Abstract
The budding and fission of vesicles during membrane trafficking requires many proteins, including those that coat the vesicles, adaptor proteins that recruit components of the coat, and small GTPases that initiate vesicle formation. In addition, vesicle formation in vitro is promoted by the hydrolysis of acyl-CoA lipid esters. The mechanisms by which these lipid esters are directed to the appropriate membranes in vivo, and their precise roles in vesicle biogenesis, are not yet understood. Here, we present the first report on membrane associated ACBP domain-containing protein-1 (MAA-1), a novel membrane-associated member of the acyl-CoA–binding protein family. We show that in Caenorhabditis elegans, MAA-1 localizes to intracellular membrane organelles in the secretory and endocytic pathway and that mutations in maa-1 reduce the rate of endosomal recycling. A lack of maa-1 activity causes a change in endosomal morphology. Although in wild type, many endosomal organelles have long tubular protrusions, loss of MAA-1 activity results in loss of the tubular domains, suggesting the maa-1 is required for the generation or maintenance of these domains. Furthermore, we demonstrate that MAA-1 binds fatty acyl-CoA in vitro and that this ligand-binding ability is important for its function in vivo. Our results are consistent with a role for MAA-1 in an acyl-CoA–dependent process during vesicle formation.
INTRODUCTION
Acyl-CoA–binding proteins (ACBPs) are a family of proteins isolated by virtue of their ability to bind acyl-CoA molecules with high affinity (Mogensen et al., 1987). The archetypal ACBP, which was first isolated from bovine liver, is a protein of 86 residues that binds long-chain acyl-CoA. The protein is thought to help render acyl-CoAs soluble in the cytoplasm and to protect them from hydrolysis by thioesterases (Knudsen et al., 2000). The protein has been very highly conserved in evolution and has been isolated from all eukaryotic cells examined to date (Burton et al., 2005). The yeast orthologue Acb1p is an 86-residue protein that, like its mammalian counterparts, forms stable complexes with fatty acyl-CoAs in vitro (Knudsen et al., 1994). Yeast cells lacking Acb1p activity display a variety of defects, including reduced growth rate, membrane perturbations, vacuolar fragmentation, and decreased ceramide synthesis (Gaigg et al., 2001; Faergeman et al., 2004).
In addition to the Acb1p orthologue, multicellular eukaryotes harbor several other proteins containing a highly conserved ACBP domain (Burton et al., 2005). These proteins contain a region with high sequence similarity to the entire Acb1p protein within a longer polypeptide sequence. The function of these larger ACBPs in vivo, however, is presently not known. Two such ACBP domain-containing proteins, predicted to be membrane associated, were isolated from human and bovine brain tissue. However, their function still remains elusive (Webb et al., 1987).
Acyl-CoAs themselves participate in several different types of biochemical reactions; they are substrates for β-oxidation, they are required for the synthesis and remodeling of lipids, and they are substrates for protein acylation reactions. Experiments with cell-free extracts aimed at understanding how transport vesicles are generated from membranous organelles in eukaryotic cells have revealed that acyl-CoAs are required for the budding of vesicles from donor organelles (Glick and Rothman, 1987; Ostermann et al., 1993). The formation of COPI vesicles from Golgi cisternae in cell-free extracts revealed that, in addition to GTP, Arf1p, and coat proteins, this process also requires long-chain acyl-CoA (Glick and Rothman, 1987; Ostermann et al., 1993). In the absence of acyl-CoA, coated buds accumulate but fail to pinch off to form vesicles. When acyl-CoA is added to the reaction, fission occurs, releasing functionally active transport vesicles. Presently, the precise role of acyl-CoA in the fission process is not completely understood. Recent results, however, suggest that one possibility is that acyl-CoAs are substrates for brefeldin-A ADP-ribosylated substrate (BARS) to allow the recruitment of the Arf1 GTPase-activating protein (Yang et al., 2005). However, how acyl-CoA is directed to the appropriate membranes is not yet known.
Important insights into the mechanisms of vesicle trafficking in metazoans have come from studies with the nematode Caenorhabditis elegans (Fares and Grant, 2002). Recently, Grant et al. and Fares and Greenwald have developed assays for the identification of new factors involved in endocytic vesicle transport (Grant and Hirsh, 1999; Fares and Greenwald, 2001a). These studies have led to the discovery of several new key regulators such as rme-1 (Grant et al., 2001), rme-6 (Sato et al., 2005), rme-8 (Zhang et al., 2001), cup-5 (Fares and Greenwald, 2001b), and cup-4 (Patton et al., 2005). Although most of the genes are absent from yeast, they are conserved from nematodes to humans. Mutations in rme-1 cause a marked reduction in the efficiency of endocytic recycling. In oocytes, this defect results in a reduction in the amount of the yolk receptor RME-2 present on the surface of oocytes and consequently a reduction in the efficiency with which yolk is taken up by oocytes (Grant et al., 2001). In the intestine, rme-1 null mutations cause large vacuoles to form that receive input from the basolateral endocytosis pathway and seem to result from the enlargement of the endosomal recycling compartment (ERC) (Grant et al., 2001). RME-1 is an ATPase that in wild-type worms is associated with the ERC (Grant et al., 2001; Lee et al., 2005a). It is thought that RME-1 promotes recycling by promoting vesicle transport between the ERC and the plasma membrane. The molecular mechanism by which it acts, however, is presently not known. A mammalian orthologue of RME-1, EHD1 (Eps15 homology domain-containing protein 1) has been isolated and shown to affect endocytic recycling in mammalian cells grown in tissue culture (Lin et al., 2001; Caplan et al., 2002).
Here, we have analyzed the function in C. elegans of maa-1, a gene predicted to encode a membrane-associated protein containing an ACBP domain. We describe experiments showing that membrane associated ACBP domain-containing protein-1 (MAA-1) in the worm modulates vesicle trafficking in the intestine, hypodermis, and oocytes. The ability to bind acyl-CoAs is important for MAA-1 function. We propose that MAA-1 acts to recruit acyl-CoAs to certain intracellular membranous organelles in order to facilitate trafficking.
MATERIALS AND METHODS
General Methods and Strains
Methods for handling and culturing C. elegans were essentially as described previously (Brenner, 1974). All strains were grown at 20°C unless otherwise stated. The wild-type parent for all strains was the Bristol N2 strain. Mutations used in this study were linkage group I, arIs37 (pmyo-3::ssGFP) (Fares and Greenwald, 2001a) LGIII maa-1(sv37 and sv38); LGV, rme-1(b1045, b1020) (Grant et al., 2001); and LGX, bIs1 (Grant and Hirsh, 1999).
Molecular Cloning of maa-1
Total RNA was extracted using TRIzol reagent according to the protocol supplied by the manufacturer (Invitrogen, Carlsbad, CA). mRNA was purified from total RNA using Poly-A-Tract mRNA isolation system (Promega, Madison, WI). The maa-1 cDNA was amplified by reverse transcription-polymerase chain reaction (RT-PCR) by using SuperScript (Invitrogen) with the primers 5′-ATG TCG GTG TTT GAA ACT GCT GT-3′ and 5′-TCA CAT CAG TAA TCG AAG AAA TC-3′. The amplified PCR product was subcloned into pCRII-Topo (Invitrogen) and sequenced with T7 and SP6 primers, by using standard techniques. The 5′ part of the transcript was amplified from total mRNA with a gene-specific primer 5′-TCG GAG TAG AAA CTT CGT GAC A-3′ and the SL1 or SL2 trans-spliced leader by using single tube SuperScript RT-PCR (Invitrogen) according to the manufacturer's instructions. A nested PCR was necessary to avoid unspecific products. The nested product was subcloned into pCRII-Topo (Invitrogen) and sequenced to identify the 5′ untranslated region.
To generate the maa-1::gfp fusion gene, the maa-1 genomic DNA together with 5 kilobases (kb) of upstream sequence was amplified by PCR by using the primers 5′-GCC GAA AAA CTC AAA TTT TCA TTT AAA AAT CCA C-3′ and 5′-ATG GCT CCG ACG ATT TCT TCG ATT CCC GGG-3′ and subcloned into pCRII-Topo. The promotor and coding region were subcloned into pPD95.77 (a kind gift from A. Fire, Departments of Pathology and Genetics, Stanford University, Stanford, CA) by using PstI and XmaI fusing the coding sequence in-frame to (S65C) green fluorescent protein (GFP) to generate pMAA-1::GFP. The mutant forms of the maa-1::GFP (mtMAA-1::GFP and tmMAA-1::GFP) were constructed by site-specific mutagenesis (Fisher and Pei, 1997). The plasmid expressing cyMAA-1::GFP was constructed by inserting a PstI-SmaI fragment from pMAA-1::GFP into pPD49.26 digested with PstI-EcoRV. Subsequently, an EcoRV fragment containing GFP from pPD102.33 was inserted into the EcoRV site found within the maa-1 gene. The transgenic lines were generated by coinjecting the reporter constructs together with pRF4 into N2 hermaphrodites. Both plasmids were injected at a concentration of 80 ng/μl.
