Abstract
The role of WASP-interacting protein (WIP) in the process of F-actin assembly during chemotaxis of Dictyostelium was examined. Mutations of the WH1 domain of WASP led to a reduction in binding to WIPa, a newly identified homolog of mammalian WIP, a reduction of F-actin polymerization at the leading edge, and a reduction in chemotactic efficiency. WIPa localizes to sites of new pseudopod protrusion and colocalizes with WASP at the leading edge. WIPa increases F-actin elongation in vivo and in vitro in a WASP-dependent manner. WIPa translocates to the cortical membrane upon uniform cAMP stimulation in a time course that parallels F-actin polymerization. WIPa-overexpressing cells exhibit multiple microspike formation and defects in chemotactic efficiency due to frequent changes of direction. Reduced expression of WIPa by expressing a hairpin WIPa (hp WIPa) construct resulted in more polarized cells that exhibit a delayed response to a new chemoattractant source due to delayed extension of pseudopod toward the new gradient. These results suggest that WIPa is required for new pseudopod protrusion and prompt reorientation of cells toward a new gradient by initiating localized bursts of actin polymerization and/or elongation.
INTRODUCTION
Many eukaryotic cells undergo chemotaxis, directed movement toward a soluble ligand. This process is necessary for many biological functions, including the migration of macrophage during wound healing in vertebrates (Devreotes and Zigmond, 1988; Martinez-Quiles et al., 2001), axonal outgrowth to target cells (Korey and Van Vactor, 2000), and the aggregation of Dictyostelium cells to form a multicellular organism (Devreotes and Parent, 1999; Chung et al., 2001). Chemotaxis also plays a role in disease states such as arthritis, cancer, and multiple sclerosis (Bagiolini, 2001). Directional cell movement requires a defined cell polarity in which components of the cytoskeleton are differentially localized at the leading edge of a migrating cell as well as its retracting posterior (Firtel and Chung, 2000; Chung et al., 2001). This polarization can be initiated by the chemoattractant binding of heterotrimeric G protein–coupled membrane-bound receptors, and subsequent activation of downstream signal transduction pathways direct reorganization of the actin and myosin cytoskeleton (Chung et al., 2001; Van Haastert and Devreotes, 2004). A significant focus of current research involves the elucidation of the pathways that mediate both cell polarization and directional sensing, the result of which makes up a cell's proper chemotactic response.
Wiskott-Aldrich syndrome (WAS) is a human X-linked immunodeficiency characterized by recurrent infections, hematopoietic malignancies, eczema, and thrombocytopenia (Aldrich et al., 1954; Derry et al., 1994; Snapper et al., 1998). These defects are caused by mutations in the gene encoding the Wiskott-Aldrich syndrome protein (WASP), a key adaptor protein that connects multiple signaling pathways to F-actin polymerization, a process essential for chemotaxis. The WASP family of proteins includes WASP, N-WASP, and SCAR or WAVE (Miki et al., 1996; Bear et al., 1998). We have previously confirmed the necessity of WASP during the chemotactic motility of Dictyostelium via targeted knockdown of this gene (Myers et al., 2005). Lowered expression of WASP significantly impaired the ability of cells to polarize, polymerize F-actin, and migrate toward the chemoattractant cAMP. Both WASP and N-WASP possess a WH1 (WASP homology 1) domain, a basic (B) domain, a Cdc42/Rac binding (GBD) domain, a polyproline (SH3-binding) domain, a G-actin binding WH2 (WASP homology 2) domain (N-WASP has two WH2 domains), a central domain, C, and a C-terminal acidic, A, segment (Symons et al., 1996; Zigmond, 2000). The importance of the WH1 domain has been suggested by the frequency of mutations found to cause WAS (Rong and Vihinen, 2000). Despite comprising only one-fifth of WASP, more than half of the known missense mutations causing WAS are present in the WH1 domain (Derry et al., 1995). The WH1 domain has been implicated in the recruitment of WASP family members to sites of actin polymerization during motility of vaccinia virus (Moreau et al., 2000), WASP interacting protein (WIP)-induced membrane protrusions (Vetterkind et al., 2002), and PI(4,5)P2-induced intracellular vesicles (Benesch et al., 2002).
WIP has been shown to interact with human N-WASP, a ubiquitously expressed homolog, at the WH1 domain (Volkman et al., 2002). WIP is known to bind to both G- and F-actin (Martinez-Quiles et al., 2001). Depending on the system examined, WIP and other verprolin family members have been found to stimulate F-actin polymerization (Ramesh et al., 1997; Kinley et al., 2003), initiate filopodia formation (Martinez-Quiles et al., 2001; Vetterkind et al., 2002), and prevent F-actin depolymerization (Martinez-Quiles et al., 2001; Kato et al., 2002). WICH, a protein related to WIP, has been shown to induce microspike formation through cooperation with N-WASP (Kato et al., 2002). We have identified a homolog of mammalian WIP, WIPa, in Dictyostelium and examined the roles of WASP and WIP in the regulation of F-actin assembly and chemotaxis. The WH1 domain of WASP was found to bind WIPa. Overexpression of WIPa leads to an increase in microspike formation that is dependent on its WH2 domain, WASP, and VASP. WIPa translocates to the cortex of cells uniformly stimulated with cAMP in a time course that parallels F-actin polymerization. Lower levels of WIPa expression reduced prompt chemotactic responses of cells to a changing chemoattractant gradient. We conclude that WIPa is important for proper regulation of actin polymerization at sites of new pseudopod formation, which allows a chemotaxing cell the flexibility to respond to a spatially dynamic chemotactic gradient.
MATERIALS AND METHODS
Cell Culture and Development
Dictyostelium cells were cultured axenically in HL5 medium supplemented with 60 U of penicillin and 60 μg of streptomycin per ml. All cell lines shown were obtained using G418 to select stable-expressing clones. Equivalent protein expression levels were determined via quantitation of YFP signal in vivo using Metamorph software (Universal Imaging, West Chester, PA). For examining developmental phenotypes, cells were washed twice with 12 μM Na/K phosphate buffer and plated on nonnutrient agar plates.
Molecular Biology
WASP and WIP expression constructs were produced by PCR amplification of Dictyostelium cDNA library or genomic DNA and subcloned into the pEXP4(+) vector in frame with eYFP at the N-terminus separated by the flexible linker GSGSG. The YFP fusion proteins were expressed under control of the actin-15 promoter. Point mutants were produced by cloning genes into pBS, using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), and subcloned into pEXP4(+). All mutants were confirmed by sequence analysis. Protein expression constructs were produced using pGEX6P-1 (GST-tag) or pET-32a+ (6xHis-tag). Hairpin WIPa RNA construct was made by amplifying full-length WIPa and C-terminal half of WIPa and cloning them into pEXP4(+) in a tail to tail manner.
In Vitro Pulldown Assay
Protein expression constructs produced using pGEX6P1 or pET-32a+ were transformed into BL21 cells. Cultures were grown to a cell density of OD600 = 0.8, and 0.1 mM IPTG was added to induce protein expression for 1 h at 37°C. Proteins were purified according to the manufacturers' instructions (Sigma-GST proteins; Qiagen-His proteins). Equal quantities of recombinant protein were added to 500 μl TBS-T containing 60 μl of GST-tagged protein bound to glutathione-conjugated agarose beads and incubated at 4°C for 1 h. The agarose beads were pelleted by centrifugation at 14,000 rpm for 5 min and washed three times with 1 ml TBS-T. Protein was then stripped off the beads in 2× sample buffer by boiling for 4 min. Samples were resolved on a 12% SDS-PAGE gel, and proteins were identified via Western analysis using a mouse anti-6x His antibody (Cell Signaling, Beverly, MA). In vivo pulldown experiments were similarly performed, but agarose beads were incubated in 100 μl cell lysate, and proteins were identified using a mouse anti-GFP antibody.
