Abstract
We identified a human orthologue of tRNA:m5C methyltransferase from Saccharomyces cerevisiae, which has been previously shown to catalyse the specific modification of C34 in the intron-containing yeast . Using transcripts of intron-less and intron-containing human genes as substrates, we have shown that m5C34 is introduced only in the intron-containing tRNA precursors when the substrates were incubated in the HeLa extract. m5C34 formation depends on the nucleotide sequence surrounding the wobble cytidine and on the structure of the prolongated anticodon stem. Expression of the human Trm4 (hTrm4) cDNA in yeast partially complements the lack of the endogenous Trm4p enzyme. The yeast extract prepared from the strain deprived of the endogenous TRM4 gene and transformed with hTrm4 cDNA exhibits the same activity and substrate specificity toward human pre-tRNALeu transcripts as the HeLa extract. The hTrm4 MTase has a much narrower specificity against the yeast substrates than its yeast orthologue: human enzyme is not able to form m5C at positions 48 and 49 of human and yeast tRNA precursors. To our knowledge, this is the first report showing intron-dependent methylation of human and identification of human gene encoding tRNA methylase responsible for this reaction.
INTRODUCTION
Maturation of eukaryotic cytoplasmic tRNAs is a multistage process that includes the processing of 5′ and 3′ ends, intron splicing in the case of intron-containing pre-tRNAs, transport from the nucleus to the cytoplasm and numerous nucleoside modifications that take place both in the nucleus and in the cytoplasm. Three classes of intron-containing tRNA genes are present in the human nuclear genome: (8 genes with intron lengths ranging from 16 to 21 bp), (5 genes with intron lengths ranging from 22 to 25 bp) and (1 gene containing a 15 bp intron) (Genomic tRNA database, http://lowelab.ucsc.edu/GtRNAdb). In all cases, introns are located one nucleotide downstream from the anticodon, which is a typical feature of nuclear intron-containing tRNA genes.
In the case of genes, it has been shown that the introduction of pseudouridine in the middle position of the anticodon (Ψ35) is intron dependent in yeast, plants, animals and humans (1–3). Moreover, it has been demonstrated that the introduction of Ψ35 strictly depends on the nucleotide sequence surrounding the U35 to be modified and depends rather on the length of the intron than on its structure (4). Transfer genes contain introns in yeast and vertebrates (5,6). However, in plants, there are no introns in these genes (7). The first position of the yeast anticodon sequence—cytosine—in is methylated to 5-methylcytosine (m5C)34. In yeast, this methylation is intron dependent and its introduction strictly depends on the intron structure (5).
The first functional evidence of the importance of Ψ35 in tRNATyr and m5C34 in tRNALeu came from the Abelson laboratory (1,5). The construction of mutant yeast tRNATyr and tRNALeu genes without introns resulted in the production of mature tRNA molecules without appropriate modified bases. When the suppressor activity of a tRNATyr product derived from intron-containing tRNA (SUP 6 coding for ochre suppressor tRNA) with the ochre suppressor tRNA transcribed from the gene without intron was compared, it turned out that tRNATyr derived from the mutated gene without intron exhibited a strong reduction in the suppressor activity. In an analogous experiment, a mutant, intron-less yeast tRNALeu gene encoding amber suppressor tRNA (SUP 53) also gave a product with a weak suppressor activity when compared with the intron-containing amber tRNALeu suppressor gene product. In both cases, the decreased suppressor activity correlated with the absence of Ψ35 and m5C34, respectively. Similarly, Ψ35 is required in the anticodon (GΨA) of cytoplasmic tRNATyr from Nicotiana rustica for UAG and UAA suppression in the translation of the tobacco mosaic virus (TMV) RNA (8). Summarizing the afore-mentioned experiments, it is clear that both Ψ35 and m5C34 are necessary to stabilize anticodon–codon pairing leading to the correct decoding of mRNA. In this paper, we show that intron in the human is indispensable for the C5-methylation of cytosine in the first position of the anticodon. We also show that the modification of C34 depends on the nucleotide sequence surrounding the position to be modified and on the structure of intron-containing prolongated anticodon stem.
Enzymes that introduce Ψ35 and m5C34 in the intron-containing pre-tRNATyr and pre-tRNALeu, respectively, have been identified in yeast (9,10). However, our knowledge about the corresponding enzymes in multicellular eukaryotes remains obscure. The first human m5C methylase Dnmt2 has been reported very recently and is responsible for the modification of C38 in tRNAAsp in mice, Drosophila melanogaster and Arabidopsis thaliana. This MTase was found to be localized in cytoplasm and acts at the level of a mature tRNA molecule (11). Here, we present the characterization of a human gene encoding a methyltransferase (MTase) that is involved in the formation of m5C34 in . The product of the gene, hTrm4, acts at the level of the intron-containing tRNA precursor and is localized in the nucleoplasm and nucleolus. During the preparation of this paper, it was found that this MTase is a novel downstream Myc-target which mediates Myc-induced cell proliferation and growth. Therefore, the characterization of hTrm4 substrate specificity can be essential since this MTase is a potential target for cancer therapies (12).
MATERIALS AND METHODS
Enzymes and reagents
The restriction enzymes, T4 DNA polymerase and T4 DNA ligase were from Roche, Fermentas and Promega. RNase T2 and SAM (S-adenosyl-l-methionine) were obtained from Sigma. [α-32P]NTPs were from Amersham/Pharmacia Biotech. Taq polymerase, T7 RNA polymerase, Calf Intestine Alkaline Phosphatase (CIAP), dNTPs and NTPs were from Fermentas. P1 nuclease was obtained from Roche. Marathon-Ready cDNA library was obtained from the Clontech, TAKARA BIO company. DNA sequencing was carried out using fmol DNA Cycle Sequencing System from Promega and CEQ DTCS Quick Start Kit from Beckman Coulter. Oligonucleotides for amplification and mutagenesis were synthesized by oligo.pl. Cellulose plates for TLC were purchased from Merck. Kits for plasmid isolation and for DNA isolation from agarose were obtained from A&A Biotechnology. The QuickChange Site-Directed Mutagenesis Kit was from Stratagene. All other reagents were from Amersham/Pharmacia Biotech, Fermentas and Sigma.
