Abstract
Mutation of TOP3 in Saccharomyces cerevisiae causes poor growth, hyperrecombination, and a failure to fully activate DNA damage checkpoints in S phase. Here, we report that overexpression of a dominant-negative allele of TOP3, TOP3Y356F, which lacks the catalytic (decatenation) activity of Top3, causes impaired S-phase progression and the persistence of abnormal DNA structures (X-shaped DNA molecules) after exposure to methylmethanesulfonate. The impaired S-phase progression is due to a persistent checkpoint-mediated cell cycle delay and can be overridden by addition of caffeine. Hence, the catalytic activity of Top3 is not required for DNA damage checkpoint activation, but it is required for normal S-phase progression after DNA damage. We also present evidence that the checkpoint-mediated cell cycle delay and persistence of X-shaped DNA molecules resulting from overexpression of TOP3Y356F are downstream of Rad51 function. We propose that Top3 functions in S phase to both process homologous recombination intermediates and modulate checkpoint activity.
INTRODUCTION
Topoisomerases are highly conserved proteins that catalyze topological rearrangements in the structure of DNA and are required for many aspects of DNA metabolism. Of the three topoisomerases in yeast, the function(s) of the sole type IA topoisomerase, Top3, remains most poorly understood. However, it is likely that Top3 fulfills important roles in vivo, because deletion of TOP3 in Saccharomyces cerevisiae causes hyperrecombination, sensitivity to genotoxic agents, meiotic defects, and poor growth due to accumulation of cells with a late S/G2 content of DNA (Wallis et al., 1989; Gangloff et al., 1994, 1999; Chakraverty et al., 2001). Similarly, deletion of top3+ in Schizosaccharomyces pombe causes lethality due to chromosome missegregation and accumulation of DNA double-strand breaks (Goodwin et al., 1999; Maftahi et al., 1999; Oh et al., 2002; Win et al., 2004). Whereas lower eukaryotes generally contain only one type IA topoisomerase, human cells, like most vertebrates, possess at least two Top3 homologues, hTOPIIIα and hTOPIIIβ (Hanai et al., 1996; Ng et al., 1999). In mice, mutation of TOP3α causes embryonic lethality (Li and Wang, 1998), whereas mutation of TOP3β causes a shortened life span (Kwan et al., 2003). Interestingly, in S. cerevisiae and S. pombe, deletion of SGS1 or rqh1+ can largely suppress the phenotypes caused by deletion of TOP3 or top3+, respectively (Gangloff et al., 1994; Goodwin et al., 1999; Maftahi et al., 1999; Chakraverty et al., 2001). Because both SGS1 and rqh1+ belong to the same family of proteins, the RecQ DNA helicases, this intriguing genetic interaction has led to the suggestion that type IA topoisomerases may be functionally associated with RecQ helicases. Indeed, both Sgs1 and Rqh1 physically interact with Top3 in their respective organisms (Gangloff et al., 1994; Bennett et al., 2000; Fricke et al., 2001; Onodera et al., 2002; Laursen et al., 2003; Ahmad and Stewart, 2005; Ui et al., 2005), and a close association between a RecQ helicase and type IA topoisomerase has now been demonstrated in a number of other organisms (Harmon et al., 1999; Wu et al., 2000; Kim et al., 2002).
RecQ helicases are evolutionarily conserved 3→5′ DNA helicase enzymes that are important for the maintenance of genomic stability in all organisms (Hickson, 2003). Of particular interest, defects in three (of the five so far identified) human RecQ helicases cause Bloom's syndrome (BLM), Rothmund-Thomson syndrome (RECQL4), and Werner's syndrome (WRN) (Ellis et al., 1995; Yu et al., 1996; Kitao et al., 1999). Each of these disorders shows genomic instability associated with a predisposition to the development of various cancers. Additionally, Rothmund–Thomson syndrome and Werner's syndrome possess features resembling premature aging. RecQ helicases have, therefore, received much interest due to their putative roles in suppressing cancer and/or aging in humans.
Consistent with an evolutionarily conserved functional interaction between RecQ helicases and a type IA topoisomerases, BLM has been demonstrated to interact with, and stimulate the DNA strand passage activity of, hTOPIIIα in vitro (Johnson et al., 2000; Wu et al., 2000; Wu and Hickson, 2002). This association is likely to be physiologically important, because BLM and hTOPIIIα act together in vitro to resolve homologous recombination (HR) intermediates containing two Holliday junctions in a process termed “double junction dissolution” (Wu and Hickson, 2003). The physiological consequence of this in vivo is likely to be in the resolution of HR repair intermediates without potentially deleterious crossing over of genetic material. Indeed, elevated levels of sister chromatid exchanges, which arise due to crossing over during or soon after S phase, are currently the most reliable diagnostic feature of Bloom's syndrome (German, 1993). Sgs1 and Top3 have also been demonstrated to function in the same HR repair pathway to prevent crossing over during double-strand break repair in S. cerevisiae (Ira et al., 2003). Therefore, current thinking suggests that RecQ helicases act in concert with type IA topoisomerases in an evolutionarily conserved pathway to process recombination intermediates that arise during DNA replication without the crossing over of genetic material.
Further evidence for a role for Top3 in the late stages of HR repair also comes from the fact that mutation of genes involved in the early steps of HR repair (e.g., RAD51 in S. cerevisiae; rhp51+ in S. pombe) partially suppresses various top3 phenotypes in S. cerevisiae or S. pombe (Fabre et al., 2002; Oakley et al., 2002; Shor et al., 2002). Together with the very low viability of yeast top3 strains, it has been proposed that DNA intermediates formed by the HR repair pathway are otherwise toxic if Top3 function is impaired (Gangloff et al., 1994; Fabre et al., 2002). Moreover, it would seem that Top3 catalyzes a step in a late stage of recombination for which there are no redundant activities, because the top3 mutant phenotype is far more severe than that of cells lacking HR repair altogether.
In addition to a putative role in processing late-stage HR intermediates, Top3 has also been implicated in DNA damage checkpoint signaling in S. cerevisiae (Chakraverty et al., 2001). Whereas wild-type cells activate the DNA damage checkpoint and slow DNA replication after DNA damage (Paulovich and Hartwell, 1995), top3 cells fail to fully activate Rad53, and, as a consequence, progress more rapidly through S phase after DNA damage (Chakraverty et al., 2001). This defect is apparent only when the damage occurs during S phase, because G1/S and G2/M checkpoints are intact in top3 cells (Chakraverty et al., 2001). Interestingly, deletion of RMI1, which encodes a Top3-interacting protein, causes a top3-like phenotype and also results in a similar failure to fully activate Rad53 in response to DNA damage (Chang et al., 2005; Mullen et al., 2005). These findings suggest that the Sgs1–Top3–Rmi1 complex (either directly or indirectly) fulfills important DNA damage checkpoint signaling functions after DNA damage. However, it remains to be determined whether this proposed role for Top3 is related to its function in HR repair.
To further investigate the putative role(s) of Top3 in yeast, we have used an allele of TOP3, TOP3Y356F, that fails to complement a top3 strain (Bennett and Wang, 2001) and causes an inducible, dominant-negative, top3-like phenotype when overexpressed in wild-type cells (Oakley et al., 2002). Because a top3 mutant strain is very poor growing and readily acquires suppressor mutations (most notably in SGS1), use of this dominant-negative system allowed us to investigate the acute effects of Top3 impairment and permitted more technically challenging experiments than would be possible with a top3 strain. Additionally, because the TOP3Y356F allele used in this study possesses a mutation in the active site tyrosine residue that abolishes the catalytic activity, but not the DNA-binding capability, of hTOPIIIα (Goulaouic et al., 1999), we reasoned that overexpression of catalytically dead Top3Y356F protein might reveal selective, separable functions of Top3 that could otherwise be overlooked in a top3 strain.