In Vitro Actin Polymerization Assay
Experiments were performed as described by Zigmond, (1997). High-speed supernatants (HSS) were prepared by pulsing cells with 30 nM cAMP at 6-min intervals for 5 h, shaking with 3 mM caffeine for 30 min, lysing cells by passage through a 5-μm pore filter in PM buffer (12 mM Na/K phosphate buffer, pH 6.1, 2 mM MgSO4, plus 20 mM KCl and 1 mM EGTA), spinning for 20 min at 2.8e5 × g, and freezing in liquid nitrogen. One hundred nanomolar purified RacC was charged with either 100 μM GTPγS or 100 μM GDPβS for 10 min at 30°C. Supernatants were then stimulated at room temperature with the nucleotide-bound RacC at time 0. Aliquots of 50 μl were taken at each time point and stopped by dilution into 850 μl of actin buffer containing 6% formaldehyde, 0.15% Triton X-100, and 1 μM TRITC-phalloidin. Actin was stained for 1 h at room temperature and spun down at 14,000 rpm for 10 min. Rhodamine-phalloidin was extracted with 300 μl of 100% methanol, and fluorescence was measured (540 excitation/575 emission).
For visualization of actin filaments, supernatants were allowed to polymerize in the presence or absence of 0.4 μM phalloidin on a slide coverslip for 5 min at 37°C before stopping reactions with actin buffer containing formaldehyde and TRITC-phalloidin. Images were taken using fluorescent microscopy with a 60× objective lens and Metamorph software (Universal Imaging).
F-Actin Pulldown Assay
Experiments were performed the same as in vivo actin polymerization assays, except 100 μl of cells were mixed at each time point in Dictyostelium lysis buffer (25 mM Tris, pH 7.6, 100 mM NaCl, 1 mM EDTA, 1% NP-40, 10% glycerol, 1 mM DTT, 3 μM phalloidin). Lysates were spun down at 14,000 rpm for 10 min. Pellets were boiled in 2× loading buffer, resolved on 10% SDS-PAGE, and blotted onto nitrocellulose, and YFP-WIPa was detected using an anti-GFP antibody.
F-Actin Staining
For phalloidin staining, cells were pulsed with 30 nM cAMP at 6-min intervals for 5 h and placed on glass coverslips in 12 mM sodium phosphate buffer (pH 6.2) + 200 μM each of MgCl2 and CaCl2 for at least 30 min. Cells were fixed with 3.7% formaldehyde for 10 min. Cells were permeabilized with 0.5% Triton X-100, washed, and incubated with TRITC or Alexa594-phalloidin (Sigma) in PBS containing 0.5% BSA and 0.05% Tween-20 for 30 min. Cells were washed in PBS containing 0.5% Tween-20. Images were captured with a Roper Coolsnap camera (Tucson, AZ) and Metamorph software (Universal Imaging).
For labeling barbed-ends, cells were permeabilized with 100 mM PIPES, pH 6.9, 1% Triton X-100, 4% PEG, 1 mM EGTA, 1 mM MgCl2, 3 μM phalloidin for 3 min and 0.4 μM rhodamine-labeled G-actin in 1 μM ATP solution was added. After 5 min staining, cells were washed three times with PIPES buffer and fixed with 3.7% paraformaldehyde. For quantitation of leading edge F-actin content, the amount of F-actin at the leading edge of these cells was quantitated by measuring the integrated phalloidin signal (area × average intensity) at the leading edge divided by the average intensity of phalloidin staining throughout the cell body.
Chemotaxis Assay
Cells were pulsed with 30 nM cAMP at 6-min intervals for 5 h. These conditions maximally induce expression of aggregation-stage genes, including the cAMP receptor cAR1 and the coupled G protein α subunit Gα2. The chemotaxis assays were performed as previously described (Chung and Firtel, 1999; Meili et al., 1999). After pulsing, cells were plated on glass-bottomed microwell dishes (MarTek, Ashland, MA). A micropipette filled with 100 μM cAMP was positioned to stimulate cells by using a micromanipulator (Eppendorf, Hamburg, Germany), and the response and movement of cells were recorded by using Metamorph software (Universal Imaging; 1 image per every 5 s). Cell movement was examined by tracing the movement of a single cell in a stack of images.
Subcellular Localization and Fluorescent Microscopy
Cells were pulsed as normal and plated onto glass-bottomed microwell dishes. Cells were stimulated with 100 μM cAMP either using a micropipette or globally with an Eppendorf pipetman. YFP localization and translocation were recorded by using Metamorph software (Universal Imaging; 1 image per every 3 s) and a YFP emission filter. Quantitation of YFP signal was performed using line scan of lines transecting the cell body for each time point before and after stimulation.
RESULTS
WH1 Mutations Lead to Aberrant Regulation of Actin Polymerization and Chemotaxis
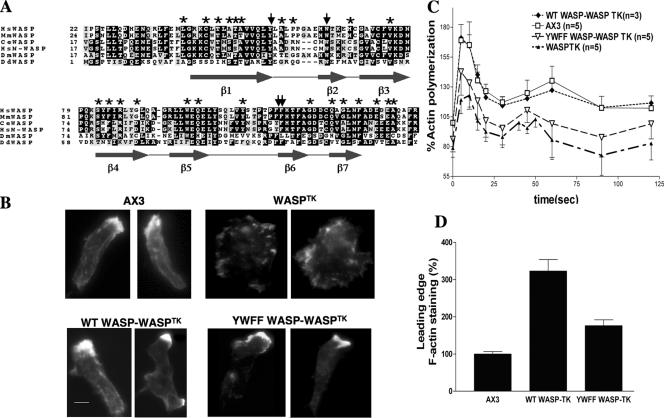
More than one-half of the known missense mutations causing WAS are found in the WH1 domain (Figure 1A). Though many of these mutations lead to lowered protein expression (Snapper and Rosen, 1999), some have been shown to affect WASP–WIP interaction (Stewart et al., 1999). A previous study identified an aromatic triad of residues within the EVH1 domain of Mena necessary for its interaction with a ligand containing the consensus binding sequence FPPPP (Prehoda et al., 1999). The WH1 domain of N-WASP has been shown to bind a proline-rich motif (DLPPPEP) of WIP similar to that recognized by the EVH1 domain of Mena (Volkman et al., 2002). Sequence alignment of the WH1 domain of Dictyostelium WASP with other EVH1/WH1-containing proteins revealed that the triad of residues is conserved (Figure 1A). To investigate the role of the WH1 domain in the function of WASP, we chose to mutate the three aromatic residues that correspond to the aromatic triad: tyrosine 33, tryptophan 39, and phenylalanine 91 (Figure 1A) to alanine. A fourth residue, phenylalanine 90, was mutated because of its proximity to F91. A construct for full-length WASP carrying all four mutations (WASPYWFF) was made. This construct and wild-type WASP were expressed in WASPTK cells. This strain has been shown to express very low levels of WASP and exhibits strong chemotaxis defects (Myers et al., 2005), although random movement appears not to be significantly affected (Table 1). Western analysis revealed equivalent expression of each protein, and a lack of degradation indicated that the aromatic residue mutations do not affect protein stability (unpublished data). Expression of either wild-type or mutant WASP rescued the ability of cells to polarize in response to cAMP pulsing; however, the WASP mutant exhibited a reduced ability to polymerize F-actin at the leading edge (Figure 1B). Figure 1D shows that the leading edge F-actin content of WASPTK cells expressing wild-type WASP (WTWASP) is threefold greater than that of wild-type cells alone, presumably because of ectopic overexpression of WASP. However, YWFFWASP mutant exhibit less than a twofold increase in F-actin accumulation over wild type. Consistent with this data, YWFFWASP-expressing WASPTK cells exhibited a reduced level of F-actin polymerization in response to global cAMP stimulation, indicating that the aromatic triad residues are required for full WASP function (Figure 1C). Chemotactic parameters such as mean velocity, angular deviation, and chemotactic index were measured. In each measurement, WASPTK cells expressing YWFFWASP exhibited an intermediate level of stimulation between that of WTWASP-expressing cells and WASPTK cells alone, indicating WH1 domain is required for the full function of WASP (Table 1).
Figure 1.
Conserved WH1 domain aromatic residues are important for WASP function. (A) Comparison of the amino acid sequence of DdWASP WH1 domain with other species: Hs, Homo sapiens; Mm, Mus musculus; Ce, C. elegans; Dm, Drosophila melanogaster; Dd, D. discoideum. Asterisks indicate residues with naturally occurring mutations in WAS patients. Arrows indicate residues mutated to alanine. Predicted secondary structure is indicated below sequence. (B) In vivo F-actin organization as determined by Alexa594-phalloidin staining. WH1 domain mutations (YWFF) reduce F-actin levels at leading edge of WASPTK cells rescued via ectopic WASP expression. Equivalent expression levels of WASP were determined via quantitation of YFP fluorescence and Western analysis. Bar, 5 μm. (C) In vivo actin polymerization assay. Full rescue of WASPTK phenotype by WASP overexpression requires WH1 aromatic residues. (D) Quantitation of F-actin at the leading edge. F-actin levels were determined by measuring the integrated Alexa594-phalloidin signal intensity (area × average intensity) at the leading edge divided by the average intensity of phalloidin staining throughout the cell body. A minimum of 100 cells of each type was taken from at least three independent experiments.
