Abstract
PDZ proteins usually contain multiple protein–protein interaction domains and act as molecular scaffolds that are important for the generation and maintenance of cell polarity and cell signaling. Here, we identify and characterize TIP-1 as an atypical PDZ protein that is composed almost entirely of a single PDZ domain and functions as a negative regulator of PDZ-based scaffolding. We found that TIP-1 competes with the basolateral membrane mLin-7/CASK complex for interaction with the potassium channel Kir 2.3 in model renal epithelia. Consequently, polarized plasma membrane expression of Kir 2.3 is disrupted resulting in pronounced endosomal targeting of the channel, similar to the phenotype observed for mutant Kir 2.3 channels lacking the PDZ-binding motif. TIP-1 is ubiquitously expressed, raising the possibility that TIP-1 may play a similar role in regulating the expression of other membrane proteins containing a type I PDZ ligand.
INTRODUCTION
PDZ proteins are generally characterized by the presence of multiple protein–protein interaction modules, which allows them to act as molecular scaffolds that recruit membrane proteins, cytoskeletal elements, trafficking machinery, and signal transduction molecules into multimeric complexes at distinct membrane domains (Brone and Eggermont, 2005; Campo et al., 2005). Many play important roles in the generation and maintenance of epithelial and neuronal polarity. A prototypical example is provided by a group of PDZ proteins, Lin-7 (mLin-7/Veli/MALS), Lin-2 (CASK) and Lin10 (Mint-1/X11), that were first discovered in Caenorhabditis elegans (Kaech et al., 1998). In vulvar precursor cells, these molecules form a tripartite protein complex that interacts with the Let-23 receptor to coordinate its expression on the basolateral membrane (Simske et al., 1996; Kaech et al., 1998; Rongo et al., 1998). Studies in C. elegans and mammalian systems have revealed a probable mechanism. Lin-2 (CASK) interacts with cell structural elements, anchoring the complex to the membrane (Cohen et al., 1998). Lin-10 offers a means of delivering the complex via its interactions with microtubule motors (Setou et al., 2000) and Munc-18 docking machinery (Hata et al., 1993; Borg et al., 1998; Butz et al., 1998). The smaller Lin-7 protein acts as the bridge between receptor and scaffold complex, binding the receptor through a type I PDZ interaction and the scaffold via a L27 domain interaction (Doerks et al., 2000) with CASK (Kaech et al., 1998). In C. elegans, depletion of any one of these three PDZ proteins causes mislocalization of Let-23 to the apical membrane, illustrating the functional significance of the complex.
Orthologues of this C. elegans PDZ protein complex have been identified in mammalian tissues (Borg et al., 1998; Butz et al., 1998; Straight et al., 2001b; Olsen et al., 2002). Although renal epithelia do not express a Lin-10 orthologue (Borg et al., 1998), the mLin-7 and CASK complex is evolutionarily conserved and localizes exclusively at the basolateral membrane (Olsen et al., 2005; Straight et al., 2001). Recently, we demonstrated that mLin-7 binds the inwardly rectifying potassium channel Kir 2.3 through a PDZ interaction and that the mLin-7/CASK complex plays an important role in coordinating polarized expression of the channel at the basolateral membrane by preventing the channel from traveling in the endocytic pathway (Olsen et al., 2002). The mLin-7/CASK complex has also been implicated in basolateral expression of the epithelial GABA transporter BGT-1 (Perego et al., 1999), the EGF-like receptor ErbB-2/Her2 (Shelly et al., 2003), and other potassium channels (Leonoudakis et al., 2004b), suggesting that interaction with the mLin-7/CASK complex represents a general mechanism for the polarized expression of basolateral membrane proteins containing a PDZ-binding motif.
While investigating the mechanism for polarized expression of Kir 2.3, we discovered a novel PDZ-binding partner for the channel, Tax-interacting Protein-1 (TIP-1). A partial TIP-1 clone was originally identified through a yeast two-hybrid screen as one of many binding partners for the viral oncoprotein Tax (Rousset et al., 1998). Here, we show that the full-length TIP-1 encodes a protein consisting of a single PDZ domain and appears to be devoid of other protein–protein interaction modules. This represents a departure from the classic PDZ protein retention/clustering scheme because PDZ proteins rely on multiple protein–protein interaction motifs to effectively scaffold membrane, cytoskeletal, and signaling proteins. Indeed, TIP-1 antagonizes the retention function of the mLin-7/CASK complex by competing for interaction with Kir 2.3. This molecular switch results in internalization of Kir 2.3 channels. The competitive PDZ interaction described here represents a novel mechanism for regulating the surface expression of Kir 2.3 channels and may provide a more general means for regulating other PDZ protein interactions.
MATERIALS AND METHODS
Yeast Two-Hybrid Interaction Trap Library Screen
The yeast two-hybrid interaction trap system was used according to established methods (Gyuris et al., 1993; Golemis and Khazak, 1997) to capture proteins that interact with the PDZ-binding motif in Kir 2.3. A sequential, two-step library screen with two different baits was performed. In the first stage of the screen, a Lex A fusion protein of the Kir 2.3 C-terminal targeting domain (amino acids 417-445) was used as the bait (pJK202 containing a HIS3 selectable marker) to screen a conditionally expressed kidney cDNA library (Clontech, Mountain View, CA) in the pJG4-5 plasmid (TRP4 selectable marker) as before (Olsen et al., 2002). In the second stage of the screen, positive library clones were screened with a lexA fusion of a mutant Kir 2.3 C-terminal domain that lacks the PDZ-binding motif (443×).
ONPG Solution β-galactosidase Assay
β-galactosidase activity was measured at Vmax using 2-nitrophenyl-β-d- galactopyranoside (ONPG), according to established methods (Mendelsohn and Brent, 1994). Transfected yeast were grown (A600 of 0.9–1.2) under conditions that induce or repress the promoter. Yeast cultures were pelleted then resuspended in buffer and diluted 1:10 in buffer containing 0.001% SDS and 2% chloroform. Tubes were equilibrated at 30°C (15 min), and ONPG was added. The reaction was stopped with 1 M Na2CO3, and product was measured by spectrofluorometry. Miller units were calculated as previously described (Reynolds et al., 1997).
Antibodies
Anti-TIP-1 antibodies were raised in rabbits against a unique epitope in TIP-1 (39-DQDPSQNPFSEDKTC) and affinity-purified. The epitope is conserved across mammalian species, including dog (Supplementary Data I). Anti-Kir 2.3 antibody was previously described (Olsen et al., 2002). Because the Kir 2.3 immunogen is not available, preimmune serum of matched IgG concentration was used as a control for the Kir 2.3 localization studies. All other antibodies were purchased from commercial suppliers: rabbit anti-His, rabbit anti-myc, and mouse anti-myc (9E10; Santa Cruz Biotechnologies, Santa Cruz, CA); mouse anti-FLAG M2 and anti-VSV monoclonal antibodies (Sigma, St. Louis, MO); mouse anti-CASK and anti-NaK/ATPase α1 (Upstate Cell Signaling Solutions, Waltham, MA); rabbit anti-Veli3 (aka Lin-7C; Zymed Laboratories, South San Francisco, CA); mouse anti-HA (Boehringer Mannheim, Indianapolis, IN); rabbit anti-Rab11 (BD Transduction Laboratories, Lexington, KY); horseradish peroxidase (HRP)-conjugated goat anti-mouse (Jackson Laboratory, Bar Harbor, ME); HRP-conjugated goat anti-Rabbit and mouse anti-GST (Amersham Biosciences, Piscataway, NJ); AlexaFluor-conjugated secondary antibodies (Molecular Probes, Eugene, OR).
DNA Constructs
FLAG and myc epitope tags were engineered into the N- or C- terminus of TIP-1, respectively, by PCR and subcloned into the pcDNA3.1 vector for cell studies or into the pShuttleCMV vector (Stratagene, La Jolla, CA) for creation of adenoviral constructs. Mutagenesis was done by PCR with PfuTubo DNA polymerase (QuikChange, Stratagene). All sequences were verified by dye termination DNA sequencing. Methods for the construction of the Kir 2.3 COOH-terminal lexA fusions and the VSV(Y2F form)-tagged wild-type Kir 2.3 have been described previously (Le Maout et al., 2001; Olsen et al., 2002).
Synthesis of Recombinant Proteins
FLAG-tagged full-length TIP-1 cDNAs were subcloned into the pRSET vector (Invitrogen) in-frame with an N-terminal His-tag to produce pRSET-TIP-1/WT, K20A, H90A, and K20A/H90A double mutant. The final 25 residues of the C-terminal tail of WT and ΔPDZ Kir 2.3 were subcloned into the pGEX 5x-1 vector (Amersham) in-frame with an N-terminal GST tag to produce pGEX-Kir 2.3 and -Kir 2.3ΔPDZ. These expression vectors were transformed into BL21(DE3)pLysS (Invitrogen) competent Escherichia coli. Recombinant His-TIP-1 proteins were purified using Ni-NTA spin columns (Qiagen, Chatsworth, CA). Recombinant GST-Kir 2.3 proteins were purified using GS4B-sepharose (Amersham) and dialyzed against PBS. Total protein concentration was determined by Bradford Assay.
GST Pulldown
GST-Kir 2.3 and GST-Kir 2.3ΔPDZ were incubated with GS4B beads for 1 h, washed with PBS, and incubated overnight with recombinant His-TIP-1. Beads were washed with TEE (50 mM Tris-HCl, pH 7.5), 1 mM EDTA, 1 mM EGTA) and then eluted by boiling in SDS-sample buffer.
Northern Analysis
Northern blot derived from multiple human tissues (Origene, Rockville, MD) containing 2 μg poly(A)-RNA per lane was probed with a 591-base pair cDNA fragment of TIP-1 (bases 400–991) and labeled by random priming with [α-32P]dCTP (Amersham) according to manufacturer's instructions (Boehringer Mannheim). The blot was stripped and reprobed with [α-32P]dCTP-labeled human β-actin cDNA fragment as a control for loading.
