Abstract
Microtubule-associated protein 1B (MAP1B) is essential for neural development. Besides the abundant expression in neurons, MAP1B recently was found in myelinating oligodendroglia. Moreover, MAP1B deficiency causes delayed myelin development, suggesting the functional importance of MAP1B in oligodendroglia. However, molecular mechanisms that control MAP1B expression in oligodendroglia remain elusive. We report here that MAP1B mRNA is markedly up-regulated in the oligodendroglia cell line CG4 upon induced differentiation, leading to elevated MAP1B protein production. A coordinated regulation of homeoprotein transcription factors was observed during CG4 cell differentiation, which recapitulates the regulation in neurons that promotes MAP1B transcription. Hence, transcriptional regulation of MAP1B appears to be a common mechanism in both neurons and oligodendroglia. In addition, we found posttranscriptional regulation of MAP1B mRNA by the selective RNA-binding protein QKI in oligodendroglia. The 3′UTR of MAP1B mRNA interacts with QKI, and oligodendroglia-specific QKI-deficiency in the quakingviable mutant mice resulted in reduced MAP1B mRNA expression. Moreover, RNAi-mediated QKI-knockdown caused destabilization of the MAP1B mRNA in CG4 cells. Furthermore, forced expression of exogenous QKI was sufficient for promoting MAP1B expression. Because QKI is absent in neurons, QKI-dependent stabilization of MAP1B mRNA provides a novel mechanism for advancing MAP1B expression specifically in oligodendroglia during brain development.
INTRODUCTION
Microtubule-associated proteins (MAPs) control the dynamic organization of microtubule cytoskeleton, which in turn governs normal cell growth and development (Takemura et al., 1992; Hirokawa, 1994). Among these MAPs, MAP1B is predominantly expressed in the nervous system and is the earliest MAP detected in the embryonic brain (Tucker et al., 1989; Ma et al., 1997; Ohyu et al., 1997). Historically, MAP1B, which has been studied mainly in the developing neurons, plays essential roles in neurite outgrowth, axonal extension, and path finding (Gonzalez-Billault et al., 2001, 2002, 2004; Bouquet et al., 2004). MAP1B expression is markedly up-regulated during neurite outgrowth in various types of neurons (Gordon-Weeks and Fischer, 2000), and MAP1B knockout mice exhibit a range of abnormalities in axonal extension and path finding (Meixner et al., 2000; Gonzalez-Billault et al., 2001; Bouquet et al., 2004). More recent studies indicated that MAP1B expression is not restricted in neurons, but also is detected in oligodendrocytes and Schwann cells that produce myelin in the central and peripheral nervous system (CNS and PNS), respectively (Fischer et al., 1990; Ma et al., 1999). In particular, MAP1B expression is elevated in oligodendrocytes that initiate ensheathment of neuronal axons during normal brain development (Wu et al., 2001) as well as in Schwann cells during nerve regeneration (Ma et al., 1999). The functional importance of MAP1B in CNS myelination is further reinforced by the defects of myelin development in the MAP1B knockout mice (Meixner et al., 2000).
Despite the important function of MAP1B in the development and function of the nervous system, molecular mechanisms that promote MAP1B expression in neurons remain poorly understood and have not been investigated in oligodendrocytes. In neurons, a coordinated action of several transcription factors are thought to control MAP1B gene transcription (Montesinos et al., 2001; Foucher et al., 2003), which up-regulates MAP1B expression during neuronal differentiation. However, whether similar or distinct mechanisms are used in regulating MAP1B expression in neurons and oligodendrocytes remain elusive. In this study, we show that the level of MAP1B mRNA is markedly elevated, which is accompanied by the increased expression of the MAP1B protein upon induced differentiation of CG4 cells, a well-established cell model system for studying oligodendrocyte development (Louis et al., 1992). A coordinated regulation of the engrailed homeoprotein (EN1) and Foxa2 was observed during CG4 cell differentiation, recapitulating the regulation in neurons that promotes MAP1B gene transcription (Montesinos et al., 2001; Foucher et al., 2003). In addition, we found that the selective RNA-binding protein QKI governs MAP1B mRNA expression in oligodendrocytes by posttranscriptional mechanisms. QKI binds to the 3′untranslated region (3′UTR) of the MAP1B mRNA in vitro and associates with the MAP1B mRNA in the developing brain. Moreover, QKI is necessary for maintaining the stability and the expression levels of the MAP1B mRNA in oligodendroglia. Furthermore, forced expression of exogenous QKI is sufficient for promoting MAP1B expression. Because QKI is abundantly expressed in oligodendrocytes but absent in neurons, QKI-mediated mRNA stabilization offers a unique means to regulate MAP1B expression specifically in oligodendroglia.
MATERIALS AND METHODS
Animals and RNA Analysis
The qkv colony was purchased from Jackson Laboratory (Bar Harbor, ME). qkv/wt and qkv/qkv littermates were produced as described previously (Li et al., 2000). Total RNA was extracted from CG4 cells or from dissected corpus callosum using Trizol reagent (Invitrogen, Carlsbad, CA). The quantity of RNA from each sample was determined by OD260 reading and further confirmed by ethidium bromide–staining of agarose gel after electrophoresis. The RNase protection assay (RPA) for MAP1B, β-actin, and GAPDH transcripts was performed as described previously (Lu et al., 2004).
Cell Culture, Transfection, and Immunodetection
CG4 cells were propagated and differentiated as described in our previous report (Wang et al., 2004). Transfection of plasmids into CG4 cells was performed using cell line Nucleofector kit V following manufacturer's protocol (Amaxa, Gaithersburg, MD). For indirect immunofluorescent staining, cells grown on coverslips were fixed with 4% (wt/vol) paraformaldehyde (Sigma, St. Louis, MO), permeabilized with 0.1% (vol/vol) Triton X-100 in PBS for 20 min, blocked by 0.1% (wt/vol) BSA in PBS, and incubated overnight at 4°C with the following primary antibodies: anti-MBP (1:1000, Chemicon, Temecula, CA), anti-MAP1B (1:1000, a gift generously provided by Dr. I. Fisher, Drexel University, Philadelphia, PA), and anti-α-tubulin (1:1000, Sigma). After washing, corresponding secondary antibodies conjugated with either FITC or Texas red were incubated with the cells for 1 h at room temperature. Fluorescence signals were detected using the Zeiss LMS510 confocal microscopic imaging system (Thornwood, NY). For immunoblot analysis, the protein quantity of each sample was estimated by Bradford assay following manufacturer's protocol (Bio-Rad, Hercules, CA) before being subjected to SDS-PAGE. After overnight transfer, the blots were subjected to Ponceau S staining (Sigma) to confirm equal protein loading before incubation with corresponding antibodies: anti-eIF5a (1:5000) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA), anti-Flag M2 (1:1000) was purchased from Sigma, anti-MAP1B (1:50,000) antibody was provided by Dr. I. Fisher. The polyclonal anti-QKI antibody for immunoprecipitation was generated as described previously (Zhang et al., 2003).
