Abstract
Mannose 6-phosphate receptors (MPRs) deliver newly synthesized lysosomal enzymes to endosomes and then recycle to the Golgi. MPR recycling requires Rab9 GTPase; Rab9 recruits the cytosolic adaptor TIP47 and enhances its ability to bind to MPR cytoplasmic domains during transport vesicle formation. Rab9-bearing vesicles then fuse with the trans-Golgi network (TGN) in living cells, but nothing is known about how these vesicles identify and dock with their target. We show here that GCC185, a member of the Golgin family of putative tethering proteins, is a Rab9 effector that is required for MPR recycling from endosomes to the TGN in living cells, and in vitro. GCC185 does not rely on Rab9 for its TGN localization; depletion of GCC185 slightly alters the Golgi ribbon but does not interfere with Golgi function. Loss of GCC185 triggers enhanced degradation of mannose 6-phosphate receptors and enhanced secretion of hexosaminidase. These data assign a specific pathway to an interesting, TGN-localized protein and suggest that GCC185 may participate in the docking of late endosome-derived, Rab9-bearing transport vesicles at the TGN.
INTRODUCTION
Mannose 6-phosphate receptors (MPRs) bind newly synthesized lysosomal hydrolases in the Golgi complex and deliver them to prelysosomes (Ghosh et al., 2003). There, the enzymes are released from MPRs, which are then recycled to the trans-Golgi network (TGN) for another round of enzyme delivery. There are two MPRs in most cell types: the cation-independent (CI)-MPR of ∼300 kDa and the cation-dependent (CD)-MPR, a dimer of ∼45 kDa. Both MPRs seem to require similar cellular components for their transport between compartments within mammalian cells (Ghosh et al., 2003).
This laboratory has studied the process by which MPRs are transported from late endosomes to the Golgi. We have shown that MPR recycling requires Rab9 GTPase and its effector p40 (Lombardi et al., 1993; Riederer et al., 1994; Diaz et al., 1997), a cargo adaptor named TIP47 (for tail-interacting protein of 47 kDa; Diaz and Pfeffer, 1998; Burguete et al., 2005) and several other general transport factors (Itin et al., 1997). Using live cell video microscopy, we were able to track yellow fluorescent protein (YFP)-Rab9–containing transport vesicles and watch them fuse with a cyan fluorescent protein-labeled trans-Golgi compartment (Barbero et al., 2002). That study showed that Rab9 is present on transport vesicles en route to the Golgi complex, but it did not provide additional clues to the mechanisms by which such vesicles are targeted.
In an effort to understand the full scope of Rab9 function, we sought to identify additional partners for the active form of this GTPase. We report here the discovery that a Golgi-localized, putative tether of the Golgin protein family binds specifically to Rab9. Golgins contain long, predicted coiled-coil motifs and are important for Golgi organization, vesicle tethering, and secretory protein transport (for reviews, see Gillingham and Munro, 2003; Gleeson et al., 2004; Short et al., 2005). Several Golgins bind Rab GTPases, but the precise functional significance of Rab–Golgin interactions remains poorly defined. Rab1 binds to the well-characterized Golgin p115 (Allan et al., 2000) and to Golgin-84 (Diao et al., 2003); Rab6a recruits the coiled-coil, Bicaudal-D protein to the TGN (Matanis et al., 2002) and also binds to a Golgin named TATA element modulatory factor (Fridmann-Sirkis et al., 2004); Rab2 interacts with the medial Golgi matrix protein Golgin-45 (Short et al., 2001).
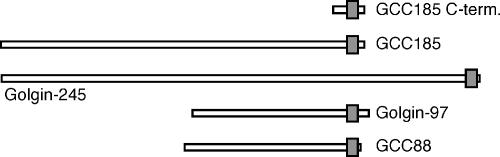
Four mammalian Golgins, named Golgin-97, Golgin-245, GCC88, and GCC185, contain so-called GRIP domains, which are ∼50 amino acids in length and have each been shown to be sufficient to target green fluorescent protein (GFP) to the trans-Golgi (Munro and Nichols, 1999; Barr, 1999; Kjer-Nielsen et al., 1999a,b; Luke et al., 2003; Figure 1). Certain GRIP domains bind to the Arf-like GTPase Arl1 (Van Valkenburgh et al., 2001; Lu and Hong, 2003), which itself is Golgi localized via Arl3 (Setty et al., 2003; Panic et al., 2003b). Interaction with Arl1 at the TGN is the basis for the localization of several GRIP domain proteins.
Figure 1.
Schematic comparison of GRIP domain-containing proteins. The shaded box shows the location of the ∼50 residue GRIP domain; the remaining sequences show a high propensity for coiled-coil formation. GCC185 C-term. represents the 110 C-terminal amino acid fragment used in these studies as a GST-fusion and by others (as a His-tagged protein; Panic et al., 2003a). The C-terminal–most residues 1343–1684 were initially identified in the yeast two-hybrid screen.
We show here that the TGN localized, GCC185 protein binds to Rab9 with strong preference for its active, GTP-bound conformation. Importantly, the Rab9-dependent, retrograde trafficking of CD-MPRs to the TGN requires GCC185 but not the related Golgin-97. Unlike Golgin-245, GCC185 binds poorly (if at all) to Arl1 GTPase in vitro, distinguishing this Golgin from other GRIP domain-containing proteins.
MATERIALS AND METHODS
Yeast Two-Hybrid Screen
To construct plasmids pRB2510-Rab9Q66LΔcc and pRB2510-Rab9S21NΔcc, Rab9 reading frames were excised from pASI-CYH2-Rab9Q66LΔcc and pASI-CYH2-Rab9S21NΔcc, respectively (Diaz et al., 1997) with NcoI and BamHI and subcloned into plasmid pRB2510 (Schwartz et al., 1997). A human placental cDNA library was amplified according to the manufacturer (Clontech, Mountain View, CA). Yeast strain Y190 (MATa gal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3,112 + URA3::GAL-lacZ,LYS2::GAL(UAS)-HIS3 cyhr) was transformed sequentially with pRB2510-Rab9Q66LΔcc and placental cDNA library by using the alkali cation treatment method (Elble, 1992) and plated on minimal media containing 10 μg/ml adenine and 50 mM 3-amino-1,2,4-triazole (Sigma-Aldrich, St. Louis, MO). Plates were incubated for 10 d at 25°C. His+/Leu+/Trp+ colonies that grew to a diameter larger than 2 mm were tested for β-galactosidase activity by 5-bromo-4-chloro-3-indolyl-β-d-galactoside colony filter assay (Bai and Elledge, 1996). The Rab9Q66LΔcc bait plasmid was lost by plating cells on minimal media lacking leucine +10 μg/ml cycloheximide (Sigma-Aldrich). Library plasmids were isolated and retransformed into strain Y190 containing either pRB2510-Rab9Q66LΔcc or pRB2510-Rab9S21NΔcc to check for plasmid linkage of the β-galactosidase–positive phenotype and to identify colonies demonstrating nucleotide preference. All such clones were sequenced.
Plasmids for Antibody Generation and Protein Expression
An N-terminal 6xHis tag was added onto the C-terminal–most 342 residues of GCC185 by polymerase chain reaction (PCR). The PCR product was then digested with NcoI-XhoI and ligated into NcoI-XhoI digested pET14b (Novagen, Madison, WI) to create plasmid pHis-GCC185. A glutathione S-transferase (GST)-tagged, 110 C-terminal amino acid fragment of GCC185 was generated by PCR using pHis-GCC185 as a template, followed by digestion with BamHI-EcoRI and ligation into BamHI-EcoRI–digested pGEX 4T-1 (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) to create pGST-GCC185. Alanine was introduced in place of GCC185 Tyr1618 by site-directed mutagenesis of pGST-GCC185 using the QuikChange II mutagenesis kit (Stratagene, La Jolla, CA) to generate pGST-GCC185(Y1618A). A GST-tagged, 123 C-terminal amino acid fragment of Golgin-245 was generated by PCR by using an expressed sequence tag clone (accession no. AA626910) as a template, followed by digestion with BamHI and EcoRI and ligation into BamHI-EcoRI digested pGEX 4T-1 (GE Healthcare) to create pGST-Golgin-245. A His-tagged Arl1 Q71L encoding plasmid was a gift of Rick Kahn (Emory University, Atlanta, GA).
Antibodies and Recombinant Proteins
Recombinant Rab1, Rab1Q70L, Rab5, Rab9, Rab9Q66L, and His-Arl1Q71L were purified as per Aivazian et al. (2006). Either pGST, pHis-GCC185, pGST-GCC185, or pGST-GCC185(Y1618A) was transformed into Escherichia coli BL21(DE3) cells. For pHis-GCC185, cells (O.D.600 of 0.6) were induced for 1 h at 37°C with 1 mM isopropyl β-d-thiogalactoside (IPTG). In pGST, pGST-GCC185, and pGST-GCC185(Y1618A), cells (O.D.600 of 0.6) were induced for 16 h at room temperature with 0.5 mM IPTG. Protein purification was as described previously (Diaz et al., 1997) by using a modified buffer for cell lysis and gel filtration (20 mM HEPES and 250 mM KCl). Gel filtration was done on a Superdex 200 column (GE Healthcare). For GST-Golgin-245, cells (O.D.600 of 0.6) were induced for 3 h at 30°C with 1 mM IPTG. Rabbit anti-His-GCC185 antibodies were prepared using 100 μg of antigen per immunization (Josman Laboratories, Napa Valley, CA). Mouse anti-Rab9 (Soldati et al., 1993) and mouse and rabbit anti-CI-MPR (Lombardi et al., 1993) have been characterized previously. Mouse anti-p115 was a gift of G. Waters (Merck Research Laboratories, Rahway, NJ). Mouse anti-Golgin-245 and anti-early endosomal antigen (EEA)1 (BD Biosciences, San Jose, CA), mouse anti-hemagglutinin (HA) 12CA5 antibody (Roche Diagnostics, Indianapolis, IN), mouse anti-Golgin-97, rabbit anti-GFP, Alexa Fluor 488 goat anti-mouse, Alexa Fluor 594 goat anti-rabbit, and Alexa Fluor 647 goat anti-mouse (Invitrogen, Carlsbad, CA), and goat anti-furin (Santa Cruz Biotechnology, Santa Cruz, CA) were purchased.