Table 1.
Chemotactic parameters
| Mean velocity (μm/min)a | Angular deviation (°)b | Chemotactic index (cosine)c | Cell number | |
|---|---|---|---|---|
| Strain | ||||
| AX3 wild type | 8.40 ± 2.10 | 42.91 ± 7.14 | 0.58 ± 0.19 | 53 |
| WTWIPa/AX3 | 6.21 ± 1.75d | 55.57 ± 8.82d | 0.35 ± 0.38d | 56 |
| K36AWIPa/AX3 | 8.58 ± 3.20 | 46.90 ± 7.35 | 0.42 ± 0.05 | 31 |
| WTWIPa/WASPTK | 1.23 ± 0.31d | 67.12 ± 7.38d | 0.03 ± 0.02d | 20 |
| hpWIPa | 9.65 ± 3.54d | 40.28 ± 7.21d | 0.66 ± 0.13d | 40 |
| WTWASP/WASPTK | 6.94 ± 2.92 | 54.92 ± 11.77 | 0.44 ± 0.21 | 40 |
| YWFFWASP/WASPTK | 4.51 ± 2.64d | 62.27 ± 11.15d | 0.10 ± 0.26d | 50 |
| WASPTK | 2.71 ± 1.10d | 77.06 ± 2.02d | −0.02 ± 0.08d | 25 |
| Random movement | ||||
| AX3 | 3.27 ± 0.71 | 75.67 ± 2.86 | nd | 20 |
| WASPTK | 2.90 ± 1.15 | 72.88 ± 10.5 | nd | 20 |
| WTWIPa/AX3 | 3.91 ± 0.48 | 69.07 ± 3.30 | nd | 12 |
| K36AWIPa/AX3 | 4.13 ± 0.55 | 66.6 ± 3.11 | nd | 12 |
nd, not determined.
a Mean velocity indicates speed of cell's centroid movement along the total path.
b Angular deviation indicates an average of the deviations of the angle of the path taken by cells from one frame to the next.
c Chemotaxis index is calculated as described in Futrelle et al. (1982). If the cell moves directly toward the gradient source it is 1, if directly away it is −1. If movements are indifferent to gradient (random movement), it is 0.
d p values were computed by Student's t test, and values below 0.05 were considered significant.
WIPa, a Dictyostelium Homolog of Mammalian WIP, Binds to the WHI Domain of WASP
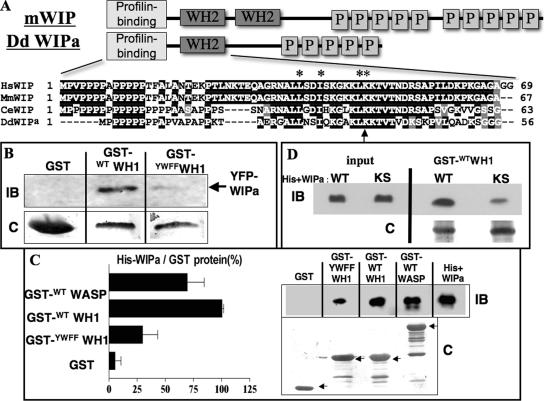
An attempt to identify homologues of WIP was made using the Dictyostelium genome database. A gene encoding a 197 amino acid protein exhibited the highest sequence homology to mammalian WIP and was designated WIPa (accession no. DDB0191055). A comparison of the derived amino acid sequence of Dictyostelium WIPa with WIP family members from various metazoans shows a moderately conserved primary sequence (32.2% identity with human WIP, 31.1% to human WICH, and 29.9% to rat CR16) and domain structure. WIPa is enriched in proline residues and has a conserved WASP Homology 2 domain (WH2) at its N-terminus (Figure 2A). This domain has previously been reported to bind G- and F-actin (Ramesh et al., 1997; Vaduva et al., 1997; Ho et al., 2001; Martinez-Quiles et al., 2001).
Figure 2.
DdWIPa is structurally and functionally similar to mammalian Wasp Interacting Protein (WIP). (A) Schematic representations of mammalian WIP and Dictyostelium WIPa. Sequence alignment of Wasp Homology 2 (WH2) domain shown with conserved residues in black. Residues essential for binding to G-actin are indicated with asterisks. Lysine 36 is indicated with an arrow. (B) YFP-WIPa was precipitated from AX3 cell lysates with either GST-fusion wild type or mutant WH1 bound to glutathione beads. Representative of three experiments is shown. IB, immunoblot; C, Coomassie staining. (C) Purified His-tagged WIPa was pulled down from solution using GST-WH1 or -WASP bound to beads. Representative of three experiments is shown. Each pulldown was quantitated and normalized to the quantity of GST-tagged protein used in the experiment. (D) Interaction between WH1 domain and WIPa depends partially on conserved residues K93 and S95 of WIPa. Purified His-tagged WT or mutant KS WIPa was pulled down from solution using GST-WH1. Three representative experiments are shown.
To determine whether WIPa interacts with the WH1 domain of WASP, we performed pulldown experiments with GST-WH1–bound glutathione beads incubated with the lysate of cells expressing YFP-WIPa. Figure 2B shows that WTWH1, but not YWFFWH1, pulls down WIPa from cell lysate. Coomassie staining of the membrane revealed equal amounts of the GST fusion proteins present. To confirm that this interaction was direct, pulldown experiments were performed using purified His-tagged WIPa. GST-WTWH1- and GST-WTWASP beads pulled down significantly more His-WIPa than GST-YWFFWH1 beads (Figure 2C).
We next attempted to identify the region of WIPa that interacts with the WH1 domain. Unlike human WIP, WIPa does not have a prominent WH1-binding domain (Volkman et al., 2002; Zetti and Way, 2002) at its C-terminus. One potential binding region lies in the middle of the protein. This region has preserved the “LPPPP”-binding motif, as well as conserved residues that have been suggested, using modeling data, to form electrostatic interactions with conserved charged residues within the WH1 domain (Volkman et al., 2002). We mutated two of these conserved residues (K93 and S95) to alanine and performed GST pulldown experiments. Figure 2D shows that the mutant WIPa (KSWIPa) exhibits a significant reduction in affinity to the WTWH1 domain. This reduction in binding averaged to 41.7 ± 8.9% of wild type over three independent experiments.
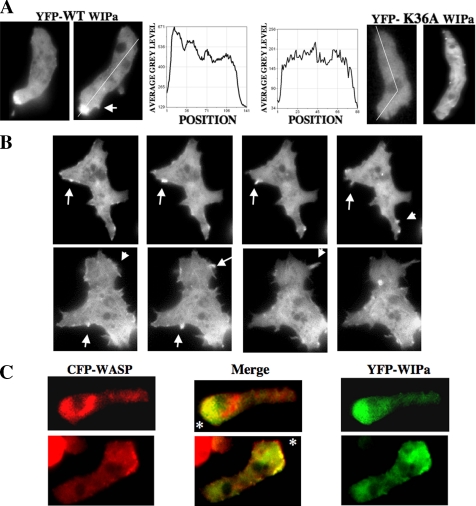
WIPa Is Enriched at the Leading Edge and Membrane Protrusions
We previously showed localization of WASP at the leading edge of chemotaxing cells (Myers et al., 2005). YFP-WIPa expressed in vivo also localized to the leading edge of polarized, chemotaxing cells (Figure 3A). YFP-WIPa colocalizes with CFP-WASP at the leading edge (Figure 3C), although they do not colocalize within intracellular compartments. WIPa is transiently enriched at membrane protrusions, which appears to be an early stage of pseudopod formation (Figure 3B). WIPa enrichment, however, seems to be lost during retraction of these protrusions, indicating that WIPa might be required for the onset of extension or maintenance of protrusion (Figure 3, A and B).
Figure 3.
