 day of differentiation, high levels of Cx43 were still located in the nuclear periphery and at regions of cellular apposition. Myogenin, the muscle-specific basic helix-loop-helix transcription factor, began to be expressed and was located in the nucleus as evidenced by its co-localization with the nuclear stain DAPI. At day 1 there were still no detectable multinucleated cells.
Figure 5.
Connexin43 is downregulated during myoblast differentiation. Arrows denote positive immunostaining for Cx43 at membrane appositions. Cells were incubated with 1:100 dilution of anti-connexin43 rabbit polyclonal antibody (Sigma) and 1 μg of anti-myogenin mouse monoclonal antibody (Sigma) overnight at 4°C in 5% HS in PBS. Dishes were examined by fluorescence and phase contrast microscopy using FITC, rhodamine and DAPI filters.
By day 2, expression of Cx43 had become undetectable in the perinuclear region and between cells, whereas myogenin expression had increased significantly. At this time, there were some multinucleated myotubes and some mononucleated myoblasts, none of which expressed Cx43. At day 3, most of the mononucleated cells had disappeared, there was no detectable Cx43 and myogenin was very highly expressed in all nuclei. These results indicate that C2C12 differentiation recapitulates the Cx43 regulatory pathway seen in developing skeletal muscle and should be a good model for dissecting the mechanism of Cx43 downregulation.
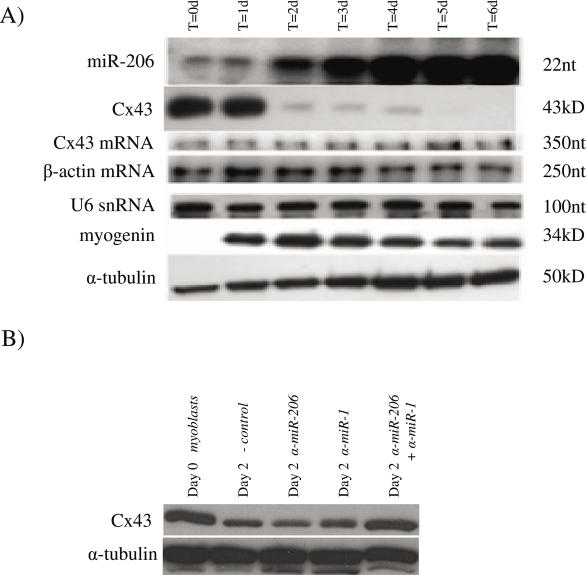
In order to further validate the in vivo correlations, we investigated the expression of Cx43 protein and mRNA as well as miR-206 during C2C12 myoblast differentiation. These cells were cultured in growth media and then induced to differentiate. Extracts were prepared at the time of induction and every 24 h thereafter. As shown in Figure 6A, Cx43 was highly expressed in proliferating myoblasts and continued its high levels of expression through day 1 of differentiation. At this time there was little expression of miR-206. By day 2 of differentiation there was a drastic reduction in the levels of Cx43 protein, and this level remained low until day 5, at which point Cx43 was undetectable. Expression of miR-206 was first clearly detectable on day 2 and came to a plateau around day 4. This is similar to the study by Chen et al. which showed that miR-1 expression was first detectible on day 3 post induction (23). The control northern blot to detect U6 showed that purification of the small RNA fraction of total RNA was consistent. The level of Cx43 mRNA was unchanged throughout the experiment, confirming that the downregulation of Cx43 protein occurred post-transcriptionally. Levels of β-actin mRNA remained unchanged throughout the experiment and served as an internal control. In fact, extended experiments have shown that Cx43 mRNA levels were largely unchanged through 12 days of differentiation in C2C12 cells (data not shown). Myogenin, the marker for myoblast differentiation, was not detectable in proliferating C2C12 cells but was rapidly upregulated in response to the medium change.
Figure 6.
(A) C2C12 cells were grown in growth medium and induced to differentiate at ∼70% confluency by switching to DMEM supplemented with 5% HS and 1× ITS. Cells were harvested prior to induction (t = 0) and at 24 h intervals thereafter. miR-206 was detected by RNase protection assay on 1 μg of total RNA. Cx43 mRNA and β-actin mRNA were detected by RNase protection assay on 10 μg of total RNA. Western blots against Cx43, myogenin and α-tubulin were performed on 10 μg of total protein. (B) C2C12 cells were grown in growth medium and induced to differentiate at ∼50% confluency by switching to DMEM supplemented with 5% HS and 1× ITS. Cells were harvested prior to induction (t = 0) or transfected with negative control microRNA inhibitor, anti-miR-206, anti-miR-1 or anti-miR-206 and anti-miR-1 and induced to differentiate. Transfected cells were harvested 48 h later. Western blots against Cx43 and α-tubulin were performed on 10 μg of total protein.
