Abstract
Accurate chromosome segregation is controlled by the spindle checkpoint, which responds to the lack of microtubule–kinetochore attachment or of tension across sister kinetochores through phosphorylation of kinetochore proteins by the Mps1, Bub1, BubR1, Aurora B, and Plk1/Plx1 kinases. The presence of the 3F3/2 phosphoepitope on kinetochores, generated by Plk1/Plx1-mediated phosphorylation of an unknown protein, correlates with the activation of the tension-sensitive checkpoint pathway. Using immunodepletion approach and a rephosphorylation assay in Xenopus extracts, we report here that not only the formation of the 3F3/2 phosphoepitope is dependent on the checkpoint activation but also the loading of the 3F3/2 substrate to kinetochores requires the prior assembly of Mps1, Bub1 and BubR1 onto kinetochores. Interestingly, generation of the 3F3/2 epitope in checkpoint extracts requires the kinase activities of Mps1 and Bub1 but not that of BubR1. Furthermore, we demonstrate that checkpoint proteins in Xenopusextracts are assembled onto kinetochores in a highly ordered pathway consisting of three steps. Mps1 and Bub1 are loaded first, and BubR1 and Plx1 second, followed by Mad1 and Mad2. The characterization of this ordered assembly pathway provides a framework for the biochemical mechanism of the checkpoint signaling and will aid in the eventual identification of the 3F3/2 substrate.
INTRODUCTION
The spindle assembly checkpoint ensures the fidelity of chromosome segregation by delaying the onset of anaphase until all chromosomes are properly aligned on the metaphase plate. This checkpoint monitors the attachment of microtubules to kinetochores and the tension across sister kinetochores exerted by microtubules in the bipolar spindle. The lack of either attachment or tension at kinetochores results in the activation of the checkpoint and cell cycle arrest. The checkpoint signals originate from unattached or untensed kinetochores, and then they are transduced into cytosol to inhibit anaphase-promoting complex/cyclosome, a ubiquitin ligase whose activation is required for the separation of sister chromatids and exit from mitosis (for review, see Shah and Cleveland, 2000; Chan and Yen, 2003).
Several proteins involved in checkpoint responses have been characterized in vertebrates. These include five protein kinases, Mps1, Bub1, BubR1, Aurora B, and Plk1/Plx1, and three additional proteins, Mad1, Mad2, and Bub3 (Millband et al., 2002; Musacchio and Hardwick, 2002; Lens and Medema, 2003; Taylor et al., 2004; Ahonen et al., 2005; Wong and Fang, 2005). Because cellular responses to the checkpoint activation include not only mitotic arrest but also correction of mitotic defects, which, in turn, shut off the checkpoint signals, several checkpoint proteins have been demonstrated to be dual functional in these responses. For example, Aurora B is required not only for the recruitment of other checkpoint proteins to kinetochores but also for correcting improper kinetochore–microtubule attachments (for review, see Lens and Medema, 2003; Vagnarelli and Earnshaw, 2004). Although the precise molecular mechanism of how the checkpoint signals are transduced on kinetochores is not clear, protein phosphorylation plays an important role. Indeed, unattached or untensed kinetochores are hyperphosphorylated (Gorbsky and Ricketts, 1993; Nicklas et al., 1998), and these phosphorylations are essential for the recruitment of Mad2 to unattached kinetochores (Waters et al., 1999; Ahonen et al., 2005; Wong and Fang, 2005). Importantly, the 3F3/2 epitope, an unknown phosphoantigen recognized by the monoclonal 3F3/2 antibody (Cyert et al., 1988), is associated with kinetochores that are not under tension, and the presence of this phosphoepitope correlates with the activation of the tension-sensitive checkpoint pathway (Gorbsky and Ricketts, 1993; Nicklas et al., 1995, 1998; Logarinho et al., 2004). Microinjection of the 3F3/2 antibody into mitotic cells delays anaphase onset, probably by preserving the 3F3/2 epitope, indicating a direct role of the 3F3/2 substrate in tension-response (Campbell and Gorbsky, 1995).
We and others have recently identified Polo-like kinase 1 (Plk1 in human and Plx1 in Xenopus) as the 3F3/2 kinase (Ahonen et al., 2005; Wong and Fang, 2005). In both human cells and in Xenopus checkpoint extracts, Polo-like kinase 1 targets Mad2 and BubR1 to the kinetochores (Ahonen et al., 2005; Wong and Fang, 2005). Moreover, Plx1 is also required for the recruitment of structural components Ndc80 and Nuf2 to kinetochores in Xenopus egg extracts (Wong and Fang, 2005). These observations indicate a role of Plk1/Plx1 in the assembly of mature kinetochores required for checkpoint responses (Wong and Fang, 2005). In contrast, knockdown of Plk1 in mammalian cells causes defects in mitotic structures and activates the spindle checkpoint rather than abolishes it (Sumara et al., 2004; van Vugt et al., 2004), suggesting a lack of a requirement of Plk1 in the checkpoint arrest, although it is possible that the residual Plk1 protein in those knockdown cells is sufficient to mediate the checkpoint arrest. A clear understanding of Plk1 in checkpoint responses awaits the identification of the 3F3/2 epitope and the analysis of the 3F3/2 phosphorylation in the checkpoint responses. Toward this end, we set out to investigate the pathway by which the 3F3/2 antigen is targeted to kinetochores. Using a combination of immunodepletion, immunofluorescence staining and an in vitro rephosphorylation assay, we demonstrate that kinetochore localization of both the 3F3/2 kinase and the protein antigen recognized by the 3F3/2 antibody (the 3F3/2 substrate) are tightly regulated by spindle checkpoint proteins. Our data also suggest an ordered pathway for the assembly of checkpoint proteins onto kinetochores, and this ordered assembly is required for the association of the 3F3/2 substrate and its kinase with kinetochores.
MATERIALS AND METHODS
Antibodies
The following Xenopus proteins were expressed and purified as recombinant proteins: His6-Mad1 (aa 321–562), His6-Mad2, GST-Bub1 (aa 1–670), GST-BubR1 (aa 1–197), GST-Mps1 (kinase-dead mutant), and His6-Plx1 (aa 225–598). All recombinant proteins were expressed in Escherichia coli, except for Mps1, which was expressed in Sf9 cells. Rabbit antibodies were raised against above-mentioned recombinant proteins and affinity purified. CenpA antibody was generously provided by A. Straight (Stanford University, Stanford, CA). Commercial antibodies were obtained as follows: 3F3/2 ascite was from Boston Biologicals (Wellesley, MA) and monoclonal Plk1 antibody (for Western blotting) was from Zymed Laboratories (South San Francisco, CA).