Immunoblot Detection of GCC185 Binding to Rab GTPases
Purified His-GCC185 (residues 1343–1684) (∼20 μM) was bound to 16 μl of nickel-nitrilotriacetic acid (Ni-NTA) agarose (QIAGEN, Valencia, CA) and rotated for 1 h at 22°C with limiting amounts (700 ng = 100 nM) of purified Rab9 or Rab5 in 300 μl binding buffer (20 mM HEPES, 250 mM KCl, 25 mM imidazole, 10 μg/ml bovine serum albumin [BSA], 1 mM MgCl2, and 100 μM guanosine 5′-O-(3-thio)triphosphate [GTPγS]). Beads were washed three times in 150 μl of binding buffer on a support and the amount of bound Rab protein was determined using standard curves by immunoblot.
Nucleotide Detection of GTPase Binding to GCC185
Rab protein (2.6 μM) was incubated with 3 μM GTPγS or 3 μM GDP, 5 μCi of [35S]GTPγS (MP Biomedicals, Irvine, CA), or 5 μCi [3H]GDP (GE Healthcare), 5 mM EDTA, 4.5 mM MgCl2, 1 mM dithiothreitol (DTT), 0.1 mg/ml BSA, 150 mM KCl, and 20 mM HEPES/KOH, pH 7.4, for 30 min at 37°C in a total volume of 1 ml. Free nucleotide was removed on a PD-10 column (GE Healthcare) equilibrated in binding buffer (5 mM MgCl2, 0.1 mg/ml BSA, 150 mM KCl, and 50 mM HEPES/KOH, pH 7.4). The specific activity of GTPγS- or GDP-bound Rab protein was determined in a filter-binding assay, and the fraction GTPγS or GDP bound Rab was calculated. Labeled Rab protein was bound to GST-GCC185 for 1 h at room temperature. Complexes were collected using 50 μl of a 50% slurry of glutathione beads (GE Healthcare) and washed with 3 × 1 ml binding buffer on a support. The fraction of Rab protein that bound GCC185 was determined after counting the beads in a scintillation counter and subtracting background binding to GST. Full-length His-Arl1 Q71L (2.6 μM) was incubated with 3 μM GTPγS, 5 μCi of [35S]GTPγS (MP Biomedicals) in 1 mM EDTA, 0.5 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA, 100 mM NaCl, and 20 mM HEPES/NaOH, pH 7.4, for 3 h at 30°C in a total volume of 1 ml. Free nucleotide was removed on a PD-10 column (GE Healthcare) equilibrated in binding buffer (5 mM MgCl2, 0.1 mg/ml BSA, 150 mM NaCl, and 50 mM HEPES/NaOH, pH 7.4). Labeled Arl1 Q71L protein or Rab9 protein was bound to GST-GCC185 or GST-Golgin-245 for 90 min at room temperature and processed as described above.
Small Interfering RNA (siRNA) Depletion of GCC185 and Rab9
Human Rab9 was targeted using the duplex: GUUUGAUACCCAGCUCUUC. Human GCC185 was targeted using two independent duplexes: GGAGUUGGAACAAUCACAU or GCUACUGUAACCUCUGAAU (Dharmacon, Lafayette, CO). HeLa or human embryonic kidney (HEK)-293 cell transfections were performed according to the manufacturer using Oligofectamine or Lipofectamine 2000, respectively (Invitrogen). Human Golgin-97 was targeted precisely as described previously (Lu et al., 2004).
Microscopy and Immunoblots
Posttransfection (48–72 h), HeLa, or BS-C-1 cells were fixed and processed for immunofluorescence (Warren et al., 1984). For Figure 3C and Supplemental Figure 1, cells were imaged using an Axiovert 200 inverted microscope (Carl Zeiss MicroImaging, Thornwood, NY) fitted with a 63×/1.4 Plan-Apochromat objective, and a charge-coupled device (CCD) camera (CoolSNAP HQ; Photometrics, Tucson, AZ). For Figures 4, top, and 6C, cells were imaged using an Axiophot 2 fluorescence microscope (Carl Zeiss MicroImaging) fitted with a Plan-Apochromat 63×/1.40 oil objective (Carl Zeiss MicroImaging), and a CCD camera (AxioCamHRc; Carl Zeiss MicroImaging) and controlled by Axiovision 4.2 software (Carl Zeiss MicroImaging). Figures 5, 6A, and Supplemental Figure 2 were acquired using a deconvolution microscope setup (Spectris; Applied Precision, Issaquah, WA), with an inverted epifluorescence microscope (IX70; Olympus America, Melville, NY), a PlanApo 60×, 1.40 numerical aperture oil immersion objective (Olympus America), a CCD camera (CoolSNAP HQ) and acquisition and deconvolution software (Delta Vision; Applied Precision). The quantitative analysis of fluorescence intensity in Figure 5 was done using ImageJ (National Institutes of Health, Bethesda, MD). All images were processed using ImageJ and Adobe Photoshop (Adobe Systems, Mountain View, CA).
Figure 3.
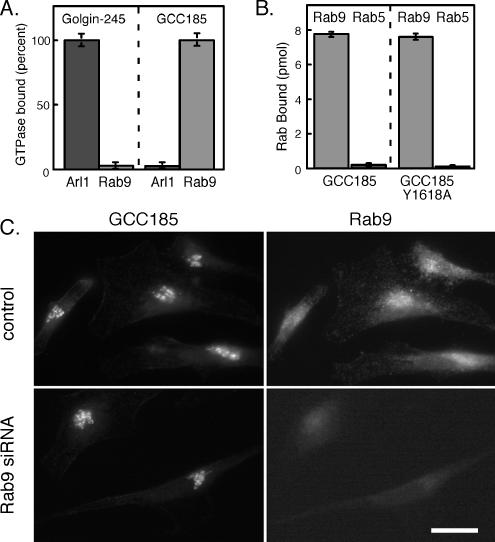
(A) Binding of His-tagged Arl1 Q71L or untagged Rab9 to GST-Golgin-245 (123 residues) or GST-GCC185 (110 residues) C termini. Reactions contained 30 pmol of Rab9 or 11 pmol of Arl1 Q71L. Golgin-245 bound 9647 cpm of Arl1 protein; GCC185 bound 6334 cpm of Rab9 protein. (B) Binding of GST-GCC185 or GST-GCC185 Y1618A C-terminal fragments to Rab9 (light bars) or Rab5 (dark bars). Reactions contained 140 pmol of Rab9 or Rab5. Experiments were carried out using [35S]GTPγS-preloaded Rab GTPases. Rab9 binding represents ∼15,000 cpm. (C) Cellular depletion of Rab9 does not influence GCC185 localization. Cells were treated with Rab9 siRNA for 72 h; the localization of endogenous GCC185 and Rab9 were then monitored using rabbit anti-GCC185 and mouse anti-Rab9 IgG. Bar, 20 μm.
Figure 4.
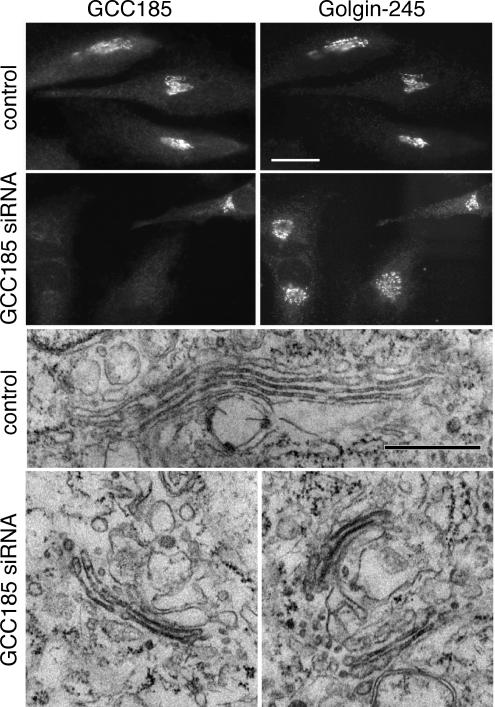
GCC185 depletion leads to minimal disruption of Golgin-245 localization and Golgi stacking. GCC185 (top left) and Golgin-245 (top right) in control, mock transfected or GCC185-depleted cells. A single cell in the second row, top right quadrant, was not depleted of GCC185; three other cells in the field were depleted and labeled for Golgin-245 (second row, right). Bar, 20 μm. Electron microscopy of control or GCC185-depleted HeLa cells (bottom). Bar, 500 nm.