Cell Culture and Transfection
All mammalian cell lines were cultured in a humidified atmosphere at 37°C in 5% CO2. COS, HEK, and type II MDCK cells were cultured as previously described (Olsen et al., 2002). Cells were transfected using LipofectAMINE transfection reagent (Invitrogen). Transfection medium was replaced 24 h after transfection with maintenance medium supplemented with 2 mM sodium butyrate (Sigma). The MDCK/TIP-1-myc stable cell line was created by a neomycin-selection protocol as previously described (Le Maout et al., 1997).
Production of TIP-1 Adenoviruses
Adenoviral vectors containing wild-type and H90A C-myc-TIP-1 were produced using AdEasy Adenoviral Vector System according to manufacturer's instructions (Stratagene). After viral amplification in low passage HEK cells, virus was harvested in Dulbecco's phosphate-buffered saline containing 1 mM MgCl2 (DPBS-M, Sigma) and purified on a cesium chloride gradient. The viral band was then loaded onto a PD-10 column (Amersham) to remove cesium chloride and eluted in DPBS-M.
Adenoviral Infection of MDCK-Kir 2.3VSV Cells
MDCK/Kir2.3-VSV cells were plated 1.5 × 105 cells/well on 12-mm-diameter Transwell polycarbonate filters (0.4-μm pore size, Corning Costar, Corning, NY) 24–48 h before infection. A “calcium switch” infection protocol was used as previously described (Henkel et al., 1998). Briefly, wells were filled with DPBS-M one time to wash, refilled with DPBS-M, and incubated for 15–30 min at 37°C and 5% CO2. Purified adenovirus was added to the apical chamber at an MOI of 100 while DPBS-M was added to the basolateral chamber. Mock-infected cells were treated identically, except that virus was omitted during the incubation period. Cells were incubated 1–2 h at 37°C and 5% CO2 before replacing complete growth medium in both the apical and basolateral chambers. Cells were incubated for 26 h after infection to permit protein expression. Epithelial integrity and cell confluence were assessed by transepithelial resistance measurements (EVOHM, WPI).
Cell Lysis
COS cells were lysed as described previously (Olsen et al., 2002). Adenovirus-infected MDCK/Kir 2.3-VSV cells were washed twice in ice-cold Ringer's solution (5 mM HEPES, 144 mM NaCl, 5 mM KCl, 1.2 mM NaH2PO4, 5.5 mM glucose, 1 mM MgCl2, 1 mM CaCl2, pH 7.4), harvested in ice-cold HEENG buffer (20 mM HEPES, pH 7.6, 25 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10% glycerol; Leonoudakis et al., 2004b), and resuspended in HEENG containing 1.0% Triton and protease inhibitors. Both COS and MDCK/Kir 2.3-VSV cell lysates were then passed though a 27-gauge needle, rotated at 4°C for 1 h, and centrifuged at max speed to pellet insoluble material. Protein concentrations were assessed by Bradford Assay Reagent (Bio-Rad, Richmond, CA).
Rat Kidney Homogenization
Isolated whole kidneys were resuspended in TSE buffer (20 mM Tris, 150 mM NaCl, 5 mM EDTA, pH 7.5) containing protease inhibitor cocktail and homogenized using a glass tissue homogenizer.
Immunoprecipitation and Immunoblotting
Cell lysates were precleared with Sepharose B (Sigma) or Fast Flow protein A Sepharose (Sigma) for 2 h at 4°C. The supernatants were than rotated overnight at 4°C with appropriate antibody and protein A. Beads were washed three times with lysis buffer containing 0.1% Triton and then eluted for 30 min at room temperature (RT) with SDS sample buffer. Eluates were separated by SDS-PAGE and transferred to nitrocellulose (Amersham). Membranes were probed with appropriate primary and HRP-conjugated secondary antibodies and developed with SuperSignal West Pico Chemiluminescent Substrate (Pierce Chemical, Rockford, IL). Densitometric measurements were made using NIH image software.
In Vitro TIP-1/Lin-7 Competition Binding
VSV-Kir 2.3/Lin 7 was recovered from COS cells cotransfected with VSV-Kir 2.3 and HA-Lin 7 (50 μl cell lysate, ∼4× cell volume) using anti-VSV antibodies (3 μg) bound to protein G-Sepharose after incubation overnight at 4°C. Subsequently, lysates from COS cells, which were separately transfected with vector (pcDNA) or myc-TIP-1 cDNA, were prepared and incubated with the immunopurified VSV-Kir 2.3/Lin 7 on anti-VSV protein G beads. Three different lysates were compared: 1) pcDNA alone (100 μl); 2) a mixture of equal volumes of vector (50 μl) and myc-TIP-1 (50 μl), and 3) myc-TIP-1 alone (100 μl). After the incubation, immunoprecipitates were washed, eluted, and immunoblotted as described above.
Surface Biotin Labeling
MDCK/Kir 2.3-VSV cells, grown on filters, were placed on ice and washed twice with ice-cold Ringer's solution. Impermeable NHS-SS-biotin (Pierce; 1.5 mg/ml) in Ringer's was added to the basolateral compartment. Cells were incubated for 30 min at 4°C, before quenching by incubation with 50 mM Tris-HCl (pH 7.5) in Ringer's for 30 min at 4°C. Cells were immediately fixed and permeabilized as described below. AlexaFluor-conjugated streptavidin was added to secondary antibody solution to visualize biotin labeling.
Immunocytochemistry
Cells were washed twice with ice-cold Ringer's and then fixed in 3% paraformaldehyde for 15 min at 4°C. Adenovirus-infected MDCK/Kir2.3-VSV cells were permeabilized/blocked for 1 h at RT with 0.4% saponin and 1.0% BSA in Ringer's solution, and 0.4% saponin was subsequently included in all wash and antibody solutions. For all other experiments, cells were permeabilized in 0.1% Triton X-100 for 30 min at 4°C, washed in ice-cold Ringer's, and blocked for 15 min at RT in 1.0% BSA. Next, cells were incubated overnight at 4°C with the primary antibodies, washed, and then incubated with appropriate AlexaFluor-conjugated secondary antibodies (Molecular Probes). Labeled cells were mounted onto slides with VectaShield glycerol mounting medium (Vector Laboratories, Burlingame, CA) and sealed with nail polish.
Fixation of male 129/SvEv mouse and Sprague Dawley rat kidneys was achieved by arterial perfusion. PBS was perfused 2 min before 30-min perfusion with paraformaldehyde (2%) and a 2-min perfusion with a cryoprotectant (10% EDTA in 0.1 M Tris). Kidney sections (12 μm) were cut with a cryostat, placed on HistoGrip-coated coverslips (Zymed), and stored at −80°C. To perform immunolocalization, sections were first rehydrated with PBS and then treated with 0.5% SDS to unmask protein epitopes. Kidney sections were then washed three times in a high-salt buffer (PBS containing 1% BSA and 385 mM NaCl), blocked (PBS containing 1% BSA and 50 mM glycine), and incubated overnight with primary antibodies (10 μg/μl) at 4°C in PBS supplemented with 0.1% BSA and 0.02% NaN3. Samples were washed in high-salt buffer and incubated with AlexaFluor-488 conjugated goat anti-rabbit secondary antibodies amplified by an additional 2-h incubation with donkey anti-goat Alexa488 before washing and mounting in VectaShield medium.
Imaging Studies
To determine cellular localization of proteins, cells were visualized using the Zeiss 410 confocal laser-scanning microscope (Carl Zeiss, Thornwood, NY) under a 63× oil immersion lens (NA = 1.40). Images were processed with Adobe Photoshop (San Jose, CA).
To assess intracellular expression of Kir 2.3 in stable TIP-1+ MDCK cells and after adenoviral mediated delivery of TIP-1 constructs into MDCK cells stably expressing Kir 2.3, images of cells were scored for intracellular localization of Kir 2.3 (2 = strong, 1 = moderate, 0 = none) by an observer who was blinded to knowledge of the treatment groups. In the adenoviral experiments, cells were colabeled with anti-VSV antibodies to detect Kir 2.3, with streptavidin to mark the basolateral membrane and anti-myc antibodies to detect myc-TIP-1–infected cells. In all cases, only cells that costained positively for TIP-1 protein expression were scored. Scores for equal numbers of cells from three separate experiments were pooled and then averaged together to produce an “Intracellular Localization Score” for each experimental condition.
Statistical Analysis
Statistical significance was calculated by one-way randomized ANOVA followed by Dunnett's post hoc test. p < 0.05 was considered significant.
RESULTS
Identification of an Unusual PDZ Protein, TIP-1, as a Binding Partner of the Kir 2.3 Channel
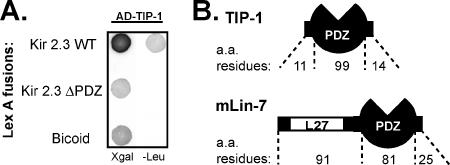
The extreme COOH-terminal tail of the Kir 2.3 channel, containing a basolateral targeting determinant (Le Maout et al., 2001) and a PDZ-binding motif (Olsen et al., 2002), was used as a bait to screen for interacting proteins in a yeast two-hybrid system. Five different human kidney cDNA library clones scored positively for interaction but did not interact with a mutant Kir 2.3 COOH-terminal tail lacking the PDZ-binding motif. Two of these encoded mLin-7 as previously reported (Olsen et al., 2002). The other three clones were overlapping cDNAs, encoding a PDZ protein called Tax-interacting protein-1 (TIP-1; Figure 1).
Figure 1.
Kir 2.3 C-terminus specifically interacts with the novel PDZ protein TIP-1. (A) Yeast were cotransformed with AD-TIP-1 and LexA DNA-binding domain fusions of wild-type (WT) Kir 2.3 C-terminus (top), a mutant Kir 2.3 C-terminus, lacking the last 3 amino acids (ΔPDZ; middle), or bicoid, an unrelated Drosophila protein (bottom). Transfected yeast were diluted and spotted onto selection/reporter plates, X-gal or -Leu. Transcriptional activation of the lacZ reporter produces blue yeast on X-gal plates; Activation of leu2 reporter allows yeast to grow on “leucine drop-out” plates (-Leu). (B) Schematic of the TIP-1 protein compared with mLin-7. Atypical of PDZ proteins, TIP-1 contains a single protein–protein interaction domain.