WIPa localizes to leading edge, sites of membrane protrusion, and colocalizes with WASP. (A) WIPa localization in chemotaxing cells. YFP-tagged WIPa, but not actin-binding domain mutant K36A. WIPa, is enriched at the leading edge of polarized cells. (B) Transient WIPa localization at sites of membrane protrusion. Images were taken at 6-s intervals. (C) YFP-tagged WIPa colocalizes with CFP-tagged WASP at the leading edge of chemotaxing cells. Asterisk indicates location of the micropipette tip releasing 100 μM cAMP.
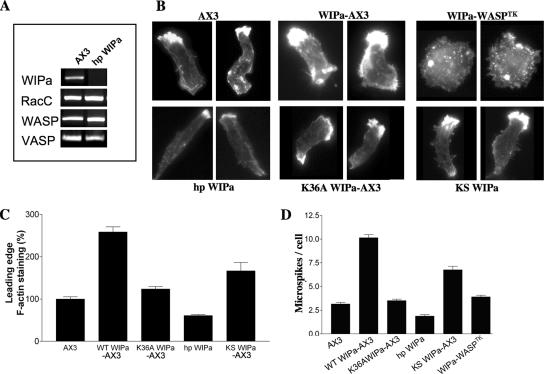
Reduced WIPa Expression Lowers Leading Edge F-actin and Microspike Formation
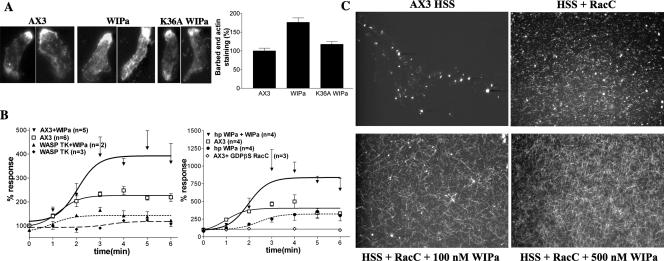
Alexa594-phalloidin was used to visualize the F-actin distribution in polarized cells expressing WIPa. Both the average steady state number of microspikes per cell, as well as the accumulation of F-actin staining at the leading edge, was significantly increased in cells overexpressing wild-type WIPa (Figure 4, B–D). WIPa expression in WASPTK cells did not stimulate microspike formation, indicating that WIPa binding to the WH1 domain is required to stimulate microspike formation (Figure 4, B and D). To investigate the role of the WASP Homology 2 (WH2) domain of WIPa in its activity, the lysine residue 36 was mutated to alanine. This residue is conserved among WH2 domain-containing proteins (Figure 2A) and has been shown to be critical for binding to G- or F-actin (Paunola et al., 2002). The K36AWIPa mutant lost its ability to localize to the leading edge of chemotaxing cells. Overexpression of K36AWIPa did not exhibit a significant increase in either microspike formation or F-actin accumulation (Figure 4, B–D), suggesting that actin binding is also required for WIPa activity.
Figure 4.
F-actin accumulation and filopodia formation are induced by WIPa. (A) Expression of hairpin WIPa hp WIPa construct reduces WIPa mRNA level. RT-PCR reactions were performed using mRNA purified from 5 h pulsed Dictyostelium cells. Expression of other genes essential for F-actin regulation was unaffected. (B) F-actin organization in cells expressing hpWIPa or WIPa mutants. At least 150 cells taken from three independent experiments were used for quantitation of leading edge F-actin content (C) and filopodia number (D). Quantitation of in vivo YFP signal revealed equivalent levels of WIPa expression in each cell type. WIPa-WASPTK refers to the overexpression of wild-type WIPa in the WASP TK background.
We used RNAi technology to reduce WIPa expression (Martens et al., 2002). A construct designed to express hairpin WIPa (hp WIPa) RNA was electroporated into AX3 cells to reduce endogenous WIPa expression. RT-PCR analysis revealed significant reduction in the expression of WIPa mRNA, but not the messages of unrelated genes involved in F-actin regulation such as WASP, RacC, and VASP (Figure 4A). Northern analysis showed similar results (unpublished data). Phalloidin staining of hp WIPa cells demonstrated moderately reduced F-actin accumulation at the leading edge; however, these cells are more polarized and elongated (average length 18 ± 0.49 μm) compared with wild type (14.83 ± 0.68 μm; Figure 4B). These cells exhibit a lower steady state number of microspikes along the cell body (Figure 4D), indicating that these cells may be defective in their ability to extend lateral membrane protrusions.
WIPa Facilitates F-actin Polymerization via Promoting Elongation of Actin Filaments
Increased level of F-actin and number of microspikes in cells overexpressing WIPa suggest that WIPa stimulates F-actin polymerization. To address more specifically the role of WIPa in actin regulation during membrane extension, the barbed ends of newly formed F-actin were labeled with TRITC-G actin in cells expressing WIPa. Figure 5A shows that wild-type WIPa, but not K36AWIPa, induces an almost twofold increase in leading edge barbed-end staining, suggesting that WIPa stimulates new actin filament formation. To support this data, in vitro F-actin polymerization assays were performed using HSS prepared from cAMP-pulsed Dictyostelium cells as described previously (Zigmond et al., 1997). GTP-bound RacC binds the GTPase-binding domain (GBD) domain of WASP and relieves its autoinhibitory interaction with the WASP acidic (A) domain (Han et al., 2006), presumably allowing WASP to activate the Arp2/3 complex. GTP-bound RacC initiated a roughly threefold stimulation of actin polymerization in wild-type supernatants (AX3), but not in supernatants prepared from WASPTK cells (Figure 5B). Consistent with Zigmond et al. (1997), this small G protein activation of actin polymerization depended on its binding to GTP, as the inactive GDP-bound conformation of RacC failed to stimulate polymerization (AX3+GDPβS RacC). Addition of 100 nM recombinant WIPa increased F-actin levels in wild-type supernatant (AX3+WIPa), but not in supernatants of WASPTK cells (WASPTK+WIPa). WIPa knockdown cell supernatants (hp WIPa) exhibited a decrease and delay in the polymerization of F-actin relative to wild-type supernatant. Addition of 100 nM recombinant WIPa to hp WIPa supernatants (hp WIPa+WIPa) rescued this defect (Figure 5B). To visualize assembled F-actin, these experiments were repeated on glass coverslips and viewed with fluorescent microscopy. RacC appears to increase the number of F-actin nucleation sites, presumably via activation of WASP and the Arp2/3 complex (Figure 5C). Addition of recombinant WIPa increases the length of these actin filaments (HSS+RacC + 100 nM WIPa), suggesting that WIPa promotes the elongation of actin filaments. We considered the possibility that WIPa stabilizes F-actin by preventing depolymerization of F-actin. If this were true, addition of WIPa in the presence of phalloidin stabilizing F-actin would be unlikely to lead to a further increase in the length of actin filaments. These reactions were repeated in the presence of 0.4 μM unlabeled phalloidin. The same WIPa-induced increase in the length of actin filaments was observed (unpublished data), suggesting that WIPa's activity does not depend solely on prevention of F-actin depolymerization. WIPa addition to the supernatant in the absence of GTPγS-charged RacC does not stimulate filament formation, indicating WIPa alone does not stimulate nucleation of F-actin (unpublished data).
Figure 5.
WIPa stimulates de novo F-actin polymerization. (A) Barbed-end staining of cells expressing wild-type WIPa or mutant (K36A WIPa). At least 60 cells taken from three independent experiments were used for quantitation. (B) In vitro F-actin polymerization assay. Supernatants of either wild-type AX3, WASPTK, or hp WIPa cells were stimulated with GTPγS-bound RacC at time 0 (or with GDPβS-bound RacC as indicated ◇). WIPa, 100 nM, was added 15–30 s after RacC addition where indicated. Reactions were stopped at the indicated time points with actin buffer containing TRITC-phalloidin, and F-actin was pelleted by centrifugation. The amount of F-actin at time 0 was standardized as 100%. (C) Visualization of polymerized F-actin. Supernatants were incubated with or without 100 nM GTPγS-charged RacC or WIPa for 5 min on a poly-l-lysine–treated glass coverslip and then fixed.