Figure 6.
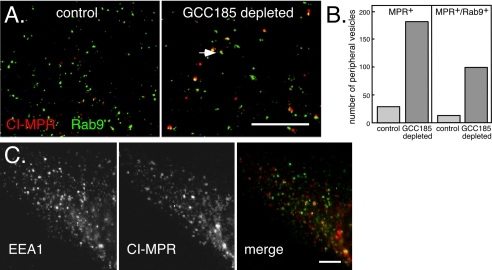
CI-MPR-containing vesicles in GCC185-depleted cells contain Rab9 but not EEA1. (A) Deconvolution microscopic image of the periphery of control (left) or GCC185-depleted (right) BS-C-1 cells expressing GFP-Rab9 (green) and stained with mouse anti-CI-MPR (red). The arrow indicates an example of a peripheral, MPR-containing vesicle colocalizing with Rab9. The image shown is a total summation of all the Z-sections. Control cells contain much less peripheral MPR staining as in Figure 5. (B) Quantitation of peripheral vesicles containing MPR or MPR and Rab9 in control (light bars) and GCC185-depleted (darker bars) cells. (C) EEA1 (left) and CI-MPR (middle) do not colocalize upon depletion of GCC185 from HeLa cells, as seen in the merged image on the right. CI-MPR is represented in red and EEA1 in green. Antibodies were zenon-labeled mouse anti-CI-MPR and mouse anti-EEA1. Bars, 20 μm.
Figure 5.
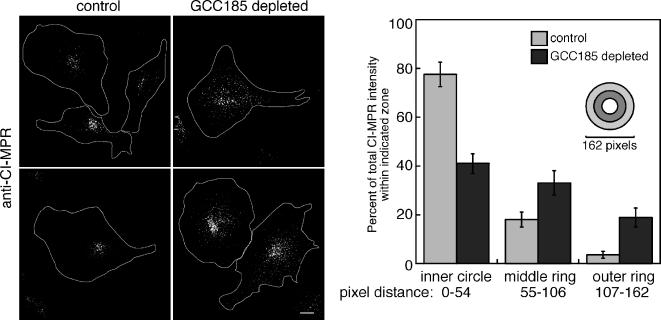
Dispersal of CI-MPR vesicles in cells depleted of GCC185. Left, perinuclear localization of CI-MPR in control HeLa cells. Right, dispersed localization of CI-MPR in cells of similar size, depleted of GCC185. The images were acquired on a deconvolution microscope and a total summation of the Z-sections is shown. Approximate cell outlines are drawn in white. The fluorescence intensity of CI-MPR in 10 GCC185-depleted and five control cells was quantified using ImageJ. Circles of different radii, centered around the peak fluorescent signal were drawn, and the intensity within the circle was quantified relative to the total intensity in the whole cell; data are presented in relation to the concentric circle pattern shown. Bar, 5 μm.
When staining with anti-Rab9 or anti-furin, cells were washed once in ice-cold glutamate buffer [25 mM HEPES, pH 7.4, 25 mM KCl, 2.5 mM Mg(Oac)2, 5 mM EGTA, and 150 mM K glutamate]. After removal of excess buffer, coverslips were frozen in liquid nitrogen, incubated for 1 min in glutamate buffer, washed twice on ice with ice-cold glutamate buffer, fixed, and then processed as described above but without detergent treatment. Mouse anti-MPR was labeled with the Zenon IgG labeling kit (Invitrogen) when costaining required mouse anti-EEA1 antibody. For electron microscopy, GCC185 siRNA-transfected HeLa cells were grown on 12-mm glass coverslips for 48 h and processed as described previously (Ganley et al., 2004). For immunoblots, 72 h after transfection with GCC185 siRNA, cells were washed three times with phosphate-buffered saline (PBS), followed by incubation with radioimmunoprecipitation assay (RIPA) buffer for 15 min. Lysates were centrifuged at 100,000 × g for 10 min, and equal amounts of protein were analyzed by immunoblot. Signals were detected by chemiluminescence (PerkinElmer Life and Analytical Sciences, Boston, MA).
Metabolic Labeling
HeLa cells seeded onto six-well plates were transfected with GCC185 siRNA. G protein of the vesicular stomatitis virus (VSVG)-YFP was transfected into HeLa cells 56 h later using FuGENE 6 (Roche Diagnostics). After a total of 72 h of transfection, cells were metabolically labeled as described previously (Burguete et al., 2005). Cells were washed three times with ice-cold DMEM, followed by binding of cell surface VSVG-YFP with anti-GFP for 1 h at 4°C. Unbound antibody was washed away with PBS, and cells were lysed in RIPA buffer containing protease inhibitors for 15 min. Antibody–protein complexes were immunoprecipitated with protein A-agarose (Roche Diagnostics) and processed. MPR turnover was determined as described previously (Ganley et al., 2004). For CD-MPR maturation, HEK-293 HeLa cells stably expressing a modified CD-MPR (Itin et al., 1997) were transfected with siRNA for 48 h as described above. Cells were metabolically labeled for 30 min, washed, and chased (Burguete et al., 2005). At the indicated times, cells were transferred to ice, washed once, and lysed in RIPA buffer (Itin et al., 1997) containing protease inhibitors for 30 min. The cell lysate was centrifuged for 10 min at 100,000 × g, at 4°C, and the supernatant was incubated with 40 μl of prewashed Ni-NTA agarose beads for 1 h at 4°C. The beads were washed three times in 500 μl of RIPA and eluted in 100 μl RIPA containing 25 mM EDTA. The CD-MPR was detected by SDS-PAGE and autoradiography.
Transport and Enzyme Assays
The in vivo transport assay was adapted to HEK-293 cells as described previously (Sincock et al., 2003), except that cells were transfected with GCC185 or Golgin-97 siRNA for 72 h before initiation of 2 h of transport. The in vitro transport assay was also adapted to HEK-293 cells (Itin et al., 1997). In some cases, purified GST or GST-GCC185 protein was added at 94 μg/ml before 90 min of transport at 37°C. To examine tyrosine sulfation of endogenous proteins, in vivo transport was conducted with cells transfected with either GCC185 siRNA or Golgin-97 siRNA for 72 h before initiation of 2 h of transport. Equal quantities of whole cell lysates were run on a 6% SDS-PAGE gel and the signal quantified by autoradiography. Transferrin uptake was measured in HeLa cells seeded onto six-well plates, transfected with GCC185 siRNA for 72 h. Cells were washed three times with PBS followed by incubation with 5 μg/ml Alexa Fluor 594 transferrin in PBS buffer A (1 mM MgCl2, 1 mM CaCl2, and 0.2% BSA in PBS) for 90 min at 4°C. Cells were washed a further three times with PBS and then incubated in DMEM media containing 7.5% fetal bovine serum for either 5 or 15 min at 37°C. Surface bound transferrin was washed away with ice-cold citrate buffer containing 25.5 mM citric acid, 24.5 mM Na citrate, and 280 mM sucrose. After a final wash in PBS buffer A, the cells were processed for immunofluorescence as described above. Images were quantified using ImageJ software. Hexosaminidase was measured according to Riederer et al. (1994) except that cells were incubated in 1 ml of DMEM media lacking phenol red plus 10 mM mannose 6-phosphate.
RESULTS
We carried out a two-tiered, yeast two-hybrid screen to identify novel Rab9 effectors. Clones (1 × 106) were selected for interaction with an activated, Rab9-GTP protein (Rab9 Q66L) in preference to a Rab9-GDP–preferring mutant, Rab9 S21N. In the past, this approach has yielded physiologically relevant binding partners, such as a protein named p40 (Diaz et al., 1997). One of the positive clones obtained in the present screen encoded 342 C-terminal residues of a protein named GCC185 (Figure 1; Luke et al., 2003). GCC185 contains a GRIP domain at its C terminus, and the remainder of the 185,000 Mr protein is predicted to form a long, extended coiled-coil (Figure 1).