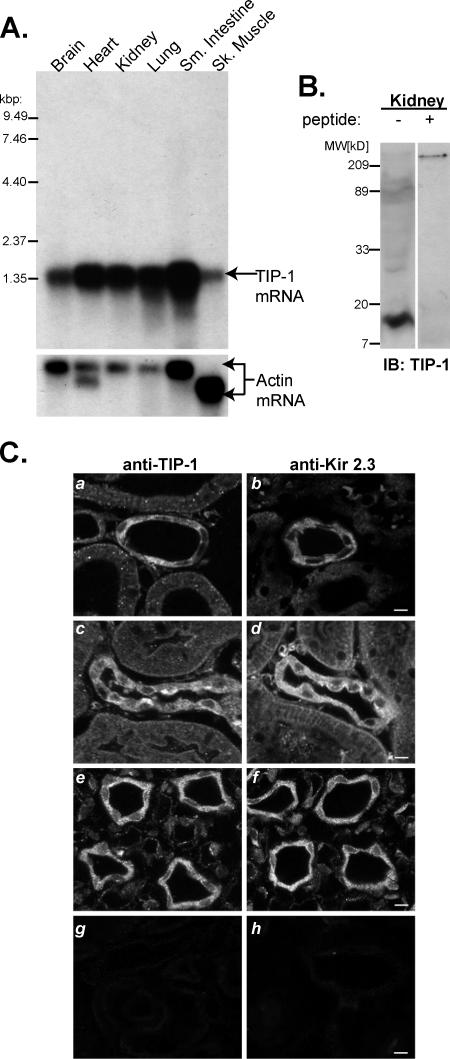
Because the original report of the TIP-1 cDNA sequence did not include the initiation of translation site (Rousset et al., 1998), we first sought to define the complete sequence of the TIP-1 gene product more precisely. In silico analysis of the human genome sequence suggested that the 1.39-kb TIP-1 cDNAs isolated here contains the entire TIP-1 open reading frame. To corroborate these observations, 200 base pairs of the predicted 3′ ends of TIP-1 were independently amplified from human kidney poly (A+) RNA by RT-PCR, subcloned, and sequenced. All sequences were identical and contained a Kozack sequence and upstream missense codon, corroborating 5′-RACE studies (Olalla et al., 2001) and verifying assignment of the open reading frame as presently annotated in the GenBank (NM014604). To further verify that the entire transcript had been isolated, Northern blot analysis of human tissue poly (A+) RNA was performed. As predicted, a single 1.4-kB transcript was detected in all tissues examined (Figure 2). These studies also revealed that TIP-1 is widely expressed but is especially enriched in epithelial tissues such as small intestine, kidney, and lungs.
Figure 2.
TIP-1 expression. (A) Northern blots of poly(A+) RNA from human tissues hybridized with TIP-1 cDNA probes. Actin probe control is shown in the bottom panel. (B) Anti-TIP-1 antibody specifically recognized a 14-kDa band in immunoblots of rat kidney lysate. Excess antigen (+peptide) blocked antibody binding with the kidney 14-kDa protein. (C) Immunolocalization of TIP-1 (a, c, and e) corresponds to sites of Kir 2.3 expression (b, d, and f) in the kidney. Both proteins localize in distal convoluted tubule (a and b), connecting tubule (c and d), and collecting duct (e and f). Controls show that immunolabeling for TIP-1 is blocked by an excess of the immunizing peptide (g) and preimmune serum shows minimal nonspecific Kir 2.3 label (h). Scale bar, 17.5 μm.
Translation of the TIP-1 cDNA sequence predicts a small, 124-amino acid protein, encoding a single PDZ domain flanked by only 11 amino acids at the N-terminus and 14 amino acids at the C-terminus. The residues bordering the PDZ domain are devoid of other known protein–protein interaction motifs or trafficking signals. Antibodies generated against a unique peptide sequence of TIP-1 specifically recognized the TIP-1 protein (Supplementary Data II). In immunoblots of rat kidney, the TIP-1 antibodies exclusively detected the expected sized (14 kDa) protein (Figure 2B), offering experimental verification of the short TIP-1 open reading frame and the first evidence of TIP-1 protein expression in native tissue. Immunohistochemical analysis of rat kidney revealed a predominately cytoplasmic distribution of TIP-1. This localization pattern was most intense in distal segments of the nephron, including distal convoluted tubule, connecting tubule, and collecting duct (Figure 2C, a, c, and e). Importantly, the Kir 2.3 channel is also predominately expressed in these segments (Figure 2C, b, d, and f), offering the opportunity for the two endogenous proteins to engage in a physiologically relevant interaction in vivo. TIP-1 staining at the apical membrane of connecting tubule intercalated cells was evident as well (Figure 2Cc). The cytoplasmic localization of TIP-1 in most of the cell types is consistent with the notion that TIP-1 lacks endogenous targeting information and instead relies on the targeting determinants of its binding partners.
The simple structure of TIP-1 predicts that it would function as a negative regulator of PDZ complexes. Interaction of Kir 2.3 with TIP-1 is predicted to displace the channel from the mLin-7/CASK complex and consequently induce endosomal targeting. Here we test this hypothesis.
Coimmunoprecipitation of TIP-1 and Kir 2.3
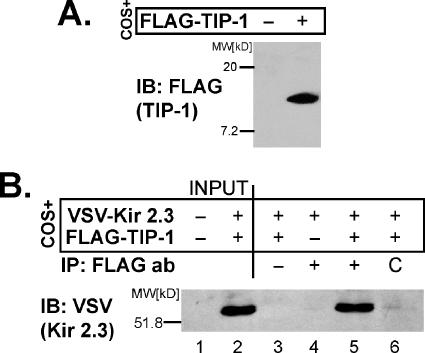
As an initial test of the scaffolding antagonist function, we verified that TIP-1 interacts with Kir 2.3 in mammalian cells using coimmunoprecipitation analysis. For this purpose, the N-termini of Kir 2.3 and TIP-1 were engineered with the VSV G protein epitope and FLAG tag, respectively. Transfection of COS cells with these constructs results in expression of the predicted proteins as detected by immunoblot analysis; anti-FLAG antibodies specifically detected a 16-kDa protein, corresponding to FLAG-tagged TIP-1 (Figure 3A), and anti-VSV antibodies recognize a 59-kDa band, corresponding to VSV-Kir 2.3 (Figure 3B, lane 2). The Kir 2.3 channel could be specifically coimmunoprecipitated with TIP-1 from COS cells cotransfected with VSV-Kir 2.3 and Flag-TIP-1, using anti-FLAG antibodies (Figure 3B, lane 5). As controls, the FLAG antibody failed to immunoprecipitate cells transfected with VSV-Kir 2.3 alone (Figure 3B, lane 4). Immunoprecipitation of the VSV-Kir 2.3 channel with FLAG-TIP-1 also required the FLAG antibody; reactions without antibody (Figure 3B, lane 3) or with a control rabbit antibody (Figure 3B, lane 6) did not immunoprecipitate VSV-Kir 2.3 from COS cells cotransfected with VSV-Kir 2.3 and FLAG-TIP-1. Collectively, these results establish that the full-length TIP-1 is capable of interacting with full-length Kir 2.3 in mammalian cells.
Figure 3.
Kir 2.3 and TIP-1 interact in mammalian cells. COS cells transiently transfected with FLAG-TIP-1– and/or VSV-Kir 2.3–expressed proteins of the predicted size as detected with either anti-FLAG (A) or anti-VSV (B, lane 2) antibodies. (B) COS cells expressing VSV-Kir 2.3 alone or coexpressed with myc-TIP-1 were solubilized, and lysates were immunoprecipitated with anti-FLAG antibodies and immunoblotted with anti-VSV antibody. The FLAG antibody, but not an irrelevant control antibody (C), specifically coimmunoprecipitated Kir 2.3, verifying a bona fide interaction.
Kir 2.3 and TIP-1 Interact in a Type I PDZ-dependent Manner
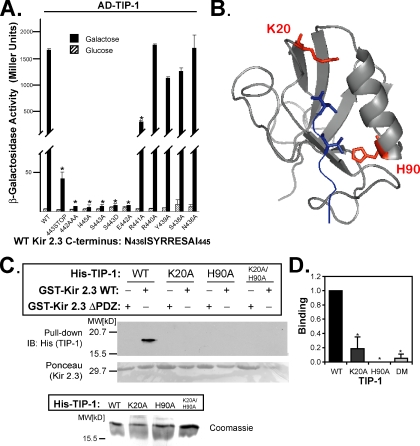
We next explored the structural basis for Kir 2.3/TIP-1 interaction. The extreme C-terminus of Kir 2.3 contains a type I PDZ-binding motif (Olsen et al., 2002) as well as a neighboring or overlapping sorting signal that directs the channel to the basolateral membrane within the biosynthetic pathway (Le Maout et al., 2001). To determine the relative contribution of the upstream sorting domain (requiring amino acids 431–441) and the canonical PDZ-binding motif for TIP-1 interaction, the target sequence in the Kir 2.3 C-terminus for TIP-1 binding was first delineated in the yeast two-hybrid system. Alanine substitution mutations were created in the lexA-Kir 2.3 C-terminus and tested for their ability to interact with the activation domain (AD) tagged-TIP-1 protein. Relative strength of interaction was inferred from reporter activity (β-galactosidase). Counting from the extreme C-terminal amino acid of Kir 2.3, residues at positions 0, -2, and -3 were found to be absolutely required for interaction with TIP-1, indicative of a type I PDZ interaction (Songyang et al., 1997). Alanine substitutions at these sites specifically rendered the channel incompetent to interact with TIP-1 (Figure 4A). Additionally, mutation of the arginine at -4 to alanine reduced the interaction strength by more than 50%. In contrast, point mutations beyond the -4 residue had no significant effect on the interaction, suggesting the sorting signal does not contribute to TIP-1 interaction. Together these observations indicate that TIP-1 may have the capacity to affect localization of the Kir 2.3 channel in a PDZ-dependent manner.
Figure 4.
