Abstract
The budding yeast protein, Chl1p, is required for sister-chromatid cohesion, transcriptional silencing, rDNA recombination and aging. In this work, we show that Chl1p is also required for viability when DNA replication is stressed, either due to mutations or if cells are treated with genotoxic agents like methylmethane sulfonate (MMS) and ultraviolet (UV) rays. The chl1 mutation caused synthetic growth defects with mutations in DNA replication genes. At semi-permissive temperatures, the double mutants grew poorly, were less viable and showed nuclear fragmentation. They were, however, not limited in their bulk DNA synthesis. When chl1 cells were treated with relatively low levels of MMS in S-phase, they lost viability. The S-phase DNA damage checkpoint pathway, however, remained active in these cells. Agarose gel electrophoresis of genomic DNA isolated from wild-type and chl1 cells, after recovery from MMS treatment, suggested that the wild-type was more proficient in the repair of DNA damage than the mutant. Our work suggests that Chl1p is required for genome integrity when cells suffer endogenously or exogenously induced DNA damage.
INTRODUCTION
High-fidelity transmission of genetic material from parent cell to daughter cells requires accurate replication and segregation of chromosomes. Cells having unreplicated, partially replicated or damaged DNA, if allowed to continue with cell cycle, would generate aneuploidy and genetic instability. Cells have devised safety mechanisms, the DNA damage/replication checkpoint machinery, which can sense the damage or replication blocks and transmit signals to its components that temporarily halt the progression of the cycle till the damage is repaired, or cause apoptosis. In the budding yeast Saccharomyces cerevisiae, the DNA damage checkpoint pathway involves the two PI3K-like kinases Mec1 and Tel1, the RFC (replication factor C)-like complex consisting of RFC1-like protein Rad24p with four small RFC subunits (Rfc2–Rfc5), the PCNA-like heterotrimeric ring consisting of Rad17, Ddc1 and Mec3 proteins, and the MRX complex of proteins, consisting of Mre11, Rad50 and Xrs2. The MRX complex, along with Tel1p, is mainly required to sense and process double-strand breaks. All the proteins function in concert as DNA damage sensors by binding DNA at the site of DNA damage [reviewed in (1–6)]. They transmit the signal to the adaptor/mediator molecule, Rad9, which is activated by phosphorylation in a Mec1/Tel1-dependent fashion. Activated Rad9p binds to the effector kinase Rad53 (Chk2) that then gets activated by Mec1/Tel1-dependent phosphorylation (7–9). Activation of Rad53p, which is a major effector kinase in S-phase, turns on the transcription of genes required for damage repair, prevents late origins from firing so that DNA synthesis in S-phase is regulated, helps in stabilizing the existing forks and restrains spindle elongation in early S-phase (10–20).
Methylmethane sulfonate (MMS), a DNA alkylating drug, can cause nicks and gaps in DNA, resulting in cell death and defective S-phase regulation in checkpoint mutants mec1, rad53, rad24, rad17 and rad9 (21). Unrepaired single-strand breaks can give rise to double-strand breaks (22). When DNA is alkylated by MMS, S-phase progression is slowed down, presumably due to the stalling of forks when they encounter alkylated DNA. The checkpoint proteins Mec1 and Rad53 are required to prevent these stalled forks from collapsing irreversibly, thus permitting continuation of DNA replication and preserving cell viability (16,17). At low levels of drug concentrations, DNA damage activates Rad53p only in S-phase and requires the formation of replication forks (23).
mcm mutations of the budding yeast were isolated to identify genes required for the initiation of DNA replication (24). mcm1–mcm7 and mcm10 have identified genes, which affect the initiation of DNA replication at yeast chromosomal replication origins [reviewed in (25–27)]. In this work, we have cloned a hitherto uncharacterized MCM gene, MCM12, and show that it is the same as Chl1p. In the previous studies, Chl1p has been shown to be required for chromosome segregation (28,29) and for sister-chromatid cohesion in mitosis and meiosis (30–32). Mating-type interconversion studies have suggested its possible role in chromatin structure (33). Recent work has documented its involvement in transcriptional silencing, rDNA recombination and aging (34). CHL1 has 23% identity to the nucleotide excision repair gene RAD3, contains all seven consensus motifs found in helicases belonging to DEAH helicase family (29), has an essential ATP-binding site and is localized in the nucleus (30,35). It also has three highly related human homologs, BACH1, hChlR1 and hChlR2. Of these, hChlR1 shows in vitro DNA helicase activity and binds to both single- and double-stranded DNA (30,36,37). BACH1 is a member of the DEAH helicase family and binds to the tumor suppressor protein BRCA1, contributing towards its DNA repair activity (38).
In this work, we show that budding yeast Chl1p is required in S-phase for preserving cell viability when the DNA damage occurs by endogenous causes such as mutations, or by exogenous agents like MMS and UV rays. We also show that chl1 cells lose viability because they are deficient in the repair of MMS-induced DNA damage.
MATERIALS AND METHODS
Media and chemicals
Media has been described before (39). Restriction enzymes and other modifying enzymes were from New England Biolabs (USA), Bethesda Research Laboratories (BRL), USA and Bangalore Genei Pvt. Ltd (India). Proteinase K was from Boehringer Mannheim, Germany. Glusulase was from DuPont Company, USA, and Zymolyase 100T was from Seikagaku Kogyo Company Limited, Japan. Low melting agarose, propidium iodide, 4′,6-diamidino-2-phenylindole (DAPI), alpha-factor and goat anti-rat alkaline phosphatase-conjugated secondary antibody were from Sigma. Rat monoclonal antibody, YOL1/34 (40) directed against alpha-tubulin from yeast was from Serotec. Rad53 goat polyclonal antibody, raised against a C-terminus peptide of yeast Rad53p, and secondary alkaline phosphatase-conjugated anti-goat antibody were from Santa Cruz Biotechnology, USA. NBT/BCIP (Nitro-Blue Tetrazolium Chloride/5-Bromo-4-Chloro-3′-Indolyphosphate p-Toluidine Salt) was from Bangalore Genei Pvt Ltd. MMS was from SRL (India). For irradiation with UV rays, the source was a germicidal UV lamp (NIS G30T8, 30 W, length 86 cm). The plates were kept at a distance of 95 cm from the UV source.
Strains and plasmids
The plasmid YIplac211 is described in (41). YCp1 (carrying ARS1) and YCp121 (carrying ARS121) have been described in (24). YCp101 (ARS1 CEN5 LEU2) was from B.-K. Tye (42). Table 1 gives the list of strains used. Strain 699 and all the strains listed below it were in W303 background while the parent strains of the remaining were from G. Fink. The construction of double mutants and deletions of SGS1, CHL1 and BAR1 are described under Supplementary Data.
Table 1.