Preparation of Xenopus Egg Extracts, Immunodepletion, and Translation in Extracts
Meiotic metaphase extracts (cytostatic factor [CSF] extracts) from Xenopus eggs, and demembranated sperm nuclei were prepared as described previously (Minshull et al., 1994).
For immunodepletion, 75 μg of affinity-purified antibodies was coupled to 8 μl of Affi-prep protein A beads (Bio-Rad, Hercules, CA) by dimethyl pimelimidate (Pierce Chemical, Rockford, IL). The antibody beads were then incubated with 50 μl of CSF extracts for 1 h at 4°C and then pelleted at 4°C. The depletion conditions were as follows: one round of immunodepletion for Mps1 and two successive rounds for Bub1 and BubR1. For Mad1 and Mad2 double depletion, extracts were depleted one round with Mad2 antibody beads followed by two rounds with Mad1 antibody beads.
For in vitro translation in extracts, mRNAs were transcribed from linearized plasmids encoding Mps1, Bub1, and BubR1 by using mMessage mMachine transcription kit (Ambion, Austin, TX). Translation reactions were performed in CSF extracts that had been depleted of the protein of interest. Proteins translated in depleted extracts were used in all rescue experiments. The kinase-dead mutants of Mps1, Bub1, and BubR1 used in this article were Mps1 D685A (provided by D. Cleveland, University of California, San Diego, La Jolla, CA) and Bub1 K872R and BubR1 K788R (gifts from R. H. Chen, Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan), respectively (Abrieu et al., 2001; Sharp-Baker and Chen, 2001; Chen, 2002).
Spindle checkpoint extracts were prepared as described previously (Minshull et al., 1994) with the following modifications: CSF extracts (fresh or depleted of the protein of interest) were incubated with demembranated sperm nuclei and nocodazole. For rescue experiments, in vitro-translated proteins were added to depleted extracts and incubated for 10 min at room temperature before the addition of sperm nuclei and nocodazole.
Immunofluorescence Microscopy
Spindle checkpoint extracts were fixed in fixation buffer (80 mM PIPES, pH 6.8, 2 mM MgCl2, 1 mM EGTA, 30% glycerol, 0.5% Triton X-100, 0.5 μM microcystin-LR, 2% formaldehyde). Each sample was then layered onto a centrifuge tube filled with CSF-XB (10 mM HEPES, pH 7.8, 50 mM sucrose, 100 mM KCl, 2 mM MgCl2, 1 mM EGTA) plus 0.5% Triton X-100 and 40% glycerol. A poly-l-lysine–coated coverslip was placed at the bottom of each tube, and nuclei were spun onto the coverslip. Coverslips were recovered, fixed with 2% formaldehyde in CSF-XBT (CSF-XB with 0.1% Triton X-100), blocked by CSF-XBT plus 3% bovine serum albumin (BSA), incubated with primary antibody at 4°C overnight and then with secondary antibodies for 1 h at room temperature (Invitrogen, Carlsbad, CA). Images were captured on an Axiovert 200M microscope (Carl Zeiss MicroImaging, Thornwood, NY) equipped with a 100× 1.4 numerical aperture oil immersion objective, a digital charge-coupled device camera (Hamamatsu Photonics, Hamamatsu City, Japan), and OpenLab 4.0.2 (Improvision, Lexington, MA). For quantitative comparison of fluorescence intensities, antibody concentrations were titrated to ensure a linear response of immunofluorescence signals to the antigen concentrations, and images were acquired and processed identically. For Figures 1C, 2, C and F, and 3, H, J, and L, the intensity of each kinetochore 3F3/2 signal was determined relative to that of CenpA obtained from the same kinetochore. The mean fluorescence intensity values and their SEMs were calculated from ≥15 kinetochores taken from multiple nuclei in different microscope fields. Subsequently, all mean fluorescence intensities were normalized to the corresponding values derived from mock-depleted extracts.
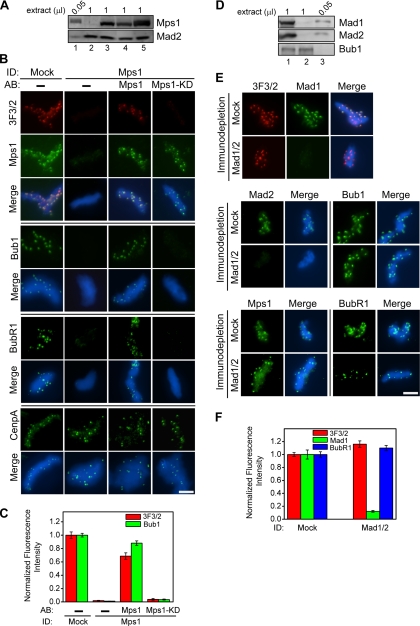
Figure 1.
Mps1, but not Mad1 or Mad2, is required for the formation of kinetochore 3F3/2 epitope and for the kinetochore association of Bub1 and BubR1. (A) Immunodepletion of Mps1 from CSF extracts. Extracts were mock-depleted (1 μl; lane 5) or depleted of Mps1 (1 μl; lanes 2–4). The Mps1-depleted extracts were then supplemented with translated Mps1 (lane 3) or Mps1-KD (lane 4). A volume of 0.05-μl input CSF extract was included in lane 1 to quantify the degree of depletion. Mad2 was shown here as a loading control. (B) Nuclei purified from checkpoint extracts that had undergone immunodepletion (ID) and add-back (AB) of the indicated proteins were stained for 3F3/2 in red and Mps1, Bub1, BubR1 and CenpA in green. (C) Mean kinetochore fluorescence intensities of 3F3/2 (red) and Bub1 (green) signals were quantified from checkpoint extracts that had undergone immunodepletion and add-back of the indicated proteins. The fluorescence intensity was calculated from ≥15 kinetochores taken from multiple nuclei in different fields and normalized to the corresponding value derived from mock-depleted extracts (see Materials and Methods for details). (D) Double depletion of Mad1 and Mad2 from CSF extracts. The indicated volumes of the mock-depleted extract (1 μl; lane 1), the Mad1-/Mad2 Ndepleted extract (1 μl; lane 2) as well as the input CSF extract (0.05 μl; lane 3) were analyzed by Western blotting to determine the depletion efficiency. Bub1 served as a loading control. (E) Nuclei purified from checkpoint extracts that had undergone immunodepletion were stained for 3F3/2 in red and Mps1, Mad1, Mad2, Bub1, and BubR1 in green. (F) Mean kinetochore fluorescence intensities were quantified and normalized as described in C for 3F3/2 (red), Mad1 (green), and BubR1 (blue) from Mad1/Mad2-depleted samples. Error bars represent SEM. Bar, 5 μm.