Kir 2.3 and TIP-1 interact in a PDZ-dependent manner. (A) Interaction of AD-TIP-1 and LexA-Kir 2.3 wild-type (WT) or mutant Kir 2.3 C-termini was inferred from β-galactosidase activity in the yeast two-hybrid system. The GAL1 promoter induces the expression of the AD-tagged TIP-1 in the presence of galactose (■) but not in the presence of glucose ( ). Reporter activation is observed only in the presence of galactose. Mean ± SE; n = 3; * p ≤ 0.01; statistical significance as measured by ANOVA. (B) Three-dimensional structure of the TIP-1-binding pocket as modeled on the crystal structure of the related third PDZ domain of PSD-95 (PDB,1BFE; Doyle et al., 1996). Two residues, K20 and H90, predicted to interact with the PDZ ligand (blue) are shown in red. (C) GST pulldown assays using GST-Kir 2.3 C-terminus wild-type (WT) or a mutant GST-Kir 2.3 C-terminus lacking the PDZ interaction motif (ΔPDZ) and different purified recombinant His-tagged TIP-1 proteins (WT, wild-type TIP-1, and TIP-1-bearing K20A, H90A, or double K20A/H90A mutations). His-TIP-1 proteins specifically bound to the GST-Kir 2.3 proteins were detected by immunoblotting with anti-His antibodies (“pull-down”). In bottom panels, Ponceau S stain of GST-Kir 2.3 input and Coomassie stain of His-TIP-1 input are shown as loading controls. (D) The amount of each TIP-1 construct bound to GST-Kir 2.3 was assessed by densitometry, background subtracted, and analyzed relative to the WT TIP-1 band intensity. The mean ± SE relative densitometry of four pulldown experiments repeated in triplicate (p ≤ 0.01) is shown.
). Reporter activation is observed only in the presence of galactose. Mean ± SE; n = 3; * p ≤ 0.01; statistical significance as measured by ANOVA. (B) Three-dimensional structure of the TIP-1-binding pocket as modeled on the crystal structure of the related third PDZ domain of PSD-95 (PDB,1BFE; Doyle et al., 1996). Two residues, K20 and H90, predicted to interact with the PDZ ligand (blue) are shown in red. (C) GST pulldown assays using GST-Kir 2.3 C-terminus wild-type (WT) or a mutant GST-Kir 2.3 C-terminus lacking the PDZ interaction motif (ΔPDZ) and different purified recombinant His-tagged TIP-1 proteins (WT, wild-type TIP-1, and TIP-1-bearing K20A, H90A, or double K20A/H90A mutations). His-TIP-1 proteins specifically bound to the GST-Kir 2.3 proteins were detected by immunoblotting with anti-His antibodies (“pull-down”). In bottom panels, Ponceau S stain of GST-Kir 2.3 input and Coomassie stain of His-TIP-1 input are shown as loading controls. (D) The amount of each TIP-1 construct bound to GST-Kir 2.3 was assessed by densitometry, background subtracted, and analyzed relative to the WT TIP-1 band intensity. The mean ± SE relative densitometry of four pulldown experiments repeated in triplicate (p ≤ 0.01) is shown.
To further elucidate the mechanism by which TIP-1 interacts with Kir 2.3, we investigated the predicted TIP-1 PDZ-binding domain. By projecting the TIP-1 sequence onto the atomic resolution structure of known type I PDZ domains (Doyle et al., 1996; Figure 4B), we identified residues in TIP-1 that are likely to participate in Kir 2.3 binding. Lysine 20 is predicted to coordinate interaction with the C-terminal carboxyl moiety of type I PDZ ligands (Songyang et al., 1997). Histidine 90, highly conserved among all type I PDZ proteins, is predicted to coordinate the hydroxyl moiety of the penultimate serine residue in type I PDZ ligands (Ser 443 in Kir 2.3). To test these predictions, we substituted K20 and H90 with alanine and studied the consequences in an in vitro binding assay. In these studies, purified, recombinant His-tagged wild-type, mutant K20A, and mutant H90A TIP-1 proteins were tested for their ability to interact with a GST fusion protein of the wild-type Kir 2.3 C-terminus (residues L416-I445). Parallel reactions were performed with a GST fusion protein of a mutated Kir 2.3 C-terminus, lacking the PDZ-binding motif (GST-Kir 2.3ΔPDZ, residues L416-I441), as a negative control. As shown in Figure 4C, wild-type TIP-1 interacted with wild-type GST-Kir 2.3, confirming that the interaction between the two proteins is direct and requires no additional protein mediators (Figure 4C). In agreement with the yeast two-hybrid data, GST-Kir 2.3ΔPDZ failed to bind to wild-type TIP-1 protein, further verifying that interaction requires the PDZ ligand motif in Kir 2.3. As expected for a PDZ domain interaction, binding was significantly abrogated by the single mutations of K20A or H90A or by the double mutation, confirming that this interaction is mediated through the TIP-1 PDZ domain (Figure 4, C and D).
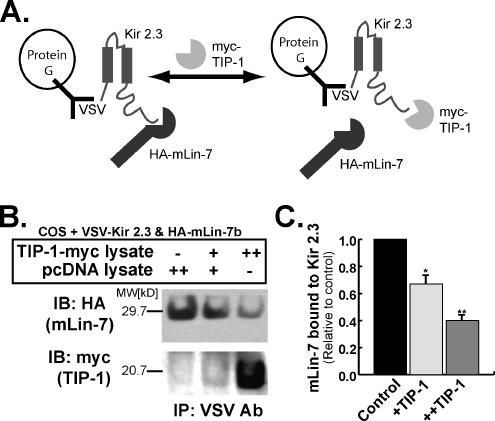
TIP-1 Can Compete with mLin-7 for Kir 2.3 Interaction
Our observations that TIP-1 binds to Kir 2.3 in a PDZ-dependent manner suggest that TIP-1 could potentially compete with the basolateral scaffolding protein, mLin-7, for channel interaction. To test this idea directly, in vitro competition binding assays were performed (Figure 5A). In these studies, Kir 2.3-mLin-7 complexes were recovered on protein G-Sepharose beads with anti-VSV antibodies from COS cells expressing VSV-tagged Kir 2.3 and HA-tagged mLin-7. Beads were then incubated at RT for 3 h with COS lysates containing either myc-tagged TIP-1 or vector (pcDNA). To assess the relative extent of channel interaction with myc-TIP-1 and HA-mLin-7, the Kir 2.3 precipitates were resolved by SDS-PAGE and immunoblotted with either anti-myc or anti-HA antibodies. As shown in Figure 5B and summarized in Figure 5C (n = 3), addition of the TIP-1 lysate reduced the amount of mLin-7 associated with Kir 2.3 in a concentration-dependent manner (Figure 5B, top panel). This was paralleled by an increase in TIP-1 binding to the channel (Figure 5B, bottom panel). Thus, TIP-1 has the capacity to uncouple Kir 2.3 from the mLin-7/CASK protein scaffold.
Figure 5.
TIP-1 competes with mLin-7 for binding to Kir 2.3. (A) Diagram illustrating the procedure for competitive immunoprecipitation experiments as described below and in Materials and Methods. (B) Kir 2.3-mLin-7 complexes were recovered on protein G-Sepharose beads with anti-VSV antibodies from COS cells expressing VSV-tagged Kir 2.3 and HA-tagged mLin-7. Three separate competition reactions were run in parallel. Lysate from COS cells transfected with pcDNA 3.1 was added (pc DNA lysate++) to one sample as a control. Another sample contained an equal volume of COS cell lysate containing TIP-1 (TIP-1-myc++). The other sample contained an equal volume of a 1:1 mix of the TIP-1-myc and pcDNA lysates (TIP-1-myc+/pcDNA+). After incubation at RT, HA-mLin-7, and TIP-1-myc bound to Kir 2.3 was recovered and detected by immunoblot. (C) Amount mLin-7 bound to Kir 2.3 relative to control (pC DNA++) was evaluated by densitometry of experiments shown in B. Mean ± SE; n = 3; p < 0.005.
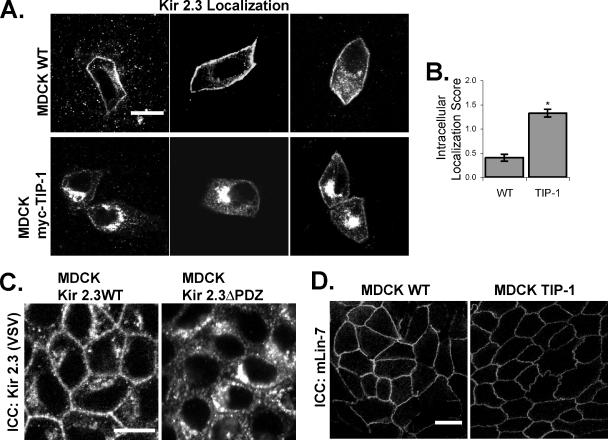
TIP-1 Causes Intracellular Accumulation of Kir 2.3 in MDCK Cells
MDCK cells provide a model system for studying the effects of TIP-1 on channel trafficking because these cells do not express TIP-1; it is not detectable in these cells by RT-PCR or Western blot. To examine the effects of TIP-1 on Kir 2.3 localization, we created a MDCK cell line stably expressing TIP-1 (TIP-1+ MDCK) and compared the steady-state localization of the Kir 2.3 channel in the TIP-1+ MDCK cells to wild-type MDCK cells. Typical examples are shown in Figure 6A, and the results from quantification of 100 cells/group are summarized in Figure 6B. In wild-type MDCK cells (Figure 6A), the transiently transfected channel localizes predominately to the basolateral membrane, identical to the expression pattern in MDCK cells stably transfected with Kir 2.3 as reported previously (Le Maout et al., 1997, 2001; Olsen et al., 2002) and shown in Figure 6C. By contrast, the Kir 2.3 channel becomes localized to a vesicular compartment in the TIP-1+ MDCK cells (78% of TIP-1+ MDCK cells exhibit significant intracellular localization of Kir2.3 compared with 26% of wild-type cells), reminiscent of the mistrafficking phenotype of mutant Kir 2.3 channels, lacking the PDZ ligand motif (Olsen et al., 2002; Figure 6C). General disruption of epithelial integrity or the basolateral mLin-7 scaffold cannot explain the effects of TIP-1; TIP-1+ MDCK monolayers exhibit a transepithelial resistance that is comparable to wild-type monolayers (unpublished data) and mLin-7 properly localizes to the basolateral membrane in the TIP-1+ MDCK cells. Together, these observations are consistent with the hypothesis that TIP-1 specifically uncouples PDZ-binding partners, such as Kir 2.3, from PDZ scaffolding complexes.
Figure 6.