Strains used in this study
| Strain | Genotype | Sources/References |
|---|---|---|
| A3 | MATaleu2-3, 112 his3-11, 15 | (24) |
| AP22 | MATα leu2-3, 112 his3-11, 15 ura3-52 trp1 | (34) |
| R67 | MATα leu2 ura3 his4 mcm12-1 | From B.-K. Tye |
| M11 | MATα leu2 ura3 his4 mcm12-2 | (24) |
| 8534-8C | MATα his4Δ34 ura3-52 leu2-3,112 | (24) |
| A3Dchl1 | MATa leu2-3, 112 his3-11, 15 chl1::HIS3 | This study |
| AP22Δ22 | MATα leu2-3, 112 his3-11, 15 ura3-52 trp1 mcm22-Δ1::TRP1 | By deleting AP22 (44) |
| AP22Dchl1 | MATα leu2-3, 112 his3-11, 15 ura3-52 trp1 chl1::HIS3 | (34) |
| M46-3C | MATα leu2-3, 112 his3-11, 15 ura3-52 mcm2-1 | (24) |
| M46-3CDchl1 | MATα leu2-3, 112 his3-11, 15 ura3-52 mcm2-1 chl1::HIS3 | This study, by disrupting CHL1 in M46-3C |
| M46-3CΔ21 | MATα leu2-3, 112 his3-11, 15 ura3-52 mcm2-1 mcm21-Δ2::LEU2 | This study |
| M46-3CDchl4 | MATα leu2-3, 112 his3-11, 15 ura3-52 mcm2-1 chl4::HIS3 | This study |
| SL13 | MATaleu2-3, 112 his3-11, 15 ura3-52 mcm2-1 | By crossing M46-3CDchl1 with A3 |
| SL13Dchl1 | MATaleu2-3, 112 his3-11, 15 ura3-52 mcm2-1 chl1::HIS3 | This study, by disrupting CHL1 in SL13 |
| 699 | MATaade2-1 trp1-1 leu2-3, 112 his 3-11, 15 ura3 can1-100 | Uttam Surana |
| 699Dchl1 | MATaade2-1 trp1-1 leu2-3, 112 his 3-11, 15 ura3 can1-100 chl1::HIS3 | This study, by disrupting CHL1 in 699 |
| SL14 | MATaade2-1 trp1-1 leu2-3, 112 his 3-11, 15 ura3 can1-100 bar1::LEU2 | This study, by disrupting BAR1 in 699 |
| SL14Dchl1 | MATaade2-1 trp1-1 leu2-3, 112 his 3-11, 15 ura3 can1-100 chl1::HIS3 bar1::LEU2 | This study, by disrupting BAR1 in 699Dchl1 |
| 699Δsgs1 | MATaade2-1 trp1-1 leu2-3, 112 his 3-11, 15 ura3 can1-100 sgs1Δ::LEU2 | This study, by deleting SGS1in 699 |
| 699Δsgs1Dchl1 | MATaade2-1 trp1-1 leu2-3, 112 his 3-11, 15 ura3 can1-100 sgs1Δ::LEU2 chl1::HIS3 | This study, by disrupting CHL1 in 699Δsgs1 |
| US456 | MATα leu2 his3 trp1 ade2 rad24::URA3 | Uttam Surana |
| SL3 | MATaleu2 his3 trp1 ade2 rad24::URA3 | By crossing US456 with 699Dchl1 |
| SL3Dchl1 | MATaleu2 his3 trp1 ade2 rad24::URA3 chl1::HIS3 | From SL3, by disrupting the CHL1gene |
| SL4 | MATaleu2 his3 trp1 ade2 rad24::URA3 chl1::HIS3 | By crossing US456 with 699Dchl1 |
| SS1 | MATaleu2 his3 trp1 ade2 rad24::URA3 bar1::LEU2 | From SL3, by disrupting the BAR1gene |
| SS1Dchl1 | MATaleu2 his3 trp1 ade2 rad24::URA3 chl1::HIS3 bar1::LEU2 | From SL3Dchl1, by disrupting the BAR1gene |
| US354 | MATα leu2 his3 trp1 ade2 ura3 rad53-21 | Uttam Surana |
| SL7 | MATaleu2 his3 trp1 ade2 ura3 rad53-21 | By crossing US354 with 699 |
| SL7Δchl1 | MATaleu2 his3 trp1 ade2 ura3 rad53-21 chl1Δ::TRP1 | This study, by deleting CHL1in SL7 |
| US3138 | MATaade2-1leu2-3,112 trp1-1 his3-11,15ura3 can1-100 bar1 GAL psi+mec1-1 | Uttam Surana |
| YB0297 | MATaade2 leu2 trp1 his3 can1 GAL psi+cdc6-3 | From B.-K. Tye |
| YB0297Dchl1 | MATaade2 leu2 trp1 his3 can1 GAL psi+cdc6-3 chl1::HIS3 | This study, by disrupting CHL1 in YB0297 |
| JRY4490 | MATacan1-100 his3-11 leu2-3, 112 lys2Δ trp1-1 ura3-1 orc2-1 | From B.-K. Tye |
| JRY4245 | MATaade2-1 can1-100 his3-11 leu2-3, 112 trp1-1 ura3-1 orc5-1 | From B.-K. Tye |
| JRY4245Dchl1 | MATaade2-1 can1-100 his3-11 leu2-3, 112 trp1-1 ura3-1 orc5-1 chl1::HIS3 | This study, by disrupting CHL1 in JRY4245 |
Cell synchronization, flow cytometry, cell viabilities and nuclear staining
Cells were synchronized in G1 phase using alpha-factor as described in (43) and processed for flow cytometry according to (44). For measuring cell viabilities, aliquots of cells were removed at indicated times, counted, appropriately diluted and plated in duplicate on YEPD plates. Cell viability was the fraction of plated cells which gave visible colonies after 3 to 4 days of growth at permissive temperatures on YEPD plates. Viabilities were normalized with respect to initial values at 0 time points. Cells were fixed for 30 min using 70% ethanol, washed with water and nuclei were stained using DAPI (45). Around 150–200 cells were counted for data involving cell cycle arrests and nuclear morphologies, using a fluorescence microscope (Leica fitted with DC 300F camera).
Protein extractions, western blots
For western blot analysis, protein extracts were prepared according to (8) from cells synchronized in G1 and released in YEPD medium containing 0.035% MMS. Proteins were separated on 8% SDS–PAGE containing an acrylamide to bis-acrylamide ratio of 80:1 and transferred to polyvinylidene difluoride (PVDF) membrane (Schleicher and Schuell). Rad53 was detected using anti-Rad53 goat polyclonal antibody at 1:1000 dilution in TBS (50 mM Tris buffer pH 7.5, 150 mM NaCl) containing 0.5% BSA for 12–16 h. Secondary alkaline phosphatase-conjugated anti-goat antibody was incubated with the membrane for 2 h at 1:2500 dilution.
Preparation of DNA in agarose plugs and agarose gel electrophoresis for DNA repair
Overnight cultures of cells were re-inoculated in liquid YEPD (at temperatures described in the text for different strains) and grown for 3–4 h to an OD610 of ∼0.8. Cells were synchronized in G1 using alpha-factor, released from arrest in the original volumes of YEPD, divided into two halves, one containing the indicated concentration of MMS and the other was left untreated. After 30 min of shaking at 28°C, equal volume of 10% sodium thiosulphate was added to each culture and cells were washed quickly at room temperature. The cells were then released in original volumes of fresh YEPD medium and aliquots were withdrawn at various time intervals. DNA from these cells was prepared in agarose plugs (inserts) according to (46). Briefly, cell pellets were washed twice with 50 mM EDTA (pH 8.0) and resuspended in 400 μl of the same solution to a concentration of ∼0.5 × 1010 cells/ml. Equal volume of 1% low melting agarose at 37°C was added to the cell suspension and zymolyase 100T was added to this mixture to a final concentration of 20 μg/ml. This mixture was distributed into 100 μl moulds (Bio-Rad mould chamber) and allowed to solidify for 30 min on ice. The agarose plugs were kept overnight at 37°C in 0.5 M EDTA pH 8.0 and 7.5% β-mercaptoethanol (2 to 3 plugs per ml) for spheroplasting. Next day the plugs were transferred to ESP (0.5 M EDTA pH 9.0, 1% laurylsarcosine and 1 mg/ml proteinase K) and kept at 50°C for 48 h. The plugs were stored at 4°C in 0.5 M EDTA pH 8.0. DNA in these plugs was electrophoresed by conventional one-dimensional agarose gel (0.5%) electrophoresis at 2.5 V/cm for 28 h in TBE (0.089 M Tris borate, 0.089 M boric acid and 0.002 M EDTA) buffer.
RESULTS
The chl1 mutation shows synergistic growth defects with mutations in genes required for DNA replication
Cloning and sequencing of the MCM12 gene showed it to be the same as CHL1 (Supplementary Data). Therefore, in this work MCM12 and Mcm12p will be referred to as CHL1 and Chl1p, respectively. The chl1 null mutant carries the chl1::HIS3 disruption mutation made in this study (Supplementary Figure 1s).
The mcm mutants have been found to affect chromosome replication or segregation (25). chl1 is known to cause chromosome missegregation due to defects in sister-chromatid cohesion (30,31). To study the role, if any, of Chl1p/Mcm12p in chromosome replication, genetic interactions of chl1 with mutations in genes required for DNA replication were studied. MCM2, CDC6, ORC2 and ORC5 are all essential genes that are required for the initiation of DNA replication while MCM2 is also required for elongation (25,27). It was reasoned that synthetic lethality of chl1 with a mutation in any of these genes would indicate the involvement of Chl1p with some aspect of DNA replication.