Finally, to show that miR-206 and miR-1 are indeed responsible for the downregulation of Cx43 in C2C12 cells, the cells were transfected with chemically modified antisense oligonucleotide microRNA inhibitors either individually or in combination immediately prior to induction of differentiation. Extracts were harvested 48 h after induction and levels of Cx43 were measured. At 2 days of differentiation, there was significant cell fusion in all experimental conditions. There was no observable difference in cell number, morphology or gross myotube size (data not shown). Prior to differentiation, Cx43 was highly expressed as shown in Figure 6B. After 48 h, the C2C12 cells transfected with a negative control oligonucleotide showed significant downregulation of Cx43. In addition, cells transfected with either anti-miR-1 or anti-miR-206 RNA alone still downregulated Cx43 expression. However, cells transfected with both anti-miR-1 and anti-miR-206 RNA demonstrated a high level of Cx43 expression, similar to the levels seen in undifferentiated cells.
DISCUSSION
Skeletal muscle development in vertebrates is an evolutionarily conserved process that involves myoblast fusion into multinucleated myotubes. Prior to and during myoblast fusion, and during the development of the motor neuron input system, muscle precursors are electrically coupled via gap junctions. In mammals, as well as in zebrafish, frogs and chickens, Cx43 is downregulated during late embryogenesis and early post-natal life. Conservation of the Cx43 expression pattern suggests the existence of a conservation of mechanism. The work presented here demonstrates the role of two related microRNAs in the downregulation of Cx43 during differentiation of skeletal muscle.
The prediction that miR-206 and to a lesser degree miR-1 would bind to the Cx43 mRNA by Lewis et al. (20) was one of many computer algorithms that predicted that a microRNA binding site was present in the Cx43 3′-UTR. Others, such as the DIANA MicroT algorithm, predict the binding of other microRNAs (24). These sites, however, are not conserved between species making them unlikely candidates to be involved in a conserved developmental mechanism. We, therefore, focused this study on the miR-206/miR-1 interaction with Cx43. An alignment of the 3′-UTR of Cx43 mRNAs from mouse, rat, human, dog, cow and chicken shows complete conservation of the ‘seed’ region of the miRNA:mRNA target interaction for both predicted targets (see Figure 1). This region corresponds to the 5′ end of the microRNA sequence and generally consists of 6–8 consecutive Watson–Crick base pairs with the mRNA target. The seed region has been repeatedly demonstrated to be important for target recognition by an miRNA (20,25–28). The conservation of these sequences increases the likelihood that Cx43 is indeed regulated by miR-206/miR-1, particularly since the overall rate of 3′-UTR sequence identity between these species is ∼36% (data not shown).
Data presented in Figure 3 show that miR-206 expression from pSUPER 206pri is capable of inhibiting the expression of a luciferase reporter bearing the 3′-UTR of Cx43. This experiment demonstrates that even with overexpression of both components of the system, the microRNA is capable of reducing luciferase expression despite the fact that the mRNA levels are not significantly affected (data not shown). As shown in Figure 3A, miR-206 inhibits the expression of Cx43 protein significantly as compared to the empty pSUPER control. From the results shown in Figure 3B and C, one can conclude that miR-206 binds to the Cx43 3′-UTR through two independent binding sites, and that it is this binding that mediates its ability to inhibit the translation of the Cx43 message. It is also shown that miR-1 is capable of regulating Cx43, albeit to a lesser degree, by virtue of its inability to regulate expression when either binding site is mutated alone. The results provided in Figure 3 present compelling evidence that miR-206, and possibly miR-1, have the potential to regulate Cx43 expression.