Bacterial, yeast and HeLa strains and plasmids
Escherichia coli TG1, CJ236 and JM109 were used as hosts for the propagation of pUC19 plasmid and M13mp19 phage derivatives, respectively. The recombinant plasmids used in this study were: pHLIVS2 carrying the human gene and pYLIVS carrying the yeast gene, respectively. Both tRNA genes were inserted under T7 RNA polymerase promoter and contained the MvaI restriction site to linearize plasmids and terminate the in vitro transcription. Human HeLa cells were used for nuclear cell-free extract preparations. Saccharomyces cerevisiae haploid strain BY4742 (wild type) and its deletion derivative with the open reading frame YBL024w (corresponding to the TRM4 gene) replaced by kanMX4, a kanamycin resistance gene mediating G418 resistance, were obtained from EUROSCARF. Yeast strains were used for transformation and tRNA:m5C methyltransferase activity assays. pYES2 (Invitrogen) vector was utilized for protein expression in yeast.
Construction of tRNALeu gene mutants
The recombinant plasmid pHLIVS2 harbouring human tRNALeu gene that contains the 22 bp intron was mutagenized using the Kunkel method (13) or QuickChange Site-Directed Mutagenesis Kit according to the manufacturer's instructions. The introduced mutations were confirmed by DNA sequencing. The mini-helix consisting of the prolongated anticodon stem of gene was synthesized according to the Milligan's method (14) with modifications described in ref. (15) using two DNA oligonucleotides: one (58 nt long) including the 17 nt long sequence of T7 RNA polymerase promoter followed by the coding sequence of prolongated anticodon stem containing 22 nt long intron and the other with the sequence complementary to the T7 RNA promoter.
T7 RNA transcription of gene and its mutants
Plasmids carrying the yeast tRNALeu gene and the human gene or its mutants (with the exception of mutant pHLIVS2 C35) were linearized by digestion with MvaI before transcription. Plasmid pHLIVS2 C35 contains a mutation that introduces a new MvaI restriction site into the tRNALeu mutant gene and was linearized with BamHI. In vitro transcription was carried out in Fermentas T7 RNA polymerase buffer, 500 μM each of CTP, GTP, UTP, 12 μM ATP, 50 μCi of [α-32P]ATP, 1 μg of linearized plasmid and 20 U of T7 RNA polymerase in total volume of 20 μl at 37°C for 1.5 h. Radiolabelled ATP was added in the case of the wild-type gene and all the mutant transcription reactions with the exception of pHLIVS2 C35, pHLIVS2 G35 and pHLIVS2 C35 where [α-32P]CTP, [α-32P]GTP or [α-32P]UTP were added, respectively. For the visualization of m5C localized at position 48/49 in tRNAs plasmids carrying the yeast , and the human genes were transcribed using [α-32P]GTP. Transcripts were purified on 10% denaturating polyacrylamide gels, identified by autoradiography, excised from the gels and eluted with 0.3 M sodium acetate (pH 5.2), 0.5 M EDTA, 0.1% (w/w) SDS. After phenol/chloroform extraction and ethanol precipitation, the RNA was resuspended in water. The average amount of the recovered labelled tRNA transcripts varied between 1 and 2 pmol per reaction. The transcription of mini-helix was performed as described in (14,15).
In vitro 5-methylcytosine formation assays in HeLa and yeast extract
A nuclear cell-free extract was prepared from HeLa cells according to Dignam et al. (16). Yeast cells used for protein extract preparations were grown as follows: single yeast colony was picked from YPD agar plate and inoculated into 15 ml SD-U synthetic medium containing 0.67% nitrogen base, 2% glucose, amino acids and no uracil. The culture was grown overnight at 30°C with shaking. The OD600 of the overnight culture was determined and the amount of culture necessary to obtain an OD600 of 0.4 in 50 ml of induction medium was calculated and taken for further experiments. Cells were pelleted at 1500 g for 5 min at 4°C, resuspended in 2 ml of induction medium (SD-U medium containing 2% galactose instead of 2% glucose), inoculated into 50 ml of the induction medium and grown overnight at 30°C with shaking. Yeast S-10 extract was prepared by the following procedure: 5 ml of cell suspension were centrifuged, collected, washed twice in 500 μl of distilled water and resuspended in twice their volume of lysis buffer [50 mM Tris–HCl, pH 7.5, 100 mM KCl, 0.1 mM EDTA, 10 mM MgCl2, 10% glycerol, 2% Protease Inhibitor Cocktail for yeast (Sigma)]. One volume of glass beads was added. The mixture was frozen in liquid nitrogen and thawed at room temperature. The suspension was vigorously vortexed 5–7 times for 1 min with 5 min pause between each vortexing when the suspension was kept on ice. The resulting homogenate was centrifuged at 12 000 g for 20 min to remove cell debris. The resulting supernatant was aliquoted, quickly frozen in liquid nitrogen and kept frozen at −70°C for later use.
Quantification of m5C34 in
About 3000 c.p.m. of 32P-labelled RNAs, obtained as described above, were digested with 0.5 U of RNase T2 for 5 h at 37°C in 10 μl of 5 mM ammonium acetate, pH 4.6. The labelled nucleotides were identified by a two-dimensional (2D) chromatographic analysis on cellulose TLC plates as described by Grosjean et al. (17). The efficiency of m5C34 formation was measured by cutting out the labelled spots from the TLC plates and counting the radioactivity by the liquid scintillation technique. Otherwise, radioactivity in the spots was evaluated by ImageQuant 5.1 software after exposure to a PhosphorImager screen (Typhoon, Molecular Dynamics, USA) and scanning. Taking into account the amount of label in each of the nucleotide spots (AMP, UMP, GMP, CMP and/or m5CMP) and knowing the relative number of labelled nucleotides per RNA substrate, the number of moles of m5C per mole of pre-tRNA was easily calculated.
