Abstract
This paper shows that the protein-primed DNA polymerases encoded by bacteriophages Nf and GA-1, unlike other DNA polymerases, do not require unwinding or processivity factors for efficient synthesis of full-length terminal protein (TP)-DNA. Analysis of their polymerization activity shows that both DNA polymerases base their replication efficiency on a high processivity and on the capacity to couple polymerization to strand displacement. Both enzymes are endowed with a proofreading activity that acts coordinately with the polymerization one to edit polymerization errors. Additionally, Nf double-stranded DNA binding protein (DBP) greatly stimulated the in vitro formation of the TP-dAMP initiation complex by decreasing the Km value for dATP of the Nf DNA polymerase by >20-fold. Whereas Nf DNA polymerase, as the φ29 enzyme, is able to use its homologous TP as well as DNA as primer, GA-1 DNA polymerase appears to have evolved to use its corresponding TP as the only primer of DNA synthesis. Such exceptional behaviour is discussed in the light of the recently solved structure of the DNA polymerase/TP complex of the related bacteriophage φ29.
INTRODUCTION
The inability of DNA polymerases to start de novo DNA synthesis imposes in most organisms the necessity of an RNA molecule to provide the 3′-OH group needed to initiate DNA elongation. This requirement creates a dilemma for the replication of the ends of linear genomes since, once the last RNA primer for the lagging strand synthesis is removed, a portion of ssDNA at the end of the genome will remain uncopied. In order to avoid the continuous shortening of the linear genomes in subsequent replication rounds, several mechanisms have evolved, most of them making use of the presence of repetitive sequences at the ends of the chromosomes that allow to create long concatemers, to circularize, or to form hairpin loops to fill the incomplete 5′ ends. In higher eukaryotes, telomerase prevents chromosome ends shortening by elongating the 3′-OH group of the ssDNA end using as template its own RNA (1).
Several phages, animal viruses as adenovirus and hepadnaviruses, mitochondrial plasmids, and linear chromosomes and plasmids of Streptomyces have solved such a quandary by using a protein as primer, called terminal protein (TP). The OH group of a specific serine, threonine or tyrosine of TP is used by the replicative DNA polymerase to start DNA synthesis from both ends of the linear genome, the TP remaining covalently linked to such 5′ ends (2).
The development of in vitro replication systems with purified proteins, mainly in the case of bacteriophage φ29 and adenovirus, has allowed the elucidation of the general bases of the protein-priming mechanism of DNA replication (2–4). Specific initiation proteins interact with the replication origins at both 5′ ends of the genome, partially opening the double helix, exposing a region of ssDNA. The complex formed by a free TP and the replicative DNA polymerase interacts with the replication origins at both ends of the genome by specific recognition of the parental TP and DNA sequences. DNA polymerase catalyses the incorporation of a specific dNMP onto the priming OH group of the TP, in a reaction directed by an internal dNMP in the template strand (initiation reaction). The initiation complex thus formed slides-back (in the case of bacteriophages φ29, GA-1, PRD1 and Cp1) or jumps-backs (as in adenovirus) to recover the terminal nucleotides, by virtue of the presence of repetitive sequences at the replication origins (5–9). Finally, the same DNA polymerase catalyses chain elongation via a strand displacement mechanism to fulfil TP-DNA replication (2).
In addition, the protein-priming mechanism of replication of the linear genomes solves the requirement of a functionally asymmetric replisome, as the placement of the two replication origins at both ends of the duplex DNA allows both strands to be replicated continuously (10) by two molecules of DNA polymerase in a processive fashion and coupled to strand displacement.
Bacteriophage φ29 DNA polymerase is the only member of the protein-priming subgroup of DNA polymerases whose structure has been crystallographically solved, giving the insights into the structural basis that confer both processivity and strand displacement capacities to the enzyme (11). The main structural difference with respect to other family B DNA polymerases is the presence of two new subdomains corresponding to the two sequence insertions specifically present in the protein-priming DNA polymerases subgroup, called Terminal Protein Region 1 and 2 (TPR1 and TPR2) (12,13). The specific TPR2 insertion, together with the exonuclease, thumb and palm subdomains, forms two tunnels capable of interacting with DNA. The major one would encircle upstream duplex DNA, conferring the DNA binding stability required to replicate processively. In addition, this tunnel also surrounds the priming domain of TP during the first phases of TP-DNA replication, confirming that both TP and DNA occupy, in a sequential manner, the same binding cleft (14). The narrow dimensions of the minor tunnel would preclude the passage of dsDNA through it, enclosing exclusively the downstream template and forcing the unwinding of both strands before the template enters such a tunnel (11). These hypotheses have recently been demonstrated by biochemical characterization of a φ29 DNA polymerase mutant lacking the TPR2 insertion (15).
Bacteriophages Nf and GA-1 belong to the group of phages that infect Bacillus. This group has been subclassified into three serological classes (2). The first class includes phages φ29, PZA, φ15 and BS32; the second one comprises phages B103, Nf and M2Y; and the third one contains phage GA-1 as the only member. As in the case of φ29, these phages possess a double-stranded linear DNA with a TP covalently linked at both 5′ ends (TP-DNA) that is replicated by a protein-priming mechanism. As in φ29, the product of bacteriophages Nf and GA-1 gene 2 is the replicative DNA polymerase. Nf DNA polymerase contains 572 amino acids (66.4 kDa), showing 81.8% of sequence identity with respect to φ29 DNA polymerase (91.3% similarity) (16). GA-1 DNA polymerase is a polypeptide of 578 amino acids (67.1 kDa) which shares 54% of sequence identity and 67.3% of similarity when compared with φ29 DNA polymerase (6,17).
In this work, we describe the catalytic properties of the Nf and GA-1 DNA polymerases responsible for efficient and accurate synthesis of full-length TP-DNA. In addition, we present data showing GA-1 DNA polymerase as the first example of a protein-primed DNA polymerase whose structure is specifically adapted to use exclusively its corresponding TP as primer of polymerization.
MATERIALS AND METHODS
Nucleotides and DNAs
Unlabelled nucleotides, as well as [α-32P]dATP [3000 Ci/mmol (1 Ci = 37 GBq)] and [γ-32P]ATP (3000 Ci/mmol) were obtained from Amersham Pharmacia. The 5′-p-nitrophenyl ester of thymidine monophosphate (pNP-TMP) was from Sigma. Oligonucleotides sp1 (5′-GATCACAGTGAGTAC), sp1p (5′-GATCACAGTGAGTAG), and sp1c+6 (5′-TCTATTGTACTCACTGTGATC) were supplied by Isogen. Oligonucleotides sp1 and sp1p were 5′-labelled with [γ-32P]ATP and phage T4 polynucleotide kinase and purified electrophoretically on 8 M urea–20% polyacrylamide gels. Both labelled sp1 and sp1p oligonucleotides were hybridized to oligonucleotide sp1c+6 in the presence of 0.2 M NaCl and 50 mM Tris–HCl (pH 7.5), resulting in a primer/template structure. To analyse processive DNA polymerization coupled to strand displacement by Nf and GA-1 DNA polymerases, M13mp18 single-stranded DNA (ssDNA) was hybridized to the universal primer (Isogen) in the presence of 0.2 M NaCl and 60 mM Tris–HCl (pH 7.5). TP-containing Nf and GA-1 DNAs were obtained as described for φ29 TP-DNA (18).
Proteins
Phage T4 polynucleotide kinase was obtained from New England Biolabs. φ29 DNA polymerase was purified from Escherichia coli BL21(DE3) cells harbouring plasmid pJLPM (a derivative of pT7-4w2) as described (19). Nf and GA-1 DNA polymerase genes were cloned and overproduced in E.coli strain BL21(DE3) (20). For this, phages Nf and GA-1 were obtained from infected Bacillus subtilis cells and purified in a cesium chloride density gradient. Phage DNA was isolated by proteinase K treatment (21). Nf and GA-1 DNA polymerase genes were amplified by PCR and digested with EcoRI and BamHI (Nf) and HindIII and BamHI (GA-1) prior to cloning in an EcoRI–BamHI digested pT7-3 (Nf) and HindIII–BamHI digested pT7-4 (GA-1) expression vectors, under the control of the T7 RNA polymerase-specific φ10 promoter (22). E.coli BL21 (DE3) cells were transformed and the cloned genes were sequenced entirely. Cells containing the DNA polymerase genes were grown overnight at 37°C (Nf) and 25°C (GA-1) in LB medium, in the presence of 100 mg/l ampicillin. Under these conditions, overexpressed Nf and GA-1 DNA polymerases were soluble. Further DNA polymerase purification steps were carried out essentially as described (16,19). DNA polymerases purity was estimated to be >90% by SDS–PAGE followed by Coomassie blue staining. φ29 TP was purified as described (18). Nf and GA-1 TP genes were cloned and overproduced in E.coli strain BL21(DE3), by previous PCR amplification of the genes from the corresponding TP-DNA and further digestion with EcoRI and BamHI. After cloning both genes into an EcoRI–BamHI digested pT7-3 vector, E.coli BL21 (DE3) cells were transformed, and the cloned genes were entirely sequenced. Cells containing the TP genes were grown overnight at 28°C (Nf) and 22°C (GA-1) in LB medium, in the presence of 100 mg/l ampicillin. Under these conditions, overexpressed Nf and GA-1 TPs were soluble. Further TP purification steps were carried out essentially as described (18). The protein was >95% homogeneous as estimated by SDS–PAGE and Coomassie blue staining. Nf DBP was purified from B.subtilis strain 110NA, infected with phage Nf, as described (23).