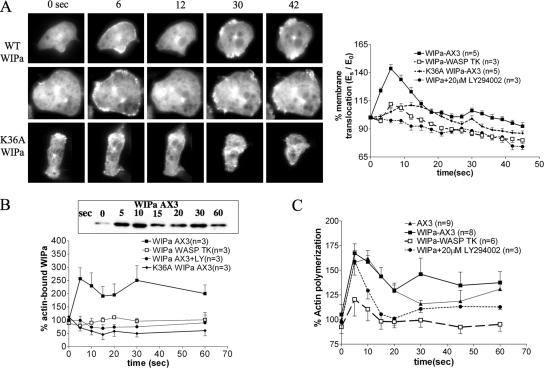
WIPa Translocates to the Cell Cortex upon cAMP Stimulation
Because F-actin polymerization and subsequent membrane protrusion and chemotaxis are processes downstream of cAMP receptor activation in Dictyostelium, WIPa localization was monitored before and after uniform stimulation of cells with cAMP (Figure 6A). A biphasic, cortical translocation of the F-actin marker coronin and the PHCRAC domain, a marker for PI(3,4,5)P3/PI(3,4)P2, has been observed in response to uniform cAMP stimulation previously (Chen et al., 2003). Similarly, two phases of WIPa translocation to the cell cortex after cAMP stimulation were observed. The first phase of translocation appeared to be more uniform along the cortex and peaked after 6 s (Figure 6A). In contrast, the smaller second peak of WIPa translocation is less uniform and more localized to sites of membrane protrusions as PHCRAC domain does. WIPa translocation appears to require its WH2 domain, as K36AWIPa showed a significantly lower level of translocation (Figure 6A). This is consistent with the inability of this mutant to localize to the F-actin–enriched leading edge of chemotaxing cells (Figure 3A). Similarly, WIPa expressed in WASPTK cells exhibited a reduced ability to translocate to the cortex. These data suggest that new F-actin polymerization at the cell cortex drives WIPa translocation. To confirm this, we determined the amount of WIPa that cosedimented with the detergent-insoluble F-actin pellet after centrifugation of lysate from cells stimulated with cAMP. Consistently, two peaks of WIPa translocation to F-actin pellet were observed. However, either mutation of the WIPa WH2 domain or WASP deletion significantly reduced the levels of WIPa in the F-actin pellet (Figure 6B).
Figure 6.
WIPa translocates to the cell cortex upon global cAMP stimulation. (A) Images of cells expressing YFP-WT WIPa or K36A mutant globally stimulated with 100 μM cAMP are shown. Data points were taken at 3-s intervals. Two linescan measurements from at least 24 cells of each cell type were used for quantitation of YFP intensity at the membrane. (B) F-actin pulldown of YFP-WIPa. WIPa interaction with F-actin depends on WH2 domain and PI3 kinase activity. (C) In vivo F-actin polymerization assay. WT WIPa overexpression leads to an increase in actin polymerization 30 s after global cAMP stimulation.
We then measured the effects of WIPa overexpression on the polymerization of F-actin in response to uniform stimulation of cAMP. Interestingly, WIPa leads to a significant increase in F-actin polymerization 30 s after cAMP stimulation, presumably at sites of membrane protrusions, whereas it does not significantly increase the first peak of F-actin polymerization at 5–10 s. This WIPa-induced increase in actin polymerization is dependent on endogenous WASP expression (Figure 6C). PI3 kinase activity also appears to be necessary for WIPa cortical translocation, as LY294002, a PI3 kinase inhibitor, but not vehicle alone (unpublished data) eliminated translocation (Figure 6, A and B). This appears to occur via a mechanism independent of actin polymerization, because LY294002 did not significantly hamper the peak increase in actin polymerization (Figure 6C).
WIPa Is Important for the Ability of Cells to Promptly Respond to a New Gradient
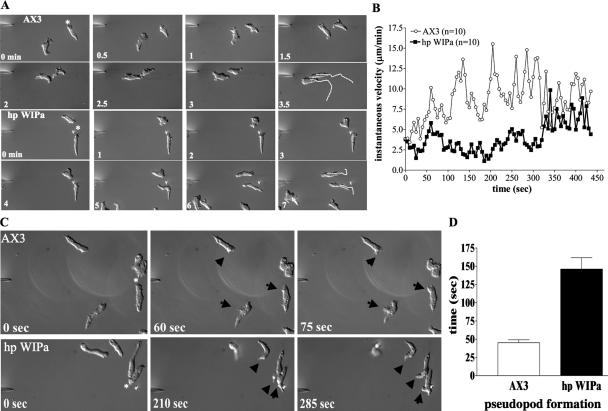
To determine the role of WIPa in controlling cell motility, we performed micropipette chemotaxis assays (Chung and Firtel, 1999). Polarized cells were plated on a glass slide and allowed to chemotax toward a cAMP-releasing micropipette for at least 5 min before taking images. Table 1 shows that WIPa overexpression reduces the mean velocity and chemotactic index of cells migrating toward the micropipette, whereas angular deviation is significantly increased, indicating these cells make more directional changes presumably due to the increase of microspike/pseudopod protrusion. These chemotactic defects are not seen in cells overexpressing K36AWIPa (Table 1). Reduction of endogenous WIPa message, however, appears to have little effect on chemotaxis efficiency. Cells expressing the hp WIPa construct are well polarized and exhibit little change in mean velocity and chemotactic index when migrating in a single direction toward a stably localized micropipette (Table 1), suggesting that WIPa may not be essential for the stabilization or maintenance of a mature leading edge.
Because WIPa overexpression caused frequent directional changes, we examined whether WIPa might be responsible for the ability of a cell to sense and respond to changes in the chemoattractant gradient, the micropipette was moved to a new location during chemotaxis. The instantaneous velocity of cells was measured immediately after the pipette was shifted to a location approximately perpendicular to the cell. Figure 7A shows that wild-type cells redirect themselves toward the new chemoattractant source quickly. However, hp WIPa cells exhibit a delayed response to the new direction. Although wild-type cells resume normal velocity in <60 s, hp WIPa cells took more than 300 s to resume normal speed (Figure 7B). We speculated that the delayed response might be due to a delay in making a new pseudopod toward the new directional cue. To test this, we measured the time required for each cell type to initiate pseudopod protrusion, in clear response to the new chemoattractant source, which eventually developed into the new leading edge (Figure 7C). The average time required for new pseudopod formation toward the new cAMP source was 48 s for wild-type cells, whereas hp WIPa cells took 145 s (Figure 7D). These results suggest that WIPa stimulates lateral membrane protrusion and prompt formation of a new pseudopod when cells respond to a change in the direction of a chemoattractant source.
Figure 7.
WIPa is necessary for prompt response to a changing chemoattractant gradient. (A) Pulsed cells were allowed to chemotax toward a cAMP-releasing micropipette (*) for ∼5 min. Time 0 indicates the first frame after which the micropipette was moved to its new position. Time-lapsed images were taken at 5 s intervals for a total of ∼90 frames. Chemotactic parameters of each cell type were averaged from three independent experiments (AX3 = 35 cells total; hp WIPa = 38 cells total). (B) Instantaneous velocity of each cell type after micropipette shift as tracked by Metamorph. Averages of 10 cells of each type from three independent experiments are shown. (C) hp WIPa cells require more time to initiate membrane protrusion and subsequently pseudopodia formation, toward the direction of the new gradient. (D) The average time required for forming new pseudopodia toward the new cAMP source.
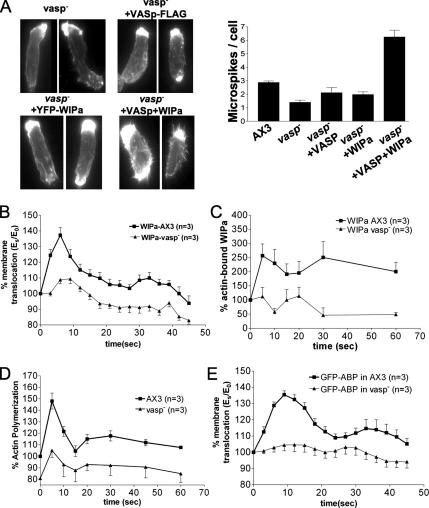
VASP Is Important for WIPa-induced Microspike Formation
Vasodilator-stimulated phosphoprotein (VASP) is a member of the Ena/VASP family, and targeted knockout of this gene in Dictyostelium was shown to reduce filopodia/microspike formation (Han et al., 2002; Schirenbeck et al., 2006). It is of great interest to determine if both WIPa and VASP are required for microspike formation. We expressed WIPa in a vasp null cell line. F-actin was stained to determine the steady state level of microspikes in each cell type. Consistent with a previous study (Han et al., 2002), vasp null cells showed a significantly lower number of microspikes. Overexpression of VASP without a membrane-targeting myristoylation tag does not significantly increase the number of fliopodia/microspikes per cell. vasp− cells overexpressing WIPa failed to exhibit an increase in the steady state level of microspikes as seen in the wild-type background (Figure 8A). Coexpression of VASP and WIPa in vasp null cells restored increased microspike formation, suggesting that VASP might be required for microspike formation by WIPa.