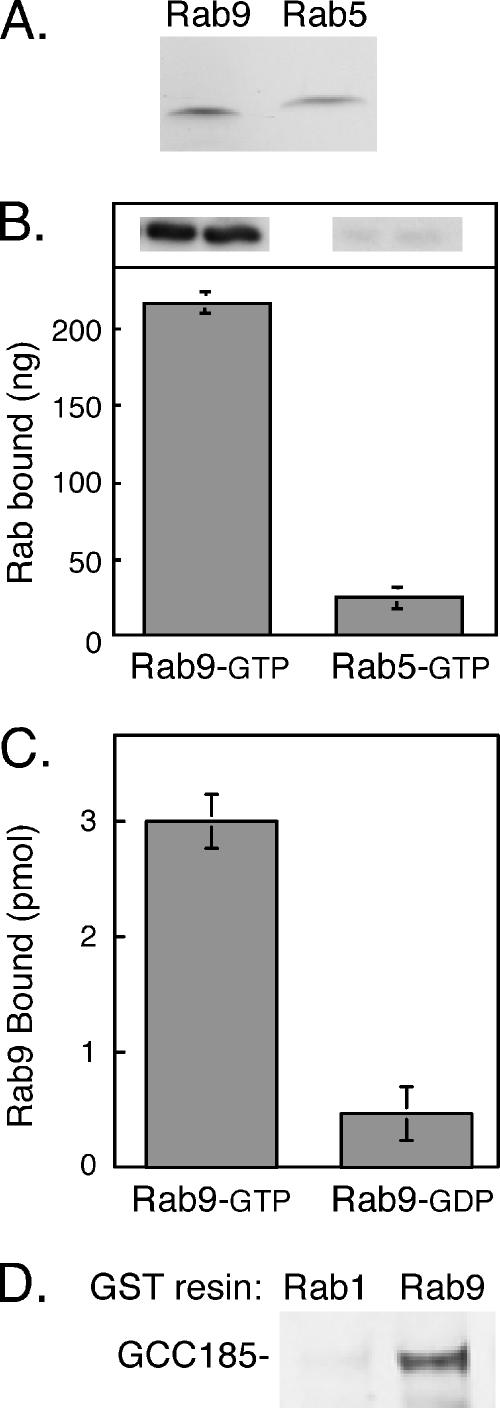
Two independent methods were used to confirm that the interaction between Rab9 and GCC185 was direct. In the first, the 342 C-terminal residues of GCC185 (1343-1684) were expressed in bacteria as a His-tagged protein. This protein was incubated with untagged Rab9 or Rab5 (Figure 2A); bound proteins were collected by Ni-NTA column chromatography and detected by immunoblot. As shown in Figure 2B, Rab9 showed significant binding to the 342 C-terminal residues using this method, whereas Rab5 did not. The second approach used Rab proteins preloaded with [35S]GTPγS or [3H]GDP and thus only measures the binding of nucleotide binding-competent GTPases to effector proteins. These experiments used a shorter Golgin construct comprised of GST fused to the C-terminal 110 residues of GCC185 (Figure 1, GCC185 C-term.). The C-terminal 110 residues of GCC185 showed specific binding to Rab9-GTP in preference to Rab9-GDP (Figure 2C).
Figure 2.
GCC185 is a Rab9 effector. (A) Purified proteins used in this study. One microgram of nontagged Rab9 and Rab5 were analyzed by 15% SDS-PAGE and Coomassie blue staining. (B) Immunoblot detection of Rab binding to His-tagged GCC185 (residues 1343–1684) by using untagged Rab9 or Rab5. Top inset, anti-Rab protein immunoblot quantified in B. (C) Binding of Rab9 to GST-GCC185 C terminus (110 residues) shows preference for GTP. Reactions contained 56 pmol of Rab9 and were carried out using [35S]GTPγS or [3H]GDP-preloaded Rab GTPases. Rab9 GTP bound corresponds to 8269 cpm. (D) Full-length GCC185 from rat liver cytosol binds GST-Rab9 but not GST-Rab1. Immunoblot of the bound fraction is shown; binding reactions were for 1.5 h at room temperature according to Aivazian et al. (2006).
Importantly, full-length GCC185 could be shown to bind to immobilized Rab9 (Figure 2D). In this experiment, a gel-filtered fraction of rat liver cytosol, enriched in high-molecular-weight proteins, was incubated with GST-tagged Rab9 or Rab1 proteins. GTP-preferring mutant forms (Rab9 Q66L and Rab1 Q70L) were used to facilitate detection. Bound proteins were eluted, and subsequent immunoblot analysis revealed specific interaction of Rab9 but not Rab1 with cytosolic GCC185. Thus, the Rab9 binding site is available for Rab interaction within the context of the native, full-length protein.
TGN Localization of GCC185
A hallmark of GRIP domain proteins is their ability to bind Arl1 GTPase. Indeed, the three-dimensional structure of the Golgin-245 GRIP domain bound to Arl1 has been determined previously (Panic et al., 2003a; Wu et al., 2004). But there has been some controversy regarding the capacity of GCC185 to bind Arl1. A His-tagged, 110 residue, GCC185 C-terminal construct was shown to bind GST-Arl1 GTPase with preference for the GTP-bound conformation (Panic et al., 2003a). However, Derby et al. (2004) failed to relocalize GCC185 from the TGN in cells expressing an endosome-associated Arl1-hybrid protein construct. Moreover, GCC185 was shown to occupy a domain of the TGN that is distinct from the other mammalian GRIP domain-containing Golgin proteins (Derby et al., 2004), suggesting that it may use a distinct mechanism to localize to the Golgi.
As shown in Figure 3A, we failed to detect binding of His-tagged, active Arl1 protein to glutathione agarose-immobilized GCC185. In contrast, significant binding of Arl1-GTP to immobilized Golgin-245 C terminus was readily detected. From these experiments, we conclude that Arl1 interacts much less strongly with GCC185 than with Golgin-245. It is possible that the dimeric nature of the GST-Arl1 construct used by others (Panic et al., 2003a) revealed a weaker binding interaction than we were able to detect using His-tagged Arl1 protein.
GRIP domain-containing proteins contain an invariant tyrosine residue that is key for Arl1 binding and in many cases, Golgi localization. Mutation of the corresponding residue (Tyr1618) in the GST-tagged GCC185 C terminus did not interfere with Rab9 binding (Figure 3B), suggesting that Rab9 binds GCC185 via a molecular interaction other than that used for typical Arl1 interaction. Future work will determine the precise binding interface of Rab9 GTPase with GCC185 C-terminal sequences.
If GCC185 does not bind tightly to Arl1, how is it localized to the Golgi complex? Most Rab9 is present on late endosomes, but a small amount is detected on the TGN (Lombardi et al., 1993). Therefore, we tested whether the Golgi localization of GCC185 was in any way dependent on Rab9 protein. Cellular depletion of Rab9 by using siRNA (Ganley et al., 2004) did not in any way seem to alter the TGN localization of GCC185 (Figure 3C). Unfortunately, attempts to alter the fraction of GCC185 that was membrane associated by Rab9 overexpression were not possible to interpret, because this resulted in a significant pool of unprenylated, cytosolic Rab9 that could have bound to cytosolic GCC185. Nevertheless, these data show that Rab9 is not a primary determinant of GCC185's Golgi association. It is possible that another Rab protein or another member of the large family of Arl proteins mediates GCC185 Golgi localization.
GCC185 Is Required for Endosome-to-Golgi Transport
Rab9 is required for the transport of MPRs from late endosomes to the TGN (Lombardi et al., 1993; Riederer et al., 1994). The established localization of GCC185 at the TGN was intriguing in that it suggested a possible role for GCC185 as a tether for Rab9-positive, MPR-containing transport vesicles (Barbero et al., 2002) en route to the Golgi from late endosomes. If true, cellular depletion of GCC185 would be predicted to block this transport pathway and lead to aberrant trafficking of proteins cycling between endosomes and the TGN. We therefore used two distinct siRNAs to deplete cells of GCC185 protein to investigate its function.
First, loss of GCC185 altered the morphological appearance of the Golgi ribbon (Figure 4). Golgin-245 is another TGN-associated GRIP domain-containing protein that shows labeling that is slightly distinct but overlapping with GCC185 (Luke et al., 2003; Derby et al., 2004; Figure 4). Golgin-245 staining is sometimes more extended than resident Golgi enzymes but is representative of the TGN as determined by electron microscopy (Luke et al., 2003). On depletion of GCC185, Golgin-245 labeled less elongated structures that nevertheless, remained on one side of the nucleus (Figure 4, top). This suggested a slight alteration in the Golgi ribbon, which was confirmed by electron microscopy of GCC185-depleted cells (Figure 4, bottom). In depleted but not control cells, clusters of Golgi mini-stacks were readily detected in individual sections. A similar change in Golgi morphology was recently reported in cells missing Golgin-245 (Yoshino et al., 2005), TATA modulatory factor (Fridmann-Sirkis et al., 2004) as well as the so-called COG complex, another putative Golgi tether (Zolov and Lupashin, 2005; for review see Puthenveedu et al., 2006). In all of these studies, Golgi morphology is reminiscent of what is observed when cells are treated with nocodazole; however, unlike nocodazole, the Golgi fragments remain localized on one side of the nucleus.
As shown in Figure 5, siRNA depletion of GCC185 led to a major alteration in the steady-state, cellular distribution of MPRs as determined by deconvolution light microscopy. MPRs are normally present in perinuclear late endosomes and to a lesser extent, in the TGN of HeLa cells. On depletion of GCC185, MPRs lost their perinuclear localization and were detected in significantly more dispersed, vesicular structures (Figure 5). The distribution of MPRs was quantified using a series of concentric rings centered upon the peak intensity of MPR staining. The amount of MPR staining detected within the inner circle, middle ring, and outer ring was then compared for cells of similar size. This analysis confirmed a significant difference in the distribution of MPRs between cells of similar size, in the presence or absence of GCC185 protein.
In all cases, MPRs seemed to be more broadly dispersed than any of the Golgi markers examined (compare Figure 5 with 4). For example, furin, a protein thought to cycle between endosomes and the Golgi, was not detected in structures other than those occupied by residual GCC185 (Supplemental Figure 1). A staining pattern resembling that observed for furin (and Golgin-245; Figure 4) was also seen for the Golgi protein p115 in GCC185-depleted cells (Supplemental Figure 2).
To determine whether the MPR-containing accumulated vesicles contained Rab9 protein, we depleted GCC185 from BS-C-1 cells stably expressing GFP-Rab9 (Figure 6A). In control BS-C-1 cells, a small amount of the total GFP-Rab9 is present in small structures in the periphery (Figure 6A, left), and in HeLa cells, it colocalizes there with Rab5 and EEA1 in early endosomes (Aivazian et al., 2006). In control cell peripheral zones containing 550 GFP-Rab9+ vesicles, 29 vesicles contained MPR; 13 of these vesicles (45%) also contained Rab9. As was shown in Figure 5, control cells contained little, peripheral MPR staining.