Immunoprecipitation and RT-PCR
Brain stems were dissected from mice at postnatal day 2, homogenized, and lysed in an ice-cold buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 500 mM EDTA, 0.5% Triton X-100, protease inhibitor, and RNase inhibitor). The postnuclear supernatant was precleared with protein A-Sepharose, incubated with or without the primary anti-QKI antibody for 1 h at 4°C. The complexes were extensively washed before being subjected to RNA extraction using Trizol. RT was performed using SuperScript II RNaseH-Reverse Transcriptase (Invitrogen), followed by PCR analysis. The following primers were used for the analysis of MAP1B: 5′-tgctgggaaactcaagg-3′ (forward) and 5′-ccttgagtttccagcag-3′ (reverse). To further confirm the identity of MAP1B mRNA in QKI immunoprecipitates, the RT-PCR product run on agarose gel was transferred to Hybond-N membrane (Amersham, Arlington Heights, IL) and hybridized overnight to a 32P-labeled probe derived from a MAP1B cDNA fragment, followed by phosphorimager analysis. The primers used to detect the GAPDH mRNA were described previously (Huang et al., 1999).
For quantitative analysis of the EN1 and Foxa2 transcripts, total RNA extracted from CG4 cells at various differentiation stages were subjected to semiquantitative RT-PCR for 25 cycles using the following 32P-labled primers: EN1, 5′-gaatgagaaggaagacaagc-3′ (forward) and 5′-actcgctctcgtctttgtcc-3′ (reverse); and Foxa2, 5′-gctgagcgagatctatcagtgg-3′ (forward) and 5′-gctcgctcaggccacctcgcttg-3′ (reverse). The MBP primers were described in our previous report (Wang et al., 2004). The PCR products were then analyzed by PAGE followed by phosphorimager quantification.
In Vitro RNA Binding
An MAP1B cDNA construct including the 5′UTR was generously provided by Dr. Gordon-Weeks (University of London, London, United Kingdom). The construct of MAP1B 3′UTR (Meixner et al., 1999) was generously provided by Dr. Propst (University of Vienna, Vienna, Austria). [35S]methionine-labeled QKI-6 was generated as described previously (Li et al., 2000). Biotinylated RNAs were derived from in vitro transcription using T7 polymerase and incubated with 35S-QKI-6. The RNA-bound 35S-QKI was captured by streptavidin-conjugated Dynabeads (Li et al., 2000) and fractionated on SDS-PAGE, followed by phosphorimager analysis or were directly subjected to scintillation counting.
QKI RNAi and Quantification of mRNAs
A predesigned small interfering RNA (siRNA) specific for QKI was purchased from Ambion (Austin, TX), and transfected into proliferating CG4 cells using Nucleofector kit (Amaxa) following the manufacturer's protocol. A negative control siRNA that has no sequence homology to any mammalian mRNA (Ambion) was used in parallel experiments. Two hundred picomoles of each siRNA was used to transfect 4 × 106 cells. The EGFPN3 plasmid was included in the transfection to mark transfected cells. The estimated plasmid transfection rate is between 50 and 70%. Cells were harvested 24 h after transfection, and total RNA was prepared with Trizol extraction (Invitrogen). Real-time quantitative RT-PCR (qRT-PCR) was performed with DyNAmo SYBR Green qPCR Kits (New England Biolabs, Beverly, MA). The QKI primers flanking the siRNA cleavage site were used to monitor the QKI siRNA effect: 5′-aaataatggtccgaggcaaaggctc (forward); and 5′-agccgcaggtaccagtaacttctt (reverse). A pair of MAP1B-specific primers 5′-ccaggacaaaagatcctcca (forward) and 5′-tggaaggagaaagagcctga (reverse) was used in parallel qRT-PCR reactions to examine the influence of QKI knockdown on MAP1B mRNA. The qRT-PCR readout for each specific transcript was normalized to that of the GAPDH housekeeping gene mRNA detected by the following primers: 5′-cacagtcaaggctgagaatgggaag (forward) and 5′-gtggttcacacccatcacaaacatg (reverse). For MAP1B mRNA decay, transcription inhibition was performed as described previously (Zhang et al., 2003). The levels of MAP1B mRNA and the 18S ribosome RNA at various time points after transcription inhibition were measured by qRT-PCR with the aforementioned MAP1B primers and the 18S rRNA primers: 5′-gtgatggggatcggggattg (forward) and 5′-ggcggtgtgtacaaagggcag (reverse).
RESULTS
Increased MAP1B Expression and Regulation of Homeoprotein Transcription Factors in CG4 Cells upon Induced Differentiation
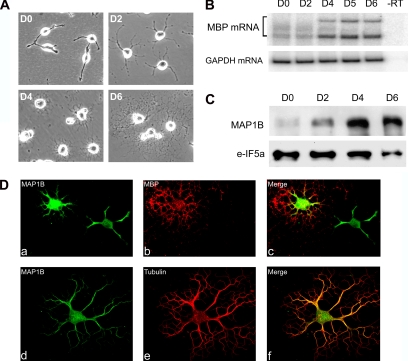
Because differentiation-induced neuritogenesis triggers vigorous up-regulation of MAP1B in neurons (Gordon-Weeks and Fischer, 2000; Gonzalez-Billault et al., 2004), we first questioned whether similar regulation of MAP1B also occurs during oligodendroglia differentiation. The CG4 cell line can be induced to differentiate in a synchronized manner and sequentially express oligodendroglia maturation markers, recapitulating that in primary cultured oligodendrocytes (Louis et al., 1992). This cell model system offers sufficient materials for biochemical analysis of MAP1B regulation during oligodendroglia differentiation. As shown in Figure 1A, proliferating cells typically harbor two primary processes. Vigorous process outgrowth was observed in cells that were differentiated for 2 d (D2), which continuous to develop sophisticated arboreous structures. The mRNA for myelin basic protein (MBP), a major myelin structural protein expressed in mature oligodendrocytes, was up-regulated in cells that had undergone 4 d of differentiation (D4, Figure 1B), recapitulating that observed in primary cultured oligodendrocytes (Kumar et al., 1989). A robust up-regulation of MAP1B protein expression was detected starting from D2 (Figure 1C), preceding the up-regulation of MBP and remained throughout the differentiation profile. Immunofluorescent staining further revealed MAP1B expression in both MBP-positive cells (mature) and MBP-negative cells (immature), with significantly higher levels of MAP1B detected in the mature MBP-positive cells (Figure 1D, a–c). The majority of the MAP1B protein was localized in the cell soma and the proximal main processes, colocalizing with microtubules (Figure 1D, d–f) but not in the finely branched distal processes enriched of microfilaments (Song et al., 2001). In contrast, MBP was predominantly detected in the periphery processes (Figure 1D, b and c). This result is consistent with the predicted role of MAP1B in modulating microtubule stability during the extension and branching of cell processes.