We report that, unlike top3 mutants (Chakraverty et al., 2001), cells overexpressing TOP3Y356F fully activate Rad53 after DNA damage. Therefore, the checkpoint defect previously reported in top3 cells (Chakraverty et al., 2001) is likely due to loss of Top3 function(s) other than loss of catalytic activity. Moreover, cells overexpressing TOP3Y356F demonstrate a persistent DNA damage checkpoint-mediated cell cycle delay in the presence of MMS. Therefore, although the catalytic activity of Top3 is not required for checkpoint activation, it is required for some aspect of S-phase progression after DNA damage. We also demonstrate that overexpression of TOP3Y356F in wild-type cells causes the accumulation of HR repair intermediates after DNA damage. Because phenotypes caused by overexpression of TOP3Y356F are downstream of Rad51 activity, our results further verify a late role for Top3 in HR repair. We propose a model in which Top3 acts late in HR repair to both process repair intermediates and (either directly or indirectly) modulate checkpoint activation/maintenance.
MATERIALS AND METHODS
S. cerevisiae Strains and Plasmids
All the strains used in this study are isogenic derivatives of T344 (Hovland et al., 1989). The rad51 and sgs1 deletion strains were constructed using a polymerase chain reaction (PCR)-based gene disruption method (Wach et al., 1994). Plasmids pYES2-TOP3 and pYES2-TOP3Y356F have been described previously (Oakley et al., 2002).
Growth Conditions, Cell Synchronization, and Flow Cytometry Analysis
Strains were grown at 30°C in CSM-Ura medium (Formedium) containing glucose [2% (wt/vol)]. For overexpression of TOP3 or TOP3Y356F from the pYES2 plasmid, 2% galactose was added. Cell cycle synchronization in the G1 phase was performed using α-factor mating pheromone (Cancer Research UK peptide synthesis laboratory, Clare Hall, United Kingdom) at 30°C for 4–5 h. Release from α-factor arrest was achieved by centrifugation, washing, and resuspension of cells in fresh medium. Cell cycle progression was monitored at 25°C using flow cytometry (fluorescence-activated cell sorting; FACS), as described previously (Chakraverty et al., 2001).
Protein Extraction and Western Blot Analysis
Protein extracts from yeast cells were prepared using a modified trichloroacetic acid (TCA) protein extraction technique (Foiani et al., 1994). Approximately 108 cells were harvested, washed once, and resuspended in 20% TCA. An equal volume of glass beads was added, and cells were disrupted using a BIO101/Savant FastPrep FP120 cell disrupter (4 × 20 s cycles at full speed; Thermo Electron Corporation, Waltham, MA). The supernatant was then removed, and the glass beads were washed twice with 200 μl of 5% TCA. Extracts were then clarified by centrifugation at 5700 rpm for 5 min, and the pellet was resuspended in 2× Laemmli buffer (Bio-Rad, Hercules, CA) diluted with an equal volume of 0.5 M Tris-HCl, pH 8.0. Samples were boiled for 3 min and then further clarified by centrifugation. Proteins were separated by SDS-PAGE using 3–8 or 7% NuPAGE Tris-acetate gels (Invitrogen, Paisley, United Kingdom). Rad53 phosphorylation status was analyzed using a mouse monoclonal antibody (EL7; kindly provided by Dr. Marco Foiani, FIRC Institute of Molecular Oncology, Milan, Italy) at a final dilution of 1:10. Top3 and Top3Y356F proteins were detected using a mouse anti-FLAG M2 antibody (Sigma-Aldrich, St. Louis, MO) at a final concentration of 10 μg/ml. Horseradish peroxidase-linked secondary antibody (Sigma-Aldrich) was used at 1:4000, and chemiluminescent detection was performed with an ECL kit (GE Healthcare, Little Chalfont Buckinghamshire, United Kingdom).
Two-Dimensional (2D) Gel Electrophoresis
The hexadecyltrimethylammonium bromide (CTAB) method of DNA extraction and two-dimensional gel procedures were described previously (Brewer and Fangman, 1987; Allers and Lichten, 2000; Lopes et al., 2003). DNA was digested with NciI and NcoI before running the first-dimension gels.
RESULTS
Overexpression of TOP3Y356F Causes Poor Growth
Mutation of the conserved tyrosine residue in the active site of hTOPIIIα abolishes catalytic activity, but not an ability to bind DNA (Goulaouic et al., 1999). The corresponding mutation in the S. cerevisiae TOP3 gene results in a TOP3 allele (TOP3Y356F) that fails to complement a top3 strain (Bennett and Wang, 2001). We confirmed that a construct, pYES2-TOP3, could complement the poor growth and DNA damage sensitivity of a top3 strain, whereas pYES2-TOP3Y356F could not (our unpublished data; Oakley, 2001). Therefore, complementation of the phenotype of a top3 mutant requires the catalytic (decatenase) activity of Top3.
When overexpressed from the GAL1 promoter of the high copy vector pYES2, TOP3Y356F (but not TOP3) causes poor growth in the wild-type S. cerevisiae YP1 strain background (Oakley, 2001; Oakley et al., 2002). In the present study, we used the T344 strain, which contains mutations in the reg101 and gal1 genes, permitting the use of galactose as a gratuitous inducer and allowing 1,500-fold induction of genes regulated by the GAL1 promoter, even in the presence of 2% glucose (Hovland et al., 1989). We confirmed that TOP3 and TOP3Y356F were overexpressed to an equivalent level in the wild-type T344 strain after exposure to 2% galactose (Supplemental Figure 1). Consistent with previous analysis (Oakley et al., 2002), overexpression of TOP3Y356F also caused poor growth in the wild-type T344 strain, whereas overexpression of TOP3 had no effect (Supplemental Figure 2). Furthermore, the poor growth caused by overexpression of TOP3Y356F was prevented in a T344 strain lacking SGS1 (Supplemental Figure 2). Western blotting again revealed that TOP3 and TOP3Y356F were similarly overexpressed in the presence of 2% glucose in our sgs1 strain (our unpublished data).
Overexpression of TOP3Y356F Causes Impaired S-Phase Progression after DNA Damage
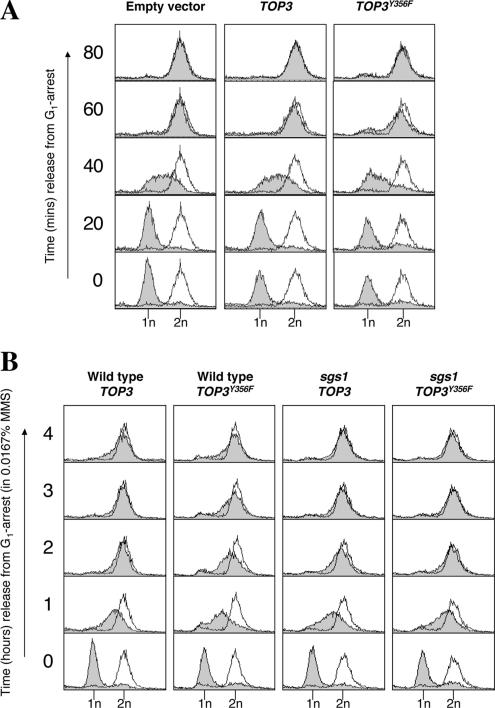
To analyze the effects of TOP3Y356F overexpression on DNA replication, wild-type strains transformed with pYES2, pYES2-TOP3, or pYES2-TOP3Y356F plasmids were synchronized in G1 with α-factor, and overexpression of TOP3 or TOP3Y356F was induced simultaneously by the addition of 2% galactose (Supplemental Figure 1). Cultures were then released from G1 arrest, and samples were taken at fixed intervals to analyze DNA content by flow cytometry (FACS). Wild-type strains transformed with empty vector or those overexpressing TOP3 traversed S phase and apparently completed DNA replication (as measured by a doubling of DNA content) within ∼60 min (Figure 1A). Wild-type cells overexpressing TOP3Y356F consistently demonstrated a slight delay in the rate of S-phase progression (Figure 1A, note DNA content at 40 min), but they nevertheless successfully completed DNA replication with only marginally delayed kinetics. Therefore, we conclude that overexpression of TOP3Y356F mildly affects some aspect of S-phase progression in unperturbed cells, but it does not noticeably affect bulk DNA synthesis per se.
