Abstract
TOPOFIT-DB (T-DB) is a public web-based database of protein structural alignments based on the TOPOFIT method, providing a comprehensive resource for comparative analysis of protein structure families. The TOPOFIT method is based on the discovery of a saturation point on the alignment curve (topomax point) which presents an ability to objectively identify a border between common and variable parts in a protein structural family, providing additional insight into protein comparison and functional annotation. TOPOFIT also effectively detects non-sequential relations between protein structures. T-DB provides users with the convenient ability to retrieve and analyze structural neighbors for a protein; do one-to-all calculation of a user provided structure against the entire current PDB release with T-Server, and pair-wise comparison using the TOPOFIT method through the T-Pair web page. All outputs are reported in various web-based tables and graphics, with automated viewing of the structure-sequence alignments in the Friend software package for complete, detailed analysis. T-DB presents researchers with the opportunity for comprehensive studies of the variability in proteins and is publicly available at http://mozart.bio.neu.edu/topofit/index.php.
INTRODUCTION
Protein structure comparison plays an essential role in understanding the similarities and differences between proteins, locating distantly related homologs/analogs, revealing functionality from similarity, and elucidating the often cryptic biological role in metabolic pathways. Protein comparison is both a complex and multidimensional problem, and while there are numerous structural alignment programs, structural comparison remains an active area of research. A review is outside of the scope of this paper and reviews can be found elsewhere.
Presently, there is a vast and rapidly growing quantity of publicly available protein structures, with the number of structures dramatically increasing since the advent of the Structural Genomics Initiative (1). Currently, one-to-all alignments to all other available structures must be calculated when a researcher is interested in a relation of a particular protein, be it published or private. The task becomes even more challenging when an analysis of all-to-all relations is required, for instance in structural classification or functional annotation. And while both computer speeds and heuristics have decreased the amount of time needed for such calculations, the sheer quantity of alignments (2 512 578 783) and size of data make such a calculation cumbersome (estimate based on 74 613 chains as of July 25, 2006). Therefore, there are needs to have pre-calculated datasets of structural alignments between representative protein structures available to researchers for quick and easy access by request from a public database.
Although currently there are several popular protein structural alignment databases (2–14) there is still no uniformly accepted gold standard; moreover the alignment methods behind the databases produce different results, for example, the FSSP and CE databases overlap in only 40% of the cases (15). Additionally, a recent study found the best alignment, coined ‘Best-of-All’ from a combination of six methods, is missed in 10–50% of the cases by many of the commonly used methods (16). Furthermore, the attempts to classify protein structures into hierarchical levels, albeit extremely functional and useful, do face an emerging alternative view in which protein space is not uniformly discrete, but more continuous and multidimensional (17). One such study has shown similarities between folds belonging to different levels of classification, bringing into question whether or not a fold designation should be viewed too rigidly (18). With this alternative point existing, along with the lack of support establishing ‘one’ structural alignment method as the most effective for all alignment pairs, we have developed TOPOFIT-DB (T-DB).
BACKGROUND OF THE DATABASE
T-DB is a public web-based relational database of structural alignments based on our recently developed TOPOFIT method (19). The approach in which TOPOFIT alignments are produced is considerably different from the alignments produced by other popular methods. The majority of methods attempt to balance between lower RMSD (root mean square deviation) and a higher number of aligned positions (Ne). The approach implemented in the TOPOFIT method employs a different strategy, one in which equivalent nearest neighbors are exploited instead of better fit. TOPOFIT identifies the largest group of residues which have the same neighbors in the same locations common in both compared structures, defined mathematically as a topological invariant. The nearest neighbors have been defined uniquely for each structure by simple, well known Delaunay tessellation (DT) (20). Therefore, TOPOFIT does not use any heuristics based on RMSD, gap penalty, or alignment length (Ne) or any combination of the three as input parameters to produce alignments. The procedure is reversed: first a saturation point in the spatial tessellation graph is detected (topomax point) and then the corresponding Cα atoms and corresponding values between the aligned structures are reported. Such an objective methodology provides unambiguous identification and separation of the structurally invariant parts from the variable parts by identifying a precise border between the two. Studying such conserved invariant regions often reveal functionally critical areas of conserved tertiary structure.