Figure 1.
MAP1B expression is up-regulated upon induced differentiation of the CG4 oligodendrocyte cell line. (A) Morphogenesis of CG4 cells during induced differentiation. Days for differentiation are marked on each panel. (B) A representative phosphorimage showing increased MBP mRNA expression as a marker of oligodendrocyte maturation during CG4 cell differentiation. Total RNA isolated from CG4 cells that had undergone various days of differentiation as indicated on the top of the corresponding lanes was subjected to a semiquantitative RT-PCR analysis using 32P-labled primers specific for MBP. RT-PCR of the housekeeping GAPDH mRNA was performed in parallel as an input reference. (C) Immunoblot analysis detects marked up-regulation of MAP1B protein upon induced differentiation of CG4 cells. The blot was reprobed with the housekeeping protein eIF5a to provide a loading control. (D) a–c, localization of MAP1B (green) and MBP (red) in differentiating CG4 cells; d–f, localization of MAP1B (green) and α-tubulin (red) in differentiating CG4 cells.
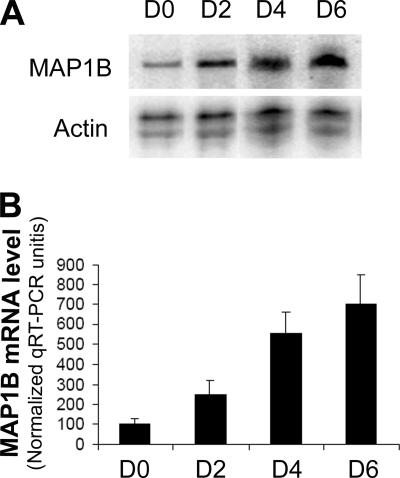
To delineate molecular mechanisms that underlie MAP1B up-regulation during oligodendrocyte differentiation, we analyzed the MAP1B mRNA levels in CG4 cells at various differentiation stages by an RNase protection assay (RPA). As shown in Figure 2A, induced differentiation triggered a significant up-regulation of MAP1B mRNA in CG4 cells, which displayed a similar profile as that observed for the up-regulation of MAP1B protein (Figure 1C). In contrast, the actin mRNA was expressed at steady levels as indicated by the RPA signals in simultaneous reactions. Quantification of the MAP1B mRNA by real-time RT-PCR (qRT-PCR) further revealed that the MAP1B mRNA level was increased approximately sevenfold in cells that had undergone 6 d of differentiation (Figure 2B). Apparently, the elevated MAP1B mRNA expression, due to up-regulation of transcription and/or mRNA stability, is a major mechanism underlying the MAP1B protein up-regulation during oligodendrocyte differentiation.
Figure 2.
Elevated MAP1B mRNA expression during CG4 cell differentiation. (A) Representative RPA gel showing MAP1B mRNA levels in CG4 cells that had undergone various days of differentiation as indicated on the top of the corresponding lanes. RPA of β-actin mRNA was performed in the same reaction to provide a loading control. (B) Real-time RT-PCR (qRT-PCR) quantification of the MAP1B mRNA levels during CG4 cell differentiation. Total RNA was extracted from four independent sets of differentiated CG4 cells (n = 4). The qRT-PCR reading of MAP1B mRNA in each sample was normalized to that of the housekeeping mRNA GAPDH. The normalized MAP1B mRNA level in undifferentiated cells (D0) was set as 100% in each set of sample. p < 0.01 based on one-way ANOVA.
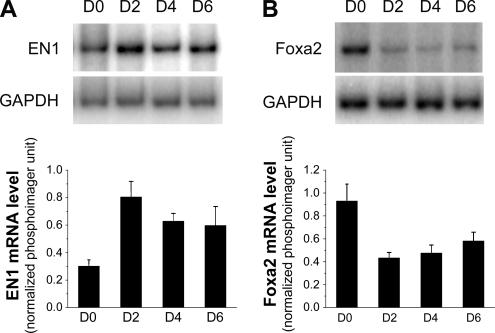
The homeoprotein transcription factors Engrailed 1 (EN1) and Foxa2 have been shown to bind the MAP1B promoter and act in concert to regulate neuronal MAP1B transcription (Foucher et al., 2003). To explore whether these transcription factors are regulated in concordance with the increased expression of the MAP1B mRNA upon induced oligodendrocyte differentiation, we examined the expression levels of these mRNAs. Semiquantitative RT-PCR analysis revealed a significant up-regulation of EN1 accompanied by a down-regulation of Foxa2 (Figure 3, A and B) in cells that had undergone 2 d of differentiation. This is associated with the initial up-regulation of MAP1B (Figure 2). The level of EN1 slightly decreased during later differentiation, but still above that in proliferating CG4 cells, whereas Foxa2 remained at low level through the differentiation profile examined. Because EN1 drives MAP1B transcription and Foxa2 antagonizes EN1 at the MAP1B promoter (Foucher et al., 2003), the cooperative regulation of EN1 and Foxa2 in CG4 cells suggests that these factors are likely involved in up-regulation of MAP1B transcription during oligodendrocyte differentiation.
Figure 3.
Expression of the homeoprotein transcription factor EN1 and Foxa2 during CG4 differentiation. Increased EN1 expression (A) and reduced Foxa2 expression (B) upon induced differentiation of CG4 cells. Total RNA isolated from CG4 cells that had undergone various days of differentiation was subjected to a semiquantitative RT-PCR analysis using 32P-labeled primers specific for EN1 and Foxa2. RT-PCR of the housekeeping GAPDH mRNA was performed in parallel as an input control (top panel). Corresponding bottom panels show phosphorimager quantification of EN1 and Foxa2 expression upon CG4 cell differentiation. The signal at each time point was quantitatively measured and was normalized to that of GAPDH (n = 3). p < 0.05 based on one-way ANOVA.
