Abstract
Rapid and long-distance secretion of membrane components is critical for hyphal formation in filamentous fungi, but the mechanisms responsible for polarized trafficking are not well understood. Here, we demonstrate that in Candida albicans, the majority of the Golgi complex is redistributed to the distal region during hyphal formation. Randomly distributed Golgi puncta in yeast cells cluster toward the growing tip during hyphal formation, remain associated with the distal portion of the filament during its extension, and are almost absent from the cell body. This restricted Golgi localization pattern is distinct from other organelles, including the endoplasmic reticulum, vacuole and mitochondria, which remain distributed throughout the cell body and hypha. Hyphal-induced positioning of the Golgi and the maintenance of its structural integrity requires actin cytoskeleton, but not microtubules. Absence of the formin Bni1 causes a hyphal-specific dispersal of the Golgi into a haze of finely dispersed vesicles with a sedimentation density no different from that of normal Golgi. These results demonstrate the existence of a hyphal-specific, Bni1-dependent cue for Golgi integrity and positioning at the distal portion of the hyphal tip, and suggest that filamentous fungi have evolved a novel strategy for polarized secretion, involving a redistribution of the Golgi to the growing tip.
INTRODUCTION
The establishment of axes of polarization is crucial for the growth and division of both unicellular and multicellular organisms. A striking example of polarized morphogenesis is the process of hyphal formation in Candida albicans. In response to a number of inductive signals, C. albicans switches from an ovoid yeast form to a highly elongated hyphal form, and its capacity to switch between these forms is postulated to be related to its virulence as a fungal pathogen (Lo et al., 1997; Gow et al., 2002). Within minutes of encountering the inducing signal, growth is restricted to the germ tube, which rapidly extends into a filament that becomes >10 times the length of the cell body within hours.
The mechanism that establishes and maintains rapid apical growth during hyphal formation in C. albicans is not well understood. However, based on our knowledge of how polarized growth occurs in other systems, it is generally accepted to involve the asymmetric deposition of membrane and cell wall at the growing tip. Studies from Saccharomyces cerevisiae have established that in response to certain spatial or positional cues, Golgi-derived secretory vesicles that fuse with the plasma membrane at random sites during isotropic growth of the mother cell are retargeted to fuse at a specified site, resulting in bud emergence and apical growth (for review, see Lew and Reed, 1995; Finger and Novick, 1998). The process of budding requires many proteins that regulate site selection, reorganization of the actin cytoskeleton, and polarization of the secretory apparatus (for review, see Pruyne et al., 2004b). Hyphal growth in C. albicans presents an additional challenge owing to the requirement to rapidly deliver materials over a long distance. Rapid hyphal formation and elongation are crucial for the success of C. albicans as a pathogen, so mechanisms that regulate efficient secretion are likely to be vital for its fitness and pathogenicity in the host environment.
Several conserved proteins that organize the actin cytoskeleton to orient polarized secretion in other organisms are known to be required for hyphal formation in C. albicans. Among these, the formins play a key role as downstream effectors that link Rho-GTPase signaling to the remodeling of the actin cytoskeleton (Kohno et al., 1996; Tominaga et al., 2000; Evangelista et al., 2002; Sagot et al., 2002). Formins function in vivo and in vitro as actin cable nucleators (Evangelista et al., 2002; Pruyne et al., 2002) and this actin nucleating activity is triggered by Rho-GTPases (see Evangelista et al., 2003). The S. cerevisiae formin Bni1 binds activated forms of Rho1 and Cdc42 through its N terminus (Kohno et al., 1996; Evangelista et al., 1997). Among other proteins, Bni1 also binds Spa2 (Fujiwara et al., 1998), which is the scaffolding component of a multiprotein structure termed the polarisome that localizes to the growing tip in a cell cycle-dependent manner (Sheu et al., 1998). These regulated interactions that recruit Bni1 to the growing tip and coordinate its activity with the cell cycle ensure that actin nucleation, and hence polarized secretion, occurs at the right time and place. As in S. cerevisiae, C. albicans encodes two partially redundant formins, CaBni1 and CaBnr1. Simultaneous loss of both formins leads to lethality, and even though both proteins can be found at the hyphal tip, Bni1, but not Bnr1, is required for normal hyphal progression (Li et al., 2005; Martin et al., 2005).
Although budding in S. cerevisiae has proved to be a useful paradigm for studying polarized growth, several observations suggest that there are some fundamental differences between the regulation of budding in S. cerevisiae and hyphal formation in C. albicans (for review, see Sudbery et al., 2004). First, budding in S. cerevisiae is tightly coupled to the cell cycle (Lew and Reed, 1995), whereas in C. albicans the initiation and elongation of hyphae is regulated independently of the cell cycle (Hazan et al., 2002). Second, unlike budding yeast, the apex of C. albicans hyphal cells contains a high density of small vesicles and other unidentified membranous structures, defined almost 80 years ago as the Spitzenkörper (“tip body”), which is thought to act as the supply center of secretory vesicles whose localization and directed deposition are essential for tip growth (Howard, 1981; Reynaga-Pena et al., 1997). These membranous structures are thought to be Golgi-derived secretory vesicles poised to fuse at the growing hyphal tip, but the composition of the Spitzenkörper remains largely uncharacterized. Studies of several filamentous fungi suggest that Bni1 in hyphal cells colocalizes with the Spitzenkörper and not the polarisome, highlighting yet another difference between yeast and hyphal cells (Sharpless and Harris, 2002; Crampin et al., 2005; Martin et al., 2005). Finally, in S. cerevisiae, polarization and the targeting of post-Golgi vesicles to the selected growth site is an actin-based process and does not require microtubules. Although the actin cytoskeleton is essential for hyphal formation in C. albicans, there are conflicting reports of the role of microtubules in this process (Yokoyama et al., 1990; Akashi et al., 1994). In other filamentous fungi, microtubules are important for rapid hyphal growth (Howard and Aist, 1977; That et al., 1988; Temperli et al., 1990; Raudaskoski et al., 1994; Steinberg et al., 2001), and it has been proposed that they are responsible for the long-distance transport of post-Golgi secretory vesicles to the Spitzenkörper, whereas actin filaments control short-range vesicle transport from the Spitzenkörper to the plasma membrane (Crampin et al., 2005; Harris et al., 2005).
To examine the role of the secretory pathway and the cytoskeleton during hyphal formation in C. albicans, we analyzed several different epitope-tagged reporters of the Golgi, endoplasmic reticulum (ER), and vacuole, in live cells and by indirect immunofluorescence. We report that in contrast to other polarized cells that achieve long-distance transport of post-Golgi secretory vesicles on cytoskeletal tracks, C. albicans has evolved an alternative and additional means of establishing polarity, whereby the majority of the Golgi complex is redistributed to and maintained at the distal portion of the hyphae near the growing apical tip. Our studies also demonstrate an additional, previously unrecognized role for the actin cable-nucleating formin Bni1, in localizing the Golgi complex at the hyphal tip, and in maintaining the structural integrity of the Golgi during the yeast-to-hyphal transition.
MATERIALS AND METHODS
Strains, Media, Growth Conditions, and Transformation Protocols
The C. albicans strains used in this study are listed in Table 1. For the characterization of Vrg4-GFP in S. cerevisiae, the chromosomal locus in W303a (MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3112 can1-100) was tagged with green fluorescent protein (GFP) by polymerase chain reaction (PCR)-mediated recombination by using the pFA6a-GFP-HIS3 plasmid as template (Longtine et al., 1998). Strains were routinely grown on YPAD (1% yeast extract, 2% Bacto Peptone, 50 mg/l adenine sulfate, 2% glucose) or a minimal (SD) medium (0.67% yeast nitrogen base without amino acids supplemented with 2% glucose) supplemented appropriately. Growth of cells used for immunofluorescence was in YPAD, because we found that addition of uridine to the growth medium resulted in an increased level of autofluorescence.
Table 1.
C. albicans strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| CAI4 | ura3Δ:: λimm434/ura3Δ:: λimm434 | Fonzi and Irwin |
| BWP17 | ura3Δ:: λimm434/ura3Δ:: λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | Wilson et al. (1999) |
| WYL2 | BWP17 bni1Δ::ARG4/bni1Δ::HIS1 | Li et al. (2005) |
| PRC1 | WYL2 bni1Δ/bni1Δ rp10::VRG4-GFP-URA3 | This study |
| PRC2 | WYL2 bni1Δ/bni1Δ rp10::ALG1-HA3-URA3 | This study |
| PRC3 | WYL2 rp10::ER-GFP-URA3 | This study |
| WYZ2 | BWP17 spa2Δ::ARG4/spa2Δ::HIS1 | Zheng et al. (2003) |
| PRC4 | WYZ2 spa2Δ/spa2Δ rp10::VRG4-GFP-URA3 | This study |
| ANC5 | CAI4 rp10::VRG4-GFP-URA3 | This study |
| ANC6 | CAI4 rp10::VRG4-HA3-URA3 | This study |
| ANC7 | CAI4 rp10::VRG4-myc3-URA3 | This study |
| ANC8 | CAI4rp10::ALG1-HA3-URA3 | This study |
| ANC9 | CAI4 rp10::ER-GFP-URA3 | This study |
| ANC10 | CAI4 rp10::GDA1-HA3-URA3 | This study |
| ANC11 | CAI4 rp10::GDA1-GFP-URA3 | This study |
| ANC14 | CAI4 rp10::TUB1-GFP-URA3 | This study |
| PRC5 | CAI4 TUB2-GFP-URA3 | This study |
| YMG7139 | BWP17 MLC1/MLC1-YFP::URA3 | Crampin et al. (2005) |
| YGM6709 | BWP17 ABP1/ABP1-YFP::URA3 | J. Berman, University of Minnesota, (St. Paul, MN) |
To induce hyphal formation, cultures were grown overnight at 30°C in YPAD to stationary phase, diluted the following day to an OD600 of 0.4 in YPAD containing 20% bovine calf serum, and incubated at 37°C for various times. For longer time-course experiments, cells were grown overnight in YPAD into stationary phase and diluted to an OD600 of 0.5–1.0 before seeding them on coverslips. This protocol enabled the growth of individual, long hyphal cells that were otherwise too tangled to view individually. Seeded coverslips were placed in 24-well plates containing prewarmed YPD + 20% bovine calf serum and incubated at 37°C for the durations indicated.
For induction of pseudohyphae, cultures were grown overnight at 30°C in YPAD to stationary phase. They were diluted the following day to an OD600 of 0.4 in YPAD buffered at pH 6.0 with citric acid and incubated at 36°C (Sudbery et al., 2004). Transformation of C. albicans to integrate linearized plasmids into chromosomal loci was carried out as described previously (Walther and Wendland, 2003).