Cloning of yeast and human TRM4 cDNA
Yeast Trm4 protein was used as a query in searching for sequences of homologous proteins in the human expressed sequence tags (ESTs) database using tBLASTn (18). For the localization of the gene in human genome and the determination of the chromosomal localization, BLAT software was used.
hTrm4 cDNA was amplified from the human cDNA library using following primers: 5′-ATGGTGGTCAACCATGATGCCTCCAGCATAC-3′ and 5′-TCACCGGGGTGGATGGACCCCCGCCGG-3′. PCR was performed in two step ‘touchdown’ manner, with the following conditions: 30 s at 96°C, followed by 25 cycles of 5 s at 96°C and 4 min at 72°C. The PCR product of expected size was excised from the agarose gel and eluted. The cDNA product was re-amplified with primers harbouring sequences for XbaI and EcoRI restriction enzymes and cloned into the pUC-19 vector (giving hTRM4/pUC construct) and sequenced. Yeast genomic DNA was isolated using QIAGEN Genomic DNA kit according to manufacturer's instructions. Primers used for the amplification of yeast Trm4p cDNA were: 5′-AAAGGATCCACCATGGCTAGAAGAAAGAATTTCAA-3′ (Kozak translation initiation sequence is marked in italics, start codon is underlined and the BamHI restriction site is in boldface) and 5′-AAAGAATTCTCAATTAGCAGCGCTAGGAG-3′ (the stop codon is underlined and EcoRI restriction sequence is in boldface). After amplification, a product of expected size was excised and eluted from the EtBr/agarose gel. Yeast and human TRM4 cDNA were cloned into pYES2 yeast expression vector (constructs TRM4/pYES2 and hTRM4/pYES2, respectively). After each cloning procedure, the correctness of insert-vector ligation was assessed by sequencing or restriction digestion.
Yeast cells transformation
Yeast strain ΔYBL024w was made competent by lithium acetate/PEG 3350 treatment according to procedures described in the User Manual for pYES2 vector system (Invitrogen). Competent cells were then transformed by purified pYES2 plasmid or its derivative constructs carrying the sequence corresponding to the yeast TRM4 gene or human TRM4 cDNA, respectively. Transformed yeast cells were grown on a synthetic selective medium as required to maintain plasmids.
Preparation of low-molecular weight RNA from S.cerevisiae
Yeast RNA was extracted as described in (19). RNA products were purified on 8% denaturing polyacrylamide gel. After EtBr staining, bands corresponding to approximate tRNA size (80–110 nt) were eluted from the gel, ethanol precipitated, carefully washed with ethanol/water (70:30), resuspended in DEPC-treated water and kept frozen at −70°C for later use.
HPLC chromatography
Low-molecular RNA (10–20 μg) was digested by P1 nuclease according to the manufacturer's manual (Roche). Nucleotides were dephosphorylated by CIAP phosphatase. The volume of the reaction was reduced to 10 μl by vacuum centrifugation. HPLC separation was performed on the reversed phase SUPELCOSIL™ LC-318 HPLC Column, 25 cm × 4.6 mm (Supelco) attached to ÄKTAexplorer 10S (Amersham Pharmacia Biotech). Chromatographic solvents were as follows: (A) 100 mM ammonium acetate and (B) 100% acetonitrile. HPLC conditions were: 4°C, 1 ml/min flow rate and absorbance measuring at λ = 277 nm. The gradient method consisted of 2 CV (Column Volume) solvent A, a gradient >10 CV to 6% solvent B, held 2 CV solvent B and column re-equilibration. To determine retention times of standards, 10 μl mix of six ribonucleosides (pseudouridine, cytidine, uridine, 5-methylcytidine, guanosine and adenosine), each at the concentration of 1 mM, was separated.
RESULTS
m5C34 in is formed only in intron-containing precursors
Green et al. (20) isolated five genes that slightly differ in intron sequence. Inspection of the Genomic tRNA database revealed that there are five human genes containing introns with nucleotide sequences identical or nearly identical to those published by Green et al (20) and Karwowska and Szweykowska-Kulinska (6).
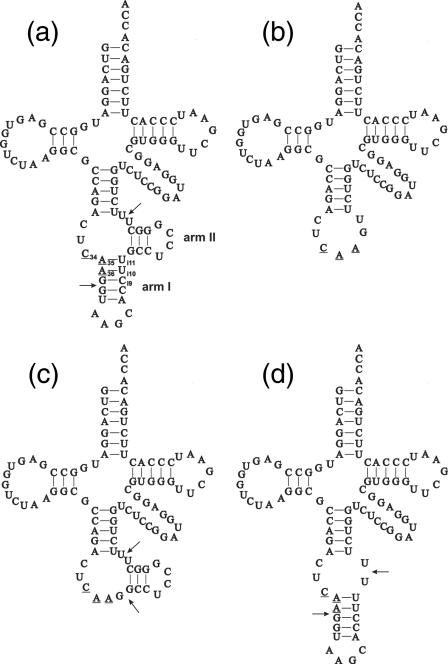
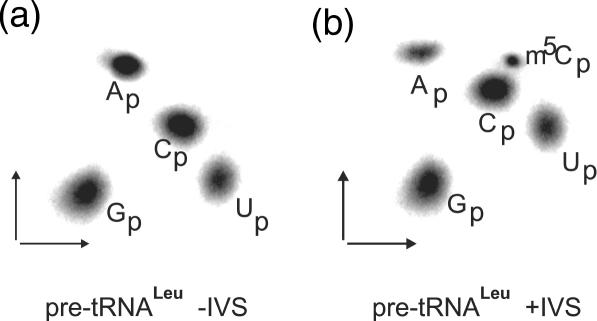
The only human sequence with the identified modified nucleosides is known from HeLa cells (21). Its nucleotide sequence corresponds exactly to the nucleotide sequence of the mature domain of the isolated human genes from HeLa cells with the exception of one additional G nucleotide in the tRNALeu gene extra arm coding sequence (6). However, Harada et al. (21) was not able to identify the nature of modified cytidine in position 34. Storbel and Abelson (5) have shown that this modification in the yeast tRNA corresponds to m5C and that its formation is intron-dependent. Using PCR, we have previously isolated two intron-containing genes from human blood (6). One of the isolated human genes was studied further. It contains 22 bp intron. The putative structure of pre-tRNALeu derived from this gene is shown in Figure 1a. The structure of the prolongated anticodon stem containing intron is proposed according to the model of yeast intron-containing (5). The putative model of the extended anticodon stem shows that (i) the second (A35) and third (A36) anticodon nucleotides and G37 can be base-paired with the complementary nucleotides of the intron; (ii) the majority of the intron can be arranged in two hairpin-stem and loop structures (arm I and arm II, see Figure 1); (iii) the 5′ splice site junction is located at the boundary of the stem–loop of the first arm in the prolongated anticodon stem; (iv) the 3′ splice site junction is located in the unpaired region. To test whether C34 in human is modified to m5C and whether its introduction depends on the presence of 22 nt long intron, the tRNALeu gene with the intron and a mutant of the tRNALeu gene without (Figure 1b) were transcribed in vitro using T7 RNA polymerase and [5′-α32P]ATP. Radiolabelled transcripts were then incubated in the presence of nuclear, cell-free HeLa extract which contained the activity necessary for m5C synthesis. The modified transcripts were gel-purified and analysed for their m5C content after a complete hydrolysis of RNA with RNase T2. Since RNase T2 produces nucleoside-3′-monophosphates, the labelled phosphate is exclusively derived from the nearest adenosine. This ensures that only five cytidine nucleotides will be labelled in the tRNALeu without intron and six in the intron-containing precursor (Figure 1), since they have 3′-flanking adenosines. The only m5C that was found in the sequenced human was C48, but this nucleoside is accompanied at the 3′ side by guanosine and thus C48 is not labelled (21). Nucleotides were resolved in a thin-layer two-dimensional chromatographic system that separates modified nucleotides from their unmodified counterparts. Figure 2 shows the autoradiograms of T2 hydrolysates derived from intron-less and intron-containing pre-tRNALeu. Only pre-tRNA containing intron gives rise to 3′-m5CMP (Figure 2a and b). It shows that the only cytosine that can be methylated in the intron-containing pre-tRNALeu is C34. Thus, the first cytosine of the anticodon of human is methylated in intron-dependent manner.
