Abstract
The Ca2+/calmodulin (CaM)-dependent protein kinase II (CaMKII)β has morphogenic functions in neurons not shared by the α isoform. CaMKIIβ contains three exons (v1, v3, and v4) not present in the CaMKIIα gene, and two of these exons (v1 and v4) are subject to differential alternative splicing. We show here that CaMKIIβ, but not α, mediated bundling of F-actin filaments in vitro. Most importantly, inclusion of exon v1 was required for CaMKIIβ association with the F-actin cytoskeleton within cells. CaMKIIβe, which is the dominant variant around birth and lacks exon v1 sequences, failed to associate with F-actin. By contrast, CaMKIIβ′, which instead lacks exon v4, associated with F-actin as full-length CaMKIIβ. Previous studies with CaMKIIβ mutants have indicated a role of nonstimulated kinase activity in enhancing dendritic arborization. Here, we show that F-actin–targeted CaMKIIβ, but not α, was able to phosphorylate actin in vitro even by nonstimulated basal activity in absence of Ca2+/CaM. In rat pancreatic islets and in skeletal muscle, the actin-associated CaMKIIβ′ and βM were the predominant variants, respectively. Thus, cytoskeletal targeting may mediate functions of CaMKIIβ variants also outside the nervous system.
INTRODUCTION
Changes in synaptic connectivity between neurons are widely thought to underlie higher brain functions such as learning and memory and are also important during development. Long-lasting changes in connectivity are often associated with morphological plasticity in structure or number of synaptic connections (for reviews, see Huntley et al., 2002; McGee and Bredt, 2003; Lamprecht and LeDoux, 2004; Segal, 2005). The α isoform of Ca2+/calmodulin(CaM)-dependent protein kinase II (CaMKII) has been studied extensively because of its prominent role in regulating the strength of individual synaptic connections (for examples, see Malinow et al., 1989; Silva et al., 1992; Giese et al., 1998; for reviews, see Malenka and Nicoll, 1999; Lisman et al., 2002; Griffith, 2004). Much less is known about the other major brain isoform, CaMKIIβ. However, CaMKIIβ has specific morphogenic functions in regulating dendritic arborization and synapse density not shared by the α isoform (Fink et al., 2003). This isoform specificity is thought to be mediated by the specific binding of CaMKIIβ, but not α to F-actin (Shen et al., 1998; Fink et al., 2003).
The four CaMKII isoforms encoded by different genes (α, β, γ, and δ) are highly homologous to each other and can phosphorylate and regulate a variety of substrate proteins in response to Ca2+ signals (for reviews, see Soderling et al., 2001; Hudmon and Schulman, 2002; Lisman et al., 2002; Colbran and Brown, 2004). The multimeric CaMKII holoenzymes are composed of 12 subunits (Kolodziej et al., 2000; Morris and Torok, 2001; Rosenberg et al., 2005; Rosenberg et al., 2006) of a single isoform or combinations of different isoforms (Bayer et al., 1998; Shen et al., 1998; Brocke et al., 1999; Lantsman and Tombes, 2005). All CaMKII isoforms contain an N-terminal kinase domain and a C-terminal association domain, connected by a variable linker region that is subject to alternative splicing (see Figure 1). Within a holoenzyme, each subunit is activated separately by Ca2+/CaM; however, binding of Ca2+/CaM to two neighboring subunits initiates an intersubunit autophosphorylation at T286 (or T287 in β, γ, and δ) that renders the kinase autonomous (active in the absence of Ca2+/CaM) (Hanson et al., 1994; Rich and Schulman, 1998). This autophosphorylation may have a molecular memory function (for review, see Lisman and McIntyre, 2001) and enables Ca2+-spike frequency decoding by CaMKII (De Koninck and Schulman, 1998; Bayer et al., 2002). At least one CaMKII isoform is present in every tissue examined (Tobimatsu and Fujisawa, 1989; Bayer et al., 1999). However, CaMKII is especially enriched in the brain, where the α and β isoforms can constitute up to 1–2% of total protein (Erondu and Kennedy, 1985). Although CaMKIIα is exclusively expressed in neurons, CaMKIIβ is additionally found in skeletal muscle and in endocrine cells such as pituitary, adrenal glands, and pancreatic β cells (Tobimatsu and Fujisawa, 1989; Urquidi and Ashcroft, 1995; Bayer et al., 1998; Rochlitz et al., 2000; Tabuchi et al., 2000). CaMKII is thought to regulate Ca2+ homeostasis in muscle (Wang and Best, 1992; Xu et al., 1993; Hain et al., 1995) and insulin secretion and production in pancreatic beta cells (Wasmeier and Hutton, 1999; Tabuchi et al., 2000; Osterhoff et al., 2003; for review, see Easom, 1999).