3′–5′ exonuclease assays
3′–5′ Exonuclease activity on ssDNA
The assay was performed essentially as described (24) in the presence of 1 ng of either Nf or GA-1 DNA polymerase and 0.075 ng of 5′-labelled sp1 oligonucleotide ssDNA substrate.
Hydrolysis of pNP-TMP
The incubation mixture contained, in 300 μl, 50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1 mM DTT, 1 mM MnCl2, 3 mM pNP-TMP (dissolved in 50 mM Tris–HCl, pH 8.0 and 150 mM NaCl) and 33 μg of either GA-1 or Nf DNA polymerase. Hydrolysis was studied by monitoring p-nitrophenol production at 420 nm with a Hitachi U-2000 spectrophotometer at 25°C, as described (25). Linearity in the production of p-nitrophenol was obtained in the 5–350 s time range. Slopes obtained by linear regression adjustments of those points allowed us to calculate the catalytic efficiency for the hydrolysis of the phosphoester bond (s−1).
3′–5′ Exonuclease assay on matched and mismatched primer-terminus
The reaction mixture was the same as described above for the use of ssDNA as substrate, but using 0.18 ng of either the hybrid molecule sp1/sp1c+6 (matched) or splp/splc+6 (mismatched), and 20 ng of either Nf or GA-1 DNA polymerase. Samples were incubated at 25°C for 2 min (Nf DNA polymerase) and 5 min (GA-1 DNA polymerase) and quenched by adding EDTA up to a final concentration of 10 mM. Each reaction was analysed as described when ssDNA was used as substrate of the 3′–5′ exonuclease.
DNA gel retardation assay
The interaction of both Nf and GA-1 DNA polymerases with a primer/template structure was assayed using the 5′-labelled sp1/sp1c+6 (15/21mer) DNA. The incubation mixture contained, in a final volume of 20 μl, 12 mM Tris–HCl (pH 7.5), 1 mM EDTA, 20 mM ammonium sulphate, 0.1 mg/ml BSA, 10 mM MgCl2, 0.18 ng of sp1/sp1c+6 and the indicated amounts of DNA polymerases. Binding to ssDNA was assayed under the same conditions described above, in the absence of MgCl2, using 0.075 ng of sp1 oligonucleotide. After incubation for 5 min at 4°C, the samples were processed and analysed as described (26,27).
Polymerase/3′–5′ exonuclease (pol/exo) coupled assay
The DNA molecule sp1/sp1c+6 (15mer/21mer) contains a 6 nt 5′-protruding end that can be used as substrate for the exonuclease activity (dsDNA) and also for DNA-dependent DNA polymerization. The assay was performed as described (28) in the presence of 0.18 ng of 5′-labelled 15/21mer, 25 ng of either Nf or GA-1 DNA polymerase and the indicated increasing concentrations of the four dNTPs. After incubation for 5 min at 25°C, the reaction was stopped and samples were analysed as described (28). Polymerization or 3′–5′-exonucleolysis was detected as an increase or decrease, respectively, in the size (15mer) of the 5′-labelled primer.
TP-primed initiation assay
The capacity to carry out the initiation step of TP-DNA replication was analysed as described (29), in the presence of 5 ng of the homologous TP, the specified amount of either φ29, Nf or GA-1 DNA polymerase, 1 mM (for GA-1 and φ29 DNA polymerases) and 2 mM (for Nf DNA polymerase) MnCl2, either 20 mM (for φ29 and GA-1 DNA polymerases) or 40 mM (Nf DNA polymerase) ammonium sulphate, 0.5 μg of the corresponding TP-DNA and 0.1 μM [α-32P]dATP (1 μCi). After incubation for the indicated time at 30°C, samples were processed and analysed as previously described (18).
Processivity and strand displacement assays
Replication of primed M13 DNA
The incubation mixture contained, in 25 μl, 50 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 40 μM each dCTP, dGTP, dTTP and [α-32P]dATP (1 μCi), 250 ng of primed M13mp18 ssDNA and 100 ng of DNA polymerase. After incubation for the indicated times at 30°C, the reaction was stopped by adding 10 mM EDTA and 0.1% SDS, and the samples were filtered through Sephadex G-50 spin columns. To determine the DNA elongation rate, samples were taken at different reaction times and the DNA replication products were analysed by alkaline agarose gel electrophoresis (30) in the presence of size markers. The average size of the newly synthesized DNA was estimated after densitometric scanning of the autoradiograms. To determine processivity during DNA synthesis, the same replication assays were performed using the indicated polymerase dilutions. After incubation for 15 min at 30°C, the reactions were stopped and processed as indicated above.
Replication assay (protein-primed initiation plus elongation) with TP-DNA as template
The assay was carried out as described (24) in the presence of 10 mM MgCl2, either 20 mM (in the case of GA-1 DNA polymerase) or 40 mM (Nf DNA polymerase) ammonium sulfate, 20 μM each of the four dNTPs, 0.5 μg of the corresponding TP-DNA, and either 10 ng of both Nf TP and DNA polymerase or 5 ng of GA-1 TP and DNA polymerase. Nf TP-DNA replication was assayed in the absence or presence of 10 μg of Nf DBP, as indicated. After incubation for the indicated times at 30°C, samples were processed and the replication rate was analysed as described above for the M13 DNA replication assay. To determine processivity during TP-DNA synthesis the same replication assays were performed using the indicated polymerase dilutions. After incubation for 10 min at 30°C, the reactions were stopped and processed as indicated above. For analysis of the transition products generated during the first steps of Nf TP-DNA replication, the indicated concentration of the corresponding dNTPs were used, as well as the metal activator and the absence or presence of 10 μg of Nf DBP. After incubation for 10 min at 30°C, samples were subjected to a 12% SDS–PAGE gel (360 mm × 280 mm × 0.5 mm) to obtain enough resolution to distinguish TP bound to the first elongation products.
RESULTS
Nf and GA-1 DNA polymerases are provided with a 3′–5′ exonuclease activity
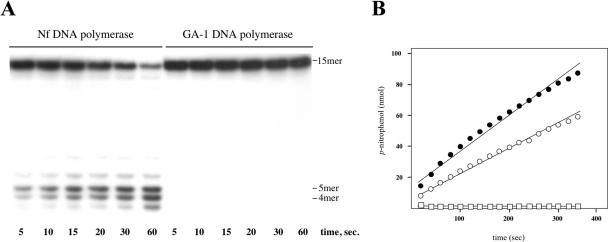
The N-terminal domain of both Nf and GA-1 DNA polymerases contains the residues predicted to be responsible for the proofreading activity (6,16). The 3′–5′ exonuclease activity of both DNA polymerases was evaluated by analyzing their capacity to degrade a 15mer (sp1) single-stranded oligonucleotide (see Materials and Methods), the preferred substrate for this activity (28). As shown in Figure 1A, Nf DNA polymerase was able to degrade the sp1 oligonucleotide to give very short products (3–6mer), without changes in the degradation pattern as time increased. The absence of intermediate degradation products indicated that the 3′–5′ exonuclease activity of Nf DNA polymerase behaved processively, without dissociation of the DNA polymerase/ssDNA complex until the length of the substrate was too short to remain stably bound to the enzyme (4/5mer). Below this size, catalysis was severely decreased and dissociation became dominant, rendering distributive exonuclease activity. The absence of activity displayed by GA-1 DNA polymerase (Figure 1A), even at a 10-fold excess of enzyme (data not shown) could be due to a reduced capacity to bind ssDNA. Indeed, analysis by gel shift assays showed a defective ssDNA binding capacity of the GA-1 DNA polymerase (data not shown).