The expressed sequence tag (EST) clone yk1249h02.5 extends 57 base pairs downstream of the gene to the poly-A tail. However, the upstream sequence was not covered by any clones in the database. To rule out any possible splice variants in the upstream region, RT-PCR was used to amplify the 5′ end of the transcript. The mRNA is trans-spliced 59 base pairs upstream of the predicted start site, and no longer splice products could be detected.
Recombinant Expression and Purification of MAA-1
Full-length MAA-1 was found to be toxic when expressed in bacteria. We therefore purified a portion of the protein containing the ACBP domain. To construct histidine- and Factor Xa-tagged MAA-1 without the transmembrane domain, the DNA sequence corresponding to amino acid residues 1–226 of MAA-1 was amplified from cDNA obtained by reverse transcription of random-hexamer primed C. elegans N2 total RNA by using the following primers: upstream, 5′-GGAATTCCATATGGGACATCACCATCACCATCACGGTAGTGGTAGTATCCAGGGGAGGTCGGTGTTTGAAACTGCTGTT-3′; and downstream, 5′-CGCCTCGAGTCACGCTTTTTTCATGAGTCCAATCAG-3′, where the restriction sites are underlined, the sequence encoding the histidine tag is in italics, and the sequence encoding the Factor Xa site is in bold. The PCR amplicon was digested with NdeI and XhoI and ligated into an NdeI/XhoI-restricted pACYC-DUET-1 expression vector (Novagen, Madison, WI).
Expression and Purification of the Truncated MAA-1.
The construct was transformed into competent BL21(DE3) cells and grown overnight in Luria Broth containing 50 μg/ml chloramphenicol. The following day, cells were diluted 100 times in fresh media and grown to an optical density at 600 nm of 1. Expression of truncated MAA-1 was induced by incubating cells with 1 mM isopropyl β-d-thiogalactoside for 3 h. MAA-1 was purified by Ni2+-Sepharose chromatography as described by the manufacturer (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) followed by Q-Sepharose anion-exchange chromatography (GE Healthcare).
Ligand Binding by Native Gel Electrophoresis.
Purified MAA-1 (2 nmol) was incubated with or without ligand (C16:0 acyl-CoA, 6 nmol) in the presence of 6 nM Triton X-100 for 10 min in H2O at room temperature and loaded on to a 12.5% native Phast-gel (GE Healthcare) equilibrated in alanine/Tris, pH 6.6. After separation, proteins were visualized by Coomassie blue staining. The human ACBP control was processed in the same manner.
Complementation of ACB1p-depleted Yeast Cells
The cDNAs encoding full-length MAA-1 or MAA-1 without its transmembrane domain were obtained by RT-PCR by using total RNA isolated from a mixed stage of the Bristol N2 strain. PCR amplicons were purified and digested with either XbaI and BamHI (full-length) or XbaI and XhoI (without transmembrane domain) and inserted into p413-ADH vector carrying the ADH promoter. Mutations were introduced by site-directed mutagenesis as described above. Plasmids were transformed into the pGAL-ACB1 strain in which expression of the ACB1 open reading is controlled by the GAL1 promoter and hence can be repressed by inoculating cells in glucose-supplemented media. The wild-type strain (Y700) was only transformed with the empty vector. Transformed cells were kept on selective YNB plates supplemented with galactose (2%). Yeast cells were inoculated at 30°C in supplemented minimal YNB medium without histidine containing glucose (2%). Cells were grown for 24 h, diluted into fresh medium to an OD600 of 0.07, and growth was followed for at least 14 h.
Isolation of maa-1 Deletion Alleles
The deletion alleles of maa-1 were isolated by screening a C. elegans deletion library generated with UV light and trimethyl-psoralen. The sv37 and sv38 deletion alleles were detected by PCR by using the primers EL 5′-TGG CCA GAA TAC GAA AAA GAT T-3′ and ER 5′-AAA CCCGTA CCT TTT GTT GTT G-3′ for the first round of PCR. A second reaction was carried out using the nested primers IL 5′-TTT GTT TGT CGG GCT CTA GAC T-3′ and IR 5′-ACT TGA CGA ATC AGC TGA CGA C-3′. The animals isolated from this screen were outcrossed nine times to the Bristol N2 strain in order to clean up the genetic background. Northern blotting was performed to confirm the absence of the MAA-1 transcript. Sequencing determined the sv37 allele to be a 1899 base pairs deletion starting 450 base pairs upstream of the ATG into exon 2 (C18D11 coordinates: 16361–18260). The sv38 allele is a 1582-base pair deletion (C18D11 coordinates: 17019–18602). It removes 787 base pairs upstream of the ATG, all of exon 1, and most of intron 1. In subsequent genotyping experiments, the mutant alleles were identified using the primers R1 5′-ACC GCA AAA ATT TTC GAT TTT A-3′, F1 5′-GGG GGG GCC TTC TTC ACT ACT C-3′ and F2 5′-TTG CGA CGA AAA TAC AGT AG-3′ in a duplex PCR reaction.
Immunostaining, Dye Labeling, and Microscopy
Intestinal dissection, fixation, and staining were performed as described previously (Grant and Hirsh, 1999). The affinity-purified rabbit antibody against RME-1 (gift from B. Grant, Department of Molecular Biology and Biochemistry, Rutgers University, Piscataway, NJ) was used at a 1:200 dilution, and a mouse anti-GFP monoclonal antibody (mAb) (Clontech, Mountain View, CA) was used at a 1:200 dilution. The mouse anti-HDEL (a gift from M. Lewis, Laboratory of Molecular Biology-Medical Research Council, Cambridge, United Kingdom) was diluted 1:10 and goat anti-GRASP55 (Santa Cruz Biotechnology, Santa Cruz, CA), 1:50. These samples were costained with a rabbit anti-GFP (Clontech) diluted 1:50. The secondary antibodies Alexa488-conjugated goat anti-rabbit (Invitrogen) and Cy3-conjugated goat anti-mouse (Jackson ImmunoResearch Laboratories, West Grove, PA) or Cy3-conjugated mouse anti-goat were used at a 1:500 dilution. N-(3-Triethylammoniumpropyl)-4-(p-diethylaminophenyl-hexatrienyl) pyridinium dibromide (FM4-64) labeling was carried out by soaking animals in 0.4 mM FM4-64 (Invitrogen) dissolved in egg salts (118 mM NaCl, 48 mM KCl, 2 mM MgCl2, 2 mM CaCl2, and 10 mM HEPES, pH 7.4) for 1 h. Animals were allowed to recover on fresh plates and mounted for microscopy. To label with LysoTracker (Invitrogen), the dye was added to the NGM medium at a final concentration of 2 μM. Animals were grown on these plates for 15 h, transferred to fresh plates, and mounted for microscopy. Fluorescent confocal micrographs were obtained on a laser scanning microscope (TCS SP2; Leica, Deerfield, IL).
Worm Whole-Cell Extracts and Fractionation
The extrachromosomal arrays wtMAA-1::GFP or tmMAA-1::GFP were integrated using standard techniques (Epstein and Shakes, 1995). Synchronized cultures of young adult worms were washed twice in extraction buffer (50 mM Tris, pH7.2, 250 mM sucrose, 2 mM EDTA, and protease inhibitors). Worms were lysed by vortexing with glass beads, and the carcasses were spun down at 2900 × g at 4°C for 20 min. The low-speed supernatant was split in two fractions and mixed with 2 volumes of either extraction buffer or sodium carbonate and incubated on ice for 1 h. The mixtures were then centrifuged at 100,000 × g at 4°C for 1 h. After this time, the supernatants were removed, and the pellets were washed with 4 ml of buffer, spun again, and dissolved in SDS-PAGE loading buffer. The supernatants were precipitated with trichloroacetic acid (13% final concentration) and dissolved in SDS-PAGE loading buffer. Fractions were run on 10% Tris-glycine gels, blotted onto polyvinylidene diflouride membranes, and probed with either anti-GFP or anti-RME-1 antibodies.
Phenotypic Analysis
The number of intestinal vacuoles was scored using Nomarski optics on a Leica DMRB compound microscope equipped with PL Fluotar 100× objective. The intestinal morphology was inspected in 40 L4 hermaphrodites for each genotype unless otherwise stated.
Accession Numbers
A list of accession numbers is available in Supplemental Table S1.
