Abstract
U12-type introns are spliced by the U12-dependent spliceosome and are present in the genomes of many higher eukaryotic lineages including plants, chordates and some invertebrates. However, due to their relatively recent discovery and a systematic bias against recognition of non-canonical splice sites in general, the introns defined by U12-type splice sites are under-represented in genome annotations. Such under-representation compounds the already difficult problem of determining gene structures. It also impedes attempts to study these introns genome-wide or phylum-wide. The resource described here, the U12 Intron Database (U12DB), aims to catalog the U12-type introns of completely sequenced eukaryotic genomes in a framework that groups orthologous introns with each other. This will aid further investigations into the evolution and mechanism of U12-dependent splicing as well as assist ongoing genome annotation efforts. Public access to the U12DB is available at http://genome.imim.es/cgi-bin/u12db/u12db.cgi.
INTRODUCTION
Two pathways for the removal of eukaryotic spliceosomal introns exist: a major pathway, that is dependent on the normal U2 snRNA-containing spliceosome and a minor pathway, that is dependent on the more recently discovered U12 snRNA-containing spliceosome [see Refs (1,2) for an in-depth review of U12-dependent splicing]. The U11 and U12 snRNAs were first discovered and characterized as low-abundance members of the Sm class of snRNAs and shown to form a complex (3,4). They were subsequently proposed (5) and then shown (6–8) to splice, along with U4atac and U6atac (7), the minor class of introns. These introns were first identified due to the presence of non-canonical AT–AC terminal dinucleotides but later found to comprise both GT–AG and AT–AC subtypes (9,10). In fact, the majority (∼70%) are GT–AG in human.
While the major pathway is present in all eukaryotes, evidence for the minor pathway only exists for plants and animals, and even within these phyla, some lineages such as nematodes appear to have lost the pathway (11). While the two spliceosomes possess distinctly different complements of snRNAs (U1, U2, U4 and U6 versus U11, U12, U4atac and U6atac), they share one snRNA, U5, and have many of the same protein components in common (12) suggesting a common evolutionary origin. Burge et al. have proposed a fission–fusion hypothesis that reconciles the observed distribution of extant U12-type introns with the divergent components of the two spliceosomes (13).
The two spliceosomes are also functionally divergent, particularly in their recognition of splice sites. The U12 consensus sequences for the donor site, RTATCCTTT, and branch point, TTCCTTRAY, are highly conserved and distinct from the U2 consensi (9). The acceptor site is unique in that the 3′-most nucleotide of the U12 acceptor site is more tolerant of substitutions, especially in those introns that begin with AT; AT–AC, AT–AA, AT–AG and AT–AT combinations have been observed (14,15). The two spliceosomes also differ in the order of spliceosomal assembly. U11 and U12 form a dimer which then recognizes the donor site and branch point simultaneously (16), whereas U1 and U2 recognize these sites independently before associating.
The compilation of several dozen U12 introns from different species paved the way for the computational identification of U12 introns using weight matrices for the donor site and branch point (10,13). A computational scan of the human genome (14) resulted in the identification of 404 U12 introns, which constitute ∼0.3% of all human introns. Similar scans of the Arabidopsis thaliana (17) and Drosophila melanogaster (18) genomes resulted in 165 and 19 putative U12 introns, respectively. A more recent scan for U12 introns present in RefSeq annotations in five species (human, mouse, D.melanogaster, Caenorhabditis elegans, and A.thaliana) resulted in 671, 625, 18, 0 and 191 U12-type introns, respectively (19). Due to their low frequency and non-canonical nature, U12 introns are frequently overlooked and often ignored by gene predictors and genome annotation pipelines. In order to remedy this systematic bias against the correct annotation of U12 introns, we have carried out a semi-automated annotation of U12 introns in the genomes of 20 species. Both the breadth of phylogenetic space that we explore and the inclusion of novel intron predictions (verified by alignment to expressed sequence) distinguish our study from previous ones. Moreover, we link orthologous introns into clusters so that they may be easily visualized in their evolutionary context. Our results are made available through the relational database described here [the U12 Intron Database (U12DB)], which is designed to catalog all U12 introns in the genomes of sequenced species. Specifically, the U12DB aims to fuel further investigations into the evolution and mechanism of U12-dependent splicing. It is also intended as a resource for gene prediction method development and genome annotation.