Figure 1.
3′–5′ Exonucleolytic activity of Nf and GA-1 DNA polymerases. (A) Exonucleolytic activity on ssDNA. The assay was carried out in the conditions described in Materials and Methods, in the presence of 1 ng of the corresponding DNA polymerase and 0.075 ng of 5′ labelled sp1 oligonucleotide. After incubation for the indicated times at 25°C, degradation of labelled DNA was analysed by electrophoresis in 8 M urea–20% polyacrylamide gels and autoradiography. The position of the unit length and degradation products is indicated. (B) Hydrolysis of the 5′-p-nitrophenyl ester of thymidine monophosphate (pNP-TMP). The assay was performed by using 33 μg of either Nf or GA-1 DNA polymerases and 3 mM pNP-TMP as substrate at 25°C. Catalytic efficiency for the hydrolysis of pNP-TMP catalysed by Nf (open circles) and GA-1 (full circles) DNA polymerases was determined spectrophotometrically by monitoring p-nitrophenol production at 420 nm at the indicated times with the further linear regression adjustment of those points. Non-enzymatic hydrolysis of pNP-TMP was also monitorized (open squares).
To rule out defects in the proper folding of the 3′–5′ exonuclease site, the ability of GA-1 DNA polymerase to hydrolyse the 5′-p-nitrophenyl ester of thymidine 5′-monophosphate (pNP-TMP) was analysed, as hydrolysis of this non-canonical nucleoside exclusively relies on the catalytic residues responsible of the exonuclease activity. The rate of hydrolysis of pNP-TMP catalysed by GA-1 and Nf DNA polymerases was determined spectrophotometrically by continuous monitoring of the p-nitrophenol produced. As it can be seen in Figure 1B, both GA-1 and Nf DNA polymerases were able to hydrolyse this substrate, showing a catalytic efficiency of 0.47 and 0.33 s−1, respectively. From these results, it can be concluded that the lack of 3′–5′ exonuclease activity shown by GA-1 DNA polymerase on ssDNA is due to a hindered binding.
3′–5′ Exonuclease and polymerization activities of Nf and GA-1 DNA polymerases are coordinated
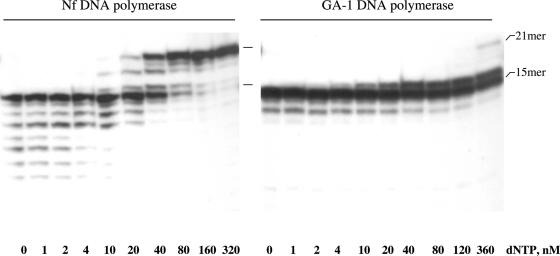
To evaluate whether both the 3′–5′ exonuclease and polymerization activities of Nf and GA-1 DNA polymerases are coordinated, degradation and extension of a fully base-paired primer/template structure (sp1/sp1c+6) was analysed (Pol/Exo assay; see Materials and Methods). As it can be seen from Figure 2, in the absence of dNTPs the only products detected in the case of Nf DNA polymerase are produced by the exonucleolytic activity. Under these conditions, the 3′–5′ exonuclease activity of GA-1 DNA polymerase produced some degradation of the primer (it has to be taken into account that in this assay the amount of polymerase was 5-fold higher and the reaction time 5-fold longer than in the 3′–5′ exonuclease assay on ssDNA), although to a much lesser extent than that of the Nf DNA polymerase, most of the DNA remaining undegraded. By adding increasing amounts of dNTP, exonucleolysis was progressively competed away by the polymerization activity, Nf DNA polymerase requiring 20–40 nM dNTPs to give a net polymerization balance. While full-length products were obtained with the Nf DNA polymerase, products longer than +1 were hardly observed, and only at very high dNTPs concentration, with GA-1 DNA polymerase. In addition, in the latter case, 50% of the initial substrate was not used (see Figure 2). The inability to elongate +1 products could result from dissociation of the GA-1 DNA polymerase/dsDNA complex or from a hindered translocation on this substrate.
Figure 2.
Coordination between the 3′–5′ exonuclease and polymerization activities of Nf and GA-1 DNA polymerases. The assay was carried out as described in Materials and Methods, using 0.18 ng of the 5′-32P-labelled hybrid molecule sp1/sp1c+6 as primer/template DNA, 25 ng of DNA polymerase, and the indicated concentration of the four dNTPs. After incubation for 5 min at 30°C, samples were analysed by 8 M urea–20% PAGE and autoradiography. The 15mer position (non-elongated primer) and 21mer position (elongated primer) are indicated. Polymerization or 3′–5′ exonuclease activities are detected as an increase and decrease, respectively, in the size of the labelled sp1 primer.
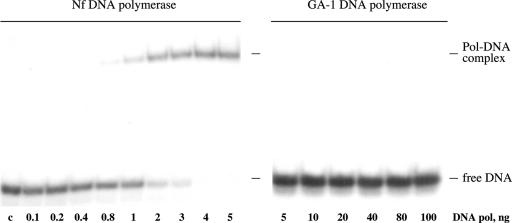
The dsDNA binding capacity of both DNA polymerases was studied by gel-shift assays (see Materials and Methods). As shown in Figure 3, Nf DNA polymerase gave rise to a single retardation band, showing a KD = 1.5 nM (considered as the concentration of DNA polymerase needed to retard 50% of the substrate molecules). Taking into account the preferential binding of a perfectly paired primer-terminus to the polymerization active site, the observed retarded band most likely corresponds to a competent polymerization complex (27). Conversely, GA-1 DNA polymerase was not capable to give either a shifted band or a smear, not even at the highest DNA polymerase concentration assayed (DNA polymerase/dsDNA ratio = 125).
Figure 3.
Gel retardation of dsDNA by Nf and GA-1 DNA polymerases. The assay was carried out as described in Materials and Methods, using 0.18 ng of 5′-labelled hybrid molecule sp1/sp1c+6 as primer/template substrate, in the presence of the indicated amount of DNA polymerase. After incubation for 5 min at 4°C, samples were subjected to gel electrophoresis, and the bands corresponding to free dsDNA and to DNA polymerase/DNA complexes were detected by autoradiography. Lane c corresponds to the mobility of the dsDNA in the absence of DNA polymerase.
To act coordinately with the polymerization activity, one of the basic criteria for an exonuclease to perform proofreading is the preferential elimination of a mismatched primer-terminus (see Materials and Methods), a physiological situation that the DNA polymerase will meet once a nucleotide is misinserted. When mispair specificity was studied by comparing the degradation efficiency of a primer with a mismatched (G:G; sp1p/sp1c+6) or matched (C:G; sp1/sp1c+6) 3′ end (see Materials and Methods), both Nf and GA-1 DNA polymerases showed a clear preference for excision of the mismatched primer/template molecule. Thus, the catalytic efficiencies (Kcat/Km) displayed by Nf DNA polymerase for degrading the matched and mismatched molecules were 0.041 and 0.068 s−1, respectively, being 0.0075 and 0.013 s−1 those obtained by GA-1 DNA polymerase. This fact, together with the dynamic equilibrium observed between the synthetic and degradative activities, allow us to conclude that both DNA polymerases are proofreading enzymes.
Nf DNA polymerase displays the capacity to couple strand displacement to polymerization
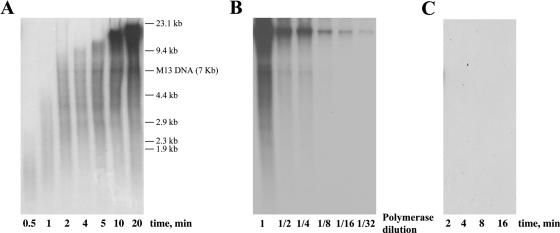
To analyse the capacity of Nf DNA polymerase to couple polymerization to strand displacement, primed M13 DNA rolling circle replication assays were performed (see Materials and Methods). As shown in Figure 4A, alkaline analysis of the DNA synthesized indicates that Nf DNA polymerase replicates M13 DNA in 4 min, proceeding further through strand displacement, displaying a replication rate of 2400 nt/min. In addition, Nf DNA polymerase carried out replication in a processive manner, as the length of the replication products remained invariable upon dilution of the enzyme up to 32-fold (see Figure 4B). As expected from previous results, GA-1 DNA polymerase did not give any detectable activity with M13 DNA (Figure 4C).
Figure 4.