The availability of myoblast cell lines that can recapitulate the myogenic process in vitro has provided a convenient method to study associated developmental processes. In C2C12 cells, myotubes begin to appear at day 2 of differentiation, concurrent with the decline in Cx43 levels. Cx43 protein levels become undetectable by day 5 of culture in differentiation medium. The downregulation of Cx43 protein is coincident with the upregulation of miR-206 which begins before day 2. The levels of Cx43 mRNA remain unchanged during the course of this experiment, consistent with the hypothesis that microRNAs are indeed responsible for the post-transcriptional inhibition of Cx43 expression. Further confirmation that miR-206 and miR-1 regulate Cx43 expression comes from the inactivation of these microRNAs by antisense RNA during C2C12 maturation. As shown in Figure 6, both microRNAs must be inactivated for Cx43 expression to remain at its undifferentiated levels. Although there is a near total recovery of Cx43 levels in those cells transfected with anti-miR-1 and anti-miR-206 together, the magnitude of decrease in Cx43 levels during differentiation seen in Figure 6B was less than what was seen in Figure 6A. This is most likely due to the difference in experimental methods between the two experiments. In Figure 6A, the cells were induced to differentiate at 70% confluence whereas in the antisense inhibition experiment the cells were transfected and induced to differentiate at only 50% confluence. This difference in methodology was necessary because of the recommended lower optimal cell density for tranfection (Lipofectamine 2000, Invitrogen). It is also possible that there is a generalized effect of transfection with small RNA molecules on the downregulation of Cx43 by miR-206/1 in these cells or a generalized inhibition of differentiation by the liposome-mediated transfection process. The differentiation of C2C12 cells in culture is highly dependent on cell–cell contact and cell density; notably, these cells were subcloned to be highly myogenic (12). The finding that Cx43 protein expression during myoblast differentiation is inversely correlated with expression of miR-206 along with the finding that the inhibition of miR-206 and its family member miR-1 relieve this downregulation, suggests that miR-206 and miR-1 perform redundant or additive functions in regulating Cx43 in C2C12 cells. This correlates with the finding that miR-206 is capable of downregulating Cx43 protein expressed from full-length Cx43 mRNA expressed in HeLa cells, and luciferase expression that bears Cx43 3′-UTR sequences. It cannot, however, be entirely ruled out that the increase in Cx43 expression at day 2 in the presence of anti-miR-1 and anti-miR-206 together is due to an inhibitory effect on myoblast differentiation that was previously described but not seen by these experimental methods (23). This correlation must be further studied in vivo in developing skeletal muscle in order to further delineate the timing and quantitative levels of each microRNA during mouse development.
A number of publications have implicated miR-1 in the regulation of skeletal muscle differentiation in both vertebrates and invertebrates as well as during cardiogenesis in vertebrates (23,29,30). Our observation that miR-1 downregulates Cx43 during C2C12 differentiation was somewhat surprising because of the previous reports that both Cx43 and miR-1 are present in adult cardiac tissue. Further studies are required to investigate whether miR-1 and Cx43 actually are coexpressed within cardiomyocytes in the adult and whether miR-1 plays a role in regulating Cx43 in the developing and/or adult heart.
The observation that Cx43 levels decrease during mammalian perinatal skeletal muscle development was first seen decades ago (1,2,9,11,14,31,32). Data from only one group have suggested that the downregulation may occur at the post-transcriptional level, although this possibility was not pursued in their work (11). In whole skeletal muscle extracts, isolated 2 days prior to birth (E16), Cx43 protein is highly expressed (Figure 4). This is in agreement with previous reports that primary and secondary myotubes, as well as undifferentiated myoblasts, are highly coupled before birth (9). In contrast, on the day of birth and thereafter, Cx43 expression is barely detectable whereas levels of miR-206 are greatly increased. Cx43 mRNA levels are not significantly altered during this period. Maximum levels of miR-206 occur around day 3 after birth and are ∼4-fold higher than adult levels (data not shown). Other reports have demonstrated that miR-1 is expressed in a similar manner during development. A quantitative comparison of miR-206 and miR-1 levels was beyond the scope of this publication.
The miR-206 gene is located between the polycystic kidney and hepatic disease 1 gene and the interleukin-17 gene in mouse (chr 1), rat (chr 9) and human (chr 6). The genomic site where the pre-miR-206 sequence is located contains another microRNA precursor sequence, miR-133b, located a mere ∼3.8 kb downstream, suggesting that miR-206 and miR-133b might be part of the same primary transcript (33). Microarray data published by other laboratories have shown that miR-133b, like miR-206, is expressed in skeletal muscle (21,22). Apparent tissue-specific co-expression of miR-206 and miR-133b supports the idea that they may be produced from the same primary transcript. In fact a novel noncoding RNA, 7H4, has been isolated and shown to be enriched in sub-synaptic nuclei of innervated muscle (34). This RNA corresponds to the genomic locations of both pre-miR-206 and pre-miR-133b. This suggests a possible correlation between NMJ maturation and specific microRNA expression. During the preparation of this manuscript, Rao et al. (35) published data demonstrating the upregulation of miR-1, miR-133 and miR-206 in C2C12 cells during differentiation. However, these authors identified no targets for miR-206, and there was no mention of redundancy or cooperation between these two microRNAs during muscle development. There has been one study which was able to show that a mutation in the 3′-UTR of the myostatin gene creates a cryptic microRNA binding site for miR-1/miR-206. This mutation was responsible for creating a phenotype that included hypertrophic skeletal muscle (36).