Plasmid Constructions
Plasmids and their relevant features used in this study are listed in Table 2. The construction of all plasmids used to express either GFP-, myc-, or influenza hemagglutinin (HA)-tagged proteins was based on CIp10, a URA3 integration plasmid that allows efficient integration of the target gene into the chromosomal RP10 locus (Murad et al., 2000). CIp10-HA3 and CIp10-myc3 allow C-terminal protein tagging with three tandem copies of the HA or myc epitope. These plasmids were made by ligating the SacI/NotI (blunted) fragment of either pSK-HA3(P/X) or pSK-Myc3(P/X) (Neiman et al., 1997) to the SacI/EcoRV fragment of CIp10.
Table 2.
Plasmids used in this study
| Plasmid | Relevant features | Source |
|---|---|---|
| CIp10-HA3 | pBS polylinker upstream HA3 epitope in URA3 integration vector | This study |
| CIp10-myc3 | pBS polylinker upstream myc3 epitopein URA3 integration vector | This study |
| CIp-ADHp-GFP | yEGFP driven by ADH1 promoter in URA3 integration vector | This study |
| CIpV1-GFP | GFP-tagged CaVRG4, driven by endogenous promoter in URA3 integration vector | This study |
| Clp-V1-HA3 | HA3-tagged CaVRG4, driven by endogenous promoter in URA3 integration vector | This study |
| Clp-V1-myc3 | myc3-tagged CaVRG4, driven by endogenous promoter in URA3 integration vector | This study |
| CIp-ER-GFP | HDEL-tagged KAR2-GFP fusion, driven by ADH1 promoter in URA3 integration vector | This study |
| CIp-ALG1-HA | HA3-tagged CaALG1, driven by endogenous promoter in URA3 integration vector | This study |
| CIp-GDA1-GFP | GFP-tagged CaGDA1, driven by endogenous promoter in URA3 integration vector | This study |
| CIp-OCH1-myc3 | myc3-tagged CaOCH1, driven by endogenous promoter in URA3 integration vector | This study |
| CIp-GDA1-HA | HA3-tagged CaGDA1, driven by endogenous promoter in URA3 integration vector | This study |
| CIp-TUB1-GFP | GFP-tagged CaTUB1, driven by endogenous promoter in URA3 integration vector | C. Gale, University of Minnesota |
| CIp-TUB2-GFP | GFP-tagged CaTUB2, driven by endogenous promoter in URA3 integration vector | W. Yue, IMCB, Singapore |
Genes encoding the various reporter proteins (CaVRG4, CaKAR2, CaGDA1, CaALG1, and CaOCH1) used in this study were generated by PCR by using CAI4 genomic DNA as template. Primers included restriction enzyme sites for in-frame ligation of each of these genes to the sequences encoding HA3, myc3, or yEGFP, in CIp10-HA3, CIp10-myc3 CIp10-GFP. Primers and their sequences are available on request. The correct sequence of all of these fusion genes was verified by DNA sequencing. Integration of these fusion genes after linearization of the plasmid with Stu1 was confirmed by PCR, and protein expression was determined by Western blotting. The functionality of each reporter was also assessed by complementation of the corresponding S. cerevisiae or C. albicans mutant (Nishikawa et al., 2002; our unpublished data).
pER-GFP encodes a yeast codon-optimized GFP tagged with the ER retention signal HDEL at the COOH terminus and the CaKar2 signal sequence at the NH2 terminus. To construct this plasmid, the CaKAR2 open reading frame (ORF) (lacking a stop codon) was amplified by PCR from genomic DNA purified from CAI4. This Xho1/Cla1 fragment was cloned into the same sites of pSK P/X HA3 (Neiman et al., 1997), which results in the in-frame fusion of the KAR2 ORF and its promoter to sequences encoding three copies of the HA epitope. Sequences encoding HDEL sequences were appended at the 3′ terminus by incorporating these sequences into a reverse primer used to amplify this fragment. This entire KAR2-HA-HDEL fragment was amplified by PCR as an Xho1/Mlu1 fragment and cloned into CIp10-ADH2p to generate CIp-CaKar2-HA-HDEL, which places this gene under the transcriptional control of the ADH2 promoter. After confirming the sequence, expression and ER localization of this fusion protein, the KAR2 ORF in CIp-CaKar2-HA-HDEL was replaced with yEGFP amplified by PCR from pUC19 containing yEGFP (Cormack et al., 1997). This generates an HDEL-tagged GFP that is targeted to the ER by the Kar2 signal sequence and whose expression is driven by the ADH2 promoter. The correct sequence of this KAR2-GFP-HDEL fragment was verified by DNA sequence analysis. The KAR2-GFP-HDEL fusion gene (“pER-GFP”) was integrated at the RP10 locus after linearization with Stu1.
Treatment of Cells with Brefeldin A (BFA), Nocodazole (NZ), and Cytochalasin A (CA)
Wild-type cells expressing CaVRG4-GFP were grown overnight in YPAD to stationary phase and induced to form hyphae for 1.5 h to allow the redistribution of the Golgi to the distal region of hyphae. BFA (Sigma-Aldrich, St. Louis, MO) was added to a final concentration of 80 μg/ml (from a 5 mg/ml stock solution in ethanol) along with 0.06% SDS, whose addition renders the cells more permeable to BFA (Pannunzio et al., 2004). Corresponding volumes of ethanol and SDS were added to the control culture, and both were incubated for an additional 30 min at 37°C with shaking. Samples were harvested and CaVrg4-GFP localization was visualized by fluorescence microscopy.
Microtubules were depolymerized by NZ (Sigma-Aldrich) treatment of yeast or hyphal cells. For examination of yeast cells, overnight cultures of a wild-type strain (BWP17) and TUB2-GFP were diluted to an OD600 of 0.2, and NZ was added (from a 10 mg/ml stock in dimethyl sulfoxide [DMSO]) at the indicated concentrations. Cells were incubated in the presence or absence of NZ for 2 h and 15 min and directly viewed to quantitate visible microtubules. An aliquot of cells was also fixed in 3.7% formaldehyde and stained with 4′, 6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA) to visualize nuclei by fluorescence microscopy. Titration of NZ in both yeast and hyphal cells demonstrated that 5 μM NZ inhibited both nuclear division and microtubule formation. At concentrations higher than 5 μM, cells showed evidence of lysis that increased with increasing NZ concentration in the culture.
For the examination of NZ effects on microtubules in hyphal cells, cells expressing GFP-tagged CaVRG4, CaTUB1, or CaTUB2 were grown overnight in YPAD to stationary phase, diluted, and induced to form hypha in the presence of 5 μM NZ for 1.5 h. Cells were harvested and immediately processed for imaging of CaVrg4-GFP, Tub1-GFP, and Tub2-GFP or fixed with 3.7% formaldehyde and stained with DAPI to visualize nuclei after NZ treatment.
To inhibit actin cable formation, C. albicans or S. cerevisiae cells were treated with CA (Fisher Scientific, Pittsburgh, PA). Overnight C. albicans cell cultures (OD600 of 30–35) were diluted and induced to form hyphae in the presence or absence of various concentrations of CA (stock solution of 5 mg/ml in DMSO) ranging from 1 to 100 μg/ml CA and 0.06% SDS to facilitate the uptake of CA into the cells. For the treatment of S. cerevisiae cells, overnight cultures (OD600 of ∼10) were diluted to 0.5 OD600 units/ml and grown for 2 h before the addition of 5 μg/ml CA. S. cerevisiae VRG4-GFP cells were incubated for 30 min before visualization of GFP. Corresponding volumes of SDS and DMSO were added to the untreated cells. C. albicans strains were incubated for variable times as indicated. It should be noted that our experiments demonstrated that CA is unstable in YPAD over hours. Thus, in all CA experiments, cells were harvested and replenished with fresh CA-containing medium at the appropriate concentration every 30 min, during which time CA retains activity (our unpublished data).
Microscopy, Indirect Immunofluorescence, and Fluorescent Staining Techniques
We used a fluorescence microscope (Zeiss Axioskop model 2 plus; Carl Zeiss, Thornwood, NY) equipped with a 100×/1.3 oil objective (Plan-neofluor), a high-performance charge-coupled device camera (DAGE-MTI, Michigan City, IN), and Scion Image software version 4.50 (Carl Zeiss) for image capture. Single focal plane images were processed using Adobe Photoshop CS, version 8 (Adobe Systems, Mountain View, CA) or Canvas, version 9 (ACD Systems of America, Miami, FL).
Indirect immunofluorescence microscopy was performed as described previously (Dean et al., 1997) with the following modifications. After inducing hyphal growth for various periods as indicated, cells in medium were fixed using formaldehyde for 30 min at 30°C, followed by an additional 3-h incubation in 100 mM potassium phosphate buffer containing 3.7% formaldehyde. Fixed spheroplasts were incubated with rabbit polyclonal anti-HA antibody (Y-11; Clontech, Mountain View, CA), diluted 1:200, or with mouse 9E10 monoclonal anti-myc antibody, diluted 1:10. After incubation with the primary antibody cells were washed and incubated with Alexa 488-conjugated anti-rabbit or anti-mouse IgG antibody (Invitrogen, Carlsbad, CA) diluted 1:300. After washing extensively with phosphate-buffered saline (PBS) containing 0.2% bovine serum albumin, cells were resuspended in mounting media and applied to slides before imaging with a fluorescein isothiocyanate (FITC) filter.
Cell membranes were stained with the lipophilic dicarbocyanine dye DiOC6 (Eastman Kodak) at a final concentration of 10–100 ng/ml, which results primarily in mitochondrial membrane staining (Koning et al., 1993 Pringle et al., 1989; Weisman et al., 1990, Walther and Wendland, 2004). A stock solution of 3,3-dihexyloxacarbocyanine iodide (DiOC6) (10 mg/ml in ethanol) was added directly to cells (107 cells/ml in YPAD) and imaged using an FITC filter set. The lipophilic fluorescent probe MDY-64 (Invitrogen) was used as a fluorescent marker to visualize yeast vacuolar membrane (Cole et al., 1998; Fratti et al., 2004) and used at a final concentration of 10 μM, according to the manufacturer's protocol (Invitrogen).