Figure 1.
Overexpression of TOP3Y356F causes impaired S-phase progression after DNA damage. (A) Wild-type strains transformed with pYES2, pYES2-TOP3, or pYES2-TOP3Y356F were arrested in G1 with α-factor and simultaneously treated with 2% galactose (to induce overexpression from the pYES2 GAL1 promoter). Cultures were released into fresh medium under normal growth conditions, and DNA content was analyzed by flow cytometry at the indicated times. The shaded peaks represent experimental data, whereas the unshaded peak is a reference to indicate a normal G2/M peak (at 2-h release from G1 arrest). The positions of the 1n (G1) and 2n (G2/M) peaks are indicated below. (B) Wild-type and sgs1 strains overexpressing TOP3 or TOP3Y356F were released from G1 arrest into fresh medium containing 0.0167% MMS, and DNA content was analyzed by flow cytometry at the indicated times.
Previous studies demonstrated that top3 cells are sensitive to the DNA-damaging agent methylmethanesulfonate (MMS) (Chakraverty et al., 2001; Shor et al., 2002). We also observed that cells overexpressing TOP3Y356F demonstrated sensitivity to MMS, whereas overexpression of TOP3 had no effect (our unpublished data). Therefore, we analyzed cell cycle progression in wild-type cells overexpressing TOP3 or TOP3Y356F in the presence of 0.0167% MMS. Under these conditions, wild-type cells overexpressing TOP3 completed DNA replication by ∼2 h (Figure 1B). Similar kinetics of DNA replication were also observed for wild-type strains transformed with empty pYES2 vector (our unpublished data). This increased S-phase duration (relative to untreated cells; Figure 1A) is a consequence of MMS-induced replication fork stalling and subsequent activation of the DNA damage checkpoint (Paulovich and Hartwell, 1995; Santocanale and Diffley, 1998; Shirahige et al., 1998; Tercero and Diffley, 2001). Wild-type cells overexpressing TOP3Y356F demonstrated much more severely delayed S-phase progression, completing DNA replication only by ∼4 h (Figure 1B). Therefore, we conclude that overexpression of TOP3Y356F increases the duration of S phase in the presence of MMS, suggesting that Top3 is required for some aspect of S-phase progression after DNA damage.
Mutation of SGS1 Suppresses the Impaired S-Phase Progression Phenotype Caused by Overexpression of TOP3Y356F
It has been reported previously that mutation of SGS1 suppresses all known top3 mitotic phenotypes (Gangloff et al., 1994; Chakraverty et al., 2001). Therefore, we examined whether this also holds true for the novel phenotype of impaired S-phase progression caused by overexpression of TOP3Y356F. In agreement with a previous report (Liberi et al., 2005), we found that mutation of SGS1 alone did not noticeably affect unperturbed S-phase progression, but it did cause a slight delay in S-phase progression in the presence of MMS (our unpublished data). Similar to what was observed in wild-type cells, sgs1 cells overexpressing TOP3 traversed S phase and apparently completed DNA replication ∼2 h after G1 release in the presence of 0.0167% MMS (Figure 1B). Interestingly, in contrast to wild-type cells overexpressing TOP3Y356F, sgs1 cells overexpressing TOP3Y356F completed DNA replication with kinetics indistinguishable from that of sgs1 cells overexpressing TOP3. We conclude, therefore, that overexpression of TOP3Y356F does not cause impaired S-phase progression after DNA damage when SGS1 is mutated. Furthermore, this suggests that impaired S-phase progression after DNA damage caused by TOP3Y356F overexpression, like previously reported top3 mitotic phenotypes (Gangloff et al., 1994; Chakraverty et al., 2001), is likely to be the consequence of Sgs1 activity uncoupled from that of Top3.
The Impaired S-Phase Progression Caused by Overexpression of TOP3Y356F after DNA Damage Is Due to a Persistent Checkpoint-mediated Cell Cycle Delay
The finding that TOP3Y356F cells show an impaired ability to progress through S phase is in contrast with previous data demonstrating that top3 cells traverse S phase more rapidly than wild-type cells in the presence of MMS due to a failure to fully activate Rad53 (Chakraverty et al., 2001). This intriguing finding suggested that the previously reported top3 phenotype could be a consequence of lack of Top3 protein rather than abolition of Top3 catalytic activity per se. We investigated, therefore, if the impaired S-phase progression observed in TOP3Y356F cells is a consequence of proficient checkpoint activation in the presence of catalytically-dead Top3Y356F protein.
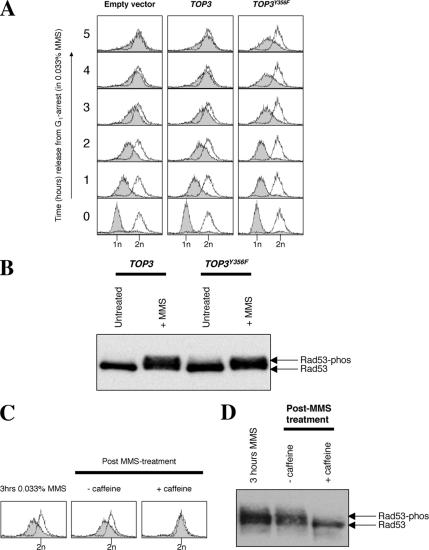
DNA damage checkpoint activation was assessed in cells exposed to a slightly higher concentration of MMS (0.033%) than used above in order to permit robust activation of the DNA damage checkpoint. Under these conditions, wild-type strains transformed with empty vector or those overexpressing TOP3 complete DNA replication by ∼5 h, whereas cells overexpressing TOP3Y356F still only demonstrate a mid-S-phase DNA content by this time (Figure 2A). DNA damage checkpoint activation was assessed 1 h after MMS treatment by an analysis of the phosphorylation status of the Rad53 checkpoint effector kinase. We observed that DNA damage-induced Rad53 phosphorylation (as indicated by a slower migrating species on protein gels) occurred to a similar extent in cells overexpressing TOP3 or TOP3Y356F (Figure 2B). This activation of Rad53 was dependent on DNA damage, because overexpression of TOP3 or TOP3Y356F did not promote activation of Rad53 in the absence of MMS (Supplemental Figure 1 and Figure 2B). Analysis of a time course of Rad53 activation also verified that Rad53 phosphorylation occurred with similar kinetics and to a similar level in cells transformed with empty pYES2 vector or cells overexpressing TOP3Y356F after exposure to MMS (Supplemental Figure 3). We conclude that, unlike in top3 cells (Chakraverty et al., 2001), Rad53 becomes fully activated in response to DNA damage in cells overexpressing TOP3Y356F.
Figure 2.