The Selective RNA-binding Protein QKI Interacts with the MAP1B mRNA
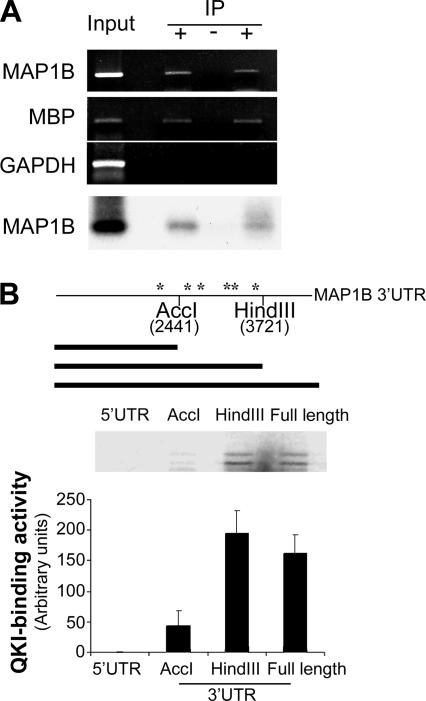
We next questioned whether posttranscriptional mechanisms may also contribute to MAP1B up-regulation in oligodendrocytes, because regulation at the level of mRNA stability plays critical roles in oligodendrocyte development and myelination (Kumar et al., 1989; Mathisen et al., 1997). In particular, the selective RNA-binding protein QKI is essential in maintaining mRNA stability during oligodendrocyte and myelin development (Li et al., 2000; Larocque et al., 2005). Moreover, simultaneous up-regulation of QKI and MAP1B was observed in actively myelinating oligodendrocytes (Wu et al., 2001). We first questioned whether QKI associates with the MAP1B mRNA in the developing brain. We immunoprecipitated (IP) QKI from the neonatal mouse brain and found that MAP1B mRNA was clearly detected in the immunoprecipitated QKI-mRNA complexes, based on RT-PCR analysis using MAP1B-specific primers (Figure 4A). Southern blot hybridization with a MAP1B-specific cDNA probe further confirmed the identity of the RT-PCR product of MAP1B. As a negative control, MAP1B mRNA was undetectable in parallel IP reactions using the protein A beads alone or using normal IgG without the QKI antibody (unpublished data). The MBP mRNA, a known target mRNA of QKI, was also detected in the immunoprecipitated QKI complexes (Figure 4A). In contrast, the GAPDH mRNA that is highly abundant in the brain lysate, was not detected in immunoprecipitated QKI-RNA complexes (Figure 4A). These data suggest that MAP1B mRNA is one of the selective ligands for QKI in the developing brain.
Figure 4.
MAP1B mRNA associates with QKI. (A) Coimmunoprecipitation of MAP1B mRNA with QKI from the neonatal brain. Cytoplasmic extracts prepared from P2 mouse brainstems was immunoprecipitated with (+) or without anti-QKI antibody (−) as indicated on top of the corresponding lanes. Coprecipitated RNA was subjected to RT-PCR as indicated on the left and Southern hybridization using a 32P-labled MAP1B probe showed in the bottom panel. Immunoprecipitation was also performed using normal IgG, which does not pull down any mRNA detected by RT-PCR. (B) Top, schematic representation of the MAP1B 3′UTR (base pairs 1752–3813 in the 3′UTR) with the restriction sites (positions in MAP1B 3′UTR marked underneath) used to generate truncated transcripts. Six putative QKI-binding elements recapitulating the consensus sequence identified by SELEX were marked by asterisks (*). Bottom, QKI-binding activity by the MAP1B 5′UTR and various truncated MAP1B 3′UTR. Inset, a representative phosphorimage of captured 35S-QKI by the corresponding truncated MAP1B transcripts. Scintillation counting for RNA-bound QKI is graphically displayed in the bottom panel.
RNA-binding proteins often interact with their mRNA ligands in the UTRs. Indeed, QKI has been reported to interact with UTRs of other mRNAs (Li et al., 2000; Larocque et al., 2005). To identify the QKI-binding sequence in the MAP1B mRNA, we performed in vitro RNA-binding analysis using various regions of the UTRs in the MAP1B transcript. As shown in Figure 4B, QKI binds the 3′UTR but not the 5′UTR of the MAP1B mRNA. Furthermore, using truncated MAP1B 3′UTR, we mapped QKI-binding activity to a 1.5-kb fragment between the AccI and HindIII restriction sites in the MAP1B 3′UTR (Figure 4B). Interestingly, this fragment contains several copies of the bipartite ACUAAY-N1–20-UAAY consensus QKI-binding element (QRE) identified by SELEX in a recent report (Galarneau and Richard, 2005). In contrast, no QREs were found in other regions of MAP1B mRNA that do not bind QKI (Figure 4B).
QKI Is a Necessary Factor for Maintaining MAP1B mRNA Stability in Oligodendrocytes
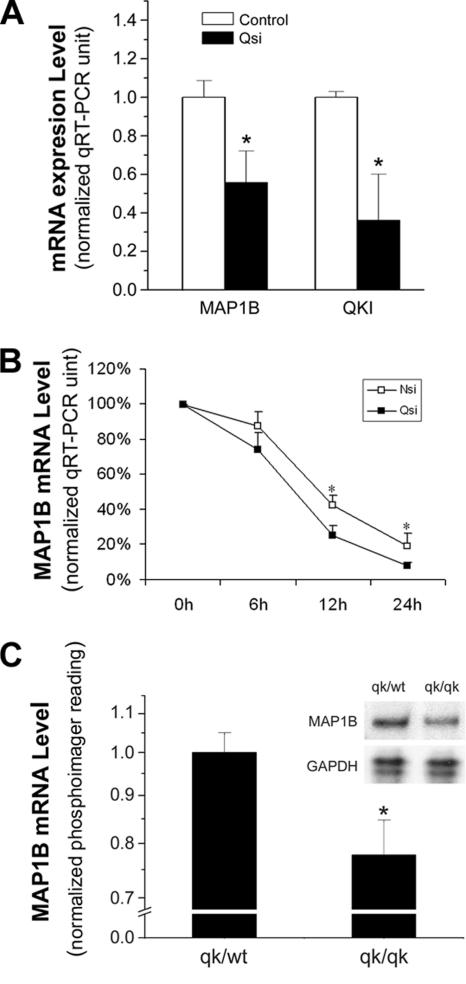
To directly test whether QKI is required for maintaining the normal expression levels of MAP1B mRNA in oligodendrocytes, we developed an siRNA to knockdown endogenous QKI expression in the CG4 cells. As expected, QKI mRNA level was significantly reduced when normalized to the GAPDH mRNA (∼64% reduction) in QKI siRNA-treated cells compared with that in control siRNA-treated cells, based on qRT-PCR analysis (Figure 5A). Consequently, the MAP1B mRNA was significantly reduced in QKI siRNA-treated cells compared with that in control siRNA-treated cells (Figure 5A). In contrast, the expression level of EN1 mRNA was not affected by the QKI siRNA (unpublished data). Therefore, knocking down QKI selectively reduced MAP1B mRNA, which is unlikely due to EN1-mediated MAP1B transcription. To further test whether the reduction of MAP1B mRNA in response to QKI knockdown is due to the loss of QKI-mediated stabilization of the MAP1B mRNA, we analyzed the decay of MAP1B mRNA in QKI siRNA- and control siRNA-treated cells when transcription was blocked. As shown in Figure 5B, MAP1B mRNA decayed more rapidly in QKI siRNA-treated cells compared with that in control siRNA-treated cells, indicating that QKI functions to stabilize the MAP1B mRNA in oligodendrocytes.