For imaging of GFP-tagged proteins, cells were washed once with PBS, resuspended in PBS, and placed on ice for 10–30 min before visualization by fluorescence microscopy. GFP fluorescent patterns were the same in the presence or absence of the PBS wash or incubation on ice, except for a higher level of background fluorescence that is presumably due to the presence of YPAD. For longer time-course experiments CaVRG4-GFP–expressing cells were induced to form hyphae by seeding on coverslips, as described above. For each time point, cells were washed twice in PBS, stained with DAPI for 5 min in the dark, and washed again in PBS before staining with 1 μg/ml calcofluor white (CW; fluorescent brightener 28, Sigma-Aldrich) solution for 1 min in the dark. The coverslips were washed with PBS and mounted on slides for fluorescence microscopy. An image was captured in a representative focal plane. In images where there was no apparent fluorescence in a particular portion of the cell, the cells were visually examined thoroughly in all focal planes to ensure the absence of any fluorescent signal in that region of the cell. For time points where the hyphal length exceeded the dimensions of the field that could be captured on the microscope, two to three images were acquired to cover the entire length of the hypha and were overlaid using Adobe Photoshop, version 7.0.
For phalloidin staining of actin, cells in medium were fixed by the addition of formaldehyde to a final concentration of 3.7%. After a 30-min incubation cells were resuspended in 50 mM potassium phosphate buffer, pH 6.8, and fixed for an additional hour in this buffer containing 3.7% formaldehyde. Cells were harvested, washed once in potassium phosphate buffer and resuspended in this buffer containing 0.1% Triton X-100, and incubated for an additional 30 min. Fixed cells were washed twice with PBS and incubated with a 1:10 dilution of Alexa 546-phalloidin (Invitrogen) in PBS overnight at 4°C. Cells were washed twice with PBS, mounted on slides, and visualized with a Texas Red filter.
Subcellular Fractionation of HA-tagged Proteins
CaVrg4-HA-containing membrane organelles in wild-type or bni1Δ/bni1Δ mutant cells were analyzed by a combination of differential centrifugation, sucrose equilibrium density, and velocity sedimentation. Twenty-five milliliters of YPAD was inoculated with overnight cultures and induced to form hyphae for 1.5 h. Cells were collected by centrifugation at 3000× g, washed once with JB buffer (HEPES-KOH, pH 6.8, 150 mM KCl, 0.1 M sorbitol, 1 mM EDTA, and 1 mM dithiothreitol [DTT]), resuspended in 500 μl of JB buffer containing a cocktail of protease inhibitors, and broken by glass bead lysis. Unlysed cells and debris were removed by centrifugation at 3000× g. Heavier membranes (ER, vacuole, and plasma membrane) were removed by centrifugation at 14,000× g for 20 min at 4°C, and the low-density membranes that remain in the supernatant (S14), including Golgi and vesicles, were isolated by centrifugation at 100,000× g (P100) for 30 min at 4°C. The resulting pellet (P100) that was greatly enriched for CaVrg4-HA was resuspended in 200 μl of JB buffer lacking sorbitol and analyzed by either sucrose equilibrium density or velocity gradients. Then, 150 μl of the P100 fraction was layered onto 2 ml of 22–60% sucrose gradient (wt/vol in JB buffer lacking sorbitol) for equilibrium density centrifugation as described previously (Vida et al., 1990) and centrifuged at 135,00× g for 18 h at 4°C. Then, 200-μl fractions were collected from the top. Aliquots of each fraction (20 μl) were analyzed directly by SDS-PAGE and immunoblotted with 12CA5 anti-HA antibody.
Alternatively, for velocity sedimentation analysis, the P100 fraction was layered onto 2-ml sucrose gradients of 0.2-ml steps of 5–25% sucrose (wt/vol in JB buffer lacking sorbitol) with a 60% sucrose cushion. Sucrose gradients were centrifuged at 135,000× g in a TLS55 rotor for 30 min at 4°C. Then, 200-μl fractions were collected from the top, and 20 μl fractions were analyzed by SDS-PAGE and immunoblotted with anti-HA antibody.
Whole cell protein extracts were prepared by the trichloroacetic acid/ β-mercaptoethanol procedure or by lysis with glass beads, exactly as described previously (Nishikawa et al., 2002), resolved by SDS-PAGE, transferred to Immobilon-polyvinylidene difluoride membranes (Millipore, Billerica, MA) and detected using anti-HA antibody. Primary antibody incubation was followed by incubation with a secondary anti-mouse antibody conjugated to horseradish peroxidase (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) followed by chemiluminescence (ECL; GE Healthcare).
RESULTS
The Golgi Complex Relocates to the Distal Hyphal Tip during Hyphal Formation
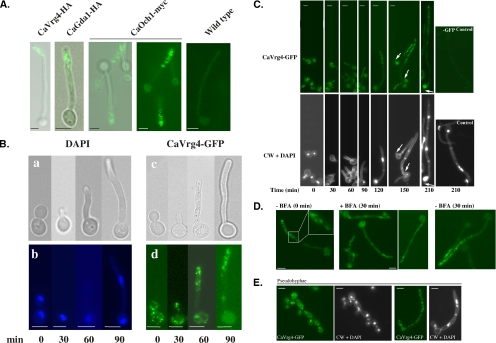
The C. albicans GDP-mannose transporter encoded by the CaVRG4 gene is a Golgi-localized protein (Nishikawa et al., 2002). Although its enrichment in a particular cisternae has not been established, owing to its abundance and its essential role in providing sugar substrates for cis-, medial-, and trans-localized Golgi glycosyltransferases, Vrg4 is a useful reporter for visualizing the yeast Golgi (Dean et al., 1997; Gao and Dean, 2000; Losev et al., 2006). Using epitope-tagged CaVrg4 as a Golgi reporter, we previously showed that in C. albicans yeast cells, the Golgi seems morphologically similar to that of S. cerevisiae, consisting of distinct, rod-like spots that are randomly distributed throughout the cell (Nishikawa et al., 2002). To examine the morphology of the Golgi during filamentous growth, the localization of CaVrg4-HA was analyzed by indirect immunofluorescence after the induction of hyphae. These cells expressed a single copy of the CaVRG4-HA gene, driven by its own promoter, and integrated at the RP10 locus. This allele complements C. albicans vrg4 null mutants, demonstrating its functionality (Nishikawa et al., 2002). After hyphal induction, aliquots of cells were removed at various times and processed for indirect immunofluorescence. In yeast cells, an average of 10 randomly distributed spots were observed in each cell (Nishikawa et al., 2002; our unpublished data). In stark contrast to this random distribution, much of the fluorescence seemed concentrated at the distal tip of the germ tube after 90 min of hyphal induction (Figure 1A). A similar pattern of Golgi distribution was seen in hyphal cells expressing CaVRG4-myc (our unpublished data), and other resident Golgi proteins, including CaGda1-HA, the Golgi guanosine diphosphatase (Herrero et al., 2002), and CaOch1-myc, a Golgi mannosyltransferase (Bates et al., 2006) (Figure 1A). The fluorescence remained associated with the distal portion of the tip and was largely absent from the cell body (compare CaOch1-myc viewed by phase contrast with that viewed by fluorescence; Figure 1A). These results demonstrate that the polarized pattern of Golgi distribution at the distal portion of the germ tube is not an artifact of indirect immunofluorescence arising from a particular antibody or due to a peculiarity of a particular Golgi protein.
Figure 1.
Polarized localization of the Golgi to the apical tip of hyphal cells. (A) Indirect immunofluorescence of Golgi proteins in hyphal cells. Cells (BWP17) expressing Ca-VRG4-HA, CaGDA1-HA, or CaOCH1-myc were induced to form hyphae and processed for indirect immunofluorescence with anti-HA antibodies as described in Materials and Methods. The last panel shows the background signal in an untagged control strain. (B) Localization of CaVrg4-GFP in hyphal cells. Cells expressing CaVRG4-GFP were induced to form hyphae, removed at the indicated times, and viewed directly for CaVrg4-GFP localization or stained with DAPI to visualize nuclear division. (C) Branching is accompanied by accumulation of Vrg4-GFP. Cells expressing CaVRG4-GFP were induced to form hyphae on coverslips. At the indicated time points, cells were stained with DAPI for visualization of nuclei and with calcofluor white for visualization of cell walls and septa. CaVrg4-GFP was visualized by fluorescence microscopy. The last panel shows the background fluorescence in a strain where Ca-VRG4 is not GFP-tagged. (D) Brefeldin A treatment of hyphal cells expressing CaVrg4-GFP. Cells expressing CaVRG4-GFP were induced to form hypha for 1.5 h to allow the redistribution of the Golgi to the distal region of hyphae. An aliquot was removed (−BFA; 0 min), and cells were incubated in the presence or absence BFA for a further 30 min before harvesting to visualize CaVrg4-GFP by fluorescence microscopy. The inset shows some of the cisternae in a wild-type hyphal cell at higher magnification. (E) Random distribution of Golgi puncta in pseudohyphal cells. For induction of pseudohyphae, cells expressing CaVRG4-GFP were incubated at 36°C in YPD buffered at pH 6.0 with citric acid. For each time point, cells seeded on coverslips were stained with DAPI and then with calcofluor white. The coverslips were mounted on slides for fluorescence microscopy. Bars, 5 μm.
The localization of GFP-tagged Vrg4 protein was also analyzed in live cells. A strain was constructed that expressed a single copy of CaVRG4-GFP whose expression was driven by the VRG4 promoter and integrated at the RP10 locus. CaVRG4-GFP–expressing cells were removed at various times after hyphal induction and imaged by microscopy. Similar to the pattern of CaVrg4-HA seen using indirect immunofluorescence, we observed characteristic Golgi “spots” that were randomly distributed in yeast cells but that clustered and remained largely associated with the extending tip during hyphal growth. In live cells, after 90–120 min of hyphal induction, in 80% of the cells that had undergone at least one nuclear division, the punctate fluorescence was localized within the top one-third of the extended germ tube (Figure 1B). As observed by indirect immunofluorescence, a notable feature of the observed Vrg4-GFP fluorescence pattern in these hyphal cells was the near absence of punctate staining in the cell body, although we occasionally observed some diffuse background staining in this region of the cell. To determine whether this absence of punctate staining in the cell body may be attributable to the quiescence of the proximal cells, we examined the Vrg4-GFP Golgi distribution in cells that were induced to form hyphae for sufficiently long times (up to 210 min) to allow branch formation. After hyphal induction, aliquots of cells were removed at various times and stained with CW to examine cell wall and septa, and with DAPI to examine nuclei (Figure 1C). The result of this experiment demonstrated that the Vrg4-GFP signal was restricted to the distal portion of the hyphae, even in cells that had undergone two nuclear divisions. However, concomitant with branch initiation, we always observed an abundance of Vrg4-GFP signal in the cell that the new branch arose from (n > 80), suggesting a strong correlation between branch formation and Golgi presence in the branching cell.