The TOP3Y356F-induced impaired S-phase progression phenotype is due to a persistent checkpoint-mediated cell cycle arrest. (A) Wild-type strains transformed with pYES2 or overexpressing TOP3 or TOP3Y356F were released from G1 arrest into fresh medium containing 0.033% MMS, and S-phase progression was monitored by FACS. (B) Cells overexpressing TOP3 or TOP3Y356F were collected before treatment with α-factor and 2% galactose (untreated), and 1 h after cells were released from α-factor arrest into medium containing 0.033% MMS. Protein extracts were prepared, and Rad53 phosphorylation status was monitored by Western blotting. The positions of the unphosphorylated Rad53 and slower migrating phosphorylated forms are shown on the right. (C and D) After 3-h treatment with 0.033% MMS, cells overexpressing TOP3Y356F were harvested and resuspended in fresh medium ± 5 mg/ml caffeine. Samples were taken 1 h later for analysis of DNA content by flow cytometry (C) and Rad53 phosphorylation status (D).
Next, we analyzed whether a persistent checkpoint-mediated cell cycle delay might be responsible for the impaired S-phase progression observed in cells overexpressing TOP3Y356F. Caffeine has been demonstrated to override DNA damage checkpoints in a number of organisms by inhibiting the DNA damage checkpoint transducer ATR (Homo sapiens)/Mec1 (S. cerevisiae)/Rad3 (S. pombe) (Schlegel and Pardee, 1986; Osman and McCready, 1998; Hall-Jackson et al., 1999; Moser et al., 2000; Vaze et al., 2002; Liberi et al., 2005). Therefore, we compared the effects of caffeine addition to a culture overexpressing TOP3Y356F after 3 h of MMS treatment. We found that cells resuspended in fresh medium lacking caffeine for 1 h failed to show any significant recovery and remained with a mid-S DNA content (Figure 2C). In contrast, cells treated with 5 mg/ml caffeine seemed to successfully traverse S phase, because a high proportion of cells possessed a 2n DNA content 1 h after the addition of caffeine (Figure 2C). To verify that this effect was due to override of the DNA damage checkpoint by caffeine, we analyzed Rad53 phosphorylation in cells before and after caffeine treatment. Consistent with the above-mentioned proposal, Rad53 was phosphorylated after 3 h of MMS treatment and remained phosphorylated 1 h later in cells released into fresh medium lacking caffeine (Figure 2D). In contrast, Rad53 became dephosphorylated in cells released into fresh medium for 1 h in the presence of 5 mg/ml caffeine (Figure 2D). Therefore, 5 mg/ml caffeine promotes DNA damage checkpoint override. Interestingly, the “resetting” of Rad53 by caffeine addition implies that Rad53 is dynamically phosphorylated and dephosphorylated with rapid kinetics. We conclude that the impaired S-phase progression observed in cells overexpressing TOP3Y356F is a consequence of a persistent DNA damage checkpoint-mediated delay caused (either directly or indirectly) by the catalytically dead Top3Y356F protein.
Overexpression of TOP3Y356F Causes Abnormal DNA Replication Intermediates (X-Molecules) to Persist after Exposure to MMS
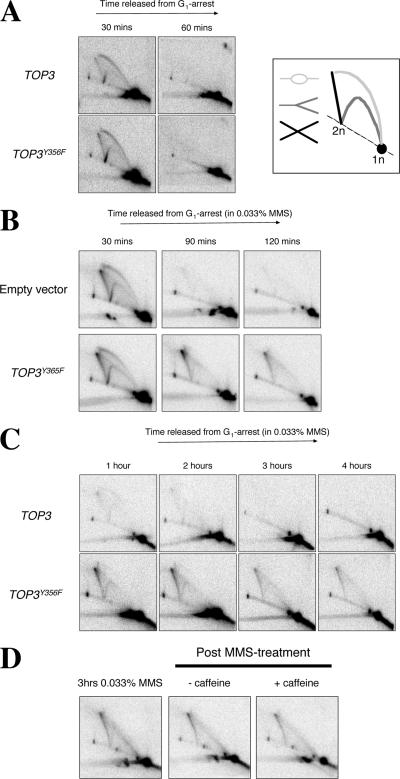
Next, we sought to determine the cause of the persistent checkpoint-mediated cell cycle delay observed in cells overexpressing TOP3Y356F. For this, we used the neutral-neutral 2D gel electrophoresis method to monitor DNA replication fork progression (Brewer and Fangman, 1987; Lopes et al., 2003). To monitor DNA replication under normal (unperturbed) growth conditions, wild-type strains overexpressing TOP3 or TOP3Y356F were released from G1 arrest into fresh medium, and samples were taken at fixed intervals to observe DNA replication intermediates on 2D gels originating from an early firing replication origin, ARS305. Genomic DNA was prepared using the CTAB method of DNA extraction to restrain branch migration of joint (X-shaped) molecules (Lopes et al., 2003). We observed that origin firing at ARS305 was detectable after 30 min in wild-type cells overexpressing either TOP3 or TOP3Y356F by the appearance of bubbles, Y-molecules, and origin-associated X-spikes (Figure 3A). Previous studies have indicated that the origin-associated X-spikes are normal DNA replication intermediates that are not dependent on Rad51 or Rad52 for their formation and are not, therefore, HR intermediates (Lopes et al., 2003). After 60 min, all of the ARS305 replication intermediates detectable at 30 min had disappeared in both strains, consistent with the completion of bulk DNA replication by this time (Figure 1A). We conclude, therefore, that overexpression of TOP3Y356F does not noticeably affect early replication origin firing or replication fork progression in the region adjacent to ARS305 in unperturbed cells.
Figure 3.
Overexpression of TOP3Y356F causes abnormal DNA replication intermediates (X-molecules) to persist after exposure to MMS. (A–C) Wild-type strains transformed with pYES2, or overexpressing TOP3 or TOP3Y356F were released from G1 arrest into fresh medium under normal growth conditions (A) or containing 0.033% MMS (B and C). DNA replication intermediates were analyzed by 2D gel electrophoresis at the times indicated. DNA samples were analyzed with a probe for the early firing ARS305 replication origin. The key in A (right) denotes DNA structures that can be identified by the 2D gel technique. (D) After 3-h treatment with 0.033% MMS, TOP3Y356F-overexpressing cells were harvested and resuspended in fresh medium ± 5 mg/ml caffeine. Samples were taken 1 h later for analysis of DNA replication intermediates by 2D gel electrophoresis.
Next, we analyzed DNA replication in the presence of 0.033% MMS. We observed that origin firing at ARS305 was again detectable at 30 min after G1 release in wild-type strains harboring either the pYES2 plasmid or pYES2-TOP3Y356F (Figure 3B). In MMS-treated cells harboring the empty pYES2 vector, all of the replication intermediates detectable at 30 min at ARS305 had largely disappeared by 90 min, consistent with completion of DNA replication in this particular region of the genome (in the presence of 0.033% MMS) by this time. In contrast, whereas cells overexpressing TOP3Y356F also revealed comparable kinetics of ARS305 origin activation (at 30 min) and bubble and Y-molecule disappearance (at 90 min), a structure corresponding to a joint molecule (X-molecule) persisted for at least 120 min. These MMS-induced X-molecules are not merely transient DNA replication intermediates, because they were still detectable in cells overexpressing TOP3Y356F (but not TOP3) 4 h after release from G1 arrest (Figure 3C). Furthermore, once formed, persistence of these X-molecules was not dependent on DNA damage checkpoint activity or cell cycle stage/progression, because driving cells into G2/M with caffeine did not promote their resolution (Figure 3D). We conclude that overexpression of TOP3Y356F causes abnormal DNA replication intermediates (X-molecules) to persist and/or accumulate during S phase in MMS-treated cells.