RESULTS
MAA-1 is the C. elegans Member of a Conserved Protein Family
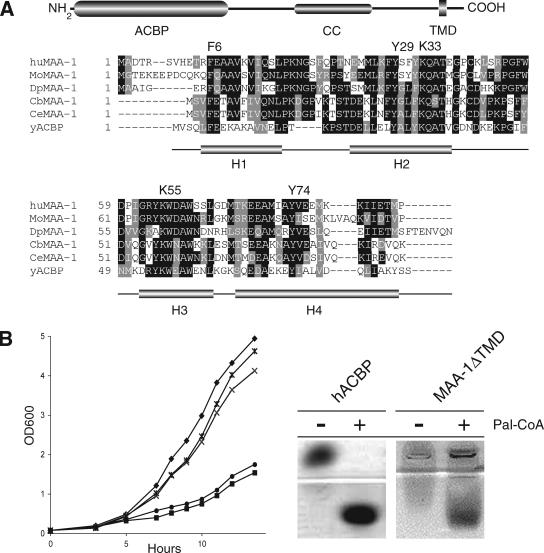
A BLAST search of the C. elegans genome by using the yeast Acb1p as query sequence identified eight predicted genes with the potential to encode proteins containing an ACBP domain. Among these predicted genes, one member (C18D11.2) encoded a potential membrane-associated (MA) isoform, MA-ACBP (Figure 1A). We have named the C18D11.2 protein MAA-1. Subsequent tblastn searches by using C. elegans, fruit fly, and bovine MA-ACBP as query identified 22 sequences with the same predicted domain structure. Phylogenetic analysis confirmed that these sequences are conserved from nematodes to humans (Supplemental Figure S1 and Table S1).
Figure 1.
Alignment, domain structure, and in vivo activity of MAA-1. (A) Alignment of the human, mouse, Drosophila, and nematode MAA-1 ACBP domains with yeast ACBP (Acb1p). Residues involved in ligand binding are shown above the alignment with numbering referring to the yeast ACBP sequence (Ce, C. elegans; Cb, Caenorhabditis briggsae; Dp, Drosophila pseudoobscura; Mo, Mus musculus; Hu, Homo sapiens). The positions of the four α helical domains (H1–4) in the yeast ACBP protein (Acb1p) are depicted below the alignment. At top is shown the overall domain structure of the C. elegans MAA-1 protein. The ACBP domain is located between residues 1–88, the coiled-coil domain (CC) between 186 and 214, and the transmembrane domain (TMD) between 228 and 250. (B) Functional analysis of the MAA-1 acyl-CoA–binding activity. Left, ability of various plasmids harboring either MAA-1 or ACB1 cDNA fragments to rescue the growth defect observed in yeast cells lacking ACB1 (Gaigg et al., 2001). (♦, reference strain; ■, Δacb1 + empty vector; ×, Δacb1 + MAA-1 fl; *, Δacb1 + MAA-1ΔTMD; •, Δacb1 + MAA-1 fl Y31F K35A). Right, both human ACBP and MAA-1 form complexes with palmitoyl-CoA in vitro.
The predicted proteins have the same overall domain organization. A cytosolic N-terminally conserved ACBP domain, containing all residues necessary for ligand binding (Kragelund et al., 1999), is followed by a more divergent region, which in most organisms contains no known motifs (Figure 1A). In C. elegans MAA-1, this region contains a coiled-coil motif between amino acids 186–214, indicating that C. elegans MAA-1 may form multimeric complexes or interacts with heterologous partners (Lupas, 1996). All members of the protein family have a predicted transmembrane domain (TMD) located close to the C terminus. The start of the TMD varies slightly in position between species but has a conserved distal sequence consisting of a WPF(I/L/V) motif.
Disruption of the only ACBP gene in yeast, ACB1, causes a slow growth phenotype (Gaigg et al., 2001). A truncated form of the C. elegans protein, MAA-1ΔTMD, lacking the transmembrane domain, fully rescued the yeast phenotype (Figure 1B). Full-length MAA-1 also rescued the growth defect to some extent but not as efficiently as the truncated protein. A form of yeast Acb1p in which two conserved residues in the protein, Y29F and K33A, are mutated has a 1000-fold lower affinity for acyl-CoA in vitro (Kragelund et al., 1999) and fails to rescue the growth defect in vivo of yeast cells lacking Acb1p (Figure 1B). A mutated C. elegans maa-1 gene, encoding a protein with the analogous amino acid sequence changes, Y31F and K35A, also failed to rescue the yeast growth defect (Figure 1B).
To examine whether the ACBP domain of the C. elegans MAA-1 was able to bind directly to long-chain acyl-CoA esters in vitro, we tested the ability of a soluble form of MAA-1 to induce a mobility shift in a native gel assay. Purified, recombinant MAA-1 caused a shift of C16:0 acyl-CoA in a manner similar to recombinant human ACBP (Figure 1B). MAA-1 was not as efficient as human ACBP at inducing a shift of the C16:0 ester, possibly because the worm protein produced in bacteria is not as soluble as human ACBP (our unpublished data). Together, these results demonstrate that MAA-1 can complement lack of Acb1p in vivo and has affinity for long-chain acyl-CoA esters in vitro; it is thus a member of the ACBP family. These results are consistent with published results using other ACBP proteins. All such proteins tested for their ligand interaction properties show high-affinity binding to long-chain acyl-CoA (Rasmussen et al., 1990; Knudsen et al., 1994; van Aalten et al., 2001).
Characterization of the C. elegans maa-1 Transcript
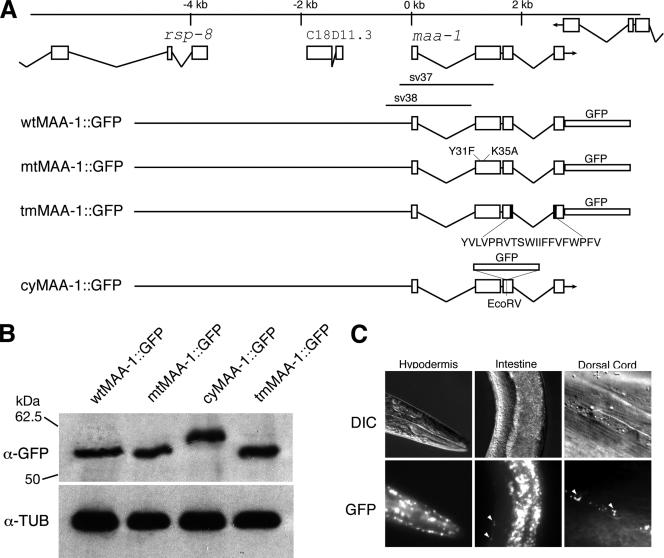
To characterize the maa-1 gene in more detail, a full-length cDNA was obtained from total RNA by RT-PCR. Sequence analysis of the amplicon and that of the published sequences of the EST clones yk510b1 and yk1249h02 confirmed the positions of the splice sites predicted by Wormbase (Chen et al., 2005). To determine in which cells MAA-1 is expressed, an 8.2-kb genomic fragment spanning the maa-1 promoter and coding region was fused in-frame to GFP (Figure 2A), and the construct generated was injected into worms. The construct encodes a protein containing GFP fused at the C terminus to full-length MAA-1. Western blotting of protein extracts from worms harboring the maa-1::gfp transgene indicated that a protein of the predicted molecular mass, 57 kDa, was expressed (Figure 2B). The fusion protein rescues maa-1 defects (see below), indicating that it is functional. In transgenic lines harboring the reporter, GFP fluorescence is detected in the hypodermis and intestine (Figure 2C). A punctuate GFP signal is also apparent close to the positions of the ventral and dorsal nerve cords (Figure 2C, intestine and nerve cord, arrowheads). No fluorescence is seen in other tissues of the worm. Within the hypodermis, the fusion protein seems to be widely expressed. Strong signals are detected in the head region and in hyp7. No signal is detected, however, in the seam cells. The fluorescent signal close to the nerve cords may be nonneuronal, because it is abolished by RNA interference (RNAi) against GFP (our unpublished data). In most genetic backgrounds, neurons in C. elegans are relatively refractory to RNAi (Timmons et al., 2001).
Figure 2.
maa-1 genomic organization and expression pattern. (A) Structure of the genomic region around maa-1. The boxes represent exons and the lines, introns. Three genes are shown, rsp-8, C18D11.3, and maa-1. The regions deleted in the sv37 and sv38 alleles are indicated below the maa-1 gene. Below are shown the transgenes encoding different MAA-1::GFP fusion proteins. wtMAA-1::GFP encodes a fusion protein in which GFP is fused to the last amino acid of full-length MAA-1. mtMAA-1::GFP encodes a mutant protein harboring the Y31F and K35A substitutions. tmMAA-1::GFP encodes a protein lacking the MAA-1 transmembrane domain (indicated below). cyMAA-1::GFP encodes a fusion protein in which GFP is N-terminal to the transmembrane domain. The GFP cloning cassette used to make the cyMAA-1::GFP transgene encodes some extra amino acids before and after GFP coding sequences. The fusion protein, therefore is slightly larger than the other MAA-1::GFP fusion proteins. (B) Western blot analysis of protein extracts from worms harboring the different transgenes. Top, signal obtained with an antibody against GFP. Bottom, same blot probed with an antibody against β-tubulin, which serves as a control for protein loading. (C) Tissue distribution of the wild-type MAA-1::GFP fusion protein. The three top panels show photomicrographs of worms viewed with Nomarski differential interference contrast (DIC) optics. The three bottom panels show the same worms viewed with fluorescence optics. Strong expression is seen in the anterior epidermis (left), the intestine (middle), and in two bands running dorsally (right) and ventrally (data not shown). The position of the two bands is consistent with localization in neurons or in hypodermal cells lying adjacent to the neurons.