Figure 1.
Cloverleaf structure of precursor containing intron (a), mature tRNA (b) and two mutants with partially deleted introns (derived from plasmids pHLIVS2 ΔI1, pHLIVS2 ΔI2) (c and d). Anticodon sequence is underlined, arrows point to splice sites.
Figure 2.
Analysis of m5C34 content in intron-less (a) and intron-containing (b) human by two-dimensional thin layer chromatography. Pre-tRNAs were incubated in the cell-free nuclear HeLa extract. tRNALeu precursors were synthesized and treated as described in Materials and Methods.
The structure of a prolongated anticodon stem and nucleotide sequence that surrounds the position to be modified are crucial for m5C34 formation in
To learn more about the sequence and structure specificity of m5C34 methylation in tRNALeu precursor, a number of mutants were generated where wild-type nucleotides occupying positions 32, 33, 35, 36 and 37 were exchanged for other nucleotides. Moreover, two mutants were constructed with partially deleted intron sequences (Figure 1c and d) and four mutants in which either the base-pairing in the prolongated anticodon stem was disrupted or base pairs were reversed. All mutant pre-tRNAs listed in Table 1 were examined for their capacity to be substrates for m5C34 formation in HeLa cell-free nuclear extract.
Table 1.
Efficiency of C34 to m5C34 conversion in HeLa extract
| Mutant name | Mutation site | Relative amount of m5C34 (%) |
|---|---|---|
| pHLIVS2 | wt | 100 |
| Mini-helix | pre-tRNALeu anticodon stem-containing intron | 0 (undetectable) |
| pHLIVS2 Δ I | Intron deleted | 0 |
| pHLIVS2 Δ I 1 | Intron arm I deleted | 0 |
| pHLIVS2 ΔI 2 | Intron arm II deleted | 0 |
| pHLIVS2 A32 | C32 → A32 | 85 |
| pHLIVS2 G32 | C32 → G32 | 0 |
| pHLIVS2 U32 | C32 → U32 | 85 |
| pHLIVS2 A33 | U33 → A33 | 85 |
| pHLIVS2 C33 | U33 → C33 | 10 |
| pHLIVS2 G33 | U33 → G33 | 10 |
| pHLIVS2 C35 | A35 → C35 | 0 |
| pHLIVS2 G35 | A35 → G35 | 0 |
| pHLIVS2 U35 | A35 → U35 | 0 |
| pHLIVS2 C36 | A36 → C36 | 0 |
| pHLIVS2 G36 | A36 → G36 | 0 |
| pHLIVS2 U36 | A36 → U36 | 0 |
| pHLIVS2 A37 | G37 → A37 | 0 |
| pHLIVS2 C37 | G37 → C37 | 0 |
| pHLIVS2 U35A11i | A35 → U35, U11i → A11i | 0 |
| pHLIVS2 U37 | G37 → U37 | 0 |
| pHLIVS2 G36Ci10 | A36 → G36, Ui10 → Ci10 | 85 |
| pHLIVS2 C37Gi9 | G37 → C37, Ci9 → Gi9 | 0 |
| pHLIVS2 Gi9Ai10Ai11 | Ci9 → Gi9, Ui10 → Ai10, Ui11 → Ai11 | 0 |
32P-labelled pre-tRNAsLeu were incubated in cell-free nuclear extract for 20 min at 30°C. The intron-containing pre-tRNALeu and its mutants were analysed for the presence of m5C34 after T2 digestion by thin layer chromatography as described in Materials and Methods. The efficiency of m5C34 formation was calculated as described in Materials and Methods.
A mini-helix consisting of prolongated anticodon stem was not methylated at the C34 position. The removal of the intron arm I or II resulted in the complete abolishment of C34 methylation. Thus, m5C34 formation depends on the structure of the intron-containing prolongated anticodon stem. The exchange of C32 to A or U had no major effect on the efficiency of m5C34 formation (85% of the wild-type efficiency). However, the exchange of C32 to G resulted in the inhibition of m5C34 methylation. The transversion of U33 to A did not significantly influence the formation of m5C34. However, the transversion of U33 to G and transition to C strongly reduced the level of m5C34 formation. All mutations in positions 35, 36 and 37 resulted in m5C34-deficient pre-tRNAsLeu. From these experiments, a consensus sequence for m5C34 methylation in pre-tRNALeu was established: C/A/U32-U/A33-C34-A35-A36-G37. Careful inspection of the prolongated anticodon stem secondary structure reveals that A35, A36 and G37 can base pair with intron nucleotides Ui11Ui10 and Ci9, respectively. To learn more about the sequence and structure dependence of m5C34 methylation within the prolongated anticodon stem, additional mutations were introduced in which potential base pairs were reversed or base-pairing was disrupted. The reversion of base pair A35–Ui11 to U35–Ai11 did not promote m5C34 formation. The reversion of base pair A36–Ui10 to G36–Ci10 resulted in 85% efficiency of m5C34 formation, showing that no A36 per se is indispensable for modification but N36–Ni10 base-pairing within the moiety of the prolongated anticodon stem is crucial. However, like in the case of the base pair A35–Ui11, the reversion of G37–Ci9 to C37–Gi9 resulted in a complete abolishment of m5C34 formation. In this case, not the base-pairing but the identity of guanosine in position 37 (and adenosine in the position 35), and possibly their pairing with Ci9 and Ui10, respectively, is crucial for the modification to occur. Disruption of all three base pairs A36–Ui11, A37–Ui10, and G37–Ci9 by the introduction of noncomplementary nucleotides within the intron sequence, i.e. Ai11, Ai10 and Gi9, leads to the inhibition of m5C34 formation. Thus, both the sequence and structure of the prolongated anticodon stem are crucial for the introduction of methyl group on C34 of pre-tRNALeu.