In GCC185-depleted cells (6A, right), peripheral zones contained many more MPR-positive structures: among 470 GFP-Rab9+ vesicles, we counted 181 MPR-positive vesicles. Of these MPR-positive vesicles, 99 (55%) also contained GFP-Rab9 protein (Figure 6B). The MPR-positive vesicles seemed slightly larger in the GCC185-depleted cells (Figure 6A, right) compared with MPR- or GFP-Rab9-positive vesicles in control cells. This is likely because these vesicles are much brighter than the few MPR- or more abundant, GFP-Rab9–positive peripheral vesicles in control cells. In summary, more than half of the peripheral MPR-containing vesicles in GCC185-depleted cells also contain Rab9.
The MPR positive, peripheral vesicles in GCC185-depleted cells were not early endosomes, by the criterion that they lacked EEA1 protein (Figure 6C, HeLa cells). The vesicles did not resemble late endosomes or lysosomes in terms of their small, apparent uniform size or distribution in these cells; the residual, perinuclear MPR staining (Figure 5) suggested that significant numbers of late endosomes remained intact. Depletion of a protein needed for vesicle consumption but not formation would be expected to result in the accumulation of transport intermediates. Together, these data are consistent with GCC185 depletion leading to a block in recycling, and possible accumulation of transport intermediates that are unable to dock at the Golgi.
GCC185 Is Needed for MPR Recycling
The dispersal of MPRs in cells lacking GCC185 (Figure 5) indicated that this protein may be needed for MPR recycling to the TGN. Because the Golgi complex was functional under these conditions (see below), we could use biochemical tests to determine whether MPR transport to the TGN was blocked—either in vivo or in vitro (Itin et al., 1997; Carroll et al., 2001). Briefly, our assays take advantage of a TGN-specific enzyme, tyrosine sulfotransferase. We use cells expressing CD-MPRs that contain an N-terminal 6-histidine tag and a consensus sequence for tyrosine sulfation derived from the cholecystokinin precursor. For in vivo analysis, late endosome-localized MPRs are accumulated in a nonsulfated form by incubation of cells in sulfate-free media containing a sulfotransferase inhibitor. The inhibitor is then removed, and transport is measured by monitoring the action of tyrosine sulfotransferase on CD-MPRs (in the presence of [35S]sulfate) that occurs upon their arrival at the TGN. This process is time, temperature, and cytosol-dependent and requires Rab9 and other components needed for a typical vesicular transport process (Itin et al., 1997).
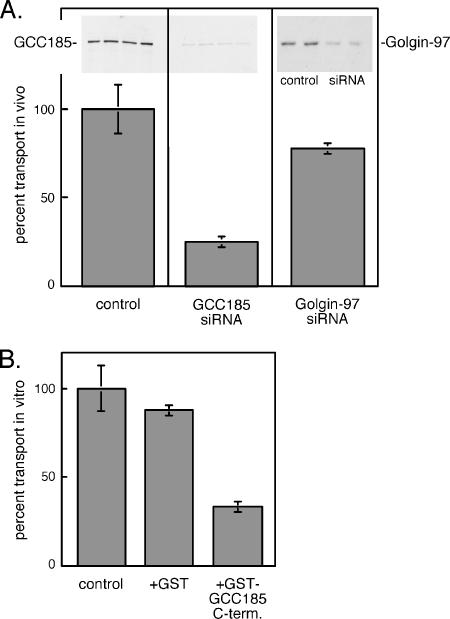
Intact cells depleted of ∼90% GCC185 (Figure 7A, top, inset) showed significant inhibition (∼75%) in the transport of CD-MPRs to the TGN (Figure 7A). In contrast, siRNA depletion of the related Golgin-97 inhibited transport only 23%. These data demonstrate that GCC185 is required for the transport of MPRs from late endosomes to the TGN in living cells. As an independent confirmation of a role for GCC185 in MPR transport, we used wild-type cell extracts to test whether excess, soluble GCC185 C terminus would function as a dominant-negative inhibitor of in vitro transport. In this case, we used an assay very similar to that described for intact cells, except that the cells are broken and reactions are supplemented with cytosol and an ATP-regenerating system (Itin et al., 1997). Addition of a polypeptide representing the 110 C-terminal–most amino acids of GCC185 led to significant inhibition of MPR transport to the TGN in vitro, relative to control reactions to which GST alone was added (Figure 7B). The inhibition observed may be due to titrating out an available binding partner on the TGN or to masking available Rab9 sites so that endogenous GCC185 cannot interact. In either case, these combined data strongly support a role for GCC185 in MPR transport to the TGN.
Figure 7.
GCC185 is needed for MPR transport to the Golgi complex in vivo and in vitro. (A) In vivo assay for MPR transport in control, mock-transfected cells or cells depleted of 90% of endogenous GCC185 protein or 77% of Golgin-97. The amounts of GCC185 or Golgin-97 were determined by loading equal protein on a 7.5% SDS-PAGE gel followed by immunoblot, as shown in the insets at the top. Transport was for 120 min at 37°C. Complete (100%) transport represents 721 cpm for the GCC185 siRNA experiment and 348 cpm for the Golgin-97 experiment, normalized per milligram of protein in each sample. (B) In vitro transport of CD-MPRs in reactions containing no addition (control), GST alone or GST-GCC185 C-terminal 110 residues (94 μg/ml). Transport was at 37°C for 90 min; 100% transport corresponded to 763 cpm.
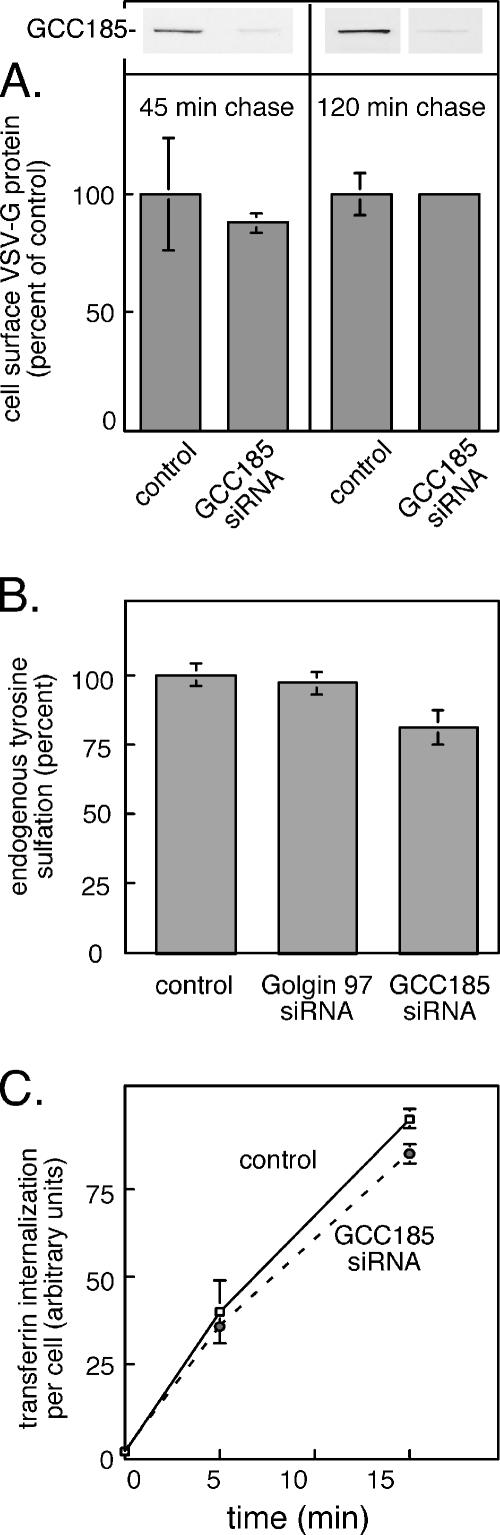
It was important to confirm the biochemical functionality of the Golgi complex after depletion of GCC185, to ensure the reliability of our conclusions. First, the Golgi complex supported the transport and processing of secretory cargo. We detected no difference in the acquisition of complex oligosaccharides by newly synthesized CD-MPRs in control and GCC185-depleted cells (Supplemental Figure 3). More importantly, to confirm that TGN export of proteins was not defective, we monitored the plasma membrane arrival of a newly synthesized hybrid protein comprised of VSV-G fused to YFP. Cells were pulse labeled with [35S]methionine for 30 min and chased for either 45 or 120 min at 37°C. Cell surface proteins were then immunoprecipitated by incubation of intact cells with antibody at 4°C, washing away unbound antibody, followed by detergent solubilization, protein A-agarose incubation, SDS-PAGE, and PhosphorImager analysis. As shown in Figure 8A, the rate with which VSV-G-YFP arrived at the surface was essentially indistinguishable between control and GCC185-depleted cells. No cell surface protein was detected in cells held on ice rather than at 37°C, as would be expected for a vesicular transport process. Thus, despite a slight dispersion of Golgi markers and generation of Golgi mini-stacks, Golgi function did not seem compromised in cells depleted of GCC185.
Figure 8.