Kir 2.3 is mislocalized to a vesicular compartment in MDCK/TIP-1 cells. (A) Confluent monolayers of wild-type (WT, top panels) or MDCK cells stably transfected with myc-TIP-1 (MDCK myc-TIP-1, bottom panels), were transiently transfected with VSV-Kir 2.3 and stained with anti-VSV antibody. Images were chosen to document the full spectrum of phenotypes observed for each cell type. Scale bar, 10 μm. (B) Cells were scored for intracellular localization of Kir 2.3 (2, strong; 1, moderate; 0, none) by a blinded observer. Data are reported as the average Kir 2.3 intracellular localization score for cells from three separate infections (n = 100; p < 0.001). (C) MDCK cells stably expressing a mutant Kir 2.3 protein lacking the PDZ ligand (ΔPDZ) display a similar mislocalization phenotype as caused by TIP-1 expression. Cells are labeled with anti-VSV antibodies, detecting Kir 2.3. (D) MDCK WT and myc-TIP-1 cells were stained with anti-mLin-7 antibodies for endogenous mLin-7. Scale bar, 20 μm.
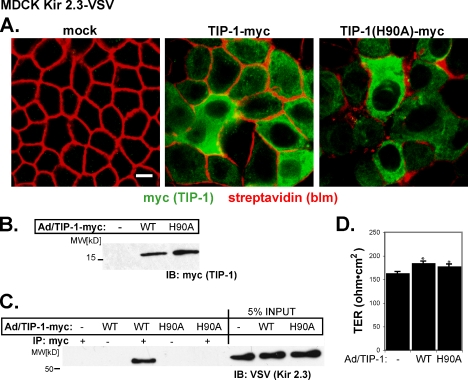
The scaffolding antagonist hypothesis predicts that the activity of TIP-1 should be carried by its PDZ-binding capacity. To test this idea directly, we compared steady-state channel distribution in cells expressing the PDZ-binding–incompetent mutant TIP-1(H90A) to those expressing wild-type TIP-1. In these studies, adenoviral-mediated cDNA transfer was used to deliver wild-type myc-tagged TIP-1 or TIP-1(H90A) to fully polarized MDCK cells, which stably express the VSV-tagged Kir 2.3 channel. The approach allows reliable, high-efficiency delivery of the TIP-1 constructs into Kir 2.3–positive MDCK monolayers on permeable supports (Figure 7). Indeed, we observed consistently that more than 70% of cells stain positively for myc-tagged TIP-1 within a day of infection (Figure 7A). Typical for adenovirus delivery systems, expression levels of TIP-1 were somewhat heterogeneous from cell to cell, yet as assessed by immunoblot analysis (Figure 7B), similar amounts of wild-type and H90A TIP-1 protein were expressed at the expected molecular weight (16 kDa; Figure 7B), allowing a reasonable comparison of the effects of the two TIP-1 constructs.
Figure 7.
TIP-1 is efficiently expressed in MDCK Kir 2.3-VSV cells by an adenoviral delivery system. Confluent, filter-grown MDCK cells stably expressing Kir 2.3-VSV were infected with wild-type (WT) or H90A TIP-1-myc adenoviruses. (A) After infection with TIP-1 adenoviruses, MDCK Kir2.3-VSV cells were decorated with biotin on the basolateral membrane and then costained with streptavidin (red) to mark the basolateral membrane and anti-myc antibodies (green) to visualize TIP-1. Scale bar, 10 μm. (B) Lysates from these cells were immunoblotted with anti-myc antibodies to detect WT and H90A TIP-1. (C) TIP-1 was immunoprecipitated with anti-myc antibodies and then immunoblotted with anti-VSV antibodies for Kir 2.3-VSV. (D) Transepithelial resistance (TER) of infected cells. Neither WT nor H90A viruses reduce the TER. Instead infection of either virus causes a small, reproducible increase in TER (n = 4; p ≤ 0.01). Mean ± SE.
Similar to the endogenous protein, the myc-tagged wild-type TIP-1 and the H90A TIP-1 mutant localize diffusely within cytoplasm (Figure 7A). Immunoprecipitation studies revealed that wild-type TIP-1 but not TIP-1(H90A) interacts with Kir 2.3, confirming that the H90A point mutation attenuates interaction of TIP-1 with the channel in MDCK cells. Importantly, adenoviral infection did not alter Kir 2.3 protein abundance (Figure 7C, input) nor did it disrupt the transepithelial monolayer resistance (Figure 7D).
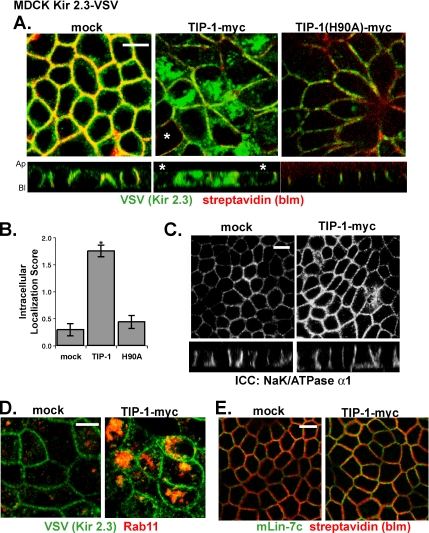
Figure 8A shows the effects of wild-type TIP-1 and the H90A mutant TIP-1 on Kir 2.3 channel localization in representative MDCK monolayers stably expressing the channel. Quantitative evaluations of three separate infections are summarized in Figure 8B. Expression of the wild-type TIP-1 caused Kir 2.3 to distribute into an intracellular vesicular compartment (Figure 8A, middle panel). By contrast, the TIP-1(H90A) mutant protein did not produce a significant change in channel localization (Figure 8A, right panel). The extent and percentage (25% of mock, 96% of TIP-1–, and 39% of H90A TIP-1–expressing cells exhibit significant intracellular localization) of wild-type TIP-1–expressing cells exhibiting intracellular localization of Kir 2.3 in these studies is remarkably similar to the results with the stable TIP-1+ MDCK cells.
Figure 8.
Adenoviral-mediated expression of TIP-1 in MDCK Kir2.3-VSV cells redistributes the channel to a vesicular compartment. (A) Shown are confluent monolayers of MDCK-Kir2.3 VSV cells grown on permeable supports after mock infection or infection with TIP-1-myc adenoviruses. Cells are colabeled with anti-VSV antibodies to detect Kir 2.3 (green) and with streptavidin to mark the basolateral membrane (red). All cells in the middle and right panels express TIP-1 except those identified with the asterisk (∗). (B) Z-plane images of cells were scored for intracellular localization of Kir 2.3 (2, strong; 1, moderate; 0, none) by a blinded observer. For both TIP-1 groups, only cells that were positively identified as expressing myc-TIP-1 by anti-myc labeling were scored. Data are reported as average Kir 2.3 intracellular localization score for cells from three separate infections. Expression of wild-type TIP-1 but not H90A TIP-1 resulted in a statistically significant increase in intracellular Kir 2.3. Mean ± SE; n = 24; p < 0.01. (C) Cells stained with anti-NaK/ATPase α1 antibodies. TIP-1 expression did not effect the localization of this endogenous basolateral marker. (D) Cells labeled with anti-VSV antibodies to detect Kir 2.3 (green) and anti-Rab11 antibodies (red) in mock and wild-type TIP-1–infected MDCK Kir 2.3-VSV cells. (E) Cells stained with anti-mLin-7c antibodies (green) and with streptavidin to mark the basolateral membrane (red). Scale bar, (A, C, D, and E) 10 μm.
The scaffold antagonist model also predicts that TIP-1 should induce mislocalization of Kir 2.3 to an endocytic compartment without altering basolateral membrane localization of Lin-7. Adenoviral-mediated expression of the wild-type TIP-1 in MDCK cells grown on filter supports caused Kir 2.3 to distribute to an intracellular, vesicular compartment that was somewhat more diffuse than observed in the stable TIP-1 cells grown on glass, presumably reflecting different experimental conditions. Nonetheless, Kir 2.3 largely accumulated into an expanded Rab11 vesicular compartment (Figure 8D), consistent with targeting to recycling and subapical endosomes (Casanova et al., 1999; Sheff et al., 1999; Wang et al., 2000). As observed in the TIP-1+ MDCK cells, adenoviral-mediated expression of TIP-1 did not change the basolateral localization of either endogenous mLin-7 protein (Figure 8E) or the Na/K-ATPase-α1 (Figure 8C), demonstrating that TIP-1 does not cause a global disruption of basolateral membrane traffic. Together, these observations indicate that TIP-1 alters steady-state localization of Kir 2.3 in a PDZ-dependent manner, consistent with a PDZ scaffolding antagonist.
TIP-1 Uncouples Kir 2.3 from the Basolateral mLin-7/CASK Complex
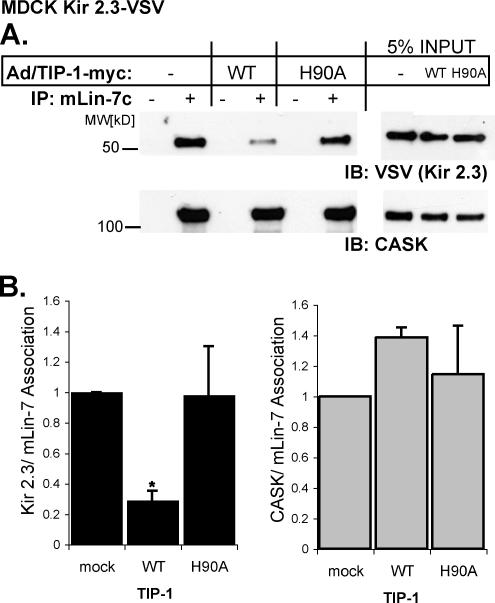
To test whether TIP-1 induces mistrafficking of Kir 2.3 by uncoupling the channel from the mLin-7 scaffolding protein, we measured the extent of mLin-7/Kir 2.3 interaction in Kir 2.3VSV+MDCK cells after adenoviral-mediated delivery of TIP-1. The effects of wild-type TIP-1 and the H90A TIP-1 mutant were compared. In these studies, endogenous mLin-7 was immunoprecipitated with an anti-mLin-7 antibody and the amount of coimmunoprecipitated Kir 2.3 and CASK were assessed by immunoblotting. As shown in Figure 9A, infection with wild-type TIP-1, but not the H90A mutant, reduced the amount of mLin-7 associated with Kir 2.3 without affecting mLin-7/CASK association. Densitometric measurements of immunoblots from three separate experiments verified that the effect is consistent and statistically significant (Figure 9B). These data provide strong evidence that TIP-1 can prevent association of Kir 2.3 with mLin-7/CASK without altering the integrity of the basolateral membrane scaffold complex.