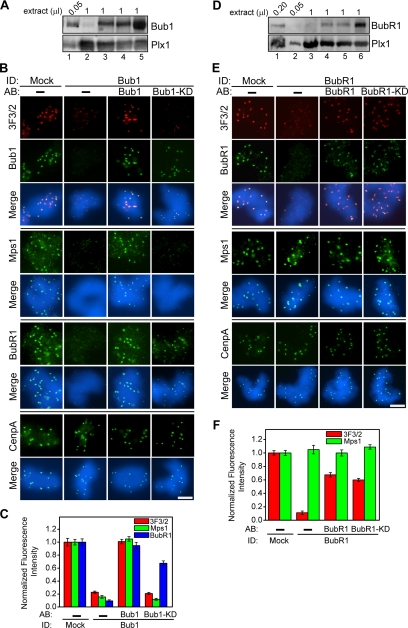
Figure 2.
Bub1 and BubR1 are required for the generation of the kinetochore 3F3/2 epitope. (A) Bub1 depletion efficiency was determined by Western analysis of the indicated volumes of the mock-depleted extract (lane 5), the Bub1-depleted extract (lane 2), the Bub1-depleted extracts with the add-back of translated Bub1 (lanes 3) or Bub1-KD (lane 4) as well as the input CSF extract (lane 1). Plx1 was shown here as a loading control. (B) Nuclei purified from checkpoint extracts that had undergone immunodepletion and add-back of the indicated proteins were stained in red for 3F3/2 and in green for Bub1, Mps1, BubR1, and CenpA. (C) Mean kinetochore fluorescence intensities of 3F3/2 (red), Mps1 (green), and BubR1 (blue) were quantified, as described in Figure 1C, from samples that were depleted of Bub1 and then added back with the indicated proteins. (D) The indicated volumes of the mock-depleted extract (lane 6), the BubR1-depleted extract (lane 3), the BubR1-depleted extracts with the add-back of translated BubR1 (lane 4) or BubR1-KD (lane 5) as well as the input CSF extract (lanes 1 and 2) were analyzed by Western blotting to determine the depletion efficiency and the add-back level. Plx1 was shown as a loading control. (E) Nuclei purified from checkpoint extracts that had undergone immunodepletion and add-back of the indicated proteins were stained for 3F3/2 (red) and for BubR1, Mps1, and CenpA (green). (F) Mean kinetochore fluorescence intensities of 3F3/2 (red) and Mps1 (green) were quantified, as described in Figure 1C, from samples that were depleted of BubR1 and then added back with the indicated proteins. Error bars represent SEM. Bar, 5 μm.
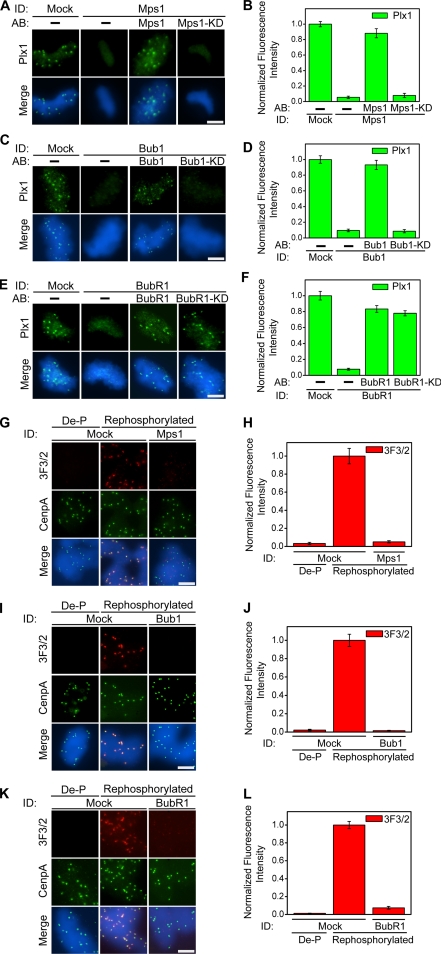
Figure 3.
Kinetochore localization of the 3F3/2 kinase and substrate is controlled by Mps1, Bub1, and BubR1. (A, C, and E) Nuclei purified from checkpoint extracts that had undergone immunodepletion and add-back of the indicated proteins were stained for Plx1 (green). (B, D, and F) Mean kinetochore fluorescence intensities of Plx1 were quantified, as described in Figure 1C, from samples that had undergone immunodepletion and add-back of the indicated proteins. (G, I, and K) Nuclei purified from checkpoint extracts that had undergone immunodepletion of the indicated proteins were dephosphorylated (De-P) with λ-phosphatase and subsequently rephosphorylated by incubation with CSF extracts. Red, 3F3/2; green, CenpA. (H, J, and L) Mean kinetochore fluorescence intensities of 3F3/2 were quantified, as described in Figure 1C, from samples that had undergone the indicated immunodepletion, dephosphorylation, and rephosphorylation. The fluorescence intensity was normalized to the corresponding value derived from mock-depleted extracts after rephosphorylation. Error bars represent SEM. Bar, 5 μm.
Dephosphorylation and Rephosphorylation Reactions
CSF extracts were either mock-depleted or depleted of Mps1, Bub1, or BubR1. Demembranated sperm nuclei were incubated with depleted extracts in the presence of nocodazole and subsequently purified onto coverslips through a glycerol cushion as described above. To remove the endogenous 3F3/2 phosphoepitope, coverslips were incubated with λ-phosphatase (New England Biolabs, Ipswich, MA) at 2 U/μl for 15 min, rinsed twice with CSF-XBT, and then blocked by CSF-XBT plus 3% BSA. To rephosphorylate, coverslips with the dephosphorylated nuclei were incubated with 50 μl of CSF extracts supplemented with 2 mM ATP and 1 μM microcystin LR for 30 min at room temperature. Finally, coverslips were processed for immunofluorescence staining as described above.