The Golgi complex in cells lacking GCC185 is functional. (A) Arrival of VSV-G-YFP protein at the surface of control (mock transfected) or GCC185 siRNA–treated cells. Cells were pulse labeled for 30 min and then chased for the times indicated; surface VSV-G-YFP was determined using a surface antibody binding procedure. Data shown are from two separate experiments in which cells were chased for 45 or 120 min, respectively. No surface VSV-G protein was detected in reactions held on ice. Values for surface VSV-G from PhosphorImager scans were normalized for protein content and are presented as percentage of control reactions; background at time 0 chase was subtracted. Examples of the GCC185 depletion blots are presented in the inset at the top. (B) Tyrosine sulfation of endogenous acceptors was determined in equal quantities of control (mock-transfected) and GCC185-depleted cells. PhosphorImager scans yielded 3280 arbitrary units for the GCC185 control and 4360 arbitrary units for the Golgin-97 control reaction. Data are normalized to 100% to ease comparison. (C) Cellular content of fluorescent transferrin internalized for 5 and 15 min determined by ImageJ software, with (dashed line) or without (solid line) GCC185 depletion.
In addition, the ability of cells to add sulfate to endogenous tyrosine acceptors was only slightly inhibited upon GCC185 depletion (Figure 8B). Finally, the specificity of the inhibition of endosome to Golgi transport was further confirmed by analyzing the ability of cells to endocytose fluorescent transferrin. As shown in Figure 8C, GCC185- depleted cells displayed wild-type transferrin endocytic capacity. Thus, it seems that only endosome-to-Golgi transport is blocked upon loss of GCC185 protein.
Several methods have been previously used to block MPR transport to the TGN in living cells. This includes expression of a Rab9 S21N dominant-negative mutant (Riederer et al., 1994), antisense oligonucleotide, or siRNA depletion of Rab9 or TIP47 proteins (Diaz and Pfeffer, 1998; Ganley et al., 2004), and expression of various TIP47 dominant-negative mutants (Carroll et al., 2001; Hanna et al., 2002; Sincock et al., 2003). These blocks in transport are usually accompanied by compensatory up-regulation of MPRs, enhanced MPR turnover, and in some cases, increased levels of lysosomal enzymes. We therefore investigated the broader consequences of GCC185 depletion, as an independent clue to the function of this protein.
In support of a role for GCC185 in MPR transport, absolute cellular levels of CI-MPRs increased approximately twofold after 72 h of GCC185 siRNA treatment (Figure 9A). Under these conditions, the levels of Rab9 were unchanged (our unpublished data). The effect was specific for GCC185 depletion, because Golgin-97 depletion did not influence the levels of CI-MPRs in these cells (Figure 9B). It is important to note that Golgin-97 depletion decreased GCC185 levels 37% (Figure 9B). This may in part explain the 23% inhibition of MPR transport detected upon Golgin-97 depletion in living cells (Figure 7A).
Figure 9.
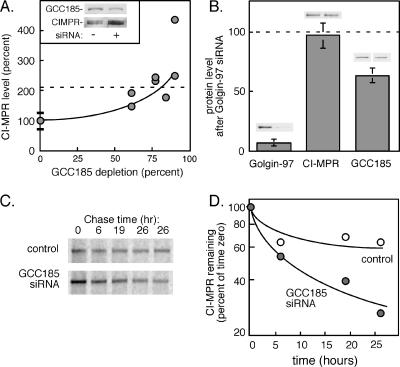
GCC185 depletion alters the levels of CI-MPRs. (A) CI-MPR protein levels determined by immunoblot of equal amounts of control (mock transfected) or GCC185 siRNA-treated cells. Values are graphed as a function of the percentage of GCC185 depleted in four separate experiments; the data are presented in relation to matched control samples (100 ± 27%; n = 9). The inset shows an example blot from one GCC185 depletion experiment with ∼60% depletion. (B) Levels of Golgin-97, CI-MPR and GCC185 in Golgin-97 siRNA-treated cells relative to control (mock-transfected) cells, determined by immunoblot. The small gels above each bar show the control and depleted levels for each distinct protein. Equal amounts of extract protein were analyzed in each case. (C) CI-MPR is unstable in GCC185-depleted cells. HeLa cells treated with GCC185 siRNA were labeled with [35S]methionine/cysteine and chased for the indicated times along with control cells. Equal amounts of cells were harvested (as determined by protein assay), and the CI-MPR was immunoprecipitated and subjected to SDS-PAGE and autoradiography. (D) Quantitation of the experiment shown in C, plotted on a semilog plot.
GCC185-depleted cells displayed an increase in the amount of newly synthesized MPRs and the rate of their turnover (Figure 9, C and D). When cells were metabolically labeled for 1 h, chased in complete medium for 3 h to allow CI-MPRs to fold and transit completely through the Golgi complex, and then chased for additional time to monitor receptor turnover (Figure 9C), we noted that at time 0, GCC185-depleted cells contained more newly synthesized and processed CI-MPR protein. In addition, the MPRs were more rapidly degraded in cells depleted of GCC185 protein (Figure 9D). Together, our data show that GCC185 is intimately involved in the normal life cycle of MPRs in vivo.
We estimate an overall difference in the CI-MPR degradation rate of about twofold (Figure 9D) and an increase of ∼2.8-fold for CI-MPR synthesis (Figure 9C). Because of the long half-life of MPRs, we had to start the metabolic labeling after only 24 h of GCC185 depletion. The steady-state levels (Figure 9A) are measured after 72 h of depletion. We think it is likely that these rates change between 24 and 72 h of GCC185 depletion, yielding an overall twofold change in CI-MPR levels at steady state.
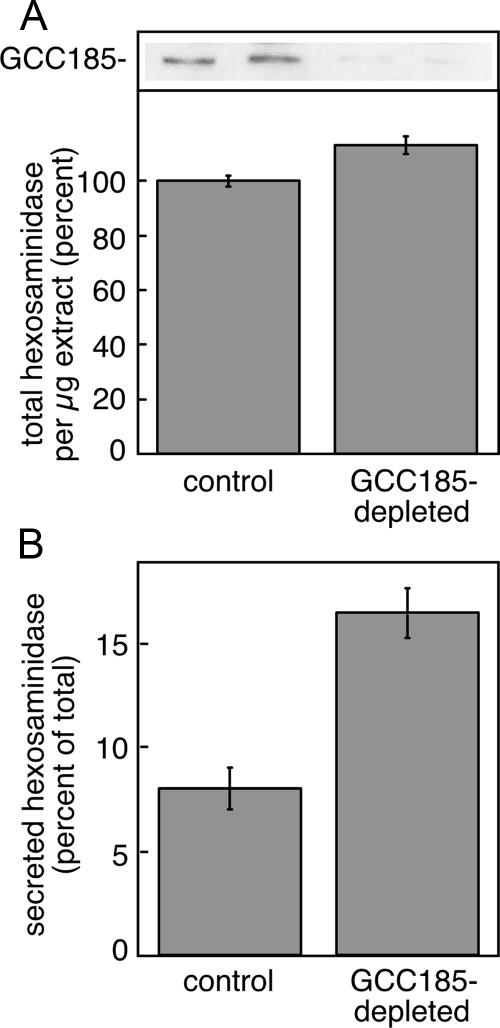
Finally, we noted that cells lacking GCC185 displayed increased secretion of hexosaminidase (Figure 10). Control and depleted cells contained similar total amounts of hexosaminidase, yet GCC185-depleted cells secreted twice as much hexosaminidase as the controls. Hexosaminidase is normally captured by mannose 6-phosphate receptors in the Golgi for transport to prelysosomes. The inability of MPRs to sort this lysosomal enzyme in the Golgi is consistent with MPR sequestration in a post-Golgi compartment. In summary, these data confirm our conclusion that GCC185 is needed for MPRs to accomplish efficient lysosomal enzyme delivery.
Figure 10.
Hexosaminidase secretion in cells depleted of GCC185. HeLa cells grown in 6-cm dishes were either mock transfected or transfected with GCC185 siRNA for 72 h. Subconfluent cells were washed and incubated for 6 h in fresh media containing 10 mM mannose 6-phosphate to block reinternalization of secreted hexosaminidase. (A) Percentage of total hexosaminidase (cellular + secreted) in control and GCC185-depleted cells. Inset at the top shows an immunoblot for GCC185 protein. (B) Percentage of total hexosaminidase secreted in the presence of 10 mM mannose 6-phosphate in control and GCC185-depleted cells, corrected for the efficiency of depletion (85%). Values represent the average of duplicate determinations.
DISCUSSION
We have shown here that the Golgi-associated coiled-coil protein GCC185 is a specific binding partner of the Rab9 GTPase and is needed along with Rab9, both in vitro and in vivo, for the transport of MPRs from late endosomes to the TGN. GCC185 is a member of the Golgin family of putative tethering factors. These proteins all contain long, extended stretches of coiled-coil forming sequences, and soluble members of this family associate with the Golgi complex via membrane-associated binding partners. Golgin tethers have been implicated in the process of vesicle docking; they are also proposed to play a role in Golgi cisternal assembly (for review, see Short et al., 2005). The close relationship between the subcellular localizations of membrane-bound GCC185 and sialyltransferase (Luke et al., 2003) is consistent with GCC185 being localized to just the right place to receive MPRs returning to the site of sialyltransferase at the TGN where they are known to become resialylated (Duncan and Kornfeld, 1988).
