Abstract
Using nuclear magnetic resonance spectroscopy, we establish that the N-terminal domain of the yeast vacuolar R-SNARE Nyv1p adopts a longin-like fold similar to those of Sec22b and Ykt6p. Nyv1p is sorted to the limiting membrane of the vacuole via the adaptor protein (AP)3 adaptin pathway, and we show that its longin domain is sufficient to direct transport to this location. In contrast, we found that the longin domains of Sec22p and Ykt6p were not sufficient to direct their localization. A YXXΦ-like adaptin-dependent sorting signal (Y31GTI34) unique to the longin domain of Nyv1p mediates interactions with the AP3 complex in vivo and in vitro. We show that amino acid substitutions to Y31GTI34 (Y31Q;I34Q) resulted in mislocalization of Nyv1p as well as reduced binding of the mutant protein to the AP3 complex. Although the sorting of Nyv1p to the limiting membrane of the vacuole is dependent upon the Y31GTI34 motif, and Y31 in particular, our findings with structure-based amino acid substitutions in the mu chain (Apm3p) of yeast AP3 suggest a mechanistically distinct role for this subunit in the recognition of YXXΦ-like sorting signals.
INTRODUCTION
The endomembrane system of eukaryotic cells is composed of a complex network of membrane-enclosed organelles, each of which contains a characteristic repertoire of lipid and protein constituents. The functional and compositional integrity of each organelle is maintained by a continued addition to and selective removal of proteins from these compartments, in a process driven by the selectivity of transport step-specific coat protein complexes and mediated by membrane-bound vesicular carriers (Rothman, 1994). Vesicle coat proteins contain binding sites, which recognize sorting motifs on their select protein cargos and direct the recruitment of these proteins into nascent transport vesicles (Lee et al., 2004; Owen et al., 2004). Subsets of vesicle cargo proteins, the soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs), are required for successful targeting and fusion of transport vesicles with their target organelles (Jahn et al., 2003). In general, SNARE proteins are differentially localized in cells, and trafficking between endomembrane subcompartments requires specific combinations of SNARE proteins. Presumably then, for a transport vesicle to be recognized by and fuse with its target organelle, the vesicle must contain the correct complement of SNARE proteins. Such a requirement implies that vesicle coat protein complexes should be able to selectively recruit the correct repertoire of SNAREs into newly forming transport vesicles. Recent experimental evidence in yeast suggests that this is indeed the case. The COPII coat, which is required for the formation of transport vesicles that deliver proteins from the endoplasmic reticulum (ER) to the Golgi apparatus, contains binding sites for the SNAREs Sed5p, Bet1p, Bos1p, and Sec22p—proteins required for fusion of ER-derived vesicles with the Golgi. Similarly, the vacuolar SNARE Vam3p contains a sorting signal that is required for its sorting to the limiting membrane of the vacuole by the coat/adaptor protein (AP) complex—AP3 (Darsow et al., 1998)—and the endosomal SNARE Pep12p contains a motif required for its sorting from the Golgi to prevacuolar endosomes, in a process presumably mediated by the GGA proteins (Black and Pelham, 2000). In addition, some SNARE proteins contain sorting signals that direct recognition by more than one coat protein complex. For example, the yeast Golgi syntaxin Sed5p cycles between the ER and Golgi (Wooding and Pelham, 1998); thus, presumably, in addition to binding to the COPII coat protein complex, Sed5p also contains a COPI-dependent sorting signal that directs its retrieval from the Golgi back to the ER.
SNARE-mediated fusion typically results in the formation a complex consisting of three Q-SNAREs and one R-SNARE. The Q/R-SNARE nomenclature is based on the x-ray structures of SNARE complexes and denotes whether a conserved glutamine (in Q-SNAREs) or arginine (in R-SNAREs) is located in the so-called zero layer of the complex (Sutton et al., 1998; Antonin et al., 2002). The genome of the yeast Saccharomyces cerevisiae encodes five R-SNAREs: Snc1/2p, Ykt6p, Sec22p, and Nyv1p. Of these five R-SNAREs, Sec22p, Ytk6p, and Nyv1p have N-terminal extensions of their SNARE-motifs greater than 100 amino acids. The structures of the N-terminal domains of Sec22b (the orthologue of yeast Sec22p) and Ytk6p have revealed that both proteins share a similar profilin-like fold (Gonzalez et al., 2001; Tochio et al., 2001). R-SNAREs that contain a profilin-like fold in their N-terminal domains have been termed longins (Filippini et al., 2001; Rossi et al., 2004), and we have adopted this nomenclature here. The longin domain (LD) is not restricted to R-SNAREs and has been identified in a variety of proteins (Rossi et al., 2004), some of which are also involved in vesicular transport. These include subunits of the adaptin complexes (Collins et al., 2002; Heldwein et al., 2004) and SEDL/Trs20p (Jang et al., 2002), a common subunit of the transport protein particle (TRAPP)I and TRAPPII multisubunit complexes, which are required for traffic between the ER and Golgi and within the Golgi (Oka and Krieger, 2005). The function of the longin fold of Sec22b/Sec22p is unknown, but in Ykt6p the longin domain folds back and binds to the SNARE-motif of the protein (Tochio et al., 2001), and this conformation is likely to be important for chaperoning the lipid modified C terminus of the cytoplasmic form of Ykt6p in cells as well as for its localization (Fukasawa et al., 2004; Hasegawa et al., 2004).
Although there is no significant amino acid similarity between the N-terminal domain of Nyv1p and the Ykt6p and Sec22b N termini, structure prediction suggests that the fold of the N-terminal domain of Nyv1p should also resemble those of Sec22p and Ykt6p. Here, we present the structure of the N-terminal domain of Nyv1p, which reveals that Nyv1p is indeed a longin SNARE, and we establish that the longin domain is chiefly responsible for the AP3-dependent sorting of Nyv1p to the limiting membrane of the vacuole via a YXXΦ-like adaptin binding motif.
MATERIALS AND METHODS
Expression and Purification of Proteins for Nuclear Magnetic Resonance (NMR) Studies
DNA fragments corresponding to the 149 residue NH2-terminal domain of Nyv1p (Nyv1p-LD) and its amino acid substitution mutants (nyv1p-LDY31Q;I34Q and nyv1p-LDL73Q;I74Q) and to the 230 residue Nyv1pΔTMD (Nyv1p minus its transmembrane domain) were amplified by the polymerase chain reaction (PCR). DNA fragments were cloned into a modified pET vector, pETH (Feng et al., 2002) to generate an in-frame fusion of a hexa-histidine [(His)6]-tag to the C terminus of the proteins. Protein expression was induced in Escherichia coli BL21(DE3) cells by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG), and the recombinant proteins were purified on Ni2+-nitrilotriacetic acid (NTA) affinity columns. After affinity purification, the (His)6-tag was removed by digesting the fusion proteins with thrombin. Purification of untagged Nyv1p proteins was accomplished by a second Ni2+-NTA affinity column purification step, followed by gel filtration chromatography. Uniformly 15N- and 15N/13C-labeled Nyv1p proteins were prepared by growing bacteria in M9 minimal medium containing 15NH4Cl (1 g/l) with or without 13C6-glucose (1 g/l).
NMR Experiments
Four NMR samples were prepared for structural determination of Nyv1p-LD with a protein concentration of ∼1.0 mM (15N-labeled protein in 90% H2O/10% D2O, two 15N/13C-labeled samples: one sample in 99.9% D2O and one sample in 90% H2O/10% D2O and unlabeled protein in 99.9% D2O). Protein samples were dissolved in 20 mM potassium phosphate buffer, pH 7.0, containing 6 mM d10-dithiothrietol.
All NMR experiments were carried out at 35°C using Varian Inova 500- and 750-MHz spectrometers. NMR spectra were processed with the nmrPipe software package (Delaglio et al., 1995) and analyzed using PIPP (Garrett et al., 1991) and Sparky (www.cgl.ucsf.edu/home/sparky/). Sequential backbone resonance assignments of the proteins were obtained by standard heteronuclear correlation experiments, including HNCO, HNCACB, and CBCA(CO)NH, and confirmed by a three-dimensional 15N-separated nuclear Overhauser effect spectroscopy (NOESY) experiments (Kay and Gardner, 1997). The nonaromatic, nonexchangeable side chain assignments were obtained using HCCH-total correlation spectroscopy (TOCSY) experiments. The side chains of aromatics were assigned by 1H two-dimensional (2D) TOCSY/NOESY experiments with an unlabeled protein sample in D2O. The stereospecific assignment of the Val and Ile methyl groups was obtained by using a 10% 13C-labeled sample (Neri et al., 1989). The -NH2 side chains of Asn and Gln residues were assigned by a three-dimensional 15N-separated NOESY experiment in which the 15N-labeled protein was dissolved in H2O.
NMR Structural Calculations
Approximate interproton distance restraints were derived from NOESY spectra (a 1H 2D homonuclear NOESY, a 15N-separated NOESY, and a 13C-separated NOESY, each with a mixing time of 100 ms). Nuclear Overhauser effects (NOEs) were grouped into three distance ranges, 1.8–2.9, 1.8–3.5, and 1.8–5.0 Å, corresponding to strong, medium, and weak NOEs, respectively. For NOEs involving NH protons, a range of 1.8–5.9 Å was added, corresponding to ultraweak NOEs. Hydrogen bonding restraints (2 per hydrogen bond, where rNH–O = 1.8–2.2 Å and rN–O = 2.2–3.3 Å), and backbone dihedral angle restraints (Φ and Ψ angles) were generated from the standard secondary structure of the protein on the basis of the NOE patterns and the backbone secondary chemical shifts. Structures were calculated by using the program CNS (Brunger et al., 1998). The coordinates of Nyv1p-LD have been deposited in the Protein Data Bank under the accession code 2FZ0.
Yeast Strains and Methods
Yeast cells lacking VPS27 or PEP12 were grown at 25°C; all other yeast strains were grown at 30°C. Cells were propagated in yeast extract peptone dextrose medium (YEPD), synthetic dextrose media (SD), or synthetic galactose (SG) media lacking the appropriate amino acids. Yeast transformations were performed using lithium acetate by the method of Elble (1992). The yeast strains used in this study are listed in Table 2.
Table 2.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf |
| SARY580 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0pep12::KanMX | Euroscarf |
| SARY1619 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0apm3::KanMX | Euroscarf |
| SARY1621 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0apl5::KanMX | Euroscarf |
| SARY1825 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0vps27::KanMX | Euroscarf |
| SARY1622 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0apl6::KanMX | Euroscarf |
| SARY1618 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0apl6::KanMX pGALAPL6-HA | This study |
| SARY1401 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0APM3::TAP | Open Biosystems |
Plasmids Used in Protein Localization and Biochemical Studies
DNA fragments encoding Nyv1p-LD and nyv1p-LDY31Q;I34Q (amino acids 1–149) were amplified by PCR and cloned into pGEX2T as BamHI–EcoRI fragments to generate glutathione S-transferase (GST) fusion proteins pGST-Nyv1-LD and pGST-nyv1-LD YI/QQ, respectively. Plasmids encoding green fluorescent protein (GFP)-Nyv1p and GFP-Yck3p were based on the yeast/bacterial shuttle vectors pRS413 (CEN6, HIS3) or pRS416 (CEN6, URA3). The PCR-generated Yck3p- and Nyv1p-encoding derivatives were cloned as EcoRI–BamHI fragments behind sequences expressing the GFP mut2 variant (Cormack et al., 1996) and the triose phosphate isomerase promoter to generate the plasmids pGFP-Nyv1p, pGFP-nyv1pYI/QQ (Y31Q, L34Q), pGFP-nyv1pLI/QQ (L73Q, I74Q), and pGFP-Yck3p, respectively. The plasmid expressing hemagglutinin antigen (HA)-tagged APL6 under control of the GAL1 promoter [pGAL1-APL6-(His)6-HA-protein A, in the plasmid BG1805] was purchased from Open Biosystems (Huntsville, AL). The plasmid expressing GFP-Pho8p, under the control of the PHO8 promoter, was a gift from Rob Piper (Department of Physiology, University of Iowa, Iowa City, IA). The plasmid expressing GNS (Reggiori et al., 2000) was a gift from Hugh Pelham (Medical Research Council-Laboratory of Molecular Biology, Cambridge, United Kingdom). The plasmid used as template for site-directed mutagenesis of the mu chain of AP3 (pRS416-APM3) was purchased from Euroscarf (Institute of Microbiology, Johann Wolfgang Goethe-University Frankfurt, Frankfurt, Germany) (pYCG_YBR288c). Plasmids expressing membrane-anchored GFP-longin domains were generated as follows. DNA fragments encoding amino acids 1–162 of Nyv1p, amino acids 1–126 of Sec22p, and amino acids 1–134 of Ykt6p were amplified by the PCR and cloned as EcoRI–HindIII fragments. The DNA encoding the Snc1p transmembrane spanning sequence (amino acids 93–117, together with a stop codon immediately after residue 117) was amplified by the PCR and cloned as a HindIII–BamHI fragment. The recombinant EcoRI–BamHI fragments were then used to replace the corresponding region of pGFP-Nyv1p. Amino acid substitutions in Nyv1p and Apm3p were generated using plasmids encoding the wild-type genes and the Kunkel method of site-directed mutagenesis (Kunkel, 1985).