We begin by outlining the pipeline that was used to populate the database. We then give an overview of the representation of U12 introns from different species in the database. Finally, we describe how to search the U12DB using the online query system and how to navigate the results pages.
DATABASE POPULATION
We employed a strategy that combined manual annotation with computational prediction for the curation of U12 introns and their orthologs in 20 species. A high-confidence set of U12 introns was compiled for each of several reference genomes. These introns were then mapped via spliced alignment to orthologous introns in each of the other genomes. This mapping procedure has two clear benefits. It conveniently constructs clusters of orthologous introns, which constitutes one of the principle relationships represented in the U12DB. Moreover, the necessity for laborious manual curation of U12 introns in each genome is reduced. On the other hand, U12 introns unique to particular non-reference species or lineages will be missed with this approach. Therefore, we chose to begin with four reference species instead of only one. More may be added in the future (see Future Directions section).
Primary data from reference genomes
Reference U12 introns were obtained from four species: Homo sapiens, Ciona intestinalis, D.melanogaster, and A.thaliana. These species were primarily chosen for their phylogenetic placement and the quality of their gene annotations. Human and A.thaliana were of particular interest due to the availability of previous compilations of U12 introns. Several sources of U12 introns were used, depending on availability: introns from known gene annotations that we classified as U12-type (D.melanogaster, A.thaliana, and H.sapiens), known U12 introns compiled from the literature (H.sapiens) and novel predicted U12 introns filtered for alignment with expressed sequence tag/cDNA sequences across the predicted exon junction (all species). In the case of annotated introns, introns were extracted from the UCSC ‘Known’ gene annotation (20) on hg17/NCBI35 (H.sapiens), FlyBase annotations (21) on BDGP Release 4 (D.melanogaster) and TAIR6 annotations (22) (A.thaliana). For annotated and predicted introns, splice junctions were scored and classified according to U12 versus U2-type using GeneID (23,24), which we modified to use positional weight matrices for both U12 and U2 donor, branch and acceptor sites. U12 training data was derived primarily from published human U12 intron sets (13,14). These same introns were mapped to the NCBI35 human genome assembly using SSAHA (25).
Intron mapping
The reference introns were mapped into each of the target species (Ensembl v37 species plus A.thaliana). A version of Exonerate (26) that we modified to better model U12 splice sites was used to align a concatenated sequence including 100 bp of sequence flanking each side of the reference intron to the genomic gene sequence of each of its Ensembl orthologs [or Inparanoid (27) homologs, in the case of A.thaliana]. If a gapped alignment to a target sequence was found and the position of the gap was identical to that of the reference intron, the target intron was considered orthologous. If an ungapped alignment or a gapped alignment where the gap was shifted relative to the reference intron, an ‘exon hit’ was recorded. If no alignment was found, an intron loss was recorded. In the case where no orthologous gene was identified for a species through Compara or Inparanoid, no record was kept. In other words, the absence of an intron record reflects the fact that no orthologous gene could be assigned. All orthologous introns were then classified according to type (U2 or U12) and subtype (GT–AG or AT–AC) using the same method as described above for scoring annotated introns. Ambiguous classifications were assigned a U12/U2 type.