Strand displacement coupled to M13 DNA replication by Nf DNA polymerase. (A) Replication of primed M13 DNA was carried out as described in Materials and Methods, using 100 ng of Nf DNA polymerase in the presence of 40 μM each of the four dNTPs. After incubation at 30°C for the indicated times, length of the synthesized DNA was analysed by alkaline 0.7% agarose gel electrophoresis alongside DNA length markers and autoradiography. M13 ssDNA unit length is also indicated. (B) Processive synthesis by Nf DNA polymerase. The assay was performed as described in Materials and Methods in the presence of 250 ng of primed M13 DNA, 40 μM each of the four dNTPs and the indicated decreasing amounts of Nf DNA polymerase, starting with 100 ng (dilution 1). After incubation at 30°C for 16 min, samples were processed as described above. (C) Absence of M13 DNA replication products by GA-1 DNA polymerase. The assay was carried out under the conditions described in (A).
Protein-primed TP-DNA replication performed by Nf and GA-1 DNA polymerases
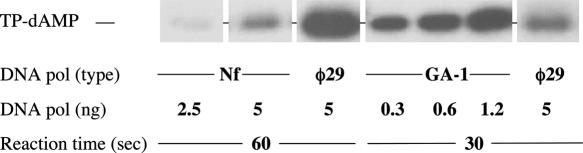
To ascertain whether both Nf and GA-1 DNA polymerases are able to perform in vitro the TP-dAMP initiation reaction, each DNA polymerase was incubated with its homologous TP and TP-DNA (see Materials and Methods), in the presence of dATP. As a control, the well-studied system of phage φ29 was included. When Mg2+ was used as metal activator, GA-1 DNA polymerase gave initiation products with an efficiency 40-fold higher than φ29 DNA polymerase. Contrarily, Nf DNA polymerase was unable to render any detectable product (data not shown). This fact compelled us to use Mn2+ as a catalyst for the reaction, an ion described to greatly stimulate such a reaction in φ29 (31). Under these conditions, both Nf and GA-1 DNA polymerases were able to accomplish the deoxyadenilylation of their homologous TPs (Figure 5), displaying a 20-fold lower and 30-fold higher efficiency than φ29 DNA polymerase, respectively. In addition, both GA-1 and Nf DNA polymerases showed a great specificity for their corresponding TP and TP-DNA, since the heterologous systems did not give any detectable reaction (data not shown), in agreement with previous results that showed a specific recognition of parental TP by DNA polymerase (16).
Figure 5.
Formation of TP-dAMP complex catalysed by Nf and GA-1 DNA polymerases. The assays were performed as described under Materials and Methods in the presence of 1 mM (for GA-1 and φ29 DNA polymerases) and 2 mM (for Nf DNA polymerase) MnCl2, 5 ng of TP, 500 ng of TP-DNA, 0.1 μM [α-32P]dATP (1 μCi), and the indicated amounts of DNA polymerase. After incubation at 30°C for the indicated times, samples were analysed by SDS–PAGE and autoradiography. The position of the TP-dAMP initiation complex is indicated. Quantification was by densitometry of the band corresponding to the labelled TP-dAMP complex, detected by autoradiography.
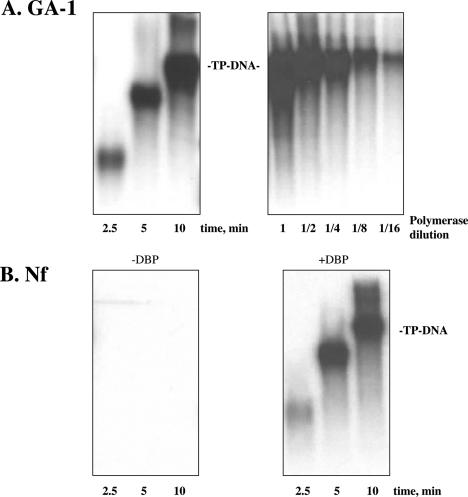
To study the elongation stage of TP-DNA replication, each DNA polymerase was incubated in the presence of TP, TP-DNA, 20 μM dNTPs and Mg2+ as metal activator (see Materials and Methods). By using this minimal replication system, GA-1 DNA polymerase was very efficient in elongating the initiation products (Figure 6A, left panel). The time needed for GA-1 TP-DNA full-length synthesis (21 129 bp) was ∼10 min, rendering a replication rate of 2260 nt/min [close to the 2280 nt/min reported for φ29 TP-DNA replication (10)], and demonstrating an efficient capacity to couple polymerization to strand displacement in a processive fashion (Figure 6A; right panel). Additionally, these results allow us to rule out a misfolding of the polymerization domain of GA-1 DNA polymerase as being responsible for the deficient polymerization activity observed when a template/primer or M13 DNA was used. As expected, and considering the lack of initiation reaction displayed by Nf DNA polymerase in the presence of Mg2+ as metal activator (see above), no replication activity was detected (Figure 6B, left panel), the presence of Nf DBP being essential to allow Nf DNA polymerase to fulfil TP-DNA replication, showing a replication rate of 2260 nt/min (Figure 6B, right panel).
Figure 6.
GA-1 and Nf TP-DNA replication. (A) Replication of GA-1 TP-DNA (left panel). The assay was carried out as described in Materials and Methods in the presence of 5 ng of GA-1 DNA polymerase, 5 ng of GA-1 TP, 500 ng of GA-1 TP-DNA and 20 μM each of the four dNTPs. After incubation for the indicated times at 30°C, the length of the synthesized DNA was analysed by alkaline 0.7% agarose gel electrophoresis. The migration position of unit length GA-1 TP-DNA is indicated. Processive synthesis of GA-1 TP-DNA by the GA-1 DNA polymerase (right panel). The replication assay was performed as described under Materials and Methods in the presence of 10 mM MgCl2, 20 mM ammonium sulfate, 20 μM each of the four dNTPs, 0.5 μg of the corresponding TP-DNA, 10 ng of GA-1 TP and the indicated amounts of GA-1 DNA polymerase, dilution one corresponding to 12.5 ng of DNA polymerase (primer-terminus/DNA polymerase ratio = 1/2.5). After incubation for 10 min at 30°C, the samples were processed as described above. (B) Efficient in vitro Nf TP-DNA replication requires the presence of Nf DBP. The assay was carried out as described in Materials and Methods, in the presence of 10 ng Nf DNA polymerase, 10 ng of Nf TP, 500 ng Nf TP-DNA, 20 μM each the four dNTPs and in the absence (left panel) or presence (right panel) of 10 μg of Nf DBP. After incubation for the indicated times at 30°C, samples were processed as described above.
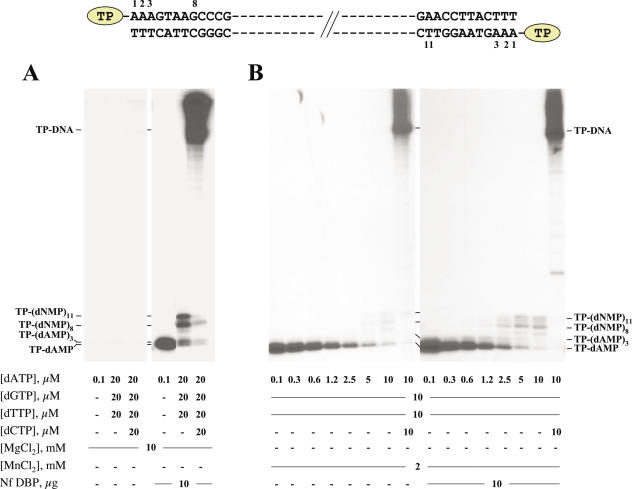
To study the effect of Nf DBP during the transition from initiation to elongation during the first steps of Nf TP-DNA replication, a truncated elongation assay was performed (see Materials and Methods). As can be observed in Figure 7A, when dATP was provided as the only nucleotide, the presence of Nf DBP greatly stimulated the initiation reaction (78-fold) performed by Nf DNA polymerase, giving rise to TP-dAMP and TP-(dAMP)2. The absence of the longer TP-(dAMP)3 product could be explained by its degradation to TP-(dAMP)2 by the 3′–5′ exonuclease activity of the DNA polymerase. Elongation of the initiation products was also very efficient in the presence of dATP, dGTP and dTTP, with the main synthesis of TP-(dNMP)8 and TP-(dNMP)11, the expected sizes according to the sequences of the replication origins (see top of Figure 7), >80% of the initiation products being elongated by the enzyme during the transition step (Figure 7A). In the presence of the four dNTPs, most of the transition products described above were elongated by the DNA polymerase synthesizing full-length DNA. The lack of detectable transition products in the absence of DBP did not allow us to study the extent of the stimulatory effect of DBP in such a replication stage. As mentioned above, Mn2+ is essential to obtain the initiation reaction in the absence of Nf DBP. Under these conditions, and using dATP as the only nucleotide, measurement of the initiation product (TP-dAMP) reflected a 20-fold reduction of the Km for this initiating nucleotide (from 0.6 to 0.03 μM) when Nf DBP was present (data not shown). Using this metal activator, we analysed also the formation of elongation products longer than TP-(dAMP)2 as a function of dATP concentration in the presence of 10 μM of both dGTP and dTTP. Thus, in the absence of DBP, TP-(dAMP)3 product starts to be detectable at 2.5 μM dATP (Figure 7B), while truncated elongation products 8–11 bases long were only observed at 10 μM dATP. In comparison, from the lowest dATP concentration assayed (0.1 μM), DBP promoted the appearance of TP-(dAMP)3 and its elongation to give detectable TP-(dNMP)8–11 molecules from 2.5 μM dATP. Bands corresponding to TP-(dNMP)9–10 most probably are generated by misaddition of one or two nucleotides to the TP-(dNMP)8 product, because of the use of Mn2+ ions. When the percentages of the elongated initiation products were plotted against dATP concentration, a 5-fold stimulatory effect of DBP in the transition stage could be estimated.