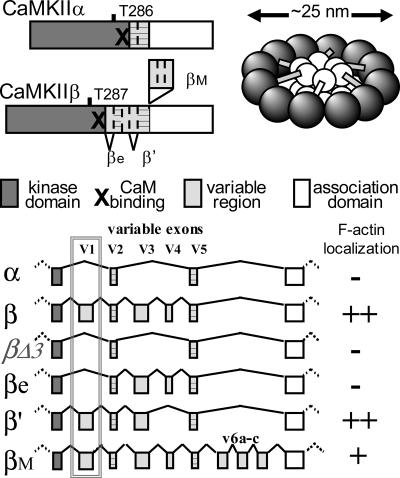
Figure 1.
CaMKII variants and their F-actin localization based on results of this study. Schematic representation of individual isoforms (left) and holoenzyme structure (right). CaMKIIα and β differ most in the variable region; however, they are products of different genes and thus also have minor differences in the kinase and association domain. Bottom, exon/intron structure of the CaMKIIα and β gene loci coding for the variable regions, with alternative splicing indicated. βΔ3 is a deletion mutant, not a splice variant found in brain (see Supplemental Figure 2). Subcellular localization of the kinase variants (see Figure 3) implicated necessity of exon v1 (boxed) for association with the F-actin cytoskeleton.
Structurally, the most striking difference between the CaMKII isoforms is within the variable linker region that connects the kinase and association domains (for review, see Hudmon and Schulman, 2002; Tombes et al., 2003). For example, the CaMKIIα gene does not contain any sequences homologous to the CaMKIIβ variable exons v1, v3, and v4 (Figure 1), whereas the CaMKIIβ gene does not contain a homologue to an insert that mediates nuclear targeting of the CaMKIIα splice variant αB (Srinivasan et al., 1994; Heist et al., 1998). Thus, we hypothesized that sequences encoded by the exons v1, v3, and/or v4 are involved in CaMKIIβ-specific targeting to the F-actin cytoskeleton. At least CaMKIIβ exons v1 and v4 are subject to alternative splicing in brain. Although expression of CaMKIIβ dominates in mature brain, expression of CaMKIIβe, which lacks exon v1, dominates in brain around and before birth (Brocke et al., 1995). Interestingly, alternative splicing of exons v1 (lacking in βe and βe′) and v4 (lacking in β′ and βe′) is differentially regulated even among individual mature hippocampal CA1 pyramidal neurons, with most neurons expressing exclusively one splice variant (Brocke et al., 1999). Thus, tightly regulated alternative splicing may control expression of differently actin-associated CaMKIIβ variants, thereby possibly affecting the morphogenic functions of CaMKIIβ in dendritic arborization and/or synapse density. Results of this study summarized in Figure 1 show that CaMKIIβ and β′, but not βe, associated with the F-actin cytoskeleton, demonstrating a developmental switch between differently targeted splice variant.