MAA-1 Is Associated with Membrane Organelles in the Golgi Apparatus and Endocytic Pathway
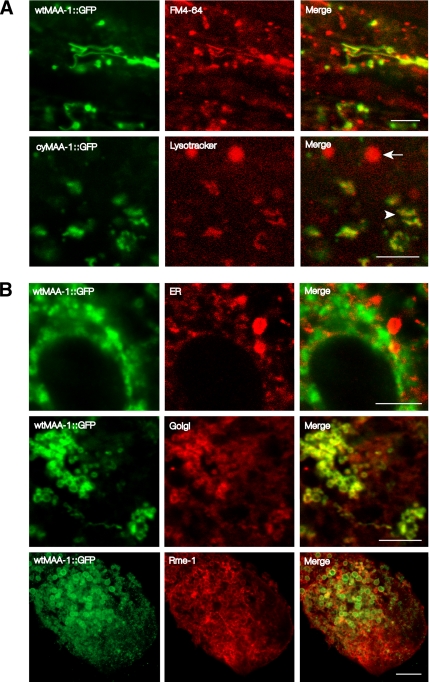
Within the hypodermis and the intestine, wtMAA-1::GFP is present in the cytoplasm and gives a punctate pattern of fluorescence. Staining is equally strong on the basal and apical sides of the cells. No signal is detected in nuclei. To determine which cellular compartment was labeled with the fusion protein, we first stained components of the endocytic pathway with fluorescent dyes and compared the staining pattern to that observed with wtMAA-1::GFP. FM4-64 is a membrane-intercalating styryl dye that is taken up by cells by endocytosis. In yeast and in the C. elegans intestine, it preferentially labels early and late endosomes and lysosomes (Vida and Emr, 1995; Grant et al., 2001). When FM4-64 is delivered to the apical plasma membrane by feeding worms the dye, endosomes on the apical side of the intestine are labeled. The organelles labeled with the wtMAA-1::GFP fusion protein were also stained by FM4-64 implying that MAA-1 is associated with a cellular organelle in the endocytic pathway (Figure 3A and Supplementary Figure 1). The pH-sensitive dye LysoTracker preferentially stains late endosomes and lysosomes. Lysosomes can be distinguished from late endosomes in C. elegans by their rounded shape and larger size. When worms harboring wtMAA-1::GFP were labeled by LysoTracker, the smaller irregularly shaped late endosomes showed GFP fluorescence and were stained by the dye. However, the larger, rounded lysosomes were stained by the dye but showed no GFP fluorescence, suggesting that MAA-1 might not be present on lysosomes. To rule out the possibility that MAA-1::GFP is in fact present on lysosomes but becomes nonfluorescent because the region C terminal to the transmembrane domain is exposed to the acid environment of the lysosome, we generated worms containing a transgene encoding a fusion protein in which GFP was N-terminal to the transmembrane domain cyMAA-1::GFP (Figure 2A). The pattern of cyMAA-1::GFP expression in these worms (Figure 2A) was the same as that in those expressing the original MAA-1::GFP fusion protein (in which GFP was fused to the C terminus). Furthermore, when worms expressing the cyMAA-1::GFP were stained with LysoTracker, cyMAA-1::GFP labeled acidic endosomes but was undetectable on lysosomes. Thus, MAA-1 seems not to be present at high levels on lysosomes.
Figure 3.
MAA-1 localizes to endosomal and Golgi compartments. Photomicrographs of the intestines of worms viewed with confocal fluorescence microscopy. The five panels on the left show GFP fluorescence in worms expressing MAA-1::GFP fusion proteins. The five middle panels show staining of the same worms with markers for different membrane organelles. FM4-64 is a membrane-intercalating dye that labels early and late endosomes. LysoTracker labels late endosomes and lysosomes. An antibody that recognizes the ER retention signal HDEL was used to label the ER. An antibody that recognizes the Golgi protein GRASP55 was used to label Golgi. Staining with this antibody substantially overlaps that obtained with a marker for Golgi in C. elegans, α-mannosidase (Chen et al., 2006) (Supplemental Figure S2). Because the epitope from human GRASP55 used to raise the antibody is almost completely conserved in the worm orthologue, it is likely that the antibody recognizes worm GRASP55. RME-1 labels the ERC. The five panels on the right show merged images of the respective left and middle panels. Bar, 5 μm. Selected images of worms at lower magnification are shown in Supplemental Figure S3.
To characterize further the subcellular distribution of MAA-1::GFP, we examined whether the organelles labeled with the fusion protein could be stained with antibodies to RME-1. RME-1 is an Eps15 homology (EH) domain protein known to be associated with the ERC in the C. elegans intestine (Grant et al., 2001) and in mammalian cells (Lin et al., 2001). RME-1 localizes to both vesicular and tubular structures close to the cell membrane (Grant et al., 2001). RME-1 staining is seen predominantly on the basal side of the cell although some staining is seen on the apical side. Immunostaining of dissected intestines from animals expressing the wtMAA-1::GFP fusion protein with an antibody against RME-1 revealed extensive overlap between the localization of the two markers in both vesicular and tubular compartments (Figure 3B and Supplemental Figure 1). Thus, MAA-1::GFP is associated with the ERC. Some MAA::GFP fluorescence was seen in organelles not stained by the anti-RME-1 antibodies. Therefore, in order to characterize the distribution of MAA-1::GFP further, we stained worms harboring the transgene with antibodies specific for other membrane organelles. To determine whether MAA-1::GFP is present on the endoplasmic reticulum (ER), we stained worms expressing the fusion protein with an antibody that recognizes the ER retention signal HDEL (Poteryaev et al., 2005). Organelles labeled with the antibody did not express MAA-1::GFP (Figure 3B) indicating that MAA-1 is not present at high levels on the ER. To determine whether MAA-1::GFP was associated with the Golgi, we stained worms with an antibody to GRASP55, a Golgi-specific protein (Kuo et al., 2000). Colocalization was observed (Figure 3B) showing that wtMAA-1::GFP is associated with at least part of the Golgi system.
MAA-1 Is a Transmembrane Protein
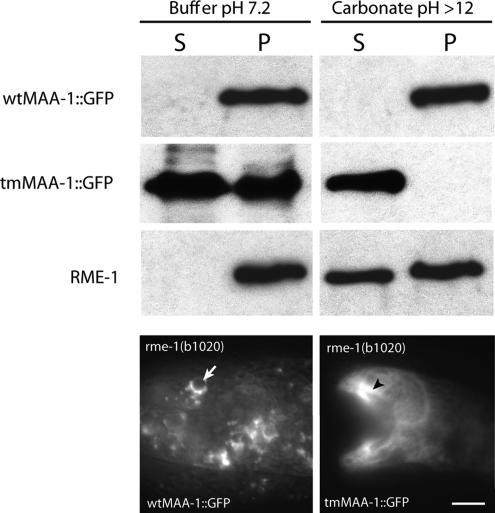
Computer analysis of the primary structure of MAA-1 predicts that the protein contains a transmembrane domain (www.cbs.dtu.dk/services/TMHMM). Consistent with this analysis, we found that wtMAA-1::GFP, but not a mutant lacking the transmembrane domain, tmMAA-1::GFP, stains the periphery of membrane organelles (Figure 4). Worms harboring tmMAA-1::GFP show diffuse staining of the cytosol. To examine further whether MAA-1 is indeed a membrane protein or is associated with membranes, we isolated membrane fractions from worms expressing the wtMAA-1::GFP or tmMAA-1::GFP fusion proteins and examined by Western blotting whether they contained GFP. Figure 4 shows that wtMAA-1::GFP is present in the pellet fraction and could not be extracted by sodium carbonate (which extracts membrane-associated proteins but not those that are membrane spanning). Strikingly, the tmMAA-1::GFP was exclusively found in the soluble fraction after extraction with carbonate buffer showing that the MAA-1 transmembrane domain is required for membrane localization (Figure 4). As a control, we also probed the different fractions with an antibody against RME-1, which is associated with membranes but does not itself contain a transmembrane domain. As expected, RME-1 was present in the membrane fraction but could be extracted by carbonate. Together, these results suggest that MAA-1 is an integral membrane protein. Consistent with a requirement for membrane localization for MAA-1 function in C. elegans, the tmMAA-1 mutant did not efficiently rescue the maa-1 phenotype (Figure 5).
Figure 4.