Mutation of RAD51 Causes Impaired S-Phase Progression after Exposure to MMS
Because Top3 has been implicated in HR repair (Gangloff et al., 1994; Fabre et al., 2002; Oakley et al., 2002; Shor et al., 2002), we considered the possibility that the X-molecules detected in MMS-treated cells overexpressing TOP3Y356F might represent unprocessed/aberrant HR repair intermediates. We analyzed, therefore, whether the phenotypes caused by TOP3Y356F overexpression were suppressed by deletion of RAD51. Rad51 catalyzes the early strand invasion step of HR and has been demonstrated to partially suppress the growth defect of a top3 mutant (Fabre et al., 2002; Oakley et al., 2002; Shor et al., 2002). In agreement with previous reports, we also observed that mutation of RAD51 in the T344 strain background partially suppressed the growth defect caused by TOP3Y356F overexpression (Supplemental Figure 4).
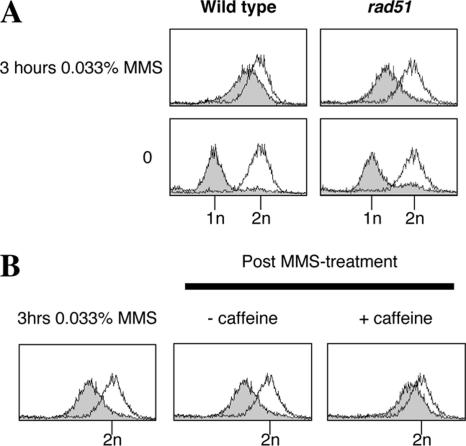
Next, we investigated whether the impaired S-phase progression caused by overexpression of TOP3Y356F is suppressed by mutation of RAD51. We observed that unperturbed rad51 cells completed bulk DNA replication by ∼60 min (our unpublished data), which are kinetics similar to that observed for wild-type cells (Figure 1A). Unlike wild-type cells, however, rad51 cells were unable to successfully traverse S phase in the presence of 0.033% MMS (Figure 4A). This finding was also verified in an independent (BY4741) strain background (our unpublished data) and suggests that the Rad51-dependent HR repair pathway is important for the repair/tolerance of MMS-lesions during S phase.
Figure 4.
Mutation of RAD51 causes impaired S-phase progression after DNA damage. (A) Wild-type and rad51 strains transformed with the pYES2 plasmid were released from G1 arrest into fresh medium containing 0.033% MMS, and DNA content was analyzed by flow cytometry at the indicated times. (B) After 3-h treatment with 0.033% MMS, the rad51-pYES2 culture was harvested and resuspended in fresh medium ± 5 mg/ml caffeine. Samples were taken 1 h later for analysis of DNA content by flow cytometry.
We note that the impaired S-phase progression observed in MMS-treated rad51 cells is actually more severe than that observed in cells overexpressing TOP3Y356F (Figure 5). Nevertheless, like TOP3Y356F cells, it seems that the impaired S-phase progression observed in MMS-treated rad51 cells is also, at least partially, caused by the DNA damage checkpoint, because addition of caffeine (after 3 h of MMS treatment) could largely override this phenotype (Figure 4B). Analysis of Rad53 phosphorylation status also confirmed that the DNA damage checkpoint is proficient and that addition of caffeine overrides the DNA damage checkpoint in rad51 cells (our unpublished data). These findings are consistent with a previous report demonstrating that Rad53 becomes fully activated earlier in rad51 cells than in wild-type cells after treatment with MMS (Liberi et al., 2005). Taken together, these data suggest that, in the absence of Rad51, the DNA damage checkpoint is both rapidly and robustly activated after exposure to MMS.
Figure 5.
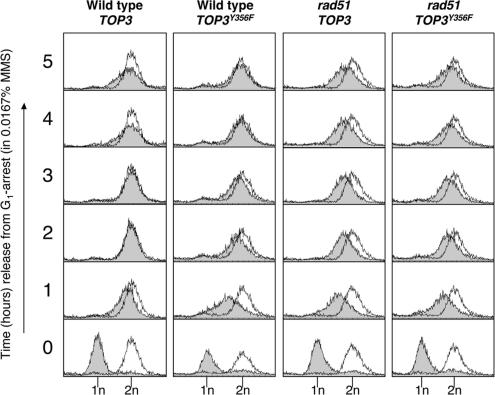
Impaired S-phase progression in rad51 cells predominates over that caused by overexpression of TOP3Y356F. Wild-type and rad51 cells overexpressing TOP3 or TOP3Y356F were released from G1 arrest into fresh medium containing 0.0167% MMS, and DNA content was analyzed by flow cytometry at the times indicated.
Impaired S-Phase Progression in rad51 Cells after Exposure to MMS Predominates Over That Caused by Overexpression TOP3Y356F
To determine whether the impaired S-phase progression phenotypes observed in rad51 mutants and cells overexpressing TOP3Y356F are epistatic or additive, we compared the effects of overexpressing TOP3 or TOP3Y356F on S-phase progression in rad51 cells in the presence of 0.0167% MMS. Under these conditions, wild-type cells overexpressing TOP3 complete DNA replication by ∼2 h, whereas wild-type cells overexpressing TOP3Y356F complete DNA replication by ∼4 h (Figures 1B and 5). In contrast, rad51 cells overexpressing TOP3 still failed to complete DNA replication by 5 h in medium containing 0.0167% MMS, confirming that the impaired S-phase progression phenotype of rad51 cells is more severe than that observed in wild-type cells overexpressing TOP3Y356F. Interestingly, we observed that the FACS profile of rad51 cells overexpressing TOP3Y356F was essentially indistinguishable from that of rad51 cells overexpressing TOP3. Therefore, overexpression of TOP3Y356F did not further impair S-phase progression in a rad51 mutant, suggesting that the impaired S-phase progression phenotypes observed in rad51 mutants or cells overexpressing TOP3Y356F are epistatic. Taken together, these data suggest that Rad51 and Top3 function in the same pathway and that the (more severe) impaired S-phase progression phenotype conferred by mutation of RAD51 predominates over that caused by overexpression of TOP3Y356F. This finding is consistent with previous reports that RAD51 and TOP3 are epistatic and that Top3 acts downstream of Rad51 in HR repair (Fabre et al., 2002; Oakley et al., 2002; Shor et al., 2002).
Mutation of RAD51 Prevents MMS-induced X-Molecules from Persisting in Cells Overexpressing TOP3Y356F
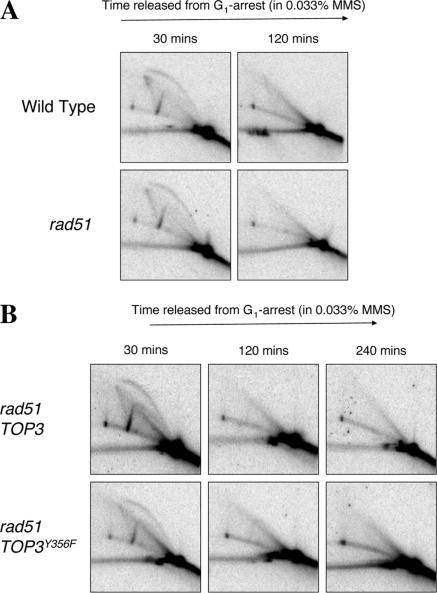
Next, we analyzed whether RAD51 mutation also prevents X-molecules from forming in cells overexpressing TOP3Y356F after exposure to MMS. We found that, despite an inability of rad51 cells to complete bulk DNA replication in the presence of 0.033% MMS (Figure 4A), these cells usually completed DNA replication (as measured by disappearance of replication intermediates on 2D gels) around the site of the ARS305 origin under routine analysis within 2 h (Figure 6A). Therefore, mutation of RAD51 does not noticeably affect early origin firing, the formation or subsequent disappearance of the (HR-independent) origin-associated X-spike, or replication fork progression around ARS305. We propose that the impaired S-phase progression after DNA damage observed in rad51 cells arises only once a (checkpoint-activating) threshold level of unrepaired, MMS-induced discontinuities in DNA synthesis is attained.