MATERIALS AND METHODS
F-Actin Bundling Assays by Centrifugation and Electron Microscopy
Purified actin from chicken muscle was a kind gift by Dr. R. Rock (Department of Biochemistry, Spudich Lab, University of California, Stanford, CA). CaMKIIα or β was purified from a baculovirus/Sf9 expression system on a phosphocellulose column followed by gel filtration chromatography, as described previously (Singla et al., 2001; Bradshaw et al., 2002; Fink et al., 2003). Actin (4 μM) was polymerized on ice in F-buffer (25 mM HEPES, pH 7.4, 50 mM KCl, 2 mM MgCl2, 1 mM EGTA, 0.5 mM dithiothreitol [DTT], and 0.2 mM ATP). CaMKII (170 nM subunits), 3 μM CaM, and/or 2 mM CaCl2 were added as indicated and binding was assessed as described previously (Fink et al., 2003). However, sedimentation was carried out at lower centrifugation speed (10,000 × g) for 20 min. Both F-actin and CaMKII were spun under the same condition before the binding assay. Supernatants and pellets were analyzed for actin and CaMKIIα or β by Western blot analysis with the specific antibodies anti-actin 20–33 (rabbit polyclonal; 1:500 in 2% bovine serum albumin [BSA]; Sigma-Aldrich, St. Louis, MO), CBα2 or CBβ1 (mouse monoclonals; 1:2000 and 1:1000 in 2.5% milk), respectively (Bayer et al., 1998; Fink et al., 2003).
For electron microscopy, actin was polymerized, and kinase was added as described above. Copper grids were coated with Formvar and carbon and then glow discharged. Approximately 10 μl of the actin/kinase mix was applied to the copper grids for 2 min and excess liquid was wicked away with blotting paper. Grids were stained with 1–2% uranyl acetate for 2 min and then briefly rinsed in distilled water. Excess liquid was removed, and grids were allowed to dry. Electron microscopy was performed on a Tecnai G2 BioTwin (at 80 kV and 49,000 magnification) in the electron microscopy core facility of University of Colorado Health Sciences Center (Aurora, CO).
Reverse Transcription-Polymerase Chain Reaction (RT-PCR) Expression Analysis
Pancreatic islets from Sprague Dawley rats (Harlan, Indianapolis, IN) were isolated at the islet core facility of the Diabetes and Endocrine Research Center at University of Colorado Health Sciences Center. Total cellular RNAs were prepared using RNAqueous-4PCR (Ambion, Austin, TX). SuperScriptIII reverse transcriptase (Invitrogen, Carlsbad, CA) was used for cDNA synthesis, primed with both random decamers and 12–18mer oligo(dT). PCR amplification for expression analysis was done with Platinum-Taq (Invitrogen) and CaMKIIβ-specific primers flanking the variable region (bvf, GACAGGAGACTGTGGAATGTC and bvr, TCAAAGTCGCCGTTGTTGAC) for 40 cycles with 15-s denaturation at 94°C for 20 s, annealing at 55°C, and 90-s elongation at 72°C (shorter elongation times favored shorter PCR products from skeletal muscle cDNA, corresponding in length to β instead of βM; our unpublished data). RT-PCR products were separated on 2% agarose gels in TAE-buffer, and stained with ethidium bromide (0.1 μg/ml gel). For direct sequencing, RT-PCR products were purified using QIAquick (QIAGEN, Valencia, CA).
Cell Culture, Transfection, and Constructs
Cos-7 cells were cultured on glass-bottomed dishes (30 mm with 12-mm glass bottom; MatTek, Ashland, MA) and transfected by the calcium phosphate method as described previously (Bayer et al., 1998, 2001, 2006). An enhanced green fluorescent protein [EGFP] A207K mutant with reduced dimerization was used to create membrane-associated form of green fluorescent protein (mGFP)-CaMKII constructs (Zacharias et al., 2002; Bayer et al., 2006). Vectors for expression of unlabeled or hemagglutinin (HA)-tagged CaMKII splice variants were described previously (Brocke et al., 1995; Bayer et al., 2002); SacI/PmlI fragments were exchanged to create the mGFP fusion protein (the SacI site in the multiple cloning site of the original mGFP-CaMKIIβ was deleted by religation after XhoI/EcoRI cut). mGFP-CaMKIIβ K43R, A303R, and T287D were created by exchanging cyan fluorescent protein in previously described constructs (Bayer et al., 2002) with mGFP. mGFP-CaMKIIβΔ3 was created by PCR mutagenesis by using mGFP-CaMKIIβe as a template and primer combination that flanked the exon v3 and v4 sequences. After sequencing of a positive clone, a SacI/PmlI fragment was exchanged with the original vector as described above to avoid possible PCR-generated mutations in the vector sequence.