Live Cell Imaging
Yeast cells were grown to mid-log phase, harvested by centrifugation, and resuspended in water. Aliquots of yeast cell suspensions (∼0.5–1.0 μl) were placed onto slides and examined immediately. Cells were visualized and photographed through a Zeiss Plan-Neofluar 100×/1.30A oil objective lens by using a Zeiss Axioskop microscope (Carl Zeiss, Jena, Germany) equipped with a SPOT-RT KE monochrome charge-coupled device camera (Diagnostic Instruments, Sterling Heights, MI). Digital images were processed with Photoshop (Adobe Systems, Mountain View, CA). The staining of yeast cells with N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenyl-hexatrienyl) pyridinium dibromide (FM4–64) (Invitrogen, Carlsbad, CA) was carried out as described by Vida and Emr (1995).
In Vivo Protein Copurification
BY4741 cells (Table 2) transformed with pGFP-Nyv1p and SARY1401 cells (expressing Apm3-TAP; Table 2) transformed with pGFP-Nyv1p, pGFP-nyv1pYI/QQ, or pRS416 were grown with constant shaking at 30°C in SD media lacking leucine (SD-Leu) until an OD660 of 0.8 was reached. Cells were collected by centrifugation at 5000 × g for 20 min at 4°C, resuspended in NP-40 buffer [15 mM Na2HPO4, 10 mM NaH2PO4·H2O, 150 mM NaCl, 1% (vol/vol) NP-40] containing 1% (vol/vol) Triton X-100 and a cocktail of protease inhibitors (EDTA-free Complete and Pefabloc; Roche Diagnostics, Mannheim, Germany), and lysed using a Mini-Bead Beater-8 (BioSpec Products, Bartlesville, OK). Unlysed cells and cellular debris were removed by centrifugation at 16,000 × g for 20 min at 4°C. IgG-beads (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) were added to yeast cell extracts, and the mixture was incubated, with constant mixing, at 4°C for 2 h. After incubation, IgG-beads were collected by centrifugation at 8000 × g for 1 min and washed four times with 10 bead volumes of NP-40 buffer containing 1% Triton X-100. After washing, IgG-beads were collected by centrifugation and resuspended in SDS-PAGE sample buffer. Proteins bound to the IgG-beads were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and immunostained with an anti-GFP antibody (Clontech, Mountain View, CA).
Purification of Proteins for In Vitro Mixing Studies
Plasmids encoding GST-Nyv1-LD, GST-nyv1-LD(Y31Q,I34Q), or GST (pGEX2T) were transformed into E. coli BL21 cells. Protein expression was induced with 0.2 mM IPTG for 2 h at 30°C after which cells were collected by centrifugation at 5000 × g for 20 min at 4°C. Bacterial cell pellets were resuspended in NP-40 buffer containing 0.5% Triton X-100 and a protease inhibitor cocktail and lysed by sonication. Bacterial cell lysates were cleared of unbroken cells and cellular debris by centrifugation at 4°C.
Equal volumes of bacterial cell lysates (corresponding to ∼10 ml of bacterial culture) were incubated with 20 μl of glutathione (GSH)-Sepharose beads (GE Healthcare) for 2 h at 4°C. After incubation, beads were washed three times with NP-40 buffer (containing 0.5% Triton X-100) and finally resuspended in NP-40 buffer before use in in vitro pull-down assays (see below).
SARY1618 cells (Table 2) were grown in SD-Ura medium at 30°C to an OD660 of 0.8. Yeast cells were collected by centrifugation, washed twice with distilled water to remove the glucose-containing medium, and resuspended in SG-Ura (2% galactose) to an OD660 of 0.1. Cells were grown in SG-Ura for 12–14 h; collected by centrifugation; resuspended in 200 mM Tris-HCl, pH 8.0, 20 mM EDTA, and 1% (vol/vol) β-mercaptoethanol; and incubated at 30°C for 15 min. After incubation, yeast cells were collected by centrifugation and resuspended in minimal medium containing 1 M sorbitol and 0.15 mg/ml Zymolyase 100T (Seikagaku, Tokyo, Japan) and incubated for 40 min at 30°C. After treatment with Zymolyase 100T, cells were washed twice with minimal medium containing 1 M sorbitol and resuspended in NP-40 buffer containing 0.5% Triton X-100. Cells were lysed in the presence of a protease inhibitor cocktail (EDTA-free Complete and Pefabloc) with 20 strokes of a Dounce homogenizer. Cell lysates were cleared by centrifugation at 16,000 × g for 20 min at 4°C.
In vitro mixing studies were performed as follows. GSH-beads containing GST-Nyv1-LD, GST-nyv1-LD(Y31Q;I34Q), or GST were mixed with ∼100 OD660 equivalents of whole cell extracts from SARY1618 cells, prepared as described above. The mixtures were incubated with constant mixing for 2 h at 4°C. After incubation, the beads were washed four times with NP-40 buffer [containing 0.5% (vol/vol) Triton X-100] for total of 0.5 h. After washing, beads were collected by centrifugation, boiled in SDS sample buffer, and processed for immunostaining with an anti-HA (Roche Diagnostics) antibody.
RESULTS
Nyv1p Is a Longin Domain-containing SNARE Protein
The structures of the N-terminal domains have been determined for Ykt6p (Tochio et al., 2001) and for the mammalian orthologue of Sec22p (Sec22b; Gonzalez et al., 2001). The structures of the N-terminal domains of Sec22b and Ykt6p are similar and bear overall fold similarity to that of the actin-binding protein profilin. To establish whether the third member of the longer R-SNARE family in yeast, Nyv1p, also contained a so-called longin domain, we determined the high-resolution solution structure of N-terminal domain of Nyv1p (residues 1–149) by NMR spectroscopy (Table 1).
Table 1.
Structural statistics for the family of the final 20 structuresa
| Distance restraints | |
| Intraresidue (i − j = 0) | 627 |
| Sequential (i − j = 1) | 617 |
| Medium range (2 ≤ i − j ≤ 4) | 525 |
| Long range (i − j > 5) | 832 |
| Hydrogen bonds | 74 |
| Total | 2675 |
| Dihedral angle restraints | |
| Φ | 47 |
| Ψ | 40 |
| Total | 87 |
| Mean r.m.s. deviations from the experimental restraints | |
| Distance (Å) | 0.017 ± 0.000 |
| Dihedral angle (°) | 0.134 ± 0.023 |
| Mean r.m.s. deviations from idealized covalent geometry | |
| Bond (Å) | 0.003 ± 0.000 |
| Angle (°) | 0.424 ± 0.007 |
| Improper (°) | 0.327 ± 0.008 |
| Mean energies (kcal mol−1) | |
| ENOEb | 56.36 ± 2.69 |
| Ecdihb | 0.10 ± 0.03 |
| EL–J | −344.99 ± 10.76 |
| Ramachandran plotc | |
| Residues 1–149 % residues in the most favorable regions | 61.6 |
| Additional allowed regions | 31.4 |
| Generously allowed regions | 5.5 |
| Disallowed regions | 1.5 |
| Atomic r.m.s. difference (Å)d (residues 5–26, 35–60, 65–141 in protein) | |
| Backbone heavy atoms (N, Cα, and C′) | 0.36 |
| Heavy atoms | 0.78 |
a None of the structures exhibits distance violations >0.3 Å or dihedral angle violations >4°.
b The final values of the square-well NOE and dihedral angle potentials were calculated with force constants of 50 kcal mol−1 Å−2 and 200 kcal mol−1 rad−2, respectively.
c The program Procheck (Laskowski et al., 1996) was used to assess the overall quality of the structures.
d The precision of the atomic coordinates is defined as the average root mean square (r.m.s.) difference between 20 final structures and the mean coordinates of the protein.
Other than the two intrinsically flexible loops (designated βB/αA- and αB/αC-loop) characterized by the low 1H, 15N heteronuclear NOE values and lack of medium- and long-range NOEs of the residues within these two loops and a few residues in the two termini, the structure of Nyv1p N-terminal domain is well defined (Figure 1A). The N-terminal domain of Nyv1p contains five β-strands and three α-helices in which the five β-strands form an antiparallel β-sheet with a βB-βA-βE-βD-βC topology. The β-sheet is sandwiched by αA on one side and by αB/αC on the other side (Figure 1B). The overall fold of the N-terminal domain of Nyv1p is similar to the structures of the N-terminal domains of the longin SNAREs Ykt6p and Sec22b (Figure 2A), and we therefore propose that Nyv1p be considered as the third yeast longin SNARE. Hereafter, we refer to the N-terminal domain of Nyv1p as Nyv1p-LD.
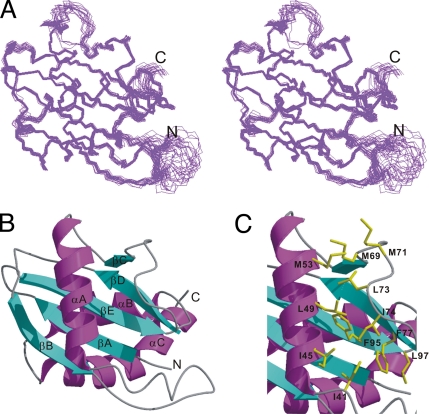
Figure 1.
Solution structure of the NH2-terminal longin domain of Nyv1p (Nyv1p-LD). (A) Stereo view of 20 superimposed NMR-derived structures of the Nyv1p-LD. The residues 4 N142 are shown in the drawing. The structures are superimposed against the averaged structure using residues 5 N26, 35 N60, and 65 N141. (B) Ribbon diagram representation of the Nyv1p-LD structure. (C) Close-up view of the hydrophobic surface of the Nyv1p-LD. The amino acid residues forming the hydrophobic surface are shown in the explicit atomic model. Molecular models in this and subsequent figures were generated using MOLMOL (Koradi et al., 1996), MOLSCRIPT (Kraulis, 1991), Raster3D (Merritt and Murphy, 1994), and DINO (www.dino3d.org).
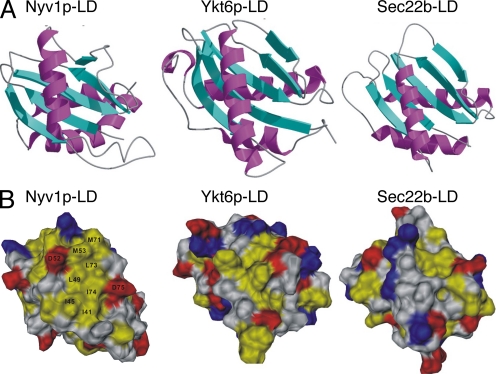
Figure 2.
Structural features of SNARE longin domains. (A) Ribbon diagram showing the longin domain structures from Nyv1p (Nyv1p-LD), Ykt6p (Ykt6p-LD), and Sec22b (Sec22b-LD). (B) The longin domain of Nyv1p contains a large surface exposed hydrophobic surface. In the surface representations of the longin domains from Nyv1p (Nyv1p-LD), Ykt6p (Ykt6p-LD), and Sec22b (Sec22b-LD), K and R are shown in blue; D and E are in red; F, V, L, I, A, M, W, and P are in yellow; and the rest of residues are shown in white. The orientation of the Nyv1p-LD is the same as that shown in Figure 1B and is similar to those of Ykt6p-LD and Sec22b-LD.
