Abstract
In Escherichia coli, proteins GidA and MnmE are involved in the addition of the carboxymethylaminomethyl (cmnm) group onto uridine 34 (U34) of tRNAs decoding two-family box triplets. However, their precise role in the modification reaction remains undetermined. Here, we show that GidA is an FAD-binding protein and that mutagenesis of the N-terminal dinucleotide-binding motif of GidA, impairs capability of this protein to bind FAD and modify tRNA, resulting in defective cell growth. Thus, GidA may catalyse an FAD-dependent reaction that is required for production of cmnmU34. We also show that GidA and MnmE have identical cell location and that both proteins physically interact. Gel filtration and native PAGE experiments indicate that GidA, like MnmE, dimerizes and that GidA and MnmE directly assemble in an α2β2 heterotetrameric complex. Interestingly, high-performance liquid chromatography (HPLC) analysis shows that identical levels of the same undermodified form of U34 are present in tRNA hydrolysates from loss-of-function gidA and mnmE mutants. Moreover, these mutants exhibit similar phenotypic traits. Altogether, these results do not support previous proposals that activity of MnmE precedes that of GidA; rather, our data suggest that MnmE and GidA form a functional complex in which both proteins are interdependent.
INTRODUCTION
GidA-like proteins are widely distributed in nature. They are conserved among Bacteria and Eukarya and have been classified into two groups, based on the size of the protein (1). One group includes proteins of about 600 amino acid residues (GidAL), whereas the other comprises proteins of approximately 450 residues (GidAS) that are truncated at the C-terminal end compared to the larger ones. Alignment of both the large and small forms of GidA revealed a conserved, dinucleotide-binding motif at the N-terminus. In fact, Myxococcus xanthus GidAS was shown to bind FAD (1). Thus, it was suggested that GidA proteins either catalyse oxidation–reduction reactions or act as sensors for the redox state of the cell (1).
Recently, it has been shown that proteins GidAS correspond to a novel class of bacterial site-specific tRNA methyltransferases (tRNA:m5U-54 Mtases), and they have been renamed TrmFO (2). These enzymes catalyse the site-specific formation of 5-methyluridine in position 54 (m5U54) of tRNA using N5, N10-methylenetetrahydrofolate (CH2H4folate) as a source of one-carbon unit and a combination of coenzymes NAD(P)H/FAD as reductant. Therefore, they differ from TrmA enzymes, which also catalyse methylation of U54 but using S-adenosylmethionine as the methyl donor. Curiously, the folate-dependent TrmFO proteins and S-AdoMet-dependent TrmA/Trm2p enzymes appear to have mutually exclusive phylogenetic distributions. Thus, the Bacteria Bacillus subtilis, Aquifex aeolicus and Thermotoga maritima lack TrmA but posses TrmFO.
Phylogenetic analyses also support that paralogous TrmFO and GidAL(hereafter designated GidA) proteins are two distinct families of proteins that probably evolved from a common ancestor but acquired different, non-overlapping cellular functions during evolution (2).
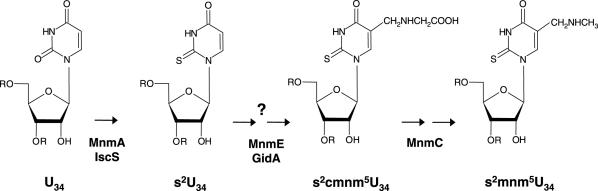
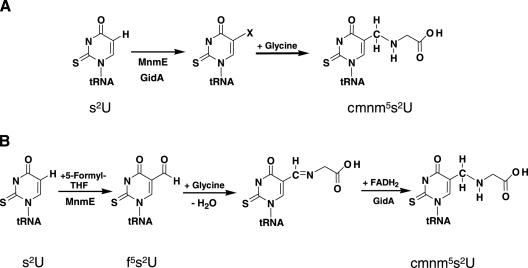
GidA was first described in Escherichia coli, where TrmFO is absent (note that methylation of U54 in E.coli relies on TrmA). It was shown that disruption of gidA in E.coli affects cell division, but only when cells are grown on glucose (3). More recent data support that gidA is allelic with trmF (also called mnmG), a gene involved in tRNA modification (4,5). According to this, GidA controls together with MnmE the addition of the carboxymethylaminomethyl (cmnm) group in position 5 of the U34 of tRNAs that read codons ending with A or G, in the mixed codon family boxes, i.e. tRNALys(UUU), tRNAGlu(UUC), tRNAGln(UUG), tRNALeu(UAA) and tRNAArg(UCU) but the precise role of both proteins in the reaction modification is hitherto unknown (4–10). In tRNALys(UUU) and tRNAGlu(UUC) (see Figure 1), MnmC transforms the cmnm5 group into the final 5-methylaminomethyl (mnm5) modification, meanwhile MnmA, together with IscS and proteins TusA-E, carries out thiolation in the 2-position of the wobble uridine (6,11–13). Modifications in the 2- and 5-positions occur independently of each other; thus, thiolation may precede or follow the synthesis of the side chain at position 5. When selenium is available in the growth medium, sulfur at position 2 may be replaced by selenium in a reaction that is dependent on SelD and YbbB (14). It should be noticed that non-thiolated derivatives are present in tRNALeu(UAA) and tRNAArg(UCU), (which contain mainly cmnm5 and mnm5, respectively), whereas tRNAGln(UUG) contains mainly cmnm5s2 (15). Moreover, it has been recently found that tRNAGly(UCC) contains mainly mnm5 (16), which suggests that this tRNA is also a substrate for MnmE and GidA. Modification at position 5 mediated by GidA and MnmE appears crucial for appropriate decoding of mRNA (4,16–20).
Figure 1.
Modification pathway of U34. Only steps relevant to this work are shown as understood. The first stage in the modification of U34 at position 5 is controlled by MnmE and GidA, but it is unclear how many steps precede the formation of cmnm5U. The cmnm5 group is then rearranged in two steps catalyzed by MnmC to synthesize the mnm5 side chain in some tRNAs. U34 may undergo thiolation at position 2 mediated by MnmA, IscS and other proteins (see text). It is worthy to mention that the mnmC gene is not evolutionarily conserved, which explains for the absence of mnm5s2U in yeast mitochondrial tRNAs.
GidA and mnmE mutations are pleiotropic, affecting diverse phenotypic traits. This may result from a translational control exerted by GidA and MnmE throughout modification of specific tRNAs. Thus, GidA has been reported to be a global regulator in the plant pathogen Pseudomonas syringae since, mutations in gidA affects antibiotic production, swarming, presence of fluorescent pigment, and virulence (21). In the human pathogen Aeromonas hydrophila, disruption of gidA reduces hemolytic and cytotoxic activity associated with enterotoxin Act (22). Since this effect was shown to be due to modulation of the act translation by GidA, it was concluded that GidA regulates the most-potent virulence factor of A.hydrophila, Act (22).
On the other hand, E.coli mnmE has been involved in oxidation of certain heterocyclic substrates, such as thiophene and furan (23), and resistance to acidic pH (24). Moreover, loss-of-function mnmE mutations are lethal in combination with mutations in some other genes involved in the mRNA decoding process [(7,8,10) M. Villarroya, L. Yim and M.-E. Armengod, in preparation]. In this respect, gidA mutations have also been reported to be incompatible with mutations disabling the normal decoding capability of certain tRNAs (25). In addition, gidA and mnmE have been shown to be essential in some human pathogens, such as Staphylococcus aureus and Helicobacter pylori (26,27), supporting the notion that tRNA modification by MnmE and GidA is crucial for survival of these species.
Interestingly, proteins GidA and MnmE are conserved between Bacteria and Eukarya. MTO1 and MSS1/GTPBP3 were found to be the respective homologues of the gidA and mnmE genes in both yeast and human (28–31). In yeast, the protein products of MTO1 and MSS1 localize in mitochondria, and their mutants are associated with respiratory defects (28,29,32). Also in yeast, it has been recently reported that mitochondrial tRNALys molecules isolated from MTO1 and MSS1 deletion strains contain s2U instead of the cmnm5s2U found in the wild-type tRNALys molecules; this indicates that MTO1 and MSS1 genes are both involved in the biosynthesis of the 5-carboxy-methylaminomethyl group of cmnm5s2 of mitochondrial tRNALys (32). Moreover, the human cDNAs of GTPBP3 and MTO1 are able to complement yeast MSS1 and MTO1 mutants, respectively (30,31). Therefore, the function of GidA and MnmE seems to be evolutionarily conserved.
The general purpose of this work was to gain further insights into the GidA cellular function by studying its location, self-assembly capability, interaction with MnmE and relationships of its putative FAD-binding motif with tRNA modification and cell viability.
MATERIALS AND METHODS
Bacterial strains, phages, plasmids and DNA manipulations
E.coli strains and plasmids used in this study are listed in Table 1, unless specified otherwise. Genetic techniques for the construction of strains were performed as described previously (33). For DNA manipulations, standard procedures were followed. DH5α strain was used as host for in vitro modified plasmids. pIC1153 expresses glutathione S-transferase (GST)-GidA fusion controlled by the Ptac promoter and LacIq repressor, and was constructed as follows: gidA was PCR-amplified from the chromosome of E.coli MC1000 strain, using Herculase DNA polymerase (Stratagene) and primers gidA5′ (5′-GGGGAATTCTCATGTTTTATCCGGATCCTTTT-3′) and gidA3′ (5′-AAAGAATTCCGTTATGCGCTACGACGCA-3′), incorporating EcoRI restriction sites (underlined). The resulting 1.9 kb fragment was digested with EcoRI and ligated to EcoRI digested pGEX-2T. A recombinant plasmid with proper orientation of gidA gene was selected. The same 1.9 kb fragment was ligated to EcoRI digested pJF119EH, rendering pIC1154. This plasmid carries gidA under the control of Ptac and LacIq. Again, a plasmid with proper orientation of insert was selected. Plasmid pIC1180 produces a FLAG–GidA fusion under control of promoter PB and protein AraC. It was constructed as follows: the FLAG epitope coding sequence (DYKDDDDK) was added in frame to 5′ end of gidA by PCR amplification of gidA with primers LY30 (5′-GACTACAAGGACGACGATGACAAGATGTTTTATCCGGATCCTTTTG-3′) and LY31 (5′-GCTCTAGATTATGCGCTACGACGCAGC-3′), coding sequence for FLAG in cursive and XbaI restriction site underlined. The resulting 1.9 kb fragment was cut with XbaI and inserted into pBAD22 previously digested with NcoI filled with Klenow and then cut with XbaI. Note that the AUG start codon of the fusion protein is included into the NcoI site, CCATGG, on the plasmid. Derivatives of pIC1154 and pIC1180 carrying gidA mutations G13A, G15A and G13A/G15A were obtained by site-directed mutagenesis (QuickChange™, Stratagene) with appropriate PCR primers. All constructs were verified by DNA sequencing. Strain carrying insertion of Tn10 in position 51 of gidA was a gift from D. Brégeon [see reference (4), mutant number 10].