Figure 6.
Mutation of RAD51 suppresses the MMS-induced X-spikes caused by overexpression of TOP3Y356F. (A) Wild-type and rad51 strains transformed with pYES2 were released from G1 arrest into fresh medium containing 0.033% MMS. DNA replication intermediates were analyzed by 2D gel electrophoresis at the times indicated. (B) rad51 cells overexpressing TOP3 or TOP3Y356F were released from G1 arrest into fresh medium containing 0.033% MMS, and DNA replication intermediates were analyzed by 2D gel electrophoresis at the times indicated.
Similar kinetics of ARS305 origin firing (at 30 min) and disappearance of replication intermediates (at 120 min) were observed in MMS-treated rad51 mutants overexpressing either TOP3 or TOP3Y356F (Figure 6B). However, we found that the persistent X-molecules normally caused by overexpression of TOP3Y356F were not evident in rad51 strains. We conclude that mutation of RAD51 prevents MMS-induced X-molecules from persisting and/or accumulating in cells overexpressing TOP3Y356F, suggesting that Rad51 is required for their formation and/or stabilization. Together, these data are consistent with the proposal that unprocessed HR repair intermediates exist in MMS-treated cells overexpressing TOP3Y356F.
DISCUSSION
To investigate the role(s) of Top3 in the maintenance of genome stability, we have used a dominant-negative allele of TOP3, TOP3Y356F, that causes poor growth when overexpressed in wild-type cells (Oakley et al., 2002). The TOP3Y356F construct used in this study possesses a mutation in the active site tyrosine that abolishes the catalytic (decatenation) activity of Top3 and fails to complement the poor growth of a top3 mutant (Bennett and Wang, 2001). Interestingly, TOP3Y356F overexpression did not cause an identical phenocopy of a top3 mutation, because cells overexpressing TOP3Y356F exhibited impaired S-phase progression after DNA damage. Conversely, top3 cells fail to fully activate Rad53 in the presence of MMS, and, as a consequence, progress more rapidly through S phase than wild-type cells (Chakraverty et al., 2001). We propose that the checkpoint-signaling defect in top3 cells arises due to absence of Top3 protein, rather than loss of Top3 catalytic activity, because Rad53 activation seemed normal in cells overexpressing TOP3Y356F after exposure to MMS. Moreover, the impaired S-phase progression observed in cells overexpressing TOP3Y356F is due to a persistent checkpoint-mediated cell cycle delay, which can be overridden by caffeine. Therefore, although the catalytic activity of Top3 is apparently not required for normal DNA damage checkpoint activation, it is required for normal S-phase progression after DNA damage. We note, however, that although cells overexpressing TOP3Y356F demonstrate a persistent checkpoint-mediated cell cycle delay, Rad53 activation seemed qualitatively similar (as detected by Western blotting) in cells overexpressing TOP3 or TOP3Y356F after MMS treatment. However, we cannot exclude the possibility that subtle alterations to Rad53 activity, such as an altered phosphorylation pattern and/or differences in Rad53 turnover rate (dephosphorylation) in cells overexpressing TOP3Y356F might not have been evident during our analyses.
In addition to causing impaired S-phase progression, we have demonstrated that overexpression of TOP3Y356F also causes abnormal DNA structures (X-shaped DNA molecules) to persist after MMS treatment. During the course of our study, Liberi et al. (2005) also demonstrated that MMS-induced X-molecules persist in sgs1, sgs1top3, and top3 cells (Liberi et al., 2005). However, because top3 mutants readily acquire suppressor mutations in SGS1 (Gangloff et al., 1994), and sgs1top3 cells exhibit X-molecule accumulation (Liberi et al., 2005), we reasoned that these findings required independent verification by using the dominant-negative TOP3Y356F system. Our data demonstrate conclusively that impairment of Top3 function does indeed cause X-molecules to persist in MMS-treated cells. Furthermore, these MMS-induced X-molecules are Rad51-dependent and therefore likely represent unprocessed recombination intermediates. It should be noted, however, that these X-molecules differ from those normally observed at firing origins. Unlike origin-associated X-molecules (Lopes et al., 2003), the X-molecules observed in cells overexpressing TOP3Y356F are both MMS and Rad51 dependent. However, it remains to be determined whether the persistent X-molecules we detect in cells overexpressing TOP3Y356F arise independently after origin X-molecule resolution, or whether they arise due to the interconversion of the origin-associated X-molecules into bona fide HR repair intermediates via the action of Rad51/Rad52 etc.
Because overexpression of TOP3Y356F does not cause any (additive) impaired S-phase progression in rad51 cells, it is likely that the checkpoint-mediated cell cycle delay caused by overexpression of TOP3Y356F, like the persistent X-molecules, is also downstream of Rad51 activity. We propose, therefore, that the futile engagement of catalytically-dead Top3Y356F protein in HR repair is inhibitory to repair and causes a more persistent and/or robust activation of the DNA damage checkpoint (Figure 7). Because mutation of the active site tyrosine of hTOPIIIα abolishes the catalytic activity, but not an ability to bind DNA (Goulaouic et al., 1999), one possibility is that catalytically dead Top3Y356F remains bound to unprocessed HR intermediates, and that the DNA damage checkpoint then recognizes this as incomplete HR repair. Conversely, it is possible that, in top3 cells, unprocessed HR intermediates persist (Liberi et al., 2005), but they cannot be recognized by the checkpoint machinery (Chakraverty et al., 2001). Therefore, we speculate that the extent of checkpoint activation after MMS treatment varies in cells depending on whether Top3 is absent (as in top3 cells), catalytically active with normal turnover (as in wild-type cells), or catalytically dead and unable to turnover (as in cells overexpressing TOP3Y356F). Although this model is consistent with our data, we cannot rule out the possibility that the effects we observed could also be explained either by Top3 influencing events at stalled replication forks (e.g., the processing of stalled forks; Hishida et al., 2004), or the stabilization of DNA polymerases at stalled forks (Bjergbaek et al., 2005)], or by the fact that persistent X-molecules in top3 cells represent different DNA structures from those in cells overexpressing TOP3Y356F.
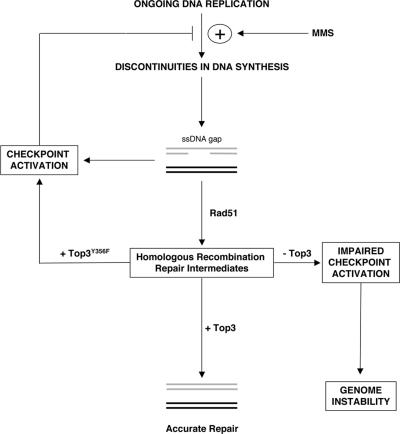
Figure 7.
Model for role of Top3 in resolving homologous recombination intermediates and modulating checkpoint activity after DNA damage. Replication forks stall when encountering MMS lesions, leading to discontinuous DNA synthesis and the accumulation of single-stranded DNA (ssDNA) gaps. Replication protein A (RPA) binds to ssDNA and then activates the DNA damage checkpoint, which promotes the stabilization of stalled replication forks at MMS lesions and inhibits further origin firing. By doing this, the DNA damage checkpoint limits further formation of recombinogenic ssDNA gaps caused by ongoing DNA replication in the presence of DNA damage. Rad51 catalyzes the early strand invasion step of homologous recombination repair of ssDNA gaps, and, through its ability to displace RPA, deactivates the checkpoint signal. Top3 acts downstream of Rad51 to resolve homologous recombination repair intermediates via its catalytic activity. Additionally, the checkpoint machinery (either directly or indirectly) senses the engagement of Top3 in homologous recombination repair and modulates checkpoint activity accordingly. Absence of Top3 protein causes unprocessed/unresolved HR intermediates to persist, and a failure to adequately delay S-phase progression after DNA damage. Conversely, catalytically dead Top3Y356F protein inhibits resolution of homologous recombination repair intermediates and promotes a persistent checkpoint-mediated cell cycle delay. DNA replication resumes at stalled replication forks once MMS-induced lesions have been removed/repaired or due to checkpoint activity falling below a threshold level.