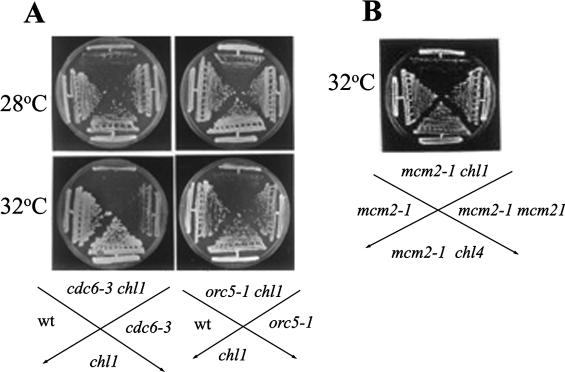
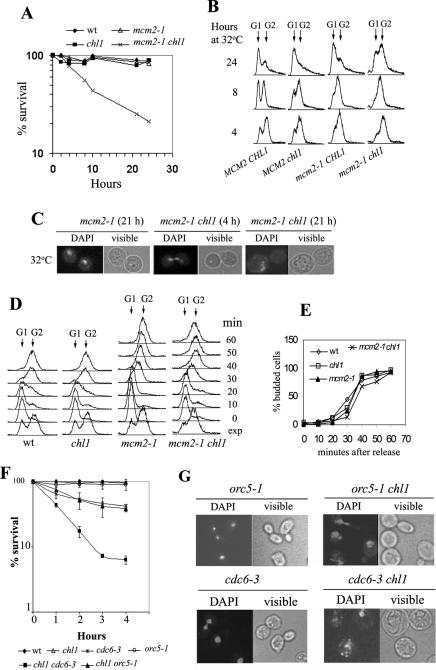
Double mutants, mcm2-1 chl1::HIS3, orc5-1 chl1::HIS3 and cdc6-3 chl1::HIS3 were tested for growth at various temperatures. All these double mutants exhibited synergistic growth defects at 32°C (Figure 1A and B). To show that these genetic interactions were specific to the chl1 mutation, double mutants mcm2-1 chl4::HIS3 and mcm2-1 mcm21-Δ2::LEU2 were also constructed, where chl4::HIS3 and mcm21-Δ2::LEU2 are mutations in genes coding for kinetochore proteins (47,48). However, mcm2-1 chl4::HIS3 and mcm2-1 mcm21-Δ2::LEU2 did not display any synergistic growth defects (Figure 1B), suggesting that mcm2-1 shared a specific interaction with chl1/mcm12 and not with mcm mutations affecting kinetochore proteins. orc2-1 was found to be synthetically lethal with chl1 null mutation (Supplementary Data). A study of cell viabilities showed that, when compared with the single mutant mcm2-1 (SL13), the double mutant mcm2-1 chl1 (SL13Dchl1) displayed a steady fall in viability at 32°C (Figure 2A). Similarly, double mutants orc5-1 chl1::HIS3 and cdc6-3 chl1::HIS3 also lost viability more rapidly than the single mutants (orc5-1, cdc6-3 and chl1::HIS3) when shifted from 23°C to 32°C for 4 h (Figure 2F).
Figure 1.
(A and B) Genetic interactions of chl1::HIS3 with mutations in DNA replication genes. Cells were streaked on YEPD plates, which were incubated at indicated temperatures for 2 days. The strains used were wild-type (wt), AP22; chl1, AP22Dchl1; cdc6-3, YB0297; orc5-1, JRY4245; mcm2-1, M46-3C; Double mutants were constructed by disrupting/deleting CHL1, CHL4 or MCM22 in these strains as described in Supplementary Data.
Figure 2.
Cell viability, G2/M arrest and nuclear fragmentation in wild-type, chl1, mcm2-1 and mcm2-1 chl1 cells. A3 (MCM2 CHL1), A3Dchl1 (MCM2 chl1), SL13 (mcm2-1 CHL1) and SL13Dchl1 (mcm2-1 chl1) cells were grown in YEPD at 23°C to log-phase and re-inoculated at 32°C in pre-warmed YEPD medium. Aliquots were removed at various times for determining cell viabilities (A), for fluorescence activated cell sorting (FACS) analysis (B) and stained with DAPI for nuclear morphology (C). Samples from a few representative time points are shown for (B) and (C) h, hours. (D and E) The same strains were arrested in G1 using alpha-factor and released at 32°C in fresh YEPD. Aliquots were removed at time intervals for monitoring S-phase progression using flow cytometry and for budding index (fraction of cells that had initiated budding after release from G1 arrest). (F) Cell viabilities of wild-type, chl1, orc5-1, orc5-1 chl1, cdc6-3 and cdc6-3 chl1 grown at 32°C. 699 (wild-type), 699Dchl1 (chl1), JRY4245 (orc5-1), JRY4245Dchl1 (orc5-1 chl1), YB0297 (cdc6-3) and YB0297Dchl1 (cdc6-3 chl1) were grown to exponential phase in liquid YEPD at 23°C, re-inoculated into pre-warmed liquid YEPD at 32°C and processed for the determination of cell viabilities as in (A). (G) Nuclear fragmentation in orc5-1 chl1 and cdc6-3 chl1 cells. JRY4245 (orc5-1), JRY4245Dchl1 (orc5-1 chl1), YB0297 (cdc6-3) and YB0297Dchl1 (cdc6-3 chl1) were grown to exponential phase in liquid YEPD at 23°C, re-inoculated into pre-warmed liquid YEPD at 32°C and grown for 4.5 h. Thereafter, cells were stained with DAPI and analyzed for their morphologies using the fluorescence microscope. Abbreviations: h, hours; min, minutes; exp, exponential.
At non-permissive temperatures, the mcm2-1 mutation is known to cause DNA damage and cell cycle (late S/G2/M) arrest which is RAD9 dependent (46,49,50). This arrest is characterized by the accumulation of large-budded uni-nucleated cells with the nucleus mostly at the neck or stretched through it. An examination of the mcm2-1 and mcm2-1 chl1 cell cultures showed that a higher percentage of mcm2-1 chl1 cells were present as large-budded cells arrested with single nucleus at the neck (Table 2). Fluorescence-activated cell sorting (FACS) analysis of cells growing exponentially at 32°C (4 and 8 h; Fig 2B) showed that most of mcm2-1 chl1 cells had a near 2n DNA content, suggesting that these cells were spending more time at G2/M. At 24 h, wild-type, mcm2-1 and chl1 cells proceeded towards stationary phase, as G1 (single) cells started to accumulate in the cultures. At this time point, mcm2-1 chl1 cells still showed a high percentage of arrested large-budded cells (Table 2). It may be mentioned that chl1 cells also pause at G2/M due to defects in sister-chromatid cohesion and independent of the DNA damage checkpoint (29,30). However, the fraction of arrested cells in mcm2-1 chl1 culture was more than additive of the single mutants, especially at later time points, suggesting that factors other than sister-chromatid cohesion were involved in causing G2/M arrest in the double mutant. The simplest explanation for the synergistic effects observed between mcm2-1 and chl1 mutations is that the double mutant cells were accumulating higher DNA damage than mcm2-1 cells and were arrested at G2/M for longer times to repair the damage.
Table 2.
chl1enhances the cell cycle arrest phenotype of mcm2-1cells
| Strain | Hours at 32°C | |||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 4 | 8 | 24 | |||||
| % Large-budded | % Arrested | % Large-budded | % Arrested | % Large-budded | % Arrested | % Large-budded | % Arrested | |
| (MCM2 CHL1) | 11 | 0 | 7.4 | 0.7 | 3 | 0 | 4.7 | 0.8 |
| (MCM2 chl1) | 27 | 7 | 15 | 6.4 | 17 | 9.2 | 2.3 | 0.6 |
| (mcm2-1 CHL1) | 17 | 1 | 42 | 23 | 35 | 16 | 27 | 10 |
| (mcm2-1 chl1::HIS3) | 30 | 15 | 65 | 43 | 77 | 47 | 60 | 26 |
Cells from Figure 2 were analyzed for cell cycle arrest (percent large-budded cells in the culture) and nuclear morphology (percent large-budded cells in the culture which had single nucleus at the neck or stretched through it). About 150–200 cells were analyzed in each case.
The double mutant culture was also marked by the presence of cells with fragmented nuclei after prolonged growth at 32°C (Figure 2C). Nuclear fragmentation could be seen in all cell types—singles, large-budded with single nucleus (arrested) or large-budded with segregated nuclei. Perhaps this last category of cells were entering catastrophic mitosis, overcoming the DNA damage checkpoint without repair of DNA damage. The fraction of cells with fragmented nuclei increased progressively with continued incubation at 32°C (Table 3). The extent of fragmentation also increased during this time interval in that cells with highly fragmented nuclei started to appear at later time points. The fraction of such cells in cultures of other strains (wild-type and the single mutants) remained <5% at all times during growth at 32°C.
Table 3.
mcm2-1 chl1 cells show increased fragmentation of nuclei at 32°C
| Strain | % Cells with fragmented nuclei | |||
|---|---|---|---|---|
| 0 h | 4 h | 10 h | 24 h | |
| MCM2 CHL1 | 3.1 | 0.7 | <0.6 | 0.8 |
| MCM2 chl1 | 1.7 | 2.0 | <0.6 | 0.6 |
| mcm2-1 CHL1 | 1.2 | 3.5 | 5.1 | 1.3 |
| mcm2-1 chl1 | 6.6 | 13 | 24 | 36 |
Cells from Figure 2A were analyzed for fragmented nuclei by staining DNA with DAPI.; ∼150–200 cells were analyzed in each case. ‘Hours’ refer to incubation time at 32°C.