Table 1.
E.coli strains and plasmids used in this study
| Strain or plasmid | Description | Origin and/or reference |
|---|---|---|
| E.coli strains | ||
| DEV16 | F−thi-1 rel-1 spoT1 lacZ105UAGmnmEC3676T [MnmEQ192X, ValR] | (5) |
| DH5α | F−endA1 hsdR17 supE44 thi1 recA1 gyrA relA1 Δ(lacZYA-argF) U169(Φ80lacZΔM15) | (42) |
| MC1000 | ||
| MG1655 | F− | D. Touati |
| IC4639 | DEV16 mnmE+ bgl (Sal+) | M. Villarroya |
| IC5241 | MG1655 gidA::Tn10 [TetR] | This work |
| IC5242 | IC5241 carrying pIC1154 [GidA+, TetR, ApR] | This work |
| IC5244 | IC5241 carrying pIC1177 [GidA G13A, TetR, ApR] | This work |
| IC5245 | IC5241 carrying pIC1178 [GidA G15A, TetR, ApR] | This work |
| IC5246 | IC5241 carrying pIC1179 [GidA G13A/G15A, TetR, ApR] | This work |
| IC5287 | XL1 carrying pIC931 [ApR] | This work |
| IC5332 | DH5α carrying pIC1154 [ApR] | This work |
| IC5358 | MG1655 mnmE::kan [MnmE−, KanR] | (10) |
| IC5550 | IC4639 gidA::Tn10 [TetR] | This work |
| V5701 | bgl (Sal+) | (7,43) |
| Plasmids | ||
| pBAD22 | Expression vector with PB and AraC control (ApR) | (44) |
| pGEX-2T | Cloning vector for GST fusions (ApR) | Pharmacia Biosciences |
| pGroESL | groES and groEL genes cloned under Ptac (CmR) | (45) |
| pJF119EH | Expression vector with Ptac and LacIq control (ApR) | (46) |
| pIC684 | GST fusion of mnmE (cloned in pGEX-2T) | (7) |
| pIC931 | mnmE cloned under Ptac[ApR] | M. Villarroya |
| pIC1153 | GST fusion of gidA (GST-GidA+ fusion, cloned in pGEX-2T) | This work |
| pIC1154 | gidA inserted into EcoRI site of pJF119EH | This work |
| pIC1177 | pIC1154 derived plasmid containing gidAG13A | This work |
| pIC1178 | pIC1154 derived plasmid containing gidAG15A | This work |
| pIC1179 | pIC1154 derived plasmid containing gidAG13AG15A | This work |
| pIC1180 | FLAG-gidA inserted between sites NcoI and XbaI of pBAD22 | This work |
| pIC1181 | pIC1180 derived plasmid containing FLAG-gidAG13A | This work |
| pIC1182 | pIC1180 derived plasmid containing FLAG-gidAG15A | This work |
| pIC1183 | pIC1180 derived plasmid containing FLAG-gidAG13AG15A | This work |
Media, growth conditions, cellular fractionations and enzyme assays
LBT (Luria–Bertani broth containing 40 μg/ml thymine) and LAT (LBT containing 20 g of Difco agar per litre) were used for routine cultures and plating of E.coli, unless otherwise specified. When required, antibiotics were added at the following concentrations: 100 μg/ml of ampicillin, 35 μg/ml of chloramphenicol, 12.5 μg/ml of tetracycline and 80 μg/ml of kanamycin. Cell growth was monitored by measuring the optical density (OD) of the cultures at 600 nm. Subcellular fractions of cells were prepared essentially as described (8). Glucose-6-phosphate dehydrogenase activity, which was used as a cytoplasmic marker, was determined as described previously (34).
Protein techniques and production of antisera
Overproduction of GST-fused GidA was done in DEV16 cells transformed with pIC1153 and pGroESL, grown during 14 h at 25°C with 10 μM isopropyl-β-d-thiogalactopyranoside (IPTG). GidA purification was carried out using affinity chromatography with glutathione-agarose and further thrombin cleavage, essentially as described (7). Cleavage of the chimera with thrombin rendered two isolated proteins; the 69 kDa expected one and another of lower molecular weight, probably due to internal protease cleavage. The 69 kDa protein was excised from SDS–acrylamide gels and used to inoculate New Zealand rabbits. The resulting antiserum was affinity purified with PVDF-bound GidA before use. SDS–PAGE and immunoblotting were carried out essentially as described (7). Purification of GST-GidA for pull down assays was done as mentioned above except that no thrombin digestion was performed and elution of the protein was achieved with free glutathione buffer. Purification of GST-MnmE or recombinant MnmE (rMnmE, 53 kDa) for pull down assays was done from DEV16 transformed with pIC684 as described previously (7). Overproduction of FLAG-GidA and mutant derivatives was achieved by growing strain DH5α transformed with pIC1180, pIC1181, pIC1182 or pIC1183, during 3 h at 37°C in LBT containing 0.2% arabinose and ampicillin. In these conditions, FLAG-GidA and derivatives were almost completely soluble so there was no need of chaperone co-expression. Cells were collected, disrupted by sonication, centrifuged and the supernatant (cleared lysate) applied over anti-FLAG M2 agarose (Sigma). The lysate was passed four times through the resin. After collecting the flow through, the resin was washed with 3 × 8 column volumes of TBS [50 mM Tris–HCl (pH 7.5) and 150 mM NaCl]. FLAG-GidA proteins were eluted with five column volumes of TBS containing 0.1 mg/ml of FLAG peptide (Sigma). Normally, two eluates were collected. Proteins were quantified by Bradford assay using BSA as standard. Overproduction of FLAG-GidA used in pull down assays was performed in DEV16 transformed with pIC1180, to avoid carrying endogenous MnmE protein with purified FLAG-GidA. Production of the anti-MnmE and anti-CyoA antibodies was described previously (7,8).
Gel filtration chromatography, native PAGE and in vitro cross-linking
Gel filtrations were performed using a Superdex 200 HR10/30 column (Amersham Biosciences) in 100 mM NaPO4 (pH 7.0), 150 mM NaCl, containing or not 5 mM DTT, at a flow rate of 0.2 ml/min. Gel filtration markers were used to calibrate the column. Proteins were detected by ultraviolet (UV) absorbance at 280 nm. Aliquots of selected fractions were analysed by SDS–PAGE and, in some cases, native PAGE. Gels and buffers used for native PAGE were made according to the standard Laemmli SDS protocol omitting the SDS. Native gels (10% polyacrylamide) were run at 4°C for 3.5–4 h at 15 mA, and stained with Coomassie blue or analysed by immunoelectroblotting using appropriate antibodies. To determine the stoichiometry of the MnmE•GidA complex, proteins FLAG-GidA and rMnmE were mixed, at 5 μM each, and incubated for 2 h at room temperature in phosphate-buffered saline (PBS) containing 2 mM MgCl2 and 5 mM DTT. Samples of the mix and each protein alone were analysed by gel filtration, in the presence of 5 mM DTT, or native PAGE. In vitro cross-linking experiments were performed as described previously (7).
GST-pull down assays
GST-GidA, GST-MnmE or GST were expressed from plasmids pIC1153, pIC684 and pGEX-2T, respectively, and bound to glutathione-agarose as described previously (7). Total cleared lysates of cells transformed with plasmids pIC931 (expressing mnmE) or pIC1154 (expressing gidA), induced during 2 h with 0.5 mM IPTG, were obtained, pre-incubated with glutathione-agarose and added to resin-bound GST-GidA or GST-MnmE, respectively. As a control, equal amounts of both lysates were added to resin-bound GST. Reactions were incubated 2 h at room temperature with gentle agitation and after collecting the unbound fraction, and washing extensively the beads with PBS containing 0.05% Triton X-100, the bound material was eluted with 50 mM Tris–HCl (pH 8), 10 mM glutathione. Equal amounts of samples from each reaction were analysed by immunoblotting using appropriate antibodies.
FLAG-pull down assays
Proteins FLAG-GidA and rMnmE, at a final concentration of 5 μM each, were incubated together for 2 h in PBS, at room temperature. Then, anti-FLAG agarose was added, and the mixture incubated overnight at 4°C with gentle agitation. As a control, rMnmE was incubated with anti-FLAG agarose without FLAG-GidA added. After collecting the unbound fraction, beads were extensively washed with PBS and the remained material eluted with PBS containing 0.1 mg/ml FLAG peptide. Equal amounts of samples from each experiment were analysed by SDS–PAGE, and the proteins detected with Coomassie blue stain.
Cofactor analysis
UV-visible spectral analysis of purified proteins (at a concentration ∼0.3 mM) were performed in a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Rockland), scanning from 220 to 750 nm. Flavine was released from FLAG-GidA by heating at 75°C for 15 min in the dark and analysed by high-performance liquid chromatography (HPLC) as described (35) with minor modifications. Briefly, HPLC separation was achieved with a LichrospherR 100 RP-18 (5 μm) using a linear gradient of water [5 mM NH4Oac (pH 6.0)] methanol (10%) to water [5 mM NH4Oac (pH 6.0)] methanol (70%) developed over 35 min at a flow rate of 1.2 ml/min. Flavins were detected by fluorescence emission using a Waters 2475 multi λ fluorescence detector. Detector wavelengths were set at 450 nm for excitation and 525 nm for emission.