RESULTS
Formation of the Kinetochore 3F3/2 Epitope Depends on Mps1, but Not Mad1 and Mad2
We reasoned that if the 3F3/2 substrate is a component in the tension-sensitive pathway, its kinetochore association is probably regulated by the spindle checkpoint proteins. To test this prediction, the checkpoint protein Mps1 was immunodepleted from CSF extracts to >95% (Figure 1A) and the checkpoint extracts were then assembled by the addition of sperm nuclei and nocodazole. Nuclei purified from Mps1-depleted extracts were stained for the 3F3/2 phosphoepitope and various kinetochore proteins as described previously (Wong and Fang, 2005). Localization of CenpA, an inner centromere protein, was not affected by Mps1 depletion, indicating that Mps1 does not control the structural integrity of inner centromeres. Interestingly, depletion of Mps1 completely removed the 3F3/2 epitope from kinetochores, indicating a lack of either the 3F3/2 kinase and/or the 3F3/2 substrate on these kinetochores (Figure 1B). Other checkpoint proteins, such as Bub1, BubR1, Mad1, and Mad2, were also absent from kinetochores in Mps1-depleted extracts, suggesting that Mps1 is a checkpoint protein acting early in the assembly pathway (Figure 1B; our unpublished data; Abrieu et al., 2001). To ensure the specificity of the Mps1 depletion, we performed the following rescue experiments. Recombinant Mps1 was translated, using in vitro-transcribed Mps1 mRNA, in Xenopus CSF extracts that had been depleted of endogenous Mps1 to >95%. Addition of in vitro-translated Mps1 to the depleted extracts efficiently restored the 3F3/2 signals and kinetochore localization of all checkpoint components tested (Figure 1B). In contrast, addition of in vitro-translated kinase-dead Mps1 mutant (Mps1-KD) restored neither the 3F3/2 epitope nor the Bub1 and BubR1 signals onto kinetochores, even though Mps1-KD was efficiently targeted to kinetochores (Figure 1B). Quantification of the fluorescence intensities of 3F3/2 and Bub1 signals at the kinetochores further supports these conclusions (Figure 1C). Therefore, the kinase activity of Mps1 is required for recruiting checkpoint proteins onto kinetochores.
To test whether the formation of the 3F3/2 epitope requires components further downstream in the checkpoint signaling pathway, Mad1 and Mad2 were simultaneously depleted from CSF extracts to >95% (Figure 1D). In sharp contrast to the results from the Mps1 depletion, the 3F3/2 epitope, Bub1, BubR1, and Mps1 all remained on the kinetochores prepared from doubly depleted extracts (Figure 1, E and F). The lack of a requirement of Mad2 for targeting of the 3F3/2 substrate is consistent with the previous observation that it is the phosphorylated kinetochores that recruit Mad2 (Waters et al., 1999). Together, our data indicate that the generation of the kinetochore 3F3/2 epitope requires Mps1 but not Mad1 or Mad2.
3F3/2 Epitope Is Under the Control of the Bub1 Kinase
Because depletion of Mps1 removed Bub1 from kinetochores (Figure 1B; Vigneron et al., 2004), we next asked whether the lack of 3F3/2 epitope in Mps1-depleted extracts is due to the absence of Bub1. On depletion of Bub1 from CSF extracts to >95% (Figure 2A), 3F3/2 signals on kinetochores were substantially reduced, although not abolished (Figure 2, B and C). Similarly, Mps1 and BubR1 also failed to localize to kinetochores in the depleted extracts. These observations were not due to a general disruption of the kinetochore structure as the CenpA localization was not affected by Bub1 depletion (Figure 2B). Furthermore, all the phenotypes resulted from specific depletion of Bub1, because addition of Bub1 to ∼5–10% of the endogenous level efficiently restored the kinetochore localization of 3F3/2, Bub1, Mps1, and BubR1 (Figure 2, A–C). Interestingly, although the kinase-dead Bub1 mutant (Bub1-KD) was able to target to the kinetochores and to restore the BubR1 staining to ∼70% of the mock-depleted level, it rescued neither the Mps1 nor the 3F3/2 signals at kinetochores (Figure 2, B and C). Thus, the kinase activity of Bub1 is required for the generation of the 3F3/2 epitope and for the kinetochore localization of Mps1, but it is not absolutely required for the recruitment of Bub1 and BubR1 to kinetochores.
Generation of the 3F3/2 Epitope Is Dependent on the BubR1 Protein but Independent of Its Kinase Activity
Results from the Bub1 depletion experiments suggested that kinetochore localization of BubR1, in the absence of Bub1 kinase activity, was not sufficient to support the formation of the 3F3/2 epitope. To directly determine whether BubR1 contributes to the generation of the 3F3/2 epitope, BubR1 was depleted from CSF extracts to ∼95% before the assembly of checkpoint extracts (Figure 2D). Similar to the results of the Bub1 depletion, the 3F3/2 epitope was greatly reduced but not abolished in the BubR1-depleted extracts, whereas the depletion had no effect on the kinetochore localization of Mps1 and CenpA (Figure 2, E and F). Thus, Mps1 is upstream of BubR1 in the kinetochore assembly pathway for the checkpoint proteins. Interestingly, addition of either wild-type BubR1 or kinase-dead BubR1 (BubR1-KD) mutant to BubR1-depleted extracts restored both BubR1 and 3F3/2 signals at kinetochores, indicating a kinase-independent function of BubR1 in the generation of the 3F3/2 epitope (Figure 2, E and F). The <100% recovery of the 3F3/2 intensities at the kinetochores, when normalized against the CenpA levels (Figure 2F), is likely due to the fact that BubR1 and BubR1-KD were only added back to ∼20% of the endogenous level (Figure 2D). We conclude that the BubR1 protein, but not its kinase activity, is required for the formation of 3F3/2 epitope. This is in sharp contrast to the requirement of the kinase activities of Mps1 and Bub1 for the generation of the 3F3/2 epitope.
Mps1, Bub1, and BubR1 Control the Kinetochore Localization of the 3F3/2 Kinase Plx1
The dependence of the 3F3/2 epitope on Mps1, Bub1, and BubR1 can result from an absence of the 3F3/2 substrate on kinetochores in these depleted extracts, from an absence of the 3F3/2 kinase Plx1 on kinetochores, or both. These possibilities were distinguished in the following experiments. Mps1, Bub1, or BubR1 was individually depleted from extracts, and the spindle checkpoint was activated in the depleted extracts. Nuclei were purified from such depleted extracts and stained for Plx1. Consistent with our previous observations, in the absence of the Mps1 kinase activity, Plx1 failed to target to kinetochores (Figure 3, A and B; Wong and Fang, 2005). Similarly, Plx1 was absent from kinetochores in the Bub1-depleted extracts, and addition of the wild-type Bub1, but not Bub1-KD, recruited Plx1 to kinetochores (Figure 3, C and D). Interestingly, the BubR1 protein, but not its kinase activity, was required for the localization of Plx1 to kinetochores, because addition of either wild-type or BubR1-KD was sufficient to restore the kinetochore localization of Plxl to a similar extent (Figure 3, E and F). In all cases, depletion of Mps1, Bub1, or BubR1 did not affect the levels of the Plx1 protein in extracts (Figure 2, A and D; Wong and Fang, 2005). Thus, Mps1, Bub1, and BubR1 are required for targeting the 3F3/2 kinase Plx1 to kinetochores.