Figure 8.
VASP is required for WIPa-induced microspike formation and membrane translocation. (A) Overexpression of WIPa fails to significantly increase the steady state level of microspikes in a vasp null background. The number of microspikes from at least 50 cells was counted in each of three independent experiments. (B) YFP-WIPa translocation. Significantly less WIPa translocates to the cell cortex upon cAMP stimulation in the absence of VASP. (C) F-actin pulldown of YFP-WIPa is decreased in VASP null cells. (D) In vivo F-actin polymerization in response to cAMP is defective in VASP-null cells. (E) Cortical translocation of the F-actin marker GFP-ABP in response to global cAMP stimulation is defective in VASP null cells.
As VASP has been shown to translocate to the cell cortex upon global cAMP stimulation and membrane targeting appears to be required for filopodia formation (Han et al., 2002), we considered the possibility that VASP may be involved in the translocation of WIPa to the cortex where it could then initiate microspike formation. Figure 8B shows a reduced WIPa translocation in the vasp null background, raising the possibility that a lack of WIPa at the plasma membrane partially explains the lack of microspikes in the vasp null background. VASP has been shown to nucleate and bundle actin filaments in vitro (Schirenbeck et al., 2006), and as we observed a dependence of WIPa translocation upon F-actin, we measured the actin polymerization response of VASP null cells to cAMP. An increase in whole cell levels of F-actin in response to cAMP was diminished in vasp null cells (Figure 8D). Consistently, expression of the F-actin–binding protein ABP in vasp null cells revealed a reduced level of actin accumulation at the cell cortex (Figure 8E). Significantly less WIPa was pelleted with F-actin after cAMP stimulation of vasp null cells, presumably due to a decrease in levels of actin polymerized and/or bundled (Figure 8C). These data suggest that, like WASP, VASP plays a role in the actin polymerization response to global cAMP. Diminished cortical actin levels in these cells lead to a decrease in the translocation of WIPa to the plasma membrane, where it might normally facilitate microspike formation with VASP.
DISCUSSION
Regulation of the actin cytoskeleton is a key event for directed cell migration. Our study suggests a role for WIPa in the regulation of the actin cytoskeleton and motility during chemotaxis. WIPa plays a role in lateral membrane protrusion and pseudopod extension that is important for prompt response to a dynamic chemoattractant gradient, whereas WASP appears to be responsible for cell polarity and the majority of actin polymerization necessary to establish the bulk protrusive force against the leading edge plasma membrane.
Our data suggests that WIPa stimulates microspike formation and lateral membrane protrusion in a polarized cell. In these processes, WIPa may facilitate elongation of actin filaments already nucleated by WASP's activation of the Arp2/3 complex as shown in our in vitro polymerization assay. It is also possible that WIPa may provide WASP with an additional actin monomer during Arp2/3 complex activation and enhances its rate of filament assembly. The number of WH2 domains in a complex of proteins including the Arp2/3 complex may determine the degree to which the complex enhances actin polymerization. WIP enhances F-actin polymerization of cortactin, which has no WH2 domain (Kinley et al., 2003). Similarly, WASP, which has one WH2 domain and is known to be a better Arp2/3 activator than cortactin, is also stimulated by WIP in a manner that depends on WIP's own WH2 domain (Ramesh et al., 1997). Consistently, the VCA domain of N-WASP, which possesses two WH2 domains, was found to be a better activator of the Arp2/3 complex and microspike formation than VCA domains possessing a single WH2 domain (Yamaguchi et al., 2000).
NMR spectroscopy and computer modeling studies have provided insight into the pathogenesis of WAS by revealing structural details of the WASP (or N-WASP) WH1 interaction with WIP (Rong and Vihinen, 2000; Volkman et al., 2002; Kim et al., 2004). Consistently, we showed that Dictyostelium WIPa binds to the WASP WH1 domain and that the aromatic triad residues are essential for this interaction. We identified a potential WH1-binding region of WIPa that lies within the middle of the protein. This region has preserved a polyproline binding motif, as well as conserved residues that have been suggested to form electrostatic interactions with conserved aspartic acid residues within the WH1 domain (Volkman et al., 2002). Mutation of two of these residues reduced the affinity of WIPa for the WH1 domain. This region, however, is oriented in a C- to N-terminal manner. As proline-rich binding modules like the WH1 domain are known to isoenergetically dock with PPII ligands in two possible orientations due to the twofold rotational pseudosymmetry of the ligand, it is plausible that this region of WIPa interacts with the WH1 domain of WASP.
When cAMP-pulsed Dictyostelium cells are globally stimulated with chemoattractant, two distinct phases of actin polymerization are triggered. Both phases have been shown to correspond to a translocation of an F-actin marker and a PI(3,4,5)P3 marker to the cell cortex (Chen et al., 2003). Cytosolic WIPa also appears to translocate to the cortical membrane in two distinct phases that parallel this actin polymerization. The reduction in translocation caused by the K36A mutation would imply that WIPa translocation is partially mediated by a WH2-F-actin interaction. Interestingly, the second phase of translocation of WIPa is localized to sites of pseudopod/microspike formation. Overexpression of WIPa would result in increased WIPa translocation to the cortex, leading to more F-actin assembly (Figure 6C) and more lateral membrane protrusion and thus lower chemotactic efficiency (Table 1). WIPa translocation to the membrane also appears to require PI3 kinase activity, suggesting that PI(3,4,5)P3 might trigger WIPa translocation. We do not fully understand how this translocation is dependent on PI3 kinase yet. In our previous study, WASP localization was significantly changed upon the inhibition of PI3 kinase by LY294002 (Myers et al., 2005). Sequestration of WIPa by WASP in LY294002-treated cells might result in defective translocation of WIPa to the cortical membrane. We suggest that upon cAMP stimulation, membrane-bound WASP stimulates actin polymerization at the cell cortex presumably via activation by PI(3,4,5)P3 and RacC. The polymerized actin recruits cytosolic WIPa to the cortex, where it facilitates elongation/polymerization of F-actin with WASP, resulting in membrane extension and protrusion. Cell cortical actin accumulation in response to cAMP also appears to require VASP, perhaps via its nucleation or bundling activity (Schirenbeck et al., 2006). A loss of this actin accumulation in VASP null cells appears to lead to a reduction in WIPa translocation similar to that seen in WASPTK cells. The reduction in cortical WIPa may be responsible for the reduced ability of WIPa overexpression to increase the steady state level of microspikes in the absence of VASP.
We have provided evidence that WIPa regulates chemotaxis. Although WIPa overexpression increases microspike formation, decreases speed, and increases angular deviation of a cell toward a chemoattractant source, reduction of WIPa expression decreases microspike formation, slightly improves the chemotactic index, but decreases the ability of cells to reorient toward a new chemoattractant source. The effects of the Dictyostelium formin dDia2 on cell migration are similar (Schirenbeck et al., 2005). Overexpression of dDia2 increases filopodium formation and leads to a decrease in cell motility. Deletion of the dDia2 gene resulted in cells with fewer filopodia but higher cell velocity. Because WIPa stimulates onset of early pseudopod at the plasma membrane, WIPa may allow cells to respond to new gradient promptly by initiating localized bursts of actin polymerization and/or elongation, whereas WASP may provide the main driving force of lamellipodia protrusion by stimulation of F-actin polymerization at the leading edge. The severe chemotactic defects exhibited by the WH1 domain WASP mutant and the lack of significant defects of WIPa knockdown cells during chemotaxis toward a single direction would imply that another polyproline-containing protein might participate in the stabilization or maintenance of WASP-generated actin assembly during lamellipodia protrusion at the leading edge. We have shown in vitro that WIPa increases the length of RacC-induced, WASP-dependent actin filaments and, in vivo, WIPa localizes to sites of newly forming pseudopodia on the cortical membrane. When a chemotaxing cell senses a change in direction in cAMP input, a localized increase in WIPa activity might provide the cytoskeletal framework for the formation of a new pseudopod and, eventually, a new leading edge by facilitating actin polymerization. A decrease in WIPa expression, then, would result in cells with a reduced ability to change direction promptly upon change of chemoattractant gradient due to delayed formation of pseudopodia/microspikes. It is likely that chemotaxis of these cells already committed toward a single chemoattractant source would not be altered or even faster due to less angular deviation. An increase in WIPa expression would produce more lateral pseudopodia resulting in a corresponding increase in the number of turns, and thus a greater angular deviation. A role for both WIP and WASP in Dictyostelium chemotaxis is consistent with the severe impairment of T-cell homing to spleen and lymph nodes from WASP−/−WIP−/− double knockout mice (Gallego et al., 2006).