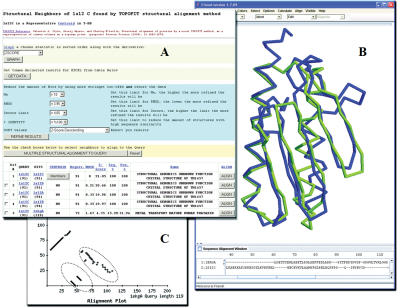
One of the intrinsic values of the TOPOFIT method is the ability to produce non-sequential alignments, from circular permutations to complex and completely reverse alignments. Many single examples of proteins with non-sequentially aligned regions have been reported (21,22); therefore, the ability to determine non-sequential alignments will permit more extensive analysis of core protein structure topology. TOPOFIT has been integrated into the Friend software (23) and is capable of reproducing and visualizing alignments stored in T-DB. To assist in the corroboration of a non-sequential alignment the user can visualize the corresponding alignment plot (Figure 1C), and display the structural superimposition in the Friend applet or stand alone application. Figure 1 displays the retrieved data from T-DB, along with the structural superimposition and the corresponding alignment plot from the alignment of Human Frataxin (PDB-code ‘1ekg’ chain A) and Hypothetical Protein TM1457 (PDB-code ‘1s12’ chain C). A precise structural match (RMSD < 2 Å) expands almost entirely over both polypeptide chains with the alignment consisting of four fragments, with three fragments aligned in reverse order. It should be mentioned, this structural relation is not present in existing structural alignments databases. TOPOFIT is capable of calculating non-sequential alignments since the method does not rely on backbone extension to produce structural alignments, i.e. segments do not have to be sequentially ordered.
Figure 1.
(A) Initial output page (web-browser) shown from a search using protein 1s12 chain C, all values from T-DB are shown in table format, by selecting the ‘ALIGN’ button, a new browser is opened showing the alignment plot, sequence alignment, and initializing the Friend Applet. (B) Friend Applet demonstrating the TOPOFIT superimposition (backbone representation) between Human Frataxin (PDB-code ‘1ekg’ chain A, blue) and Hypothetical Protein TM1457 (PDB-code ‘1s12’ chain C, green), Ne 74, RMSD 1.8 Å. (C) An example of a non-sequential alignment found by the TOPOFIT method, the alignment plot between 1ekg chain A (x-axis) and 1s12 chain C (y-axis) is shown, notice the three reverse fragments (dotted circles).
Using the TOPOFIT method, we have developed the TOPOFIT database (T-DB) for public use; along with T-Server for one-to-all comparisons with known structures from the PDB, and T-Pair for the comparison of any two protein structures. To provide users with an effective way to utilize the data from T-DB, the database has been linked to the Friend software package. This software package allows a user to conveniently and simultaneously visualize, and analyze multiple structural superimpositions and sequence alignments. The software package includes both an applet for straightforward online viewing of the structure-sequence alignments, and a stand-alone version, which must be locally installed.
DATABASE CONTENT
T-DB release (0.9) currently contains 86 033 950 TOPOFIT structural alignments, 66 161 PDB protein chains and 9209 representatives (centroids) from the PDB (24) (July 2005). The database has been initiated using the CE clusters of the structural neighbors from the CE database (February 2002 release) (generously provided by Dr I. Shindyalov). Protein structures inside each CE cluster have been aligned by TOPOFIT all-to-all and a new representative structure has been chosen by the criteria of maximum sum of Z-scores to all other structures in the cluster and named ‘centroid’. Thus, an initial set of centroids and their clusters have been created. All the centroids have been compared to each other by TOPOFIT and the clusters with ΔNe (<15 for up to 100 residues, and <30 for over 100 residues in protein length) between centroids have been joined together resulting in 3579 initial clusters.