Cultures of orc5-1 chl1 grown for 4.5 h at 32°C also had a greater fraction of arrested cells with fragmented nuclei when compared with the single mutants (Table 4,Figure 2G). Under same conditions of growth, cdc6-3 and cdc6-3 chl1 cells displayed similar fractions of large-budded uni-nucleated (arrested) cells in their respective cultures (Table 4). cdc6-3 chl1 cells were, however, abnormally large, looked sick and displayed extensive nuclear fragmentation as opposed to cdc6-3 cells (Figure 2G, Table 4).
Table 4.
Cell and nuclear morphology in orc5-1, orc5-1 chl1, cdc6-3 and cdc6-3 chl1 mutant cells after 4.5 h of growth at 32°C
| Strain | Cell and nuclear morphology | ||
|---|---|---|---|
| % Large-budded | % Arrested | % Fragmented nuclei | |
| orc5-1 | 25 | 7.3 | 3.3 |
| orc5-1 chl1 | 54 | 33 | 19 |
| cdc6-3 | 72 | 62 | 5.3 |
| cdc6-3 chl1 | 69 | 53 | 33 |
Thus, replication defects in these mutants were exacerbated by chl1 and suggested a role of Chl1p in the maintenance of DNA integrity in replication mutants. One possibility was Chl1p's involvement in the initiation or for some other aspect of DNA replication, due to which the double mutants discussed above could be severely compromised in the synthesis of their DNA. A comparison of S-phase progression of wild-type and mutant cells at 32°C showed that chl1 cells progressed at the same rate as the wild-type, reaching G2 within 40 min (Figure 2D). The budding index (Figure 2E) showed that both the strains entered S-phase at similar timings. This shows that the absence of Chl1p does not affect bulk DNA synthesis. As expected, the replication mutant mcm2-1 synthesized DNA more slowly, taking 60 min to reach G2. If chl1 affected the efficiency of replication origin usage and mcm2-1 chl1 cells were growing poorly because of synergistic deficiency in origin utilization, then the double mutant was expected to synthesize DNA more slowly than mcm2-1. This, however, was not the case. On the contrary, mcm2-1 chl1 cells progressed somewhat faster than the mcm2-1 cells. The budding index (Figure 2E) showed that mcm2-1 and mcm2-1 chl1 cells entered S-phase at about the same time. This result was reproducible in three experiments. Similar results were obtained with orc5-1 and orc5-1 chl1 (not shown). Therefore, chl1 cells were not limited in the utilization of their replication origins nor in bulk DNA synthesis. This was further corroborated by the observations that chl1 mutation did not show ARS-specificity nor ARS-dosage-dependence in the maintenance of minichromosomes (51). Furthermore, chl1 enhanced chromosome loss rate but did not significantly elevate genetic recombination frequency, making it unlikely that Chl1p has a major role in DNA replication (29,51,52).
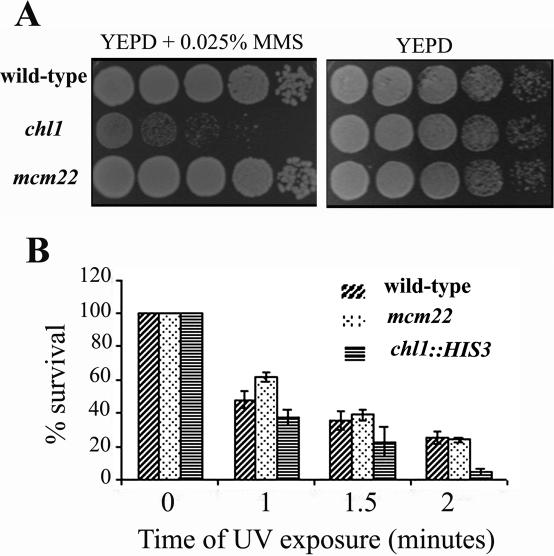
The chl1 null mutant is more sensitive than the wild-type to MMS and UV rays
Cells carrying defects in DNA damage repair or checkpoint pathways are more sensitive to killing by DNA damaging agents like UV rays and the DNA alkylating drug MMS. In genome-wide screens, it has been reported that chl1 deletion mutant is hypersensitive to MMS (53,54). Our own results showed the same. AP22Dchl1 (chl1), AP22Δ22 [deleted for MCM22 that codes for a kinetochore protein (44)] and the wild-type strain, AP22, were all tested for their sensitivities towards MMS. While the chl1 null mutant showed higher sensitivity towards this drug (Figure 3A), the kinetochore mutant mcm22 grew like the wild-type. Cell viabilities of chl1 and wild-type cells in the presence of 0.035% MMS confirmed these results (not shown). MMS-treated cultures of mutant and wild-type cells showed >80% accumulation of G2/M-arrested large-budded cells, each with a single nucleus at the neck or stretched through it (not shown), suggesting that the G2/M DNA damage checkpoint pathway is functional in the mutant.
Figure 3.
The chl1 mutation confers growth sensitivity in the presence of MMS and UV rays. (A) Spot assay for MMS sensitivity of chl1::HIS3 (AP22Dchl1), AP22Δ22 (mcm22-Δ1::TRP1), and wild-type (AP22) strains. Growing cells were serially diluted and spotted on YEPD plates containing 0.025% MMS or no MMS (control). The YEPD plate was incubated at 30°C for 2 days while the MMS-containing plate was incubated at the same temperature for 3–4 days. (B) chl1 mutation confers hypersensitivity to UV rays. Exponentially growing cells, appropriately diluted, were plated on YEPD plates, exposed to UV rays for various times and incubated in the dark at 30°C. Control plates were also kept which were not irradiated and were used to calculate total number of cells plated. Colonies were counted after 2–3 days of incubation. Results are the mean values of four independent experiments and error bars are SD values from the mean.
To determine whether chl1 disruption mutation was more susceptible to killing by UV damage, we compared the cell-viabilities of the wild-type AP22, the chl1 disruption mutant AP22Dchl1 and the kinetochore mutant AP22Δ22, in response to different dosages of UV irradiation. Figure 3B shows that AP22Dchl1 was more sensitive towards killing by UV irradiation than the wild-type or the kinetochore mutant AP22Δ22.
The chl1 mutant is proficient in DNA damage checkpoint pathway
The observations that the chl1 null mutation gives synergistic growth defects with DNA replication mutants, shows hypersensitivity to genotoxic agents like UV rays and MMS, causes chromosome loss without significant increase in genetic recombination rates, suggests that Chl1p could be involved in DNA damage repair or in checkpoint function or both. Although the presence of arrested cells in MMS-treated chl1 cultures and in double mutants described above suggested that the DNA damage checkpoint pathway was active in these cells, we decided to check on this pathway by more direct experiments, as described below.
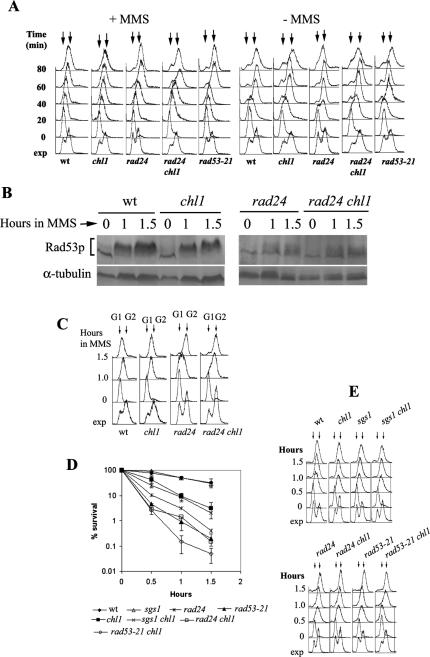
When replication forks stall in the presence of MMS, S-phase is slowed down (16,21). However, when DNA is damaged in some S-phase checkpoint mutants such as mec1, rad9, rad17, rad24 and rad53, S-phase appears to progress faster because late origins fire inappropriately, causing additional DNA synthesis, which can be detected by flow cytometry (16,21). To test whether Chl1p had an intra S-phase checkpoint function, the progression of the cell cycle was studied in cells synchronized with alpha-factor. rad24 (SL3) and rad53-21 (SL7) mutants were also included in these studies, as they are known to be defective in regulating their DNA synthesis when it is damaged. After their release from alpha-factor, the cells were resuspended in fresh medium containing 0.035 % MMS and the progression of cells across S-phase were monitored by flow cytometry. After 80 min of treatment with MMS, both chl1 and wild-type strains were still in S-phase (Figure 4A). In contrast to chl1, rad24 and rad53-21 mutants continued to synthesize DNA whose content reached G2 within 60 min of release from G1 arrest. The double mutant rad24 chl1 was not very different from the single mutant rad24, in so far as the time taken to reach G2 DNA content was concerned. Since the S-phase progressions in chl1 and wild-type cells were similar in the presence of MMS, it suggests that the DNA damage checkpoint pathway was mostly active in these cells.