Figure 5.
QKI deficiency causes reduced expression of MAP1B in vitro and in vivo. (A) Knocking down QKI expression by siRNA significantly reduced MAP1B mRNA expression in CG4 cells. RNA extracted from QKI siRNA-treated cells and control siRNA-treated cells was used for qRT-PCR analysis of the mRNAs for QKI, MAP1B, and GAPDH. qRT-PCR reading for each sample was normalized to that of the GAPDH housekeeping mRNA, and results shown as the mean ± SD of four independent experiments are graphically displayed. (B) The decay of MAP1B mRNA in QKI siRNA- and control siRNA-treated CG4 cells. Twenty-four hours after siRNA-treatment, parallel cultures of cells were treated with actinomycin D to block transcription before isolating total RNA at the indicated time points of transcription inhibition. The qRT-PCR reading of MAP1B mRNA was normalized to that of the 18S rRNA before being plotted against time to generate the decay curve. (C) Reduced MAP1B mRNA expression in the qkv corpus callosum. RPA was performed using total RNA isolated from the dissected corpus callosum derived from qkv/qkv and the nonphenotypic qkv/wt littermate control. Inset, a representative RPA image of MAP1B and the loading control GAPDH. MAP1B mRNA level was quantitatively measured by a phosphorimager, normalized to the GAPDH signal and graphically displayed with SD (n = 5). * p < 0.05 by standard t test.
To further test whether QKI is a necessary factor for maintaining the MAP1B mRNA levels in oligodendrocytes in vivo, we took the advantage of the well-known quakingviable (qkv) dysmyelination mutant mice in which QKI is specifically diminished in oligodendrocytes (Hardy et al., 1996; Lu et al., 2003). Although MAP1B is most abundantly expressed in neurons, the corpus callosum is enriched of oligodendrocytes and neuronal axons that harbor less mRNA than that in the neuronal cell body, hence allowing us to evaluate the influence of QKI on MAP1B expression in oligodendrocytes in vivo. Indeed, quantitative RPA revealed reduced MAP1B mRNA in the corpus callosum isolated from the homozygous qkv mutant compared with that in the nonphenotypic littermate controls (Figure 5C), suggesting that QKI is necessary for MAP1B expression in oligodendrocytes in vivo. However, the actual reduction level of MAP1B mRNA in this experiment is likely underestimated because of the existence of neuronal MAP1B mRNA in the corpus callosum, which is unlikely affected in the qkv mutant because QKI is absent in neurons (Hardy et al., 1996).
QKI Is Sufficient for Promoting MAP1B Expression
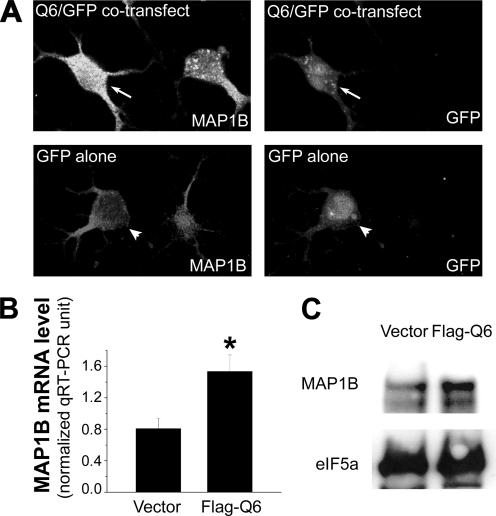
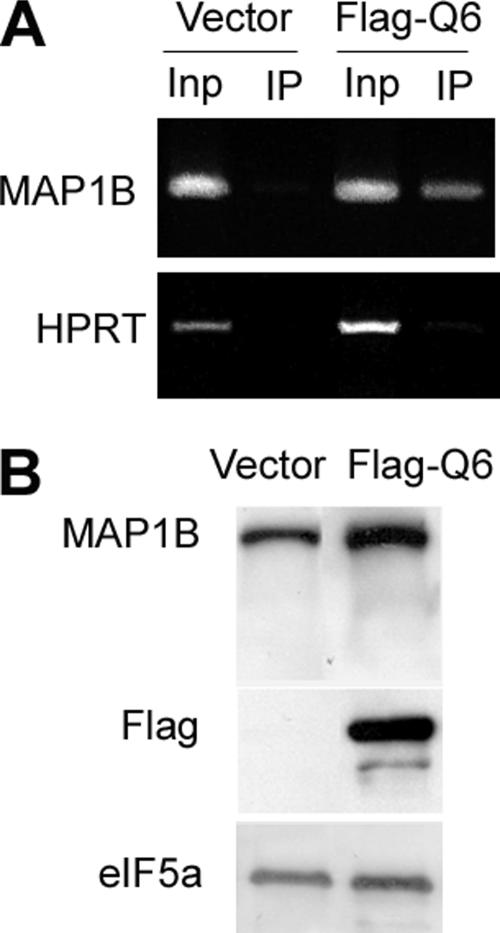
We next questioned whether exogenous QKI is sufficient for promoting MAP1B expression. Three major QKI isoforms (QKI-5, -6, and -7) are expressed in oligodendrocytes, among which QKI-6 is the most abundant (Lu et al., 2003). In CG4 cells transfected with Flag-QKI-6, the expression level of endogenous MAP1B protein was markedly elevated, as evidenced by the increased intensity of immunofluorescent staining of MAP1B (Figure 6A), the increased MAP1B mRNA measured by qRT-PCR (Figure 6B), and the increased MAP1B protein on immunoblot (Figure 6C). We further tested whether forced expression of QKI-6 in neurons that do not express endogenous QKI can also promote MAP1B expression. The immortalized brain neuronal cell line CAD does not express QKI (Zhang et al., 2003), but harbors abundant MAP1B mRNA that can be immunoprecipitated with Flag-QKI-6 (Figure 7A). In contrast, we found that the mRNA encoding the housekeeping protein HPRT did not associate with Flag-QKI-6. Moreover, the MAP1B level in CAD cells that express Flag-QKI-6 is elevated compared with that in control CAD cells transfected with the vehicle plasmid (Figure 7B). These results indicate that QKI harbors the ability to promote MAP1B expression in both the oligodendroglia and the neuronal lineage. However, because QKI expression is eliminated from neurons during neural cell fate specification (Hardy, 1998), QKI-mediated MAP1B mRNA stabilization offers a mechanism for promoting MAP1B expression specifically in oligodendroglia.
Figure 6.