We have shown here that the interaction of the putative TGN tether GCC185 with the Rab9 GTPase is not used to localize the majority of GCC185 protein in cells. Indeed, siRNA depletion of Rab9 did not change the Golgi association of GCC185. Thus, Rab9 interaction with GCC185 may represent an interaction important for transport vesicle targeting, rather than Golgin localization. There are several examples of specific Rab effectors, including Golgins, that do not require a Rab protein for their steady-state localization. For example, a mutant form of EEA1 that binds phosphatidylinositol 3-phosphate but not Rab5 is targeted to early endosomes as efficiently as the wild-type protein (Lawe et al., 2002). Likewise, Golgin-45 uses GRASP55 for Golgi association, yet it is a Rab2 effector (Short et al., 2001). Moreover, the integral membrane protein Golgin-84 interacts with Rab1 (Diao et al., 2003; Satoh et al., 2003). Thus, there are numerous examples of Rab interactions that do not necessarily drive protein localization.
Models for GCC185 Function in MPR Transport
The simplest model for a TGN-localized, Rab9 binding partner would be for the Golgi-bound protein to engage a Rab9-decorated transport vesicle. Yet, the Rab9 binding site we have identified is located near the C terminus of GCC185 that is likely to be oriented adjacent to the TGN membrane. It is difficult to imagine how a membrane-proximal binding site would be of benefit in an initially, long-distance tethering process. Several alternative models resolve this dilemma.
First, GCC185 may have an additional binding site for Rab9 at its N terminus. A second possibility is equally plausible: there is a significant pool of cytosolic GCC185 protein (∼50%) that seems much fainter in fluorescence micrographs because it is not concentrated in the perinuclear region. Because Rab9 does not localize GCC185 to the Golgi, we favor a model in which Rab9 transport vesicles interact first with cytosolic GCC185. In yeast exocytosis, for example, a cytosolic exocyst tether is thought to bind transport vesicles and participate in their docking at the plasma membrane. Moreover, p115 is thought to act as a tether from a soluble pool. Thus, such a model is not without precedent.
If cytosolic GCC185 first engages the transport vesicle-associated, Rab9 protein, the GCC185-decorated transport vesicle may engage another tethering protein at the TGN—either GCC185 that is already localized there, or another protein. All tethers must release their bound vesicles, to enable a vesicle to fuse with its target. Nothing is known about tether release, and it will be of interest to determine where GCC185 engages Rab9, and how this pairing is released.
Our data support a model in which GCC185 uses a protein other than Rab9 to achieve its localization at the TGN. Because Rab9 binding to GCC185 did not require a conserved Tyr residue that is an important determinant of typical Arl1 binding, Rab9 may bind GCC185 by interaction with different amino acids than those used by Arl1. The protein construct we have used in our studies contains 110 residues of GCC185, and experiments are in progress to map the precise Rab9 interaction domain within this construct. It remains possible that another member of the large family of Arl proteins uses the GRIP domain of GCC185 to mediate TGN binding. Perhaps this interaction competes with Rab9, allowing release of GCC185 from a docked vesicle, and subsequent fusion.
Interference with the Rab9-dependent pathway for MPR transport from late endosomes to the Golgi increases the total number of MPRs within such cells (Riederer et al., 1994; Ganley et al., 2004). Similar changes were seen here in cells depleted of GCC185, consistent with a role for this protein in MPR recycling from late endosomes. In contrast, MPR levels remained constant in cells depleted of Golgin-97. Independent evidence that GCC185 is needed for efficient lysosomal sorting comes from our finding that cells lacking GCC185 showed hypersecretion of normally lysosomally targeted hexosaminidase.
Recently, Lu et al. (2004) and Yoshino et al., (2005) implicated Golgin-97 and Golgin-245, respectively, in early/recycling endosome-to-TGN transport of Shiga toxin. Unlike the Rab9-dependent MPR transport from late endosomes that we study, endosome to TGN transport of Ricin is independent of Rab9 (Iversen et al., 2001), and early endosome-to-TGN transport of Shiga toxin requires Rab6A (Mallard et al., 2002) and Rab11 instead (Wilcke et al., 2000). In their very thorough biochemical study, Lu et al., (2004) showed that purified Golgin-97 GRIP domain inhibited Shiga toxin transport to the Golgi in vitro, and expression of that GRIP domain inhibited transport in vivo. siRNA depletion of >90% of Golgin-97 led to a ∼35% block in Shiga toxin transport from early endosomes to the TGN in those cells. Yoshino et al. (2005) used morphological localization as a measure of Shiga toxin transport to the TGN, which was inhibited in cells lacking Golgin-245.
In our experiments, GCC185 depletion led to >75% inhibition of MPR recycling from late endosomes in living cells under conditions in which Golgin-97 depletion only inhibited MPR transport 23%. Although GCC185 depletion in the HEK-293 cells used for transport assays did not influence Golgin-97 levels (our unpublished data), it should be noted that Golgin-97 depletion in this cell line decreased GCC185 levels 37% (Figure 9B). This decrease could be responsible for the 23% transport inhibition that we observed under these conditions (Figure 7A). Nevertheless, the residual GCC185 remaining in the Golgin-97–depleted reactions was sufficient to support MPR transport to the TGN. We conclude that most MPR transport represents a GCC185-mediated process; a small proportion may use Golgin-97.
In summary, we have shown that GCC185 acts in concert with Rab9 to ensure efficient transport of MPRs from endosomes to the Golgi complex. An important challenge for the future will be to determine how GCC185 is able to bind to Rab9 in a manner that enables transport vesicles, but not entire late endosome compartments, to dock productively at the trans-Golgi network.
Supplementary Material
ACKNOWLEDGMENTS
We are especially indebted to Dr. Bill Balch for encouraging us to pursue another two-hybrid screen and to Dr. Monica Calero for participating in the secondary screen of all two-hybrid clones for lack of binding to Rab9S21N. This research was funded by the Ara Parseghian Medical Research Foundation and National Institutes of Health Grant DK-37332 (to S.P.). K.S. was the recipient of a fellowship from the Leukemia and Lymphoma Society.
Abbreviations used:
- CD-MPR
cation-dependent mannose 6-phosphate receptor
- CI-MPR
cation-independent mannose 6-phosphate receptor
- MPR
mannose 6-phosphate receptor
- siRNA
small interfering RNA
- TIP47
tail-interacting protein of 47 kDa
- VSVG
G protein of the vesicular stomatitis virus.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-02-0153) on August 2, 2006.
REFERENCES
- Aivazian D., Serrano R. L., Pfeffer S. TIP47 is a key effector for Rab9 localization. J. Cell Biol. 2006;173:917–926. doi: 10.1083/jcb.200510010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan B. B., Moyer B. D., Balch W. E. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 2000;289:444–448. doi: 10.1126/science.289.5478.444. [DOI] [PubMed] [Google Scholar]
- Bai C., Elledge S. J. Gene identification using the yeast two-hybrid system. Methods Enzymol. 1996;273:331–347. doi: 10.1016/s0076-6879(96)73029-x. [DOI] [PubMed] [Google Scholar]
- Barbero P., Bittova L., Pfeffer S. R. Visualization of Rab9-mediated vesicle transport from endosomes to the trans-Golgi in living cells. J. Cell Biol. 2002;156:511–518. doi: 10.1083/jcb.200109030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F. A. A novel Rab6-interacting domain defines a family of Golgi-targeted coiled-coil proteins. Curr. Biol. 1999;9:381–384. doi: 10.1016/s0960-9822(99)80167-5. [DOI] [PubMed] [Google Scholar]
- Burguete A. S., Sivars U., Pfeffer S. Purification and analysis of TIP47 function in Rab9–dependent mannose 6-phosphate receptor trafficking. Methods Enzymol. 2005;403:357–366. doi: 10.1016/S0076-6879(05)03031-4. [DOI] [PubMed] [Google Scholar]
- Carroll K. S., Hanna J., Simon I., Krise J., Barbero P., Pfeffer S. R. Role of Rab9 GTPase in facilitating receptor recruitment by TIP47. Science. 2001;292:1373–1376. doi: 10.1126/science.1056791. [DOI] [PubMed] [Google Scholar]
- Derby M. C., van Vliet C., Brown D., Luke M. R., Lu L., Hong W., Stow J. L., Gleeson P. A. Mammalian GRIP domain proteins differ in their membrane binding properties and are recruited to distinct domains of the TGN. J. Cell Sci. 2004;117:5865–5874. doi: 10.1242/jcs.01497. [DOI] [PubMed] [Google Scholar]
- Diao A., Rahman D., Pappin D. J., Lucocq J., Lowe M. The coiled-coil membrane protein Golgin-84 is a novel rab effector required for Golgi ribbon formation. J. Cell Biol. 2003;160:201–212. doi: 10.1083/jcb.200207045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz E., Pfeffer S. R. TIP 47, a cargo selection device for mannose 6-phosphate receptor trafficking. Cell. 1998;93:433–443. doi: 10.1016/s0092-8674(00)81171-x. [DOI] [PubMed] [Google Scholar]