Figure 4.
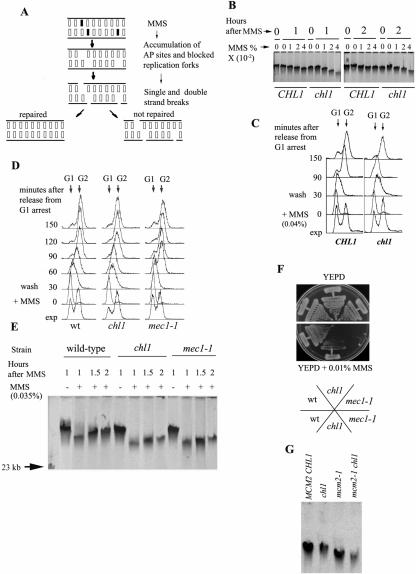
DNA damage checkpoint is active in chl1 mutant cells. (A) S-phase progression of mutant and wild-type cells in the presence of MMS. 699 (CHL1 RAD24 RAD53), 699Dchl1 (chl1), SL3 (rad24), SL4 (rad24 chl1) and SL7 (rad53-21) cells were all synchronized with alpha-factor, washed free of alpha-factor, resuspended in pre-warmed YEPD medium at 30°C, divided into two parts and MMS was added to a final concentration of 0.035% to one part. Both the cultures were kept shaking at 30°C. Aliquots were removed at various times for FACS analysis. Arrows indicate G1 and G2 DNA contents. (B) chl1 are proficient in Rad53p phosphorylation in response to MMS treatment in S-phase. SL14 (CHL1 RAD24), SL14Dchl1 (chl1), SS1 (rad24) and SS1Dchl1 (rad24 chl1) were arrested in G1 phase and released in fresh YEPD medium containing 0.035% MMS at 30°C. Rad53p phosphorylation was detected by western blot analysis of proteins extracted from aliquots of cells removed at indicated times, using antibodies directed against the Rad53 protein. (C) DNA content of cells from the same aliquots analyzed by flow cytometry. (D) chl1 cells are hypersensitive towards killing by MMS in S-phase. 699 (wild-type), 699Dchl1 (chl1), SL3 (rad24), SL3Dchl1 (rad24 chl1), 699Δsgs1 (sgs1), 699Δsgs1Dchl1 (sgs1 chl1), SL7 (rad53-21), SL7Δchl1 (rad53-21 chl1) cells were arrested by alpha-factor in G1 and released in fresh YEPD containing 0.035% MMS. Aliquots were removed for cell viabilities and for DNA content. Data are averages of two independent experiments which gave very similar results. Bars are deviations from the average values and are not shown in rad24 and rad24 chl1 series for the sake of clarity in rad53 and rad53 chl1 series. (E) DNA content of cells in (D) measured by flow cytometry. Arrows indicate G1 and G2 DNA contents. Abbreviations are as in Figure 2.
To confirm this, Rad53p activation was studied directly by assaying for its phosphorylation in MMS-treated cells. Cells were synchronized with alpha-factor and released in YEPD in the presence of 0.035% MMS. Aliquots were withdrawn at indicated times. Figure 4B and C show that chl1 cells had Rad53p phosphorylated to the same levels as the wild-type in S-phase. rad24 cells, as expected, showed much lower levels of Rad53p phosphorylation while rad24 chl1 mutant was no different than rad24. Thus, Chl1p is not required to activate the DNA damage checkpoint pathway when cells are treated with MMS in S-phase.
Finally, it was found that S-phase chl1 cells were significantly more sensitive towards killing by MMS than the wild-type. Mutant and wild-type cells were synchronized in G1, released in S-phase and exposed to 0.035% MMS for varying times. Figure 4D and E show the viability curve. chl1 cells were more prone than the wild-type towards killing by the drug while rad24 and rad53-21 displayed very low viabilities. The fall in the viability of chl1 cells was additive with both rad24 and rad53-21 mutants in that rad24 chl1 and rad53-21 chl1 cells showed even lower viabilities than the single mutants. rad53-21 is a null mutation in so far as its role in DNA replication/damage checkpoint pathways is concerned (55). Higher cell death in rad53-21 chl1 cells, as compared with rad53-21 cells, suggested that Chl1p was required in addition to the DNA damage checkpoint pathway to maintain viability when cells were treated with MMS in S-phase.
Sgs1p is a member of the RecQ family of helicases which are involved in maintaining genome stability (56,57). In yeast, Sgs1p is a component of the DNA replication checkpoint pathway which gets activated upon a block in replication (58). sgs1 chl1 double mutant was included to see if there was a functional overlap between the two helicases, Chl1p and Sgs1p, that would give synergistic loss in cell viability. Figure 4D shows that the sgs1 mutant displayed the same viability as the wild-type and did not display any synthetic interactions with chl1. Therefore, unlike Chl1p, Sgs1p does not play a major role in maintaining genome integrity when cells suffer limited DNA damage in S-phase by low concentrations (0.035%) of MMS. The DNA damage checkpoint pathway, involving Rad24p, appears to be the major route for the maintenance of genome stability under these conditions.
Chl1p is required in S-phase for MMS-induced DNA damage repair
To determine if Chl1p was required for the repair of DNA damage inflicted in S-phase by MMS, mutant and wild-type cells were synchronized with alpha-factor and released in YEPD containing MMS. After a period of treatment with MMS, the cells were released in fresh YEPD medium without the drug and grown to repair the damage. We argued that if the mutant DNA had more unrepaired nicks and gaps than the wild-type DNA, double-strand breaks would be generated (22,59) and the average size of mutant DNA would be lesser than that of the wild-type (Figure 5A). This difference would manifest itself during the movement of DNA through agarose.
Figure 5.
Chl1p is required for DNA damage repair in S-phase. (A) A schematic representation of the mechanism by which alkylated DNA can give rise to nicks, gaps and double-strand breaks, resulting in higher mobility of genomic DNA during agarose gel electrophoresis. Alkylated bases are cleaved by DNA glycosylases creating apurinic or apyrimidinic (AP) sites which are cleaved by AP endonuclease/lyase giving rise to single-strand breaks. Closely opposed and unrepaired single-strand breaks (SSB), coupled with the stalling of replication forks at alkylated bases can give rise to double-strand breaks in DNA (59) which are repaired in the wild-type but not in the mutant. (B and C) Overnight cultures of wild-type (SL14) and mutant strains chl1 (SL14Dchl1) grown at 30°C in liquid YEPD were re-inoculated in YEPD at 30°C till mid-log-phase (OD610 ≅ 0.8–1) and synchronized in G1 using alpha-factor. Cells were released from arrest in liquid YEPD at 30°C in the presence or absence of varying concentrations of MMS. At indicated times, aliquots were taken out for FACS analysis (C) and to isolate DNA in agarose plugs as described in Methods section. Agarose gel (0.5%) electrophoresis of DNA was carried out for 28 h at 2.5 V/cm, as described under Methods (B). (D) Overnight cultures of wild-type (SL14) and mutant strains chl1 (SL14Dchl1) and mec1-1 (US3138), grown at 23°C in liquid YEPD were re-inoculated in YEPD at 28°C till mid-log phase (OD610 ≅ 0.8–1) and synchronized in G1 using alpha-factor. Cells were released from arrest in liquid YEPD at 28°C in the presence or absence of 0.035% MMS. At indicated times, aliquots were taken out for FACS analysis (D) and to isolate DNA in agarose plugs as described in Methods. (E) Agarose gel electrophoresis of the DNA in plugs from (D) was carried out as described above. There were no differences in the mobilities of genomic DNA isolated from cultures not exposed to MMS at all time points in the experiment. DNA from untreated cells (−MMS) corresponds to genomic DNA isolated 1.5 h after release from arrest of untreated cells, that is, 1 h after MMS wash. ‘−’ indicates no MMS was added while ‘+’ indicates MMS added. (F) Growth of wild-type SL14 (CHL1 MEC1), SL14Dchl1 (chl1 MEC1) and US3138 (mec1-1) on YEPD plates with or without 0.01% MMS. Plates were incubated for 3 days at 23°C. (G) Agarose gel electrophoresis (as described above) of DNA isolated in plugs from wild-type (A3), chl1 (A3Dchl1), mcm2-1 (SL13) and mcm2-1 chl1 (SL13Dchl1) cells grown in liquid YEPD at 32°C for 24 h.