A species-specific difference, however, between the importance of the WASP family of proteins may exist. Although lamellipodia and filopodia formation appear to be more dependent on SCAR in Drosophila BG2 cells (Biyasheva et al., 2004) and WAVE2 in mammalian cells (Yan et al., 2003), WASP deficiency appears to have a greater effect on membrane protrusion and actin assembly than SCAR deficiency in Dictyostelium. SCAR deficiency does lead to decreased actin assembly at the leading edge (Bear et al., 1998), but these cells polarize well and form visible streams, unlike WASPTK cells, suggesting the possibility of rather normal chemotactic movement despite abnormal F-actin organization. It has been suggested that SCAR is not required for the generation of actin protrusions, but may be more involved in extending and reshaping them (Blagg et al., 2004). This would imply the evolutionary acquisition of WASP-dependent roles of actin assembly during chemotaxis by WAVE/SCAR family members sometime after the divergence of the protozoans. Alternatively, it is conceivable that WASP function is more essential in highly motile cells such as Dictyostelium, neutrophils, and macrophage to provide highly dynamic regulation of the actin cytoskeleton than in slower moving cells like fibroblasts. It has been demonstrated that the VCA domain of SCAR shows different potency in stimulating Arp2/3 than that of WASP or N-WASP, at least in in vitro systems. The VCA domain of N-WASP and WASP induce rapid actin polymerization, whereas SCAR1 induces the slowest rate of nucleation (Zalevsky et al., 2001). It is conceivable to expect WASP or N-WASP to be the primary regulators of dynamic F-actin assembly. Thus, lack of WASP function might cause more severe phenotypes in highly motile cells.
In conclusion, WIPa translocates from the cytoplasm to the plasma membrane upon cAMP stimulation. The regulation of the F-actin cytoskeleton by WIPa at sites of membrane protrusion and pseudopod formation appears necessary to allow chemotaxing cells the flexibility of responding to new sources of chemoattractant and to prevent premature commitment to a single chemoattractant source.
ACKNOWLEDGMENTS
We thank Dr. Alissa Weaver and Scott Gruver for useful discussions and critical reading of the manuscript. We also thank Jihyun Moon for excellent technical assistance. This work was supported, in part, by grants from National Institutes of Health (GM68097 to C.Y.C.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-05-0376) on August 9, 2006.
REFERENCES
- Aldrich R. A., Steinberg A. G., Campbell D. C. Pedigree demonstrating a sex linked recessive condition characterized by draining ears, eczematoid dermatitis, and bloody diarrhea. Pediatrics. 1954;13:133–139. [PubMed] [Google Scholar]
- Bagiolini M. Chemokines in pathology and medicine. J. Intern. Med. 2001;250:91–104. doi: 10.1046/j.1365-2796.2001.00867.x. [DOI] [PubMed] [Google Scholar]
- Bear J. E., Rawls J. F., Saxe C. L. SCAR, a WASP-related protein, isolated as a suppressor of receptor defects in late Dictyostelium development. J. Cell Biol. 1998;142:1325–1335. doi: 10.1083/jcb.142.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benesch S., Lommel S., Steffen A., Stradal T. E., Scaplehorn N., Way M., Wehland J., Rottner K. Phosphatidylinositol 4,5-Biphosphate (PIP2)-induced vesicle movement depends on N-WASP and involves Nck, WIP, and Grb2. J. Biol. Chem. 2002;277:37771–37776. doi: 10.1074/jbc.M204145200. [DOI] [PubMed] [Google Scholar]
- Biyasheva A., Svitkina T., Kunda P., Baum B., Borisy G. Cascade pathway of filopodia formation downstream of SCAR. J. Cell Sci. 2004;117:837–848. doi: 10.1242/jcs.00921. [DOI] [PubMed] [Google Scholar]
- Blagg S. L., Stewart M., Sambles C., Insall R. H. PIR121 regulates pseudopod dynamics and SCAR activity in Dictyostelium. Curr. Biol. 2003;13:1480–1487. doi: 10.1016/s0960-9822(03)00580-3. [DOI] [PubMed] [Google Scholar]
- Blagg S. L., Insall R. H., et al. Solving the WAVE function. Nat. Cell Biol. 2004;6:279–281. doi: 10.1038/ncb0404-279. [DOI] [PubMed] [Google Scholar]
- Chen L., Janetopoulos C., Huang Y. E., Iijima M., Borleis J., Devreotes P. N. Two phases of actin polymerization display different dependencies on PI(3,4,5)P3 accumulation and have unique roles during chemotaxis. Mol. Biol. Cell. 2003;14:5028–5037. doi: 10.1091/mbc.E03-05-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C. Y., Funamoto S., Firtel R. A. Signaling pathways controlling cell polarity and chemotaxis. Trends Biochem. Sci. 2001;26:557–566. doi: 10.1016/s0968-0004(01)01934-x. [DOI] [PubMed] [Google Scholar]
- Chung C. Y., Firtel R. A. PAKa, a putative PAK family member, is required for cytokinesis and the regulation of the cytoskeleton in Dictyostelium discoideum cells during chemotaxis. J. Cell Biol. 1999;147:559–575. doi: 10.1083/jcb.147.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derry J. M., Kerns J. A., Weinberg K. I., Ochs H. D., Volpini V., Estivill X., Walker A. P., Francke U. WASP gene mutations in Wiskott-Aldrich syndrome and X-linked thrombocytopenia. Hum. Mol. Genet. 1995;4:1127–1135. doi: 10.1093/hmg/4.7.1127. [DOI] [PubMed] [Google Scholar]
- Devreotes P. N., Zigmond S. H. Chemotaxis in eukaryotic cells: a focus on leukocytes and Dictyostelium. Annu. Rev. Cell Biol. 1988;4:649–686. doi: 10.1146/annurev.cb.04.110188.003245. [DOI] [PubMed] [Google Scholar]
- Devreotes P., Parent O. A cell's sense of direction. Science. 1999;284:765–770. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- Firtel R. A., Chung C. Y. The molecular genetics of chemotaxis: sensing and responding to chemoattractant gradients. BioEssays. 2000;22:603–615. doi: 10.1002/1521-1878(200007)22:7<603::AID-BIES3>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Futrelle R. P., Traut F. J., McKee W. G. Cell behavior in Dictyostelium discoideum: preaggregation response to localized cAMP pulses. J. Biol. Chem. 1982;92:807–821. doi: 10.1083/jcb.92.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M. D., Fuente M. A., Anton I. M., Snapper S., Fuhlbrigge R., Geha R. S. WIP and WASP play complementary roles in T cell homing and chemotaxis to SDF-1a. Int. Immun. 2006;18:221–232. doi: 10.1093/intimm/dxh310. [DOI] [PubMed] [Google Scholar]
- Han Y., Chung C. Y., Wessels D., Stephens S., Titus M. A., Soll D. R., Firtel R. Requirement of a vasodilator-stimulated phosphoprotein family member for cell adhesion, the formation of filopodia, and chemotaxis in Dictyostelium. J. Biol. Chem. 2002;277:49877–49887. doi: 10.1074/jbc.M209107200. [DOI] [PubMed] [Google Scholar]
- Han J. W., Leeper L., Rivero F., Chung C. Y. Role of RacC for the regulation of WASP activity and PI3 kinase localization during chemotaxis of Dictyostelium. J. Biol. Chem. 2006 doi: 10.1074/jbc.M605997200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho H. H., Rohatgi R., Ma L., Kirschner M. W. CR16 forms a complex with N-WASP in brain and is a novel member of a conserved proline-rich actin-binding protein family. Proc. Natl. Acad. Sci. USA. 2001;98:11306–11311. doi: 10.1073/pnas.211420498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M., Sun Y., Drubin D. G. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Kato M., Miki H., Kurita S., Endo T., Nakagawa H., Miyamoto S., Takenawa T. WICH, a novel verprolin homology domain-containing protein that functions cooperatively with N-WASP in actin-microspike formation. Biochem. Biophys. Res. Commun. 2002;291:41–47. doi: 10.1006/bbrc.2002.6406. [DOI] [PubMed] [Google Scholar]