NSUN2 encodes yeast TRM4 orthologue
The BLAST similarity search of human UniGene database at NCBI (22) using the yeast Trm4p (enzyme that introduces m5C into yeast in an intron-dependent manner) as a query enabled us to identify EST sequences of a putative human orthologue. The putative protein sequence encoded by the contig sequence, constructed from identified ESTs using the CAP3 program (23), shows 35% identity to the yeast sequence. A comparison of the contig sequence with the human genome revealed only one gene with an identical putative coding sequence (NCBI accession no. NM_017755), located on the human chromosome 5, with precise position 5p15.31. This gene is called NSUN according to HUGO Gene Nomenclature Committee and contains 19 exons, and encompasses ∼34 000 nt. Based on the results of the analyses reported in this article, we propose to rename it as hTRM4. Bioinformatic analysis of amino acid sequence of putative m5C34 MTases and the alignment of regions forming the active site in yeast and human Trm4 enzymes were already published by Bujnicki et al. (24). Sequence analysis using the programs predictNLS (25), SignalP (26) and PSORT II (http://psort.hgc.jp) revealed 60–70% probability of nuclear localization of hTrm4. However, we did not find any classical nuclear localization signals (NLS) within the protein sequence.
Using the cDNA library from HeLa cells, we isolated a 2304 bp cDNA encoding hTrm4 protein. Its nucleotide sequence was confirmed to be identical to the virtually constructed cDNA contig from EST libraries and to exon sequences of the identified human gene.
Human Trm4 catalyses the formation of m5C34 in human
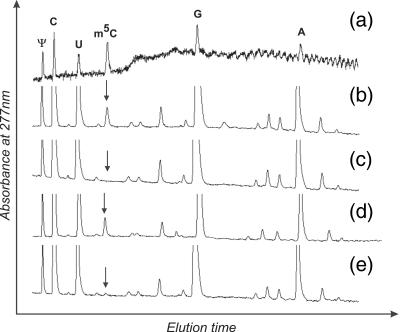
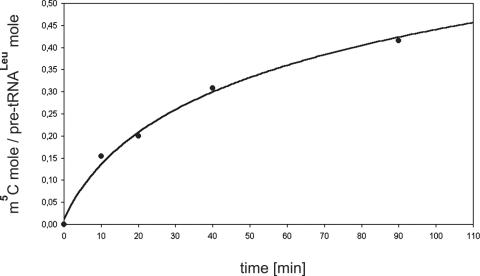
Since our efforts to overexpress the hTrm4 protein in different prokaryotic expression systems were unsuccessful (data not shown), we decided to investigate hTrm4 activity in yeast strain BY4742 with the deleted endogenous TRM4 gene. The deletion of this gene has no phenotypic effect on cell growth (27). Δtrm4 yeast strain BY4742 was transformed with hTrm4 cDNA that was cloned into yeast pYES2 plasmid. The same yeast strain was also transformed with pYES2 plasmid containing yeast Trm4p cDNA. Low-molecular weight RNA was isolated from both yeast lines, digested with P1 nuclease and digestion products were dephosphorylated. Nucleosides were separated on HPLC chromatography using RPC18 column. Figure 3 presents the chromatographic elution profiles of nucleosides from yeast lines: wild type, Δtrm4, Δtrm4 transformed with plasmid carrying hTrm4 cDNA and Δtrm4 transformed with plasmid carrying yeast Trm4p cDNA. In the elution profile of nucleosides obtained from low-molecular weight RNA from Δtrm4 + yeast Trm4p, the level of m5C was restored to that achieved for m5C for wild-type yeast strain. In the Δtrm4 yeast strain, m5C nucleosides are undetectable. In the case of Δtrm4 + hTrm4 cDNA, we observed only a low level of m5C. However, this small peak appeared repeatedly in all consecutive experiments. The low level of m5C methylation may be due to several (mutually not exclusive) reasons: (i) hTrm4 protein recognizes a small group of substrates that are normally methylated by the yeast Trm4p enzyme, (ii) the expressed hTrm4 enzyme has only residual activity, e.g. because of non-native folding or post-translational modification or (iii) hTrm4 localization in yeast is not fully correct. To test whether hTrm4 exhibits activity toward human containing intron, we prepared total protein extracts from Δtrm4, Δtrm4 + hTrm4 cDNA, Δtrm4 + Trm4p cDNA and wild-type yeast lines, and conducted activity tests similar to those in the HeLa cell-free nuclear extract. Figure 4 shows the m5C34 analyses of human intron-less and intron-containing in the wild-type yeast strain (Figure 4a), Δtrm4 (Figure 4b), Δtrm4 + hTrm4 cDNA (Figure 4c) and Δtrm4 + Trm4p cDNA yeast lines (Figure 4d). In the case of the wild-type yeast strain, human pre-tRNALeu containing intron is a substrate for endogenous MTase while pre-tRNALeu without intron is not. When the protein extract from Δtrm4 yeast line was used, there was no m5C in intron-containing and intron-less pre-tRNAs. However, when we used the protein extract prepared from Δtrm4 + Trm4p cDNA, human intron-containing pre-tRNALeu was methylated at C34, demonstrating that in the wild-type yeast strain TRM4 MTase was responsible for m5C34 formation. In the case of Δtrm4 + hTrm4 cDNA, human was methylated when its precursor contained an intron. Finally, yeast containing intron is methylated at C34 in the S-10 yeast extract prepared from the Δtrm4 + hTrm4 strain (Figure 4e). Figure 5 shows the kinetics of m5C34 formation in human intron-containing in the extract prepared from the Δtrm4 + hTrm4 cDNA yeast line. The efficiency of m5C34 formation in yeast pre-tRNALeu in the same yeast line is about the same (∼45%, data not shown). Several mutants of the human were tested in yeast extracts containing the human Trm4 enzyme. In all cases, we obtained similar results as in the case of HeLa cell-free nuclear extract (data not shown). These biochemical experiments definitely prove that the human gene identified by us encodes a functional counterpart of the yeast Trm4p enzyme.