Wild-type and chl1 cells were released from alpha-factor arrest into YEPD containing 0.01%, 0.02% and 0.04% MMS. Control cells were also maintained under similar conditions without MMS. After 30 min, MMS treatment was stopped, cells were washed and grown in YEPD for further periods of 1 and 2 h to allow for recovery from DNA damage. Genomic DNA isolated in agarose plugs (to avoid shearing) from all these time points was run on a 0.5% agarose gel as described in the Methods section. Figure 5B shows that after 1 h recovery period, DNA from cells treated with progressively higher concentrations of MMS showed a progressive increase in DNA mobility, suggestive of the presence of DNA having lower average molecular weight. Thus, the extent of MMS treatment corresponded with the level of DNA damage. MMS-treated DNA from mutant cells showed higher mobility than corresponding cells from the wild-type, suggestive of the presence of additional damage in the DNA of mutant cells. After an additional hour of recovery, DNA from MMS-treated wild-type cells began to migrate close to DNA from untreated cells. However, DNA from MMS-treated mutant cells still lagged behind in upward shift, showing a slow recovery from DNA damage. The FACS profile of the cells treated with 0.04% MMS (Figure 5C) showed that most of the cells from the two strains were arrested in G2 phase of the cell cycle.
We next performed the same experiment using 0.035% MMS which we have used for viabilty and other studies in this work. The DNA replication and damage checkpoint mutant, mec1-1, was also included as a control. DNA was isolated from these cells in agarose plugs and it was run in 0.5% agarose gel. After 30 min treatment in the presence of MMS, the cells, still in S-phase (Figure 5D), were released in fresh YEPD medium and allowed to grow for 60, 90 and 120 min to repair the damage. Genomic DNA from both chl1 and mec1-1 cells, isolated 60 min after treatment with MMS (1 h repair period), moved faster than the wild-type DNA isolated under similar conditions (Figure 5E). Assuming that the level of DNA damage inflicted by MMS was the same in wild-type and mutant cells, it is reasonable to deduce that the higher mobility of mutant DNA was due to the inability of these cells to repair DNA damage at the same pace as the wild-type cells. chl1 and mec1-1 cells approached or reached G2 phase without repairing their DNA damage to the same extent as the wild-type cells. Genomic DNA isolated 90 min after MMS treatment showed some repair in all the strains, as evidenced by the decreased mobility of DNA. However, the DNA of wild-type strain moved the slowest, showing maximum repair. At this stage, ∼4.5%, 2.8% and 13% of the wild-type, chl1 and mec1-1 cells had segregated their respective DNA. The percentages of large-budded cells in the cultures were 54, 57 and 31 respectively. After 2 h of treatment, the mobilities of wild-type MMS-treated and untreated DNA were almost the same. The DNA from the mutant (chl1 and mec1-1) cells, however, still exhibited higher mobilities indicative of preponderance of lower molecular weight species due to defective repair. At this time chl1 cells were still arrested at G2/M (81% large-budded and only 3% of total cells had segregated nuclei) but wild-type had initiated nuclear segregation (77% large-budded cells and 10% of cells had segregated nuclei). The mec1-1 culture had 67% large-budded cells and 33% of the cells had segregated nuclei. This precocious segregation of damaged DNA would be one of the reasons why mec1-1 cells are much less viable than chl1 cells when challenged with MMS (Figure 5F). Thus, the FACS profile (Figure 5D) and nuclear morphologies showed that the majority of wild-type and chl1 cells were still arrested at G2/M after 2 h of recovery time. Yet there was an upward shift of DNA in this period. The simplest explanation is that while the cells were resting at G2/M, the DNA was getting continuously repaired. This repair was sluggish in the mutant cells. Thus, Chl1p is required for efficient repair of MMS-induced DNA damage.
We have shown earlier that the mcm2-1 mutant synthesizes damaged DNA when grown at semi-permissive temperatures (46). Genomic DNA isolated from mcm2-1 cells moves faster on agarose gels [(46), Figure 5G] and this phenotype is suppressed by minichromosomes carrying the MCM2 gene (60). In this work, we found that mcm2-1 chl1 mutant, when cultured at 32°C, accumulates large-budded cells containing fragmented nuclei (Figure 2C). When DNA of such cells was isolated in agarose plugs, it showed even higher mobility than mcm2-1 DNA (Figure 5G), confirming that in the absence of Chl1p, mcm2-1 cells are unable to repair DNA damage that arises due to the absence of Mcm2p activity.
DISCUSSION
Earlier work on the budding yeast protein, Chl1p, has implicated its role in sister-chromatid cohesion, chromatin structure, rDNA recombination and aging. In this work, we ascribe additional roles to Chl1p in the budding yeast cell cycle. We show that this protein is required in S-phase to maintain cell viability if DNA replication is put under stress due to mutations or by treatment with genotoxic agents like UV radiation and MMS. All these observations are consistent with a role of Chl1p in DNA replication, repair or checkpoint function. Since chl1 mutation does not elevate genetic recombination frequency significantly (29,51), nor does it slow down S-phase, Chl1p does not appear to have a role in bulk DNA synthesis under normal conditions of growth. In the presence of MMS, the absence of Chl1p led to a loss in cell viability in S-phase but the DNA damage checkpoint pathway remained fully active. The double mutant rad53-21 chl1 showed higher cell death than the single mutant rad53-21 that lacks a functional DNA damage checkpoint pathway (55). This observation is strongly suggestive of Chl1p's involvement in checkpoint-independent MMS-induced DNA damage repair. Agarose gel electrophoresis of genomic DNA isolated from MMS-treated S-phase cells, which were allowed to repair the DNA damage, showed that chl1 cells were less proficient than the wild-type in repairing the damage. Similar results were obtained with the mec1-1 mutant, which is known to be defective in the repair of damaged DNA, lending credence to the theory that Chl1p is required for DNA damage repair.
It is interesting that MMS-treated genomic DNA moved primarily as a band (with some evidence of smearing below the band, Figure 5B) and was not highly degraded into a low molecular weight smear. Each such band would actually consist of a range of high molecular weight DNA molecules which, just like untreated genomic DNA, cannot be resolved by simple agarose gel electrophoresis. The absence of prominent smears flanking the band was indicative of a relatively narrow distribution of DNA size in that band, the reason for which is not immediately clear. Nevertheless, the mobility shift experiments indicated that the average size of DNA decreased with increasing concentrations of MMS. We believe that the average molecular weight of DNA from MMS-treated cells remains high because the treatment is for a limited period of time with low concentrations of MMS. DNA could be protected against heavy damage because not all hits by MMS may result in DNA alkylation; DNA organized into chromatin within the cell could be less accessible to MMS due to the surrounding milieu which may not favor the alkylation reaction. Furthermore, a simultaneous repair process could protect DNA from extensive fragmentation during MMS treatment. This repair could help to prevent the degradation of DNA into low molecular weight species. This approach should be of use to study the response of cells in relation to DNA damage when they are exposed to different levels of this drug.
One of the reasons for the observed synthetic lethal or sick interactions of chl1 with DNA replication mutations could simply be due to the inability of the double mutants to repair damaged DNA. This was corroborated by the finding that these cells showed fragmented nuclei with loss in viability when grown at semi-permissive temperatures and that mcm2-1 chl1 cells synthesized DNA which migrated faster than mcm2-1 DNA in 0.5% agarose gels. The mutational alleles mcm2-1, orc2-1 and orc5-1, all lead to under initiations at yeast replication origins (50,61,62). In contrast, cdc6-3 is a gain of function allele which allows promiscuous initiation of DNA replication even in G2/M (63). This allele causes cell death at high temperatures, very likely due to a lack of genomic integrity caused by continued re-replications (63,64). At non-permissive temperatures, cells carrying any of these mutations do not enter mitosis and arrest as large-budded cells, each having a single nucleus. The presence of DNA damage in mcm2-1 and orc2-1 has been documented (46,49,65,66). It has been shown that the spindle checkpoint-dependent G2/M arrest observed at non-permissive temperatures in orc5-1 cells is also elicited due to DNA damage as a result of extended S-phase (67,68). We believe that in the absence of Chl1p and at semi-permissive temperatures, the DNA damage was extensive in these mutants, leading to lower cell viabilities and high incidence of nuclear fragmentation. Nuclear fragmentation in double mutants could not have occurred due to prolonged arrest at G2/M, since a high percentage of cdc6-3 cells were also arrested at G2/M but did not show significant levels of nuclear fragmentation. It remains to be seen whether this phenotype constitutes apoptosis-like death in the double mutants (66).