- Diaz E., Schimmöller F., Pfeffer S. R. A novel Rab9 effector required for endosome to TGN transport. J. Cell Biol. 1997;138:283–290. doi: 10.1083/jcb.138.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan J. R., Kornfeld S. Intracellular movement of two mannose 6-phosphate receptors: return to the Golgi apparatus. J. Cell Biol. 1988;106:617–628. doi: 10.1083/jcb.106.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- Fridmann-Sirkis Y., Siniossoglou S., Pelham H.R.B. TMF is a Golgin that binds Rab6 and influences Golgi morphology. BMC Cell Biol. 2004;5:18–27. doi: 10.1186/1471-2121-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley I., Carroll K., Bittova L., Pfeffer S. R. Rab9 GTPase regulates late endosome size and requires effector interaction for its stability. Mol. Biol. Cell. 2004;15:5420–5430. doi: 10.1091/mbc.E04-08-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P., Dahms N. M., Kornfeld S. Mannose 6-phosphate receptors: new twists in the tale. Nat. Rev. Mol. Cell Biol. 2003;4:202–212. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- Gillingham A. K., Munro S. Long coiled-coil proteins and membrane traffic. Biochim. Biophys. Acta. 2003;1641:71–85. doi: 10.1016/s0167-4889(03)00088-0. [DOI] [PubMed] [Google Scholar]
- Gleeson P. A., Lock J. G., Luke M. R., Stow J. L. Domains of the TGN: coats, tethers and G proteins. Traffic. 2004;5:315–326. doi: 10.1111/j.1398-9219.2004.00182.x. [DOI] [PubMed] [Google Scholar]
- Hanna J., Carroll K., Pfeffer S. R. Identification of residues in TIP47 essential for Rab9 binding. Proc. Natl. Acad. Sci. USA. 2002;99:7450–7454. doi: 10.1073/pnas.112198799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itin C., Rancaño C., Nakajima Y., Pfeffer S. R. A novel assay reveals a role for alpha-SNAP in mannose 6-phosphate receptor transport from endosomes to the TGN. J. Biol. Chem. 1997;272:27737–27744. doi: 10.1074/jbc.272.44.27737. [DOI] [PubMed] [Google Scholar]
- Iversen T. G., Skretting G., Llorente A., Nicoziani P., van Deurs B., Sandvig K. Endosome to Golgi transport of ricin is independent of clathrin and of the Rab9- and Rab11-GTPases. Mol. Biol. Cell. 2001;12:2099–2107. doi: 10.1091/mbc.12.7.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjer-Nielsen L., Teasdale R. D., van Vliet C., Gleeson P. A. A novel Golgi-localisation domain shared by a class of coiled-coil peripheral membrane proteins. Curr. Biol. 1999a;9:385–388. doi: 10.1016/s0960-9822(99)80168-7. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L., van Vliet C., Erlich R., Toh B. H., Gleeson P. A. The Golgi-targeting sequence of the peripheral membrane protein p230. J. Cell Sci. 1999b;112:1645–1654. doi: 10.1242/jcs.112.11.1645. [DOI] [PubMed] [Google Scholar]
- Lawe D. C., Chawla A., Merithew E., Dumas J., Carrington W., Fogarty K., Lifshitz L., Tuft R., Lambright D., Corvera S. Sequential roles for phosphatidylinositol 3-phosphate and Rab5 in tethering and fusion of early endosomes via their interaction with EEA1. J. Biol. Chem. 2002;277:8611–8617. doi: 10.1074/jbc.M109239200. [DOI] [PubMed] [Google Scholar]
- Lombardi D., Soldati T., Riederer M. A., Goda Y., Zerial M., Pfeffer S. R. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J. 1993;12:677–682. doi: 10.1002/j.1460-2075.1993.tb05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Hong W. Interaction of Arl1-GTP with GRIP Domains Recruits Autoantigens Golgin-97 and Golgin-245/p230 onto the Golgi. Mol. Biol. Cell. 2003;14:3767–3781. doi: 10.1091/mbc.E03-01-0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Tai G., Hong W. Autoantigen Golgin-97, an effector of Arl1 GTPase, participates in traffic from the endosome to the Trans-Golgi network. Mol. Biol. Cell. 2004;15:4426–4443. doi: 10.1091/mbc.E03-12-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke M. R., Kjer-Nielsen L., Brown. D. L., Stow J. L., Gleeson P. A. GRIP domain-mediated targeting of two new coiled-coil proteins, GCC88 and GCC185, to subcompartments of the trans-Golgi network. J. Biol. Chem. 2003;278:4216–4226. doi: 10.1074/jbc.M210387200. [DOI] [PubMed] [Google Scholar]
- Mallard F., Tang B. L., Galli T., Tenza D., Saint-Pol A., Yue X., Antony C., Hong W., Goud B., Johannes L. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J. Cell Biol. 2002;156:653–664. doi: 10.1083/jcb.200110081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matanis T., et al. Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat. Cell Biol. 2002;4:986–992. doi: 10.1038/ncb891. [DOI] [PubMed] [Google Scholar]
- Munro S., Nichols B. J. The GRIP domain – a novel Golgi-targeting domain found in several coiled-coil proteins. Curr. Biol. 1999;9:377–380. doi: 10.1016/s0960-9822(99)80166-3. [DOI] [PubMed] [Google Scholar]
- Panic B., Perisic O., Veprintsev D. B., Williams R. L., Munro S. Structural basis for Arl1-dependent targeting of homodimeric GRIP domains to the Golgi apparatus. Mol. Cell. 2003a;12:863–874. doi: 10.1016/s1097-2765(03)00356-3. [DOI] [PubMed] [Google Scholar]
- Panic B., Whyte J. R., Munro S. The ARF-like GTPases Arl1p and Arl3p act in a pathway that interacts with vesicle-tethering factors at the Golgi apparatus. Curr. Biol. 2003b;13:405–410. doi: 10.1016/s0960-9822(03)00091-5. [DOI] [PubMed] [Google Scholar]
- Puthenveedu M. A., Bachert C., Puri S., Lanni F., Linstedt A. D. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi enzyme distribution. Nat. Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- Riederer M. A., Soldati T., Shapiro A. D., Lin J., Pfeffer S. R. Lysosome biogenesis requires Rab9 function and receptor recycling from endosomes to the trans-Golgi network. J. Cell Biol. 1994;125:573–582. doi: 10.1083/jcb.125.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A., Wang Y., Malsam J., Beard M. B., Warren G. Golgin-84 is a rab1 binding partner involved in Golgi structure. Traffic. 2003;4:153–161. doi: 10.1034/j.1600-0854.2003.00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz K., Richards K., Botstein D. BIM1 encodes a microtubule-binding protein in yeast. Mol. Biol. Cell. 1997;8:2677–2691. doi: 10.1091/mbc.8.12.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty S. R., Shin M. E., Yoshino A., Marks M. S., Burd C. G. Golgi recruitment of GRIP domain proteins by Arf-like GTPase 1 is regulated by Arf-like GTPase 3. Curr. Biol. 2003;13:401–404. doi: 10.1016/s0960-9822(03)00089-7. [DOI] [PubMed] [Google Scholar]
- Short B., Haas A., Barr F. A. Golgins and GTPases, giving identity and structure to the Golgi apparatus. Biochim. Biophys. Acta. 2005;1744:383–395. doi: 10.1016/j.bbamcr.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Short B., Preisinger C., Körner R., Kopajtich R., Byron O., Barr F. A. A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J. Cell Biol. 2001;155:877–883. doi: 10.1083/jcb.200108079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sincock P. M., Ganley I. G., Krise J. P., Diederichs S., Sivars U., O'Connor B., Ding L., Pfeffer S. R. Self-assembly is important for TIP47 function in mannose 6-phosphate receptor transport. Traffic. 2003;4:18–25. doi: 10.1034/j.1600-0854.2003.40104.x. [DOI] [PubMed] [Google Scholar]
- Soldati T., Riederer M. A., Pfeffer S. R. Rab GDI: a solubilizing and recycling factor for rab9 protein. Mol. Biol. Cell. 1993;4:425–434. doi: 10.1091/mbc.4.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valkenburgh H., Shern J. F., Sharer J. D., Zhu X., Kahn R. A. ADP-ribosylation factors (ARFs) and ARF-like 1 (ARL1) have both specific and shared effectors: characterizing ARL1-binding proteins. J. Biol. Chem. 2001;276:22826–22837. doi: 10.1074/jbc.M102359200. [DOI] [PubMed] [Google Scholar]
- Warren G., Davoust J., Cockcroft A. Recycling of transferrin receptors in A431 cells is inhibited during mitosis. EMBO J. 1984;3:2217–2225. doi: 10.1002/j.1460-2075.1984.tb02119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcke M., Johannes L., Galli T., Mayau V., Goud B., Salamero J. Rab11 regulates the compartmentalization of early endosomes required for efficient transport from early endosomes to the trans-Golgi network. J. Cell Biol. 2000;151:1207–1220. doi: 10.1083/jcb.151.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Lu L., Hong W., Song H. Structural basis for recruitment of GRIP domain Golgin-245 by small GTPase Arl1. Nat. Struct. Mol. Biol. 2004;11:86–94. doi: 10.1038/nsmb714. [DOI] [PubMed] [Google Scholar]
- Yoshino A., et al. tGolgin-1 (p230, golgin-245) modulates Shiga-toxin transport to the Golgi and Golgi motility towards the microtubule-organizing centre. J. Cell Sci. 2005;118:2279–2293. doi: 10.1242/jcs.02358. [DOI] [PubMed] [Google Scholar]
- Zolov S. N., Lupashin V. V. Cog3p depletion blocks vesicle-mediated Golgi retrograde trafficking in HeLa cells. J. Cell Biol. 2005;168:747–759. doi: 10.1083/jcb.200412003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.