To confirm that the CaVrg4-GFP staining pattern reflected the labeling of Golgi cisternae, hyphal cells expressing VRG4-GFP were treated with BFA. BFA causes a reversible disassembly of the Golgi complex and the redistribution of Golgi enzymes into the ER. In cells treated with BFA, CaVrg4-GFP no longer seemed as distinct rod-like structures that concentrate in the distal region of the hyphae (Figure 1D). Instead, CaVrg4-GFP redistributed to a reticular pattern of membranes throughout the hypha and mother cell, in a pattern resembling the ER in hyphal cells (Figure 2C). BFA treatment also caused a substantial fluorescent signal that colocalized with vacuolar markers (our unpublished data) and interfered with the mesh-like distribution of the CaVrg4-GFP signal due to its ER distribution. Nevertheless, the BFA-dependent marked change in the distribution of the CaVrg4-GFP signal provided evidence that the fluorescent signal in cells expressing VRG4-GFP indeed reflected the in vivo distribution of the Golgi.
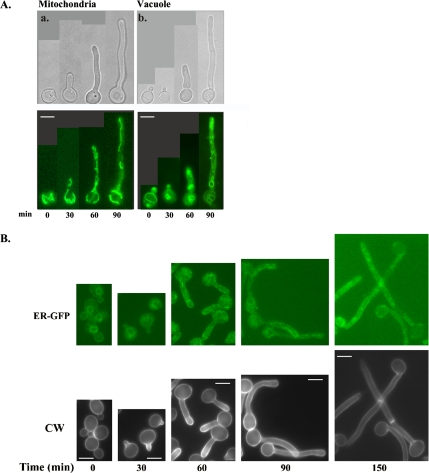
Figure 2.
ER, mitochondria, and vacuoles are randomly distributed in hyphal cells. Cells (BWP17) were induced to form hyphae by growth at 37°C and the addition of serum. Aliquots of cells were removed every 30 min and processed for fluorescence microscopy. (A) Mitochondria (a) were visualized by staining cells with DiOC6. Vacuoles (b) were visualized using MDY-64. (B) ER was visualized in a strain expressing an HDEL-tagged Kar2-GFP fusion protein (ER-GFP; see description in Materials and Methods). Cells expressing ER-GFP were induced to form hyphae, removed at the indicated times, and viewed directly for GFP localization, or stained with CW to visualize cell wall and septa as described in Materials and Methods. Bars, 5 μm.
The restricted localization of CaVrg4-GFP to distal sites of polarized growth is seen in hyphal but not budding C. albicans cells. To determine whether this Golgi localization pattern is unique to hyphal cells, we also examined the distribution of Vrg4-GFP in cells that were induced to form pseudohyphae by incubation at 36°C in media buffered at pH 6.0 (Sudbery et al., 2004). Under these conditions the pattern of the Golgi remained punctate and seemed randomly distributed throughout the cell, with no evidence of polarity in pseudohyphal cells, even very long ones that superficially resemble hyphal cells (Figure 1E). These results demonstrate that the polarized localization of the Golgi to the active growth site is a hyphal-specific event.
The spatially restricted distribution of the Golgi in the distal portion of the filament in hyphal cells was unique to Golgi membranes and not observed for other organelles (Figure 2). For example, mitochondria (DiOC6 staining; Figure 2A), vacuoles (MDY-64 staining; Figure 2A), and ER (“ER-GFP” fluorescence; Figure 2B) were distributed throughout the cell and germ tube in both yeast and in hyphal cells. Even in cells that had been induced to form hyphae for 150 min in which there was clear evidence of septation, the ER remained randomly distributed (Figure 2B). This ER pattern was in marked contrast to that of the Golgi, which was largely absent from the cell body, except for those cells that were undergoing branch formation. These results also imply that the quiescence of these unbranched proximal cells seemed selective for the Golgi but not for the ER. This same, random distribution was also observed for another ER protein, Sec12-GFP (our unpublished data). Together, these results demonstrate that there is a marked redistribution of the Golgi during the transition of yeast cells to hyphal cells, in which the Golgi is largely absent from the cell body and remains associated with the distal hypha as it extends.
Golgi Polarization to the Distal Portion of the Hyphae Is Not Dependent on Microtubules
To explore the mechanism by which the Golgi repolarizes itself during hyphal formation, we wanted to determine whether the organelle's distal localization depended on cytoskeletal components. The requirement of microtubules for hyphal formation in C. albicans was tested by treating cells with NZ, which causes microtubule depolymerization. The efficacy of inhibition of microtubules by NZ was assayed visually, by monitoring the absence of microtubule formation in a TUB1-GFP (encoding α-tubulin) and TUB2-GFP (encoding β-tubulin) strain (Figure 3), and by quantitatively monitoring inhibition of nuclear division due to the consequent activation of the spindle checkpoint (Table 3). Treatment with 5 μM NZ resulted in a 10-fold reduction in the number of cells within the population that successfully divided their nuclei. In addition, almost all of the cells in the treated samples arrested as large budded cells (97 verses 51.7% in the untreated control). These results demonstrate that this concentration of NZ efficiently inhibited microtubule function and therefore arrested nuclear division. Consistent with these results, under these conditions of NZ treatment, microtubules in TUB1-GFP and TUB2-GFP hyphal cells could not be detected visually, although they were easily detected in the untreated control cells (Table 3 and Figure 3). The only remaining fluorescent signal visible in these NZ-treated cells were spots in the cell body, which likely represent the microtubule organizing centers (Figure 3). Together, these results demonstrate that NZ efficiently inhibited microtubule function and formation in these cells. Importantly, this inhibition of microtubule formation by NZ affected neither the ability of cells to form hyphae nor their rate of formation (Figure 3).
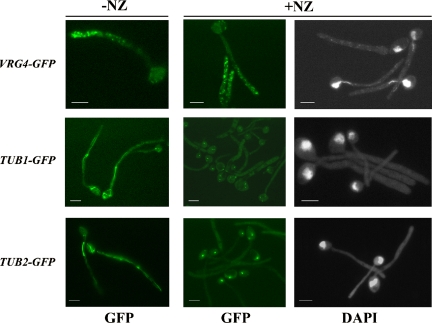
Figure 3.
Redistribution of Golgi to the distal region of hyphae does not require microtubules. Cultures of wild-type cells expressing either CaVRG4-GFP, CaTUB1-GFP, or CaTUB2-GFP were induced to form hyphae in the presence (+NZ) or absence (−NZ) of 5 μM NZ. Samples were harvested after 1.5 h of hyphal induction and stained with DAPI for the visualization of CaVrg4-GFP, Tub1-GFP and Tub2-GFP or nuclei, as described in Materials and Methods. Bars, 5 μm.
Table 3.
NZ (5 μM) depolymerizes microtubules and inhibits nuclear division in C. albicans
| Cells | % Cells with at least one visible microtubule | % Cells with divided nuclei | % Large-budded cells |
|---|---|---|---|
| (−) Nocodazole | 100.0 (n = 384) | 27.3 (n = 300) | 51.7 (n = 300) |
| (+) Nocodazole | 3.3 (n = 272) | 2.7 (n = 289) | 97.0 (n = 300) |
Cells expressing Tub2-GFP were grown overnight into stationary phase in YPD, diluted to an OD600 of 0.3, and allowed to grow in the presence or absence of 5 μM nocodazole at 30°C for 2 h 15 min before harvesting. Cells were washed once in PBS, and microtubules were visualized and quantitated directly by fluorescence microscopy. Large-budded cells were cells where the daughter cell was at least two-thirds the size of the mother cell. The status of nuclear division was assessed by staining cells with DAPI.
Although microtubules seemed to be efficiently depolymerized in the presence of 5 μM NZ, CaVrg4-GFP was still redistributed to the distal tip of hyphae in 85% (n > 300) of the NZ-treated cells (Figure 3). As in the untreated control, almost no CaVrg4-GFP signal could be observed in the body of cells treated with NZ. We also found that if cells were first allowed to form hyphae and then treated with 5 μM NZ, the CaVrg4-GFP signal was still restricted to the distal region of the hypha. Furthermore, although the Tub1- and Tub2-GFP signals in untreated control cells were sensitive to cold (i.e., the signal was lost over time if samples were kept on ice for several hours), the CaVrg4-GFP signal was not (our unpublished data). Together, these results demonstrate that microtubules do not play a role in transporting or in maintaining the Golgi near the growing hyphal tip.
Golgi Positioning and Integrity in Cells Forming Hyphae Require Actin and the Formin CaBni1
To examine the requirement of actin for Golgi polarization, we first assayed the effect of treatment of cells with CA whose effect on cell polarity in C. albicans is well established (Akashi et al., 1994; Crampin et al., 2005). CA is a fungal metabolite that binds to the barbed ends of actin filaments and to actin monomers and thereby inhibits new filament formation (Cooper, 1987). To identify a concentration of CA that was sufficient to disrupt the organization of the actin cytoskeleton in C. albicans cells, wild-type cells grown overnight into stationary phase were diluted and induced to form hyphae in serum containing various concentrations of CA ranging from 1 to 100 μg/ml CA, and 0.06% SDS to facilitate the uptake of CA into the cells. Consistent with previous reports, addition of 20 μg/ml CA was sufficient to abolish both budding and hyphal formation in C. albicans cells and >98% of cells in this culture were round and unbudded at the endpoint (our unpublished data; Akashi et al., 1994).
Because this concentration of CA abolished hyphal formation, we were unable to address the question of whether Golgi redistribution of the to the distal region of hyphae required the actin cytoskeleton. However, we could address whether an intact actin cytoskeleton is required to maintain the Golgi at the distal region once hyphae have formed. To establish the efficacy of CA as an actin inhibitor in hyphal cells, its effect on actin structure and function was assessed by 1) visualizing the actin cytoskeleton directly in Alexa 546-phalloidin–stained hyphal cells; 2) visualizing the localization pattern of yellow fluorescent protein (YFP)-tagged myosin light chain protein 1 (Mlc1-YFP), a Spitzenkörper component whose hyphal tip localization relies on an intact actin cytoskeleton (Crampin et al., 2005); and 3) visualizing actin indirectly in hyphal cells expressing YFP-tagged actin binding protein 1 (Abp1-YFP), which localizes predominantly to cortical actin patches in C. albicans yeast and hyphal cells (Berman and Gerami-Nejad, unpublished data). As reported previously (Hazan et al., 2002), actin in hyphal cells (assayed by phalloidin-stained cells and by visualization of fluorescence in the Abp1-YFP strain) was primarily detected as bright cortical patches associated with the distal region of the growing hyphae (Figure 4, A and B). Actin filaments in hyphal cells seemed much thinner than in yeast cells and although detectable by microscopy, they were difficult to capture in single focus plane images. Importantly, treatment of cells with 20 μg/ml CA abolished hyphal extension and led to a loss of the bright tip localization pattern of phalloidin-stained actin, Abp1-YFP, and Mlc1-YFP (Figure 4, A and B). Thus, although we cannot rule out the remains of some undetectable actin cytoskeleton, these results suggested that this concentration of CA is effective in inhibiting actin function in hyphal cells.