F-Actin Staining and Microscopy
mGFP-CaMKII localization in Cos-7 cells was analyzed 2 d after transfection. Scoring of actin cytoskeletal localization in live cells was done on a Nikon TE-300 inverted microscope with a 63× objective, blind of the mGFP-CaMKII variant used. Fifty or all transfected cells per dish were scored; eight dishes were analyzed for each construct. Alternatively, cells were fixed in 4% paraformaldehyde in 100 mM phosphate buffer, pH 7.2, for 10 min at room temperature, permeabilized for 20–30 min in 0.1% Triton X-100 in phosphate-buffered saline (PBS), and F-actin was stained by 165 nM Texas Red-phalloidin (Invitrogen) in PBS for 30 min. Images were taken on a Zeiss Axiovert 200M microscope (Carl Zeiss, Thornwood, NY) equipped with a 63× plan-apo/1.2 numerical aperture objective, 175-W xenon light source, independently controlled excitation and emission filter wheels, and a CoolSnapHQ camera (Roper Scientific, Trenton, NJ). Fluorescence images were acquired blind of the mGFP construct and analyzed for GFP/Texas Red colocalization correlation index by using SlideBook software (Intelligent Imaging Innovations, Denver, CO). Six images 0.5 μm apart in the z-plane were taken per cell, deconvoluted, and maximum projected.
Fluorescence Recovery after Photobleaching (FRAP) Analysis
Motility of mGFP-CaMKII variants in transfected Cos-7 cells was analyzed by FRAP on the Zeiss imaging setup described above, at 30°C in 50 mM HEPES-buffered, pH 7.4, Hank's balanced saline. Bleaching was done using a micropoint FRAP system (Photonic Instruments, St. Charles, IL), and images were acquired every 5 s; the first image acquisition possible on the setup was ∼2 s after bleach (set to t = 0 s). Background fluorescence, measured outside cells, was subtracted. The average fluorescence intensity during 60 s before bleach was rescaled to 1, and the intensity on the first image after bleach was rescaled to 0. Photobleach caused by image acquisition was minimal and was not corrected for in the data presentation. The slope (k) and maximum (ymax) of the recovery curves was determined by curve fit to the single exponential association function y = ymax * (1 − e−kx) in GraphPad Prism software (GraphPad Software, San Diego, CA). For calculating bleach efficiency, intensity before bleach was rescaled to 0%, and background intensity was rescaled to −100%. To correct the estimate of the recoverable pool for recovery occurring even before the first test image, intensity of the first live image after bleach was rescaled to apparent bleach efficiency in this image divided by the real bleach efficiency as measured in fixed cells (instead of rescaling it to 0). The estimate of the actual recoverable pool was not corrected for reduction in the total fluorescence within cells caused by the bleach.
F-Actin Phosphorylation Assay
Purified CaMKII (100 nM subunits) was added to 4 μM actin polymerized in F-buffer as described above. The kinase/actin mix was then diluted 1:5 into phosphorylation buffer (25 mM HEPES, pH 7.4, 50 mM KCl, 20 mM MgCl2, 0.2 mM [γ-32P]ATP at 0.5 Ci/mmol, and 1.2 mM EGTA) and incubated for 15 min at 30°C to measure phosphorylation by basal activity. Stimulated phosphorylation was induced for 3 min by a mix that contained 2 mM CaCl2 and 1 μM CaM instead of EGTA. CaMKII inhibitors were added as indicated. To assess phosphorylation of kinase and actin, the reaction mixes were separated on SDS-gels, blotted onto nitrocellulose membrane, and subjected to autoradiography.