The Nyv1p-LD contains a prominent hydrophobic surface formed by residues from αA and from the βC/βD loop (Ile45, Lue49, Met53, Met71, Lue73, and Ile74; Figure 1C and 2B). Due to the packing of Met71 and Leu73 from the extended βC/βD-loop, the hydrophobic surface of the Nyv1p-LD is significantly larger than that of the Ykt6p-LD (Tochio et al., 2001, Figure 2B).
Nyv1p Does Not Adopt a Folded-back Conformation
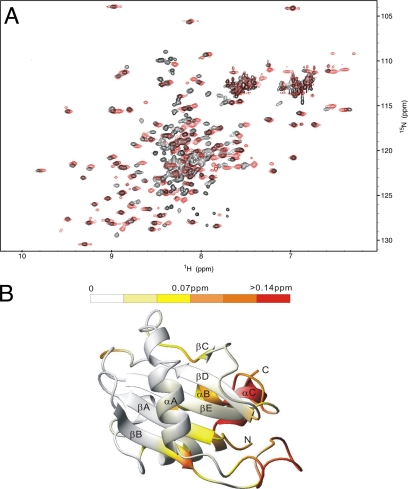
The hydrophobic surface on the Ykt6p-LD has been suggested to play an autoinhibitory role by binding to the SNARE-motif of Ykt6p in cis (Tochio et al., 2001) and may also be important for chaperoning the lipid-modified C terminus of the protein (Hasegawa et al., 2004). In contrast, the longin domain of Sec22b does not contain such a hydrophobic surface, and the SNARE-motif of Sec22b does not bind to the longin domain but rather adopts an open conformation (Gonzalez et al., 2001). Given the presence of the large solvent-exposed hydrophobic surface, we reasoned that like Ykt6p, the longin domain of Nyv1p might also be able to bind to its SNARE-motif, thereby allowing Nyv1p to adopt an autoinhibited conformation. To test this hypothesis, we compared the 1H, 15N-heteronuclear single-quantum coherence (HSQC) spectrum of the Nyv1p-LD to that of the full-length soluble form of Nyv1p, i.e., lacking its membrane spanning region (designated Nyv1pΔTMD). A subset of peaks from the Nyv1pΔTMD spectra overlaps well with the entire spectrum of Nyv1p-LD (Figure 3A). In particular, the amino acid residues that form the hydrophobic surface of Nyv1p-LD did not experience any detectable core domain-induced chemical shift changes, indicating that the SNARE-motif of Nyv1pΔTMD does not interact with its longin domain (Figure 3, A and B). The extra peaks in the Nyv1pΔTMD 1H, 15N-HSQC spectra are located in a region indicative of a random coil structure that presumably corresponds to amino acid residues of the Nyv1p SNARE-motif. In Figure 3B, the small chemical shift changes observed in the αC helix are a result of the covalent attachment of the SNARE core domain to the αC helix. The even smaller chemical shift changes to the residues in the αA/βB-loop are due to the proximity of this region with the SNARE core domain. Because one would expect to observe much greater chemical shift changes (∼0.5 ppm or even higher; Wuthrich, 2000) if the αC helix and αA/βB-loop were in direct contact with the SNARE core domain, we concluded that Nyv1pΔTMD adopts an open conformation in solution.
Figure 3.
Nyv1pΔTMD does not adopt a folded-back conformation in solution. (A) Overlay of 1H, 15N-HSQC spectra of Nyv1p-LD (red), and Nyv1pΔTMD (black). (B) Summary of 1H and 15N combined chemical shift changes in the NH2-domain of Nyv1pΔTMD induced by the SNARE-motif. The combined 1H and 15N chemical shift changes are defined as Δppm = [(ΔδHN)2 + (ΔδN × αN)2]½, where ΔδHN and ΔδN represent chemical shift differences of amide proton and nitrogen chemical shifts of Nyv1p-LD with and without the SNARE-motif. The scaling factor (αN) used to normalize the 1H and 15N chemical shifts is 0.17.
The Longin Domain Is Sufficient to Target Nyv1p to the Limiting Membrane of the Vacuole
Nyv1p localizes to the limiting membrane of the vacuole and a previous study has shown that the sorting of Nyv1p to this location requires a functional AP3 adaptin complex (Reggiori et al., 2000). AP3-dependent sorting signals have previously been identified on the vacuolar syntaxin Vam3p (Darsow et al., 1998), the yeast alkaline phosphatase Pho8p (Vowels and Payne, 1998), and on a yeast casein kinase I, Yck3p (Sun et al., 2004). The sorting signals on these proteins conform to either the so-called acidic dileucine motif D/EXXXLL (found in Pho8p and Vam3p) or to a tyrosine-containing motif designated YXXΦ, where Φ denotes a bulky hydrophobic residue (found in Yck3p). The nature of the AP3-dependent sorting signal on Nyv1p is not known. We therefore examined Nyv1p for the presence of amino acid sequences that conformed to either the acidic dileucine- or YXXΦ-based motifs and found several potential sorting signals that span the length of the protein.
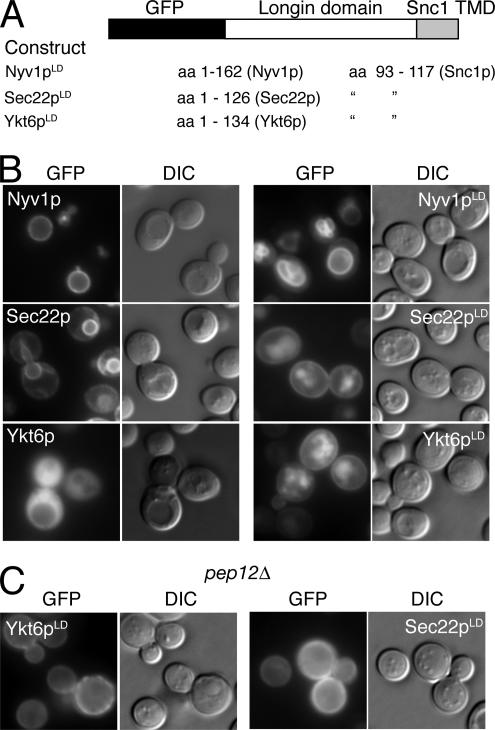
To establish whether Nyv1p, Ykt6p, or Sec22p contained localization determinants in their longin domains, we created fusion proteins in which the longin domains of these proteins were fused to GFP at their N termini and to the transmembrane domain of the R-SNARE Snc1p at their C termini (designated Nyv1pLD, Sec22pLD, and Ykt6pLD in Figure 4A). We reasoned that those membrane-anchored longin domains that lacked dominant sorting signals would be localized to the cell surface by virtue of the Snc1p transmembrane spanning sequence (Reggiori et al., 2000). Plasmids encoding GFP-tagged, full-length SNAREs or encoding membrane anchored longin domains from either Nyv1p, Sec22p, or Ykt6p were transformed into yeast cells, and the intracellular location of the fusion proteins was visualized by microscopy (Figure 4B). As expected, GFP-Nyv1p localized to the limiting membrane of the vacuole, whereas GFP-Sec22p was localized to the ER. GFP-Ykt6p was found throughout the cell, and the intensity of cytoplasmic staining effectively masked any evidence of membrane localization (Figure 4B, left). Like the full-length protein, GFP-Nyv1-LD was also localized to the limiting membrane of the vacuole (Figure 4B, right). In contrast, the membrane-anchored longin domains of Sec22p and Ykt6p localized to the cell surface as well as to the lumen of the vacuole (Figure 4B, right). The GFP-Sec22-LD and GFP-Ykt6-LD protein found in the lumen of the vacuole likely corresponds to misfolded protein targeted for degradation, because traffic to this location could be blocked in cells that lacked the prevacuolar endosomal syntaxin Pep12p (Figure 4C).
Figure 4.
The longin domain is sufficient to direct the sorting of Nyv1p to the limiting membrane of the vacuole. (A) Scheme used for the construction of GFP fusions to the longin domains of Nyv1p, Sec22p, and Ykt6p. (B) The localization of GFP-Nyv1p, GFP-Sec22p and GFP-Ykt6p and their membrane-anchored longin domains (Nyv1pLD, Sec22pLD, and Ykt6pLD) in wild-type cells. (C) Ykt6pLD and Sec22pLD localize to the plasma membrane in pep12Δ cells. Yeast cells were transformed with plasmids expressing the various GFP-fusion proteins and processed for microscopy as described in Materials and Methods
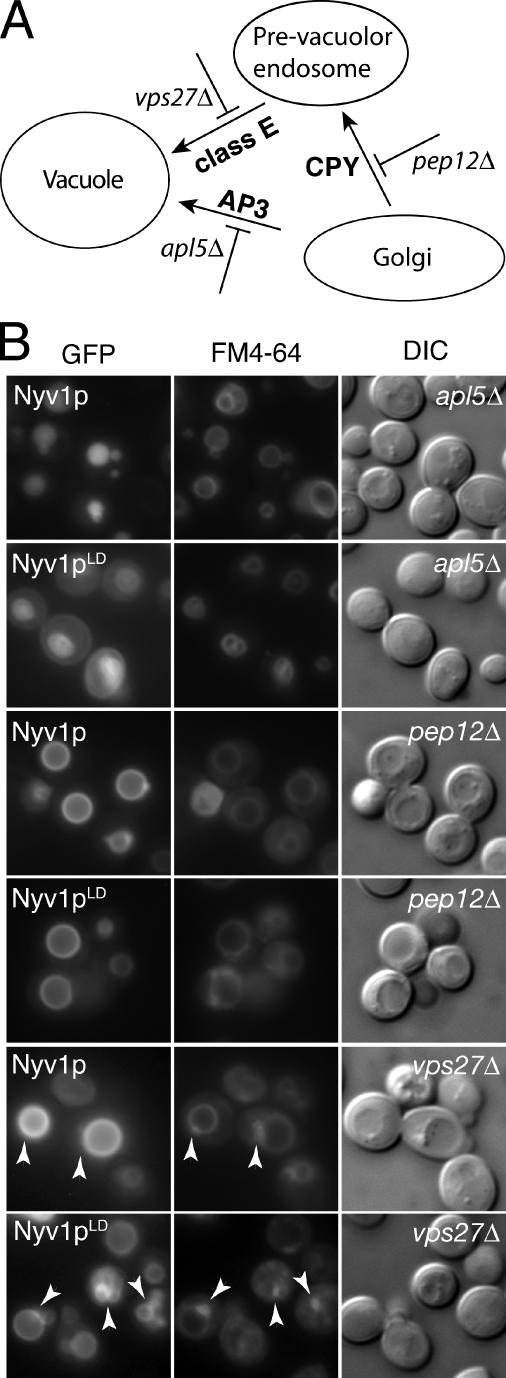
To determine whether the localization of Nyv1-LD (Figure 4B) to the limiting membrane of the vacuole was mediated via the AP3 pathway (as has been shown to be the case for Nyv1p, Reggiori et al., 2000), we examined the localization of GFP-Nyv1-LD, and for comparison, GFP-Nyv1p, in various yeast mutants defective in transport from the Golgi to the vacuole (Figure 5). In yeast, two distinct transport routes have been identified that mediate protein transport to the vacuole. The so-called carboxypeptidase Y (CPY) pathway transports select proteins from the Golgi to the vacuole via the prevacuolar endosome (Figure 5A) as well as functioning as a default transport route to the vacuole for many mislocalized proteins. The AP3 pathway mediates the transport of select proteins, bearing AP3-dependent sorting signals, directly from the Golgi to the vacuole (Figure 5A). Thus, cells lacking APL5 (apl5Δ; Figure 5A), which encodes the δ subunit of yeast AP3, are defective in transport from the Golgi directly to the vacuole, whereas cells lacking PEP12 (pep12Δ; Figure 5A), which encodes the prevacuolar resident syntaxin, are defective in transport from the Golgi to vacuole along the CPY pathway. Cells lacking VPS27 (vps27Δ; Figure 5A) accumulate proteins in a compartment adjacent to the vacuole termed the ′class E′ compartment (Piper et al., 1995), and vps27Δ cells were used here to identify proteins destined for the vacuole, which had either transited the prevacuolar endosome (via the CPY pathway) or had been endocytosed (Figure 5A). To visualize vacuoles, cells were stained with the lipophilic styryl dye FM4-64 (Vida and Emr, 1995). In principle, proteins bearing AP3-dependent sorting signals would be expected to be mislocalized in cells lacking a functional AP3 complex but to have their localization unaffected by blocking transport to the vacuole via the CPY pathway (i.e., in pep12Δ cells). Similarly, proteins transported to the vacuole via the CPY pathway would be expected to accumulate in the prevacuolar compartment in class E mutants, as would also be the case for the FM4-64 dye (Darsow et al., 1998; Reggiori et al., 2000; Teo et al., 2006). Thus, if GFP-Nyv1p and GFP-Nyv1-LD contained AP3-dependent sorting signals, their localization to the limiting membrane of the vacuole should be unaffected by deletion of PEP12 and neither protein should accumulate in the class E compartment in cells lacking VPS27. In contrast, cells lacking APL5 should show aberrant localization of both GFP-Nyv1p and GFP-Nyv1-LD.