A calculation pipeline using simple assignment procedure has been developed to automatically add new structures to the clusters. Each new structure has been compared to all other centroids using the same criteria as above: if a close relation to a centroid is present the structure was assigned to it, if not then a new cluster has been created and added to the list of centroids used for comparison there after, resulting in 9208 centroids. This stringent clustering has resulted in tight clusters, where all the members of the cluster are essentially the same, which is important when a search is initiated in T-DB, because centroids are used in all searching in an attempt to circumvent the high degree of redundancy found throughout the PDB. The calculations have been performed on two clusters: our in-house 20 node dual CPU 2.8 GHz cluster and NEU's 65 node dual 3.0 GHz processor cluster (http://opportunity.neu.edu/), approximately five CPU months of calculations were required to fill T-DB. All data has been placed into a dedicated MySQL server, with dual 2.8 GHz processors and 8 GB RAM, running on Fedora Core 5 × 64.
QUERYING THE DATABASE
T-DB has a query page with search parameters by PDB-code and chain identifier or by SCOP/ASTRAL (25) domain definitions. The results (Figure 1) of a search provide a list of structurally similar proteins initially sorted by decreasing Ne, which can be further restricted by number of alignments, by lower limit of Z-score, by upper limit of the output RMSD and by length difference between query and subject. All data is displayed inside a web-based hit-table with each structural neighbor in a single row. For each hit the table shows the hit number, PDB-codes of query and subject proteins along with lengths, parameters of the structural alignment (Ne, RMSD, Z-score, % sequence identity and positives), and the name of the subject protein. Optionally, additional values about topology correspondence between structures can be displayed. Finally, in an attempt to assist a user, vocabulary on the search and results pages has been linked to help content, which defines the terms to help aid in the analysis of the data.
After a search has been completed, there are several actions a user can conduct with the list of pre-computed structural neighbors (Figure 1). All individual alignments (hits) can be visualized over the web by using the ‘Align’ button in each row, resulting in the construction of an alignment plot, colored sequence alignment, and initialization of the Friend Applet (‘3D View’ button). The results of the search can be re-sorted by Ne, RMSD, and Z-score and hits can be removed by thresholds of Ne, RMSD, Z-score, and % identity. Results from the table can be saved as a comma delimited text format for spreadsheet analysis. A set of every numeric parameter in the table can be displayed in a graph, with the selected value in sorted order. The members in a centroid can be examined by selecting the ‘Members’ button in the Centroid column. And finally, a multiple alignment of selected protein chains can be produced. The alignment is represented in three ways: as a graph, as a text alignment with residues colored by biochemical properties and as a file in FASTA or SKY (Friend specific) formats. Along with sequence data, files in SKY format contain reference to PDB-chains corresponding to sequences and a scripting section describing structure manipulation. The locally installed version of Friend executes the script right after all sequence and corresponding structures are loaded into memory. As a result one gets the multiple structural superimpositions corresponding to the alignment with differentiated aligned and unaligned residues automatically displayed in the Friend software. The aligned areas indicate the invariant regions which can be visualized to facilitate in studies of regions which contribute to structure stability along with functionally important active/ligand binding site residues.
T-SERVER AND T-PAIR
T-Server is a web-based server allowing users to submit their structures (private or selected pieces, i.e. domains) to be compared to all currently available structures in the PDB. The user's structures are uploaded by browser, or selected from the PDB, and a link is provided to the user to check whether calculations have finished; optionally, the user can be notified by email about the calculation progress. Upon completing the one-to-all calculation the results can be downloaded from the T-Server web-page. T-Pair pairwise comparison server allows a user to align two structures, either identified by PDB-code and chain or uploaded, using the TOPOFIT method. Results are shown in similar fashion to the above ‘Align’ button, additionally a summary email can be sent to a provided email address.