MAA-1 is an integral membrane protein. The top six panels show Western blots of extracts from worms probed with antibodies to either GFP or RME-1. S and P denote supernatant and pellet fractions, respectively. wtMAA-1::GFP is present only in the pellet fraction (containing membranes) and is not extracted by carbonate. Thus, wtMAA-1::GFP is an integral membrane protein rather than a membrane-associated protein. tmMAA-1::GFP, which lacks the transmembrane domain, is not present in the membrane fraction generated in the presence of carbonate, indicating that the transmembrane domain is important for membrane localization of MAA-1. The bottom two panels show photomicrographs of worms expressing wild-type (left) or mutant (right) MAA-1::GFP with or without the transmembrane domain. Whereas the wild-type protein shows a punctate staining pattern (arrow), the staining of the mutant protein is diffuse throughout the cytoplasm (arrowheads). Bar, 10 μm.
Figure 5.
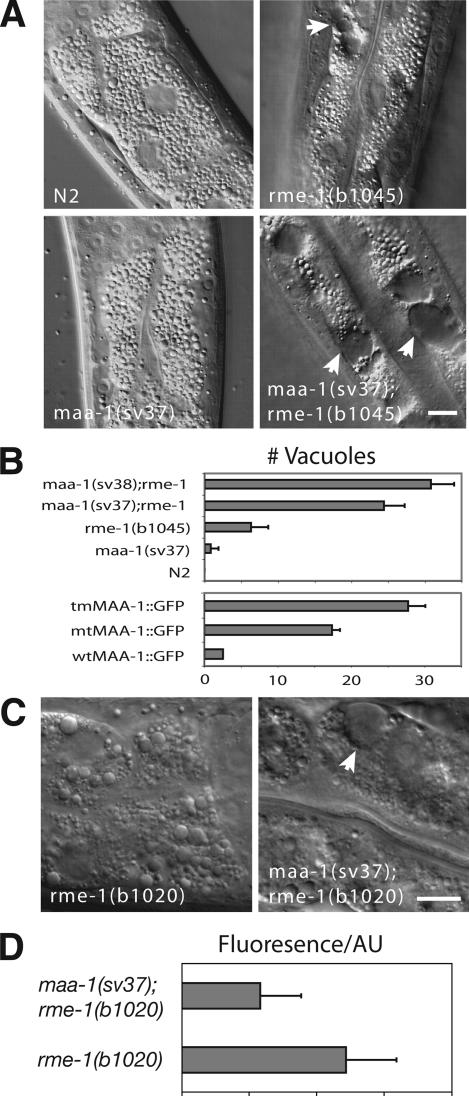
maa-1 loss-of-function mutations enhance the rme-1 intestinal phenotype. (A) Photomicrographs of L4 larval worms viewed with Nomarski DIC optics. The genotypes are indicated. Note the large vacuoles (indicated by arrowheads) present in the rme-1(b1045) single and maa-1(sv37); rme-1(b1045) double mutants. Bar, 10 μm. (B) Top box, maa-1 mutations enhance the rme-1 vacuolar phenotype. Genotypes are given on the y-axis. x-axis, average number of vacuoles in the intestines. The numbers of vacuoles present in 40 individuals of each genotype were determined. The bars represent the average number for a given genotype (±SEM). Bottom box, ability of the different maa-1::gfp transgenes to rescue the maa-1; rme-1 double mutant phenotype. The genotypes were maa-1(sv37); rme-1(b1045); svEx[tmMAA-1::GFP], maa-1(sv37); rme-1(b1045); svEx[mtMAA-1::GFP], and maa-1(sv37); rme-1(b1045); svEx[wtMAA-1::GFP]. For each of the different transgenes, three independent transgenic lines were established and assayed. The intensity of the GFP signal was similar in all transgenic strains tested. Data for each of the three transgenes represent the average number of vacuoles in a total of 40 individuals. (C) Micrographs of the intestines of worms viewed with Nomarski DIC optics. The genotypes are indicated. Note that whereas in rme-1(b1020) single mutants, small vacuoles are present but not large ones, in maa-1(sv37); rme-1(b1020) double mutants, both small and large vacuoles are present. A large vacuole is indicated with an arrowhead. Bar, 10 μm. (D) Quantification of yolk uptake in the maturing oocytes of rme-1(b1020) single and maa-1(sv37); rme-1(b1020) double mutant animals. Each bar represents the average value of fluorescence for 10 proximal oocytes from 10 animals.
We believe that the MAA-1 transmembrane domain is necessary for rescue of the maa-1 phenotype in C. elegans but not for that of the acb1 defect in yeast because yeast Acb1p and C. elegans MAA-1 have different functions. Yeast Acb1p is thought to protect acyl-CoAs from hydrolysis by thioesterases. Membrane localization is not necessary for this function. Our results are consistent with the possibility that the function of MAA-1 in C. elegans is not merely to bind acyl-CoAs but to deliver them to certain membrane organelles.
Generation of maa-1 Deletion Mutants
To gain insight into the function of maa-1 in C. elegans, we isolated two deletion alleles, sv37 and sv38 from a frozen C. elegans deletion library (see Materials and Methods). sv37 removes all of exon 1, including the start codon, and most of exon 2, which encodes the ACBP domain. It is therefore likely to eliminate maa-1 activity. sv38 removes all of exon 1 and ∼1 kb upstream of it (Figure 2A). Northern blotting confirmed that no transcript is generated by either allele (our unpublished data), suggesting that these are indeed null alleles. maa-1(sv37) or maa-1(sv38) homozygous mutant animals displayed no gross defects when observed under the dissecting microscope. The animals had normal growth rates, but the brood sizes were slightly reduced compared with the Bristol N2 wild-type strain (average 220, n = 10; cf. 300 for N2).
maa-1 Mutations Enhance rme-1 Mutant Phenotypes
rme-1 is required for efficient vesicle transport from the ERC to the plasma membrane in C. elegans (Grant et al., 2001; Lin et al., 2001). In the intestines of rme-1 null mutants, large vacuoles accumulate that are thought to be enlarged ERCs (Grant et al., 2001). rme-1 mutant hermaphrodites have a second defect associated with abnormal recycling: oocytes fail to take up yolk from the psuedocoelomic fluid. This latter defect is thought to be the result of inefficient recycling of the yolk receptor, RME-2, from the ERC to the plasma membrane (Grant et al., 2001; Lin et al., 2001). Because both RME-1 and MAA-1 localize to endosomal compartments, and because RME-1 is known to have a role in vesicle transport, we examined genetic interactions between maa-1 and rme-1. maa-1 mutations enhanced the intestinal defect caused by an rme-1 null mutation. The number of intestinal vacuoles was clearly greater in the maa-1: rme-1 double mutant animals than in the rme-1 single mutant (Figure 5B; our unpublished data). The activity of maa-1 in modulating vacuolar formation seems to be related to its ability to bind acyl-CoA. An MAA-1::GFP fusion protein containing wild-type MAA-1 fused to GFP rescued the enhancement of the rme-1 phenotype, but a fusion protein with the Y31F and K35A sequence changes, which was unable to complement the growth defect in a yeast strain depleted for Acb1p (Figure 1B), failed to rescue the synthetic phenotype in C. elegans (Figure 5B).
In an otherwise wild-type background, the phenotype caused by the rme-1 partial loss-of-function (lf) mutation, rme-1(b1020) is weaker than that caused by rme-1(b1045): although vacuoles are present in the intestines of both mutants, they are on average considerably smaller in size in rme-1(b1020) than in rme-1(b1045) mutant animals (Figure 5C). We found that maa-1(lf) enhanced the rme-1(b1020) mutant phenotype such that the intestines of maa-1; rme-1(b1020) double mutant hermaphrodites resemble those in rme-1(b1045) single mutants (Figure 5C) and contained large vacuoles. Loss of maa-1 activity also enhanced the rme-1 vacuolar defect in the hypodermis. This tissue is more sensitive to the formation of vacuoles than the intestine. In the hypodermis, large vacuoles are sometimes seen even in animals homozygous for the weaker rme-1 mutation, b1020. maa-1 null greatly enhanced the penetrance of the phenotype. Large hypodermal vacuoles were present in 2/18 rme-1(b1020) single mutant hermaphrodites examined, and in 22/25 maa-1(sv37); rme-1(b1020) double mutant hermaphrodites.
maa-1 null also enhanced the oocyte yolk uptake defect caused by rme-1(b1020) (Figure 5D). Yolk uptake in maa-1; rme-1(b1020) double mutant hermaphrodites was significantly reduced compared with that in rme-1(b1020) single mutants. Together, these results suggested that maa-1 might be involved in modulating vesicle trafficking within the endosomal system in both the intestine and oocytes. A function for MAA-1 in the germline was not suggested by the pattern of GFP fluorescence observed in transgenic worms harboring the MAA-1::GFP fusion protein. However, it should be born in mind that transgenes in C. elegans are only rarely expressed in the germline (Kelly et al., 1997; Kelly and Fire, 1998), so that absence of GFP expression does not preclude a function in this tissue.