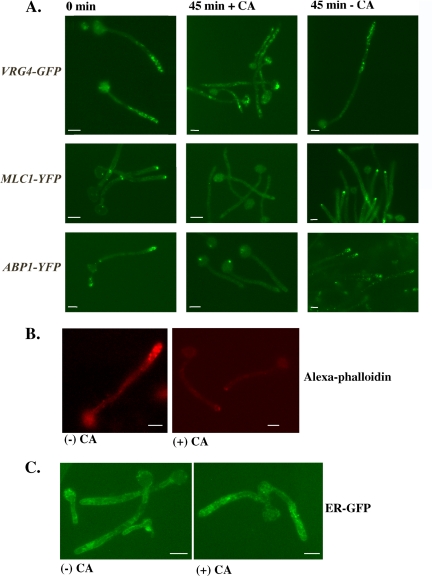
Figure 4.
Maintenance of Golgi organization near the hyphal tip requires an intact actin cytoskeleton. (A) Actin inhibition results in the disorganization of Golgi in hyphal cells. Stationary phase cultures of wild-type cells harboring CaVRG4-GFP, MLC1-YFP, or ABP1-YFP were induced to form hyphae for 1 h 45 min. Cells were treated with buffer alone (DMSO + SDS) (−CA) or buffer containing 20 μg/ml CA (+CA) for 45 min before visualization of cells by fluorescence microscopy. (B) CA inhibits actin organization in hyphal cells. Cells grown as described in A were incubated in the presence or absence of CA, fixed, and stained with Alexa 546-phalloidin as described in Materials and Methods. (C) Treatment of hyphal cells with CA has no effect on ER organization. BWP17 cells expressing ER-GFP were induced to form hyphae and then incubated in the presence or absence of 20 μg/ml CA for 45 min as described in A and visualized by fluorescence microscopy. Bars, 5 μm.
To determine whether actin is required to maintain the polarized Golgi localization at the distal portion of the hyphae, cells were induced to form hypha and then incubated in the presence or absence of CA. Cells producing either CaVrg4-GFP as a Golgi marker, or ER-GFP as an ER marker were induced to form hyphae by serum for 1.5 h. Then, 20 μg/ml CA was added, and the samples were harvested after 45 min for the visualization of the Golgi and the ER by fluorescence microscopy. Although the treatment with CA had no effect on the membrane-like distribution of the ER-GFP signal throughout the length of the hyphae and the cell body (Figure 4C), we observed that CA treatment caused a marked disorganization of the Golgi (Figure 4A). Furthermore, 63% of the CA-treated cells (n > 300) showed a region of CaVrg4-GFP-dependent bright fluorescence in the cell body (Figure 4A), which seemed qualitatively different from the puncta in branching cells and did not colocalize with the DAPI signal, vacuole or ER membranes (our unpublished data). Longer treatment with CA (up to 2 h) resulted in the same fluorescent pattern. Together, these results demonstrated that the maintenance of the Golgi at the distal portion of hyphal cells requires an intact actin cytoskeleton.
To understand better the involvement of the actin cytoskeleton in the apical redistribution of Golgi in hyphal cells, we chose a genetic approach and examined a mutant in which we expected defects in actin-dependent processes. Bni1 and its partially redundant homologue Bnr1 are required for actin nucleation and linear cable assembly at sites of polarized growth (Evangelista et al., 2002; Sagot et al., 2002). Unlike bnr1 mutants, in which hyphal formation is normal, C. albicans bni1 mutants still undergo the yeast-to-hyphal switch, but they exhibit severe polarity defects, forming short, swollen true hyphae that are kinetically delayed (Li et al., 2005; Martin et al., 2005). Despite these hyphal defects, bni1 null mutants are unaffected in budding and germination.
To examine Golgi distribution in the absence of Bni1, a bni1Δ/bni1Δ strain was constructed that expressed a single copy of CaVRG4-GFP, driven by its endogenous promoter and integrated at the RP10 locus. Overnight cultures of the isogenic wild-type parental strain (BWP17) and bni1Δ/bni1Δ VRG4-GFP cells were induced to form hyphae for 1.5 h, and the Golgi was examined in live cells by examining CaVrg4-GFP fluorescence. Unexpectedly, rather than the typical punctate, tip localized pattern of Golgi observed in wild-type cells, the fluorescent signal of CaVrg4-GFP in these bni1Δ/bni1Δ hyphal cells manifested as a haze that was evenly distributed throughout the cells (Figure 5A). In addition to its role as a nucleator of actin cables, Bni1 physically interacts with a number of different proteins, including the polarisome scaffold Spa2. C. albicans spa2Δ/spa2Δ mutants display polarity phenotypes that are similar to those of bni1Δ (Zheng et al., 2003). The loss of CaVrg4-GFP fluorescence was not seen in spa2Δ/spa2Δ mutants, demonstrating that this phenotype was due to loss of BNI1 rather than an indirect effect via the polarisome (Figure 5A). Although bni1Δ cells display polarity defects and are delayed in hyphal formation, true hyphae (i.e., parallel-sided tubes that are clearly septated and whose mother/daughter neck lacks constrictions) are eventually produced (Figure 5B). Even in bni1Δ/bni1Δ cells that had produced elongated hyphae, Vrg4-GFP fluorescence still occurred as nonresolvable haze, thus ruling out the possibility that the absence of Golgi-associated fluorescence is a consequence of the inability of these bni1Δ/bni1Δ cells to form true hyphae.
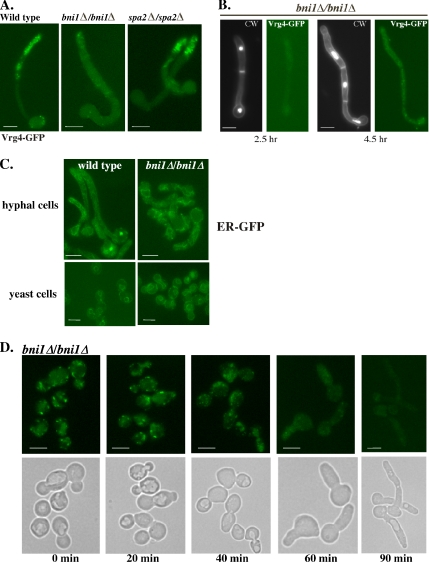
Figure 5.
Bni1 has a hyphal-specific function in the maintenance of Golgi integrity and localization. (A) Golgi disassembly in the bni1Δ/bni1Δ strain. The Golgi was visualized in wild-type (BWP17), bni1Δ/bni1Δ, or spa2Δ/spa2Δ cells harboring CaVRG4-GFP. Overnight cultures of were induced to form hyphae with serum and viewed after 1.5 h of hyphal induction (WT) and 2.5 h for bni1Δ/bni1Δ, spa2Δ/spa2Δ (B) bni1Δ/bni1Δ form true hyphae but are delayed. bni1Δ/bni1Δ cells were induced to form hyphae as in A but harvested after 4.5 h and stained with DAPI and calcofluor white to visualize the nuclei, and cell walls and septa, respectively. (C) Loss of Bni1 has no effect on ER morphology. The ER was visualized in wild-type or bni1Δ/bni1Δ yeast or hyphal cells, harboring pER-GFP. (D) Loss of Golgi puncta in hyphal but not yeast bni1Δ mutant cells. An overnight culture of the bni1Δ/bni1Δ strain harboring VRG4-GFP was diluted and grown into logarithmic phase at 30°C and harvested for microscopy (time, 0 min). These cells were induced to form hyphae and samples were collected at the indicated time points for the visualization of CaVrg4-GFP distribution. Bars, 5 μm.
This pattern of CaVrg4-GFP fluorescence in bni1Δ/bni1Δ hyphal cells was specific to the Golgi, because the ER (observed using the ER-GFP reporter) looked normal and was indistinguishable from the ER pattern in wild-type cells (Figure 5C). Remarkably, this CaVrg4-GFP fluorescent haze was not seen when bni1Δ/bni1Δ cells were growing as yeast cells (Figure 5D, 0-min hyphal induction). Examination of the CaVrg4-GFP Golgi fluorescence in bni1Δ/bni1Δ cells over time after the induction of hyphae demonstrated that the punctate, randomly distributed Golgi spots looked normal in bni1Δ/bni1Δ yeast cells and at early times after induction of hyphae (Figure 5D, times 0 and 20 min) and dispersed only gradually, after the induction of hyphae (Figure 5C, e.g., 20–90 min).
To determine whether this loss of CaVrg4-GFP fluorescence in bni1Δ/bni1Δ cells was due to Vrg4 degradation during hyphal formation, the steady-state level of Vrg4 was compared in bni1Δ/bni1Δ cells during hyphal formation. To facilitate detection of protein, extracts were prepared from bni1Δ/bni1Δ and isogenic wild-type parental strains expressing a CaVRG4-HA-tagged allele. After induction of hypha, aliquots of cells were removed, protein was extracted, and its concentration measured. Equal amounts of protein were analyzed by Western blotting with anti-HA antibody. It should be noted that C. albicans expresses an ∼50-kDa, 12CA5 cross-reacting protein (denoted by the asterisk in Figure 6A, right), which serves as a convenient loading control. In both wild-type cells (Figure 6A, left) and the bni1Δ/bni1Δ mutant strain, the relative amount of Vrg4, normalized to either total protein (Figure 6A) or to dry cell weight (our unpublished data) remained constant over 2 h of hyphal formation. These results ruled out the possibility that the attenuated, dispersed fluorescent signal in the bni1Δ/bni1Δ strain was due to degradation of Vrg4. Instead, these results suggested that the dispersed haze observed in bni1Δ/bni1Δ may be due to the fragmentation or vesiculation of the Golgi.
Figure 6.