Analysis of tRNA modification by HPLC
Strains MG1655 (wild-type) and derivatives IC5241 (gidA::Tn10) and IC5358 (mnmE::kan) were grown until late log phase in LBT, then cells were collected and processed as described previously for nucleoside analysis (10). IC5241 cells carrying plasmids pIC1154 (gidA+), pIC1177 (gidA G13A), pIC1178 (gidA G15A) and pIC1179 (gidA G13A/G15A) were grown during 3 h with or without inducer (1 mM IPTG), before being collected and processed as above. Note that LBT is deficient in selenium; thus, MnmE/GidA-specific tRNAs isolated from strains grown in this medium mostly carry sulfur at position 2.
RESULTS
GidA antibody production and subcellular localization
M.xanthus carries genes encoding both TrmFO and GidA proteins, which share about 25% identity in their N-terminal 450 amino acids (1). Subcellular fractionation followed by immunoblotting with anti-TrmFO antibody led to the idea that the M.xanthus GidA protein is localized in periplasm, or in association with the inner membrane, since the anti-TrmFO antibody reacted slightly with a protein of 75 kDa, the expected size for GidA (1). No null gidA mutant was available to be introduced as a negative control in these experiments. Moreover, it was observed that M.xanthus GidA does not have any regions that score well as membrane-spanning segments, nor does it have an N-terminal sequence that resembles a signal peptide (1). Therefore, the putative periplasmic localization of M.xanthus GidA remains to be checked.
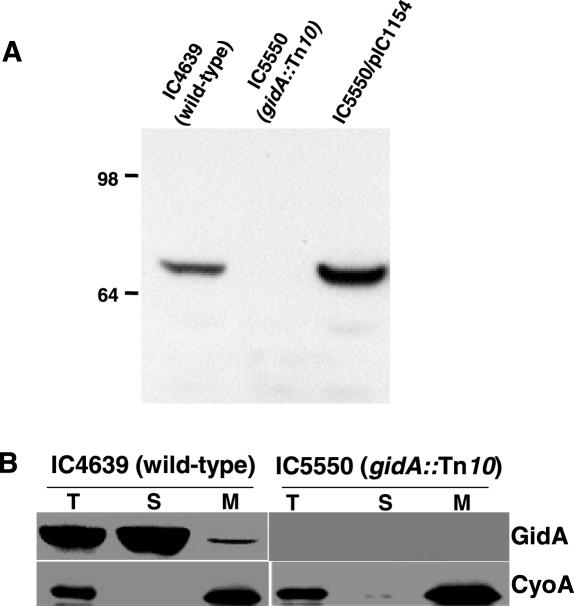
Periplasm is not the expected localization for an enzyme involved in tRNA modification. Thus, immunoelectron microscopy and subcellular fractionation revealed that the E.coli MnmE protein is a cytoplasmic protein partially associated with the inner membrane (7,8). Here, it was of interest to determine the localization of E.coli GidA, and to this end, an anti-GidA antibody was generated. GidA was expressed as a protein fused to the C-terminal region of the GST, but the GST–GidA chimera (96 kDa) was only soluble in the presence of overproduced GroES and GroEL chaperons. Cleavage with thrombin of the chimera, after purification by affinity chromatography, allowed the isolation of a recombinant GidA with the expected mass of about 69 kDa that was used to raise antibodies in rabbits. Specificity of the anti-GidA antibody was tested in strains IC4639 and IC5550, which carry the wild-type and gidA::Tn10 allele, respectively (Figure 2A).
Figure 2.
(A) Specificity of the anti-GidA antibody. Equal amounts of total extracts of strains IC4639, IC5550 and IC5550 harbouring plasmid pIC1154 (where gidA is under control of Ptac promoter and LacI repressor) were subjected to western blot analysis using anti-GidA antibody. Strain IC5550/pIC1154 was grown in the absence of inducer IPTG, but there is escape of gidA expression. Molecular weight markers (on the left) are in kDa. (B) Subcellular localization of GidA. Western blot analysis with anti-GidA and anti-CyoA antibodies of cell fractions from strains IC4639 and IC5550 is shown. Equal amounts of bulk protein (50 μg) were loaded in each lane. T, total cell lysate; S, soluble fraction; M: membrane fraction.
E.coli strains either expressing GidA at wild-type levels (IC4639) or lacking GidA (IC5550) were subjected to subcellular fractionation followed by immunoblotting to identify GidA in the fractions. As shown in Figure 2B, GidA was mostly found in the soluble fraction, although at least a portion was also detected in the membrane fraction. Activity of glucose-6-phosphate dehydrogenase, used as a cytoplasmic marker, was negligible in the membrane fraction (lanes M), indicating little, if any, cross-contamination with the soluble fraction (lanes S). These results altogether support that, in E.coli, GidA and MnmE display identical localization.
Self-association of GidA
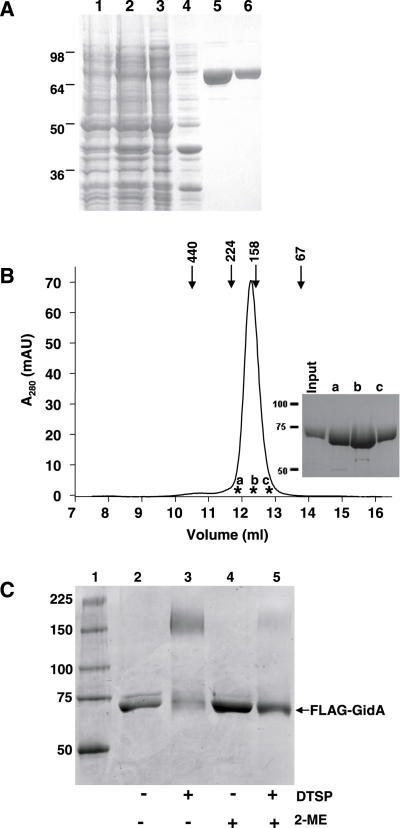
It has been recently reported that the B.subtilis TrmFO protein forms homodimers in vitro (2). Given that TrmFO and GidA proteins share about 25% identity (40% similarity) in their N-terminal homologous region, we asked whether E.coli GidA is also able to dimerize. To this end, purified FLAG-GidA protein (Figure 3A) was applied to a Superdex-200 HR gel filtration column. As shown in Figure 3B, FLAG-GidA protein eluted as a single peak at ∼168 kDa, which is consistent with a dimeric form of this protein (expected molecular weight, 141 kDa). An identical result was obtained from gel filtration experiments performed in the presence of DTT, 5 and 10 mM (data not shown), which supports that GidA homodimers may be maintained by non-covalent interactions.
Figure 3.
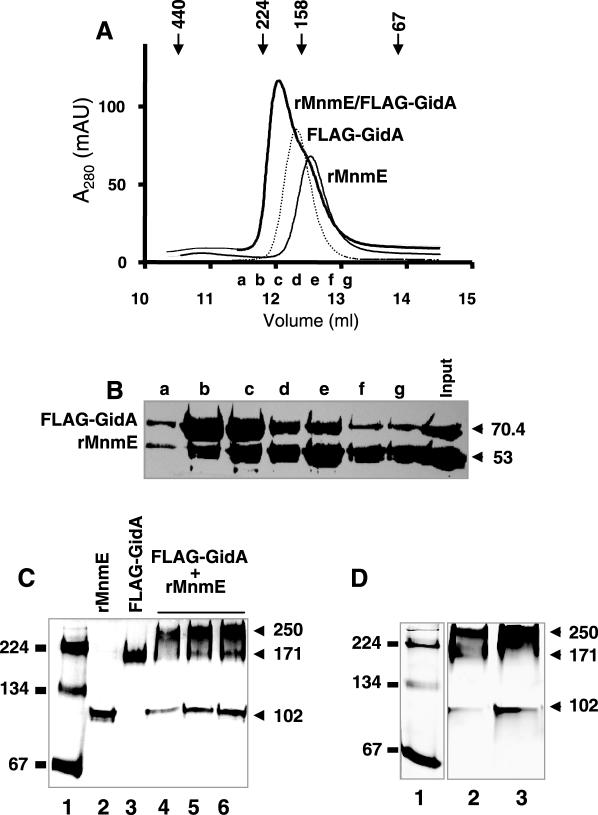
Self-assembly of FLAG-GidA protein. (A) Purification of the recombinant protein FLAG-GidA. SDS–PAGE analysis of samples from different purification steps of the FLAG-GidA protein. The gel was stained with Coomassie blue. Lane 1, crude extract from uninduced DEV16/pIC1180; lane 2, crude extract from DEV16/pIC1180 induced with arabinose; lanes 3 and 4, cleared lysate (supernatant after sonication) and insoluble material (pellet after sonication) of the induced strain, respectively; lane 5, first eluate with elution buffer (containing free FLAG peptide); lane 6, second eluate with elution buffer. (B) Gel filtration analysis of purified FLAG-GidA. A total of 100 μg of FLAG-GidA were applied on a Superdex HR 200 column as described in Materials and Methods and 0.5 ml fractions collected. Markers indicate the positions of the standards: ferritin (440 kDa), β-amylase (224 kDa), aldolase (158 kDa), albumin (67 kDa). Inset: SDS–PAGE analysis of fractions indicated with asterisks in the chromatogram and designated as a, b, and c. (C) In vitro cross-linking of FLAG-GidA with DTSP. Protein samples, untreated or cross-linked with DTSP, were analysed by non-reducing SDS–PAGE. Lanes 4 and 5 show the effect of 2-ME on the reactions. Size of mass markers, run in lane 1, is indicated on the left in kDa.
The ability of GidA to dimerize was also analysed by in vitro chemical cross-linking. Extensive cross-linking in solution is taken to be indicative of specific interactions, since random collisions are expected to produce a minimal amount of cross-linking [(7) and references therein]. Dithiobis (succinimidyl propionate) (DTSP) was used as the cross-linking molecule; it reacts primarily with ɛ-amines of lysine residues, and is cleavable under reducing conditions. Incubation of FLAG-GidA with DTSP led to the formation of a complex of around 150 kDa, (Figure 3C, compare lanes 2 and 3), whereas the subsequent addition of the strong reducing agent 2-mercaptoethanol (2-ME) induced dissociation into monomers (Figure 3C, compare lanes 3 and 5).