Figure 5.
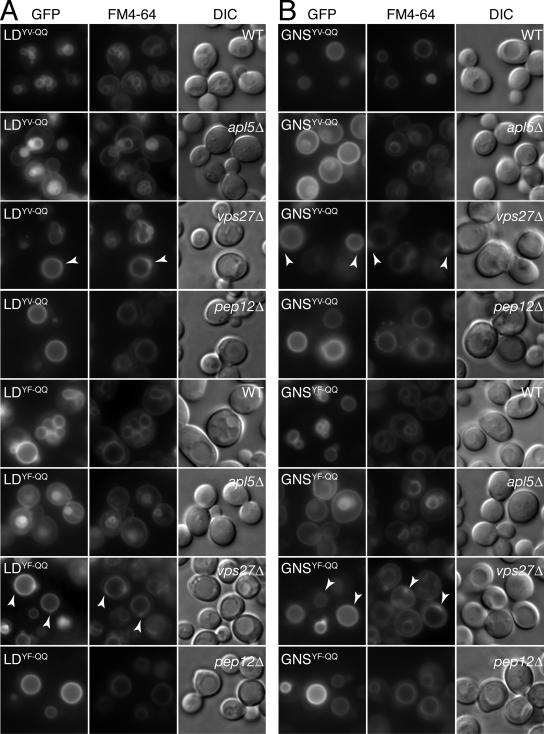
Sorting of GFP-Nyv1p and GFP-Nyv1-LD to the limiting membrane of the vacuole is mediated by AP3. (A) Schematic representation of trafficking routes from the Golgi to the vacuole in yeast. Trafficking routes are denoted by the use of arrows. Yeast gene deletion mutants (used in this study) defective in a particular trafficking route are indicated below a given arrow. Thus, cells lacking PEP12 (pep12Δ) are defective in transport from the Golgi to the prevacuolar endosome designated CPY. Cells lacking APL5 (apl5Δ) are defective in transport from the Golgi to the vacuole designated AP3, and cells lacking VPS27 (vps27Δ) accumulate proteins destined for the vacuole via endosomes, in an aberrant structure, termed the class E compartment. Yeast cells lacking APL5, VPS27, or PEP12 were used to assess trafficking routes taken by GFP-Nyv1p, GFP-Nyv1LD, and GNS (and their corresponding amino acid substitution mutants); see text for details. (B) The localization of GFP-Nyv1p (Nyv1p) and GFP-Nyv1-LD (Nyv1pLD) in wild-type cells and in the indicated yeast single gene deletion strains (see Table 2 and text for details). To visualize vacuoles, cells were stained with FM4-64 (see Materials and Methods). Arrowheads in the FM4-64 panels (vps27Δ) indicate the location of the class E compartment. Note the absence of the GFP fusion proteins from the class E compartment in the corresponding vps27Δ GFP panels.
Consistent with a previous report (Reggiori et al., 2000), GFP-Nyv1p was mislocalized to the lumen of the vacuole in cells defective in AP3-dependent transport (apl5Δ; Figure 5B). GFP-Nyv1-LD was also mislocalized in apl5Δ cells where it was found on the cell surface; in addition, some GFP-Nyv1-LD also localized to the vacuole in apl5Δ cells. In contrast, deletion of PEP12 had no discernible effect on the targeting of GFP-Nyv1-LD or GFP-Nyv1p to the limiting membrane of the vacuole (pep12Δ; Figure 5B) and unlike FM4-64, neither GFP-Nyv1p nor GFP-Nyv1-LD accumulated in the class E compartment in cells lacking VPS27 (vps27Δ; Figure 5B). Therefore, both GFP-Nyv1p and GFP-Nyv1-LD satisfy the established criteria (Darsow et al., 1998; Vowels and Payne, 1998; Reggiori et al., 2000; Sun et al., 2004) for proteins bearing AP3-dependent sorting signals. Moreover, our data suggest that an AP3-dependent sorting signal is located in the Nyv1p longin domain. The residual vacuolar staining observed for GFP-Nyv1-LD in apl5Δ cells likely represents sorting of the protein via the CPY pathway in the absence of a functional AP3 complex, a phenomena previously observed for three proteins ordinarily transported to the vacuole via the AP3 pathway: Vam3p, Pho8p, and Yck3p (Stepp et al., 1997; Darsow et al., 1998; Vowels and Payne, 1998; Sun et al., 2004).
Thus, unlike the Nyv1p-LD, when the longin domains of Sec22p and Ykt6p are tethered to membranes via the Snc1p transmembrane domain, they do not seem to convey any dominant protein sorting signals. However, we cannot exclude the possibility that the longin domain of Sec22p is required for COPII-mediated export from the ER (Miller et al., 2003; Mossessova et al., 2003; Liu et al., 2004).
The Longin Domain of Nyv1p Contains a YXXΦ-Motif That Is Required for Its AP3-dependent Sorting to the Limiting Membrane of the Vacuole
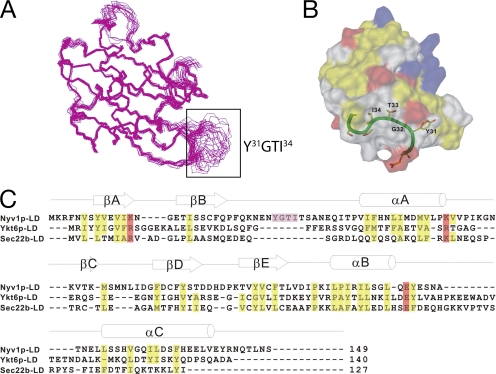
Having established that the longin domain of Nyv1p contained an AP3-dependent sorting signal, we wanted to identify the location and nature of the motif. The longin domain of Nyv1p contains three potential YXXΦ-motifs: Y8VEV11, Y31GTI34, and Y92VCF95; two of these consensus amino acid sequences map to the βA (Y8VEV11) and βE (Y92VCF95) regions of the longin fold, and both are buried in the core of the protein, whereas the third sequence, Y31GTI34, is located in a surface-exposed and highly dynamic region between βB and αA (Figure 6, A and B). A structural-based amino acid sequence alignment of the longin domains of Nyv1p, Ykt6p, and Sec22b revealed that the Y31GTI34 YXXΦ-like motif is uniquely placed in the longin domain of Nyv1p (Figure 6C). To determine whether the Y31GTI34 sequence functioned as an AP3-dependent sorting signal in Nyv1p, a double amino acid substitution of Y31 to Q and I34 to Q (nyv1pYI-QQ) was generated by site-directed mutagenesis, and the localization pattern of the mutant GFP-tagged protein was visualized in yeast by microscopy (Figure 7A).
Figure 6.
A YXXΦ-motif is located in a unique, surface exposed and highly dynamic region of the Nyv1p longin domain. (A) Superimposed NMR-derived structures of Nyv1p-LD showing the location of the surface exposed and highly dynamic loop (boxed) containing the Y31GTI34 YXXΦ-motif. (B) Surface representation of the Nyv1p-LD in which the locations of Y31, G32, T33, and I34 are prominently represented. The model shown represents a 90° clockwise rotation of the image shown in A. Lys and Arg are shown in blue; Asp and Glu are in red; Phe, Val, Leu, Ile, Ala, Met, Trp, and Pro are in yellow; and the rest of residues are shown in white. (C) A structure-based alignment of the longin domains of Nyv1p (amino acids 1–149), Ykt6p (amino acids 1–140), and Sec22b (amino acids 1–127). For reference, the secondary structure of the Nyv1p-LD is shown above the alignment. The amino acid residues of the Y31GTI34 YXXΦ-motif in the Nyv1p-LD (between βB and αA) are shaded in purple. Amino acids shaded in yellow denote hydrophobic residues that are conserved between Nyv1p-LD, Ykt6p-LD, and Sec22b-LD, whereas amino acids shaded in red denote conserved charged residues.
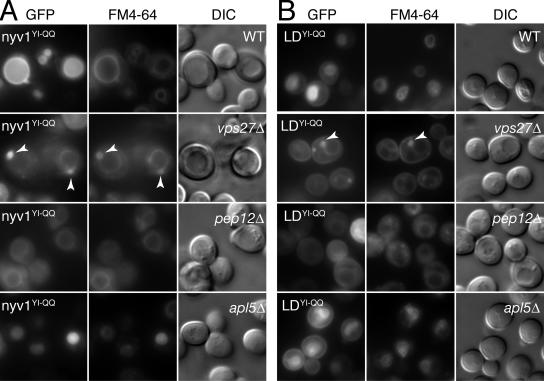
Figure 7.
Amino acid substitutions to the Y31GTI34 YXXΦ-like motif in Nyv1p and Nyv1p-LD disrupt their AP3-dependent sorting. (A) The localization and sorting requirements of GFP-nyv1p Y31Q;I34Q (designated nyv1YI-QQ), in wild type and in the indicated yeast single gene deletion strains. The arrowheads in the vps27Δ panel denote the position of the class E compartment, which is visible in both the FM4-64 and GFP images. (B) The localization and sorting requirements of GFP-nyv1-LD Y31Q;I34Q (designated LDYI-QQ), in wild type and in the indicated yeast single gene deletion strains. The arrowheads in the vps27Δ panel indicate the position of the class E compartment, which is visible in both the FM4-64 and GFP images.
In wild-type cells GFP-nyv1pYI-QQ was predominantly localized to the lumen of the vacuole (Figure 7A). A similar localization pattern was also observed for GFP-Nyv1p in an AP3 mutant (see the apl5Δ panel in Figure 5B; Reggiori et al., 2000). These observations suggested to us that when expressesd in wild-type cells, the majority of GFP-nyv1pYI-QQ followed the same trafficking route as GFP-Nyv1p in AP3 mutants—reaching the plasma membrane but then being subsequently endocytosed and transported to the vacuole via the multivesicular body sorting pathway (Reggiori et al., 2000). This conclusion is consistent with the fate of GFP-nyv1pYI-QQ in vps27Δ cells where it accumulated in the class E compartment (Figure 7A) as well as with the fate of GFP-nyv1pYI-QQ in pep12Δ cells in which sorting to the vacuole was largely defective. When expressed in apl5Δ cells, GFP-nyv1pYI-QQ, like GFP-Nyv1p, was found in the lumen of the vacuole (Figures 7A and 5B, respectively). We note that because some GFP-nyv1pYI-QQ was still observed on the limiting membrane of the vacuole in both wild-type and pep12Δ cells, it is possible that Nyv1p may use additional (albeit weaker) signals to direct this residual (and presumably) AP3-mediated sorting.
We also examined the effect of the Y31 to Q and I34 to Q double amino acid substitution on the sorting of GFP-Nyv1-LD (Figure 7B). As expected, GFP-nyv1-LDYI-QQ was localized to the cell surface, indicating that Y31 and I34 are required to direct GFP-Nyv1-LD to the vacuole. However, in addition to cell surface localization, GFP-nyv1-LDYI-QQ was also found on the limiting membrane of the vacuole as well as in the lumen of the vacuole. Consistent with this observation, GFP-nyv1-LDYI-QQ accumulated in the class E compartment of vps27Δ cells (Figure 7B). Evidently, some GFP-nyv1-LDYI-QQ is targeted for degradation in the vacuole via the CPY pathway (pep12Δ; Figure 7B), indicating that this protein is either misfolded and/or recognized as being aberrant. Regardless, some GFP-nyv1-LDYI-QQ is clearly mislocalized to the cell surface, supporting the notion that Y31GTI34 functions as an AP3-dependent sorting signal in the longin domain of Nyv1p. The vacuolar localization observed for GFP-nyv1-LDYI-QQ in pep12Δ cells may reflect the use of additional AP3-dependent sorting signals (see below) or alternatively, represent protein transported to the vacuole by default via the AP3 pathway (Bruinsma et al., 2004). In contrast, the relatively faint vacuolar localization observed for GFP-nyv1-LDYI-QQ in apl5Δ cells most likely corresponds to protein redirected to the vacuole by default via the CPY pathway (Figures 5A and 7B).