Golgi membranes from bni1Δ/Δ cells have the same density as wild-type Golgi, but differ in size. (A) Western blot of CaVrg4-HA in hyphal cells. Wild-type or bni1Δ/bni1Δ mutant cells, expressing CaVRG4-HA, were induced to form hyphae, and protein extracts prepared from aliquots collected over time of induction. Equal amounts of protein extract were separated by SDS-PAGE and immunoblotted with anti-HA antibody. (B) Sedimentation equilibrium analysis of Golgi membranes. Protein extracts were prepared from wild-type or bni1Δ/bni1Δ mutant hyphal cells expressing VRG4- HA and subjected to differential centrifugation. The Golgi-enriched P100 fraction was layered on to a 22–60% sucrose gradient, centrifuged at 135,000× g for 18 h. Fractions were removed from the top, subjected to SDS-PAGE, and analyzed by immunoblotting with anti-HA antibodies. (C) Velocity sedimentation analysis of Golgi membranes. The Golgi enriched P100 fraction from wild-type or bni1Δ/bni1Δ cells was layered on to a 5–25% sucrose gradient and centrifuged at 135,000× g for 30 min. Fractions were removed from the top, subjected to SDS-PAGE, and analyzed by immunoblotting with anti-HA antibodies.
To test this idea, the relative density and size of Golgi membranes from bni1Δ/bni1Δ hyphal cells were compared with those from wild-type cells. Golgi membranes, assayed quantitatively by following Vrg4-HA, were isolated from hyphal whole cell lysates, separated from ER, and enriched by differential centrifugation. Using protocols established for the enrichment of Golgi membranes in S. cerevisiae (see Materials and Methods), we found that Vrg4-containing Golgi membranes from wild-type and bni1Δ/bni1Δ hyphal cells remain soluble, could be separated from CaAlg1-containing ER membranes during sedimentation at 14,000× g, and were quantitatively recovered by sedimentation at 100,000× g (Figure 6B; our unpublished data). The yield of CaVrg4- HA protein recovered in the Golgi fraction from equal cell numbers of the wild-type and bni1Δ/bni1Δ mutant strain was similar, providing further evidence that the dispersed fluorescence in bni1Δ/bni1Δ strains is due to a dispersal of Golgi membranes, rather than degradation of CaVrg4-GFP (Figure 6B). The membranes that pelleted at 100,000× g (P100) were resuspended and subjected to sedimentation equilibrium analysis to compare their relative density, and sedimentation velocity analysis, to compare their size.
After equilibrium density sedimentation was performed, fractions were collected from the top, and the position of Golgi membranes within these gradients assayed by Western blotting each fraction for the presence of CaVrg4-HA. As shown in Figure 6B, the relative position of Golgi-enriched membranes from wild-type and bni1Δ/bni1Δ cells was virtually identical, with the bulk of CaVrg4-HA peaking in fractions 3 and 4. These results suggest that CaVrg4-HA–containing Golgi membranes from bni1Δ/bni1Δ hyphal cells are of a similar density, and hence composition, to those of wild type.
To compare the relative size of these membranes, velocity sedimentation was performed. Fractions were collected from the top and assayed for Vrg4-HA as described above. Under these conditions, Golgi-enriched membranes from bni1Δ/bni1Δ cells fractionated differently than wild type. The peak of Golgi membranes from the wild-type strain sedimented in fractions 7 and 8 and displayed a leading edge toward the bottom (heavier) portion of the sucrose gradient (Figure 6C). The peak of Golgi membranes from the bni1Δ/bni1Δ strain sedimented in fraction five and in the lighter portion of the gradient, and CaVrg4-HA was largely absent from the bottom of the gradient. Under these conditions, ER membranes (monitored by the presence of CaAlg1-HA) from both wild-type and bni1Δ/bni1Δ cells, pelleted at the bottom of this gradient (our unpublished data). These results suggest that Vrg4-HA-containing Golgi membranes from the bni1Δ/bni1Δ strain are of a smaller size than those from the wild-type strain. Together, we conclude that the dispersed haze observed in the bni1Δ/bni1Δ hyphal cells is due to a fragmentation or vesiculation of the Golgi cisternae and therefore that Bni1 is required for maintaining the structural integrity of the Golgi in C. albicans hyphal cells.
To address the question of whether an intact actin cytoskeleton is important for maintaining the structural integrity of the Golgi in yeast cells, we asked whether treatment of wild-type C. albicans cells with CA could mimic the bni1Δ/bni1Δ Golgi dispersal phenotype. Wild-type and the bni1Δ/bni1Δ yeast cells expressing VRG4-GFP were grown to logarithmic phase and treated with 5 μg/ml CA, an intermediate concentration that we determined prevents hyphal formation but not budding (our unpublished data). Aliquots of CA-treated cells were removed every 30 min and viewed for CaVrg4-GFP localization. The results of this experiment demonstrated that this intermediate CA treatment could indeed lead to a Golgi dispersal phenotype in wild-type cells, similar to that seen in bni1Δ mutants. As seen in Figure 7A, after exposure to CA for 30–60 min, wild-type cells displayed a gradual loss of the fluorescence associated with typical bright Golgi puncta and a concomitant increase in the fluorescent haze that is characteristic of the Golgi in bni1Δ hyphal cells. In addition, treatment of bni1Δ/bni1Δ mutant yeast cells with 5 μg/ml CA had a similar effect, but it occurred more rapidly. In bni1Δ/bni1Δ cells, loss of CaVrg4-GFP fluorescence was evident by 30 min of CA treatment (Figure 7A).
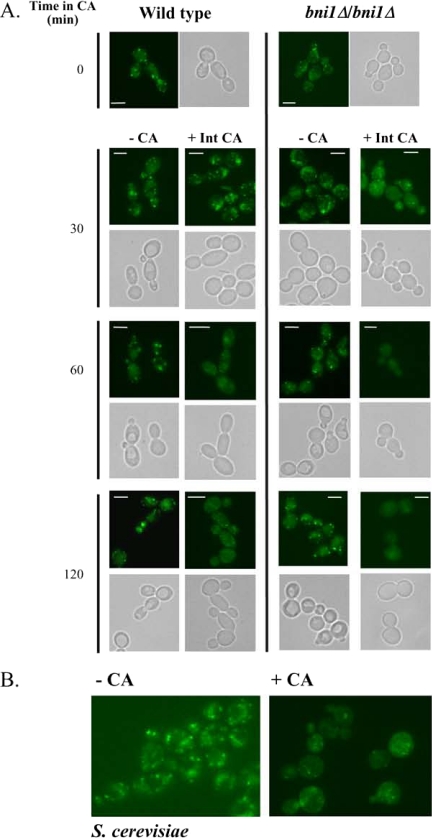
Figure 7.
An intact actin cytoskeleton is essential for preserving the structural integrity of the Golgi in yeast cells. (A) Overnight cultures of wild-type and the bni1Δ/bni1Δ strain harboring VRG4-GFP were grown to logarithmic phase and treated with 5 μg/ml CA in DMSO, 0.06% SDS (+Int CA) or with 0.06% SDS and an equivalent volume of DMSO. Aliquots were removed every 30 min and processed for microscopy. Note that the images of bni1Δ/bni1Δ cells were captured at longer exposure times (1.5- to 2-fold) than the wild type due to their decreased signal intensity. Bars, 5 μm. (B) S. cerevisiae cells expressing VRG4-GFP were grown to logarithmic phase and treated with 5 μg/ml CA in DMSO, 0.06% SDS (+CA) or with 0.06% SDS and an equivalent volume of DMSO (−CA). Cells were visualized after 30 min.
To test whether the requirement of an actin cytoskeleton is generally important for Golgi integrity in yeast cells, we also examined the morphology of the Golgi in S. cerevisiae. A strain expressing Vrg4-GFP as the sole GDP-mannose transporter was constructed (see Materials and Methods). This strain exhibited a normal phenotype, demonstrating that the addition of GFP on the C terminus of ScVrg4 does not affect its normal function. As was seen in C. albicans VRG4-GFP strain, treatment of this S. cerevisiae strain for 30 min with 5 μg/ml CA led to a similar dispersal of the Vrg4-GFP signal (Figure 7B). Together, these experiments demonstrate that the actin cytoskeleton has a previously unrecognized role in maintaining the integrity of the Golgi in C. albicans and S. cerevisiae yeast cell, although the role of BNI1 in S. cerevisiae has not yet been characterized. These data also demonstrate that in C. albicans, the absence of Bni1 results in a Golgi that is more susceptible to undergo fragmentation if the actin cytoskeleton is perturbed.
DISCUSSION
In this study, we monitored the Golgi complex in C. albicans cells undergoing the yeast-to-hyphal transition, by using fluorescent reporters. We report that hyphal formation in C. albicans is accompanied by a redistribution of most of the Golgi complex toward the growing tip, and this localization is dependent on the formin Bni1. Shortly after inducing hypha, the randomly distributed Golgi spots seen in vegetatively growing yeast cells migrate with the growing hyphal tip. Throughout hyphal extension, the Golgi remains associated with the distal portion of the hypha. Several pieces of evidence support our conclusion that this fluorescence is coincident with the Golgi. First, this pattern of localization was observed by both indirect immunofluorescence by using several different antibodies and different reporters as well as by analysis of a GFP-tagged Golgi reporter in live cells. Second, this pattern of localization was lost after treatment with brefeldin A, a compound that causes a rapid but reversible fusion of the Golgi with the ER (Figure 1). Third, this pattern is distinct from that of the ER and vacuolar membranes, which are found throughout the cell body and extended filament (Figure 2). Neither CaVrg4 nor any Golgi proteins have been precisely assigned to a particular cisternae in C. albicans, so it remains to be determined whether this phenomenon reflects the entire Golgi or just a subset of cisternae. Nevertheless, the distribution of any of the Golgi cisternae to the distal portion of the hypha implies that post-Golgi secretory vesicles in hyphae do not require long-distance transport from the cell body to reach the growing tip, but rather they are generated in proximity to the growth site.
We found that hyphal formation and the redistribution of the Golgi progressed normally in the absence of microtubules. This result is in contrast to previous studies that reported the requirement of microtubules for hyphal formation in C. albicans (Akashi et al., 1994; Crampin et al., 2005). This apparent discrepancy may be due to the different drugs that were used to inhibit microtubule formation. Although we used NZ to inhibit microtubule formation, the previous studies used benomyl, or its derivative methyl benzimidazole carbamate. We also found that benomyl does indeed inhibit hyphal growth but only when added at very high concentrations, much higher than its solubility in the medium (our unpublished data). One explanation for the different effects seen with NZ versus benomyl is that benomyl has secondary effects at such high concentrations, including an effect on actin, as was suggested by previous reports (Akashi et al., 1994). Therefore, we chose to assess microtubule requirements using NZ, whose efficacy as a microtubule-depolymerizing agent in hyphal cells was established in this work (Table 3 and Figure 3). Our NZ data also underscore the fact that hyphal formation and the redistribution of the Golgi, unlike in mammalian cells, are not coupled to the partitioning of the genetic material, because nuclear division was arrested in NZ-treated cells.