GidA and MnmE interact and form an α2β2 heterotetrameric complex
Several results indicate that both GidA as well as MnmE form homodimers in vitro (Figure 3B and C; 7–9). Previously, it was observed that when a mitochondrial extract obtained from a yeast transformant overexpressing proteins MTO1 and MSS1 was centrifuged through a sucrose gradient, the two proteins displayed identical sedimentation properties (29). Considering that MTO1 and MSS1 peaked in a region of the gradient between the lactate dehydrogenase and hemoglobin markers, it was concluded that they form a heterodimer complex with a molecular weight of around 121–128 kDa. However, because of these experiments did not include appropriate controls showing the sedimentation properties of each protein alone, and in the light of the new results, we thought that other explanations were also possible. For example, it could not be discarded that the gradient fraction supposed to contain the putative heterodimer, actually included MTO1 and MSS1 homodimers. In this respect, it is worthy to mention that the MnmE dimer has an elongated structure, which determines that it migrates in gel filtration at a molecular mass greater than expected (9). Consequently, it would not be surprising that a dimer of its homologue, MSS1, could also display an anomalous sedimentation profile, peaking at a position similar to that of a putative MTO1 dimer. We also reasoned that complexes between MTO1 and MSS1 homodimers, if exist, could not have been detected by sucrose gradient centrifugation because they might be sensitive to pressure and dissociate during high velocity centrifugation. Alternatively, it could not be either discarded the possibility that the MSS1•MTO1 complex displayed aberrant sedimentation properties, so that the putative heterodimer were actually a heterotetramer. Thus, we considered that additional experiments were required to prove and clarify interaction between the MnmE and GidA family proteins.
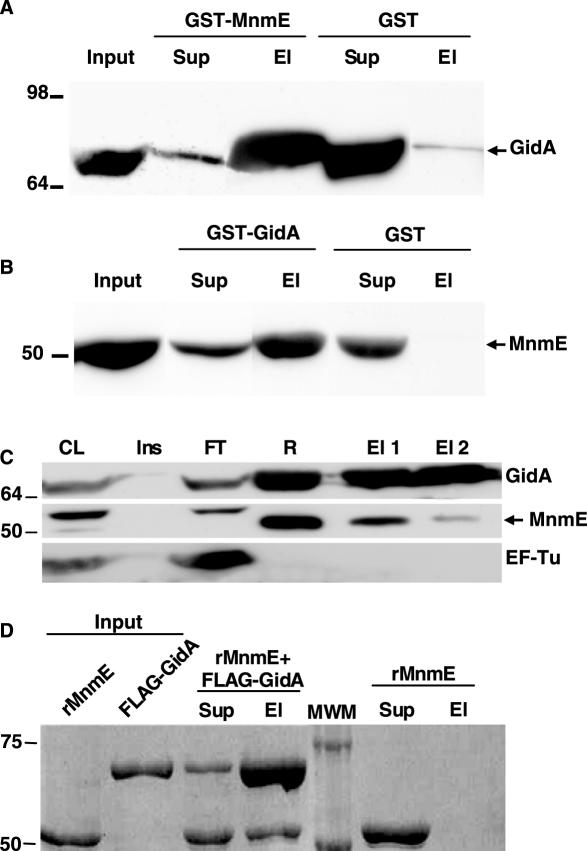
To explore whether the E.coli MnmE and GidA proteins physically interact, we performed GST-pull down analysis using purified GST-MnmE or, alternatively, GST-GidA as a bait. GST-MnmE immobilized on glutathione-agarose beads was incubated with total protein extracts of a GidA overexpressing strain. As can be seen in Figure 4A, GidA could be pulled down from the extracts by GST-MnmE but not by GST alone. Moreover, protein EF-Tu, used as a control, was detected only in the unbound supernatant fraction (data not shown). Conversely, when GST-GidA was used as a bait, MnmE was pulled down from a total extract of an MnmE overproducing strain meanwhile, in the same conditions, MnmE was not pulled down by GST alone (Figure 4B). These results suggest that MnmE and GidA interact with each other in total E.coli extracts; however, they do not inform whether such an interaction is direct or mediated by another protein. In addition, the need to co-overproduce chaperones to obtain soluble GST-GidA protein raises the possibility that some chaperone mediates interaction of MnmE with GST-GidA. Therefore, we decided to perform co-immunoprecipitation experiments using FLAG-GidA protein as bait. This protein is overproduced by adding arabinose to the medium and remains mostly soluble, eliminating the requirement for chaperone co-expression (see Figure 3A). We purified FLAG-GidA by firstly passing it through an anti-FLAG agarose column and later eluting it with excess of FLAG peptide. When analysing the fractions eluted from the column by Western blotting (Figure 4C), we observed that MnmE, but not EF-Tu, was co-eluted with FLAG-GidA, supporting that MnmE and FLAG-GidA were able to interact in total extracts of E.coli. Moreover, we used purified FLAG-GidA as bait to pull down purified recombinant MnmE (rMnmE, molecular mass, 53 kDa). As shown in Figure 4D, rMnmE is pulled down with anti-FLAG agarose only when FLAG-GidA is present, and is eluted from the resin using an excess of FLAG peptide. We therefore conclude that the E.coli MnmE and GidA proteins are able to directly interact with each other.
Figure 4.
GidA and MnmE interact with each other. (A) Western blot of GST pull down analysis using anti-GidA antibody. GST-MnmE or GST alone were used as baits for pull down analysis. Input: total lysate of strain IC5332 that was added to the corresponding bait previously immobilized to glutathione-agarose. Sup: fraction not retained by the resin-bound bait protein. El: the bound material that is eluted from the resin with glutathione buffer. Equivalent amounts of each fraction were loaded. (B) The same as in (A), but the baits were GST-GidA or GST, the lysate was from strain IC5287, and the western blot was developed with anti-MnmE antibody. (C) Coimmunopurification of FLAG-GidA and MnmE. FLAG-GidA was purified from strain DH5α/pIC1180 with anti-FLAG agarose as described in Materials and Methods, and the resulting fractions were analyzed by western blot using the antibodies indicated in the right side of each panel. Anti-EF-Tu was used as a negative control. The arrowhead marks MnmE position. The upper band detected in the MnmE western blot corresponds to an unrelated, unknown protein that is recognized by the anti-MnmE antibody and serves as a negative control. CL: cleared lysate of DH5α/pIC1180 (supernatant after sonication). Ins: pellet of cleared lysate. FT: flow through after incubation with anti-FLAG agarose. R: resin after washes. El 1: first eluate with elution buffer (free FLAG peptide). El 2: second eluate with elution buffer. (D) In vitro pull down analysis of purified recombinant MnmE (rMnmE) and FLAG-GidA proteins. After incubating both proteins together, anti-FLAG resin was added and fractions collected as described in Material and Methods. As a negative control, MnmE was processed without adding FLAG-GidA. Sup: supernatant after pelleting FLAG-agarose. El: proteins eluted with free FLAG peptide. Equivalent amounts of samples were loaded in each well. Size of molecular weight markers (MWM) is indicated on the left in kDa.
Next, considering that both MnmE and GidA alone dimerize, we decided to investigate the stoichiometry of the GidA•MnmE complex. To this end, purified rMnmE (53 kDa) and FLAG-GidA (70.4 kDa) proteins were applied, either separately or jointly, to a Superdex-200 HR gel filtration column. Figure 5A shows that rMnmE and GidA eluted as single peaks at molecular weights of 149 and 168 kDa. Note that the estimated mass of MnmE is larger than expected, but this feature has been previously described and considered to be due to the elongated shape of the MnmE dimer (9). When a mix of GidA and MnmE was chromatographed, both proteins coeluted at a peak of about 195 kDa, which is indicative of their presence in a complex (Figure 5A). Moreover, given that this analysis was performed in the presence of DTT, 5 mM, it may be concluded that the in vitro formation of the GidA•MnmE complex does not involve disulfide bridges. From these results, and considering that: (i) the apparent mass of the complex is larger than that of each homodimer, (ii) no peaks corresponding to monomeric forms are distinguishable from the elution profile of the mix, and (iii) the peak of the mix (elution fraction c) contains approximately equal amounts of GidA and MnmE (Figure 5B), we propose that the peak of 195 kDa corresponds to a GidA•MnmE heterotetrameric complex of type α2β2, whose compact conformation results in an apparent mass smaller than expected. To confirm this proposal, we analysed migration of a GidA/MnmE mix in native gels. As shown in Figure 5C, purified rMnmE and FLAG-GidA proteins migrate, when separately loaded, at positions corresponding to a molecular mass of 102 and 171 kDa, respectively. Note that in this case, the apparent mass of MnmE fits well with that expected for its dimeric form. The mix of MnmE and GidA produces bands of 102, 171 and 250 kDa. Since the last one is recognized by both anti-MnmE and anti-GidA antibody (data not shown), and its size fits well with the sum of sizes of the MnmE and GidA homodimers, as estimated from their migration in the same native gel, we conclude that it corresponds to the α2β2 heterotetrameric form of the GidA•MnmE complex. It is worthy to mention that, when samples of the elution fractions b and c from the gel filtration experiment (Figure 5A) were analysed by native PAGE, a predominant 250 kDa band was also observed (Figure 5D, lanes 2 and 3), which supports the idea that the apparent mass of the heterotetrameric GidA•MnmE complex estimated from gel filtration is smaller than that estimated from native PAGE.
Figure 5.