Exogenous Flag-QKI-6 promotes MAP1B expression in CG4 cells. (A) Representative image of increased MAP1B expression in Flag-QKI-6 transfected cells. CG4 cells were cotransfected with PC-Flag-QKI-6 and N3EGFP plasmids (arrows in top panels) or N3GFP alone (arrowheads in bottom panels). Two days after transfection, cells were stained with anti-MAP1B antibody (left panels). Transfected cells are marked by GFP (right panels). Note, MAP1B intensity is increased in Flag-QKI-6–transfected cells (arrow in top-left panel) compared with untransfected cells, whereas GFP alone does not affect MAP1B expression (arrowhead in bottom-left panel). (B) qRT-PCR analysis indicates increased MAP1B mRNA expression in Flag-QKI-6–transfected cells compared with that in pcDNA3 vector-transfected cells. MAP1B mRNA level was normalized to that of the GAPDH mRNA and graphically displayed with SD (n = 3). * p < 0.05 by standard t test. (C) Representative immunoblot (for 3 independent experiments) indicates increased MAP1B protein expression in cells transfected with PC-Flag-QKI-6 compared with that in cells transfected by the PCDNA3 parental vector.
Figure 7.
Exogenous expression of Flag-QKI-6 in the CAD neuron cell line promotes MAP1B expression. (A) Endogenous MAP1B mRNA associates with exogenous Flag-QKI in transfected CAD cells. Immunoprecipitation was performed using the anti-Flag M2 beads from cells transfected with PC-Flag-QKI-6 or the PCDNA parental vector in parallel reactions. RT-PCR was performed using RNA extracted from the input lysate (Inp) and the immunoprecipitates (IP) in parallel. Note, MAP1B mRNA is detected in immunoprecipitates in cells expressing Flag-QKI-6 but not in immunoprecipitates derived from vector-transfected control cells that lack Flag-QKI-6. The HPRT mRNA is not detected in either IPs. (B) Representative immunoblot indicates increased MAP1B protein expression in CAD cells transfected with PC-Flag-QKI-6 compared with that in control cells transfected with the PCDNA3 parental vector. The same blot is reprobed with anti-Flag and anti-eIF5a as indicated on the left. Similar results were observed from three independent experiments.
DISCUSSION
We show here that MAP1B mRNA levels are markedly up-regulated in the CG4 oligodendroglia cell line upon induced differentiation, leading to increased production of the MAP1B protein. The main goal of this study is to delineate molecular mechanisms that promote MAP1B expression in oligodendrocytes. The concurrent regulation of homeoprotein transcription factors EN1 and Foxa2 with elevated MAP1B expression in the differentiating CG4 cells suggests up-regulation of MAP1B gene transcription. In addition, we demonstrated that the selective RNA-binding protein QKI is necessary for stabilization of the MAP1B mRNA in oligodendrocytes and exogenous QKI is sufficient for promoting MAP1B expression. Because QKI is absent in neurons but abundantly expressed in oligodendrocytes, QKI-dependent mRNA stabilization provides a means for regulating MAP1B expression specifically in oligodendrocytes.
The robust up-regulation of MAP1B during induced differentiation of CG4 cells (Figure 1) suggests an increase in the functional requirement of MAP1B in supporting the vigorous process extension of oligodendrocytes, recapitulating the well-known function of MAP1B in neuritogenesis (Gordon-Weeks and Fischer, 2000). Consistent with the classical role of MAP1B in modulating microtubule stability, MAP1B is predominantly colocalized with tubulin in CG4 cells (Figure 1). The fact that MAP1B up-regulation occurs before the expression of MBP (Figure 1D) is consistent with the previous report that MAP1B functions at the early stage of oligodendrocyte development (Vouyiouklis and Brophy, 1993). Furthermore, elevated MAP1B expression sustains in mature oligodendrocytes that expression MBP (Figure 1D), suggesting that MAP1B also plays important roles in oligodendroglia maturation.
The cooperative regulation of EN1 and Foxa2 associates with MAP1B up-regulation in differentiating CG4 cells (Figure 3). Such coregulation of EN1 and Foxa2 promotes MAP1B transcription in neurons (Foucher et al., 2003). Hence, regulation of MAP1B transcription by these transcription factors appears to be a commonly shared mechanism in neurons and oligodendrocytes. On the other hand, MAP1B mRNA is associated with QKI (Figure 4A), a key player that controls mRNA stability during oligodendrocyte development (Li et al., 2000; Zhang et al., 2003; Larocque et al., 2005). Consistent with the role of QKI in protecting other mRNAs from degradation (Zhang et al., 2003; Larocque et al., 2005), RNAi-mediated QKI knockdown demonstrated that QKI is necessary for maintaining the stability of the MAP1B mRNA in oligodendroglia (Figure 5), identifying MAP1B as a new target for QKI in oligodendrocyte differentiation. The simultaneous up-regulation of MAP1B and QKI in myelinating oligodendrocytes in vivo (Wu et al., 2001) further suggests a role of QKI in controlling MAP1B expression in myelination beyond the early stage of oligodendrocyte differentiation. Noticeably, QKI is also up-regulated during oligodendroglia development (Hardy et al., 1996). Hence, QKI-mediated mRNA stabilization offers a synergistic mechanism with the transcriptional regulation of MAP1B. The functional requirement of MAP1B in myelination is reinforced by the fact that genetic disruption of MAP1B in several mouse models leads to impaired myelination (Meixner et al., 2000; Takei et al., 2000). In this regard, the reduced MAP1B expression in the qkv mutant mice due to QKI deficiency (Figure 5C) may contribute not only to the defect in oligodendrocyte differentiation, but also to the failure in myelinogenesis.
During neural cell fate specification, QKI expression is eliminated in the neuronal lineage and is up-regulated in oligodendrocytes during myelin development (Hardy et al., 1996; Hardy, 1998). Interestingly, exogenous QKI is sufficient for promoting MAP1B expression in oligodendroglia that harbors QKI as well as in the CAD neuronal cell line that lacks endogenous QKI (Figures 6 and 7). This result suggests that QKI does not require oligodendrocyte-specific cofactors to associate with MAP1B mRNA and protects it from degradation. However, because QKI is absent in neurons but is elevated in oligodendroglia during myelin development, QKI-dependent MAP1B mRNA stabilization provides a mechanism for promoting MAP1B expression specifically in oligodendrocytes in the developing brain.