An important aspect of this present work is our observation that loss of Bni1 affects Golgi organization and inhibits its localization to the hyphal tip. Remarkably, in the course of this study, we uncovered a previously unrecognized feature of the fungal Golgi; its capacity to disassemble in a manner reminiscent of the Golgi haze in mitotic animal cells. During the formation of hyphae in bni1Δ cells, the gradual disappearance of Golgi spots was accompanied by the appearance of a haze that pervaded the entire cell (Figure 5). Several lines of evidence support the idea that this haze represents Golgi vesicles that cannot be resolved by fluorescence microscopy. First, the disappearance of Vrg4 during bni1Δ hyphal induction is not due to Vrg4 degradation. Second, Vrg4-enriched Golgi membrane fractions from wild-type and bni1Δ cells display similar differential centrifugation properties and density, but they differ in their velocity sedimentation properties (Figure 6). Loss of Bni1 has no obvious affect on ER (Figure 5) or on vacuole morphology (our unpublished data), suggesting this is a Golgi-specific affect. The simplest interpretation of these data are that the Golgi in bni1Δ cells is fragmented.
This interpretation raises the pivotal question of how the loss of Bni1 can affect the structural integrity of the Golgi. We favor a model in which normal Golgi morphology and localization to the distal tip require actin filaments that are nucleated by Bni1 during hyphal formation (Figure 8). A direct role for actin in mediating Golgi localization and integrity comes from our observation that actin formation is required to maintain an organized Golgi localization at the distal portion of the hyphae, whereas inhibition of actin formation caused Golgi disassembly in wild-type yeast cells as is seen in bni1Δ mutants (Figures 4 and 7). In S. cerevisiae, the two partially redundant formins, Bnr1 and Bni1, localize to either the bud neck or the bud cortex, respectively. This difference in formin localization explains, in part, the orientation of actin cables in S. cerevisiae and the different mutant phenotypes of the bni1 and bnr1 mutants (Pruyne et al., 2004a). Loss of ScBnr1 results in cell separation defects, whereas loss of Bni1 results in budding defects (for review, see Pruyne et al., 2004b). However, spatial differences cannot fully explain the different hyphal phenotypes of C. albicans bni1Δ and bnr1Δ null mutants. CaBni1 performs unique functions during hyphal formation, because bnr1Δ null mutants do not display severe hyphal phenotypes, but bni1Δ cells do (Li et al., 2005; Martin et al., 2005). Moreover, like Bni1, Bnr1 is also localized at the hyphal tip in C. albicans and actin cables emanating from the hyphal tip persist in the bni1 null mutant (Martin et al., 2005). The conclusion from this observation is that the effect on Golgi morphology and localization is not due to a complete loss of actin cables in bni1 null hyphal cells; instead, it suggests the existence of a signal-induced actin polymerization in hyphal cells, critically dependent on Bni1, that is necessary to maintain both proper polarized growth and Golgi architecture. Studies in budding yeast of actin cables that differ in their length, location, orientation, dynamics, and sensitivity to latrunculin A (Yang and Pon, 2002) provide a precedent for the idea that hyphal cells may contain specialized actin cables required for Golgi repositioning. One can envisage that upon activation of the hyphal program, Golgi-localized proteins interact with these specialized filaments to allow migration of the Golgi to the tip hyphal cells, and they maintain its integrity while doing so. An alternative possibility is that hyphal cells demand more actin cables than yeast cells to maintain polarized growth and Golgi integrity and that the defect in both these processes in the bni1Δ strain during the yeast-to-hyphal switch represents the threshold point at which this high demand is unmet.
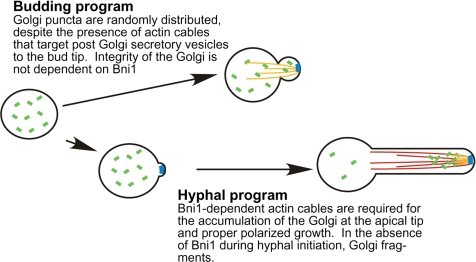
Figure 8.
Model of Golgi organization in C. albicans yeast and hyphal cells. The Golgi consists of puncta (denoted in green) that are randomly distributed throughout the cell during the cell division cycle. On induction of the hyphal program, Bni1-dependent nucleation of actin cables (denoted in red) mediates the redistribution of Golgi cisternae to the distal portion of the extending hyphae. During execution of the hyphal, but not the budding program, Bni1 is also required for the structural integrity of the Golgi (blue denotes Bni1). See text for further discussion.
Our data do not address why the effect of Bni1 on Golgi repositioning and structure is limited to the window encompassing the yeast-to-hyphal transition. This undoubtedly involves a hyphal-specific interaction of Bni1 with other proteins that could, in principle, modulate the rate of actin nucleation or elongation, or the structure of the filament. Most formins, including Bni1, are activated by GTP-bound Rho-GTPases (for review, see Higgs, 2005). Our results demonstrating the hyphal-specific effects of Bni1 are consistent with the idea that a hyphal specific Rho-GTPase effector may modulate Bni1 activity. Alternatively, hyphal specific actin-binding proteins may modulate actin structure in a Bni1-dependent way. It is noteworthy that the expression of several C. albicans genes predicted to encode small GTPase and cytoskeletal modulators is highly up-regulated in response to serum (Nantel et al., 2002). These encode homologues of the S. cerevisiae Yke2 (an actin binding protein), Rdi1 (Rho-GTPase inhibitor that is implicated in regulating Cdc42), Pfy1 (profilin), and Ybl060w (a Sec7 GEF domain-containing protein). These proteins represent attractive candidates for hyphal-specific Bni1 or actin cable modulators, and it will be of interest to investigate whether these proteins affect Golgi structure and repositioning during hyphal formation. The molecular mechanism linking Bni1 activation, Golgi repositioning, and actin remodeling is likely to involve a complex network and will require further investigations.
The Golgi complex is a highly dynamic organelle that requires a continuous influx and efflux of membrane trafficking to maintain its structure. It is notable that despite the dispersed Golgi haze observed in the bni1Δ mutant, secretion is not abolished. Cells lacking Bni1 still form hyphae (albeit slowly), display only a minor (25%) decrease in the secretion of acid phosphatase (Menzies and Dean, unpublished data), and do not seem to have any defects in glycoprotein processing (our unpublished data). Therefore the Golgi seems to remain largely functional. This raises several questions. First, what is the benefit of polarizing the Golgi to hyphal apex in C. albicans? Although purely speculative, redistribution of the Golgi to the growing tip, although not essential, may provide a selective advantage during infection when rapid hyphal growth enables efficient host tissue invasion. In support of this idea, bni1 null strains display a severe virulence defect (Li et al., 2005).
The second question raised by the bni1Δ fragmented Golgi phenotype is how such a fragmented Golgi can retain function? There is evidence that the yeast Golgi is comprised of tubulovesicular clusters, rather than fixed, large saccules and that a series of fixed structural compartments is not strictly required for transport of cargo between the ER and post-Golgi secretory vesicles (Rambourg et al., 2001). Although the role of actin cables in targeting post-Golgi vesicles to the plasma membrane is well established, our data that implicate Bni1 in the maintenance of Golgi architecture raise the novel possibility that Golgi clusters in fungi may depend on the Bni1-dependent actin cytoskeleton to remain intact. This is, to our knowledge, the first time a direct role for actin cables has been reported in the maintenance of Golgi integrity. Although Bni1-dependent actin polymerization may be required for retaining the cisternal appearance of the Golgi, residual actin cytoskeleton, nucleated by Bnr1 in the absence of Bni11, may be sufficient to allow the asymmetric targeting of post-Golgi secretory vesicles to the hyphal tip. Further experiments that rely on the development of additional reporters, C. albicans mutants, and higher resolution microscopy will be required to provide a more accurate description of this haze in bni1Δ hyphal cells.
ACKNOWLEDGMENTS
We thank Wang Yue for providing the bni1/bni1Δ and spa2Δ/spa2Δ strains and the TUB2-GFP plasmid, Cheryl Gale for providing the TUB1-GFP plasmid, Judith Berman and Maryam Gerami-Nejad for the ABP1-YFP strain, and Peter Sudbery for the MLC1-YFP strain. We acknowledge the excellent technical assistance of Sabine-Keppler Ross. We thank Nicole Averbeck and Catherine Menzies for critical reading of this manuscript. We also thank the anonymous reviewers for insightful comments that contributed to a much-improved study. This work was funded by National Institutes of Health Grant R0 GM-48467 (to N.D.).
Abbreviations used:
- CA
cytochalasin A
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- HA
influenza hemagglutinin
- NZ
nocodazole.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-02-0143) on July 19, 2006.