Figure 7.
Effect of Nf DBP on in vitro Nf TP-DNA replication. (A) Effect of Nf DBP on the formation of the TP-dAMP initiation complex and limited elongation. The assay was performed as described in Materials and Methods, using 10 mM MgCl2, 0.5 μg of Nf TP-DNA, 10 ng of Nf DNA polymerase, 10 ng of Nf TP and the indicated concentration of the corresponding dNTP. Samples were incubated either in the absence (−) or presence of 10 μg of Nf DBP. After incubation for 10 min at 30°C, the initiation and transition products were detected by high resolution SDS–PAGE. Completely replicated molecules remain at the interphase with the stacking gel. The length of the different partially elongated products and the position corresponding to full-length Nf TP-DNA are indicated on the left. (B) Effect of Nf DBP on the truncated elongation of the TP-dAMP complex. The assay was performed as described in (A), using 2 mM MnCl2 and increasing dATP concentrations in the presence of 10 μM dGTP and dTTP. When indicated, the reaction mixture contained 10 μM dCTP and/or 10 μg of Nf DBP. Samples were processed and analysed as described in (A).
DISCUSSION
Extensive studies performed both in vitro and in vivo, mainly using bacteriophage φ29 and adenovirus, have provided the general insights about the mechanism of protein-primed DNA replication (32,33). Both 5′ ends of the linear genome contain a TP covalently linked that, together with specific DNA sequences, constitute the replication origins. The replicative eukaryotic-type [family B, (34)] DNA polymerase catalyses both, the initial formation of the covalent complex between a free TP molecule and the 5′ terminal nucleotide, and its further elongation coupled to strand displacement. Such coupling can be accounted for by the polymerase itself, as in the case of bacteriophage φ29, or by the assistance of unwinding proteins as it occurs in adenovirus.
Here, we have carried out a biochemical characterization of the main properties of the DNA polymerases encoded by bacteriophages GA-1 and Nf, whose linear genome is also replicated via a protein-priming mechanism (6,35).
DNA polymerase from bacteriophages Nf and GA-1
Based on the high degree of identity between φ29, Nf and GA-1 DNA polymerases, the protein structure homology-modelling server Swiss-Model (36,37) has provided a model for GA-1 and Nf DNA polymerases, obtained by using the recently solved crystallographic structure of φ29 DNA polymerase as template (11). The predicted structures exhibit two well-structured independent domains (see Figure 8A for GA-1 DNA polymerase model). The N-terminal exonuclease domain [that is structurally conserved in the A, B and C families of DNA polymerases (38)] of both Nf and GA-1 DNA polymerases has the three universally conserved motifs Exo I, Exo II and Exo III containing the four carboxylic residues involved in binding the two metal ions responsible for the 3′–5′ exonuclease activity (38–40), as well as other residues described as primer-terminus and TP ligands at the exonuclease site (24,29,41–43). This domain also presents the Kx2h motif that contains a Lys residue which plays an auxiliary role in the exonucleolytic catalysis in family B DNA polymerases (44), and the (S/T)Lx2h motif, whose residues have been involved in making contacts with DNA and TP (29,42,45,46). The C-terminal polymerization domain shows the universal palm, fingers and thumb subdomains structured as a partially open right hand and forming a U-shaped groove predicted to bind the duplex DNA, like in other DNA polymerases cocrystallized with this substrate (47–51). Primer-terminus will lie on the palm subdomain as it includes the conserved motifs Dx2SLYP (motif A) and YxDTDS (motif C) containing the three catalytic carboxylates responsible for the polymerization catalysis (48,51–56), the KxY motif including DNA ligands at the polymerase site (52), and the YxGG/A and Tx2G/AR motifs whose residues are making contacts with the DNA and TP substrates (52,57,58). The fingers subdomain includes motifs Pre-B and Kx3NSxYG (motif B) responsible for interacting with the incoming nucleotide and the template strand (52,59–61). The thumb subdomain contains a Leu residue involved in stabilizing the primer-terminus at the exonuclease site, in addition to other positively charged residues implicated in steadying primer-terminus at the polymerization site as well as in coordinating both the exonuclease and polymerization activities (62). In addition, as the rest of protein-priming DNA polymerases, those of Nf and GA-1 also contain two specific insertions into the sequence of the polymerase domain called terminal protein regions 1 and 2 (TPR1 and TPR2) (12,13,63). TPR1 is located between motif A and fingers subdomain and contains residues involved in making contacts with both dsDNA and TP (13,63). TPR2 immediately follows the fingers subdomain, and together with the palm, fingers and thumb subdomains forms a tunnel that will embrace the DNA and TP substrates, providing the required stability to account for processive TP-DNA replication (11,14,15).
Figure 8.
(A) Ribbon representation of the structural model of GA-1 DNA polymerase. Models for both GA-1 and Nf DNA polymerases were provided by the homology-modelling server Swiss-Model, using as template the crystallographic structure of φ29 DNA polymerase (PDB code 1XHX). The 3′–5′ exonuclease domain is shown in red, the palm in pink, the fingers in dark blue and the thumb in green. Protein-primed DNA polymerases specific insertions TPR1 and TPR2 are coloured in orange and cyan, respectively. Alignment of the amino acid sequences corresponding to family B motifs of the related φ29, GA-1 and Nf DNA polymerases are shown, as well as their spatial placement. Catalytic amino acids responsible for the exonuclease and the polymerization activities are coloured in red, DNA ligand residues in blue, those interacting with both DNA and TP substrates are in orange, incoming nucleotide ligands in magenta, and residues predicted to make contacts with TP-DNA in green. The linear arrangement of these sequence motifs is shown at the bottom. (B) Structural differences in the TPR1 loop of GA-1 and φ29 DNA polymerases. Superposition of the modelled GA-1 DNA polymerase and crystal structure of φ29 DNA polymerase was obtained by fitting the catalytic amino acid residues responsible of both the exonuclease and polymerization activities by using the Swiss-PdbViewer program (http://www.expasy.org/spdbv/) and further rendering with Pymol (http://www.pymol.org). Figure is restricted to the TPR1 loop region of both DNA polymerases. The DNA has been taken from the structure of the RB69 ternary complex (50) and homology modeled by aligning the palm subdomains of RB69, GA-1 and φ29 DNA polymerases. φ29 and GA-1 DNA polymerases are coloured in cyan and green, respectively. The alignment of the β-turn-β structure of TPR1 insertion is shown at the bottom. Arg and Phe residues located at the tip of the TPR1 loop are shown in orange and yellow, respectively.
Nf and GA-1 DNA polymerases are proofreading enzymes
Most replicative DNA-dependent DNA polymerases possess an associated 3′–5′ exonuclease activity that enhances base substitution fidelity from a few fold to more than two orders of magnitude (64,65).
Efficient editing of polymerization errors requires the primer-terminus to be properly placed at the catalytic site by virtue of DNA ligand residues that form a cleft designed to place exclusively ssDNA (frayed terminus) (66–69). Whereas Nf DNA polymerase was able to degrade efficiently the ssDNA substrates, the 3′–5′ exonuclease activity of GA-1 DNA polymerase could be only detected by using pNP-TMP, a substrate used to dissociate the catalytic efficiency in hydrolysing the phosphodiester bond from DNA binding.