Additional recent studies have demonstrated regulatory pathways responsible for the expression of miR-206 during embryonic development (35,37). This work and the work of others have demonstrated an early expression pattern associated with somites in chicken and mouse embryos and with developing muscle in zebrafish (24,38,39). Recently, another microRNA, miR-181 has been shown to be required for efficient myoblast differentiation by regulating Hox-A11 (40). Clearly there are multiple microRNAs that play a role in skeletal muscle development and this tissue system will provide fertile ground to study the regulatory networks involved in differentiation.
Acknowledgments
We would like to thank C. E. Murry and H. Reinecke for the generous gift of the C2C12 subclone as well as helpful technical advice, R. Myers for critical reading of this manuscript and J. Anderson in the DNA Core Lab for DNA sequencing assistance. This work was supported by NIH grant HD34152 to R.W. Funding to pay the Open Access publication charges for this article was provided by R.W.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dennis M.J., Ziskind-Conhaim L., Harris A.J. Development of neuromuscular junctions in rat embryos. Dev. Biol. 1981;81:266–279. doi: 10.1016/0012-1606(81)90290-6. [DOI] [PubMed] [Google Scholar]
- 2.Fladby T. Postnatal loss of synaptic terminals in the normal mouse soleus muscle. Acta Physiol. Scand. 1987;129:229–238. doi: 10.1111/j.1748-1716.1987.tb08063.x. [DOI] [PubMed] [Google Scholar]
- 3.Lewis J., Chevallier A., Kieny M., Wolpert L. Muscle nerve branches do not develop in chick wings devoid of muscle. J. Embryol. Exp. Morphol. 1981;64:211–232. [PubMed] [Google Scholar]
- 4.Araya R., Eckardt D., Maxeiner S., Kruger O., Theis M., Willecke K., Saez J.C. Expression of connexins during differentiation and regeneration of skeletal muscle: functional relevance of connexin43. J. Cell Sci. 2005;118:27–37. doi: 10.1242/jcs.01553. [DOI] [PubMed] [Google Scholar]
- 5.Armstrong D.L., Turin L., Warner A.E. Muscle activity and the loss of electrical coupling between striated muscle cells in Xenopus embryos. J. Neurosci. 1983;3:1414–1421. doi: 10.1523/JNEUROSCI.03-07-01414.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Proulx A., Merrifield P.A., Naus C.C. Blocking gap junctional intercellular communication in myoblasts inhibits myogenin and MRF4 expression. Dev. Genet. 1997;20:133–144. doi: 10.1002/(SICI)1520-6408(1997)20:2<133::AID-DVG6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 7.Landmesser L.T., O'Donovan M.J. Activation patterns of embryonic chick hind limb muscles recorded in ovo and in an isolated spinal cord preparation. J. Physiol. 1984;347:189–204. doi: 10.1113/jphysiol.1984.sp015061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalderon N., Epstein M.L., Gilula N.B. Cell-to-cell communication and myogenesis. J. Cell Biol. 1977;75:788–806. doi: 10.1083/jcb.75.3.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross J.J., Duxson M.J., Harris A.J. Formation of primary and secondary myotubes in rat lumbrical muscles. Development. 1987;100:383–394. doi: 10.1242/dev.100.3.383. [DOI] [PubMed] [Google Scholar]
- 10.Gorbe A., Becker D.L., Dux L., Stelkovics E., Krenacs L., Bagdi E., Krenacs T. Transient upregulation of connexin43 gap junctions and synchronized cell cycle control precede myoblast fusion in regenerating skeletal muscle in vivo Histochem. Cell Biol. 2005;125:705–716. doi: 10.1007/s00418-004-0745-2. [DOI] [PubMed] [Google Scholar]
- 11.Balogh S., Naus C.C., Merrifield P.A. Expression of gap junctions in cultured rat L6 cells during myogenesis. Dev. Biol. 1993;155:351–360. doi: 10.1006/dbio.1993.1034. [DOI] [PubMed] [Google Scholar]
- 12.Reinecke H., Murry C.E. Transmural replacement of myocardium after skeletal myoblast grafting into the heart. Too much of a good thing? Cardiovasc. Pathol. 2000;9:337–344. doi: 10.1016/s1054-8807(00)00055-7. [DOI] [PubMed] [Google Scholar]
- 13.Mege R.M., Goudou D., Giaume C., Nicolet M., Rieger F. Is intercellular communication via gap junctions required for myoblast fusion? Cell Adhes. Commun. 1994;2:329–343. doi: 10.3109/15419069409014208. [DOI] [PubMed] [Google Scholar]