It remains to be determined whether the activation/modulation of Rad53 activity by Top3 is direct (i.e., mediated by Top3 itself) or indirect (i.e., mediated by a Top3-binding protein). Interestingly, deletion of RMI1, which encodes a Top3-interacting protein, also causes a top3-like phenotype and results in a similar failure to fully activate Rad53 in response to DNA damage (Chang et al., 2005; Mullen et al., 2005). It is worth noting that deletion of either TOP3 or RMI1 adversely affects the stability of Sgs1 and causes a reduction in Sgs1 protein levels (Chang et al., 2005). It is possible, therefore, that Sgs1 mediates the activation of Rad53 after DNA damage via its ability to physically interact with Rad53 (Frei and Gasser, 2000; Bjergbaek et al., 2005).
Consistent with previous data (Gangloff et al., 1994; Chakraverty et al., 2001), we also found that mutation of SGS1 suppresses the poor growth and impaired S-phase progression after DNA damage caused by overexpression of TOP3Y356F. This suggests that phenotypes caused by TOP3Y356F overexpression, like other previously reported top3 phenotypes (Gangloff et al., 1994; Chakraverty et al., 2001), may arise due to the uncoupling of Sgs1 activity from that of Top3. However, we could not directly test whether mutation of SGS1 abolishes the X-molecules we observed in cells overexpressing TOP3Y356F, because sgs1 cells also accumulate (MMS-induced) Rad51-dependent X-molecules (Liberi et al., 2005; our unpublished data). Given the very different catalytic activities of Sgs1 and Top3, it seems likely that X-molecules detected in sgs1 mutants or cells overexpressing TOP3Y356F could represent different types of HR intermediates. Consistent with this idea, it has been proposed that RecQ helicases act in concert with a type IA topoisomerase to resolve key HR repair intermediates containing double Holliday junctions, in a process termed double junction dissolution (Ira et al., 2003; Wu and Hickson, 2003). This process is thought to consist of a two-step process: branch migration of individual Holliday junctions by a RecQ helicase to form a hemicatenane intermediate, which a type IA topoisomerase then decatenates via its strand passage activity. We speculate that the X-molecules detectable in sgs1 cells may represent double Holliday junctions, whereas those caused by Top3 impairment represent hemicatenanes. Ongoing experiments are aimed at testing this hypothesis by attempting to discriminate between, or selectively resolve, X-molecules present in sgs1 mutants and cells overexpressing TOP3Y356F.
Of wider significance, if the proposed functions of Top3 in resolving HR repair intermediates and modulating checkpoint activity reported here also holds true for hTOPOIIIα, this suggests that a putative hTOPOIIIα inhibitor that selectively inhibits the catalytic activity of hTOPOIIIα alone, could provide a means to greatly sensitize rapidly proliferating cells to DNA damage. Therefore, we propose that hTOPOIIIα might represent an attractive anticancer drug target in human cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Cotta-Ramusino, G. Liberi, and M. Foiani for technical advice, sharing unpublished data, and the EL7 Rad53 antibody. We also thank the Cancer Research UK peptide synthesis laboratory for providing α-factor; M. Resnick for the T344 strain; S. Gasser for the SGS1::TRP1 deletion cassette; and P. McHugh, L. Wu, R. Borts, and various members of the Hickson laboratory for helpful comments and criticisms. This work was funded by Cancer Research UK.
Abbreviation used:
- HR
homologous recombination.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-06-0516) on August 9, 2006.
REFERENCES
- Ahmad F., Stewart E. The N-terminal region of the Schizosaccharomyces pombe RecQ helicase, Rqh1p, physically interacts with Topoisomerase III and is required for Rqh1p function. Mol. Genet. Genomics. 2005;273:102–114. doi: 10.1007/s00438-005-1111-3. [DOI] [PubMed] [Google Scholar]
- Allers T., Lichten M. A method for preparing genomic DNA that restrains branch migration of Holliday junctions. Nucleic Acids Res. 2000;28:e6. doi: 10.1093/nar/28.2.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R. J., Noirot-Gros M. F., Wang J. C. Interaction between yeast sgs1 helicase and DNA topoisomerase III. J. Biol. Chem. 2000;275:26898–26905. doi: 10.1074/jbc.M003137200. [DOI] [PubMed] [Google Scholar]
- Bennett R .J., Wang J. C. Association of yeast DNA topoisomerase III and Sgs1 DNA helicase: studies of fusion proteins. Proc. Natl. Acad. Sci. USA. 2001;98:11108–11113. doi: 10.1073/pnas.201387098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjergbaek L., Cobb J. A., Tsai-Pflugfelder M., Gasser S. M. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J. 2005;24:405–417. doi: 10.1038/sj.emboj.7600511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Chakraverty R. K., Kearsey J. M., Oakley T. J., Grenon M., de La Torre Ruiz M. A., Lowndes N. F., Hickson I. D. Topoisomerase III acts upstream of Rad53p in the S-phase DNA damage checkpoint. Mol. Cell. Biol. 2001;21:7150–7162. doi: 10.1128/MCB.21.21.7150-7162.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M., Bellaoui M., Zhang C., Desai R., Morozov P., Delgado-Cruzata L., Rothstein R., Freyer G. A., Boone C., Brown G. W. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J. 2005;24:2024–2033. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis N. A., Groden J., Ye T. Z., Straughen J., Lennon D. J., Ciocci S., Proytcheva M., German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- Fabre F., Chan A., Heyer W. D., Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M., Marini F., Gamba D., Lucchini G., Plevani P. The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol. 1994;14:923–933. doi: 10.1128/mcb.14.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei C., Gasser S. M. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- Fricke W. M., Kaliraman V., Brill S. J. Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J. Biol. Chem. 2001;276:8848–8855. doi: 10.1074/jbc.M009719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S., de Massy B., Arthur L., Rothstein R., Fabre F. The essential role of yeast topoisomerase III in meiosis depends on recombination. EMBO J. 1999;18:1701–1711. doi: 10.1093/emboj/18.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S., McDonald J. P., Bendixen C., Arthur L., Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German J. Bloom syndrome: a Mendelian prototype of somatic mutational disease. Medicine. 1993;72:393–406. [PubMed] [Google Scholar]
- Goodwin A., Wang S. W., Toda T., Norbury C., Hickson I. D. Topoisomerase III is essential for accurate nuclear division in Schizosaccharomyces pombe. Nucleic Acids Res. 1999;27:4050–4058. doi: 10.1093/nar/27.20.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulaouic H., Roulon T., Flamand O., Grondard L., Lavelle F., Riou J. F. Purification and characterization of human DNA topoisomerase IIIalpha. Nucleic Acids Res. 1999;27:2443–2450. doi: 10.1093/nar/27.12.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Jackson C. A., Cross D. A., Morrice N., Smythe C. ATR is a caffeine-sensitive, DNA-activated protein kinase with a substrate specificity distinct from DNA-PK. Oncogene. 1999;18:6707–6713. doi: 10.1038/sj.onc.1203077. [DOI] [PubMed] [Google Scholar]