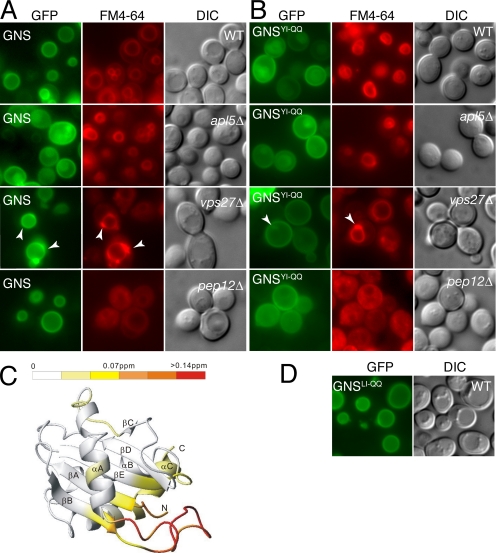
To address the potential contribution of the transmembrane domain of nyv1pYI-QQ to the localization of protein to the limiting membrane of the vacuole in wild-type and pep12Δ cells (Figure 7A) as well as to better visualize the extent of mislocalization of the Nyv1p site-directed mutant protein, we used a GFP-tagged version of Nyv1p in which the Nyv1p transmembrane spanning sequence was replaced with that of Snc1p (GFP-Nyv1-Snc1TMD) termed GNS (Reggiori et al., 2000). Yeast mutants that effect the sorting of GNS and as well as mutations in GNS that effect the localization of the protein will result in their redirection from the vacuole to the cell surface.
As expected, the localization of GNS was indistinguishable from that of GFP-Nyv1p in wild-type cells (compare Figure 4B with Figure 8A). In wild-type cells, GNS was found on the limiting membrane of the vacuole, and its sorting to this location required a functional AP3 complex; as when expressed in cells lacking the APL5 gene (which encodes the δ subunit of yeast AP3), GNS was largely mislocalized to the plasma membrane (Figure 8A). As anticipated, the sorting of GNS to the limiting membrane of the vacuole did not proceed via the CPY pathway (Figure 5A), because the protein failed to accumulate in the class E compartment of vps27Δ cells (Figure 8A) and its localization to the limiting membrane of the vacuole was unaffected in cells lacking PEP12 (pep12Δ; Figure 8A). In contrast to the double amino acid substitution mutant of Nyv1p (GFP-nyv1YI-QQ), which was found primarily in the lumen of the vacuole (Figure 7A), GNSYI-QQ was mislocalized to the plasma membrane in wild-type cells (Figure 8B). The extent of mislocalization of GNSYI-QQ in wild-type cells was essentially indistinguishable from that of GNS and GNSYI-QQ in apl5Δ cells (compare Figure 8, A and B), suggesting that the Y31GTI34 motif is the predominant AP3-dependent sorting signal in the protein. In contrast to GFP-nyv1pYI-QQ (Figure 7A), GNSYI-QQ did not localize to the limiting membrane of the vacuole in pep12Δ cells (Figure 8B). We therefore concluded that either the transmembrane domain of Nyv1p also contributed modestly to the AP3-dependent sorting of the protein in wild-type cells, or alternatively, that GFP-nyv1pYI-QQ but not GNSYI-QQ was sorted by default via the AP3 pathway in pep12Δ cells (Bruinsma et al., 2004).
Figure 8.
Localization and sorting requirements of GFP-Nyv1-Snc1TMD (GNS) and its amino acid substitution mutants GFP-Nyv1Y31Q;I34Q-Snc1TMD (GNSYI-QQ) and GFP-Nyv1L71Q;I74Q-Snc1TMD (GNSLI-QQ). (A) The localization pattern of GNS in wild-type, apl5Δ, vps27Δ, and pep12Δ cells. The arrowheads in the vps27Δ panel indicate the position of the class E compartment, which is visible in the FM4-64 panel but not in the GFP panel. (B) The localization pattern of GNS Y31Q;I34Q (designated GNSYI-QQ) in wild type, apl5Δ, vps27Δ, and pep12Δ cells. The arrowheads in the vps27Δ panel indicate the location of the class E compartment, which is visible in the FM4-64 panel and into which GNSYI-QQ also accumulates, albeit modestly (arrowhead in the corresponding GFP panel). (C) The folding of the Nyv1p-LD is not effected by the introduction of the Y31Q;I34Q amino acid substitutions. The structure perturbations caused by the Y31Q;I34Q substitutions are predominantly restricted to the loop containing the YXXΦ-like motif, indicating that the structural integrity of the longin fold is largely unaffected in GNSYI-QQ, see the text for details. (D) Amino acid substitutions that disrupt the surface exposed hydrophobic patch of the Nyv1p-LD (L71;I74Q) have no effect on the localization of GNSLI-QQ.
The residual localization of GNS to the limiting membrane of the vacuole apparent in apl5Δ cells (Figure 8B) most likely corresponds to rerouting of some GNS via the CPY pathway to the vacuole (as discussed above for GFP-Nyv1-LD; also see Stepp et al., 1997; Darsow et al., 1998; Vowels and Payne, 1998; Sun et al., 2004). In contrast, the portion of GNSYI-QQ that localized to the limiting membrane in wild-type and apl5Δ cells may represent misfolded protein targeted for degradation in the vacuole, because some colocalization of GNSYI-QQ to the class E compartment was apparent in vps27Δ cells and staining of the limiting membrane was by and large absent in pep12Δ cells (Figure 8B).
Because the introduction of amino acid substitutions can sometimes result in changes to the folding of a protein, we wanted to eliminate mutation-induced folding alterations as a possible explanation for the degree of mislocalization observed for GNSYI-QQ (Figure 8B). To address this possibility, we investigated conformation changes to the Nyv1p LD that resulted from the Y31Q, I34Q mutations, by using an NMR chemical shift perturbation approach (Figure 8C). From these data, it is clear that with the exception of the βB/αA-loop, the Y31Q, I34Q amino acid substitutions did not alter the conformation of the longin fold (Figure 8C).
Like Ykt6p-LD, Nyv1p-LD also contains a prominent hydrophobic surface (yellow in Figure 2B), and we next addressed whether this surface in the Nyv1p-LD might also be required for interaction of the longin domain with the AP3 complex. We reasoned that amino acid substitutions that disrupted the hydrophobic surface would be expected to disrupt interactions between Nyv1p/GNS and AP3, resulting in mislocalization of Nyv1p/GNS. Using site-directed mutagenesis, we introduced amino acid substitutions at L73 (L73Q) and I74 (I74Q) to generate GNSLI-QQ. Unlike GNSYI-QQ, GNSLI-QQ was localized to the limiting membrane of vacuole (Figure 8D), and we therefore concluded that L73 and I74 (and therefore the solvent-exposed hydrophobic surface) were not critical for interactions between Nyv1p/GNS and the AP3 sorting machinery.
An alternate explanation for the residual localization of GFP-Nyv1YI-QQ and GNSYI-QQ to limiting membrane of the vacuole in wild-type cells (Figures 7A and 8B) is the presence of one or more additional (albeit weaker) AP3-dependent sorting signals in these proteins. In addition to the Y31GTI34 motif, the longin domain of Nyv1p also contains two additional YXXΦ consensus sequences: Y8VEV11 and Y92VCF95. As discussed above, the structure of the Nyv1-LD has revealed that both of these motifs are buried in the protein and thus would presumably only function as AP3-dependent sorting signals if the longin domain underwent a substantial conformational change. Nonetheless, to address the possible roles of the Y8VEV11 and Y92VCF95 motifs as contributors to the AP3-dependent sorting of GNS and Nyv1-LD, double amino acid substitutions were introduced by site-directed mutagenesis, creating nyv1-LDYV-QQ (Y8Q;V11Q), nyv1-LDYF-QQ (Y92Q;F95Q), GNSYV-QQ (Y8Q;V11Q), and GNSYF-QQ (Y92Q;F95Q). The localization patterns of the mutant GFP-tagged proteins were then visualized in wild-type cells as well as in various yeast mutants (Figure 5A) by microscopy (Figure 9, A and B). The localization patterns observed for both the GFP-nyv1-LD (LD; Figure 9A) and GNS amino acid substitution mutants (GNS; Figure 9B) were essentially indistinguishable from the localization patterns observed previously for GFP-Nyv1-LD (Figure 5B) and GNS (Figure 8A). Although the degree of mislocalization of LDYV-QQ in apl5Δ is not as profound as LDYF-QQ in apl5Δ cells, the localization pattern of the corresponding GNS mutant (GNSYV-QQ, Figure 9B) is entirely consistent with the view that Y8VEV11 does not function as an AP3-dependent sorting signal. Note that, as for GNS and GFP-Nyv1-LD in apl5Δ cells (Figures 5B and 8A), some the corresponding YV-QQ and YF-QQ mutant proteins were redirected to the vacuolar membrane via the CPY pathway, presumably by default (Stepp et al., 1997; Darsow et al., 1998; Vowels and Payne, 1998; Sun et al., 2004), because no class E compartment staining was observed for any of these mutants (vps27Δ; Figure 9, A and B).
Figure 9.
Amino acid substitutions targeting the additional YXXΦ-like motifs in Nyv1p (Y8VEV11 and Y92VCF95) do not disrupt AP3-dependent sorting of the mutant proteins. (A) The localization and sorting requirements of the Y8Q;V11Q (LDYV-QQ) and Y92Q;F95Q (LDYF-QQ) longin domain amino acid substitution mutants. (B) The localization and sorting requirements of the Y8Q;V11Q (GNSYV-QQ) and Y92Q;F95Q (GNSYF-QQ) GFP-Nyv1-Snc1TMD amino acid substitution mutants. For both A and B, the arrowheads in the vps27Δ panels indicate the location of the class E compartment. Note the absence of GFP localization to the class E compartment in all vps27Δ panels.
In sum, we conclude that Y31GTI34 represents the predominant YXXΦ-motif in the Nyv1p longin domain and that this sequence functions as the principal AP3-dependent sorting signal in Nyv1p.
Amino Acid Substitutions in the Longin Domain of Nyv1p That Disrupt the Sorting of GFP-Nyv1p In Vivo Reduce Binding to the AP3 Complex
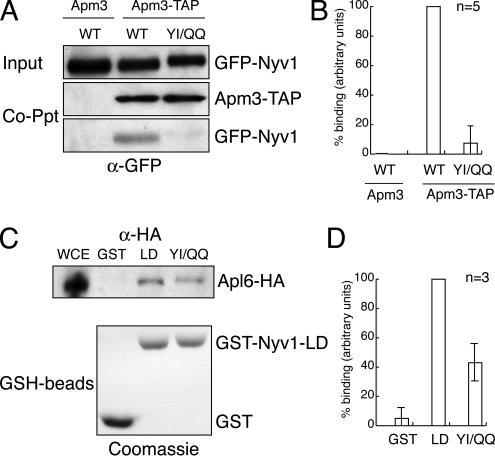
Having established that the longin domain of Nyv1p was sufficient to mediate AP3-dependent sorting via the Y31GTI34 YXXΦ-like motif, we next wanted to determine whether Nyv1p could bind to the AP3 sorting machinery in vivo. To accomplish this, we expressed GFP-Nyv1p and GFP-nyvlpYI-QQ in yeast cells (SARY1401; Table 2) in which the chromosomal copy of the mu chain of AP3 (Apm3p) had been modified through the addition of a tandem affinity purification tag (termed Apm3-TAP; Gavin et al., 2002). The localization pattern of GFP-Nyv1p in yeast cells expressing Apm3-TAP as their sole source of Apm3p was indistinguishable from that in wild-type cells (our unpublished data). Whole cell extracts were prepared from SARY1401 cells, and proteins that copurified with Apm3-TAP were isolated on IgG beads by virtue of the protein-A portion of the TAP tag to bind to immunoglobulin-coupled beads. As a control for nonspecific binding of GFP and/or GFP-Nyv1p proteins to IgG-beads, these experiments were carried out in parallel using whole cell extracts prepared from wild-type cells (BY4741; Table 2) expressing GFP-Nyv1p (Apm3; Figure 10, A and B). Proteins bound to IgG-beads were resolved by SDS-PAGE and immunostained with anti-GFP antibodies. Although both GFP-Nyv1p and GFP-nyv1p YI-QQ could be copurified with Apm3-TAP, the relative amount of GFP-nyv1pYI-QQ bound to the AP3 complex was by comparison an average of ∼13-fold less (7.42 ± 7.82%) than the amount of GFP-Nyv1p that copurified with Apm3-TAP (from 5 independent experiments). The results from one of these experiments are shown in Figure 10A. The data for the five experiments are summarized graphically in Figure 10B. For comparison purposes, the binding of GFP-Nyv1p to Apm3-TAP was arbitrarily set to 100%; hence, there are no error bars for this data set in Figure 10B (i.e., WT in Apm3-TAP). Because the binding of GFP-Nyv1p to IgG-beads was undetectable (WT in Apm3; Figure 10, A and B), we concluded that neither Nyv1p nor GFP bound nonspecifically to IgG-beads under the conditions used here.