Z-SCORE IMPROVEMENT
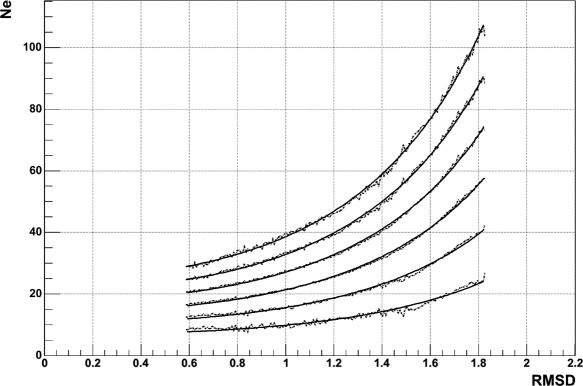
A large scale protein comparison was conducted which resulted in an improved Z-score value. The new Z-score was derived based on a more accurate description of the produced random distribution of Ne and RMSD. The same dataset of non-related proteins used in the TOPOFIT paper were used in this new analysis. The structures in the set were compared to produce the distribution of Ne and RMSD representing the random model. The distribution of Ne for each value of RMSD was approximated by Gaussian distribution with mean μ = μNe (RMSD) and σ = μNe (RMSD) depending on RMSD. The parameters μ and σ were obtained from the least-squares fit of the experimental distributions for each value of RMSD. For a given RMSD and Ne the Z-score was calculated as the deviation of Ne from the Gaussian average μ normalized to the Gaussian standard deviation σ. The dependences: μ, μ + σ, μ + 2σ, μ + 3σ, μ + 4σ, μ + 5σ were approximated by quadratic exponents (Figure 2) rather than by linear exponent as it was done in the original paper. Such an approximation allows for the calculation of Z-score analytically instead of tabulating values of μ and σ for every value of RMSD.
Figure 2.
Quadratic exponent fit (solid line) of dependencies μ, μ + σ, μ + 2σ, μ + 3σ, μ + 4σ, μ + 5σ (dashed line).
The comparison (data not shown) between the new and old way of Z-score estimation concluded that Z-score values are significantly different for RMSD < 0.5 Å and >1.5 Å, with new values being lower. This resulted in the assignment of a lower significance to protein alignments sharing similarities in only short secondary structure elements.
SUMMARY AND FUTURE WORK
Currently T-DB is useful for quick and effortless retrieval of statistically significant structural neighbors, along with automated viewing of T-DB's stored structural alignments in the Friend software package for complete, thorough analysis. Additionally, a user can use T-Server for the comparison of a structure not found in T-DB, against the entire PDB, or use T-Pair to align two structures; providing functional utilities for the study of recently determined protein structures of unknown function.
The pipeline for updating T-DB with TOPOFIT alignments for new structures has been developed, and new structures from the PDB will be compared and added once the calculations are completed. Future improvement will include: a deeper analysis between protein clusters, examination of the invariant cores in protein families, analysis of the variable regions across protein super-families, analysis of functional variations inside each cluster, downloadable similarity matrices within clusters, and graphically visualizing the distance between closely related clusters, allowing the users to browse from one cluster to neighboring clusters. TOPOFIT's non-parametric and objective way to separate the common part from the variable part, along with T-DB's accessibility will permit for these detailed studies to be shared with the scientific community.