GidA and MnmE form an α2β2 heterotetrameric complex. (A) Estimation of the molecular mass of the peak fractions in preparations of rMnmE, FLAG-GidA and a FLAG-GidA/rMnmE mix by gel-filtration chromatography. The major peaks from each preparation eluted at 149, 168 and 195 kDa, respectively. Markers indicate the positions of the standards: ferritin (440 kDa), β-amylase (224 kDa), aldolase (158 kDa), albumin (67 kDa). Elution fractions a to g from chromatography of the FLAG-GidA/MnmE mix were pooled for further analysis. (B) SDS–PAGE of elution fractions a to g from the FLAG-GidA/rMnmE mix chromatography showed in (A). Fractions (250 μl) were precipitated with trichloroacetic acid before loading. The gel was stained with Coomassie blue. Molecular masses are indicated on the right in kDa. (C) Native PAGE analysis of rMnmE, FLAG-GidA and a mix of FLAG-GidA/rMnmE. Lane 1: Molecular mass markers were β-amylase and albumin (monomer and dimer). Lane 2: 10 μg of rMnmE. Lane 3: 10 μg of FLAG-GidA. Lanes 4–6: 15, 30, and 40 μg, respectively, of total protein from a FLAG-GidA/rMnmE mix prepared as indicated in Materials and Methods. Positions of mass markers and their size in kilodaltons are indicated on the left. Positions of rMnmE, FLAG-GidA and FLAG-GidA•rMnmE specific complexes are indicated on the right, together with the apparent molecular mass of the complexes. (D) Native PAGE of fractions coming from gel-filtration of a FLAG-GidA/rMnmE mix. Lane 1: molecular mass markers were as (C), Lanes 2 and 3: elution fractions b and c, respectively, from an experiment like that shown in (A). Size of the mass markers and complexes are indicated on the right and left, respectively. Note correspondence between complexes detected here and those of (C).
The FAD domain of GidA is required for the tRNA modifying function of this protein
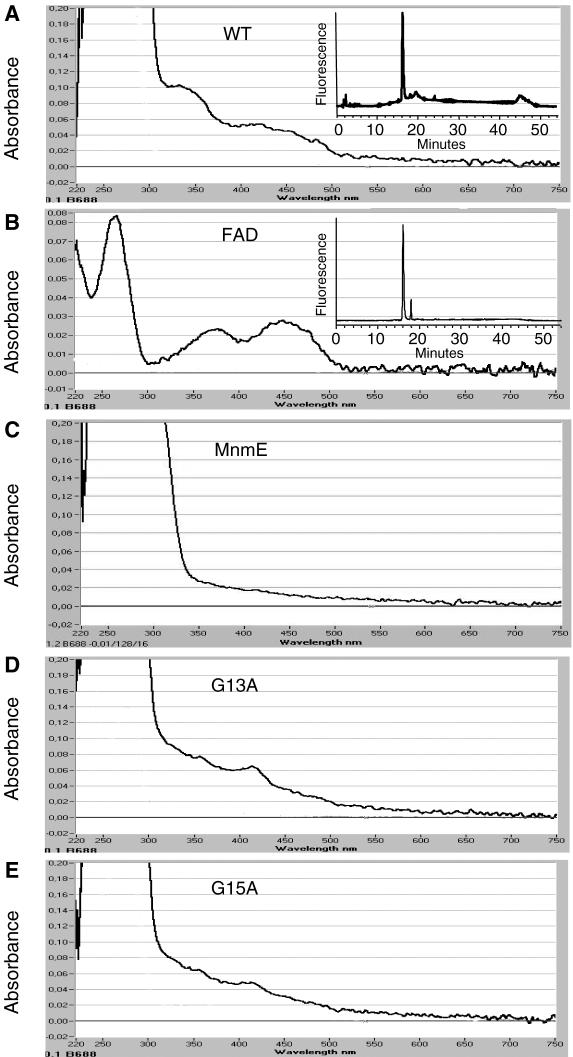
Alignment of Gid proteins in the database reveals a highly conserved dinucleotide-binding motif [xhxhxGxGxxG(x)3hxxh(x)9hxhxxE/D, where x is any residue and h is a hydrophobic residue] near the N-terminus (1,2). In fact, the TrmFO proteins from M.xanthus and B.subtilis (1,2) and GidA proteins from B.subtilis and E.coli (2,9) were reported to bind FAD. In this study, we have found that FLAG-GidA absorbs at about 340 and 430 nm (Figure 6A), in agreement with the idea that GidA binds flavin. Moreover, HPLC analysis indicated that the cofactor released from FLAG-GidA by heating the protein at 75°C for 15 min was FAD (Figure 6A, inset). In contrast, the visible spectrum of MnmE, the GidA partner in the modification reaction, reveals that this protein does not contain flavin (Figure 6C).
Figure 6.
Spectral analysis of purified FLAG-GidA and mutant derivatives, to detect the presence of bound FAD. The UV-visible spectrum of the wild type, G13A, and G15A FLAG-GidA proteins is shown in panel A, D and E, respectively. Free FAD was used to obtain a reference spectrum (panel B), and rMnmE as a negative control (panel C). Insets: HPLC analysis of the GidA-bound cofactor (A) and FAD (B), which had retention times of 16.19 and 16.05 min, respectively. FMN had a retention time of 18.61 min (data not shown).
The dinucleotide-binding motif present in the N-terminal end of GidA is known to be part of the Rossmann fold characteristic of the glutathion reductase family (36). The importance of the invariant glycine residues (GxGxxG) in this motif is well understood, with the two first glycines determining close contact of the main chain to the pyrophosphate of FAD (36). In agreement with this, when we separately changed these invariant glycines to alanine on FLAG-GidA, the UV-visible spectrum of the purified proteins G13A (from plasmid pIC1181) and G15A (from plasmid pIC1182) indicated that the absorbing peaks at around 340 and 430 nm were significantly lower than those produced by the wild-type protein (Figure 6D and E), suggesting that both mutant proteins bind FAD in less extent than the wild-type GidA (Figure 6A).
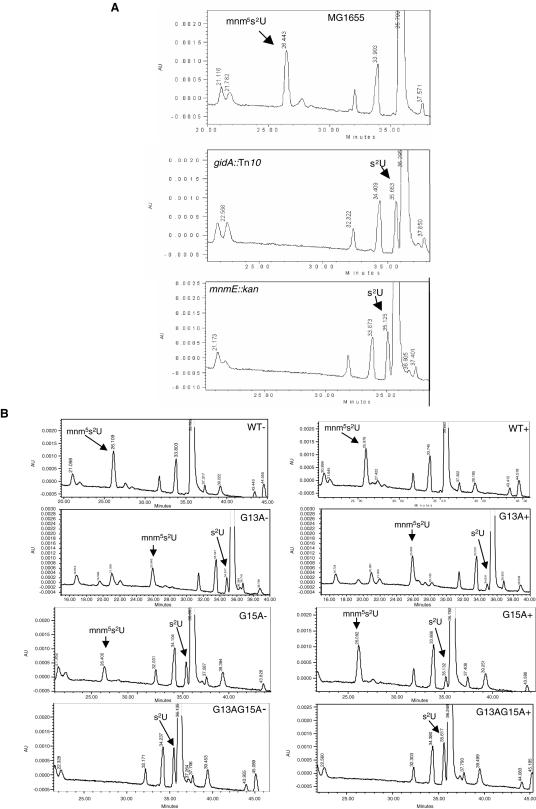
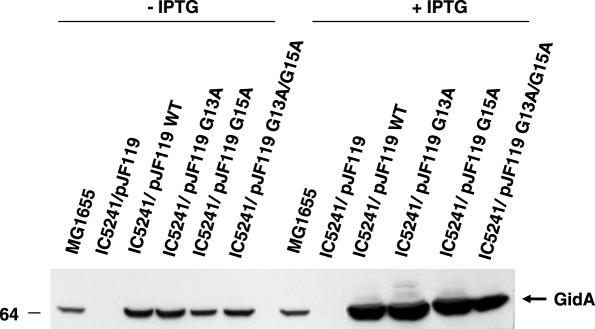
To determine whether the FAD-binding motif of GidA is involved in the tRNA modifying function of this protein, we introduced mutations G13A and G15A on plasmid pIC1154 (carrying the gidA wild-type gene under control of promoter Ptac) and proceeded to analyse the nucleoside composition of tRNA from strains producing wild-type or mutant GidA proteins. First, total tRNA from strains MG1655 (wild-type) and IC5241 (MG1655 gidA::Tn10) was hydrolysed and analysed by HPLC. As can be seen in Figure 7A, a peak corresponding to nucleoside mnm5s2U is detected in tRNA from strain MG1655, but not in tRNA isolated from IC5241. Conversely, a peak corresponding to s2U is seen in tRNA from IC5241, but not in tRNA from the wild-type strain. This indicates that in the gidA::Tn10 strain, the addition of cmnm group in position 5 of U34 is defective, rendering U34 modified only in position 2 (see Figure 1). Note that this pattern is similar to that observed in tRNAs obtained from the mnmE::kan strain (10; Figure 7A, lower panel). These results show for the first time that interruption of the gidA gene impairs formation of mnm5s2U34, and definitively prove that gidA is allelic to trmF (see Introduction). In this respect, it should be pointed out that the original proposal by Brégeon et al. (4) that trmF and gidA are allelic was mostly based on genetic mapping data, (which were not precise in the trmF case) as well as on phenotypic traits. Next, we analysed the nucleoside composition of strains producing mutant GidA proteins. Thus, total tRNA from strain IC5241 (MG1655 gidA::Tn10) transformed with pIC1154 (Ptac-gidA) or each one of its mutant derivatives (pIC1177, pIC1178 and pIC1179 carrying alleles G13A, G15A or G13A/G15A, respectively), and grown with or without IPTG, was hydrolysed and analysed by HPLC. As shown in Figure 7B (without IPTG, left panels), a peak corresponding to s2U is detected in the analysis of tRNA from strains G13A, G15A and G13A/G15A, although it is most prominent in the last one. This pattern is indicative of a defect in the addition of the cmnm group to position 5 of U34 (see Figure 1), as shown in the analysis of strain gidA::Tn10 (Figure 7A). Note, however, that a small peak corresponding to mnm5s2U34 is present in tRNA isolated from mutants G13A, and G15A but not in tRNA isolated from G13A/G15A (Figure 7B, left panels). This suggests that even in the absence of the IPTG inducer, a certain amount of protein G13A or G15A is produced, which is able to modify a fraction of tRNA molecules. When tRNA is isolated from IPTG-induced G13A and G15A strains (Figure 7B, right panels), the peak corresponding to s2U decreases whereas that corresponding to mnm5s2U increases. This behaviour is not observed with tRNA isolated from mutant G13A/G15A, which suggests that the GidA protein produced by this mutant is a full loss-of-function protein. Western blot analysis confirmed production of the mutant proteins even in the absence of the IPTG inducer, which is probably due to an incomplete repression of the strong promoter Ptac (Figure 8, −IPTG). No significant differences among cellular levels of the plasmid encoded wild-type and mutant proteins were observed both in the presence as well as in the absence of IPTG (Figure 8), indicating that the defects in tRNA modification observed in cells expressing mutant proteins (Figure 7B) are not due to a lower accumulation of these proteins into the cell. It is also interesting to note that the cellular levels of proteins G13A and G15A synthesized in the absence of IPTG were similar to the native levels of GidA in strain MG1655 (Figure 8, −IPTG). This supports that proteins G13A and G15A are not as effective as the native GidA protein in conducting tRNA modification since s2U is present in tRNA purified from strains expressing proteins G13A and G15A (Figure 7B, left panels), but not in tRNA obtained from the wild-type strain (Figure 7A). Moreover, a small but discernible peak of s2U is still detected from tRNA of strains overexpressing proteins G13A and G15A (Figure 7B, right panel), in spite of their respective cellular levels are much higher than the native levels of GidA (Figure 8, +IPTG). Therefore, it is reasonable to conclude that G13A and G15A are partial loss-of-function proteins.