Mps1, Bub1, and BubR1 Are All Required for Targeting the 3F3/2 Substrate to Kinetochores
Next, we sought to determine whether the kinetochore localization of the 3F3/2 substrate itself is under the control of Mps1, Bub1 or BubR1. To achieve this, we modified a previously developed rephosphorylation assay (Wong and Fang, 2005). Nuclei were incubated with nocodazole in the Mps1-, Bub1- or BubR1-depleted extracts and subsequently purified through a glycerol cushion. The purified nuclei were then fixed and treated with λ-phosphatase to dephosphorylate all kinetochore proteins. The resulting nuclei were then used as substrates in an in vitro phosphorylation assay using CSF extracts as the source of the 3F3/2 kinase supplied in trans. We have previously demonstrated that CSF extracts, when added in trans to the dephosphorylated and fixed nuclei, were sufficient to regenerate the 3F3/2 epitope on kinetochores (Wong and Fang, 2005). Furthermore, control experiments confirmed that the recovery of the 3F3/2 signals in the rephosphorylation assay does not require stable association of the 3F3/2 kinase Plx1 with kinetochores (our unpublished data). Consistent with our previous observations, CSF extracts were able to phosphorylate and recover the kinetochore 3F3/2 signals on nuclei purified from mock-depleted extracts (Figure 3, G–L). In contrast, none of the Mps1-, Bub1- or BubR1-depleted extracts regained the kinetochore 3F3/2 signals upon rephosphorylation with CSF extracts (Figure 3, G–L). Quantification of the kinetochore fluorescence intensities further confirms these conclusions (Figure 3, H, J, and L). These results indicate that the 3F3/2 substrate was absent on kinetochores purified from Mps1-, Bub1- or BubR1-depleted extracts and that the 3F3/2 substrate in CSF extracts was not recruited to the kinetochores during the rephosphorylation reaction under our experimental conditions. Thus, Mps1, Bub1, and BubR1 are all required for the localization of the 3F3/2 substrate to kinetochores.
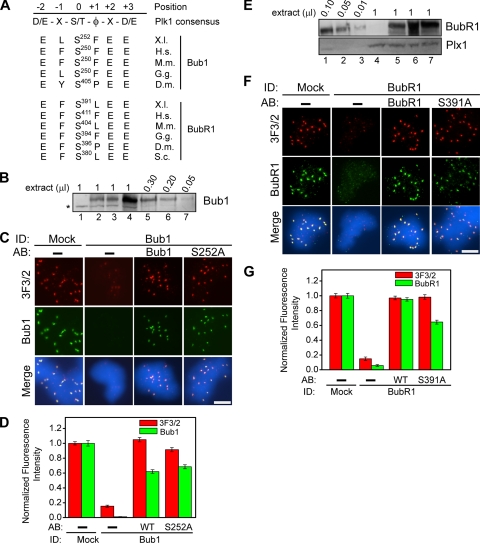
The Conserved Plx1 Sites in Bub1 and BubR1 Are Not the 3F3/2 Epitope
Our data raise the possibility that either Bub1 or BubR1 is the 3F3/2 substrate. Sequence analysis indicates that Bub1 and BubR1 each contain multiple sites that fit the Plk1 consensus phosphorylation sequence (Nakajima et al., 2003). Among these candidate sites, Bub1 and BubR1 each only has one Plk1 site (serine252 in Bub1 and serine391 in BubR1) that is also conserved among their respective homologues during evolution (Figure 4A). Given that the tension-sensitive 3F3/2 epitope on kinetochores is conserved from Drosophila to human (Bousbaa et al., 1997; Logarinho et al., 2004; Ahonen et al., 2005; Wong and Fang, 2005), these two sites represent the best candidates for the 3F3/2 epitope. Thus, we generated Bub1-S252A and BubR1-S391A mutants in which the conserved serine residues were mutated to alanines and tested the effect of the mutations on the formation of the 3F3/2 epitope in checkpoint extracts. Bub1 was first depleted from CSF extracts to >95%, and, as expected, the 3F3/2 epitope was lost in the depleted checkpoint extracts (Figure 4, B and C). However, addition of either the Bub1-S252A mutant or the wild-type Bub1 rescued the 3F3/2 signals to a similar extent (Figure 4, C and D), indicating that serine252 in Bub1 is not the 3F3/2 epitope, at least not the sole or the major 3F3/2 epitope.
Figure 4.
Conserved candidate Plx1 sites in Bub1 and BubR1 are not the 3F3/2 epitope. (A) Sequence alignment of the putative conserved Plk1 phosphorylation sites in Bub1 and BubR1 among different species. The consensus sequence for Plk1 sites was identified by Nakajima et al. (2003). φ denotes hydrophobic amino acids. X represents any amino acids. X.l., Xenopus laevis; H.s., Homo sapiens; M.m., Mus musculus; G.g., Gallus gallus; D.m., Drosophila melanogaster; and S.c., Saccharomyces cerevisiae. (B) Extracts were mock-depleted (lane 4), Bub1-depleted (lanes 1–3) with the add-back of the wild-type Bub1 (lane 2) or the Bub1-S252A mutant (lane 3). The indicated volumes of input CSF extracts (lanes 5–7) were loaded to quantify the degree of depletion and add-back. The asterisk denotes a cross-reacting band. (E) Extracts were mock-depleted (lane 7), BubR1-depleted (lanes 4–6) with the add-back of the wild-type BubR1 (lane 6) or the BubR1-S391A mutant (lane 5). The indicated volumes of input CSF extracts (lanes 1–3) were loaded to quantify the degree of depletion and add-back. (C and F) Immunofluorescence staining of nuclei purified from checkpoint extracts that had undergone immunodepletion and add-back of the indicated wild-type proteins, the Bub1-S252A mutant, or the BubR1-S391A mutant. Red, 3F3/2; green, Bub1 and BubR1. Bar, 5 μm. (D and G) Mean kinetochore fluorescence intensities of 3F3/2 (red) and Bub1 or BubR1 (green) were quantified, as described in Figure 1C, from samples that had undergone the indicated immunodepletion and add-back of the indicated wild-type (WT) or mutant (S252A and S391A) proteins. Error bars represent SEM.
Next, we tested whether serine391 in BubR1 is the 3F3/2 epitope. Depletion of BubR1 removed the 3F3/2 epitope from kinetochores. However, addition of either BubR1-S391A or wild-type BubR1 efficiently rescued the 3F3/2 signals at kinetochores (Figure 4, E–G), indicating that serine391 in BubR1 is not the site recognized by the 3F3/2 antibody, at least not the sole or the major 3F3/2 epitope. We also noted that in the depletion and add-back experiments, the BubR1-S391A mutant protein was targeted to the kinetochores with a slightly reduced efficiency compared with the wild-type BubR1 (Figure 4, E–G). This is likely due to a slightly lower protein level in the BubR1-S391A added-back extracts (Figure 4E).