Recently, a number of DNA replication checkpoint proteins have been identified which help in the maintenance of sister-chromatid cohesion (69). Damaged DNA could well affect the loading of cohesin complex, leading to cohesion defects. The physical association of Chl1p with Ctf7, which itself associates with RF-C factors when bound to Rad24p or Ctf18p (70), is strongly suggestive of the presence of Chl1p at replication forks which stall due to DNA damage. Its helicase activity may be required to unwind DNA so that alternative structures could be formed at the fork that mediate either checkpoint proteins/cohesin loading or facilitate DNA repair. In another recent study, Suter et al. (71) have found synthetic lethal or sick interactions between mutations in CTF4, CTF8, CTF18 and DCC1 (genes involved in sister-chromatid cohesion) and mutations in genes required for DNA replication initiation, suggesting that robust cohesion and the integrity of DNA being synthesized are interdependent. Furthermore, loading of fresh cohesin is needed for the repair of double-strand DNA breaks (72,73). Since Chl1p is also required for sister-chromatid cohesion, improper cohesin loading at sites of DNA damage could be instrumental in defective DNA repair leading to cell death, and thus this protein may be playing an indirect role in DNA repair. One of the ways by which the interplay between MMS sensitivity and cohesion defects can be studied is by analyzing phenotypes of a large number of chl1 point mutations to see if the two phenotypes are always linked. Experiments involving synergism between chl1 and cohesion mutations like ctf4 will also be of great help in answering the question whether Chl1p is directly involved in DNA repair or this phenotype is due to cohesion defects. These experiments are in progress.
Chl1p has a human homolog, BACH1, which is a DNA helicase and binds to the breast tumour suppressor protein BRCA1, contributing towards its DNA repair function (38,74). Germline mutations that target its helicase activity were found in the BACH1 gene of some patients having early onset of breast cancer (74). Our work shows a similar role for Chl1p. Further work on yeast Chl1p will throw more light on the molecular mechanisms which interlink sister-chromatid cohesion, transcriptional silencing and DNA damage repair.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
We are very grateful to Bik Kwoon Tye, in whose laboratory this work had originated and for providing the strains. We are also grateful to Uttam Surana for strains. We are thankful to Anuradha Lohia and our laboratory colleagues for helpful comments on the manuscript. The laboratory assistance of Md. Asraf Ali Molla is gratefully acknowledged. This work was supported by Grant SP/SO/DO3/2001 from the Department of Science and Technology, Government of India to P.S. S.P.D and S.S are supported by CSIR grants (Sanction Numbers: 9/15(254)/2002-EMR-I and 9/15(314)/2006-EMR-I). The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Melo J., Toczyski D. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 2002;14:237–245. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- 2.Nyberg K.A., Michelson R.J., Putnam C.W., Weinert T.A. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- 3.Osborn A.J., Elledge S.J., Zou L. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 2002;12:509–516. doi: 10.1016/s0962-8924(02)02380-2. [DOI] [PubMed] [Google Scholar]
- 4.Friedberg E.C. DNA damage and repair. Nature. 2003;421:436–440. doi: 10.1038/nature01408. [DOI] [PubMed] [Google Scholar]
- 5.Friedberg E.C., McDaniel L.D., Schultz R.A. The role of endogenous and exogenous DNA damage and mutagenesis. Curr. Opin. Genet. Dev. 2004;14:5–10. doi: 10.1016/j.gde.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Cobb J.A., Shimada K., Gasser S.M. Redundancy, insult-specific sensors and thresholds: unlocking the S-phase checkpoint response. Curr. Opin. Genet. Dev. 2004;14:292–300. doi: 10.1016/j.gde.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Emili A. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol. Cell. 1998;2:183–189. doi: 10.1016/s1097-2765(00)80128-8. [DOI] [PubMed] [Google Scholar]
- 8.Vialard J.E., Gilbert C.S., Green C.M., Lowndes N.F. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 1998;17:5679–5688. doi: 10.1093/emboj/17.19.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz M.F., Duong J.K., Sun Z., Morrow J.S., Pradhan D., Stern D.F. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell. 2002;9:1055–1065. doi: 10.1016/s1097-2765(02)00532-4. [DOI] [PubMed] [Google Scholar]
- 10.Kiser G.L., Weinert T.A. Distinct roles of yeast MEC and RAD checkpoint genes in transcriptional induction after DNA damage and implications for function. Mol. Biol. Cell. 1996;7:703–718. doi: 10.1091/mbc.7.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aboussekhra A., Vialard J.E., Morrison D.E., de la Torre-Ruiz M.A., Cernakova L., Fabre F., Lowndes N.F. A novel role for the budding yeast RAD9 checkpoint gene in DNA damage-dependent transcription. EMBO J. 1996;15:3912–3922. [PMC free article] [PubMed] [Google Scholar]
- 12.Navas T.A., Sanchez Y., Elledge S.J. RAD9 and DNA polymerase epsilon form parallel sensory branches for transducing the DNA damage checkpoint signal in Saccharomyces cerevisiae. Genes Dev. 1996;10:2632–2643. doi: 10.1101/gad.10.20.2632. [DOI] [PubMed] [Google Scholar]
- 13.Santocanale C., Diffley J.F.X. A Mec1- and Rad53- dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 14.Shirahige K., Hori Y., Shiraishi K., Yamashita M., Takahashi K., Obuse C., Tsurimoto T., Yoshikawa H. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 15.Lowndes N.F., Murguia J.R. Sensing and responding to DNA damage. Curr. Opin. Genet. Dev. 2000;10:17–25. doi: 10.1016/s0959-437x(99)00050-7. [DOI] [PubMed] [Google Scholar]
- 16.Tercero J.A., Diffley J.F. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- 17.Lopes M., Cotta-Ramusino C., Pellicioli A., Liberi G., Plevani P., Muzi-Falconi M., Newlon C.S., Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 18.Sogo J.M., Lopes M., Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- 19.Clarke D.J., Segal M., Andrews C.A., Rudyak S.G., Jensen S., Smith K., Reed S.I. S-phase checkpoint controls mitosis via an APC-independent Cdc20p function. Nat. Cell. Biol. 2003;5:928–935. doi: 10.1038/ncb1046. [DOI] [PubMed] [Google Scholar]
- 20.Krishnan V., Nirantar S., Crasta K., Cheng A.Y., Surana U. DNA replication checkpoint prevents precocious chromosome segregation by regulating spindle behaviour. Mol. Cell. 2004;16:687–700. doi: 10.1016/j.molcel.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Paulovich A.G., Margulies R.U., Garvik B.M., Hartwell L.H. Rad9, Rad17 and Rad24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics. 1997;145:45–62. doi: 10.1093/genetics/145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pascucci B., Russo M.T., Crescenzi M., Bignami M., Dogliotti E. The accumulation of MMS-induced single strand breaks in G1 phase is recombinogenic in DNA polymerase β defective mammalian cells. Nucleic Acids Res. 2005;33:280–288. doi: 10.1093/nar/gki168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tercero J.A., Longhese M.P., Diffley J.F.X. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell. 2003;11:1323–1336. doi: 10.1016/s1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- 24.Maine G.T., Sinha P., Tye B.-K. Mutants of S. cerevisiae defective in the maintenance of minichromosome. Genetics. 1984;106:365–385. doi: 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tye B.-K. MCM proteins in DNA replication. Annu. Rev. Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- 26.Bell S.P., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 27.Forsburg S.L. Eukaryotic MCM proteins: beyond replication initiation. Microbiol. Mol. Biol. Rev. 2004;68:109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haber J.E. Bisexual mating behavior in a diploid of Saccharomyces cerevisiae: evidence for genetically controlled non-random chromosome loss during vegetative growth. Genetics. 1974;78:843–858. doi: 10.1093/genetics/78.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerring S.L., Spencer F., Hieter P. The CHL1(CTF1) gene product of Saccharomyces cerevisiae is important for chromosome transmission and normal cell cycle progression in G2/M. EMBO J. 1990;9:4347–4358. doi: 10.1002/j.1460-2075.1990.tb07884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skibbens R.V. Chl1p, a DNA helicase-like protein in budding yeast, functions in sister-chromatid cohesion. Genetics. 2004;166:33–42. doi: 10.1534/genetics.166.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayer M.L., Pot I., Chang M., Xu H., Aneliunas V., Kwok T., Newitt R., Aebersold R., Boone C., Brown G.W., Hieter P. Identification of protein complexes required for efficient sister chromatid cohesion. Mol. Biol. Cell. 2004;15:1736–1745. doi: 10.1091/mbc.E03-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petronczki M., Chwalla B., Siomos M.F., Yokobayashi S., Helmhart W., Deutschbauer A.M., Davis R.W., Watanabe Y., Nasmyth K. Sister-chromatid cohesion mediated by the alternative RF-C Ctf18/Dcc1/Ctf8, the helicase Chl1 and the polymerase-α-associated protein Ctf4 is essential for chromatid disjunction during meiosis II. J. Cell Sci. 2004;117:3547–3559. doi: 10.1242/jcs.01231. [DOI] [PubMed] [Google Scholar]