The interaction of MAP1B mRNA with QKI is mediated by the 3′UTR but not the 5′UTR (Figure 4B), particularly to the fragment that harbors several copies of the QKI-responsible element (QRE) that contains the bipartite sequence motif of ACUAAY-N1–20-UAAY (Galarneau and Richard, 2005). Noticeably, the 5′ portion of this SELEX-identified QRE matches with the consensus QKI recognition sequence 5′NA(A>C)U(A≫C/U)A-3′ identified by competition fluorescence-polarization (Ryder and Williamson, 2004). In addition, the QKI-binding segment in the MAP1B 3′UTR contains several clusters of UAAY “half-core” QRE that is present in various QKI mRNA ligands and binds QKI with relatively lower affinity in vitro (Galarneau and Richard, 2005). Besides the aforementioned QREs, QKI has also been reported to bind the TGE sequence motif (Saccomanno et al., 1999; Lakiza et al., 2005) that contains part of the QRE identified by SELEX (Galarneau and Richard, 2005) and can suppress translation of some of its mRNA ligands (Lakiza et al., 2005). However, QKI unlikely suppresses MAP1B translation because forced expression of QKI increased MAP1B protein (Figures 6 and 7). It is important to point out that the MAP1B 3′UTR contains several putative AU-rich elements (ARE), which are well documented in controlling mRNA stability of other mRNA species (Chen and Shyu, 1995; Brennan and Steitz, 2001; Bevilacqua et al., 2003; Dean et al., 2004; Wilusz and Wilusz, 2004). What ARE-binding proteins may be involved in controlling MAP1B mRNA decay and how QKI may coordinate with these proteins to protect MAP1B mRNA from degradation in oligodendrocytes still remain elusive. Furthermore, although QKI is not expressed in neurons, other neuronal-specific RNA-binding proteins may be identified by future studies that recapitulate the role of QKI, contributing to the elevated MAP1B expression in neurons during neurite outgrowth (Larsen et al., 1998; Goold and Gordon-Weeks, 2001).
ACKNOWLEDGMENTS
We thank Dr. I. Fisher (Drexel University) for providing the anti-MAP1B antibody, Dr. Gordon-Weeks for the MAP1B cDNA clone, and Dr. F. Propst for the genomic DNA clone containing the MAP1B 3′UTR. This work is supported by National Institutes of Health Grant NS39551 and National Multiple Sclerosis Society (NMSS) Grant RG 3296 to Y.F. and NMSS Grant PP0864 to P.L.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-04-0355) on July 19, 2006.
REFERENCES
- Bevilacqua A., Ceriani M. C., Capaccioli S., Nicolin A. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J. Cell. Physiol. 2003;195:356–372. doi: 10.1002/jcp.10272. [DOI] [PubMed] [Google Scholar]
- Bouquet C., Soares S., von Boxberg Y., Ravaille-Veron M., Propst F., Nothias F. Microtubule-associated protein 1B controls directionality of growth cone migration and axonal branching in regeneration of adult dorsal root ganglia neurons. J. Neurosci. 2004;24:7204–7213. doi: 10.1523/JNEUROSCI.2254-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C. M., Steitz J. A. HuR and mRNA stability. Cell Mol. Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Y., Shyu A. B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Dean J. L., Sully G., Clark A. R., Saklatvala J. The involvement of AU-rich element-binding proteins in p38 mitogen-activated protein kinase pathway-mediated mRNA stabilisation. Cell Signal. 2004;16:1113–1121. doi: 10.1016/j.cellsig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Fischer I., Konola J., Cochary E. Microtubule associated protein (MAP1B) is present in cultured oligodendrocytes and co-localizes with tubulin. J. Neurosci. Res. 1990;27:112–124. doi: 10.1002/jnr.490270117. [DOI] [PubMed] [Google Scholar]
- Foucher I., Montesinos M. L., Volovitch M., Prochiantz A., Trembleau A. Joint regulation of the MAP1B promoter by HNF3beta/Foxa2 and Engrailed is the result of a highly conserved mechanism for direct interaction of homeoproteins and Fox transcription factors. Development. 2003;130:1867–1876. doi: 10.1242/dev.00414. [DOI] [PubMed] [Google Scholar]
- Galarneau A., Richard S. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat. Struct. Mol. Biol. 2005;12:691–698. doi: 10.1038/nsmb963. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Billault C., Avila J., Caceres A. Evidence for the role of MAP1B in axon formation. Mol. Biol. Cell. 2001;12:2087–2098. doi: 10.1091/mbc.12.7.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Billault C., Jimenez-Mateos E. M., Caceres A., Diaz-Nido J., Wandosell F., Avila J. Microtubule-associated protein 1B function during normal development, regeneration, and pathological conditions in the nervous system. J. Neurobiol. 2004;58:48–59. doi: 10.1002/neu.10283. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Billault C., Owen R., Gordon-Weeks P. R., Avila J. Microtubule-associated protein 1B is involved in the initial stages of axonogenesis in peripheral nervous system cultured neurons. Brain Res. 2002;943:56–67. doi: 10.1016/s0006-8993(02)02534-9. [DOI] [PubMed] [Google Scholar]
- Goold R. G., Gordon-Weeks P. R. Microtubule-associated protein 1B phosphorylation by glycogen synthase kinase 3beta is induced during PC12 cell differentiation. J. Cell Sci. 2001;114:4273–4284. doi: 10.1242/jcs.114.23.4273. [DOI] [PubMed] [Google Scholar]
- Gordon-Weeks P. R., Fischer I. MAP1B expression and microtubule stability in growing and regenerating axons. Microsc. Res. Tech. 2000;48:63–74. doi: 10.1002/(SICI)1097-0029(20000115)48:2<63::AID-JEMT2>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Hardy R. J. QKI expression is regulated during neuron-glial cell fate decisions. J. Neurosci. Res. 1998;54:46–57. doi: 10.1002/(SICI)1097-4547(19981001)54:1<46::AID-JNR6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Hardy R. J., Loushin C. L., Friedrich V. L., Jr, Chen Q., Ebersole T. A., Lazzarini R. A., Artzt K. Neural cell type-specific expression of QKI proteins is altered in quakingviable mutant mice. J. Neurosci. 1996;16:7941–7949. doi: 10.1523/JNEUROSCI.16-24-07941.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N. Microtubule organization and dynamics dependent on microtubule-associated proteins. Curr. Opin. Cell Biol. 1994;6:74–81. doi: 10.1016/0955-0674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- Huang Y., Myers S. J., Dingledine R. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat. Neurosci. 1999;2:867–872. doi: 10.1038/13165. [DOI] [PubMed] [Google Scholar]