Figure 3.
Detection of m5C in bulk low-molecular weight RNA extracted from yeast wild-type strain BY4742 (b), BY4742 ΔTRM4 (c), BY4742 transformed with pYES2 containing yeast Trm4p cDNA (d) and BY4742 ΔTRM4 transformed with pYES2 containing hTrm4 cDNA (e). Separation of a test nucleoside mixture (Ψ, U, C, m5C, G and A) is presented in (a). The total RNA fraction was completely hydrolysed by nuclease P1 and the resulting nucleotides dephosphorylated by CIAP. Analysis of nucleosides was performed as described in Materials and Methods.
Figure 4.
Analysis of m5C34 content in human without intron and containing intron by 2-D thin layer chromatography. Pre-tRNAs were incubated in yeast S-10 extract derived from wild-type strain (a), BY4742 ΔTRM4 (b), BY4742 ΔTRM4 transformed with pYES2-containing hTrm4 cDNA (c) and BY4742 ΔTRM4 transformed with pYES2-containing yeast Trm4p cDNA (d). (e) Incubation of yeast containing intron in S-10 extract prepared from BY4742 ΔTRM4 transformed with pYES2-containing hTrm4 cDNA. tRNALeu precursors were synthesized and treated as described in Materials and Methods.
Figure 5.
Kinetics of m5C34 formation in human intron-containing . 32P-labelled pre-tRNALeu transcripts were incubated with S-10 yeast extract prepared from BY4742 Δtrm4 + hTrm4 cDNA at 30°C for 10, 20, 40 and 90 min and analyzed for the relative amount of m5C residues at position 34. The measurements were taken as described in Materials and Methods.
Human Trm4 has narrower substrate specificity than its yeast orthologue
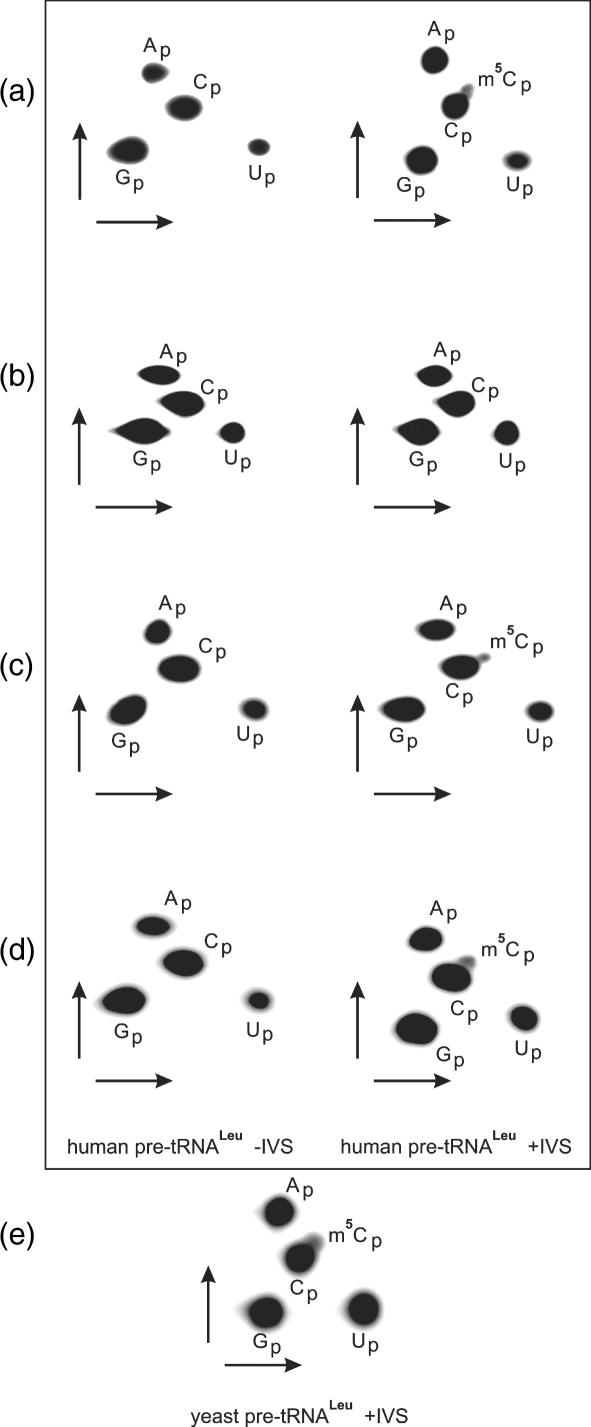
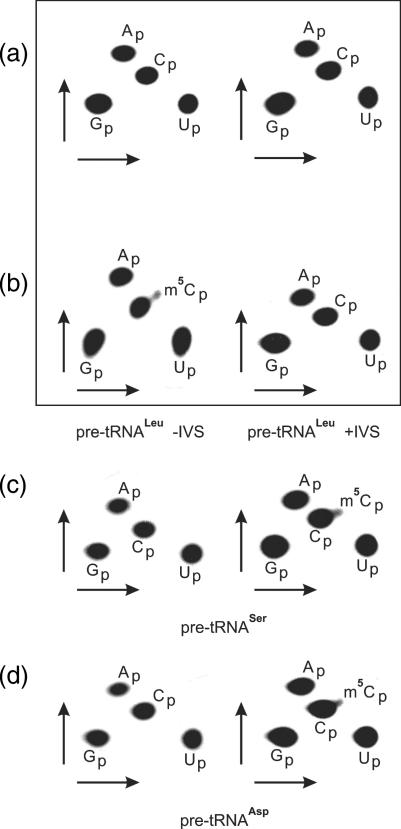
Apart from methylating C34 in yeast pre-tRNALeu containing intron, yeast Trm4p is required for C48 and C49 methylation in different yeast tRNAs. Using total protein extracts from yeast lines described in the preceding chapter, we examined hTrm4 ability to methylate cytosines at positions 48/49. hTrm4 is not able to introduce a methyl group at position C48 of human tRNALeu, both at the level of intron-containing and intron-less precursors (Figure 6a), while the yeast Trm4p methylates human intron-less precursor and does not recognize intron-containing substrate (Figure 6b). Additionally, we analysed m5C48 and m5C49 formation in yeast and , respectively. We show that neither of them is methylated at C48 or C49 by hTrm4 and both are substrates for yeast Trm4p (Figure 6c and d). Control experiments carried out in the Δtrm4 strain show no detection of m5C48/49 formation in any tRNA tested (data not shown). Thus, human Trm4 substrate specificity is narrower than in the case of its yeast orthologue.