Figure 7.
HPLC chromatograms of tRNA hydrolysates. Strains used were: MG1655 (wild-type), IC5241 (gidA::Tn10), and IC5348 (mnmE::kan), in (A), and IC5241 transformed with plasmids pIC1154, pIC1177, pIC1178 and pIC1179 (carrying the wild-type, G13A, G15A and G13A/G15A gidA allele, respectively), in (B). Strains in (B) were grown with (+, right side) or without (−, left side) IPTG. The nucleosides were monitored at 314 nm to maximize the detection of thiolated nucleosides. mnm5s2U and s2U were identified by comparing UV spectra with published spectra (41). AU, absorbance units.
Figure 8.
Western blot analysis of total extracts from strains MG1655 (wild-type) and IC5241 (gidA::Tn10) transformed with pJF119 or its derivatives pIC1154, pIC1177, pIC1178 and pIC1179 (carrying the wild-type, G13A, G15A and G13A/G15A gidA alleles, respectively), using anti-GidA antibody. The strains were grown during 2.5 h in LBT at 37°C with or without 1 mM IPTG, before processed. 50 μg of total proteins were loaded in each well. Size of the molecular weight marker (on the left) is in kDa.
Phenotype of GidA mutants
Full loss-of-function mnmE mutations are lethal in the genetic backgrounds of strains JC7623 and V5701 but not in those of MC1000, DEV16 and MG1655 (7,8,10). At present, we have demonstrated that the lethal phenotype results from incompatibility of loss-of-function mnmE mutations with mutations in genes involved in the ribosomal dynamic (M. Villarroya, L. Yim and M.-E. Armengod, manuscript in preparation). Here, we have found that the gidA::Tn10 allele cannot be recovered on the V5701 chromosome after P1 transductions unless the receptor strain carries a second copy of gidA on a plasmid that allows accumulation of the wild-type, G13A or G15A GidA protein (i.e. plasmids pIC1154, pIC1177 and pIC1178; data not shown). This indicates that the gidA::Tn10 null allele is also lethal in the V5701 background and that the single mutations G13A and G15A should be partial loss-of-function mutations, given that overexpression of the corresponding proteins allows recovery of allele gidA::Tn10 on the V5701 chromosome. It should be pointed out that overexpression of protein G13A/G15A (from plasmid pIC1179) does not lead to recovery of allele gidA::Tn10, which means that it is not able to complement for the null allele and, therefore, that mutation G13A/G15A is a full loss-of-function mutation.
Note that, similarly to full loss-of-function mnmE mutations (7,8,10), the G13A/G15A or gidA::Tn10 mutations could be recovered on the DEV16 or MG1655 chromosome even in the absence of a second copy of the wild-type gidA gene (see e.g. strains IC5241 and IC5550 in Table 1). Interestingly, the cell growth is slowed down by about 30% when mutation gidA::Tn10 is introduced into strain MG1655, and this effect can be suppressed by a plasmid overexpressing protein G13A or G15A but not by a plasmid overexpressing protein G13A/G15A (data not shown). This indicates that mutation G13/G15 affects cell growth in the same extent than a null gidA mutation does, which is in agreement with the idea that G13/G15 is a full loss-of-function mutation.
We have also found that mutations gidA::Tn10 and mnmE::kan reduce the doubling time of strain MG1655 to a similar extent in LB (doubling times: 29.9 ± 2.3, 37.8 ± 2.7 and 38.2 ± 3.1, for wild-type, mnmE and gidA strains, respectively), minimal medium supplemented with casaminoacid (doubling times: 50.3 ± 3.6, 63.7 ± 4.1 and 64.1 ± 4.2, for wild-type, mnmE and gidA strains, respectively), and minimal medium, in which both mutations seem to have a minor effect (doubling times: 90 ± 5.5, 103.1 ± 5.9 and 100.2 ± 4.5, for wild-type, mnmE and gidA strains, respectively). Curiously, Brégeon et al. (4) found that whilst the growth rate of their gidA::Tn10 and mnmE::Tn10 mutants was reduced in LB to the same level, only mutation gidA::Tn10 significantly reduced the growth rate in minimal medium. This difference of phenotype conferred by gidA and mnmE mutations led Brégeon et al. (4) to propose that the MnmE activity precedes that of GidA in the tRNA modification pathway, and that accumulation of the unknown product of MnmE in gidA mutants could be toxic for cells under slow growth conditions. If MnmE worked before GidA, we should find a reduction of the s2U levels in gidA::Tn10 mutants since the hypothetical MnmE product would be generated in these mutants at expenses of s2U (see Figure 1). However, we have found that the levels of s2U are near identical in full loss-of-function gidA and mnmE mutants (Table 2). Therefore, our results do not support the idea that the MnmE activity precedes that of GidA. Moreover, as above pointed out, we have not found significant differences between the growth rate of gidA and mnmE mutants in minimal medium. The discrepancy between these results and those reported by Brégeon et al. (4) might be due to the use of different insertion mutations. Our strains carry the Tn10 insertion at position 51 in gidA, whereas Brégeon et al. used a gidA mutant carrying the Tn10 insertion at position 707 for their experiments. In both insertions, the orientation of the minitransposon remains to be determined. Minitransposons may be polar and their effect on neighbouring genes may depend on their location and orientation within the target gene. Therefore, it is possible that the effect of each Tn10 insertion on gidA neighbouring genes (i.e. oriC and gidB, which encodes a putative methyltransferase of unknown function) were different, and this may be the true cause of the different behaviour of gidA insertion mutants in minimal medium. Since plasmid pIC1154, which just carries the structural gidA gene under control of a suitable promoter, reverted the growth rate of our gidA::Tn10 mutant to the wild-type level, we are confident that, in this strain, the polar effects of the minitransposon, if there exist, are negligible. Consequently, our data support the conclusion that loss-of-function gidA and mnmE mutations have similar effects on cell growth (including the synthetic lethality phenomenon and growth rate, see above) and the tRNA modification status (Table 2).
Table 2.
Levels of s2U in mutant strains
| Straina | Relevant genotype | s2U/s4Ub | |
|---|---|---|---|
| Chromosome | Plasmid | ||
| MG1655 | mnmE::kan | — | 0.026 |
| MG1655 | gidA::Tn10 | — | 0.022 |
| MG1655 (−) | gidA::Tn10 | GidA G13A | 0.011 |
| MG1655 (−) | gidA::Tn10 | GidA G15A | 0.016 |
| MG1655 (−) | gidA::Tn10 | GidA G13AG15A | 0.025 |
| MG1655 (+) | gidA::Tn10 | GidA G13A | 0.004 |
| MG1655 (+) | gidA::Tn10 | GidA G15A | 0.006 |
| MG1655 (+) | gidA::Tn10 | GidA G13AG15A | 0.026 |
a(+) and (−) indicate presence or absence of inducer IPTG in the growth medium.
bThe numbers are calculated as the absorbance of s2U relative to the absorbance of s4U at 314 nm (values were taken from Figure 7).
DISCUSSION
The role of tRNA in mRNA decoding is crucial to the accuracy and efficiency of protein synthesis. The post-transcriptional modifications that occur at the wobble position of tRNAs that read codons in the mixed codon boxes are particularly significant (16,37,38). E.coli mutants defective in the biosynthesis of mnm5s2U, the modified nucleoside that is present in the wobble position of tRNAs specific for Lys and Glu, were firstly isolated based on the readthrough phenotype at UAG codons (5). This allowed the characterization of the trmE and trmF loci, which were mapped to two different locations on the E.coli chromosome, although the phenotype of both type of mutants was quite similar. The isolation and study of trmC (mnmC) mutants allowed to conclude that trmF and trmE control the addition of the cmnm group in position 5 of the wobble uridine (6; see Figure 1). Later, the trmE gene was shown to be allelic with a gene encoding a 50 kDa GTPase, and renamed mnmE (7), whereas trmF was proposed to be allelic with gidA from indirect genetic evidence and phenotypic traits (4). However, no analysis of the tRNA modification status in gidA mutants has been performed up to now. In this study, we show for the first time that mutations in the E.coli gidA gene impair the biosynthesis of mnm5s2U (Figure 7) and, therefore, definitively prove that gidA is allelic to trmF. The tRNA modifying function assigned to gidA is in agreement with recent findings in yeast showing that disruption of the gidA homologous gene, MTO1, impairs the biosynthesis of cmnm5U, the nucleoside normally present in the wobble position of mitochondrial tRNALys (32), which strongly supports the idea that the function of GidA is evolutionarily conserved. Similarly, disruption of MSS1, the homologous gene of mnmE in yeast, also impairs biosynthesis of cmnm5U (32), which argue in favour of a conservative evolution of the cmnm5U biosynthesis pathway. However, the precise role of MnmE and GidA family proteins in this pathway remains undetermined. One way to gain insights into such a role is by elucidating the activities of both proteins that are required for their tRNA modifying function. Thus, we have previously found that E.coli MnmE exhibits a GTPase activity that is essential for that function (8,10). Moreover, biochemical and structural data indicated that T.maritima MnmE binds 5-formyl-tetrahydrofolate (9). This strongly supports a direct participation of MnmE in the tRNA modification reaction since formyl-tetrahydrofolate might be used as the one-carbon unit donor in the formation of the cmnm group (Figure 1). Such a proposal is in line with previous data indicating that the first carbon atom in the side chain at 5 position of cmnm5U originates from a source other than methionine (6). Furthermore, alignment of GidA proteins and their paralogues TrmFO revealed a highly conserved dinucleotide-binding motif near the N-terminus (1,2). In fact, the TrmFO proteins from M.xanthus and B.subtilis (1,2), and GidA proteins from B.subtilis and E.coli (2,9) were reported to bind FAD, although in the case of GidA proteins, data were never shown. In this respect, we have found here that GidA contains noncovalently bound FAD (Figure 6A), and that change to alanine of G13 and G15, two residues belonging to the FAD-binding motif of GidA, impairs capability of this protein to modify tRNA (Figure 7). This indicates that GidA, whichever the mechanism of U34 modification is, catalyses an FAD-depending reaction that is also required for generation of cmnm5U.