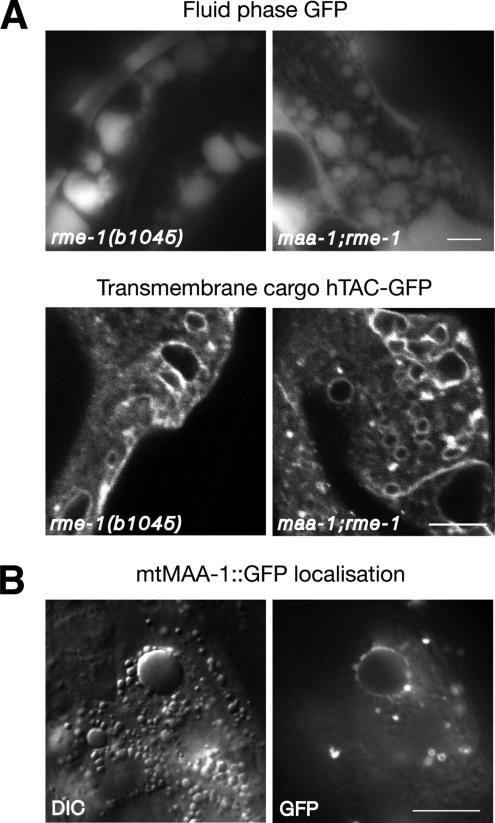
To help determine how maa-1 mutations enhance the rme-1 intestinal phenotype, we first analyzed the nature of the vacuoles in the double mutant. If the vacuoles, like those in the rme-1 single mutant (Grant et al., 2001), are formed by fusion of vesicles in the endocytic pathway, then they should receive input from basolateral fluid phase endocytosis. Consistent with this idea, a fluid phase GFP marker expressed by muscle cells and secreted into the body cavity (pseudocoelom) surrounding the basal side of the intestine (Fares and Greenwald, 2001b) accumulated in the vacuoles of the maa-1; rme-1 double mutant (Figure 6). This observation suggests that the vacuoles receive input from the basolateral endocytosis pathway. Further evidence that the enlarged vacuoles in the maa-1; rme-1 double mutants are derived from normal cellular trafficking compartments is that the same organelles are labeled by a membrane marker for endocytosis (Figure 6).
Figure 6.
The vacuoles present in maa-1; rme-1 double mutants receive input from the baso-lateral endocytosis pathway. (A) Top, photomicrographs of the intestines of worms expressing a fluid phase GFP marker in the pseudocelom, the fluid surrounding the basolateral surface of the intestine. The GFP signal is clearly visible in the intestinal vacuoles in both the single and double mutant animals. Bottom, photomicrographs of the intestines of worms expressing a membrane marker for endocytosis, hTAC::GFP (Chen et al., 2006). The peripheral membranes of the vacuoles both in the single mutant and in the maa-1; rme-1 double mutant are labeled with the marker. (B) Mutant mtMAA-1::GFP fusion protein labels the periphery of enlarged endosomal compartments in the double mutant background. Bars, 10 μm.
Loss of maa-1 Activity Perturbs Receptor Recycling
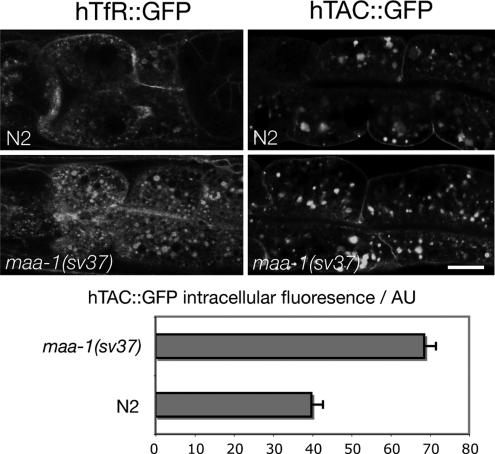
One way in which maa-1 mutations might enhance the rme-1(b1020) vacuolar phenotype could be by a further slowing of recycling from the ERC to the plasma membrane. To investigate the effect of maa-1 mutations on recycling more directly, we examined the effect of maa-1 mutations on well-established markers for endocytic recycling. The human transferrin receptor (hTfR) is a marker for clathrin-dependent endocytosis and rme-1–dependent recycling in mammalian cells (Burack et al., 2000; Lin et al., 2001). The α-chain of human interleukin-2 receptor (TAC), in contrast, is internalized via a clathrin-independent pathway but recycled in an rme-1–dependent manner (Naslavsky et al., 2004; Chen et al., 2006). In C. elegans, worms expressing hTfR-GFP or hTAC-GFP fusion proteins in the intestine, the proteins are associated with basolateral plasma membrane and with basolaterally located endosomes (Chen et al., 2006). In maa-1 null mutant worms, however, the steady-state patterns of expression of hTfR-GFP and hTAC-GFP are clearly different from those in wild type. Consistent with an effect on recycling, in both cases, maa-1 null caused a greater accumulation of the markers inside the cell (Figure 7) relative to that in wild type. Together, these results demonstrate that lack of maa-1 activity perturbs transport through the recycling pathway back to the basolateral plasma membrane in the C. elegans intestine.
Figure 7.
Transport of hTfR-GFP and hTAC-GFP is altered in maa-1 mutant animals. Top, confocal micrographs showing the steady-state localization of hTfR::GFP and hTAC::GFP in wild-type and maa-1 worms. Bottom, quantification of the fluorescence intensity of the hTAC::GFP marker. Fluorescence intensity was measured by using software associated with a Leica confocal microscope. Loss of maa-1 activity causes a greater accumulation of the receptors in intracellular compartments compared with wild type. This observation is consistent with a reduced rate of recycling to the plasma membrane. Bar, 10 μm.
maa-1 Mutants Display an Altered Organelle Morphology
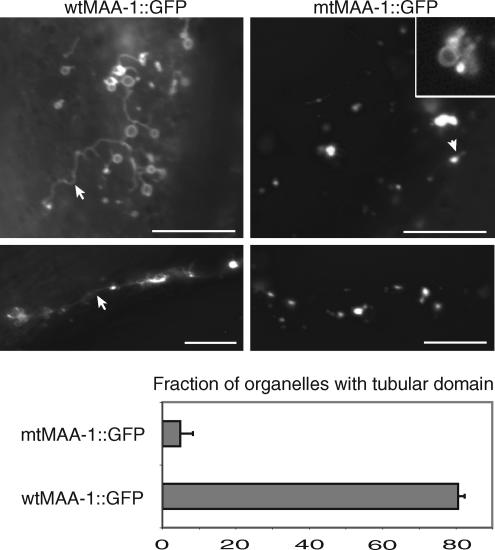
In the course of examining maa-1; rme-1 mutant worms, we noticed that the maa-1 mutation not only enhanced the vacuolar phenotype caused by rme-1 but also altered the shape and morphology of endosomes. Although the effect is seen in maa-1 null single mutants, it is most easily seen in the background of an rme-1 mutation. In particular, whereas in rme-1(b1020) worms with wild-type maa-1 activity, most MAA-1–positive organelles in the hypodermis had tubular protrusions, those in maa-1; rme-1(b1020) double mutants lacked such protrusions. Representative examples of organelles in the single and double mutants are shown in Figure 8. Quantification of the defect revealed that tubular domains were present on >80% of organelles in wild type but <10% of those in the maa-1 mutant. These results suggest that the function of maa-1 is to facilitate the generation or maintenance of tubular domains from which vesicular transport occurs.
Figure 8.
maa-1 mutations alter the morphology of membrane organelles. The top four panels show fluorescence photomicrographs of hypodermal membrane organelles in maa-1(sv37); rme-1(b1020) double mutant worms harboring either the wild-type (left) or mutant (right) transgenes. The four micrographs were obtained at identical exposure settings. The inset in the panel at top right shows an enlargement of part of a micrograph of worms harboring mtMAA-1::GFP obtained using shorter exposure times. Under these conditions, the GFP fluorescence is seen to be around the rims of the organelles. Note that in worms with wild-type maa-1 activity, the organelles have a ring-like shape from which tubules emerge. In animals lacking maa-1 activity, extensive tubulation is absent. Lower panel shows the fraction of organelles with a tubular domain (wtMAA-1::GFP, n = 62; mtMAA-1::GFP, n = 164).
DISCUSSION
In this report, we have characterized the function of C. elegans MAA-1. We have shown that MAA-1 encodes an ACBP that is membrane associated. MAA-1 has a restricted pattern of expression in the worm and is highly expressed in the hypodermis and in the intestine. Within these tissues, MAA-1 is associated with membrane organelles in the endosomal and Golgi systems. maa-1 mutations cause a reduction in the rate of endosomal recycling and enhance phenotypes caused by mutations in rme-1, a gene required for efficient recycling.