- Kumar S., Cole R., Chiappelli F., de Vellis J. Differential regulation of oligodendrocyte markers by glucocorticoids: post-transcriptional regulation of both proteolipid protein and myelin basic protein and transcriptional regulation of glycerol phosphate dehydrogenase. Proc. Natl. Acad. Sci. USA. 1989;86:6807–6811. doi: 10.1073/pnas.86.17.6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakiza O., Frater L., Yoo Y., Villavicencio E., Walterhouse D., Goodwin E. B., Iannaccone P. STAR proteins quaking-6 and GLD-1 regulate translation of the homologues GLI1 and tra-1 through a conserved RNA 3′UTR-based mechanism. Dev. Biol. 2005;287:98–110. doi: 10.1016/j.ydbio.2005.08.038. [DOI] [PubMed] [Google Scholar]
- Larocque D., Galarneau A., Liu H. N., Scott M., Almazan G., Richard S. Protection of p27(Kip1) mRNA by quaking RNA binding proteins promotes oligodendrocyte differentiation. Nat. Neurosci. 2005;8:27–33. doi: 10.1038/nn1359. [DOI] [PubMed] [Google Scholar]
- Larsen K. E., Pacheco M., Roth J., Aletta J. M. Increased MAP1B expression without increased phosphorylation in manganese-treated PC12Mn cells. Exp. Cell Res. 1998;245:105–115. doi: 10.1006/excr.1998.4222. [DOI] [PubMed] [Google Scholar]
- Li Z., Zhang Y., Li D., Feng Y. Destabilization and mislocalization of myelin basic protein mRNAs in quaking dysmyelination lacking the QKI RNA-binding proteins. J. Neurosci. 2000;20:4944–4953. doi: 10.1523/JNEUROSCI.20-13-04944.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis J. C., Magal E., Muir D., Manthorpe M., Varon S. CG-4, a new bipotential glial cell line from rat brain, is capable of differentiating in vitro into either mature oligodendrocytes or type-2 astrocytes. J. Neurosci. Res. 1992;31:193–204. doi: 10.1002/jnr.490310125. [DOI] [PubMed] [Google Scholar]
- Lu R., Wang H., Liang Z., Ku L., O'Donnell W. T., Li W., Warren S. T., Feng Y. The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proc. Natl. Acad. Sci. USA. 2004;101:15201–15206. doi: 10.1073/pnas.0404995101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Zhang Y., Ku L., Wang H., Ahmadian A., Feng Y. The quakingviable mutation affects qkI mRNA expression specifically in myelin-producing cells of the nervous system. Nucleic Acids Res. 2003;31:4616–4624. doi: 10.1093/nar/gkg635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Chow S., Obrocka M., Connors T., Fischer I. Induction of microtubule-associated protein 1B expression in Schwann cells during nerve regeneration. Brain Res. 1999;823:141–153. doi: 10.1016/s0006-8993(99)01148-8. [DOI] [PubMed] [Google Scholar]
- Ma D., Nothias F., Boyne L. J., Fischer I. Differential regulation of microtubule-associated protein 1B (MAP1B) in rat CNS and PNS during development. J. Neurosci. Res. 1997;49:319–332. doi: 10.1002/(sici)1097-4547(19970801)49:3<319::aid-jnr7>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Mathisen P. M., Johnson J. M., Kawczak J. A. Stabilization of myelin mRNAs as measured in a brain slice system. J. Neurosci. Res. 1997;50:1030–1039. doi: 10.1002/(SICI)1097-4547(19971215)50:6<1030::AID-JNR14>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Meixner A., Haverkamp S., Wassle H., Fuhrer S., Thalhammer J., Kropf N., Bittner R. E., Lassmann H., Wiche G., Propst F. MAP1B is required for axon guidance and is involved in the development of the central and peripheral nervous system. J. Cell Biol. 2000;151:1169–1178. doi: 10.1083/jcb.151.6.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meixner A., Wiche G., Propst F. Analysis of the mouse MAP1B gene identifies a highly conserved 4.3 kb 3′ untranslated region and provides evidence against the proposed structure of DBI-1 cDNA. Biochim. Biophys. Acta. 1999;1445:345–350. doi: 10.1016/s0167-4781(99)00062-7. [DOI] [PubMed] [Google Scholar]
- Montesinos M. L., Foucher I., Conradt M., Mainguy G., Robel L., Prochiantz A., Volovitch M. The neuronal microtubule-associated protein 1B is under homeoprotein transcriptional control. J. Neurosci. 2001;21:3350–3359. doi: 10.1523/JNEUROSCI.21-10-03350.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyu J., Yamanouchi H., Takashima S. Immunohistochemical study of microtubule-associated protein 5 (MAP5) expression in the developing human brain. Brain Dev. 1997;19:541–546. doi: 10.1016/s0387-7604(97)00075-2. [DOI] [PubMed] [Google Scholar]
- Ryder S. P., Williamson J. R. Specificity of the STAR/GSG domain protein Qk1, implications for the regulation of myelination. RNA. 2004;10:1449–1458. doi: 10.1261/rna.7780504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccomanno L., Loushin C., Jan E., Punkay E., Artzt K., Goodwin E. B. The STAR protein QKI-6 is a translational repressor. Proc. Natl. Acad. Sci. USA. 1999;96:12605–12610. doi: 10.1073/pnas.96.22.12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Goetz B. D., Baas P. W., Duncan I. D. Cytoskeletal reorganization during the formation of oligodendrocyte processes and branches. Mol. Cell Neurosci. 2001;17:624–636. doi: 10.1006/mcne.2001.0974. [DOI] [PubMed] [Google Scholar]
- Takei Y., Teng J., Harada A., Hirokawa N. Defects in axonal elongation and neuronal migration in mice with disrupted tau and map1b genes. J. Cell Biol. 2000;150:989–1000. doi: 10.1083/jcb.150.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura R., Okabe S., Umeyama T., Kanai Y., Cowan N. J., Hirokawa N. Increased microtubule stability and alpha tubulin acetylation in cells transfected with microtubule-associated proteins MAP1B, MAP2 or tau. J. Cell Sci. 1992;103(Pt 4):953–964. doi: 10.1242/jcs.103.4.953. [DOI] [PubMed] [Google Scholar]
- Tucker R. P., Garner C. C., Matus A. In situ localization of microtubule-associated protein mRNA in the developing and adult rat brain. Neuron. 1989;2:1245–1256. doi: 10.1016/0896-6273(89)90309-7. [DOI] [PubMed] [Google Scholar]
- Vouyiouklis D. A., Brophy P. J. Microtubule-associated protein MAP1B expression precedes the morphological differentiation of oligodendrocytes. J. Neurosci. Res. 1993;35:257–267. doi: 10.1002/jnr.490350305. [DOI] [PubMed] [Google Scholar]
- Wang H., Ku L., Osterhout D. J., Li W., Ahmadian A., Liang Z., Feng Y. Developmentally-programmed FMRP expression in oligodendrocytes: a potential role of FMRP in regulating translation in oligodendroglia progenitors. Hum. Mol. Genet. 2004;13:79–89. doi: 10.1093/hmg/ddh009. [DOI] [PubMed] [Google Scholar]
- Wilusz C. J., Wilusz J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Wu H. Y., Dawson M. R., Reynolds R., Hardy R. J. Expression of QKI proteins and MAP1B identifies actively myelinating oligodendrocytes in adult rat brain. Mol. Cell Neurosci. 2001;17:292–302. doi: 10.1006/mcne.2000.0941. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Lu Z., Ku L., Chen Y., Wang H., Feng Y. Tyrosine-phosphorylation of QKI mediates developmental signals to regulate mRNA metabolism. EMBO J. 2003;15:1801–1810. doi: 10.1093/emboj/cdg171. [DOI] [PMC free article] [PubMed] [Google Scholar]