Database implementation
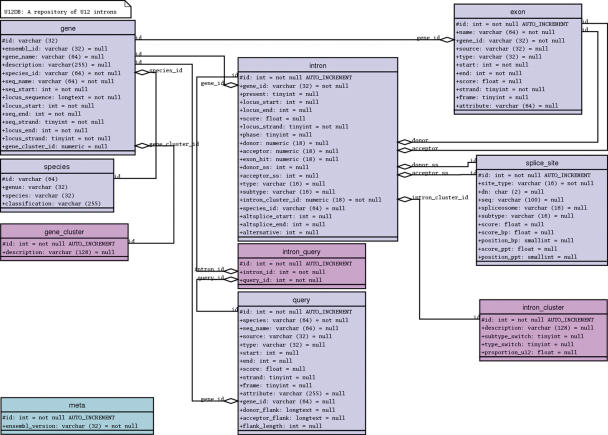
The U12DB has been implemented as a relational database in order to provide fast and flexible queries as well as to facilitate future data and schema updates. The data are stored in a MySQL database with tables for storing information on the reference introns (the query table), the target introns, the genes containing the reference and target introns, the sequence flanking the target introns, the splice sites defining the introns, orthologous gene clusters, orthologous intron clusters, taxonomy, and database version numbers. The database schema is depicted in Figure 1. Database population and updating is carried out using custom Perl scripts. The web query interface is also written in Perl.
Figure 1.
The U12DB schema. Square rectangles are tables in MySQL. The lines connecting them represent foreign key—primary key relationships. Transcript-confirmed or annotated U12 introns from reference genomes are loaded into the query table. The introns are then associated with Ensembl or TAIR6 genes and these genes are inserted into the gene table. The Ensembl Compara database or a custom-built Inparanoid human/Arabidopsis database is then searched for orthologs. The orthologs are inserted into the gene table and assigned a gene cluster id that is stored in a separate table. Each reference intron is then mapped with Exonerate to each gene in the gene cluster to which its host gene belongs. The introns are assigned an intron cluster id and stored in the intron table. Splice site and exon information are stored in separate tables. The system is designed to be updatable so that intron and gene records are not duplicated but, instead, are updated if they are the target of a new reference intron mapping from the query set. Therefore, there is a many-to-many relationship between query and intron.
DATABASE CONTENTS
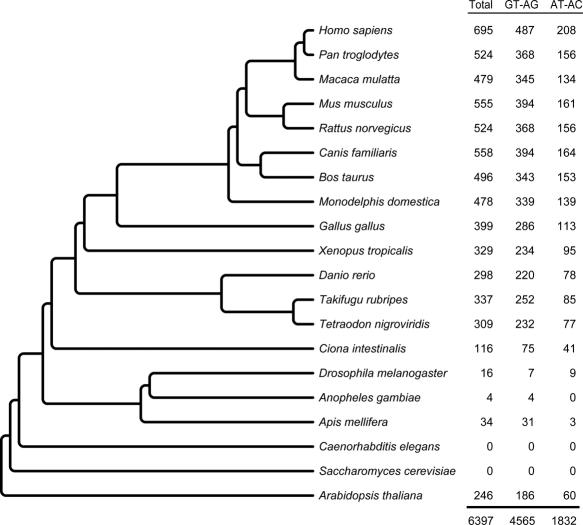
Release 1 of the U12DB contains information on the genomic location and properties of 6397 known and computationally predicted introns with U12-type characteristics in 20 species (Figure 2). The species with the largest number of U12 introns is H.sapiens (695 introns), whereas some species (e.g. Anopheles gambiae) have very few. Aside from the presence of species-specific U12 introns, several additional explanations are plausible for the discrepancies in absolute intron numbers among species in the database. First, vertebrates and plants have more genes and more introns per gene than lower eukaryotes. Another factor to consider is that the volume of expressed sequence tags and cDNAs available, the level of alternative transcript annotation and the genome sequence quality are higher in human than in other species. Finally, because our intron annotation pipeline relies on an alignment step, the number of introns successfully mapped between genomes decreases as a factor of the phylogenetic distance. To illustrate this point, if we filter out human U12 introns that are not conserved across species (those with <75% of their orthologs U12-type), only 603 human U12 introns are retained, which is more in line with the numbers found in other vertebrates. The relative abundance of U12 introns in U12DB for A.thaliana is also relatively high, given its early branching position in the phylogenetic tree, but this may also be due to its use as a reference genome in the pipeline.