That the MAA::GFP fusion protein is localized to endosomal membranes and maa-1 mutations enhance rme-1 mutant phenotypes in the intestine suggests a role for maa-1 in vesicle trafficking. The increased number of endosome-derived vacuoles in maa-1; rme-1 double mutants could be caused by increased trafficking of vesicles to the vacuoles or by a reduction in the rate at which vesicles leave. The abnormal morphology of the ERC membrane in maa-1 mutants is also consistent with a role for maa-1 in vesicle fusion or fission.
One possible model to explain our observations is that MAA-1 functions to recruit long-chain acyl-CoA esters to membrane organelles in the endosomal and Golgi systems and that these promote vesicle biogenesis or fusion. That the ACBP domain of MAA-1 binds acyl-CoA esters in vitro and the MAA-1 ACBP domain can complement the growth defect in yeast cells depleted for Acb1p in vivo is consistent with this suggestion. Long-chain acyl-CoA esters have been shown to be required for both fission and fusion of vesicles that mediate trafficking between Golgi cisternae in vitro (Pfanner et al., 1989, 1990). Nonhydrolyzable acyl-CoA analogues can inhibit the fission of Golgi-derived COPI vesicles in vitro (Glick and Rothman, 1987; Ostermann et al., 1993). It is therefore thought that COPI vesicle formation involves one or more acylation reactions of either proteins or lipids. Presently, however, no substrates for these reactions have been identified. It is noteworthy that the enzymes endophilin and C-terminal–binding protein (CtBP)/BARS are able to catalyze the acylation of lysophosphatidic acid to phosphatidic acid (PA) in vitro by using fatty acyl-CoA esters as a substrate (Schmidt et al., 1999; Weigert et al., 1999). Disruption of endophilin A function in living lamprey synapses leads to inhibition of membrane fission and the accumulation of budding intermediates in the plasma membrane of neurons (Gad et al., 2000). PA, whose biophysical properties are thought to promote membrane curvature (Kooijman et al., 2003), is able to stimulate the budding and release of vesicles from the trans-Golgi network and ERC (Siddhanta and Shields, 1998; Jovanovic et al., 2005). One speculative model consistent with our results, therefore, is that MAA-1 recruits acyl-CoA esters to membranes where they are used as substrates for reactions catalyzed by acyl-transferases that lead to the local production of PA. However, recent results question the importance of the acyltransferase activity of endophilin and CtBP/BARS in the formation of transport vesicles (Gallop et al., 2005). Alternatively, it is very interesting that the compound CI-976, a potent lysophospholipid acyltransferase inhibitor, has been reported to block transport of both the transferrin receptor from the ERC to the plasma membrane as well as Golgi transport (Chambers et al., 2005). Other models are also consistent with our results. For example, fatty acyl-CoA esters brought to the membrane by MAA-1 might be used as substrate by other enzymes, such as protein acyl transferases, for the modification of proteins that function in vesicle biogenesis.
The genetic interactions we have observed between maa-1 and rme-1 do not imply a direct biochemical connection between the MAA-1 and RME-1 proteins. Indeed, that a null mutation in maa-1 enhances a null mutation in rme-1 suggests that the two proteins do not function in precisely the same process. MAA-1 shows a rather restricted pattern of expression both within the cell and within the organism. It is highly expressed in the intestine and in some hypodermal cells, but it is absent from most other tissues, including the nervous system where vesicle trafficking also occurs. Within the intestine, it is present on some endosomal membranes and in the Golgi but seems to be absent from some other membranous organelles such as lysosomes and the ER. Fission and fusion from these other membrane compartments therefore must occur via a mechanism distinct from one involving MAA-1. It is known that different types of vesicle mediate transport between different organelles. For example, trafficking in the secretory pathway between the ER and the Golgi is mediated by COPII vesicles. No evidence exists presently for a requirement for acyl-CoA for the generation of COPII vesicles. The Sar1p GTPase together with the heterodimeric complexes Sec23p/Sec24p and Sec13p/31p are together sufficient to generate COPII vesicles from synthetic liposomes (Lee et al., 2005b).
The complete disruption of membrane trafficking is likely to be lethal to the cell and the organism. Therefore, that maa-1 null mutant worms are viable as homozygotes suggests that alternative mechanisms may exist for targeting acyl-CoA esters to the membranes of organelles in the intestine. This function could possibly be performed by one or more of the seven other ACBPs predicted to be encoded by the C. elegans genome. However, MAA-1 is the sole ACBP containing a predicted transmembrane domain. It is possible that these other ACBPs might either themselves be recruited to membranes via a second membrane-bound protein or transiently interact with membranes and thereby partly replace MAA-1 function. We have carried out RNAi for all other ACBP genes in the background of an maa-1 null mutation, but in no case did we observe a synthetic lethal phenotype (The list of genes tested is given in Supplemental Table 2). However, RNAi does not eliminate gene activity for all genes so a possible redundancy between the ACBP genes cannot be ruled out by these experiments. Another possibility is that acyl-CoA, synthesized by endosomal-associated acyl-CoA synthetases, by simple diffusion are able to bypass to some extent the requirement for MAA-1. The results presented here together with work by others suggest that C. elegans is an excellent organism to identify new factors required for vesicle trafficking. It should be possible to carry out genetic screens for mutations that are synthetically lethal in combination with mutations in maa-1 and thereby identify new factors involved in vesicular transport.
Supplementary Material
ACKNOWLEDGMENTS
We thank Carlos Chen and Barth Grant for generously providing the anti-RME-1 antibody and the hTAC-GFP and hTfR-GFP marker strains. The anti-HDEL antibody was a kind gift from M. Lewis. We thank Y. Kohara for cDNA clones. The pPD plasmids were a gift from A. Fire. M.L. was supported by a stipend from the Wenner-grenska Samfundet and by a postdoctoral fellowship from the Danish Natural Research Council. J.K. and N.F. were supported by a grant from Novo Nordisk Fonden and Danish Natural Research Council. Research in S.T.'s laboratory is supported by grants from Cancerfonden and the Wallenberg Foundation. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-01-0035) on July 26, 2006.
REFERENCES
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burack M. A., Silverman M. A., Banker G. The role of selective transport in neuronal protein sorting. Neuron. 2000;26:465–472. doi: 10.1016/s0896-6273(00)81178-2. [DOI] [PubMed] [Google Scholar]
- Burton M., Rose T. M., Faergeman N. J., Knudsen J. Evolution of the acyl-CoA binding protein (ACBP) Biochem. J. 2005;392:299–307. doi: 10.1042/BJ20050664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan S., Naslavsky N., Hartnell L. M., Lodge R., Polishchuk R. S., Donaldson J. G., Bonifacino J. S. A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 2002;21:2557–2567. doi: 10.1093/emboj/21.11.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers K., Judson B., Brown W. J. A unique lysophospholipid acyltransferase (LPAT) antagonist, CI-976, affects secretory and endocytic membrane trafficking pathways. J. Cell Sci. 2005;118:3061–3071. doi: 10.1242/jcs.02435. [DOI] [PubMed] [Google Scholar]
- Chen C. C., Schweinsberg P. J., Vashist S., Mareiniss D. P., Lambie E. J., Grant B. D. RAB-10 is required for endocytic recycling in the C. elegans intestine. Mol. Biol. Cell. 2006;17:1286–1297. doi: 10.1091/mbc.E05-08-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., et al. WormBase: a comprehensive data resource for Caenorhabditis biology and genomics. Nucleic Acids Res. 2005;33:D383–D389. doi: 10.1093/nar/gki066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein H. F., Shakes D. C. San Diego: Academic Press; 1995. Methods in Cell Biology Series. [Google Scholar]
- Faergeman N. J., Feddersen S., Christiansen J. K., Larsen M. K., Schneiter R., Ungermann C., Mutenda K., Roepstorff P., Knudsen J. Acyl-CoA. binding protein, Acb1p, is required for normal vacuole function and ceramide synthesis in Saccharomyces cerevisiae. Biochem. J. 2004;15:907–918. doi: 10.1042/BJ20031949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares H., Grant B. Deciphering endocytosis in Caenorhabditis elegans. Traffic. 2002;3:11–19. doi: 10.1034/j.1600-0854.2002.30103.x. [DOI] [PubMed] [Google Scholar]