- Kim M. K., Kim K. S., Kim D. S., Choi I. H., Moon T., Yoon C. N., Shin J. Two novel mutations of Wiskott-Aldrich syndrome: the molecular prediction of interaction between the mutated WASP L101P with WASP-interacting protein by molecular modeling. Biochim. Biophys. Acta. 2004;14:134–140. doi: 10.1016/j.bbadis.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Kinley A. W., Weed S. A., Weaver A. M., Karginov A. V., Bissonette E., Cooper J. A., Parsons J. T. Cortactin interacts with WIP in regulating Arp2/3 activation and membrane protrusion. Curr. Biol. 2003;13:384–393. doi: 10.1016/s0960-9822(03)00107-6. [DOI] [PubMed] [Google Scholar]
- Korey C. A., Van Vactor D. From the growth cone surface to the cytoskeleton: one journey, many paths. J. Neurobiol. 2000;44:184–193. [PubMed] [Google Scholar]
- Martens H., Novotny J., Oberstrass J., Steck T. L., Postlethwait P., Nellen W. RNAi in Dictyostelium: the role of RNA-directed RNA polymerases and double-stranded RNase. Mol. Biol. Cell. 2002;13:445–453. doi: 10.1091/mbc.01-04-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Quiles N., et al. WIP regulates N-WASP-mediated actin polymerization and filopodium formation. Nat. Cell Biol. 2001;3:484–491. doi: 10.1038/35074551. [DOI] [PubMed] [Google Scholar]
- Meili R., Ellsworth C., Lee S., Reddy T.B.K., Ma H., Firtel R. A. Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. EMBO J. 1999;18:2092–2105. doi: 10.1093/emboj/18.8.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H., Miura K., Takenawa T. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 1996;15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- Moreau V., Frischknecht F., Rechmann I., Vincentelli R., Rabut G., Stewart D., Way M. A complex of N-WASP and WIP integrates signaling cascades that lead to actin polymerization. Nat. Cell Biol. 2000;2:441–448. doi: 10.1038/35017080. [DOI] [PubMed] [Google Scholar]
- Myers S. A., Han J. W., Lee Y., Firtel R. A., Chung C. Y. A Dictyostelium homolog of WASP is required for polarized F-actin assembly during chemotaxis. Mol. Biol. Cell. 2005;16:2191–2206. doi: 10.1091/mbc.E04-09-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunola E., Mattila P. K., Lappalainen P. WH2 domain: a small, versatile adapter for actin monomers. FEBS Lett. 2002;513:92–97. doi: 10.1016/s0014-5793(01)03242-2. [DOI] [PubMed] [Google Scholar]
- Prehoda K. E., Lee D. J., Lim W. A. Structure of the Enabled/VASP homology 1 domain-peptide complex: a key component in the spatial control of actin assembly. Cell. 1999;97:471–480. doi: 10.1016/s0092-8674(00)80757-6. [DOI] [PubMed] [Google Scholar]
- Ramesh N., Anton I. M., Hartwig J. H., Geha R. S. WIP, a protein associated with Wiskott-Aldrich Syndrome protein, induces actin polymerization and redistribution in lymphoid cells. Proc. Natl. Acad. Sci. USA. 1997;94:14671–14676. doi: 10.1073/pnas.94.26.14671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong S. B., Vihinen M. Structural basis of Wiskott-Aldrich syndrome causing mutations in the WH1 domain. J. Mol. Med. 2000;78:530–537. doi: 10.1007/s001090000136. [DOI] [PubMed] [Google Scholar]
- Schirenbeck A., Arasada R., Bretschneider T., Stradal T. E., Schleicher M., Faix J. The bundling activity of vasodilator-stimulated phosphoprotein is required for filopodium formation. Proc. Natl. Acad. Sci. USA. 2006;103:7694–7699. doi: 10.1073/pnas.0511243103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirenbeck A., Bretschneider T., Arasada R., Schleicher M., Faix J. The diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat. Cell Biol. 2005;7:619–625. doi: 10.1038/ncb1266. [DOI] [PubMed] [Google Scholar]
- Snapper S. B., et al. Wiskott-Aldrich syndrome protein-deficient mice reveal a role for WASP in T but not B cell activation. Immunity. 1998;9:81–91. doi: 10.1016/s1074-7613(00)80590-7. [DOI] [PubMed] [Google Scholar]
- Snapper S. B., Rosen F. S. The Wiskott-Aldrich Syndrome protein (WASP): roles in signaling and cytoskeletal organization. Annu. Rev. Immunol. 1999;17:905–929. doi: 10.1146/annurev.immunol.17.1.905. [DOI] [PubMed] [Google Scholar]
- Stewart D. M., Tian L., Nelson D. L. Mutations that cause the Wiskott-Aldrich syndrome impair the interaction of Wiskott-Aldrich syndrome protein (WASP) with WASP interacting protein. J. Immunol. 1999;162:5019–5024. [PubMed] [Google Scholar]
- Symons M., Derry J. M., Karlak B., Jiang S., Lemahieu V., McCormick F., Francke U., Abo A. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- Vaduva G., Martin N. C., Hopper A. K. Actin-binding verprolin is a polarity development protein required for the morphogenesis and function of the yeast actin cytoskeleton. J. Cell Biol. 1997;139:1821–1833. doi: 10.1083/jcb.139.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haastert P. J., Devreotes P. N. Chemotaxis: signalling the way forward. Nat. Rev. Mol. Cell Biol. 2004;5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- Vetterkind S., Miki H., Takenawa T., Klawitz I., Scheidtmann K., Preuss U. The rat homologue of Wiskott-Aldrich Syndrome protein (WASp)-Interacting Protein (WIP) associates with actin filaments, recruits N-WASp from the nucleus, and mediates mobilization of actin from stress fibers in favor of filopodia formation. J. Biol. Chem. 2002;277:87–95. doi: 10.1074/jbc.M104555200. [DOI] [PubMed] [Google Scholar]
- Volkman B. F., Prehoda K. E., Scott J. A., Peterson F. C., Lim W. A. Structure of the N-WASP EVH1 domain-WIP complex: insight into the molecular basis of Wiskott-Aldrich syndrome. Cell. 2002;111:565–576. doi: 10.1016/s0092-8674(02)01076-0. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H., Miki H., Suetsugu S., Ma L., Kirschner M. W., Takenawa T. Two tandem verprolin homology domains are necessary for a strong activation of Arp2/3 complex-induced actin polymerization and induction of microspike formation. Proc. Natl. Acad. Sci. USA. 2000;97:12631–12636. doi: 10.1073/pnas.190351397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., et al. WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility. EMBO J. 2003;22:3602–3612. doi: 10.1093/emboj/cdg350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky J., Lempert L., Kranitz H., Mullins R. D. Different WASP family proteins stimulate different Arp2/3 complex-dependent actin-nucleating activities. Curr. Biol. 2001;11:1903–1913. doi: 10.1016/s0960-9822(01)00603-0. [DOI] [PubMed] [Google Scholar]
- Zetti M., Way M. The WH1 and EVH1 domains of WASP and Ena/VASP family members bind distinct sequence motifs. Curr. Biol. 2002;12:1617–1622. doi: 10.1016/s0960-9822(02)01112-0. [DOI] [PubMed] [Google Scholar]
- Zigmond S. H. How WASP regulates actin polymerization. J. Cell Biol. 2000;150:F117–F120. doi: 10.1083/jcb.150.6.f117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond S. H., Joyce M., Borleis J., Bokoch G. M., Devreotes P. N. Regulation of actin polymerization in cell-free systems by GTPγS and Cdc42. J. Cell Biol. 1997;138:363–374. doi: 10.1083/jcb.138.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]