The preference for excision of a mismatched primer/template molecule with respect to a matched one displayed by GA-1 and Nf DNA polymerases, together with the dynamic equilibrium between the polymerization and exonuclease activities (Pol/Exo assay) show that both are coupled and act coordinately to remove the misinserted nucleotides.
Nf and GA-1 DNA polymerases couple polymerization to strand displacement processively
The results presented in this paper clearly indicate that Nf and GA-1 DNA polymerases can account for their genome replication without the assistance of unwinding and processivity factors, in contrast to most replicative DNA polymerases which require their physical association to processivity factors and DNA unwinding proteins (1,70). Strand displacement capacity has also been shown for other protein-primed DNA polymerases as those of bacteriophages φ29 (10), Cp-1 (71) and PRD1 (72,73). On the contrary, adenovirus DNA polymerase, although processive, cannot couple polymerization to strand displacement, requiring the DNA unwinding activity of the adenovirus DBP to perform strand displacement (74,75).
Whereas it was possible to obtain GA-1 DNA replication by using exclusively the GA-1 TP and DNA polymerase, Nf DNA polymerase, although provided with competent strand displacement and processivity features, required the presence of Nf DBP for an effective in vitro replication of Nf TP-DNA. Results presented here show that Nf DBP strongly stimulates the formation of the TP-dAMP initiation complex by decreasing the Km for dATP and facilitates the transition from initiation to elongation, as it occurs in φ29 (76). These results point to either a specific and direct contact between DBP and DNA polymerase that promotes conformational changes at the polymerization active site or to an effect of DBP in conferring the optimal template structure to direct initiating nucleotide insertion. A similar role has been proposed for adenovirus DBP, a DNA unwinding protein (77). As in the case of φ29 and Nf DBP, this protein stimulates the rate of initiation also by decreasing the Km for the initiating nucleotide (74). The fact that an adenovirus DBP mutant defective in unwinding can still stimulate initiation precludes the unwinding role as the one responsible for such an activation (77,78). In this case, contacts between DBP and pTP/DNA polymerase complex have been reported (77).
The effect of Nf DBP in promoting elongation of the initiation products could be due to a decrease of the Km also for the incorporation of the dNMPs during the transition stage from initiation to elongation, to a different type of contact with the DNA polymerase that helps transition to elongation, or both. The similarity in replication rates when comparing M13 DNA replication, performed in the absence of DBP (2400 nt/min), with Nf TP-DNA replication in the presence of DBP (2260 nt/min), suggests that the DBP stimulatory role is restricted to the first phases of Nf TP-DNA replication.
GA-1 DNA polymerase, a paradigmatic enzyme
The ability displayed by GA-1 DNA polymerase to hydrolyse the pNP-TMP substrate, together with its high efficiency in carrying out protein-primed initiation and elongation, allowed us to rule out a global misfolding as the cause of its hindered capacity to use ssDNA as substrate of its 3′–5′ exonuclease activity, as well as of its impaired ability to elongate DNA primers. These results could indicate that GA-1 DNA polymerase has developed an extraordinary selectivity to use exclusively its natural primer, the TP, a rather unusual behaviour not shared by the rest of reported replicative protein-primed DNA polymerases that can use both types of primers, TP and DNA. The polymerization domain could be occluded somehow in the absence of TP, preventing the binding of GA-1 DNA polymerase to DNA substrates other than TP-DNA, restricting the use of the polymerase for TP-DNA replication. This fact would also explain why GA-1 DNA polymerase is highly impaired in the use of ssDNA as substrate of the exonuclease activity, taking into account that the cleft that binds the primer strand in the editing mode emanates from the polymerization active site [see Figure 8A and (11)]. Substrates such as the pNP-TMP can be exonucleolyticaly degraded since its small size would allow it to diffuse into the exonuclease site. If this hypothesis were correct, the sequestration of the DNA polymerase by DNAs other than the viral TP-DNA would be impeded, optimizing the usage of the DNA polymerase for viral replication.
The high degree of both sequence identity and similarity (54% and 67.5%, respectively) shared by GA-1 and φ29 DNA polymerases makes difficult to find out structural differences between the modelled GA-1 DNA polymerase structure and the crystallized φ29 DNA polymerase that could be responsible for the substrate specificity of the former. The major difference is found at the β-turn-β structure of the TPR1 insertion. Homology modelled φ29 DNA polymerase/DNA complex allows to predict a direct contact between the loop formed by the TPR1 β-turn-β and the DNA substrate through its major groove (11) (see also Figure 8B). Overlapping of GA-1 and φ29 DNA polymerase structures shows differences in this region of the enzyme because of one position displacement of the GA-1 sequence Arg309–Phe310 with respect to the corresponding one in φ29 DNA polymerase (see Figure 8B). This could imply that the large side chains of these two residues in GA-1 DNA polymerase were facing towards the deepest part of the DNA major groove sterically hindering an initial DNA binding, in contrast to the outer orientation showed by the corresponding residues of φ29 DNA polymerase. Structural comparison of φ29 apo polymerase and DNA polymerase/TP heterodimer structures shows that significant differences in the DNA polymerase structure occur only in the loop between residues 304 and 314 in the TPR1 subdomain (14). This region has to curve out to allow TP access to the active site of the polymerase, the TP priming domain occupying the DNA binding cleft. Similar conformational changes are predicted to occur in GA-1 DNA polymerase TPR1 loop. As the TP priming domain is elongated, the growing DNA must displace it. After the incorporation of ∼6 nt, total dissociation of the heterodimer will take place and the TPR1 loop will adopt the orientation showed in the apoenzyme, fitting into the DNA major groove to confer binding stability. The main binding difference is that in this latter case, the DNA would be already placed into the polymerase active site before the TPR1 loop adopts its final straight conformation.
Acknowledgments
We are grateful to L. Villar for the purification of bacteriophage Nf TP. This investigation was aided by research grant BFU 2005-00733 from the Spanish Ministry of Education and Science and by an institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular ‘Severo Ochoa’. E.L. was a pre-doctoral fellow of the Ministerio de Educación y Ciencia. Funding to pay the Open Access publication charges for this article was provided by research grant BFU 2005-00733 from the Spanish Ministry of Education and Science.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kornberg A., Baker T. DNA Replication. 2nd edn. New York: W.H. Freeman; 1992. [Google Scholar]
- 2.Salas M. Mechanisms of initiation of linear DNA replication in prokaryotes. Genet. Eng. (NY) 1999;21:159–171. doi: 10.1007/978-1-4615-4707-5_8. [DOI] [PubMed] [Google Scholar]
- 3.Brenkman A.B., Breure E.C., van der Vliet P.C. Molecular architecture of adenovirus DNA polymerase and location of the protein primer. J. Virol. 2002;76:8200–8207. doi: 10.1128/JVI.76.16.8200-8207.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rademaker H.J., Fallaux F.J., Van den Wollenberg D.J., De Jong R.N., Van der Vliet P.C., Hoeben R.C. Relaxed template specificity in fowl adenovirus 1 DNA replication initiation. J. Gen. Virol. 2006;87:553–562. doi: 10.1099/vir.0.81328-0. [DOI] [PubMed] [Google Scholar]
- 5.Caldentey J., Blanco L., Bamford D.H., Salas M. In vitro replication of bacteriophage PRD1 DNA. Characterization of the protein-primed initiation site. Nucleic Acids Res. 1993;21:3725–3730. doi: 10.1093/nar/21.16.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Illana B., Blanco L., Salas M. Functional characterization of the genes coding for the terminal protein and DNA polymerase from bacteriophage GA-1. Evidence for a sliding-back mechanism during protein-primed GA-1 DNA replication. J. Mol. Biol. 1996;264:453–464. doi: 10.1006/jmbi.1996.0653. [DOI] [PubMed] [Google Scholar]