- 14.Constantin B., Cronier L., Raymond G. Transient involvement of gap junctional communication before fusion of newborn rat myoblasts. C. R. Acad. Sci. III. 1997;320:35–40. doi: 10.1016/s0764-4469(99)80084-5. [DOI] [PubMed] [Google Scholar]
- 15.Araya R., Eckardt D., Maxeiner S., Kruger O., Theis M., Willecke K., Saez J.C. Expression of connexins during differentiation and regeneration of skeletal muscle: functional relevance of connexin43. J. Cell Sci. 2005;118:27–37. doi: 10.1242/jcs.01553. [DOI] [PubMed] [Google Scholar]
- 16.Tatusova T.A., Madden T.L. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 17.Jeanmougin F., Thompson J.D., Gouy M., Higgins D.G., Gibson T.J. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 18.Brummelkamp T.R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 19.Pfeifer I., Anderson C., Werner R., Oltra E. Redefining the structure of the mouse connexin43 gene: selective promoter usage and alternative splicing mechanisms yield transcripts with different translational efficiencies. Nucleic Acids Res. 2004;32:4550–4562. doi: 10.1093/nar/gkh792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 21.Baskerville S., Bartel D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Babak T., Zhang W., Morris Q., Blencowe B.J., Hughes T.R. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J.F., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M., Conlon F.L., Wang D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nature Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiriakidou M., Nelson P.T., Kouranov A., Fitziev P., Bouyioukos C., Mourelatos Z., Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 26.Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robins H., Li Y., Padgett R.W. Incorporating structure to predict microRNA targets. Proc. Natl Acad. Sci. USA. 2005;102:4006–4009. doi: 10.1073/pnas.0500775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doench J.G., Sharp P.A. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor M.V. Developmental biology: micromanaging muscle growth. Curr. Biol. 2006;16:R20–23. doi: 10.1016/j.cub.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y., Samal E., Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 31.Ontell M., Kozeka K. The organogenesis of murine striated muscle: a cytoarchitectural study. Am. J. Anat. 1984;171:133–148. doi: 10.1002/aja.1001710202. [DOI] [PubMed] [Google Scholar]
- 32.Jansen J.K., Fladby T. The perinatal reorganization of the innervation of skeletal muscle in mammals. Prog. Neurobiol. 1990;34:39–90. doi: 10.1016/0301-0082(90)90025-c. [DOI] [PubMed] [Google Scholar]
- 33.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Velleca M.A., Wallace M.C., Merlie J.P. A novel synapse-associated noncoding RNA. Mol. Cell Biol. 1994;14:7095–7104. doi: 10.1128/mcb.14.11.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rao P.K., Kumar R.M., Farkhondeh M., Baskerville S., Lodish H.F. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl Acad. Sci. USA. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clop A., Marcq F., Takeda H., Pirottin D., Tordoir X., Bibe B., Bouix J., Caiment F., Elsen J.M., Eychenne F., et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nature Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 37.Sweetman D., Rathjen T., Jefferson M., Wheeler G., Smith T.G., Wheeler G.N., Munsterberg A., Dalmay T. FGF-4 signaling is involved in mir-206 expression in developing somites of chicken embryos. Dev. Dyn. 2006;235:2185–2191. doi: 10.1002/dvdy.20881. [DOI] [PubMed] [Google Scholar]
- 38.Kloosterman W.P., Wienholds E., de Bruijn E., Kauppinen S., Plasterk R.H. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nature Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 39.Wienholds E., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 40.Naguibneva I., Ameyar-Zazoua M., Polesskaya A., Ait-Si-Ali S., Groisman R., Souidi M., Cuvellier S., Harel-Bellan A. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nature Cell Biol. 2006;8:278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]