Figure 9.
TIP-1 expression reduces amount of Kir 2.3 associated with the mLin-7/CASK complex. (A) Confluent MDCK Kir 2.3-VSV cells were infected with wild-type (WT) or H90A TIP-1-myc adenoviruses and compared with mock-infected cells (Ad/TIP-1-myc−). Lysates were immunoprecipitated with anti-mLin-7c antibodies (+) or no-antibody (−) and then immunoblotted with anti-VSV antibodies for Kir 2.3-VSV (above) or anti-CASK antibodies (below). (B) Mean ± SE densitometry (immunoprecipitated protein/input) from three separate infections; n = 3; p ≤ 0.05.
DISCUSSION
In the present study, we identified TIP-1 as a highly unusual PDZ protein that negatively modulates the localization of a binding target, the Kir 2.3 channel, on the renal epithelial cell basolateral membrane. Our results provide support for a novel mechanism whereby the small PDZ protein exhibits PDZ scaffolding antagonist activity. We found that TIP-1 directly associates with the PDZ-binding motif in Kir 2.3 and competes for interaction with mLin-7. Consequently, TIP-1 interaction prevents the channel from efficiently engaging the basolateral PDZ protein scaffold complex and, thereby, disrupts cell surface localization. Indeed, either stable expression of TIP-1 or acute adenoviral-mediated TIP-1 cDNA transfer into MDCK cells caused Kir 2.3 to accumulate in an endosomal compartment, reminiscent of the mistrafficking phenotype exhibited by mutant Kir 2.3 channels lacking their PDZ-binding motif (Olsen et al., 2002). A single point mutation in the PDZ-binding pocket of TIP-1 that reduced Kir 2.3 binding affinity was sufficient to attenuate the endosomal trafficking phenotype, indicating that the effects of wild-type TIP-1 are specific and PDZ-dependent.
Our study suggests that TIP-1 may normally operate as a natural “dominant negative” PDZ protein. This would represent the first example of a PDZ protein antagonist and expand an emerging biological paradigm of naturally “fractured” (Kroiher et al., 2001) or truncated molecules that function in a wide variety of biological processes as negative modulators. For instance, transcription by basic helix-loop-helix (bHLH) transcription factors are negatively regulated by “broken” mimics of themselves that contain a bHLH domain but lack a DNA-binding domain necessary for transcriptional activity (Yokota and Mori, 2002). Similarly, decoy receptors of the tumor necrosis factor receptor superfamily contain the extracellular ligand-binding domain but lack intracellular domains required for effective signaling (Khosla, 2001; Mantovani et al., 2004). Similar examples of “natural dominant negatives” can be found among structural proteins; the neuronal gene product, Homer1a, provides an elegant example. Proper signaling by type I metabotropic glutamate receptors (mGluRs) requires association with self-multimerizing Homer proteins (Xiao et al., 1998; Ango et al., 2001). Homer1a lacks a conserved coiled-coil domain necessary for the multimerization process, and thus up-regulation of endogenous Homer1a by excitotoxic brain injury results in decreased association of other Homer proteins with mGluRs and disregulated signaling by the receptor (Ango et al., 2001). Each of the negative modulators described above lacks a structural module critical to the normal functioning of typical proteins within the family. TIP-1 is no exception; it is composed almost entirely of a single PDZ domain and lacks any other obvious protein–protein interaction modules.
Unlike TIP-1, typical PDZ proteins contain multiple protein–protein interaction modules, allowing them to act as macromolecular scaffolds (Kim and Sheng, 2004; Brone and Eggermont, 2005). The basolateral scaffolding function of mLin-7, for example, is made possible by the presence of its two different protein–protein interaction modules. mLin-7 interacts with target proteins, such as Kir 2.3, via a PDZ interaction (Olsen et al., 2002), while simultaneously binding to CASK via a heterotypic L27 domain interaction (Borg et al., 1998; Straight et al., 2000). CASK, in turn, associates with the basolateral membrane through a web of interactions, forming a multimeric complex that has the capacity to act as a stable basolateral membrane anchor. Indeed, multiple protein–protein interactions sites allow CASK to simultaneously bind to mLin-7, extracellular matrix receptors, adhesion molecules, and the actin cytoskeleton (Cohen et al., 1998). SAP97, another Kir 2.3 PDZ-binding partner, also interacts with CASK (Leonoudakis et al., 2001) through an independent L27 domain interaction (Karnak et al., 2002; Lee et al., 2002; Roh et al., 2002). The mLin-7 and SAP97 L27 domains separately assemble with the two L27 domains of CASK, possibly as a dimer of L27 heterodimers (Feng et al., 2004; Li et al., 2004), to form a mLin-7/SAP97/CASK complex (Leonoudakis et al., 2004b). Our studies indicate that TIP-1 disrupts Kir 2.3 interaction with mLin-7 without altering the mLin-7/CASK complex.
Dominant negative mLin-7 mutants, lacking their L27 domain (mLin-7-ΔL27), also behave like TIP-1. Removal of the L27 domain in mLin-7 disrupts CASK association, causing a cytoplasmic localization pattern that is identical to TIP-1 (Straight et al., 2000). Similar effects have been observed with SAP97 mutants, lacking their L27 domain (Roh et al., 2002). Shelly et al. (2003) showed that expression of mLin-7-ΔL27 mutants alters basolateral membrane targeting of a mLin-7 PDZ-binding partner, ErbB-2, in MDCK cells, reminiscent of the effects observed here with TIP-1 on Kir 2.3. Similarly, expression of a dominant negative CASK construct that contains both L27 domains but no basolateral membrane localization determinants causes a related mLin-7 binding partner, Kir 2.2, to mislocalize into a vesicular compartment in MDCK cells (Leonoudakis et al., 2004a, 2004b). Without L27 domains, neither mLin-7 nor SAP97 can bridge their PDZ-binding target proteins to CASK and the basolateral membrane. TIP-1 only contains a PDZ-binding domain, and consequently it produces similar mistargetting defects as engineered dominant-interfering PDZ constructs.
TIP-1 appears to be well suited for its role as a natural dominant-negative PDZ protein. First, we found that TIP-1 is constitutively expressed in many tissues and is especially enriched in epithelia, suggesting that there is sufficient copy number to compete with PDZ-based scaffolding proteins, such as mLin-7. In principal cells of the renal collecting duct TIP-1 is expressed as a cytoplasmic protein, consistent with it acting as a physiologically relevant PDZ scaffold antagonist in the epithelial cells where Kir 2.3 is found. Second, we have demonstrated by competitive coimmunoprecipitation that TIP-1 can successfully vie with mLin-7 for access to the C-terminal PDZ ligand of Kir 2.3, suggesting that TIP-1 has adequate binding affinity for the substrate to compete successfully with PDZ-protein scaffolds. It is left to future studies to determine whether TIP-1 abundance or binding affinity is physiologically regulated.
The subcellular locale where TIP-1 exerts its dominant negative effects also remains to be precisely identified. Although it is well established that the mLin-7/CASK complex stabilizes mLin-7 target proteins at the basolateral membrane by limiting endocytic traffic (Perego et al., 1999; Straight et al., 2001; Olsen et al., 2002; Shelly et al., 2003), it is still uncertain if this function is carried out at the plasmalemma or within endosomes or both. Consequently, TIP-1 could disrupt any of basolateral membrane stabilization mechanisms that that have been attributed to mLin-7, including altering plasmalemma retention (Perego et al., 1999) or active recycling in the postendocytic pathway (Straight et al., 2001). Previous studies demonstrated that unrelated PDZ scaffolding proteins, NHERF (Cao et al., 1999) and syntenin (Zimmermann et al., 2005), influence surface stability of their binding targets by augmenting endocytic recycling. Importantly, the ability of these PDZ proteins to affect endocytic turnover depends on the presence of additional interaction modules that link the PDZ protein/target complex to cellular machinery or signaling molecules. For instance, expression of a syntenin mutant, lacking its PIP2-interaction site, causes syndecan, a type II PDZ-binding target of syntenin, to accumulate within engorged recycling endosomes (Zimmermann et al., 2005), similar to the effect of TIP-1 expression on Kir 2.3 localization.
TIP-1 is also poised to disrupt PDZ association with the dystrophin-associated protein complex (DPC), the other major basolateral membrane PDZ protein scaffold. The complex consists of dystroglycans, β-dystrobrevin and utrophin, and the PDZ protein, β2-syntrophin (Kachinsky et al., 1999). Syntrophin protein contains a type I PDZ-binding domain and additional domains that are required for utrophin binding. According to Kachinsky et al., steady state localization of β2-syntrophin at the basolateral membrane is dependent on utrophin binding. Because utrophin can simultaneously associate indirectly with laminin in the extracellular matrix via the dystroglycans (James et al., 2000) and can bind directly to the actin cytoskeleton (Keep et al., 1999), it is poised to provide a secure anchoring point for syntrophin at the basolateral membrane. Importantly, the association of syntrophin with utrophin and the basolateral membrane occurs independently of the syntrophin PDZ domain (Kachinsky et al., 1999). This leaves the PDZ domain free to interact with other cell components containing a type I PDZ ligand motif, such as Kir 2 channel proteins (Leonoudakis et al., 2004a). Analysis of currently known TIP-1–binding partners suggests a modified type I PDZ consensus binding motif of R(D/E)(S/T)X(V/L/I)-COOH, matching the preferred ligands for the syntrophin PDZ domain (Wiedemann et al., 2004). Thus, TIP-1 should disrupt association of type I PDZ ligands, such as Kir 2.3, to the syntrophin/DPC complex, similar to what we have observed with the mLin-7/CASK scaffold.