Figure 6.
Analysis of m5C48 (a–c) and m5C49 (d) content in human and yeast tRNAs using hTrm4 and yeast Trm4p enzymes by 2D TLC. Pre-tRNAs were labelled using [α-32P] GTP during T7 RNA polymerase transcription, gel purified and incubated in S-10 extract. (a) BY4742 Δtrm4 + hTrm4 yeast strain and human pre-tRNALeu, (b) BY4742 Δtrm4 + yeast Trm4p and human pre-tRNALeu, (c) BY4742 Δtrm4 + hTrm4 (left) and BY4742 Δtrm4 + Trm4p (right) and yeast , and (d) BY4742 Δtrm4 + hTrm4 (left), BY4742 Δtrm4 + Trm4p (right) and yeast . Yeast and genes do not contain introns.
DISCUSSION
In this article, we describe the identification, cloning and enzyme activity of the corresponding gene product encoding human Trm4 enzyme. This is the first report showing intron-dependent methylation of human and identification of human gene encoding tRNA methylase responsible for this reaction. The hTRM4 gene was identified on the basis of its amino acid sequence similarity to a known Trm4p MTase from yeast. Our studies provide experimental validation of the phylogenetic analysis of RNA:m5C MTases by Bujnicki et al. who predicted the product of the NSUN2 gene as the closest human homologue of yeast Trm4p (24). hTRM4 is a single-copy gene, located on the human chromosome 5, with the precise position 5p15.31. The hTrm4 protein is a member of an evolutionarily conserved family, which includes genuine and putative RNA:m5C MTases from all three domains of life, such as the human nucleolar proliferation-associated antigen p120 (NOL1) and its homologues. Wu et al. (27) reported that the yeast Trm4p was found to be localized in the nucleus, including the nucleolus, and was concentrated at the nuclear periphery. Our experiments have shown that its human homologue hTrm4 is always localized in the nucleoplasm and in nucleoli of HeLa cells, and in several cases we also observed a concentration of hTrm4 at the nuclear periphery (data not shown). These data have been recently confirmed by Frye and Watt (12).
The presence of an intron is required for the m5C34 formation in human as demonstrated in this paper by the maturation of intron-less and intron-containing tRNA precursor in HeLa cell-free nuclear extract. In earlier studies, Harada et al. (21) showed that this position in the mature human is occupied by a hypermodified derivative. The authors suggest that this is a 2′-O-methylated and further hypermodified cytidine derivative (21). Very similar results were obtained by Randerath et al. (28) when leucine tRNAs were isolated and sequenced from rat tumour, Morris hepatoma 5123D cells. Our studies show that the first stage of C34 modification process in HeLa cells is the m5C34 formation. However, the final chemical structure of this modification remains to be characterized. In addition to intron-dependent m5C34 formation, we have shown that the activity of hTrm4 strictly depends on a specific sequence surrounding the cytosine to be modified and on the intron structure. Similar results were obtained studying the yeast Trm4p enzyme by Storbel and Abelson (5). However, our experiments have shown that minisubstrate composed of the anticodon stem–loop extended by the intron was not methylated at C34. In contrast, Motorin and Grosjean (10) presented data that analogous minihelix of yeast was an efficient substrate for tRNA:m5C-methyltransferase. These results imply that there are differences in the ability of enzyme–substrate recognition between the human and fungi Trm4 enzymes. The nucleotide sequence that surrounds C34 in human tRNA precursor is more restrictive for m5C34 formation than in the case of yeast. The substitution of adenosine from the middle position of the anticodon to uridine did not promote m5C34 formation in human tRNA precursor while in the case of yeast amber leucine-inserting pre-tRNA containing intron (anticodon CUA), m5C34 was formed. Additionally, our experiments have shown that the reversion of base pair A35–Ui11 to U35–Ai11 also resulted in the m5C34 deficient . Moreover, human MTase responsible for the introduction of methyl group at the anticodon wooble cytosine (hTrm4) requires a consensus nucleotide sequence C/A/U32-U/A33-C34-A35-A36-G37 as a prerequisite for m5C34 formation. Thus, all nucleotides from the anticodon loop that are present upstream from the intron sequence are essential for pre-tRNALeu modification. These results again strengthen the conclusion about the differences in the ability of enzyme–substrate recognition between human and yeast Trm4 enzymes.
Motorin and Grosjean showed (10) that the yeast Trm4p enzyme catalysed m5C formation at four distinct sites in different tRNAs: 34, 40, 48 and 49. m5C was found at positions 34 and 40 only in two yeast tRNAs [ (Sup53) and , respectively], whereas most elongator tRNAs bore either m5C48 or m5C49, but never both in the same tRNA molecule. Motorin and Grosjean concluded that yeast Trm4p MTase is responsible for cytosine conversion in all yeast tRNAs and pre-tRNAs, while two other identified MTases encoded in S.cerevisiae genome are required for proper rRNA m5C modification. We transformed yeast strain BY4742 deprived of the TRM4 gene with yeast Trm4p cDNA and human Trm4 cDNA. In the Δtrm4 + Trm4p cDNA strain, the level of m5C in low-molecular RNA molecules was restored to that of the wild-type yeast strain (see Figure 3). However, in the Δtrm4 + hTrm4 cDNA strain, the level of m5C was hardly detectable. It clearly shows that human enzyme is not able to recognize the majority of yeast tRNA substrates. Since our experiments show that yeast and human Trm4 MTases both recognize mammalian containing intron, the conclusion can be drawn that the yeast enzyme has much broader substrate specificity than the human Trm4. We confirm this statement showing that hTrm4 is not able to introduce m5C48 at the level of intron-containing and intron-less human pre-tRNALeu precursor. Moreover, we show that it does not methylate different yeast tRNA precursors at positions 48 and 49. In the light of these observations, it is interesting to note that Bujnicki et al. (24) identified two additional human putative orthologues of yeast Trm4p, which have probably originated from duplications and subsequent functional specializations of the original TRM4 gene in the animal lineage. One or both of these so-far uncharacterized genes may be responsible for some of the methylations carried out by the yeast Trm4p, but not exhibited by its human counterpart. This speculation can explain a very low level of m5C in the Δtrm4 + hTrm4 strain. Summarizing our results concerning the hTrm4 enzyme, it can be concluded that the enzyme acts on intron-containing precursors, it is more sensitive to the nucleotide sequence that surrounds the position to be modified than its yeast homologue Trm4p and it is equally sensitive to the anticodon prolongated, intron-containing structure. Moreover, its sensitivity to substrate specificity is more constrained than in the case of yeast homologue—Trm4p.