- Fares H., Greenwald I. Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics. 2001a;159:133–145. doi: 10.1093/genetics/159.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares H., Greenwald I. Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat. Genet. 2001b;28:64–68. doi: 10.1038/ng0501-64. [DOI] [PubMed] [Google Scholar]
- Fisher C. L., Pei G. K. Modification of a PCR-based site-directed mutagenesis method. Biotechniques. 1997;23:570–571. doi: 10.2144/97234bm01. 574. [DOI] [PubMed] [Google Scholar]
- Gad H., et al. Fission and uncoating of synaptic clathrin-coated vesicles are perturbed by disruption of interactions with the SH3domain of endophilin. Neuron. 2000;27:301–312. doi: 10.1016/s0896-6273(00)00038-6. [DOI] [PubMed] [Google Scholar]
- Gaigg B., et al. Depletion of acyl-coenzyme A-binding protein affects sphingolipid synthesis and causes vesicle accumulation and membrane defects in Saccharomyces cerevisiae. Mol. Biol. Cell. 2001;12:1147–1160. doi: 10.1091/mbc.12.4.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallop J. L., Butler P. J., McMahon H. T. Endophilin and CtBP/BARS are not acyl transferases in endocytosis or Golgi fission. Nature. 2005;438:675–678. doi: 10.1038/nature04136. [DOI] [PubMed] [Google Scholar]
- Glick B. S., Rothman J. E. Possible role for fatty acyl-coenzyme A in intracellular protein transport. Nature. 1987;326:309–312. doi: 10.1038/326309a0. [DOI] [PubMed] [Google Scholar]
- Grant B., Hirsh D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell. 1999;10:4311–4326. doi: 10.1091/mbc.10.12.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B., Zhang Y., Paupard M. C., Lin S. X., Hall D. H., Hirsh D. Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat. Cell Biol. 2001;3:573–579. doi: 10.1038/35078549. [DOI] [PubMed] [Google Scholar]
- Jovanovic O. A., Brown F. D., Donaldson J. G. An effector domain mutant of Arf6 implicates phospholipase D in endosomal membrane recycling. Mol. Biol. Cell. 2005;17:327–335. doi: 10.1091/mbc.E05-06-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly W. G., Fire A. Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans. Development. 1998;125:2451–2456. doi: 10.1242/dev.125.13.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly W. G., Xu S., Montgomery M. K., Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen J., Faergeman N. J., Skott H., Hummel R., Borsting C., Rose T. M., Andersen J. S., Hojrup P., Roepstorff P., Kristiansen K. Yeast acyl-CoA-binding protein: acyl-CoA-binding affinity and effect on intracellular acyl-CoA pool size. Biochem. J. 1994;302:479–485. doi: 10.1042/bj3020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen J., Neergaard T. B., Gaigg B., Jensen M. V., Hansen J. K. Role of acyl-CoA binding protein in acyl-CoA metabolism and acyl-CoA-mediated cell signaling. J. Nutr. 2000;130:294S–298S. doi: 10.1093/jn/130.2.294S. [DOI] [PubMed] [Google Scholar]
- Kooijman E. E., Chupin V., de Kruijff B., Burger K. N. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic. 2003;4:162–174. doi: 10.1034/j.1600-0854.2003.00086.x. [DOI] [PubMed] [Google Scholar]
- Kragelund B. B., Poulsen K., Andersen K. V., Baldursson T., Kroll J. B., Neergard T. B., Jepsen J., Roepstorff P., Kristiansen K., Poulsen F. M., Knudsen J. Conserved residues and their role in the structure, function, and stability of acyl-coenzyme A binding protein. Biochemistry. 1999;38:2386–2394. doi: 10.1021/bi982427c. [DOI] [PubMed] [Google Scholar]
- Kuo A., Zhong C., Lane W. S., Derynck R. Transmembrane transforming growth factor-alpha tethers to the PDZ domain-containing, Golgi membrane-associated protein p59/GRASP55. EMBO J. 2000;19:6427–6439. doi: 10.1093/emboj/19.23.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. W., Zhao X., Scarselletta S., Schweinsberg P. J., Eisenberg E., Grant B. D., Greene L. E. ATP binding regulates oligomerization and endosome association of RME-1 family proteins. J. Biol. Chem. 2005a;280:17213–17220. doi: 10.1074/jbc.M412751200. [DOI] [PubMed] [Google Scholar]
- Lee M. C., Orci L., Hamamoto S., Futai E., Ravazzola M., Schekman R. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell. 2005b;122:605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Lin S. X., Grant B., Hirsh D., Maxfield F. R. Rme-1 regulates the distribution and function of the endocytic recycling compartment in mammalian cells. Nat. Cell Biol. 2001;3:567–572. doi: 10.1038/35078543. [DOI] [PubMed] [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- Mogensen I. B., Schulenberg H., Hansen H. O., Spener F., Knudsen J. A novel acyl-CoA-binding protein from bovine liver. Effect on fatty acid synthesis. Biochem. J. 1987;241:189–192. doi: 10.1042/bj2410189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naslavsky N., Weigert R., Donaldson J. G. Characterization of a nonclathrin endocytic pathway: membrane cargo and lipid requirements. Mol. Biol. Cell. 2004;15:3542–3552. doi: 10.1091/mbc.E04-02-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermann J., Orci L., Tani K., Amherdt M., Ravazzola M., Elazar Z., Rothman J. E. Stepwise assembly of functionally active transport vesicles. Cell. 1993;75:1015–1025. doi: 10.1016/0092-8674(93)90545-2. [DOI] [PubMed] [Google Scholar]
- Patton A., Knuth S., Schaheen B., Dang H., Greenwald I., Fares H. Endocytosis function of a ligand-gated ion channel homolog in Caenorhabditis elegans. Curr. Biol. 2005;15:1045–1050. doi: 10.1016/j.cub.2005.04.057. [DOI] [PubMed] [Google Scholar]
- Pfanner N., Glick B. S., Arden S. R., Rothman J. E. Fatty acylation promotes fusion of transport vesicles with Golgi cisternae. J. Cell Biol. 1990;110:955–961. doi: 10.1083/jcb.110.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N., Orci L., Glick B. S., Amherdt M., Arden S. R., Malhotra V., Rothman J. E. Fatty acyl-coenzyme A is required for budding of transport vesicles from Golgi cisternae. Cell. 1989;59:95–102. doi: 10.1016/0092-8674(89)90872-6. [DOI] [PubMed] [Google Scholar]
- Poteryaev D., Squirrell J. M., Campbell J. M., White J. G., Spang A. Involvement of the actin cytoskeleton and homotypic membrane fusion in ER dynamics in Caenorhabditis elegans. Mol. Biol. Cell. 2005;16:2139–2153. doi: 10.1091/mbc.E04-08-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen J. T., Borchers T., Knudsen J. Comparison of the binding affinities of acyl-CoA-binding protein and fatty-acid-binding protein for long-chain acyl-CoA esters. Biochem. J. 1990;265:849–855. doi: 10.1042/bj2650849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Sato K., Fonarev P., Huang C. J., Liou W., Grant B. D. Caenorhabditis elegans RME-6 is a novel regulator of RAB-5 at the clathrin-coated pit. Nat. Cell Biol. 2005;7:559–569. doi: 10.1038/ncb1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Wolde M., Thiele C., Fest W., Kratzin H., Podtelejnikov A. V., Witke W., Huttner W. B., Soling H. D. Endophilin I mediates synaptic vesicle formation by transfer of arachidonate to lysophosphatidic acid. Nature. 1999;401:133–141. doi: 10.1038/43613. [DOI] [PubMed] [Google Scholar]
- Siddhanta A., Shields D. Secretory vesicle budding from the trans-Golgi network is mediated by phosphatidic acid levels. J. Biol. Chem. 1998;273:17995–17998. doi: 10.1074/jbc.273.29.17995. [DOI] [PubMed] [Google Scholar]
- Timmons L., Court D. L., Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- van Aalten D. M., Milne K. G., Zou J. Y., Kleywegt G. J., Bergfors T., Ferguson M. A., Knudsen J., Jones T. A. Binding site differences revealed by crystal structures of Plasmodium falciparum and bovine acyl-CoA binding protein. J. Mol. Biol. 2001;309:181–192. doi: 10.1006/jmbi.2001.4749. [DOI] [PubMed] [Google Scholar]
- Vida T. A., Emr S. D. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb N. R., Rose T. M., Malik N., Marquardt H., Shoyab M., Todaro G. J., Lee D. C. Bovine and human cDNA sequences encoding a putative benzodiazepine receptor ligand. DNA. 1987;6:71–79. doi: 10.1089/dna.1987.6.71. [DOI] [PubMed] [Google Scholar]
- Weigert R., et al. CtBP/BARS induces fission of Golgi membranes by acylating lysophosphatidic acid. Nature. 1999;402:429–433. doi: 10.1038/46587. [DOI] [PubMed] [Google Scholar]
- Yang J. S., Lee S. Y., Spano S., Gad H., Zhang L., Nie Z., Bonazzi M., Corda D., Luini A., Hsu V. W. A role for BARS at the fission step of COPI vesicle formation from Golgi membrane. EMBO J. 2005;24:4133–4143. doi: 10.1038/sj.emboj.7600873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Grant B., Hirsh D. RME-8, a conserved J-domain protein, is required for endocytosis in Caenorhabditis elegans. Mol. Biol. Cell. 2001;12:2011–2021. doi: 10.1091/mbc.12.7.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.