Our observations are most consistent with TIP-1 directly uncoupling basolateral PDZ scaffolding protein targets, such as Kir 2.3, in collecting duct principal cells, yet, it is presently unknown if TIP-1 may have different functions in other renal epithelial cell types or other tissues. For example, our observation that TIP-1 localizes to both the cytoplasm and the apical membrane in intercalated cells of the renal cortical collecting duct suggests that TIP-1 may have additional activities or even different mechanisms of negative regulation. Considering that formation of PDZ protein scaffolding networks, such as the apical membrane-associated NHERF1 protein complex (Fouassier et al., 2000), can depend on PDZ–PDZ domain interactions, it will be important to determine if TIP-1 might also negatively regulate PDZ scaffolds by binding to and blocking PDZ–PDZ domain interactions.
It also remains unclear how many other additional type I PDZ functions could be negatively controlled by TIP-1. Only five other TIP-1–binding partners have been described: the Rho-activator rhotekin (Reynaud et al., 2000), glutaminase (Olalla et al., 2001), β-catenin (Kanamori et al., 2003), and the viral oncoproteins HTLV-1 Tax (Rousset et al., 1998) and HPV16 E6 (Hampson et al., 2004). Significantly, two of them, β-catenin (Perego et al., 2000; Kanamori et al., 2003) and the HTLV-1 oncoprotein Tax (Rousset et al., 1998), are also mLin-7–binding partners. At present, in is not know if TIP-1 actually antagonizes functions of these target proteins that ordinarily depend on mLin-7 interaction. In fact, the observation that TIP-1 suppresses the transcriptional activity of the cytoplasmic pool of β-catenin (Kanamori et al., 2003) suggests that TIP-1 has negative regulatory activities beyond uncoupling scaffold complexes. On the basis of this previous report and our own observations of TIP-1 as a natural PDZ scaffold antagonist, we hypothesize that TIP-1 not only functions to prevent β-catenin from interacting with mLin-7, but that TIP-1 caps the C-terminal tail of β-catenin so as to prevent formation of the C-terminal fold-back conformation that is important for nuclear signaling (Gottardi and Gumbeiner, 2004). Our model would offer an explanation for the transcriptionally inactive pool of cytoplasmic β-catenin that is also unable to associate with mLin-7/cadherin adhesive complexes (Gottardi et al., 2001). Obviously, further studies will be required to determine if TIP-1 does function as a dual-inhibitory β-catenin–sequestering factor.
In summary, TIP-1 represents a departure from classical PDZ protein structure because it contains only a single protein–protein interaction motif and appears to lack endogenous membrane–targeting information. Binding of TIP-1 antagonizes the surface membrane stabilization provided by association of a target protein with the basolateral membrane PDZ scaffolds and represents a novel means of regulating the surface density of membrane proteins containing a type I PDZ ligand motif.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Institutes of Health, National Institutes of Diabetes, Digestive and Kidney Diseases Grants DK-54231 and DK-63049 (P.A.W.) and Grant DK-32839 (J.B.W.).
Abbreviations used:
- bHLH
basis helix-loop-helix
- DPBS-M
Dulbecco's phosphate-buffered saline with 1 mM MgCl2
- DPC
dystrophin-associated protein complex
- mGluR
metabotropic glutamate receptor
- mLin-7
mammalian Lin-7
- TIP-1
Tax Interacting Protein-1.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-02-0129) on July 19, 2006.
REFERENCES
- Ango F., Prezeau L., Muller T., Tu J. C., Xiao B., Worley P. F., Pin J. P., Bockaert J., Fagni L. Agonist-independent activation of metabotropic glutamate receptors by the intracellular protein Homer. Nature. 2001;411:962–965. doi: 10.1038/35082096. [DOI] [PubMed] [Google Scholar]
- Borg J. P., Straight S. W., Kaech S. M., de Taddeo-Borg M., Kroon D. E., Karnak D., Turner R. S., Kim S. K., Margolis B. Identification of an evolutionarily conserved heterotrimeric protein complex involved in protein targeting. J. Biol. Chem. 1998a;273:31633–31636. doi: 10.1074/jbc.273.48.31633. [DOI] [PubMed] [Google Scholar]
- Brone B., Eggermont J. PDZ proteins retain and regulate membrane transporters in polarized epithelial cell membranes. Am. J. Physiol. Cell Physiol. 2005;288:C20–C29. doi: 10.1152/ajpcell.00368.2004. [DOI] [PubMed] [Google Scholar]
- Butz S., Okamoto M., Sudhof T. C. A. tripartite protein complex with the potential to couple synaptic vesicle exocytosis to cell adhesion in brain. Cell. 1998;94:773–782. doi: 10.1016/s0092-8674(00)81736-5. [DOI] [PubMed] [Google Scholar]
- Cao T. T., Deacon H. W., Reczek D., Bretscher A., von Zastrow M. A kinase-regulated PDZ-domain interaction controls endocytic sorting of the β2-adrenergic receptor. Nature. 1999;401:286–290. doi: 10.1038/45816. [DOI] [PubMed] [Google Scholar]
- Campo C., Mason A., Maouyo D., Olsen O., Yoo D., Welling P. A. Molecular mechanisms of membrane polarity in renal epithelial cells. Rev. Physiol. Biochem. Pharmacol. 2005;153:47–99. doi: 10.1007/s10254-004-0037-1. [DOI] [PubMed] [Google Scholar]
- Casanova J. E., Wang X., Kumar R., Bhartur S. G., Navarre J., Woodrum J. E., Altschuler Y., Ray G. S., Goldenring J. R. Association of Rab25 and Rab11a with the apical recycling system of polarized Madin-Darby canine kidney cells. Mol. Biol. Cell. 1999;10:47–61. doi: 10.1091/mbc.10.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A. R., Woods D. F., Marfatia S. M., Walther Z., Chishti A. H., Anderson J. M., Wood D. F. W. Human CASK/LIN-2 binds syndecan-2 and protein 4.1 and localizes to the basolateral membrane of epithelial cells. J. Cell Biol. 1998;142:129–138. doi: 10.1083/jcb.142.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerks T., Bork P., Kamberov E., Makarova O., Muecke S., Margolis B. L27, a novel heterodimerization domain in receptor targeting proteins Lin-2 and Lin-7. Trends Biochem. Sci. 2000;25:317–338. doi: 10.1016/s0968-0004(00)01599-1. [DOI] [PubMed] [Google Scholar]
- Doyle D. A., Lee A., Lewis J., Kim E., Sheng M., MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- Fouassier L., Yun C. C., Fitz J. G., Doctor R. B. Evidence for ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) self-association through PDZ-PDZ interactions. J. Biol. Chem. 2000;275:25039–25045. doi: 10.1074/jbc.C000092200. [DOI] [PubMed] [Google Scholar]
- Feng W., Long J. F., Fan J. S., Suetake T., Zhang M. The tetrameric L27 domain complex as an organization platform for supramolecular assemblies. Nat. Struct. Mol. Biol. 2004;11:475–480. doi: 10.1038/nsmb751. [DOI] [PubMed] [Google Scholar]
- Golemis E. A., Khazak V. Alternative yeast two-hybrid systems. The interaction trap and interaction mating. Methods Mol. Biol. 1997;63:197–218. doi: 10.1385/0-89603-481-X:197. [DOI] [PubMed] [Google Scholar]
- Gottardi C. J., Wong E., Gumbiner B. M. E-cadherin suppresses cellular transformation by inhibiting beta-catenin signaling in an adhesion-independent manner. J. Cell Biol. 2001;153(5):1049–1060. doi: 10.1083/jcb.153.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottardi C. J., Gumbiner B. M. Distinct molecular forms of beta-catenin are targeted to adhesive or transcriptional complexes. J. Cell Biol. 2004;167(2):339–349. doi: 10.1083/jcb.200402153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyuris J., Golemis E. A., Chertkov H., Brent R. Cdi, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- Hampson L., Li C., Oliver A. W., Kitchener H. C., Hampson I. N. The PDZ protein Tip-1 is a gain of function target of the HPV16 E6 oncoprotein. J. Oncol. 2004;25:1249–1256. [PubMed] [Google Scholar]
- Hata Y., Slaughter C. A., Sudhof T. C. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature. 1993;366:347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- Henkel J. R., Apodaca G., Altschuler Y., Hardy S., Weisz O. A. Selective perturbation of apical membrane traffic by expression of influenza M2, an acid-activated ion channel, in polarized madin-darby canine kidney cells. Mol. Biol. Cell. 1998;9:2477–2490. doi: 10.1091/mbc.9.9.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James M., Nuttall A., Ilsley J. L., Ottersbach K., Tinsley J. M., Sudol M., Winder S. J. Adhesion-dependent tyrosine phosphorylation of (beta)-dystroglycan regulates its interaction with utrophin. J. Cell Sci. 2000;113(Pt 10):1717–1726. doi: 10.1242/jcs.113.10.1717. [DOI] [PubMed] [Google Scholar]
- Kachinsky A. M., Froehner S. C., Milgram S. L. A PDZ-containing scaffold related to the dystrophin complex at the basolateral membrane of epithelial cells. J. Cell Biol. 1999;145:391–402. doi: 10.1083/jcb.145.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S. M., Whitfield C. W., Kim S. K. The LIN-2/LIN-7/LIN-10 complex mediates basolateral membrane localization of the C. elegans EGF receptor LET-23 in vulval epithelial cells. Cell. 1998;94:761–771. doi: 10.1016/s0092-8674(00)81735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori M., Sandy P., Marzinotto S., Benetti R., Kai C., Hayashizaki Y., Schneider C., Suzuki H. The PDZ protein tax-interacting protein-1 inhibits beta-catenin transcriptional activity and growth of colorectal cancer cells. J. Biol. Chem. 2003;278:38758–38764. doi: 10.1074/jbc.M306324200. [DOI] [PubMed] [Google Scholar]