Figure 2.
Representation of U12 introns in the U12DB. The total number of U12 introns and the number of each subtype are shown in the columns to the right of the species tree. Grand totals are shown at the bottom. The number of U12 introns in the database for each species does not necessarily reflect the actual number of U12 introns in its genome. Note that branch lengths are not to scale. Branch order was obtained from the NCBI taxonomy browser.
To conclude, we find that the relative abundance in plants and vertebrates is consistent with a model of U12 intron loss, with a lower rate of U12 intron depletion in plants and vertebrates than in yeast, nematodes and insects. We cannot rule out that the technical bias of our pipeline is a factor, but the low number of introns found in D.melanogaster, another of our reference genomes, would suggest that this technical bias is not great and is likely to account for only ∼15% of the difference in intron abundance.
DATABASE ACCESS
Web interface
The U12 Intron Database web interface (http://genome.imim.es/datasets/u12/) provides easy access to the data stored in the database. Queries are constructed using a combination of text fields, drop-down lists, radio buttons and check boxes. It is possible to query by intron id, intron cluster id, gene id, Ensembl gene id, gene name and gene description. Wild cards and partial matches are allowed. Results can be further restricted by species, intron type and subtype, involvement in alternative splicing (currently restricted to human), and the presence of U12-type orthologous introns in other species. It is also possible to retrieve all orthologs of introns meeting the search criteria by selecting the ‘intron cluster’ option.
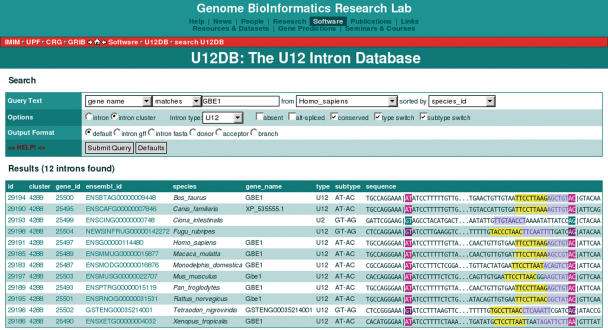
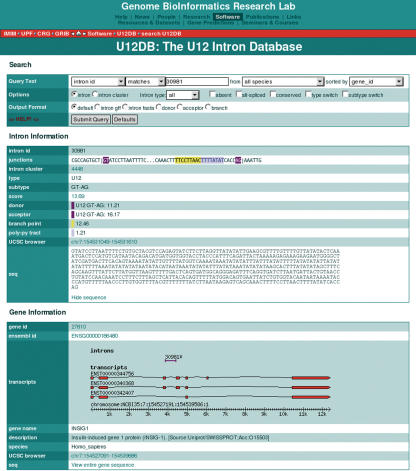
Results are displayed either in a multiple intron view or in a single intron view depending on the number of records returned. The layout of the results has been designed so that the evolutionary relationships of introns within and between species can be easily explored from any result page via hyperlinks. In the multi-intron view, information is summarized in one line such that that the splice junction sequences are aligned to each other (Figure 3). Links are provided to individual intron records, to the cluster of orthologous introns, to other introns in the same gene, and to the Ensembl gene record. In the single intron view, more detailed information is provided and includes the following: splice site and branch point scores, links to Ensembl and the Alternative Splicing Database (28), links to the intron and gene locations in the UCSC genome browser (29), a graphic of the intron location with respect to Ensembl transcripts, the gene name and the gene description (Figure 4).
Figure 3.
The U12DB multi-intron view. The search box appears on every page, including the results page, in order to allow quick navigation of the database. The donor and acceptor splice sites are color coded by type and subtype (U2: green, U12gtag: purple, U12atac: magenta). The predicted branch point consensus is highlighted in yellow and the putative polypyrimidine tract is highlighted in light blue (note that U12 introns do not generally possess a polypyrimidine tract).