In a similar assay, mGFP-CaMKIIβ wild type and K43R in extracts from transfected Cos-7 cells were used, with extracts of mock-transfected cells as control. Cos-7 cells were extracted in HB (50 mM PIPES, pH 7.2, 10% glycerol, 1 mM EGTA, 1 mM DTT, and 1X Boehringer protease inhibitor cocktail), and kinase was solubilized with 150 mM sodium perchlorate before a 20-min 16,000 × g centrifugation at 4°C. The extract supernatants were adjusted for amount of kinase (as determined by GFP fluorescence) and total Cos protein (using extracts from mock-transfected cells; determined by Bradford assay with BSA standard). Adjusted extracts were centrifuged for 60 min at 100,000 × g. The sodium perchlorate was removed from the supernatants using Microcon YM-100 spin columns (Millipore, Billerica, MA), yielding the final extracts used in the phosphorylation reactions.
RESULTS
F-Actin Bundling by CaMKIIβ, but Not α
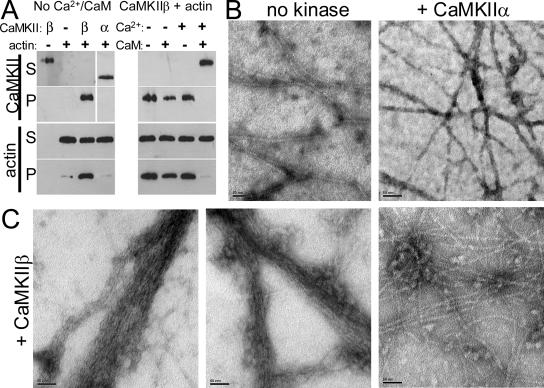
Based on the multimeric holoenzyme structure (Figure 1), we hypothesized that CaMKIIβ may be able to interact with several F-actin filaments simultaneously. Formation of such large F-actin complexes can be assessed in vitro by low-speed sedimentation assays. Although individual filaments are sedimented at 100,000 × g, presence of a cross-linking protein is required for F-actin sedimentation at 10,000 × g. Indeed, F-actin was sedimented at low speed only in presence of CaMKIIβ (Figure 2A). As previously described for binding, formation of large CaMKIIβ/F-actin complexes was disrupted by Ca2+/CaM; either Ca2+ or CaM alone was not sufficient to disrupt these complexes (Figure 2A). By contrast, CaMKIIα did not result in F-actin sedimentation under any conditions.
Figure 2.
CaMKIIβ, but not α, bundles F-actin. (A) Actin was polymerized before addition of CaMKIIβ or α, as indicated. CaMKII and actin content of supernatants (S) and pellets (P) after low-speed centrifugation (10,000 × g; 20 min) were analyzed by the Western blots shown. Left, CaMKIIβ and actin were found in the pellet only when mixed; CaMKIIα and actin did not cosediment. Right, Addition of Ca2+/CaM (2 mM/3 μM), but not Ca2+ or CaM alone, prevented cosedimentation of CaMKIIβ and actin. (B) F-actin filaments are visualized by electron microscopy and do not form bundles in absence of kinase (left) or in presence of CaMKIIα (right). (C) Addition of CaMKIIβ to polymerized actin resulted in bundling of the F-actin filaments, as assessed by electron microscopy. In some areas, F-actin cross-linking without bundling was apparent (right). Bars, 50 nm.
To examine the nature of the larger F-actin/CaMKIIβ complexes, electron microscopy was performed after negative staining with uranyl acetate. Without kinase or in presence of CaMKIIα, no formation of extensive actin filament bundles was observed (Figure 1B and Supplemental Figure 1), as expected. By contrast, addition of CaMKIIβ resulted in formation of F-actin bundles with multiple parallel filaments (Figure 2C). Additionally, in some areas CaMKIIβ seemed to cross-link filaments without bundle formation (Figure 2C, right). In these areas, particles of CaMKII holoenzyme size (∼25 nm in diameter) were observed associated with the actin filaments. Similar particles were also seen associated with the F-actin bundles. Although such particles also may be present within the tightly packed bundles, they could not be clearly distinguished in such location (Figure 2C).