Figure 10.
Amino acid substitutions that disrupt the sorting of Nyv1p in vivo reduce binding to the AP3 adaptin complex. (A) GFP-nyv1YI-QQ is defective in binding to the AP3 complex in vivo. Whole cell extracts were prepared from either BY4741 (Apm3, as a negative control) or SARY1401 cells (Amp3-TAP) transformed with GFP-Nyv1p (Apm3/WT) in the case of BY4741 cells and either GFP-Nyv1p (Apm3-TAP/WT) or GFP-nyv1p Y31Q;I34Q (Apm3-TAP/YI/QQ) in the case of SARY1401 cells. After incubation, proteins bound to IgG-beads were resolved on SDS-PAGE gels, transferred to nitrocellulose membranes, and immunostained with an anti-GFP antibody (as described in Materials and Methods). Note that Apm3-TAP binds to the secondary antibody used in these experiments (anti-IgG) and serves as a gel loading control. (B) Graphic representation of the copurification data from five independent experiments quantified by scanning densitometry. Binding of wild-type GFP-Nyv1p to AP3 was arbitrarily set as 100% and other data points are plotted relative to wild-type protein binding. The data (for n = 5) were WT (GFP-Nyv1p, 100%) and YI/QQ (GFP-nyv1pY31Q,I34Q, 7.42 ± 7.82%). Binding of GFP-Nyv1p to IgG-beads was undetectable (Apm3/WT; A and B). See text for details. (C) The longin domain of Nyv1p binds to AP3 in vitro. Equivalent amounts (∼10 μg) of bacterially expressed, purified recombinant GST, GST-Nyv1p-LD (LD), and GST-nyv1p(Y31Q, I34Q)-LD (YI/QQ) were immobilized on GSH-beads and mixed with equal amounts of whole cell extract prepared from wild-type cells expressing a galactose-inducible HA-tagged form of Apl6p from a plasmid (SARY1618 cells; see Table 2). Proteins bound to GSH-beads were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and immunostained with an anti-HA antibody. An aliquot of whole cell extract from SARY1618 cells (WCE) was also included as a gel mobility reference for HA-tagged Apl6p. Bottom, Coomassie brilliant blue-stained SDS-PAGE gel of identical amounts of recombinant proteins bound to GSH-beads that were used in the pull-down assays shown above. (D) Graphic representation of the copurification data from three independent experiments, quantified by scanning densitometry. The binding of the wild-type longin domain of Nyv1p (LD) to AP3 was arbitrarily set as 100%, and other data points are plotted relative to wild-type protein binding. The data were LD (100%), GST (4.97 ± 7.52%) and YI/QQ (42.98 ± 15.29%).
To establish whether the longin domain of Nyv1p was sufficient to interact with the AP3 sorting machinery, we performed in vitro mixing experiments. Bacterially expressed, purified recombinant GST-Nyv1p-LD immobilized on glutathione-Sepharose beads (or GST alone) was mixed with whole cell extracts prepared from yeast cells expressing an HA epitope-tagged form of the β subunit of AP3 (Apl6p-HA) as their sole source of the protein (SARY1618 cells; Table 2). Proteins bound to GST-Nyv1p-LD or GST-glutathione beads were resolved by SDS-PAGE and immunostained with an anti-HA antibody. We found that Alp6p-HA copurified with GST-Nyv1p-LD and to a lesser extent with GST-nyv1p-LD(Y31Q,I34Q)—an average of twofold less (from 3 independent experiments). The results from one of these experiments are shown in Figure 10C, and the data from all three experiments are summarized in Figure 10D. For comparison, the binding of GST-Nyv1p-LD to Apl6p-HA was arbitrarily set to 100%; hence, there are no error bars for this data set (i.e., LD in Figure 10D).
Although the reduction in binding observed for the Nyv1p YI/QQ mutant in our in vivo and in vitro experiments was modest, our data from the in vivo coprecipitation experiments (Figure 10A) are in line with another study that examined interactions between mammalian AP3 and one of its YXXΦ-containing ligands (Nishimura et al., 2002). We therefore consider our biochemical data to be consistent with our in vivo protein sorting data. Together, these experiments support the view that amino acid substitutions to the Y31GTI34 motif that disrupted sorting of Nyv1p in vivo were most likely the result of a reduced binding affinity between the Nyv1p longin domain AP3.
Amino Acid Substitutions in the mu Chain of Yeast AP3 Preferentially Effect Sorting of YXXΦ-containing Cargoes
Because GFP-Nyv1p could be copurified with AP3, we next wanted to determine which subunit(s) of AP3 was responsible for recognition of the Y31GTI34 YXXΦ-like motif in the longin domain of Nyv1p. To accomplish this, we took advantage of what had been learned about the molecular basis of the recognition and sorting of YXXΦ signal-containing cargo by the mu chain of the mammalian AP2 complex (Owen and Evans, 1998; Owen et al., 2004; Honing et al., 2005), because relatively little is known about the molecular basis of recognition of YXXΦ-containing cargo proteins by the AP3 adaptor complex. Using a structure-based amino acid sequence alignment of the C-terminal domain of the mu chain of rat AP2 as a model for YXXΦ ligand binding, amino acid substitutions were introduced into the mu chain subunit (Apm3p) of yeast AP3 (μ1–μ4, Figure 11A). The effects of these amino acid substitutions on the sorting of YXXΦ- and acidic dileucine-containing yeast AP3 cargo proteins were assessed by microscopy. In these experiments, the AP3-dependent acidic dileucine-containing cargo protein GFP-Pho8p served as a control for assessing whether any of our mu chain amino acid substitutions resulted in loss-of-function of AP3 (as in rat AP2, binding of the acidic dileucine motif occurs at a different site from that of YXXΦ-containing proteins; Honing et al., 2005). Mislocalization in individual μ-amino acid substitution mutants was assessed on the basis of how closely the GFP-fusion protein localization phenotype (Figure 11A) coincided with that observed in apm3Δ cells expressing the same GFP-tagged marker protein. For example, GNS and GFP-Yck3p were considered mislocalized if any cell surface staining was apparent, even if some protein was still localized to the vacuole (as is the case for both GNS and GFP-Yck3p in apm3Δ cells; Figure 11E; Sun et al., 2004). As discussed above, the residual localization of GNS to the limiting membrane likely represented protein that had been redirected, via the CPY pathway, to the vacuolar membrane in the absence of a functional AP3 pathway (Figures 5A and 8, A and B; Sun et al., 2004).
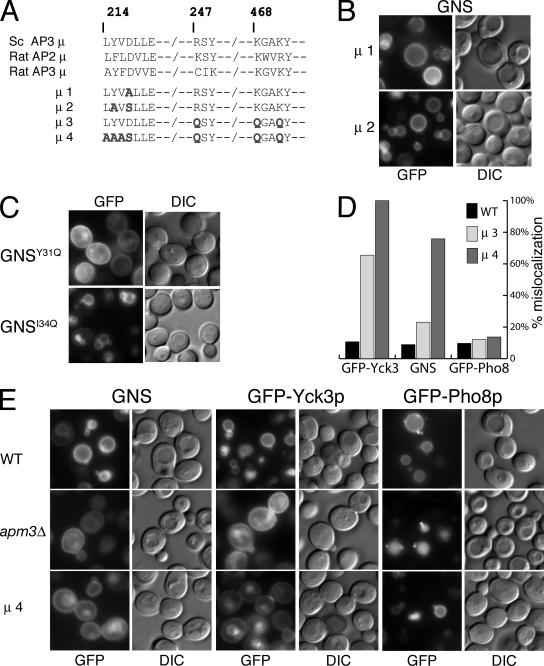
Figure 11.
Effect of amino acid substitutions in the mu chain of yeast AP3 (Apm3p) on YXXΦ-motif and acidic dileucine motif containing cargo proteins. (A) Top, an alignment of a portion of the C-terminal regions of the mu chains from yeast AP3 (Sc AP3μ), rat AP2 (Rat AP2μ), and rat AP3 (rat AP3μ). The amino acid residue numbering corresponds to that of the yeast AP3 mu chain (Apm3p). Bottom, summary of amino acid substitution mutations introduced into the mu chain of yeast AP3, the position and nature of amino acid substitutions are indicated with bold font (see text for details). (B) Effect of the Apm3p amino acid substitution mutants μ1 and μ2 on the sorting of GNS. (C) Effect of single amino acid substations in the Y31GTI34 YXXΦ-like motif on the localization of GNS. (D) Percentage of mislocalization of YXXΦ and acidic dileucine-containing cargo proteins in the μ3 and μ4 mu chain amino acid substitution mutants (see A and text for details). Yeast cells expressing the indicated APM3 mutant as their sole source of Apm3p were transformed with plasmids expressing GNS (YXXΦ signal-containing cargo protein), GFP-Yck3p (YXXΦ signal-containing cargo protein) or GFP-Pho8 (acidic dileucine signal-containing cargo protein). Mislocalization was assessed on the basis of how closely the localization phenotype of individual cells coincided with those of apm3Δ mutants expressing the same GFP-tagged marker protein (see text for details). GFP-Yck3p and GNS were considered to be mislocalized if cell surface localization was apparent. GFP-Pho8p was considered to be mislocalized if localization to the lumen of the vacuole was apparent. For wild-type cells, the data scored were GFP-Yck3p (mislocalized, n = 18; wild type, n = 151) GNS (mislocalized, n = 19; wild type, n = 194), GFP-Pho8p (mislocalized, n = 20; wild type n = 187). For the μ3 mutant the data collected were GFP-Yck3p (mislocalized, n = 148; wild type, n = 78), GNS (mislocalized, n = 51; wild type, n = 169), GFP-Pho8p (mislocalized, n = 30; wild type, n = 189). For the μ4 mutant, the data collected were GFP-Yck3p (mislocalized, n = 150; wild type, n = 0) GNS (mislocalized, n = 175; wild type, n = 56), GFP-Pho8p (mislocalized, n = 15; wild type, n = 108). (E) Mislocalization of YXXΦ-motif containing cargo proteins in apm3 cells expressing the μ4 mutant (apm34). Note that GFP-Yck3p and GNS localizes to the cell surface (as well as the vacuole) in the majority of μ4 mutant cells, whereas GFP-Pho8p localization in the μ4 mutant cells is essentially indistinguishable from that in wild-type cells.
We first examined the effect of an amino acid substitution in the C-terminal region of Apm3p (D217) that we predicted to be equivalent to D176 in the mu chain of rat AP2 (Owen and Evans, 1998). Asp176 has been shown to be a critical residue for interactions with the tyrosine of YXXΦ motifs, and substitution of A for D at amino acid 176 has been shown to significantly reduce transferrin receptor internalization in vivo (Nesterov et al., 1999). Yeast cells expressing apm3p(D217A) as their sole source of the AP3 mu chain (μ1 in Figure 11A) were transformed with plasmids expressing the AP3-dependent cargo proteins: GNS (Figure 8A), GFP-Yck3p (which contains a YXXΦ-sorting motif; Sun et al., 2004), or GFP-Pho8p (which contains an acidic dileucine-like sorting motif; Vowels and Payne, 1998), and transformants were examined by microscopy. Unexpectedly, cells expressing apm3p(D217A) were, based on our criteria, not defective in the sorting of GNS (which localized to the limiting membrane of the vacuole; μ1 in Figure 11B) or any of the other GFP-tagged AP3 cargo proteins examined, the localization pattern of which seemed indistinguishable from that in wild-type cells (our unpublished data). Following on the work of Honing et al. (2005), we generated an additional mutant of Apm3p analogous to their F174A; D176S form of the rat AP2 mu chain (μ2 in Figure 11A), which the authors described as abrogating binding to YXXΦ peptides. Cells expressing apm3p(Y215A,D217S) as their sole source of Apm3p were also not defective in the sorting of GNS (μ2 in Figure 11B) or of the other GFP-tagged AP3 cargo proteins examined (our unpublished data).