- Hanai R., Caron P. R., Wang J. C. Human TOP 3, a single-copy gene encoding DNA topoisomerase III. Proc. Natl. Acad. Sci. USA. 1996;93:3653–3657. doi: 10.1073/pnas.93.8.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon F. G., DiGate R. J., Kowalczykowski S. C. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol. Cell. 1999;3:611–620. doi: 10.1016/s1097-2765(00)80354-8. [DOI] [PubMed] [Google Scholar]
- Hickson I. D. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- Hishida T., Han Y. W., Shibata T., Kubota Y., Ishino Y., Iwasaki H., Shinagawa H. Role of the Escherichia coli RecQ DNA helicase in SOS signaling and genome stabilization at stalled replication forks. Genes Dev. 2004;18:1886–1897. doi: 10.1101/gad.1223804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovland P., Flick J., Johnston M., Sclafani R. A. Galactose as a gratuitous inducer of GAL gene expression in yeasts growing on glucose. Gene. 1989;83:57–64. doi: 10.1016/0378-1119(89)90403-4. [DOI] [PubMed] [Google Scholar]
- Ira G., Malkova A., Liberi G., Foiani M., Haber J. E. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson F. B., Lombard D. B., Neff N. F., Mastrangelo M. A., Dewolf W., Ellis N. A., Marciniak R. A., Yin Y., Jaenisch R., Guarente L. Association of the Bloom syndrome protein with topoisomerase IIIalpha in somatic and meiotic cells. Cancer Res. 2000;60:1162–1167. [PubMed] [Google Scholar]
- Kim Y. C., Lee M. H., Ryu S. S., Kim J. H., Koo H. S. Coaction of DNA topoisomerase IIIalpha and a RecQ homologue during the germ-line mitosis in Caenorhabditis elegans. Genes Cells. 2002;7:19–27. doi: 10.1046/j.1356-9597.2001.00496.x. [DOI] [PubMed] [Google Scholar]
- Kitao S., Shimamoto A., Goto M., Miller R. W., Smithson W. A., Lindor N. M., Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- Kwan K. Y., Moens P. B., Wang J. C. Infertility and aneuploidy in mice lacking a type IA DNA topoisomerase III beta. Proc. Natl. Acad. Sci. USA. 2003;100:2526–2531. doi: 10.1073/pnas.0437998100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen L. V., Ampatzidou E., Andersen A. H., Murray J. M. Role for the fission yeast RecQ helicase in DNA repair in G2. Mol. Cell. Biol. 2003;23:3692–3705. doi: 10.1128/MCB.23.10.3692-3705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wang J. C. Mammalian DNA topoisomerase IIIalpha is essential in early embryogenesis. Proc. Natl. Acad. Sci. USA. 1998;95:1010–1013. doi: 10.1073/pnas.95.3.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberi G., Maffioletti G., Lucca C., Chiolo I., Baryshnikova A., Cotta-Ramusino C., Lopes M., Pellicioli A., Haber J. E., Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M., Cotta-Ramusino C., Liberi G., Foiani M. Branch migrating sister chromatid junctions form at replication origins through Rad51/Rad52-independent mechanisms. Mol. Cell. 2003;12:1499–1510. doi: 10.1016/s1097-2765(03)00473-8. [DOI] [PubMed] [Google Scholar]
- Maftahi M., Han C. S., Langston L. D., Hope J. C., Zigouras N., Freyer G. A. The top3(+) gene is essential in Schizosaccharomyces pombe and the lethality associated with its loss is caused by Rad12 helicase activity. Nucleic Acids Res. 1999;27:4715–4724. doi: 10.1093/nar/27.24.4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser B. A., Brondello J. M., Baber-Furnari B., Russell P. Mechanism of caffeine-induced checkpoint override in fission yeast. Mol. Cell. Biol. 2000;20:4288–4294. doi: 10.1128/mcb.20.12.4288-4294.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J. R., Nallaseth F. S., Lan Y. Q., Slagle C. E., Brill S. J. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol. Cell. Biol. 2005;25:4476–4487. doi: 10.1128/MCB.25.11.4476-4487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S. W., Liu Y., Hasselblatt K. T., Mok S. C., Berkowitz R. S. A new human topoisomerase III that interacts with SGS1 protein. Nucleic Acids Res. 1999;27:993–1000. doi: 10.1093/nar/27.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley T. J. Ph.D. Thesis. Oxford, United Kingdom: University of Oxford; 2001. S. cerevisiae Topoisomerase III. [Google Scholar]
- Oakley T. J., Goodwin A., Chakraverty R. K., Hickson I. D. Inactivation of homologous recombination suppresses defects in topoisomerase III-deficient mutants. DNA Repair. 2002;1:463–482. doi: 10.1016/s1568-7864(02)00032-0. [DOI] [PubMed] [Google Scholar]
- Oh M., Choi I. S., Park S. D. Topoisomerase III is required for accurate DNA replication and chromosome segregation in Schizosaccharomyces pombe. Nucleic Acids Res. 2002;30:4022–4031. doi: 10.1093/nar/gkf531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera R., Seki M., Ui A., Satoh Y., Miyajima A., Onoda F., Enomoto T. Functional and physical interaction between Sgs1 and Top3 and Sgs1-independent function of Top3 in DNA recombination repair. Genes Genet. Syst. 2002;77:11–21. doi: 10.1266/ggs.77.11. [DOI] [PubMed] [Google Scholar]
- Osman F., McCready S. Differential effects of caffeine on DNA damage and replication cell cycle checkpoints in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 1998;260:319–334. doi: 10.1007/s004380050901. [DOI] [PubMed] [Google Scholar]
- Paulovich A. G., Hartwell L. H. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- Santocanale C., Diffley J. F. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- Schlegel R., Pardee A. B. Caffeine-induced uncoupling of mitosis from the completion of DNA replication in mammalian cells. Science. 1986;232:1264–1266. doi: 10.1126/science.2422760. [DOI] [PubMed] [Google Scholar]
- Shirahige K., Hori Y., Shiraishi K., Yamashita M., Takahashi K., Obuse C., Tsurimoto T., Yoshikawa H. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- Shor E., Gangloff S., Wagner M., Weinstein J., Price G., Rothstein R. Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics. 2002;162:647–662. doi: 10.1093/genetics/162.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero J. A., Diffley J. F. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- Ui A., Seki M., Ogiwara H., Onodera R., Fukushige S., Onoda F., Enomoto T. The ability of Sgs1 to interact with DNA topoisomerase III is essential for damage-induced recombination. DNA Repair. 2005;4:191–201. doi: 10.1016/j.dnarep.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Vaze M. B., Pellicioli A., Lee S. E., Ira G., Liberi G., Arbel-Eden A., Foiani M., Haber J. E. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell. 2002;10:373–385. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pohlmann R., Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wallis J. W., Chrebet G., Brodsky G., Rolfe M., Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- Win T. Z., Goodwin A., Hickson I. D., Norbury C. J., Wang S. W. Requirement for Schizosaccharomyces pombe Top3 in the maintenance of chromosome integrity. J. Cell Sci. 2004;117:4769–4778. doi: 10.1242/jcs.01351. [DOI] [PubMed] [Google Scholar]
- Wu L., Davies S. L., North P. S., Goulaouic H., Riou J. F., Turley H., Gatter K. C., Hickson I. D. The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- Wu L., Hickson I. D. The Bloom's syndrome helicase stimulates the activity of human topoisomerase IIIalpha. Nucleic Acids Res. 2002;30:4823–4829. doi: 10.1093/nar/gkf611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Hickson I. D. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Yu C. E., et al. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.