Figure 4.
The U12DB single-intron view. This view provides more detail on the intron returned by the query. The splice junction sequences are color-coded as in Figure 2. In addition, the user can choose to display the intronic sequence or the gene sequence. For certain species, one can also link to UCSC genome browser where a user-defined track will display the intron position. The transcript view provides a visualization of the intron with respect to the Ensembl transcripts. The transcripts are clickable and link to the corresponding ExonView in Ensembl. The U12DB introns are also clickable and link to the intron record in U12DB. This is useful in the case of two or more U12-type introns that are located in the same gene.
Additional selectable output formats include General Feature Format (http://www.sanger.ac.uk/Software/formats/GFF/) and FASTA formats for the output of intron locations and sequences, respectively, and tabular format for the output of donor, acceptor or branch point sequences. These formats are provided to aid researchers in further analysis of the data.
SQL files
Full access to the data is provided in the form of SQL commands that can be executed to reconstitute the entire database. These files are provided as downloadable links on the home page of U12DB (http://genome.imim.es/datasets/u12/).
FUTURE DIRECTIONS
Our focus will be on making U12DB updates automatic and periodic. Ideally, we would like to synchronize with new Ensembl releases. However, compatibility with the UCSC browser display will have to be manually reviewed and configured depending on the overlap in genomes available on each browser. Increased automation will also allow us to increase the number of reference genomes, thus making U12DB more comprehensive and more robust to potential mapping errors.
Additional features that we foresee implementing in the future include the ability to query by the type of evidence supporting each intron–exon junction in the database and the ability to search for sequence characteristics (homology, pattern or motif searches) in the intronic sequence or flanking sequences.
Acknowledgments
This work was carried out in the group of Roderic Guigó, who provided the original inspiration for this project and whose comments on the database interface features were particularly helpful. I also thank Enrique Blanco García for answering questions about GeneID, the members of the Guigó group for helpful discussions and Gina Alioto for comments on the manuscript. This work was fully supported by the Biosapiens Network of Excellence project. The BioSapiens project is funded by the European Commission within its FP6 Programme, under the thematic area ‘Life sciences, genomics and biotechnology for health’, contract number LSHG-CT-2003-503265. Funding to pay the Open Access publication charges for this article was provided by the BioSapiens project.
Conflict of interest statement. None declared.
REFERENCES
- 1.Patel A.A., Steitz J.A. Splicing double: insights from the second spliceosome. Nature Rev. Mol. Cell Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- 2.Will C.L., Luhrmann R. Splicing of a rare class of introns by the U12-dependent spliceosome. Biol. Chem. 2005;386:713–724. doi: 10.1515/BC.2005.084. [DOI] [PubMed] [Google Scholar]
- 3.Montzka K.A., Steitz J.A. Additional low-abundance human small nuclear ribonucleoproteins: U11, U12, etc. Proc. Natl Acad. Sci. USA. 1988;85:8885–8889. doi: 10.1073/pnas.85.23.8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wassarman K.M., Steitz J.A. The low-abundance U11 and U12 small nuclear ribonucleoproteins (snRNPs) interact to form a two-snRNP complex. Mol. Cell Biol. 1992;12:1276–1285. doi: 10.1128/mcb.12.3.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall S.L., Padgett R.A. Conserved sequences in a class of rare eukaryotic nuclear introns with non-consensus splice sites. J. Mol. Biol. 1994;239:357–365. doi: 10.1006/jmbi.1994.1377. [DOI] [PubMed] [Google Scholar]
- 6.Tarn W.Y., Steitz J.A. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell. 1996;84:801–811. doi: 10.1016/s0092-8674(00)81057-0. [DOI] [PubMed] [Google Scholar]