Immunoelectron microscopy and subcellular fractionation indicated that MnmE is localized in both the cytoplasm and, to a lesser but significant extent, the inner membrane (7,8). In this work, experiments of subcellular fractionation show that GidA is mostly found in the soluble fraction, although at least a portion was also detected in the membrane fraction (Figure 2B). We find this to be indicative that both proteins have identical subcellular distribution. A predominant cytoplasmic localization, or even a partial association with the cytoplasmic membrane inner face, is foreseeable for enzymes whose substrate is tRNA. Moreover, we show for the first time that purified MnmE and GidA proteins interacts in vitro (Figure 4). Altogether, these results suggest that both proteins form a functional complex that carries out the modification reaction. Considering that dimerization of MnmE is required to form its tetrahydrofolate binding site (9) and that GidA mostly forms dimers (Figure 3), it seems reasonable to propose that a heterotetramer of type α2β2 is the functional form of the MnmE•GidA complex. In fact, our results from gel filtration experiments and analysis in native gels strongly support this proposal (Figure 5). It is of interest to mention that whilst the apparent mass of the MnmE dimer estimated from gel filtration is larger than expected (149 versus 100 kDa), because of its elongated shape (9), the apparent mass of the α2β2 MnmE•GidA complex is smaller than expected (∼195 kDa, see Figure 5A). Since the mobility of particles on gel filtration depends on their size and their shape, our results suggest that formation of the heterotetrameric complex increases the compactness of the interacting partners. The MnmE•GidA complex runs in native gels with an apparent mass of about 250 kDa, which fits well with the expected mass for a heterotetramer of type α2β2 (Figure 5C).
MnmE and GidA are evolutionarily conserved proteins present in mitochondria of yeast and human (28–31). Interestingly, taurine is directly incorporated into human mitochondrial tRNAs in a modification process that probably involves the human MnmE and GidA homologues (32,39). Thus, it was proposed a model where both proteins catalyse the formation of an unknown intermediate, and the subsequent activity of a taurine or glycine transferase is responsible for construction of the 5-taurinomethyl group in humans, or 5-cmnm group in yeast and bacteria [(32), see Figure 9A]. On the other hand, differences in growth rate between E.coli mnmE and gidA null mutants led to the idea that the MnmE activity precedes that of GidA in the tRNA modification pathway (4). Thus, considering the ability of MnmE and GidA to bind 5-formyl-tetrahydrofolate and FAD, respectively, it was proposed a new model where MnmE catalyses the transfer of the formyl group from formyl-tetrahydrofolate onto the 5 position of the uridine base, producing a formylated uridine and, subsequently, GidA mediates transfer of glycine and catalyses a reduction reaction required for generation of the cmnm5 modification [(9), see Figure 9B]. According to this model, in the HPLC analysis from tRNA hydrolysates, we would expect to observe disappearance of s2U if a thiolated intermediate (s2f5U) were synthesized by MnmE in gidA mutants. In contrast, s2U is present in hydrolysates of tRNA isolated from mutants gidA::Tn10 and G13A/G15A (Figure 7). In fact, the relative levels of s2U found in these mutants were near identical to those found in the mnmE::kan strain (Table 2). We have observed that MnmE is pulled down by the FLAG-GidA G13A/G15A mutant protein (data not shown), which indicates that the FAD-binding ability of GidA is not required for the physical interaction with MnmE. However, MnmE does not seem to work successfully in the presence of protein G13A/G15A since s2U, as pointed out above, is found in tRNA of mutant G13A/G15A at similar levels than in tRNA from the mnmE::kan mutant (Figure 7 and Table 2). These results do not support previous proposals that activity of MnmE precedes that of GidA in the MnmE/GidA pathway (4,9); rather, our data fit better with a model where proteins GidA and MnmE form a functional complex in which both proteins are interdependent. Moreover, we have not observed differences between the growth rate of gidA and mnmE mutants in minimal medium, so that we think that differences reported by other authors (4) might be due to polar effects of the gidA::Tn10 mutation used in their experiments. The similar phenotype produced by loss-of-function gidA and mnmE mutations on cell survival and cell growth that we describe here (see subsection ‘Phenotype of GidA mutants’) also supports a model in which MnmE and GidA are interdependent proteins. Development of an in vitro assay using a recombinant MnmE•GidA complex will provide a key to clarify the molecular mechanism of the MnmE/GidA dependent pathway.
Figure 9.
Models for the biosynthetic pathway leading to cmnm5U34, summarized from references 32 (A) and 9 (B).
Phylogenetic analysis indicate that GidA and TrmFO protein families have evolved from a common ancestor but acquired different, non-overlapping cellular functions during evolution (2). Our results clearly show that GidA and MnmE form a heterocomplex, and suggest that both proteins drive the tRNA modification process by forming such a complex. In other words, whilst the TrmFO enzymes catalyze folate-dependent formation of 5-methyluridine at position 54 of tRNA by themselves (2), GidA requires association with MnmE to catalyze formation of cmnm5U at position 34. Thus, the MnmE•GidA complex seems to represent a new evolutionary way to use a folate derivative and FAD in the post-transcriptional modification of tRNA.
Interestingly, s2mnm5U was reported to be present in tRNA from Archaea (40). However, we have not found homologous of gidA and mnmE in the sequenced genomes of archaeal organisms. Search for mnmE homologues led to find proteins from Archaea that only exhibit a minor homology with MnmE in the G domain, which could just indicate that they are GTP-binding proteins. Thus, it seems that if ancient gidA and mnmE homologues there existed, divergent evolution between Bacteria and Archaea has produced proteins with a very low level of homology, even undetectable in the case of GidA. Alternatively, it might be possible that formation of mnm5 in Archaea depends on proteins that are not evolutionarily related to MnmE and GidA. If so, this would mean that a pathway for synthesis of mnm5 has been established independently at least twice during evolution.
Acknowledgments
The authors thank Dr E. Knecht, Dr M. J. Vicent-Docón, and Dr J. F. Sanz-Cervera for valuable advice, Dr D. Brégeon for the generous gift of strains, and Dr M. Vicente for the anti-EF-Tu antibody. The authors also thank Silvia Prado and Rafa Ruiz for donating purified rMnmE and FLAG-GidA proteins used in some experiments, Magda Villarroya and Elvira Cebolla for allowing us to use their data on growth rate of mutants mnmE and gidA, and Rafa Ruiz and Alfonso Benítez for discussions during preparation of this manuscript. I.M. is a Fellow of Bancaixa and Centro de Investigación Príncipe Felipe. This work was supported in part by the Ministerio de Educación y Ciencia (Grants BMC2001-1555 and BFU2004-05819), the Ministerio de Sanidad (Grants G03/203, G03/011, and PI051791) and the Generalidad Valenciana (Grant GRUPOS04/07) to M.-E.A., and by grants from the Swedish Cancer Foundation (Project 680) and the Swedish Science Research Council (Project BU-2930) to G.R.B. Funding to pay the Open Access publication charges for this article was provided by the Ministerio de Educación y Ciencia (Grant BFU2004-05819).
Conflict of interest statement. None declared.