Our findings on the sorting of AP3-dependent YXXΦ-containing cargo proteins in cells expressing the Apm3p D217 and Apm3p Y215, D217 substitution mutants were perplexing particularly as substitution of Y31 to Q was sufficient to mislocalize the majority of GNSY31Q (Figure 11C), highlighting the relative importance of the tyrosine residue in the Y31GTI34 motif. Substitution of I34 with Q, in contrast, had significantly less impact on the localization of GNSI34Q, were the majority of cells showed localization to the limiting membrane of the vacuole (Figure 11C). On the basis of these results, we concluded that D217 of Apm3p was not crucial for the binding of AP3-interacting YXXΦ-motifs in vivo. To establish whether amino acid substitutions elsewhere in the presumptive YXXΦ binding site of Apm3p would effect the localization of GNS in cells, we generated additional amino acid substitutions that (by analogy to rat μ2) would be predicted to affect the binding of YXXΦ-motifs mediated by the Φ residue (termed the Y + 3 position) as well as by the tyrosine (Owen and Evans, 1998). Yeast cells expressing the first of these substitution mutants as their sole source of Apm3p (μ3 in Figure 11, A and D) showed a significant defect in the localization of GFP-Yck3p (>60% of cells showed some mislocalization), but a more moderate effect was observed on the localization of GNS (where ∼20% of cells showed some mislocalization, Figure 11D). In contrast, cells expressing the μ3 mutant of APM3 showed a very modest defect in the sorting of the acidic dileucine cargo protein GFP-Pho8p, were the localization pattern observed was on the whole indistinguishable from wild-type cells (Figure 11D).
When the scope of the amino substitutions was extended to encompass multiple presumptive contact sites for the tyrosine and Y + 3 positions (μ4 in Figure 11, A, D, and E), substantial defects in the sorting of both GFP-Yck3p and GNS were observed. The localization pattern of GFP-Yck3p in cells expressing the apm3p μ4 substitution mutant was essentially indistinguishable from that of GFP-Yck3p in apm3Δ cells, whereas ∼80% of cells showed some cell surface localization of GNS in μ4-expressing cells (Figure 11, D and E). In contrast, the localization pattern of the AP3 cargo protein containing the acidic dileucine motif (GFP-Pho8p) was largely unaffected in μ4-expressing cells (Figure 11, D and E), where it was found on the limiting membrane of the vacuole rather than in the lumen of the vacuole (compare GFP-Pho8p in WT, apm3Δ and μ4; Figure 11E). However, we cannot rule out the possibility that the μ3 and μ4 amino acid substitution mutants represent partial loss-of-function mutations in AP3, in which the sorting of GFP-Pho8p is significantly more robust than that of either GNS or GFP-Yck3p. Nonetheless, together, the data presented in Figure 11 are consistent with the μ subunit of yeast AP3 (Apm3p) playing an important but distinct mechanistic role in the recognition of tyrosine-based YXXΦ-like motifs.
DISCUSSION
In this study, we establish that Nyv1p, like Ykt6p and Sec22b, contains a longin domain in its N terminus, although Nyv1p does not share any significant amino acid similarity to the corresponding regions of either Ykt6p or Sec22b/Sec22p (Figures 1 and 2). Thus, the genome of S. cerevisiae encodes three longin domain-containing R-SNAREs—Ykt6p, Sec22p, and Nyv1p—and we have addressed the potential role of this fold in the localization of these proteins in this report. We find that GFP-Sec22p localizes to ER membranes, whereas GFP-Ykt6p, although functioning on multiple compartments, including the Golgi, endosome and vacuole, appears as a largely diffuse cytoplasmic haze (Figure 4B). Although the basis for the localization of Sec22p and Ykt6p is not known, the requirement for AP3 in the localization of Nyv1p to the limiting membrane of the vacuole has been reported previously (Figure 5; Reggiori et al., 2000). Here, we have established that the AP3-dependent vacuolar localization of Nyv1p is mediated by a YXXΦ-like sorting motif (Y31GTI34) located in its longin domain. A membrane-anchored form of the Nyv1p longin domain (GFP-Nyv1LD) is sufficient to mediate AP3-dependent sorting (Figures 4 and 5A) and amino acid substitutions to the Y31GTI34 motif (YI-QQ) disrupt the AP3-dependent sorting of the full-length protein as well as of the longin domain construct (Figure 5, A and B). In contrast, when tethered to the membrane via the Snc1p transmembrane domain, the longin domains of Ykt6p and Sec22p seem to lack any dominant sorting signals and are primarily localized to the cell surface (Figure 4, A and B). The Ykt6p-LD result was surprising to us, because Ykt6p is thought to function as a v-SNARE in AP3-mediated Golgi–vacuole protein sorting (Kweon et al., 2003) and might therefore be expected to contain an AP3-dependent sorting signal. The fact that Ykt6p, unlike Sec22p and Nyv1p, is not an integral membrane protein, but rather associates with membranes by virtue of the addition of lipid moieties to its C terminus, suggests alternate mechanisms may be used to localize Ykt6p in cells.
Thus, in yeast the SNARE longin domain does not seem to function as a generic protein-sorting module. However, we cannot exclude the possibility that addition of GFP to the N termini of Ykt6-LD and Sec22-LD/Sec22p does not mask localization signals in these proteins. In contrast, the capacity of the longin domain to direct the sorting of Nyv1p may reflect an evolutionarily conserved feature of a subset of R-SNARE longin domains. Two recent studies have identified a role for the longin domain in the sorting of presumptive Nyv1p homologues. In mammalian cells, the longin domain-containing SNARE TiVAMP/VAMP7 has been shown to be sorted to late endosomes via AP3, and the intact longin-domain of the protein is required to mediate this sorting (Martinez-Arca et al., 2003). In Arabidopsis, the longin domain of the vacuole-localized member of the VAMP7 family, VAMP711, seems to be sufficient to mediate its sorting; moreover, the longin domains of other Arabidopsis VAMP7 family members (the genome of A. thaliana encodes 11 members) also seem to be capable of directing their compartment-specific localization (Uemura et al., 2005). However, neither the location nor the nature of the sorting signals has been determined for the plant or mammalian VAMP7 family proteins.
Like Ykt6p, the longin domain of Nyv1p contains a substantial surface-exposed hydrophobic region that likely represents a protein-binding site (Figure 2B). In Ykt6p, this hydrophobic region is involved in binding to the SNARE-motif of the protein, thereby allowing Ykt6p to adopt a so-called “closed” or “folded-back” conformation (Tochio et al., 2001). What is the function of the surface-exposed hydrophobic surface on Nyv1p? Here, we addressed the potential role for the hydrophobic region in mediating the binding of Nyv1p to its SNARE-motif as well as the role of this region in the localization and sorting of the protein. Our data revealed that Nyv1p does not seem to adopt a folded-back conformation analogous to Ykt6p (Figure 3). In addition, amino acid substitutions that disrupted the hydrophobic surface (L73Q, I74Q) did not effect the sorting of Nyv1p in wild-type cells, suggesting that this region of the longin domain does not represent a critical binding site for interactions with AP3 (Figure 8D). The role of this region of the Nyv1p longin domain therefore remains to be established.
Although GNSYI-QQ was substantially mislocalized in wild-type cells, in keeping with the requirement for the Y31GTI34 motif in the AP3-dependent sorting of the protein, some localization of the protein to the limiting membrane of the vacuole was still apparent (Figure 8B). Although this residual staining may reflect rerouting of GNSYI-QQ to the vacuolar membrane by default (via the CPY pathway; Figure 5A) as occurs for several other proteins ordinarily trafficked by AP3 (Stepp et al., 1997; Darsow et al., 1998; Vowels and Payne, 1998; and Sun et al., 2004). It was also possible that additional, albeit less effective, AP3-dependent sorting signals operated in Nyv1p. Indeed, Nyv1p contains two additional YXXΦ-like motifs in its longin domain: Y8VEV11 and Y92VCF95. However, unlike amino acid substitutions to the Y31GTI34 motif (Figure 8B) comparable substitutions at Y8VEV11 and Y92VCF95 did not have a significant effect on the AP3-dependent sorting of the respective LD- or GNS-based constructs (Figure 9, A and B). We therefore concluded that Y31GTI34 functioned as the predominant AP3-dependent sorting motif in GNS as well as Nyv1p.
In addition to the effects of amino acid substitutions to the Y31GTI34 motif on protein localization (GFP-nyv1YI-QQ and GNSYI-QQ, Figures 7, A and B, and 8B), we also established that GFP-nyv1YI-QQ (Figure 10, A and B) and GST-nyv1-LDYI-QQ (Figure 10, C and D) were defective for binding to AP3 in biochemical experiments. For both, the in vivo coprecipitation experiments (Figure 10, A and B) and the in vitro protein pull-down assays (Figure 10, C and D), we consistently obtained reduced binding to AP3, relative to the wild-type proteins, for an average ∼12-fold reduction in the copurification of GFP-nyv1YI-QQ (Figure 10B; n = 5) and an average approximately twofold reduction in binding of GST-nyv1-LDYI-QQ (Figure 10D; n = 3) to the AP3 complex. Are these data in line with the effects of the YI-QQ amino acid substitutions on the sorting of Nyv1p, Nyv1-LD, and GNS observed in vivo? It is clear from our in vivo localization studies that the YI-QQ substitution had a profound effect on the localization of Nyv1p (Figure 7) and GNS (Figure 8), with the majority of the protein being mislocalized to the lumen of the vacuole and plasma membrane, respectively. In the absence of any other prominent AP3-dependent sorting signals in Nyv1p (Figure 9) how can we reconcile the ostensibly modest reduction in binding of GFP-nyv1YI-QQ and GST-nyv1-LDYI-QQ to AP3 (Figure 10)? One rather trivial, but entirely plausible explanation, is that the coprecipitation and protein mixing experiments, although giving consistent and reproducible reduced binding, are simply not sensitive enough to mimic the effects seen on the sorting of the YI-QQ amino acid substitution mutants. Alternatively, these biochemical data do reflect the effects of the YI-QQ amino acid substitutions seen on the sorting of Nyv1p and GNS for the following reason. Presumably, the AP3 complex does not identify substrates for sorting at a distance, but rather “samples” potential candidate proteins in situ, on Golgi membranes. Such a sampling mechanism would require AP3 to bind to proteins, with varying affinities, whether they bear a sorting signal or not. Thus, outwardly modest differences in the capacity of AP3 to bind to a particular protein could, in physiological terms, translate into considerable differences in that protein's capacity to be sorted by AP3. Thus, GFP-Nyv1YI-QQ may retain a level of binding to AP3 detectable by biochemical experiments, but it is too weak to support AP3-dependent sorting of the protein. When considered in this context, we believe that our biochemical data could be regarded as being consistent with the effects we observed on the localization patterns of the Nyv1p and GNS mutant proteins in vivo.
Having established that Nyv1p is sorted to the limiting membrane of the vacuole via a YXXΦ-like motif located in its longin domain, we set out to determine which subunit of the AP3 complex mediated binding between Nyv1p (GNS in this case) and AP3 (Figure 11). In these experiments, we focused on the role of the mu chain (encoded by the APM3 gene) as the corresponding subunit of mammalian AP2 (μ2) was known to be involved in binding to YXXΦ-motifs (Owen and Evans, 1998; Owen et al., 2004; Honing et al., 2005). However, our results on the effects of amino acid substitutions to either the YXXΦ-motif of GNS (Figure 11C) or to the mu chain of yeast AP3 (Figures 11, A, B, D, and E) were unexpected. Together, these data suggested to us that significant differences likely existed in the binding mechanism of YXXΦ-motif containing proteins to yeast AP3. For example, substitution of Y31 by Q or A at the YXXΦ-motif (Y31GTI34) in Nyv1p resulted in mislocalization of the protein to the cell surface, whereas the substitution at the Φ position had no apparent effect on localization (Figure 11B). In contrast, amino acid substitutions at either the Y or Φ residues in the Yck3p YXXΦ-motif did result in mislocalization of the protein, although it was incomplete (Sun et al., 2004). The differences observed in the effects of amino acid substitutions to the YXXΦ-motifs of Nyv1p and Yck3p may partially explain the observed differences in the binding affinity of these cargos for our various AP3 mu chain amino acid substitution mutants (Figure 11, D and E). The Yck3p AP3-dependent sorting signal contains an acidic residue at the Y + 1 position (YDSI), a common finding in YXXΦ-motifs in proteins sorted to the lysosome; the Y + 1 position of the motif (YGTI) of Nyv1p is a G residue, which is a relatively uncommon feature of such motifs (Bonifacino and Traub, 2003).