DISCUSSION
The 3F3/2 phosphoepitope is involved in the tension-sensitive response upon activation of the spindle checkpoint. We report here that the presence of this epitope depends on the prior assembly of checkpoint proteins Mps1, Bub1, and BubR1 onto the kinetochores. Although the 3F3/2 epitope is directly phosphorylated by the Plk1/Plx1 kinase, the generation of this phosphoepitope also requires the kinase activities of the Mps1 and Bub1 in the extracts. Interestingly, the kinase activity of BubR1 is not required for the formation of the 3F3/2 epitope on kinetochores even though this activity is involved in the checkpoint arrest (Mao et al., 2003). In contrast, the checkpoint proteins Mad1 and Mad2 are not required for the generation of the 3F3/2 epitope, indicating that these two proteins either act downstream of the 3F3/2 substrate or function in a parallel pathway.
Ordered Assembly of Spindle Checkpoint Proteins onto Kinetochores
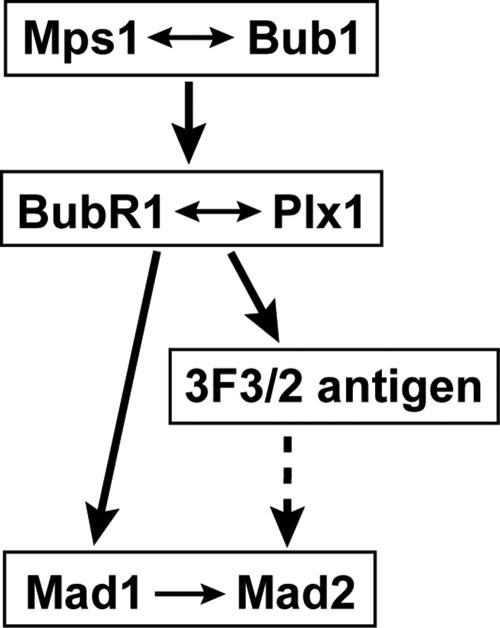
Upon activation of the spindle checkpoint, several checkpoint proteins, including Mps1, Bub1, BubR1, Mad1 and Mad2, are recruited to kinetochores. Association of these proteins with kinetochores mediates checkpoint signaling. Thus, understanding their assembly pathway will provide insight on the biochemical mechanism of the checkpoint responses. In this work, we demonstrated that in egg extracts, the assembly of checkpoint proteins onto kinetochores follows a hierarchical order, as summarized in Figure 5.
Figure 5.
Proposed model for the assembly of the spindle checkpoint proteins and the 3F3/2 antigen onto kinetochores. Arrows indicate the direction of the regulation, and double arrows indicate interdependency. Dotted line represents a possible regulation.
At the top of this checkpoint hierarchy are Mps1 and Bub1 (Figure 5), because depletion of either Mps1 or Bub1 removed BubR1 and Plx1 from kinetochores (Figures 1B, 2B, and 3, A–D). The association of Mps1 and Bub1 with kinetochores is dependent on each other (Figures 1B and 2B). Although this mutual dependence has been recently reported in egg extracts (Vigneron et al., 2004), we further demonstrated here that the kinase activities of both proteins are essential for this mutual dependence (Figures 1B and 2B). These observations differ from those in mammalian cells in which knockdown of Mps1 does not affect the kinetochore localization of Bub1 (Martin-Lluesma et al., 2002). Moreover, in mammalian cells, kinetochore targeting of BubR1 and Mps1 are not affected by Bub1 knockdown (Meraldi et al., 2004). Although we cannot exclude the possibility that this discrepancy between the mammalian cells and Xenopus extracts may result from an incomplete knockdown of checkpoint proteins in cultured cells, these results also suggest a possibility that there may exist system-dependent or species-specific kinetochore assembly pathways.
The next level in the assembly pathway is BubR1 and Plx1 (Figure 5). In our immunodepletion experiments, the kinetochore association of both BubR1 and Plx1 are dependent on Mps1 and Bub1, but not vice versa (Figures 1B, 2, B and E, and 3, A–D; our unpublished data), indicating that BubR1 and Plx1 act downstream of Mps1 and Bub1. In addition, the kinetochore localization of Plx1 and BubR1 in egg extracts is interdependent, because depletion of either one of them removed the other from kinetochores (Figure 3, E and F; Wong and Fang, 2005). Interestingly, there also exist differential requirements for kinetochore association of BubR1 and Plx1. BubR1 kinetochore localization requires the kinase activities of Mps1 and Plx1 as well as the Bub1 protein, but not its kinase activity (Figures 1B and 2B; Wong and Fang, 2005). On the other hand, the association of Plx1 with kinetochores requires the kinase activities of Mps1 and Bub1 as well as the BubR1 protein, but not its kinase activity (Figure 3, A–F; Wong and Fang, 2005), suggesting a complex regulatory network.
At the bottom of this hierarchy is the Mad1–Mad2 complex (Figure 5). We and others have previously shown that depletion of Plk1/Plx1 significantly reduces the levels of Mad2 on kinetochores, both in Xenopus egg extracts and in mammalian cells (Ahonen et al., 2005; Wong and Fang, 2005). Similarly, depletion of BubR1 abolishes the localization of Mad1 and Mad2 to kinetochores (our unpublished data; Chen, 2002). Thus, the Mad1–Mad2 complex acts downstream of BubR1 and Plx1. In contrast, simultaneous depletion of both Mad1 and Mad2, each to >95%, has essentially no effect on the kinetochore localization of other checkpoint proteins (Figure 1, D–F). Our observations are consistent with the results from small interfering RNA studies in mammalian system in which knockdown of Mad1 or Mad2 does not affect the kinetochore localization of Mps1, Bub1, or BubR1 (Martin-Lluesma et al., 2002; Meraldi et al., 2004). Although efficient depletion of Mad1 and Mad2 has no significant effect on the localization of BubR1 to kinetochores in our hands (Figure 1, E and F), a previous study in egg extracts reported a slight reduction of BubR1 at kinetochores upon depletion of Mad1 (Chen, 2002). Although we cannot exclude the possibility that Mad1 may help kinetochores to retain BubR1 more efficiently, our study shows that neither Mad1 nor Mad2 is absolutely required for the recruitment of BubR1 to kinetochores.