- 7.King A.J., van der Vliet P.C. A precursor terminal protein-trinucleotide intermediate during initiation of adenovirus DNA replication: regeneration of molecular ends in vitro by a jumping back mechanism. EMBO J. 1994;13:5786–5792. doi: 10.1002/j.1460-2075.1994.tb06917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martín A.C., Blanco L., García P., Salas M., Méndez J. In vitro protein-primed initiation of pneumococcal phage Cp-1 DNA replication occurs at the third 3′ nucleotide of the linear template: a stepwise sliding-back mechanism. J. Mol. Biol. 1996;260:369–377. doi: 10.1006/jmbi.1996.0407. [DOI] [PubMed] [Google Scholar]
- 9.Méndez J., Blanco L., Esteban J.A., Bernad A., Salas M. Initiation of Φ29 DNA replication occurs at the second 3′ nucleotide of the linear template: a sliding-back mechanism for protein-primed DNA replication. Proc. Natl Acad. Sci. USA. 1992;89:9579–9583. doi: 10.1073/pnas.89.20.9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanco L., Bernad A., Lázaro J.M., Martín G., Garmendia C., Salas M. Highly efficient DNA synthesis by the phage Φ29 DNA polymerase. Symmetrical mode of DNA replication. J. Biol. Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 11.Kamtekar S., Berman A.J., Wang J., Lázaro J.M., de Vega M., Blanco L., Salas M., Steitz T.A. Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage Φ29. Mol. Cell. 2004;16:609–618. doi: 10.1016/j.molcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 12.Blasco M.A., Blanco L., Parés E., Salas M., Bernad A. Structural and functional analysis of temperature-sensitive mutants of the phage Φ29 DNA polymerase. Nucleic Acids Res. 1990;18:4763–4770. [PMC free article] [PubMed] [Google Scholar]
- 13.Dufour E., Méndez J., Lázaro J.M., de Vega M., Blanco L., Salas M. An aspartic acid residue in TPR-1, a specific region of protein-priming DNA polymerases, is required for the functional interaction with primer terminal protein. J. Mol. Biol. 2000;304:289–300. doi: 10.1006/jmbi.2000.4216. [DOI] [PubMed] [Google Scholar]
- 14.Kamtekar S., Berman A.J., Wang J., Lázaro J.M., de Vega M., Blanco L., Salas M., Steitz T.A. The Φ29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition. EMBO J. 2006;25:1335–1343. doi: 10.1038/sj.emboj.7601027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodríguez I., Lázaro J.M., Blanco L., Kamtekar S., Berman A.J., Wang J., Steitz T.A., Salas M., de Vega M. A specific subdomain in Φ29 DNA polymerase confers both processivity and strand-displacement capacity. Proc. Natl Acad. Sci. USA. 2005;102:6407–6412. doi: 10.1073/pnas.0500597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González-Huici V., Lázaro J.M., Salas M., Hermoso J.M. Specific recognition of parental terminal protein by DNA polymerase for initiation of protein-primed DNA replication. J. Biol. Chem. 2000;275:14678–14683. doi: 10.1074/jbc.m910058199. [DOI] [PubMed] [Google Scholar]
- 17.Meijer W.J., Horcajadas J.A., Salas M. Φ29 family of phages. Microbiol. Mol. Biol. Rev. 2001;65:261–287. doi: 10.1128/MMBR.65.2.261-287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peñalva M.A., Salas M. Initiation of phage Φ29 DNA replication in vitro: formation of a covalent complex between the terminal protein, p3, and 5′-dAMP. Proc. Natl Acad. Sci. USA. 1982;79:5522–5526. doi: 10.1073/pnas.79.18.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lázaro J.M., Blanco L., Salas M. Purification of bacteriophage Φ29 DNA polymerase. Methods Enzymol. 1995;262:42–49. doi: 10.1016/0076-6879(95)62007-9. [DOI] [PubMed] [Google Scholar]
- 20.Studier F.W., Moffatt B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 21.Inciarte M.R., Viñuela E., Salas M. Transcription in vitro of Φ29 DNA and EcoRI fragments by Bacillus subtilis RNA polymerase. Eur. J. Biochem. 1976;71:77–83. doi: 10.1111/j.1432-1033.1976.tb11091.x. [DOI] [PubMed] [Google Scholar]
- 22.Tabor S., Richardson C.C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl Acad. Sci. USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freire R., Serrano M., Salas M., Hermoso J.M. Activation of replication origins in Φ29-related phages requires the recognition of initiation proteins to specific nucleoprotein complexes. J. Biol. Chem. 1996;271:31000–31007. doi: 10.1074/jbc.271.48.31000. [DOI] [PubMed] [Google Scholar]
- 24.de Vega M., Lázaro J.M., Salas M., Blanco L. Primer-terminus stabilization at the 3′-5′ exonuclease active site of Φ29 DNA polymerase. Involvement of two amino acid residues highly conserved in proofreading DNA polymerases. EMBO J. 1996;15:1182–1192. [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar J.K., Chiu E.T., Tabor S., Richardson C.C. A unique region in bacteriophage t7 DNA polymerase important for exonucleolytic hydrolysis of DNA. J. Biol. Chem. 2004;279:42018–42025. doi: 10.1074/jbc.M406103200. [DOI] [PubMed] [Google Scholar]
- 26.Carthew R.W., Chodosh L.A., Sharp P.A. An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell. 1985;43:439–448. doi: 10.1016/0092-8674(85)90174-6. [DOI] [PubMed] [Google Scholar]
- 27.Méndez J., Blanco L., Lázaro J.M., Salas M. Primer-terminus stabilization at the Φ29 DNA polymerase active site. Mutational analysis of conserved motif TX2GR. J. Biol. Chem. 1994;269:30030–30038. [PubMed] [Google Scholar]
- 28.Garmendia C., Bernad A., Esteban J.A., Blanco L., Salas M. The bacteriophage Φ29 DNA polymerase, a proofreading enzyme. J. Biol. Chem. 1992;267:2594–2599. [PubMed] [Google Scholar]
- 29.de Vega M., Blanco L., Salas M. Φ29 DNA polymerase residue Ser122, a single-stranded DNA ligand for 3′-5′ exonucleolysis, is required to interact with the terminal protein. J. Biol. Chem. 1998;273:28966–28977. doi: 10.1074/jbc.273.44.28966. [DOI] [PubMed] [Google Scholar]
- 30.McDonell M.W., Simon M.N., Studier F.W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J. Mol. Biol. 1977;110:119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- 31.Esteban J.A., Bernad A., Salas M., Blanco L. Metal activation of synthetic and degradative activities of Φ29 DNA polymerase, a model enzyme for protein-primed DNA replication. Biochemistry. 1992;31:350–359. doi: 10.1021/bi00117a006. [DOI] [PubMed] [Google Scholar]
- 32.Salas M. Protein-priming of DNA replication. Annu. Rev. Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 33.Salas M., Miller J., Leis J., DePamphilis M. Mechanisms for Priming DNA Synthesis. New York: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 34.Bernad A., Zaballos A., Salas M., Blanco L. Structural and functional relationships between prokaryotic and eukaryotic DNA polymerases. EMBO J. 1987;6:4219–4225. doi: 10.1002/j.1460-2075.1987.tb02770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.González-Huici V., Salas M., Hermoso J.M. Sequence requirements for protein-primed initiation and elongation of phage Φ29 DNA replication. J. Biol. Chem. 2000;275:40547–40553. doi: 10.1074/jbc.M007170200. [DOI] [PubMed] [Google Scholar]
- 36.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 37.Schwede T., Kopp J., Guex N., Peitsch M.C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernad A., Blanco L., Lázaro J.M., Martín G., Salas M. A conserved 3′–5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989;59:219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- 39.Derbyshire V., Freemont P.S., Sanderson M.R., Beese L., Friedman J.M., Joyce C.M., Steitz T.A. Genetic and crystallographic studies of the 3′,5′-exonucleolytic site of DNA polymerase I. Science. 1988;240:199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- 40.Beese L.S., Steitz T.A. Structural basis for the 3′–5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Vega M., Lázaro J.M., Salas M. Phage Φ29 DNA polymerase residues involved in the proper stabilisation of the primer-terminus at the 3′–5′ exonuclease active site. J. Mol. Biol. 2000;304:1–9. doi: 10.1006/jmbi.2000.4178. [DOI] [PubMed] [Google Scholar]
- 42.de Vega M., Lázaro J.M., Salas M., Blanco L. Mutational analysis of Φ29 DNA polymerase residues acting as ssDNA ligands for 3′-5′ exonucleolysis. J. Mol. Biol. 1998;279:807–822. doi: 10.1006/jmbi.1998.1805. [DOI] [PubMed] [Google Scholar]