- 33.Weiler K.S., Szeto L., Broach J.R. Mutations affecting donor preference during mating type interconversion in Saccharomyces cerevisiae. Genetics. 1995;139:1495–1510. doi: 10.1093/genetics/139.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das S.P., Sinha P. The budding yeast protein Chl1p has a role in transcriptional silencing, rDNA recombination and aging. Biochem. Biophys. Res. Commun. 2005;337:167–172. doi: 10.1016/j.bbrc.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 35.Holloway S.L. CHL1 is a nuclear protein with an essential ATP binding site that exhibits a size-dependent effect on chromosome segregation. Nucleic Acids Res. 2000;28:3056–3064. doi: 10.1093/nar/28.16.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amann J., Kidd V.J., Lahti J.M. Characterization of putative human homologues of the yeast chromosome transmission fidelity gene, CHL1. J. Biol. Chem. 1997;272:3823–3832. doi: 10.1074/jbc.272.6.3823. [DOI] [PubMed] [Google Scholar]
- 37.Hirota Y., Lahti J.M. Characterization of the enzymatic activity of hChlR1, a novel human DNA helicase. Nucleic Acids Res. 2000;28:917–924. doi: 10.1093/nar/28.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cantor S.B., Bell D.W., Ganesan S., Kass E.M., Drapkin R., Grossman S., Wahrer D.C.R., Sgroi D.C., Lane W.S., Haber D.A., Livingston D.M. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- 39.Maiti A.K., Sinha P. The mcm2 mutation of yeast affects replication, rather than segregation or amplification of the two micron plasmid. J. Mol. Biol. 1992;224:545–558. doi: 10.1016/0022-2836(92)90543-s. [DOI] [PubMed] [Google Scholar]
- 40.Kilmartin J.V., Wright B., Milstein C. Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J. Cell Biol. 1982;93:576–582. doi: 10.1083/jcb.93.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gietz R.D., Sugino A. New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 42.Fitch M.J., Donato J.J., Tye B.-K. Mcm7, a subunit of the presumptive MCM helicase, modulates its own expression in conjunction with Mcm1. J. Biol. Chem. 2003;278:25408–25416. doi: 10.1074/jbc.M300699200. [DOI] [PubMed] [Google Scholar]
- 43.Breeden L.L. Alpha-factor synchronization of budding yeast. Methods Enzymol. 1997;283:332–341. doi: 10.1016/s0076-6879(97)83027-3. [DOI] [PubMed] [Google Scholar]
- 44.Poddar A., Roy N., Sinha P. MCM21 and MCM22, two novel genes of the yeast Saccharomyces cerevisiae are required for chromosome transmission. Mol. Microbiol. 1999;31:349–360. doi: 10.1046/j.1365-2958.1999.01179.x. [DOI] [PubMed] [Google Scholar]
- 45.Ghosh S.K., Sau S., Lahiri S., Lohia A., Sinha P. The Iml3 protein of the budding yeast is required for the prevention of precocious sister chromatid separation in meiosis I and for sister chromatid disjunction in meiosis II. Curr. Genet. 2004;46:82–91. doi: 10.1007/s00294-004-0516-6. [DOI] [PubMed] [Google Scholar]
- 46.Ray A., Sinha P. The mcm2-1 mutation of yeast causes DNA damage with a RAD9 requirement for repair. Curr. Genet. 1995;27:95–101. doi: 10.1007/BF00313422. [DOI] [PubMed] [Google Scholar]
- 47.Roy N., Poddar A., Lohia A., Sinha P. The mcm17 mutation of yeast shows a size-dependent segregational defect of a mini-chromosome. Curr. Genet. 1997;32:182–189. doi: 10.1007/s002940050264. [DOI] [PubMed] [Google Scholar]
- 48.Cheeseman I.M., Drubin D.G., Barnes G. Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 2002;157:199–203. doi: 10.1083/jcb.200201052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sinha P., Chang V., Tye B.-K. A mutant that affects the function of autonomously replicating sequences in yeast. J. Mol. Biol. 1986;192:805–814. doi: 10.1016/0022-2836(86)90030-6. [DOI] [PubMed] [Google Scholar]
- 50.Yan H., Merchant A.M., Tye B.-K. Cell cycle-regulated nuclear localization of MCM2 and MCM3, which are required for the initiation of DNA synthesis at chromosomal replication origins in yeast. Genes Dev. 1993;7:2149–2160. doi: 10.1101/gad.7.11.2149. [DOI] [PubMed] [Google Scholar]
- 51.Hajra S. Kolkata: Jadavpur University; 2003. Kinetochore structure of the budding yeast Saccharomyces cerevisiae: a study using genetic and protein–protein interactions. PhD Thesis. [Google Scholar]
- 52.Hartwell L.H., Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics. 1985;110:381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang M., Bellaoui M., Boone C., Brown G.W. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc. Natl Acad. Sci. USA. 2002;99:16934–16939. doi: 10.1073/pnas.262669299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pan X., Ye P., Yuan D.S., Wang X., Bader J.S., Boeke J.D. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 55.Desany B.A., Alcasabas A.A., Bachant J.B., Elledge S.J. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes. Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watt P.M., Hickson I.D., Borts R.H., Louis E.J. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Myung K., Datta A., Chen C., Kolodner R.D. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homologous recombination. Nature Genet. 2001;27:113–116. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- 58.Frei C., Gasser S.M. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- 59.Memisoglu A., Samson L. Base excision repair in yeast and mammals. Mutat. Res. 2000;451:39–51. doi: 10.1016/s0027-5107(00)00039-7. [DOI] [PubMed] [Google Scholar]
- 60.Pal A. Kolkata: PhD Thesis, Jadavpur University; 1992. Characterization of a mutant that effects the function of autonomously replicating sequences in yeast. [Google Scholar]
- 61.Fox C.A., Loo S., Dillin A., Rine J. The origin recognition complex has essential functions in transcriptional silencing and chromosomal replication. Genes Dev. 1995;9:911–924. doi: 10.1101/gad.9.8.911. [DOI] [PubMed] [Google Scholar]
- 62.Liang C., Weinreich M., Stillman B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- 63.Liang C., Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Green B.M., Li J.J. Loss of rereplication control in Saccharomyces cerevisiae results in extensive DNA damage. Mol. Biol. Cell. 2005;16:421–432. doi: 10.1091/mbc.E04-09-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watanabe K., Morishita J., Umezu K., Shirahige K., Maki H. Involvement of Rad9-dependent damage checkpoint control in arrest of cell cycle, induction of cell death, and chromosome instability caused by defects in origin recognition complex in Saccharomyces cerevisiae. Eukaryotic Cell. 2002;1:200–212. doi: 10.1128/EC.1.2.200-212.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weinberger M., Ramachandran L., Feng L., Sharma K., Sun X., Marchetti M., Huberman J.A., Burhans W.C. Apoptosis in budding yeast caused by defects in initiation of DNA replication. J. Cell Sci. 2005;118:3543–3553. doi: 10.1242/jcs.02477. [DOI] [PubMed] [Google Scholar]
- 67.Dillin A., Rine J. Roles for ORC in M phase and S phase. Science. 1998;279:1733–1737. doi: 10.1126/science.279.5357.1733. [DOI] [PubMed] [Google Scholar]
- 68.Gibson D.G., Bell S.P., Aparicio O.M. Cell cycle execution point analysis of ORC function and characterization of the checkpoint response to ORC inactivation in Saccharomyces cerevisiae. Genes Cells. 2006;11:557–573. doi: 10.1111/j.1365-2443.2006.00967.x. [DOI] [PubMed] [Google Scholar]
- 69.Warren C.D., Eckley D.M., Lee M.S., Hanna J.S., Hughes A., Peyser B., Jie C., Irizarry R., Spencer F.A. S-phase checkpoint genes safeguard high-fidelity sister chromatid cohesion. Mol. Biol. Cell. 2004;15:1724–1735. doi: 10.1091/mbc.E03-09-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kenna M.A., Skibbens R.V. Mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different replication factor C complexes. Mol. Cell. Biol. 2003;23:2999–3007. doi: 10.1128/MCB.23.8.2999-3007.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suter B., Tong A., Chang M., Yu L., Brown G.W., Boone C., Rine J. The origin recognition complex links replication, sister chromatid cohesion and transcriptional silencing in Saccharomyces cerevisiae. Genetics. 2004;167:579–591. doi: 10.1534/genetics.103.024851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sjogren C., Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 2001;11:991–995. doi: 10.1016/s0960-9822(01)00271-8. [DOI] [PubMed] [Google Scholar]
- 73.Strom L., Lindroos H.B., Shirahige K., Sjogren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 74.Cantor S., Drapkin R., Zhang F., Lin Y., Han J., Pamidi S., Livingston D.M. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc. Natl Acad. Sci. USA. 2004;101:2357–2362. doi: 10.1073/pnas.0308717101. [DOI] [PMC free article] [PubMed] [Google Scholar]