Kinetochore Association of the 3F3/2 Substrate Depends on Spindle Checkpoint Proteins
The phenomenon that unattached or untensed kinetochores are hyperphosphorylated was observed over a decade ago (Gorbsky and Ricketts, 1993; Nicklas et al., 1998). A key phosphoepitope in the 3F3/2 substrate is recognized by the 3F3/2 antibody (Cyert et al., 1988). The observations that the 3F3/2 epitope is associated with untense kinetochores (Gorbsky and Ricketts, 1993; Nicklas et al., 1998; Logarinho et al., 2004) and that microinjection of the 3F3/2 antibody delays anaphase onset (Campbell and Gorbsky, 1995) lead to the postulation that the 3F3/2 substrate and/or its kinase are tension-sensitive components of the spindle checkpoint (Nicklas et al., 1995, 1998). Although the molecular identity of the 3F3/2 substrate remains to be discovered, our rephosphorylation data suggest that its kinetochore localization is regulated by checkpoint kinases Mps1, Bub1, and BubR1.
The Identity of the 3F3/2 Substrate
Although several biochemical antigens with 3F3/2 epitopes have been identified, their direct connections to the kinetochore biology and to the tension responses remain to be established (Daum and Gorbsky, 1998; Daum et al., 2000). Our finding that the loading of the 3F3/2 epitope onto kinetochores is dependent on the prior assembly of checkpoint proteins raises the possibility that the 3F3/2 substrate could, in fact, be a bona fide checkpoint protein. However, the fact that the 3F3/2 epitope persist in Mad1-Mad2 doubly depleted extracts excludes the possibility that either Mad1 or Mad2 is the 3F3/2 substrate, consistent with the facts that Mad1 and Mad2 are primarily involved in the attachment-sensitive branch of the kinetochore signaling and that they dissociate from kinetochores upon microtubule attachment, even in the absence of tension (Chen et al., 1996, 1998; Waters et al., 1998; Howell et al., 2000). Similarly, because the nuclei purified from the BubR1-depleted extracts retained Mps1 on kinetochores (Figure 2, E and F) but lacked the 3F3/2 signals in our rephosphorylation assay (Figure 3, K and L), Mps1 is not the 3F3/2 substrate. In addition, the kinetochore localization of the Aurora B kinase has been shown to be not affected by the depletion of either Mps1 or Bub1 (Vigneron et al., 2004), and yet the 3F3/2 signals were absent in Mps1-depleted or Bub1-depleted nuclei, even after rephosphorylation with CSF extracts (Figures 1–3). Thus, Aurora B is unlikely to be the 3F3/2 substrate as well.
Our observation that Bub1 and BubR1 are required for kinetochore localization of the 3F3/2 substrate together with the well documented roles of Bub1 and BubR1 in the tension-sensitive branch of the checkpoint responses (Skoufias et al., 2001) raises the possibility that either Bub1 or BubR1 may harbor the 3F3/2 epitope. However, mutations of the highly conserved candidate Plk1 phosphorylation sites in Bub1 and BubR1 did not abolish the 3F3/2 epitope in checkpoint extracts (Figure 4). Thus, these two conserved sites, S252 in Bub1 and S391 in BubR1, are not recognized by the 3F3/2 antibody as the sole 3F3/2 epitope, although these results do not exclude the possibility that additional, nonconserved sites in Bub1 or BubR1 may be responsible for the formation of the kinetochore 3F3/2 signals.
The kinetochore-associated microtubule motor protein CENP-E is an activator of the BubR1 kinase (Mao et al., 2003). The observations that kinetochore localization of CENP-E requires BubR1 and that CENP-E is required for silencing BubR1-dependent checkpoint signaling (Mao et al., 2003, 2005) make CENP-E another attractive candidate for the 3F3/2 substrate.
In summary, the characterization of the kinetochore assembly pathway for the checkpoint proteins and for the 3F3/2 substrate will aid the eventual identification of the 3F3/2 epitope, which, in turn, will address the functional importance of this phosphorylation in checkpoint control.
ACKNOWLEDGMENTS
We thank Drs. D. Cleveland, R. H. Chen, and A. Straight for reagents, and members of the Fang laboratory for discussions. This work was supported by National Institutes of Health Grant GM-062852 (to G.F.) and by the Burroughs-Wellcome Career Award in Biomedical Sciences.
Abbreviations used:
- Plk1/Plx1
Polo-like kinase 1.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-04-0346) on August 2, 2006.
REFERENCES
- Abrieu A., Magnaghi-Jaulin L., Kahana J. A., Peter M., Castro A., Vigneron S., Lorca T., Cleveland D. W., Labbâe J. C. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106:83–93. doi: 10.1016/s0092-8674(01)00410-x. [DOI] [PubMed] [Google Scholar]
- Ahonen L. J., Kallio M. J., Daum J. R., Bolton M., Manke I. A., Yaffe M. B., Stukenberg P. T., Gorbsky G. J. Polo-like kinase 1 creates the tension-sensing 3F3/2 phosphoepitope and modulates the association of spindle-checkpoint proteins at kinetochores. Curr. Biol. 2005;15:1078–1089. doi: 10.1016/j.cub.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Bousbaa H., Correia L., Gorbsky G. J., Sunkel C. E. Mitotic phosphoepitopes are expressed in Kc cells, neuroblasts and isolated chromosomes of Drosophila melanogaster. J. Cell Sci. 1997;110:1979–1988. doi: 10.1242/jcs.110.17.1979. [DOI] [PubMed] [Google Scholar]
- Campbell M. S., Gorbsky G. J. Microinjection of mitotic cells with the 3F3/2 anti-phosphoepitope antibody delays the onset of anaphase. J. Cell Biol. 1995;129:1195–1204. doi: 10.1083/jcb.129.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan G. K., Yen T. J. The mitotic checkpoint: a signaling pathway that allows a single unattached kinetochore to inhibit mitotic exit. Prog. Cell Cycle Res. 2003;5:431–439. [PubMed] [Google Scholar]
- Chen R. H. BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. J. Cell Biol. 2002;158:487–496. doi: 10.1083/jcb.200204048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. H., Shevchenko A., Mann M., Murray A. W. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. H., Waters J. C., Salmon E. D., Murray A. W. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- Cyert M. S., Scherson T., Kirschner M. W. Monoclonal antibodies specific for thiophosphorylated proteins recognize Xenopus MPF. Dev. Biol. 1988;129:209–216. doi: 10.1016/0012-1606(88)90175-3. [DOI] [PubMed] [Google Scholar]