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by Northeastern University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vitkup D., Melamud E., Moult J., Sander C. Completeness in structural genomics. Nature Struct. Biol. 2001;8:559–566. doi: 10.1038/88640. [DOI] [PubMed] [Google Scholar]
- 2.Shindyalov I.N., Bourne P.E. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 3.Holm L., Sander C. Dali/FSSP classification of three-dimensional protein folds. Nucleic Acids Res. 1997;25:231–234. doi: 10.1093/nar/25.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orengo C.A., Michie A.D., Jones S., Jones D.T., Swindells M.B., Thornton J.M. CATH—a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 5.Murzin A.G., Brenner S.E., Hubbard T., Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 6.Gough J., Karplus K., Hughey R., Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 7.Chen J., Anderson J.B., DeWeese-Scott C., Fedorova N.D., Geer L.Y., He S., Hurwitz D.I., Jackson J.D., Jacobs A.R., Lanczycki C.J., et al. MMDB: Entrez's 3D-structure database. Nucleic Acids Res. 2003;31:474–477. doi: 10.1093/nar/gkg086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye Y., Godzik A. FATCAT: a web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 2004;32:W582–W585. doi: 10.1093/nar/gkh430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krissinel E., Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 10.Lackner P., Koppensteiner W.A., Sippl M.J., Domingues F.S. ProSup: a refined tool for protein structure alignment. Protein Eng. 2000;13:745–752. doi: 10.1093/protein/13.11.745. [DOI] [PubMed] [Google Scholar]
- 11.Marti-Renom M.A., Ilyin V.A., Sali A. DBAli: a database of protein structure alignments. Bioinformatics. 2001;17:746–747. doi: 10.1093/bioinformatics/17.8.746. [DOI] [PubMed] [Google Scholar]
- 12.Balaji S., Sujatha S., Kumar S.S., Srinivasan N. PALI-a database of Phylogeny and ALIgnment of homologous protein structures. Nucleic Acids Res. 2001;29:61–65. doi: 10.1093/nar/29.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizuguchi K., Deane C.M., Blundell T.L., Overington J.P. HOMSTRAD: a database of protein structure alignments for homologous families. Protein Sci. 1998;7:2469–2471. doi: 10.1002/pro.5560071126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ortiz A.R., Strauss C.E., Olmea O. MAMMOTH (matching molecular models obtained from theory): an automated method for model comparison. Protein Sci. 2002;11:2606–2621. doi: 10.1110/ps.0215902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shindyalov I.N., Bourne P.E. An alternative view of protein fold space. Proteins. 2000;38:247–260. [PubMed] [Google Scholar]
- 16.Kolodny R., Koehl P., Levitt M. Comprehensive evaluation of protein structure alignment methods: scoring by geometric measures. J. Mol. Biol. 2005;346:1173–1188. doi: 10.1016/j.jmb.2004.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolodny R., Petrey D., Honig B. Protein structure comparison: implications for the nature of ‘fold space’, and structure and function prediction. Curr. Opin. Struct. Biol. 2006;16:393–398. doi: 10.1016/j.sbi.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Harrison A., Pearl F., Mott R., Thornton J., Orengo C. Quantifying the similarities within fold space. J. Mol. Biol. 2002;323:909–926. doi: 10.1016/s0022-2836(02)00992-0. [DOI] [PubMed] [Google Scholar]
- 19.Ilyin V.A., Abyzov A., Leslin C.M. Structural alignment of proteins by a novel TOPOFIT method, as a superimposition of common volumes at a topomax point. Protein Sci. 2004;13:1865–1874. doi: 10.1110/ps.04672604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voronoi G.F. Nouveles applications des parametres continus a la thorie des formes quadratiques. J. Reine Angew. Math. 1908;134:198–287. [Google Scholar]
- 21.Grishin N.V. Fold change in evolution of protein structures. J. Struct. Biol. 2001;134:167–185. doi: 10.1006/jsbi.2001.4335. [DOI] [PubMed] [Google Scholar]
- 22.Nagano N., Orengo C.A., Thornton J.M. One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol. 2002;321:741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- 23.Abyzov A., Errami M., Leslin C.M., Ilyin V.A. Friend, an integrated analytical front-end application for bioinformatics. Bioinformatics. 2005;21:3677–3678. doi: 10.1093/bioinformatics/bti602. [DOI] [PubMed] [Google Scholar]
- 24.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandonia J.M., Hon G., Walker N.S., Lo C.L., Koehl P., Levitt M., Brenner S.E. The ASTRAL Compendium in 2004. Nucleic Acids Res. 2004;32:D189–D192. doi: 10.1093/nar/gkh034. [DOI] [PMC free article] [PubMed] [Google Scholar]