- Karnak D., Lee S., Margolis B. Identification of multiple binding partners for the amino-terminal domain of synapse-associated protein 97. J. Biol. Chem. 2002;277:46730–46735. doi: 10.1074/jbc.M208781200. [DOI] [PubMed] [Google Scholar]
- Keep N. H., Winder S. J., Moores C. A., Walke S., Norwood F. L., Kendrick-Jones J. Crystal structure of the actin-binding region of utrophin reveals a head-to-tail dimer. Structure Fold Des. 1999;7:1539–1546. doi: 10.1016/s0969-2126(00)88344-6. [DOI] [PubMed] [Google Scholar]
- Khosla S. Minireview: the OPG/RANKL/RANK system. Endocrinology. 2001;142:5050–5055. doi: 10.1210/endo.142.12.8536. [DOI] [PubMed] [Google Scholar]
- Kim E., Sheng M. PDZ domain proteins of synapses. Nat. Rev. Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Kroiher M., Miller M. A., Steele R. E. Deceiving appearances: signaling by “dead” and “fractured” receptor protein-tyrosine kinases. Bioessays. 2001;23:69–76. doi: 10.1002/1521-1878(200101)23:1<69::AID-BIES1009>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Le Maout S., Brejon M., Merot J., Welling P. A. Polarized expression of an epitope tagged CCD Kir Channel, CCD-IRK3, in MDCK cells. Proc. Natl. Acad. Sci. USA. 1997;94:13329–13334. doi: 10.1073/pnas.94.24.13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Maout S., Welling P. A., Brejon M., Olsen O., Merot J. Basolateral membrane expression of a K+ channel, Kir 2.3, is directed by a cytoplasmic COOH-terminal domain. Proc. Natl. Acad. Sci. USA. 2001;98:10475–10480. doi: 10.1073/pnas.181481098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Fan S., Makarova O., Straight S., Margolis B. A novel and conserved protein-protein interaction domain of mammalian Lin-2/CASK binds and recruits SAP97 to the lateral surface of epithelia. Mol. Cell. Biol. 2002;22:1778–1791. doi: 10.1128/MCB.22.6.1778-1791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonoudakis D., Conti L. R., Anderson S., Radeke C. M., McGuire L. M., Adams M. E., Froehner S. C., Yates J. R., 3rd, Vandenberg C. A. Protein trafficking and anchoring complexes revealed by proteomic analysis of inward rectifier potassium channel (Kir2.x)-associated proteins. J. Biol. Chem. 2004a;279:22331–22346. doi: 10.1074/jbc.M400285200. [DOI] [PubMed] [Google Scholar]
- Leonoudakis D., Conti L. R., Radeke C. M., McGuire L. M., Vandenberg C. A. A multiprotein trafficking complex composed of SAP97, CASK, Veli, and Mint1 is associated with inward rectifier Kir2 potassium channels. J. Biol. Chem. 2004b;279:19051–19063. doi: 10.1074/jbc.M400284200. [DOI] [PubMed] [Google Scholar]
- Leonoudakis D., Mailliard W., Wingerd K., Clegg D., Vandenberg C. Inward rectifier potassium channel Kir2.2 is associated with synapse-associated protein SAP97. J. Cell Sci. 2001;114:987–998. doi: 10.1242/jcs.114.5.987. [DOI] [PubMed] [Google Scholar]
- Li Y., Karnak D., Demeler B., Margolis B., Lavie A. Structural basis for L27 domain-mediated assembly of signaling and cell polarity complexes. EMBO J. 2004;23:2723–2733. doi: 10.1038/sj.emboj.7600294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A., Locati M., Polentarutti N., Vecchi A., Garlanda C. Extracellular and intracellular decoys in the tuning of inflammatory cytokines and Toll-like receptors: the new entry TIR8/SIGIRR. J. Leukoc. Biol. 2004;75:738–742. doi: 10.1189/jlb.1003473. [DOI] [PubMed] [Google Scholar]
- Mendelsohn A. R., Brent R. Applications of interaction traps/two-hybrid systems to biotechnology research. Curr. Opin. Biotechnol. 1994;5:482–486. doi: 10.1016/0958-1669(94)90061-2. [DOI] [PubMed] [Google Scholar]
- Olalla L., Aledo J. C., Bannenberg G., Marquez J. The C-terminus of human glutaminase L mediates association with PDZ domain-containing proteins. FEBS Lett. 2001;488:116–122. doi: 10.1016/s0014-5793(00)02373-5. [DOI] [PubMed] [Google Scholar]
- Olsen O., Liu H., Wade J. B., Merot J., Welling P. A. Basolateral membrane expression of the Kir 2.3 channel is coordinated by PDZ interaction with Lin-7/CASK complex. Am. J. Physiol. Cell Physiol. 2002;282:C183–C195. doi: 10.1152/ajpcell.00249.2001. [DOI] [PubMed] [Google Scholar]
- Olsen O., Wade J. B., Morin N., Bredt D. S., Welling P. A. Differential localization of mammalianLin-7 (MALS/Veli) PDZ proteins in the kidney. Am. J. Physiol. Renal Physiol. 2005;288:F345–F352. doi: 10.1152/ajprenal.00235.2004. [DOI] [PubMed] [Google Scholar]
- Perego C., Vanoni C., Massari S., Longhi R., Pietrini G. Mammalian LIN-7 PDZ proteins associate with beta-catenin at the cell-cell junctions of epithelia and neurons. EMBO J. 2000;19:3978–3989. doi: 10.1093/emboj/19.15.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego C., Vanoni C., Villa A., Longhi R., Kaech S. M., Frohli E., Hajnal A., Kim S. K., Pietrini G. PDZ-mediated interactions retain the epithelial GABA transporter on the basolateral surface of polarized epithelial cells. EMBO J. 1999;18:2384–2393. doi: 10.1093/emboj/18.9.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud C., Fabre S., Jalinot P. The PDZ protein TIP-1 interacts with the Rho effector rhotekin and is involved in Rho signaling to the serum response element. J. Biol. Chem. 2000;275:33962–33968. doi: 10.1074/jbc.M000465200. [DOI] [PubMed] [Google Scholar]
- Reynolds A., Lundblad V., Dorris D., Keaveney M. Yeast vectors and assays for expression of cloned genes. In: Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K., editors. Current Protocols in Molecular Biology. Boston: John Wiley & Sons; 1997. 13.6.1–13.6.6. [DOI] [PubMed] [Google Scholar]
- Roh M. H., Makarova O., Liu C. J., Shin K., Lee S., Laurinec S., Goyal M., Wiggins R., Margolis B. The Maguk protein, Pals1, functions as an adapter, linking mammalian homologues of Crumbs and Discs Lost. J. Cell Biol. 2002;157:161–172. doi: 10.1083/jcb.200109010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongo C., Whitfield C. W., Rodal A., Kim S. K., Kaplan J. M. LIN-10 is a shared component of the polarized protein localization pathways in neurons and epithelia. Cell. 1998;94:751–759. doi: 10.1016/s0092-8674(00)81734-1. [DOI] [PubMed] [Google Scholar]
- Rousset R., Fabre S., Desbois C., Bantignies F., Jalinot P. The C-terminus of the HTLV-1 Tax oncoprotein mediates interaction with the PDZ domain of cellular proteins. Oncogene. 1998;16:643–654. doi: 10.1038/sj.onc.1201567. [DOI] [PubMed] [Google Scholar]
- Setou M., Nakagawa T., Seog D. H., Hirokawa N. Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science. 2000;288:1796–1802. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- Sheff D. R., Daro E. A., Hull M., Mellman I. The receptor recycling pathway contains two distinct populations of early endosomes with different sorting functions. J. Cell Biol. 1999;145:123–139. doi: 10.1083/jcb.145.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelly M., Mosesson Y., Citri A., Lavi S., Zwang Y., Melamed-Book N., Aroeti B., Yarden Y. Polar expression of ErbB-2/HER2 in epithelia. Bimodal regulation by Lin-7. Dev. Cell. 2003;5:475–486. doi: 10.1016/j.devcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Simske J. S., Kaech S. M., Harp S. A., Kim S. K. LET-23 receptor localization by the cell junction protein LIN-7 during C. elegans vulval induction. Cell. 1996;85:195–204. doi: 10.1016/s0092-8674(00)81096-x. [DOI] [PubMed] [Google Scholar]
- Songyang Z., Fanning A. S., Fu C., Xu J., Marfatia S. M., Chishti A. H., Chan A. C., Anderson J. M., Cantley L. C. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- Straight S. W., Chen L., Karnak D., Margolis B. Interaction with mLin-7 alters the targeting of endocytosed transmembrane proteins in mammalian epithelial cells. Mol. Biol. Cell. 2001;12:1329–1340. doi: 10.1091/mbc.12.5.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight S. W., Karnak D., Borg J. P., Kamberov E., Dare H., Margolis B., Wade J. B. mLin-7 is localized to the basolateral surface of renal epithelia via its NH(2) terminus. Am. J. Physiol. Renal. Physiol. 2000;278:F464–F475. doi: 10.1152/ajprenal.2000.278.3.F464. [DOI] [PubMed] [Google Scholar]
- Wang X., Kumar R., Navarre J., Casanova J. E., Goldenring J. R. Regulation of vesicle trafficking in madin-darby canine kidney cells by Rab11a and Rab25. J. Biol. Chem. 2000;275:29138–29146. doi: 10.1074/jbc.M004410200. [DOI] [PubMed] [Google Scholar]
- Wiedemann U., Boisguerin P., Leben R., Leitner D., Krause G., Moelling K., Volkmer-Engert R., Oschkinat H. Quantification of PDZ domain specificity, prediction of ligand affinity and rational design of super-binding peptides. J. Mol. Biol. 2004;343:703–718. doi: 10.1016/j.jmb.2004.08.064. [DOI] [PubMed] [Google Scholar]
- Xiao B., Tu J. C., Petralia R. S., Yuan J. P., Doan A., Breder C. D., Ruggiero A., Lanahan A. A., Wenthold R. J., Worley P. F. Homer regulates the association of group 1 metabotropic glutamate receptors with multivalent complexes of homer-related, synaptic proteins. Neuron. 1998;21:707–716. doi: 10.1016/s0896-6273(00)80588-7. [DOI] [PubMed] [Google Scholar]
- Yokota Y., Mori S. Role of Id family proteins in growth control. J. Cell. Physiol. 2002;190:21–28. doi: 10.1002/jcp.10042. [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Zhang Z., Degeest G., Mortier E., Leenaerts I., Coomans C., Schulz J., N′Kuli F., Courtoy P. J., David G. Syndecan recycling is controlled by syntenin-PIP2 interaction with ARF6. Dev. Cell. 2005;9:377–388. doi: 10.1016/j.devcel.2005.07.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.