- Daum J. R., Gorbsky G. J. Casein kinase II catalyzes a mitotic phosphorylation on threonine 1342 of human DNA topoisomerase IIalpha, which is recognized by the 3F3/2 phosphoepitope antibody. J. Biol. Chem. 1998;273:30622–30629. doi: 10.1074/jbc.273.46.30622. [DOI] [PubMed] [Google Scholar]
- Daum J. R., Tugendreich S., Topper L. M., Jorgensen P. M., Hoog C., Hieter P., Gorbsky G. J. The 3F3/2 anti-phosphoepitope antibody binds the mitotically phosphorylated anaphase-promoting complex/cyclosome. Curr. Biol. 2000;10:R850–R852. doi: 10.1016/s0960-9822(00)00836-8. [DOI] [PubMed] [Google Scholar]
- Gorbsky G. J., Ricketts W. A. Differential expression of a phosphoepitope at the kinetochores of moving chromosomes. J. Cell Biol. 1993;122:1311–1321. doi: 10.1083/jcb.122.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B. J., Hoffman D. B., Fang G., Murray A. W., Salmon E. D. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J. Cell Biol. 2000;150:1233–1250. doi: 10.1083/jcb.150.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lens S. M., Medema R. H. The survivin/Aurora B complex: its role in coordinating tension and attachment. Cell Cycle. 2003;2:507–510. doi: 10.4161/cc.2.6.559. [DOI] [PubMed] [Google Scholar]
- Logarinho E., Bousbaa H., Dias J. M., Lopes C., Amorim I., Antunes-Martins A., Sunkel C. E. Different spindle checkpoint proteins monitor microtubule attachment and tension at kinetochores in Drosophila cells. J. Cell Sci. 2004;117:1757–1771. doi: 10.1242/jcs.01033. [DOI] [PubMed] [Google Scholar]
- Mao Y., Abrieu A., Cleveland D. W. Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell. 2003;114:87–98. doi: 10.1016/s0092-8674(03)00475-6. [DOI] [PubMed] [Google Scholar]
- Mao Y., Desai A., Cleveland D. W. Microtubule capture by CENP-E silences BubR1-dependent mitotic checkpoint signaling. J. Cell Biol. 2005;170:873–880. doi: 10.1083/jcb.200505040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Lluesma S., Stucke V. M., Nigg E. A. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297:2267–2270. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- Meraldi P., Draviam V. M., Sorger P. K. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Millband D. N., Campbell L., Hardwick K. G. The awesome power of multiple model systems: interpreting the complex nature of spindle checkpoint signaling. Trends Cell Biol. 2002;12:205–209. doi: 10.1016/s0962-8924(02)02276-6. [DOI] [PubMed] [Google Scholar]
- Minshull J., Sun H., Tonks N. K., Murray A. W. A MAP kinase-dependent spindle assembly checkpoint in Xenopus egg extracts. Cell. 1994;79:475–486. doi: 10.1016/0092-8674(94)90256-9. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Hardwick K. G. The spindle checkpoint: structural insights into dynamic signalling. Nat. Rev. Mol. Cell Biol. 2002;3:731–741. doi: 10.1038/nrm929. [DOI] [PubMed] [Google Scholar]
- Nakajima H., Toyoshima-Morimoto F., Taniguchi E., Nishida E. Identification of a consensus motif for Plk (Polo-like kinase) phosphorylation reveals Myt1 as a Plk1 substrate. J. Biological Chem. 2003;278:25277–25280. doi: 10.1074/jbc.C300126200. [DOI] [PubMed] [Google Scholar]
- Nicklas R. B., Campbell M. S., Ward S. C., Gorbsky G. J. Tension-sensitive kinetochore phosphorylation in vitro. J. Cell Sci. 1998;111:3189–3196. doi: 10.1242/jcs.111.21.3189. [DOI] [PubMed] [Google Scholar]
- Nicklas R. B., Ward S. C., Gorbsky G. J. Kinetochore chemistry is sensitive to tension and may link mitotic forces to a cell cycle checkpoint. J. Cell Biol. 1995;130:929–939. doi: 10.1083/jcb.130.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J. V., Cleveland D. W. Waiting for anaphase: Mad2 and the spindle assembly checkpoint. Cell. 2000;103:997–1000. doi: 10.1016/s0092-8674(00)00202-6. [DOI] [PubMed] [Google Scholar]
- Sharp-Baker H., Chen R. H. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J. Cell Biol. 2001;153:1239–1250. doi: 10.1083/jcb.153.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias D. A., Andreassen P. R., Lacroix F. B., Wilson L., Margolis R. L. Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc. Natl. Acad. Sci. USA. 2001;98:4492–4497. doi: 10.1073/pnas.081076898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I., Gimenez-Abian J. F., Gerlich D., Hirota T., Kraft C., de la Torre C., Ellenberg J., Peters J. M. Roles of polo-like kinase 1 in the assembly of functional mitotic spindles. Curr. Biol. 2004;14:1712–1722. doi: 10.1016/j.cub.2004.09.049. [DOI] [PubMed] [Google Scholar]
- Taylor S. S., Scott M. I., Holland A. J. The spindle checkpoint: a quality control mechanism which ensures accurate chromosome segregation. Chromosome Res. 2004;12:599–616. doi: 10.1023/B:CHRO.0000036610.78380.51. [DOI] [PubMed] [Google Scholar]
- Vagnarelli P., Earnshaw W. C. Chromosomal passengers: the four-dimensional regulation of mitotic events. Chromosoma. 2004;113:211–222. doi: 10.1007/s00412-004-0307-3. [DOI] [PubMed] [Google Scholar]
- van Vugt M. A., van de Weerdt B. C., Vader G., Janssen H., Calafat J., Klompmaker R., Wolthuis R. M., Medema R. H. Polo-like kinase-1 is required for bipolar spindle formation but is dispensable for anaphase promoting complex/Cdc20 activation and initiation of cytokinesis. J. Biol. Chem. 2004;279:36841–36854. doi: 10.1074/jbc.M313681200. [DOI] [PubMed] [Google Scholar]
- Vigneron S., Prieto S., Bernis C., Labbâe J. C., Castro A., Lorca T. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol. Biol. Cell. 2004;15:4584–4596. doi: 10.1091/mbc.E04-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters J. C., Chen R. H., Murray A. W., Gorbsky G. J., Salmon E. D., Nicklas R. B. Mad2 binding by phosphorylated kinetochores links error detection and checkpoint action in mitosis. Curr. Biol. 1999;9:649–652. doi: 10.1016/s0960-9822(99)80287-5. [DOI] [PubMed] [Google Scholar]
- Waters J. C., Chen R. H., Murray A. W., Salmon E. D. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong O. K., Fang G. Plx1 is the 3F3/2 kinase responsible for targeting spindle checkpoint proteins to kinetochores. J. Cell Biol. 2005;170:709–719. doi: 10.1083/jcb.200502163. [DOI] [PMC free article] [PubMed] [Google Scholar]