In sum, although we have been able to establish that the longin domain of Nyv1p contains a YXXΦ-like sorting signal (Y31GTI34) that is required for localization of the protein to the limiting membrane of the vacuole, our findings suggest that the mode of recognition of this motif is likely to be distinct from that previously described for mammalian AP2 and its YXXΦ-bearing binding partners. The molecular details underlying these apparent differences between mammalian AP2 and yeast AP3 require detailed structural information on AP3. It is noteworthy that the ς chain and the N-terminal region of the mu chain of adaptins contain a longin fold (Collins et al., 2002; Heldwein et al., 2004) as apparently do the ζ and δ subunits of the COPI coat protein complex (Schlenker et al., 2006). The significance of a shared fold in the vesicle coats and cargo sorting complexes that presumably recognize and sort longin SNAREs merits investigation.
ACKNOWLEDGMENTS
We thank Hugh Pelham and Rob Piper for plasmids. This work was supported by Hong Kong Research Grants Council Grants HKUST6105/02M and HKUST6407/05M (to D.K.B.) and HKUST6125/02M, HKUST6138/03M, and HKUST6125/04M (to M.Z.). The NMR spectrometers used in this study were purchased with funds donated to the Biotechnology Research Institute by the Hong Kong Jockey Club. M.Z. was a Croucher Foundation Senior Research Fellow.
Abbreviations used:
- AP
adaptor protein
- ER
endoplasmic reticulum
- FM4-64
N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenyl-hexatrienyl) pyridinium dibromide
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- LD
longin domain
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment receptor protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-02-0128) on July 19, 2006.
REFERENCES
- Antonin W., Fasshauer D., Becker S., Jahn R., Schneider T. R. Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nat. Struct. Biol. 2002;9:107–111. doi: 10.1038/nsb746. [DOI] [PubMed] [Google Scholar]
- Bonifacino J. S., Traub L. M. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- Black M. W., Pelham H.R.B. A selective transport route from Golgi to late endosomes that requires the yeast GGA proteins. J. Cell Biol. 2000;151:587–600. doi: 10.1083/jcb.151.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruinsma P., Spelbrink R. G., Nothwehr S. F. Retrograde transport of the mannosyltransferase Och1p to the early Golgi requires a component of the COG transport complex. J. Biol. Chem. 2004;279:39814–39823. doi: 10.1074/jbc.M405500200. [DOI] [PubMed] [Google Scholar]
- Brunger A. T. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Collins B. M., McCoy A. J., Kent H. M., Evans P. R., Owen D. J. Molecular architecture and functional model of the endocytic AP2 complex. Cell. 2002;109:523–535. doi: 10.1016/s0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- Cormack B. P., Valdivia R. H., Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Darsow T., Burd C. G., Emr S. D. Acidic di-leucine motif essential for AP3-dependent sorting and restriction of the functional specificity of the Vam3p vacuolar t-SNARE. J. Cell Biol. 1998;142:913–922. doi: 10.1083/jcb.142.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- Feng W., Fan J. S., Jiang M., Shi Y. W., Zhang M. PDZ7 of glutamate receptor interacting protein binds to its target via a novel hydrophobic surface area. J. Biol. Chem. 2002;277:41140–41146. doi: 10.1074/jbc.M207206200. [DOI] [PubMed] [Google Scholar]
- Filippini F., Rossi V., Galli T., Budillon A., D'Urso M., D'Esposito M. Longins: a new evolutionary conserved VAMP family sharing a novel SNARE domain. Trends Biochem. Sci. 2001;26:407–409. doi: 10.1016/s0968-0004(01)01861-8. [DOI] [PubMed] [Google Scholar]
- Fukasawa M., Varlamov O., Eng W. S., Sollner T. H., Rothman J. E. Localization and activity of the SNARE Ykt6 determined by its regulatory domain and palmitoylation. Proc. Natl. Acad. Sci. USA. 2004;101:4815–4820. doi: 10.1073/pnas.0401183101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett D. S., Powers R., Gronenborn A. M., Clore G. M. A common sense approach to peak picking in two-, three- and four-dimensional spectra using automatic computer analysis of contour diagrams. J. Magn. Reson. 1991;95:214–220. doi: 10.1016/j.jmr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Gavin A. C., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gonzalez L. C., Jr, Weis W. I., Scheller R. H. A novel snare N-terminal domain revealed by the crystal structure of Sec22b. J. Biol. Chem. 2001;276:24203–24211. doi: 10.1074/jbc.M101584200. [DOI] [PubMed] [Google Scholar]
- Hasegawa H., Yang. Z., Oltedal L., Davanger S., Hay J. C. Intramolecular protein-protein and protein-lipid interactions control the conformation and subcellular targeting of neuronal Ykt6. J. Cell Sci. 2004;117:4495–4508. doi: 10.1242/jcs.01314. [DOI] [PubMed] [Google Scholar]
- Heldwein E. E., Macia E., Wang J., Yin H. L., Kirchhausen T., Harrison S. C. Crystal structure of the clathrin adaptor protein 1 core. Proc. Natl. Acad. Sci. USA. 2004;101:14108–14113. doi: 10.1073/pnas.0406102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honing S., Ricotta D., Krauss M., Spate K., Spolaore B., Motley A., Robinson M., Robinson C., Haucke V., Owen D. J. Phosphatidylinositol-(4,5)-bisphosphate regulates sorting signal recognition by the clathrin-associated adaptor complex AP2. Mol. Cell. 2005;18:519–531. doi: 10.1016/j.molcel.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Jahn R., Lang T., Sudhof T. C. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- Jang S. B., Kim Y. G., Cho Y. S., Suh P. G., Kim K. H., Oh B. H. Crystal structure of SEDL and its implications for a genetic disease spondyloepiphyseal dysplasia tarda. J. Biol. Chem. 2002;277:49863–49869. doi: 10.1074/jbc.M207436200. [DOI] [PubMed] [Google Scholar]
- Kay L. E., Gardner K. H. Solution NMR spectroscopy beyond 25 kDa. Curr. Opin. Struct. Biol. 1997;7:722–731. doi: 10.1016/s0959-440x(97)80084-x. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter M., Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Kraulis P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon Y., Rothe A., Conibear E., Stevens T. H. Ykt6p is a multifunctional yeast R-SNARE that is required for multiple membrane transport pathways to the vacuole. Mol. Biol. Cell. 2003;14:1868–1881. doi: 10.1091/mbc.E02-10-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–886. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Lee M. C., Miller E. A., Goldberg J., Orci L., Schekman R. Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- Liu Y., Flanagan J. J., Barlowe C. Sec22p export from the endoplasmic reticulum is independent of SNARE pairing. J. Biol. Chem. 2004;279:27225–27232. doi: 10.1074/jbc.M312122200. [DOI] [PubMed] [Google Scholar]
- Martinez-Arca S., et al. A dual mechanism controlling the localization and function of exocytic v-SNAREs. Proc. Natl. Acad. Sci. USA. 2003;100:9011–9016. doi: 10.1073/pnas.1431910100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt E., Murphy M. Raster3D version 2.0, a program for photorealistic molecular graphics. Acta Crystallogr. 1994;D50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- Miller E. A., Beilharz T. H., Malkus P. N., Lee M. C., Hamamoto S., Orci L., amd Schekman R. Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell. 2003;114:497–509. doi: 10.1016/s0092-8674(03)00609-3. [DOI] [PubMed] [Google Scholar]
- Mossessova E., Bickford L. C., Goldberg J. SNARE selectivity of the COPII coat. Cell. 2003;114:483–495. doi: 10.1016/s0092-8674(03)00608-1. [DOI] [PubMed] [Google Scholar]
- Neri D., Szyperski T., Otting G., Senn H., Wuthrich K. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry. 1989;28:7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- Nesterov A., Carter R. E., Sorkina T., Gill G. N., Sorkin A. Inhibition of the receptor-binding function of clathrin adaptor protein AP2 by dominant-negative mutant mu2 subunit and its effects on endocytosis. EMBO J. 1999;18:2489–2499. doi: 10.1093/emboj/18.9.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N., Plutner H., Hahn K., Balch W. E. The subunit of AP-3 is required for efficient transport of VSV-G from the trans-Golgi network to the cell surface. Proc. Natl. Acad. Sci. USA. 2002;99:6755–6760. doi: 10.1073/pnas.092150699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T., Krieger M. Multi-component protein complexes and Golgi membrane trafficking. J. Biochem. 2005;137:109–114. doi: 10.1093/jb/mvi024. [DOI] [PubMed] [Google Scholar]
- Owen D. J., Collins B. M., Evans P. R. Adaptors for clathrin coats: structure and function. Annu. Rev. Cell Dev. Biol. 2004;20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- Owen D. J., Evans P. R. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science. 1998;282:1327–1332. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper R. C., Cooper A. A., Yang H., Stevens T. H. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Black M. W., Pelham H.R.B. Polar transmembrane domains target proteins to the interior of the yeast vacuole. Mol. Biol. Cell. 2000;11:3737–3749. doi: 10.1091/mbc.11.11.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi V., Banfield D. K., Vacca M., Dietrich L. E., Ungermann C., D'Esposito M., Galli T., Filippini F. Longins and their longin domains: regulated SNAREs and multifunctional SNARE regulators. Trends Biochem. Sci. 2004;29:682–688. doi: 10.1016/j.tibs.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Rothman J. E. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Schlenker O., Hendricks A., Sinning I, Wild K. The structure of the mammalian SRP receptor as prototype for the interaction of small GTPases with longin domains. J. Biol. Chem. 2006;281:8898–8906. doi: 10.1074/jbc.M512415200. [DOI] [PubMed] [Google Scholar]
- Sun B., Chen L., Cao W., Roth A. F., Davis N. G. The yeast casein kinase Yck3p is palmitoylated, then sorted to the vacuolar membrane with AP3-dependent recognition of a YXXPhi adaptin sorting signal. Mol. Biol. Cell. 2004;15:1397–1406. doi: 10.1091/mbc.E03-09-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepp J. D., Huang K., Lemmon S. K. The yeast adaptor protein complex, AP-3, is essential for the efficient delivery of alkaline phosphatase by the alternate pathway to the vacuole. J. Cell Biol. 1997;139:1761–1774. doi: 10.1083/jcb.139.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton R. B., Fasshauer D., Jahn R., Brunger A. T. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Teo H., Gill D. J., Sun J., Perisic O., Veprintsev D. B., Vallis Y., Emr S. D., Williams R. L. ESCRT-I Core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell. 2006;125:99–111. doi: 10.1016/j.cell.2006.01.047. [DOI] [PubMed] [Google Scholar]
- Tochio H., Tsui M.M.K., Banfield D. K., Zhang M. An autoinhibitory mechanism for nonsyntaxin SNARE proteins revealed by the structure of Ykt6p. Science. 2001;293:698–702. doi: 10.1126/science.1062950. [DOI] [PubMed] [Google Scholar]
- Uemura T., Sato M. H., Takeyasu K. The longin domain regulates subcellular targeting of VAMP7 in Arabidopsis thaliana. FEBS Lett. 2005;579:2842–2846. doi: 10.1016/j.febslet.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Vida T. A., Emr S. D. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels J. J., Payne G. S. A dileucine-like sorting signal directs transport into an AP3-dependent, clathrin-independent pathway to the yeast vacuole. EMBO J. 1998;17:2482–2493. doi: 10.1093/emboj/17.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding S., Pelham H. R. The dynamics of Golgi protein traffic visualized in living yeast cells. Mol. Biol. Cell. 1998;9:2667–2680. doi: 10.1091/mbc.9.9.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuthrich K. Protein recognition by NMR. Nat. Struct. Biol. 2000;7:188–189. doi: 10.1038/73278. [DOI] [PubMed] [Google Scholar]