- 43.Eisenbrandt R., Lázaro J.M., Salas M., de Vega M. Φ29 DNA polymerase residues Tyr59, His61 and Phe69 of the highly conserved ExoII motif are essential for interaction with the terminal protein. Nucleic Acids Res. 2002;30:1379–1386. doi: 10.1093/nar/30.6.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Vega M., Ilyina T., Lázaro J.M., Salas M., Blanco L. An invariant lysine residue is involved in catalysis at the 3′–5′ exonuclease active site of eukaryotic-type DNA polymerases. J. Mol. Biol. 1997;270:65–78. doi: 10.1006/jmbi.1997.1093. [DOI] [PubMed] [Google Scholar]
- 45.Rodríguez I., Lázaro J.M., Salas M., de Vega M. Φ29 DNA polymerase residue Phe128 of the highly conserved (S/T)Lx2h motif is required for a stable and functional interaction with the terminal protein. J. Mol. Biol. 2003;325:85–97. doi: 10.1016/s0022-2836(02)01130-0. [DOI] [PubMed] [Google Scholar]
- 46.Rodríguez I., Lázaro J.M., Salas M., de Vega M. Φ29 DNA polymerase-terminal protein interaction. Involvement of residues specifically conserved among protein-primed DNA polymerases. J. Mol. Biol. 2004;337:829–841. doi: 10.1016/j.jmb.2004.02.018. [DOI] [PubMed] [Google Scholar]
- 47.Beese L.S., Derbyshire V., Steitz T.A. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- 48.Doubliè S., Tabor S., Long A.M., Richardson C.C., Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 49.Eom S.H., Wang J., Steitz T.A. Structure of Taq polymerase with DNA at the polymerase active site. Nature. 1996;382:278–281. doi: 10.1038/382278a0. [DOI] [PubMed] [Google Scholar]
- 50.Franklin M.C., Wang J., Steitz T.A. Structure of the replicating complex of a pol α family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 51.Kiefer J.R., Mao C., Braman J.C., Beese L.S. Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature. 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- 52.Blanco L., Salas M. Relating structure to function in Φ29 DNA polymerase. J. Biol. Chem. 1996;271:8509–8512. doi: 10.1074/jbc.271.15.8509. [DOI] [PubMed] [Google Scholar]
- 53.Joyce C.M., Steitz T.A. Function and structure relationships in DNA polymerases. Annu. Rev. Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 54.Pelletier H., Sawaya M.R., Wolfle W., Wilson S.H., Kraut J. Crystal structures of human DNA polymerase beta complexed with DNA: implications for catalytic mechanism, processivity, and fidelity. Biochemistry. 1996;35:12742–12761. doi: 10.1021/bi952955d. [DOI] [PubMed] [Google Scholar]
- 55.Saturno J., Lázaro J.M., Blanco L., Salas M. Role of the first aspartate residue of the ‘YxDTDS’ motif of Φ29 DNA polymerase as a metal ligand during both TP-primed and DNA-primed DNA synthesis. J. Mol. Biol. 1998;283:633–642. doi: 10.1006/jmbi.1998.2121. [DOI] [PubMed] [Google Scholar]
- 56.Steitz T.A. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 57.Brenkman A.B., Heideman M.R., Truniger V., Salas M., van der Vliet P.C. The (I/Y)XGG motif of adenovirus DNA polymerase affects template DNA binding and the transition from initiation to elongation. J. Biol. Chem. 2001;276:29846–29853. doi: 10.1074/jbc.M103159200. [DOI] [PubMed] [Google Scholar]
- 58.Truniger V., Blanco L., Salas M. Role of the ‘YxGG/A’ motif of Φ29 DNA polymerase in protein-primed replication. J. Mol. Biol. 1999;286:57–69. doi: 10.1006/jmbi.1998.2477. [DOI] [PubMed] [Google Scholar]
- 59.Saturno J., Lázaro J.M., Esteban F.J., Blanco L., Salas M. Φ29 DNA polymerase residue Lys383, invariant at motif B of DNA-dependent polymerases, is involved in dNTP binding. J. Mol. Biol. 1997;269:313–325. doi: 10.1006/jmbi.1997.1053. [DOI] [PubMed] [Google Scholar]
- 60.Truniger V., Lázaro J.M., Blanco L., Salas M. A highly conserved lysine residue in Φ29 DNA polymerase is important for correct binding of the templating nucleotide during initiation of phi29 DNA replication. J. Mol. Biol. 2002;318:83–96. doi: 10.1016/S0022-2836(02)00022-0. [DOI] [PubMed] [Google Scholar]
- 61.Truniger V., Lázaro J.M., Salas M. Two positively charged residues of Φ29 DNA polymerase, conserved in protein-primed DNA polymerases, are involved in stabilisation of the incoming nucleotide. J. Mol. Biol. 2004;335:481–494. doi: 10.1016/j.jmb.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 62.Pérez-Arnaiz P., Lázaro J.M., Salas M., de Vega M. Involvement of Φ29 DNA polymerase thumb subdomain in the proper coordination of synthesis and degradation during DNA replication. Nucleic Acids Res. 2006;34:3107–3115. doi: 10.1093/nar/gkl402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dufour E., Rodríguez I., Lázaro J.M., de Vega M., Salas M. A conserved insertion in protein-primed DNA polymerases is involved in primer terminus stabilisation. J. Mol. Biol. 2003;331:781–794. doi: 10.1016/s0022-2836(03)00788-5. [DOI] [PubMed] [Google Scholar]
- 64.Brutlag D., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XXXVI. A proofreading function for the 3′ leads to 5′ exonuclease activity in deoxyribonucleic acid polymerases. J. Biol. Chem. 1972;247:241–248. [PubMed] [Google Scholar]
- 65.Kunkel T.A. Exonucleolytic proofreading. Cell. 1988;53:837–840. doi: 10.1016/s0092-8674(88)90189-4. [DOI] [PubMed] [Google Scholar]
- 66.Cowart M., Gibson K.J., Allen D.J., Benkovic S.J. DNA substrate structural requirements for the exonuclease and polymerase activities of procaryotic and phage DNA polymerases. Biochemistry. 1989;28:1975–1983. doi: 10.1021/bi00431a004. [DOI] [PubMed] [Google Scholar]
- 67.Franklin M.C., Wang J., Steitz T.A. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 68.Freemont P.S., Friedman J.M., Beese L.S., Sanderson M.R., Steitz T.A. Cocrystal structure of an editing complex of Klenow fragment with DNA. Proc. Natl Acad. Sci. USA. 1988;85:8924–8928. doi: 10.1073/pnas.85.23.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J., Sattar A.K., Wang C.C., Karam J.D., Konigsberg W.H., Steitz T.A. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 70.Watson J., Baker T., Bell S., Gann A., Levine M., Losick R. Molecular Biology of the Gene. 5th edn. Plainview, New York: Cold Spring Harbor Lab. Press; 2004. [Google Scholar]
- 71.García P., Hermoso J.M., García J.A., García E., López R., Salas M. Formation of a covalent complex between the terminal protein of pneumococcal bacteriophage Cp-1 and 5′-dAMP. J. Virol. 1986;58:31–35. doi: 10.1128/jvi.58.1.31-35.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Savilahti H., Caldentey J., Lundstrom K., Syvaoja J.E., Bamford D.H. Overexpression, purification, and characterization of Escherichia coli bacteriophage PRD1 DNA polymerase. In vitro synthesis of full-length PRD1 DNA with purified proteins. J. Biol. Chem. 1991;266:18737–18744. [PubMed] [Google Scholar]
- 73.Zhu W., Ito J. Purification and characterization of PRD1 DNA polymerase. Biochim. Biophys. Acta. 1994;1219:267–276. doi: 10.1016/0167-4781(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 74.Dekker J., Kanellopoulos P.N., Loonstra A.K., van Oosterhout J.A., Leonard K., Tucker P.A., van der Vliet P.C. Multimerization of the adenovirus DNA-binding protein is the driving force for ATP-independent DNA unwinding during strand displacement synthesis. EMBO J. 1997;16:1455–1463. doi: 10.1093/emboj/16.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.King A.J., Teertstra W.R., Blanco L., Salas M., van der Vliet P.C. Processive proofreading by the adenovirus DNA polymerase. Association with the priming protein reduces exonucleolytic degradation. Nucleic Acids Res. 1997;25:1745–1752. doi: 10.1093/nar/25.9.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blanco L., Gutiérrez J., Lázaro J.M., Bernad A., Salas M. Replication of phage Φ29 DNA in vitro: role of the viral protein p6 in initiation and elongation. Nucleic Acids Res. 1986;14:4923–4937. doi: 10.1093/nar/14.12.4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.de Jong R.N., van der Vliet P.C., Brenkman A.B. Adenovirus DNA replication: protein priming, jumping back and the role of the DNA binding protein DBP. Curr. Top. Microbiol. Immunol. 2003;272:187–211. doi: 10.1007/978-3-662-05597-7_7. [DOI] [PubMed] [Google Scholar]
- 78.van Breukelen B., Brenkman A.B., Holthuizen P.E., van der Vliet P.C. Adenovirus type 5 DNA binding protein stimulates binding of DNA polymerase to the replication origin. J. Virol. 2003;77:915–922. doi: 10.1128/JVI.77.2.915-922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]