REFERENCES
- 1.White D.J., Merod R., Thomasson B., Hartzell P.L. GidA is an FAD-binding protein involved in development of Myxococcus xanthus. Mol. Microbiol. 2001;42:503–517. doi: 10.1046/j.1365-2958.2001.02659.x. [DOI] [PubMed] [Google Scholar]
- 2.Urbonavičius J., Skouloubris S., Myllykallio H., Grosjean H. Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria-evolutionary implications. Nucleic Acids Res. 2005;33:3955–3964. doi: 10.1093/nar/gki703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.von Meyenburg K., Jørgensen B.B., Nielsen J., Hansen F.G. Promoters of the atp operon coding for the membrane-bound ATP synthase of Escherichia coli mapped by Tn10 insertion mutations. Mol. Gen. Genet. 1982;188:240–248. doi: 10.1007/BF00332682. [DOI] [PubMed] [Google Scholar]
- 4.Brégeon D., Colot V., Radman M., Taddei F. Translational misreading: a tRNA modification counteracts a +2 ribosomal frameshift. Genes Dev. 2001;15:2295–2306. doi: 10.1101/gad.207701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elseviers D., Petrullo L.A., Gallagher P. Novel E. coli mutants deficient in biosynthesis of 5-methylaminomethyl-2-thiouridine. Nucleic Acids Res. 1984;12:3521–3534. doi: 10.1093/nar/12.8.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagervall T.G., Edmonds C.G., McCloskey J.A., Björk G.R. Transfer RNA(5-methylaminomethyl-2-thiouridine)-methyltransferase from Escherichia coli K-12 has two enzymatic activities. J. Biol. Chem. 1987;262:8488–8495. [PubMed] [Google Scholar]
- 7.Cabedo H., Macián F., Villarroya M., Escudero J.C., Martínez-Vicente M., Knecht E., Armengod M.E. The Escherichia coli trmE (mnmE) gene, involved in tRNA modification, codes for an evolutionarily conserved GTPase with unusual biochemical properties. EMBO J. 1999;18:7063–7076. doi: 10.1093/emboj/18.24.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yim L., Martínez-Vicente M., Villarroya M., Aguado C., Knecht E., Armengod M.E. The GTPase activity and C-terminal cysteine of the Escherichia coli MnmE protein are essential for its tRNA modifying function. J. Biol. Chem. 2003;278:28378–28387. doi: 10.1074/jbc.M301381200. [DOI] [PubMed] [Google Scholar]
- 9.Scrima A., Vetter I.R., Armengod M.E., Wittinghofer A. The structure of the TrmE GTP-binding protein and its implications for tRNA modification. EMBO J. 2005;24:23–33. doi: 10.1038/sj.emboj.7600507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martínez-Vicente M., Yim L., Villarroya M., Mellado M., Pérez-Payá E., Björk G.R., Armengod M.E. Effects of mutagenesis in the switch I region and conserved arginines of Escherichia coli MnmE protein, a GTPase involved in tRNA modification. J. Biol. Chem. 2005;280:30660–30670. doi: 10.1074/jbc.M503223200. [DOI] [PubMed] [Google Scholar]
- 11.Kambampati R., Lauhon C.T. MnmA and IscS are required for in vitro 2-thiouridine biosynthesis in Escherichia coli. Biochemistry. 2003;42:1109–1117. doi: 10.1021/bi026536+. [DOI] [PubMed] [Google Scholar]
- 12.Bujnicki J.M., Oudjama Y., Roovers M., Owczarek S., Caillet J., Droogmans L. Identification of a bifunctional enzyme MnmC involved in the biosynthesis of a hypermodified uridine in the wobble position of tRNA. RNA. 2004;10:1236–1242. doi: 10.1261/rna.7470904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeuchi I., Shigi N., Kato J., Nishimura A., Suzuki T. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol. Cell. 2006;21:97–108. doi: 10.1016/j.molcel.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Wolfe M.D., Ahmed F., Lacourciere G.M., Lauhon C.T., Stadtman T.C., Larson T.J. Functional diversity of the rhodanese homology domain: the Escherichia coli ybbB gene encodes a selenophosphate-dependent tRNA 2-selenouridine synthase. J. Biol. Chem. 2004;279:1801–1809. doi: 10.1074/jbc.M310442200. [DOI] [PubMed] [Google Scholar]
- 15.Chen P., Crain P.F., Näsvall S.J., Pomerantz S.C., Björk G.R. A ‘gain of function’ mutation in a protein mediates production of novel modified nucleosides. EMBO J. 2005;24:1842–1851. doi: 10.1038/sj.emboj.7600666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Björk G.R., Hagervall T.G. Transfer tRNA modification. In: Curtiss R. III, Böck A., Ingrahan J.L., Kaper J.B., Maloy S., Neidhardt F.C., Riley M.M., Squires C.L., Wanner B.L., editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington DC: ASM Press; 2005. [Online] http://www.ecosal.org. [Google Scholar]
- 17.Krüger M.K., Sørensen M.A. Aminoacylation of hypomodified tRNAGlu in vivo. J. Mol. Biol. 1998;284:609–620. doi: 10.1006/jmbi.1998.2197. [DOI] [PubMed] [Google Scholar]
- 18.Curran J.M. Modified nucleosides in translation. In: Grosjean H., Benne R., editors. Modification and Editing of RNA. Washington, D.C.: American Society for Microbiology; 1998. pp. 493–516. [Google Scholar]
- 19.Urbonavičius J., Quian Q., Durand J.M.B., Hagervall T.G., Björk G.R. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yarian C., Townsend H., Czestkowski W., Sochacka E., Malkiewicz A.J., Guenther R., Miskiewicz A., Agris P.F. Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem. 2002;277:16391–16395. doi: 10.1074/jbc.M200253200. [DOI] [PubMed] [Google Scholar]
- 21.Kinscherf T.G., Willis D.K. Global regulation by gidA in Pseudomonas syringae. J. Bacteriol. 2002;184:2281–2286. doi: 10.1128/JB.184.8.2281-2286.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sha J., Kozlova E.V., Fadl A.A., Olano J.P., Houston C.W., Peterson J.W., Chopra A.K. Molecular characterization of a glucose-inhibited division gene, gidA, that regulates cytotoxic enterotoxin of Aeromonas hydrophila. Infect. Immun. 2004;72:1084–1095. doi: 10.1128/IAI.72.2.1084-1095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alam K.Y., Clark D.P. Molecular cloning and sequence of the thdF gene, which is involved in thiophene and furan oxidation by Escherichia coli. J. Bacteriol. 1991;173:6018–6024. doi: 10.1128/jb.173.19.6018-6024.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong S., Ma Z., Foster J.W. The Era-like GTPase TrmE conditionally activates gadE and glutamate-dependent acid resistance in Escherichia coli. Mol. Microbiol. 2004;54:948–961. doi: 10.1111/j.1365-2958.2004.04312.x. [DOI] [PubMed] [Google Scholar]
- 25.Nakayashiki T., Inokuchi H. Novel temperature-sensitive mutants of Escherichia coli that are unable to grow in the absence of wild-type tRNA6Leu. J. Bacteriol. 1998;180:2931–2935. doi: 10.1128/jb.180.11.2931-2935.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forsyth R.A., Haselbeck R.J., Ohlsen K.L., Yamamoto R.T., Xu H., Trawick J.D., Wall D., Wang L., Brown-Driver V., Froelich J.M., et al. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 2002;43:1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- 27.Karita M., Etterbeek M.L., Forsyth M.H., Tummuru M.K.R., Blaser M.J. Characterization of Helicobacter pylori dapE and construction of a conditionally lethal dapE mutant. Infect. Immun. 1997;65:4158–4164. doi: 10.1128/iai.65.10.4158-4164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Decoster E., Vassal A., Faye G. MSS1, a nuclear-encoded mitochondrial GTPase involved in the expression of COX1 subunit of cytochrome c oxidase. J. Mol. Biol. 1993;232:79–88. doi: 10.1006/jmbi.1993.1371. [DOI] [PubMed] [Google Scholar]
- 29.Colby G., Wu M., Tzagoloff A. MTO1 codes for a mitochondrial protein required for respiration in paromomycin-resistant mutants of Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:27945–27952. doi: 10.1074/jbc.273.43.27945. [DOI] [PubMed] [Google Scholar]
- 30.Li X., Guan M.-X. A human mitochondrial GTP binding protein related to tRNA modification may modulate phenotypic expression of the deafness-associated mitochondrial 12S rRNA mutation. Mol. Cell. Biol. 2002;22:7701–7711. doi: 10.1128/MCB.22.21.7701-7711.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X., Li R., Lin X., Guan M.X. Isolation and characterization of the putative nuclear modifier gene MTO1 involved in the pathogenesis of deafness-associated mitochondrial 12 S rRNA A1555G mutation. J. Biol. Chem. 2002;277:27256–27264. doi: 10.1074/jbc.M203267200. [DOI] [PubMed] [Google Scholar]
- 32.Umeda N., Suzuki T., Yukawa M., Ohya Y., Shindo H., Watanabe K., Suzuki T. Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J. Biol. Chem. 2005;280:1613–1624. doi: 10.1074/jbc.M409306200. [DOI] [PubMed] [Google Scholar]
- 33.Miller J.H. A Short Course in Bacterial Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 34.Ujita S., Kimura K. Glucose-6-phosphate dehydrogenase, vegetative and spore Bacillus subtilis. Meth. Enzymol. 1982;89:258–261. doi: 10.1016/s0076-6879(82)89046-0. [DOI] [PubMed] [Google Scholar]
- 35.Light D.R., Walsh C., Marletta M.A. Analytical and preparative high-performance liquid chromatography separation of flavin and flavin analog coenzymes. Anal. Biochem. 1980;109:87–93. doi: 10.1016/0003-2697(80)90014-7. [DOI] [PubMed] [Google Scholar]
- 36.Dym O., Eisenberg D. Sequence-structure analysis of FAD-containing proteins. Protein Sci. 2001;10:1712–1728. doi: 10.1110/ps.12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yokoyama S., Nishimura S. Modified nucleosides and codon recognition. In: Söll D., RajBhandary U.L., editors. tRNA: Structure, Biosynthesis, and Function. Washington, D.C.: ASM Press; 1995. pp. 207–223. [Google Scholar]
- 38.Agris P.F. Decoding the genome: a modified view. Nucleic Acids Res. 2004;32:223–238. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki T., Suzuki T., Wada T., Saigo K., Watanabe K. Taurine as a constituent of mitochondrial tRNAs: new insights into the functions of taurine and human mitochondrial diseases. EMBO J. 2002;21:6581–6589. doi: 10.1093/emboj/cdf656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCloskey J.A., Graham D.E., Zhou S., Crain P.F., Ibba M., Konisky J., Söll D., Olsen G.J. Post-transcriptional modification in archaeal tRNAs: identities and phylogenetic relations of nucleotides from mesophilic and hyperthermophilic Methanococcales. Nucleic Acids Res. 2001;29:4699–4706. doi: 10.1093/nar/29.22.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gehrke C.W., Kuo K.C. Ribonucleoside analysis by reversed-phase high-performance liquid chromatography. J. Chromatogr. 1989;471:3–36. doi: 10.1016/s0021-9673(00)94152-9. [DOI] [PubMed] [Google Scholar]
- 42.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 43.Bachmann B. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F.C., et al., editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, D.C.: American Society for Microbiology; 1996. pp. 2460–2488. [Google Scholar]
- 44.Guzmán L.M., Belin D., Carson M.J., Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goloubinoff P., Gatenby A.A., Lorimer G.H. GroE heat-shock proteins promote assembly of foreign prokaryotic ribulose bisphosphate carboxylase oligomers in Escherichia coli. Nature. 1989;337:44–47. doi: 10.1038/337044a0. [DOI] [PubMed] [Google Scholar]
- 46.Furste J.P., Pansegrau W., Frank R., Blocker H., Scholz P., Bagdasarian M., Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]