REFERENCES
- Akashi T., Kanbe T., Tanaka K. The role of the cytoskeleton in the polarized growth of the germ tube in Candida albicans. Microbiology. 1994;140:271–280. doi: 10.1099/13500872-140-2-271. [DOI] [PubMed] [Google Scholar]
- Bates S., et al. Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J. Biol. Chem. 2006;281:90–98. doi: 10.1074/jbc.M510360200. [DOI] [PubMed] [Google Scholar]
- Cole L., Orlovich D.A., Ashford A. E. Structure, function and motility of vacuoles in filamentous fungi. Fungal Genet. Biol. 1998;24:86–100. doi: 10.1006/fgbi.1998.1051. [DOI] [PubMed] [Google Scholar]
- Cooper J. A. Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 1987;105:1473–1478. doi: 10.1083/jcb.105.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack B. P., Bertram G., Egerton M., Gow N. A., Falkow S., Brown A. J. Yeast-enhanced green fluorescent protein (yEGFP)a reporter of gene expression in Candida albicans. Microbiology. 1997;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- Crampin H., Finley K., Gerami-Nejad M., Court H., Gale C., Berman J., Sudbery P. Candida albicans hyphae have a Spitzenkörper that is distinct from the polarisome found in yeast and pseudohyphae. J. Cell Sci. 2005;118:2935–2947. doi: 10.1242/jcs.02414. [DOI] [PubMed] [Google Scholar]
- Dean N., Zhang Y. B., Poster J. B. The VRG4 gene is required for GDP-mannose transport into the lumen of the Golgi in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 1997;272:31908–31914. doi: 10.1074/jbc.272.50.31908. [DOI] [PubMed] [Google Scholar]
- Fratti R. A., Jun Y, Merz A. J., Margolis N., Wickner W. Interdependent assembly of specific regulatory lipids and membrane fusion proteins into the vertex ring domain of docked vacuoles. J. Cell Biol. 2004;167:1087–1098. doi: 10.1083/jcb.200409068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M., Blundell K., Longtine M. S., Chow C. J., Adames N., Pringle J. R., Peter M., Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Evangelista M., Pruyne D., Amberg D. C., Boone C., Bretscher A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 2002;4:260–269. doi: 10.1038/ncb770. [DOI] [PubMed] [Google Scholar]
- Evangelista M., Zigmond S., Boone C. Formins: signaling effectors for assembly and polarization of actin filaments. J. Cell Sci. 2003;116:2603–2611. doi: 10.1242/jcs.00611. [DOI] [PubMed] [Google Scholar]
- Finger F. P., Novick P. Spatial regulation of exocytosis: lessons from yeast. J. Cell Biol. 1998;142:609–612. doi: 10.1083/jcb.142.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi W. A., Irwin M. Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Tanaka K., Mino A., Kikyo M., Takahashi K., Shimizu K., Takai Y. Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Biol. Cell. 1998;9:1221–1233. doi: 10.1091/mbc.9.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X. D., Dean N. Distinct protein domains of the yeast Golgi GDP-mannose transporter mediate oligomer assembly and export from the endoplasmic reticulum. J. Biol. Chem. 2000;275:17718–17727. doi: 10.1074/jbc.M909946199. [DOI] [PubMed] [Google Scholar]
- Gow N. A., Brown A. J., Odds F. C. Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 2002;5:366–371. doi: 10.1016/s1369-5274(02)00338-7. [DOI] [PubMed] [Google Scholar]
- Harris S. D., Read N. D., Roberson R. W., Shaw B., Seiler S., Plamann M., Momany M. Polarisome meets Spitzenkörper: microscopy, genetics, and genomics converge. Eukaryot. Cell. 2005;4:225–229. doi: 10.1128/EC.4.2.225-229.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan I., Sepulveda-Becerra M., Liu H. Hyphal elongation is regulated independently of cell cycle in Candida albicans. Mol. Biol. Cell. 2002;13:134–145. doi: 10.1091/mbc.01-03-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero A. B., Uccelletti D., Hirschberg C. B., Dominguez A., Abeijon C. The Golgi GDPase of the fungal pathogen Candida albicans affects morphogenesis, glycosylation, and cell wall properties. Eukaryot. Cell. 2002;1:420–431. doi: 10.1128/EC.1.3.420-431.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs H. N. Formin proteins: a domain-based approach. Trends Biochem. Sci. 2005;30:342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Howard R. J. Ultrastructural analysis of hyphal tip cell growth in fungi: Spitzenkörper, cytoskeleton and endomembranes after freeze-substitution. J. Cell Sci. 1981;48:89–103. doi: 10.1242/jcs.48.1.89. [DOI] [PubMed] [Google Scholar]
- Howard R. J., Aist J. R. Effects of MBC on hyphal tip organization, growth, and mitosis of Fusarium acuminatum, and their antagonism by D2O. Protoplasma. 1977;92:195–210. doi: 10.1007/BF01279458. [DOI] [PubMed] [Google Scholar]
- Kohno H., et al. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15:6060–6068. [PMC free article] [PubMed] [Google Scholar]
- Koning A. J., Lum P. Y., Williams J. M., Wright R. DiOC6 staining reveals organelle structure and dynamics in living yeast cells. Cell Motil. Cytoskeleton. 1993;25:111–128. doi: 10.1002/cm.970250202. [DOI] [PubMed] [Google Scholar]
- Lew D. J., Reed S. I. Cell cycle control of morphogenesis in budding yeast. Curr. Opin. Genet. Dev. 1995;5:17–23. doi: 10.1016/s0959-437x(95)90048-9. [DOI] [PubMed] [Google Scholar]
- Li C. R., Wang Y. M., De Zheng X., Liang H. Y., Tang J. C., Wang Y. The formin family protein CaBni1p has a role in cell polarity control during both yeast and hyphal growth in Candida albicans. J. Cell Sci. 2005;118:2637–2648. doi: 10.1242/jcs.02393. [DOI] [PubMed] [Google Scholar]
- Lo H. J., Kohler J. R., DiDomenico B., Loebenberg D., Cacciapuoti A., Fink G. R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Losev E., Reinke C. A., Jellen J., Strongin D. E., Bevis B. J., Glick B. S. Golgi maturation visualized in living yeast. Nature. 2006;441:1002–1006. doi: 10.1038/nature04717. [DOI] [PubMed] [Google Scholar]
- Martin R., Walther A., Wendland J. Ras1-induced hyphal development in Candida albicans requires the formin Bni1. Eukaryot. Cell. 2005;4:1712–1724. doi: 10.1128/EC.4.10.1712-1724.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad A. M., Lee P. R., Broadbent I. D., Barelle C. J., Brown A. J. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast. 2000;16:325–327. doi: 10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Nantel A., et al. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell. 2002;13:3452–3465. doi: 10.1091/mbc.E02-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman A. M., Mhaiskar V., Manus V., Galibert F., Dean N. Saccharomyces cerevisiae HOC1, a suppressor of pkc1, encodes a putative glycosyltransferase. Genetics. 1997;145:637–645. doi: 10.1093/genetics/145.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa A., Poster J. B., Jigami Y., Dean N. Molecular and phenotypic analysis of CaVRG4, encoding an essential Golgi apparatus GDP-mannose transporter. J. Bacteriol. 2002;184:29–42. doi: 10.1128/JB.184.1.29-42.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannunzio V. G., Burgos H. I., Alonso M., Mattoon J. R., Ramos E. H., Stella C. A. A simple chemical method for rendering wild-type yeast permeable to brefeldin a that does not require the presence of an erg6 mutation. J. Biomed. Biotechnol. 2004;2004:150–155. doi: 10.1155/S1110724304308077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle J. R., Preston R. A., Adams A. E., Stearns T., Drubin D. G., Haarer B. K., Jones E. W. Fluorescence microscope methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Evangelista M., Yang C., Bi E., Zigmond S., Bretscher A., Boone C. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Gao L., Bi E., Bretscher A. Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol. Biol. Cell. 2004;15:4971–4989. doi: 10.1091/mbc.E04-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D., Legesse-Miller A., Gao L., Dong Y., Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 2004;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- Rambourg A., Jackson C. L., Clermont Y. Three dimensional configuration of the secretory pathway and segregation of secretion granules in the yeast Saccharomyces cerevisiae. J. Cell Sci. 2001;114:2231–2239. doi: 10.1242/jcs.114.12.2231. [DOI] [PubMed] [Google Scholar]
- Raudaskoski M., Mao W. Z., Yli-Mattila T. Microtubule cytoskeleton in hyphal growth Response to nocodazole in a sensitive and a tolerant strain of the homobasidiomycete Schizophyllum commune. Eur. J. Cell Biol. 1994;64:131–141. [PubMed] [Google Scholar]
- Reynaga-Pena C. G., Gierz G., Bartnicki-Garcia Analysis of the role of the Spitzenkörper in fungal morphogenesis by computer simulation of apical branching in Aspergillus niger. Proc Natl. Acad. Sci. USA. 1997;94:9096–9101. doi: 10.1073/pnas.94.17.9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagot I., Rodal A. A., Moseley J., Goode B. L., Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 2002;4:626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- Sharpless K. E., Harris S. D. Functional characterization and localization of the Aspergillus nidulans formin SEPA. Mol. Biol. Cell. 2002;13:469–479. doi: 10.1091/mbc.01-07-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu Y. J., Santos B., Fortin N., Costigan C., Snyder M. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol. Cell Biol. 1998;18:4053–4069. doi: 10.1128/mcb.18.7.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg G., Wedlich-Soldner R., Brill M., Schulz I. Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J. Cell Sci. 2001;114:609–622. doi: 10.1242/jcs.114.3.609. [DOI] [PubMed] [Google Scholar]
- Sudbery P., Gow N., Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Temperli E., Roos U. P., Hohl H. R. Actin and tubulin cytoskeletons in germlings of the oomycete fungus Phytophthora infestans. Eur. J. Cell Biol. 1990;53:75–88. [PubMed] [Google Scholar]
- That T. C., Rossier C., Barja F., Turian G., Roos U. P. Induction of multiple germ tubes in Neurospora crassa by antitubulin agents. Eur. J. Cell Biol. 1988;46:68–79. [PubMed] [Google Scholar]
- Tominaga T., Sahai E., Chardin P., McCormick F., Courtneidge S. A., Alberts A. S. Diaphanous-related formins bridge Rho GTPase and Src tyrosine kinase signaling. Mol. Cell. 2000;5:13–25. doi: 10.1016/s1097-2765(00)80399-8. [DOI] [PubMed] [Google Scholar]
- Vida T. A., Graham T. R., Emr S. D. In vitro reconstitution of intercompartmental protein transport to the yeast vacuole. J. Cell Biol. 1990;111:2871–2884. doi: 10.1083/jcb.111.6.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther A., Wendland J. An improved transformation protocol for the human fungal pathogen Candida albicans. Curr. Genet. 2003;42:339–343. doi: 10.1007/s00294-002-0349-0. [DOI] [PubMed] [Google Scholar]
- Walther A., Wendland J. Apical localization of actin patches and vacuolar dynamics in Ashbya gossypii depend on the WASP homolog Wal1p. J. Cell Sci. 2004;117:4947–4958. doi: 10.1242/jcs.01377. [DOI] [PubMed] [Google Scholar]
- Weisman L. S., Emr S. D., Wickner W. T. Mutants of Saccharomyces cerevisiae that block intervacuole vesicular traffic and vacuole division and segregation. Proc. Natl. Acad. USA. 1990;87:1076–1080. doi: 10.1073/pnas.87.3.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R. B., Davis D., Mitchell A. P. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. C., Pon L. A. Actin cable dynamics in budding yeast. Proc. Natl. Acad. USA. 2002;99:751–756. doi: 10.1073/pnas.022462899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K., Kaji H., Nishimura K., Miyaji M. The role of microfilaments and microtubules in apical growth and dimorphism of Candida albicans. J. Gen. Microbiol. 1990;136:1067–1075. doi: 10.1099/00221287-136-6-1067. [DOI] [PubMed] [Google Scholar]
- Zheng X. D., Wang Y. M., Wang Y. CaSPA2 is important for polarity establishment and maintenance in Candida albicans. Mol. Microbiol. 2003;49:1391–1405. doi: 10.1046/j.1365-2958.2003.03646.x. [DOI] [PubMed] [Google Scholar]