Acknowledgments
This work was supported by Badania Statutowe grant of the Faculty of Biology at Adam Mickiewicz University and by the Polish Committee for Scientific Research project no. 6 P04A 02721. We would like to thank Dr Urszula Karwowska for her technical support in mutant genes construction and for carrying out activity tests in HeLa cell extracts, and Dr Janusz Bujnicki for carefully reading the manuscript. Funding to pay the Open Access publication charges for this article was provided by Badania Statutowe of the Department of Gene Expression and Biology Faculty of Adam Mickiewicz University, Poznań.
Conflict of interest statement. None declared.
REFERENCES
- 1.Johnson P.F., Abelson J. The yeast tRNATyr gene intron is essential for correct modification of its tRNA product. Nature. 1983;302:681–687. doi: 10.1038/302681a0. [DOI] [PubMed] [Google Scholar]
- 2.Pienkowska J., Wrzesinski J., Szweykowska-Kulinska Z. A cell-free yellow lupin extract containing activities of pseudouridine 35 and 55 synthases. Acta Biochim. Polon. 1998;45:745–754. [PubMed] [Google Scholar]
- 3.Van Tol H., Beier H. All human tRNATyr genes contain introns as a prerequisite for pseudouridine biosynthesis in the anticodon. Nucleic Acids Res. 1988;16:1951–1966. doi: 10.1093/nar/16.5.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szweykowska-Kulińska Z., Beier H. Sequence and structure requirements for the biosynthesis of pseudouridine (Ψ35) in plant pre-tRNATyr. EMBO J. 1992;11:1907–1912. doi: 10.1002/j.1460-2075.1992.tb05243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Storbel M.C., Abelson J. Effect of intron mutations on processing and function of Saccharomyces cervisiae SUP53 tRNA in vitro and in vivo. Mol. Cell Biol. 1986;6:2663–2673. doi: 10.1128/mcb.6.7.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karwowska U., Szweykowska-Kulinska Z. New intron-containing human tRNALeu genes. Acta Bichom. Polon. 1997;4:791–794. [PubMed] [Google Scholar]
- 7.Fiedorow P., Szweykowska-Kulinska Z. Intergenic sequences of clustered tRNA genes: new type of genetic marker for phylogenetic studies, with application to the taxonomy of liverworts. Plant Mol. Biol. 1998;38:1257–1261. doi: 10.1023/a:1006084711003. [DOI] [PubMed] [Google Scholar]
- 8.Zerfass K., Beier H. Pseudouridine in the anticodon GΨA of plant cytoplasmic tRNA(Tyr) is required for UAG and UAA suppression in the TMV-specific context. Nucleic Acids Res. 1992;20:5911–5918. doi: 10.1093/nar/20.22.5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behm-Ansmant I., Urban A., Ma X., Yu Y-T., Motorin Y., Branlant C. The Saccharomyces cerevisiae U2 snRNA: pseudouridine-synthase Pus7p is a novel multisite-multisubstrate RNA:Ψ-synthase also acting on tRNAs. RNA. 2003;9:1371–1382. doi: 10.1261/rna.5520403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Motorin Y., Grosjean H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA. 1999;5:1105–1118. doi: 10.1017/s1355838299982201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goll M.G., Kirpekar F., Maggert K.A., Yoder J.A., Hsieh C.-L., Zhang X., Golic K.G., Jacobsen S.E., Bestor T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 12.Frye M., Watt F.M. The RNA methyltransferase Misu (Nsun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr. Biol. 2006;16:971–981. doi: 10.1016/j.cub.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 13.Kunkel T.A., Roberts J.D., Zakour R.A. Rapid and efficient site-specific mutagenesis without phenotypic effect. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 14.Milligan J.F., Uhlenbeck O. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 15.Szweykowska-Kulinska Z., Krajewski J., Wypijewski K. Mutations of Arabidopsis thaliana pre-tRNATyr affecting pseudouridylation of U35. Biochim. Biophys. Acta. 1995;1264:87–92. doi: 10.1016/0167-4781(95)00129-5. [DOI] [PubMed] [Google Scholar]
- 16.Dignam J.D., Lebowitz R.M., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grosjean H., Droogmans L., Giege R., Uhlenbeck O.C. Guanosine modifications in runoff transcripts of synthetic transfer RNA-Phe genes microinjected into Xenopus oocytes. Biochim Biophys Acta. 1990;1050:267–273. doi: 10.1016/0167-4781(90)90179-6. [DOI] [PubMed] [Google Scholar]
- 18.Altschul S. F, Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Schmitt M.E., Brown T.A., Trumpower B.L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green C.J., Sohel I., Vold B.S. The discovery of new intron-containing human tRNA genes using the polymerase chain reaction. J. Biol. Chem. 1990;265:12139–12142. [PubMed] [Google Scholar]
- 21.Harada F., Matsubara M., Kato N. Stable tRNA precursors in HeLa cells. Nucleic Acids Res. 1984;12:9263–9269. doi: 10.1093/nar/12.24.9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34(Database issue):D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang X., Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bujnicki J.M., Feder M., Ayres C.L., Redman K.L. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. 2004;32:2453–2463. doi: 10.1093/nar/gkh564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cokol M., Nair R., Rost B. Finding nuclear localization signals. EMBO Rep. 2000;1:411–415. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bendtsen J.D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 27.Wu P., Brockenbrough J.S., Paddy M.R., Aris J.P. NCL1, a novel gene for non-essential nuclear protein in Saccharomyces cerevisiae. Gene. 1998;220:109–117. doi: 10.1016/s0378-1119(98)00330-8. [DOI] [PubMed] [Google Scholar]
- 28.Randerath E., Gupta R.C., Morris H.P., Randerath K. Isolation and sequence analysis of two major leucine transfer ribonucleic acids (anticodon Mm-A-A) from a rat tumor, Morris hepatoma 5123D. Biochemistry. 1980;19:3476–3483. doi: 10.1021/bi00556a011. [DOI] [PubMed] [Google Scholar]