- 7.Tarn W.Y., Steitz J.A. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT–AC introns. Science. 1996;273:1824–1832. doi: 10.1126/science.273.5283.1824. [DOI] [PubMed] [Google Scholar]
- 8.Hall S.L., Padgett R.A. Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science. 1996;271:1716–1718. doi: 10.1126/science.271.5256.1716. [DOI] [PubMed] [Google Scholar]
- 9.Dietrich R.C., Incorvaia R., Padgett R.A. Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol. Cell. 1997;1:151–160. doi: 10.1016/s1097-2765(00)80016-7. [DOI] [PubMed] [Google Scholar]
- 10.Sharp P.A., Burge C.B. Classification of introns: U2-type or U12-type. Cell. 1997;91:875–879. doi: 10.1016/s0092-8674(00)80479-1. [DOI] [PubMed] [Google Scholar]
- 11.Collins L., Penny D. Complex spliceosomal organization ancestral to extant eukaryotes. Mol. Biol. Evol. 2005;22:1053–1066. doi: 10.1093/molbev/msi091. [DOI] [PubMed] [Google Scholar]
- 12.Will C.L., Schneider C., Reed R., Luhrmann R. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science. 1999;284:2003–2005. doi: 10.1126/science.284.5422.2003. [DOI] [PubMed] [Google Scholar]
- 13.Burge C.B., Padgett R.A., Sharp P.A. Evolutionary fates and origins of U12-type introns. Mol. Cell. 1998;2:773–785. doi: 10.1016/s1097-2765(00)80292-0. [DOI] [PubMed] [Google Scholar]
- 14.Levine A., Durbin R. A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res. 2001;29:4006–4013. doi: 10.1093/nar/29.19.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietrich R.C., Fuller J.D., Padgett R.A. A mutational analysis of U12-dependent splice site dinucleotides. RNA. 2005;11:1430–1440. doi: 10.1261/rna.7206305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frilander M.J., Steitz J.A. Initial recognition of U12-dependent introns requires both U11/5' splice-site and U12/branchpoint interactions. Genes Dev. 1999;13:851–863. doi: 10.1101/gad.13.7.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu W., Brendel V. Identification, characterization and molecular phylogeny of U12-dependent introns in the Arabidopsis thaliana genome. Nucleic Acids Res. 2003;31:4561–4572. doi: 10.1093/nar/gkg492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider C., Will C.L., Brosius J., Frilander M.J., Luhrmann R. Identification of an evolutionarily divergent U11 small nuclear ribonucleoprotein particle in Drosophila. Proc. Natl Acad. Sci. USA. 2004;101:9584–9589. doi: 10.1073/pnas.0403400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheth N., Roca X., Hastings M.L., Roeder T., Krainer A.R., Sachidanandam R. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 2006;34:3955–3967. doi: 10.1093/nar/gkl556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu F., Kent W.J., Clawson H., Kuhn R.M., Diekhans M., Haussler D. The UCSC known genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 21.Drysdale R.A., Crosby M.A. FlyBase: genes and gene models. Nucleic Acids Res. 2005;33:D390–D395. doi: 10.1093/nar/gki046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhee S.Y., Beavis W., Berardini T.Z., Chen G., Dixon D., Doyle A., Garcia-Hernandez M., Huala E., Lander G., Montoya M., et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parra G., Blanco E., Guigo R. GeneID in Drosophila. Genome Res. 2000;10:511–515. doi: 10.1101/gr.10.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guigo R. Assembling genes from predicted exons in linear time with dynamic programming. J. Comput. Biol. 1998;5:681–702. doi: 10.1089/cmb.1998.5.681. [DOI] [PubMed] [Google Scholar]
- 25.Ning Z., Cox A.J., Mullikin J.C. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slater G.S., Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Brien K.P., Remm M., Sonnhammer E.L. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:D476–D480. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamm S., Riethoven J.J., Le Texier V., Gopalakrishnan C., Kumanduri V., Tang Y., Barbosa-Morais N.L., Thanaraj T.A. ASD: a bioinformatics resource on alternative splicing. Nucleic Acids Res. 2006;34:D46–55. doi: 10.1093/nar/gkj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]