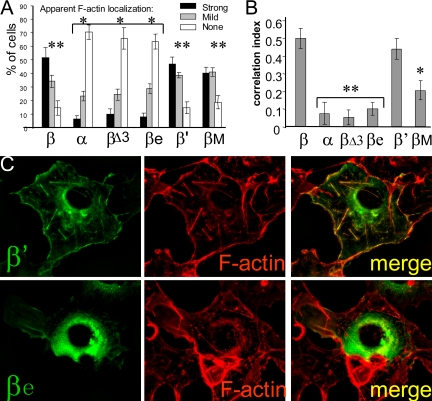
CaMKIIβ and β′, but Not βΔ3 or βe, Associated with the Actin Cytoskeleton
We hypothesized that the β variable exons v1, v3, and/or v4, which are lacking in the α isoform, are necessary for F-actin association (Figure 1). CaMKIIβe and β′ lack exons v1 and v4, respectively. By contrast, no splice variant lacking exon v3 was detected in rat brain (Supplemental Figure 2). Thus, we generated a CaMKIIβ mutant (βΔ3) that lacks exons v1, v3, and v4. mGFP fusion proteins of the CaMKIIα and β isoforms, the βΔ3 mutant, and the splice variants β′, βe, and βM (Figure 1) were expressed in Cos-7 cells, and their subcellular localization was analyzed (Figure 3). First, apparent F-actin localization (stress fibers, cortical actin, and membrane ruffles) of the GFP fluorescence in live cells was scored into three categories (strong, mild, and no apparent F-actin localization) (Figure 3A). Then, cells were fixed and F-actin was stained with phalloidin-Texas Red, and the index of GFP/Texas Red colocalization was determined on deconvoluted images (Figure 3B). Examples of phalloidin-stained transfected Cos-7 cells are shown in Figure 3C. As hypothesized, CaMKIIβΔ3 did not associate with F-actin structures. Even lack of exon v1 alone, in the splice variant βe, was sufficient to disrupt F-actin association. Localization of both βΔ3 and of βe was indistinguishable from CaMKIIα localization in both analysis methods. By contrast, lack of exon v4 in β′ did not reduce F-actin association seen for the full-length CaMKIIβ. CaMKIIβM contains an additional insert C-terminal of exon v5 (Figure 2) that generates putative binding sites for Src homology 3 (SH3) domains (Bayer et al., 1998). Localization of βM, β, and β′ were indistinguishable in the scoring analysis in live cells (Figure 3A). After phalloidin stain, the F-actin colocalization coefficient for βM was significantly higher than for α, βΔ3, and βe; however, it was lower compared with β and β′ (Figure 3B). This may reflect association of the βM variant with additional subcellular targets, as has been speculated previously based on putative SH3 binding (Urquidi and Ashcroft, 1995; Bayer et al., 1998). Together, the results clearly indicate that the variable region exon v1, which is lacking in CaMKIIβe, is necessary for localization to the actin cytoskeleton within cells (for overview, see Figure 1).
Figure 3.
Alternative splicing modulates F-actin association of CaMKIIβ. (A) Scores of apparent F-actin localization for GFP-CaMKII variants in live Cos-7 cells. Scoring was done blind of the variant. n = 8 coverslips (50 or all transfected cells scored). There are no differences within the groups labeled * or ** (p > 0.3; analysis of variance [ANOVA]), but each * is different from each ** (p < 0.00005; two-tailed t tests) (tested were “strong” and “none” as independent values). (B) F-actin/GFP-CaMKII correlation of localization values determined using SlideBook software, after staining of F-actin with phalloidin-Texas Red as shown in C. n = 9 images. **, not different from each other (p > 0.2; ANOVA), but different from β and β′ (p < 0.00001) and βM (p < 0.05). Error bars show SEM. (C) Examples of GFP-CaMKIIβ variant localization in Cos-7 cells. F-actin was stained by phalloidin-Texas Red after fixation.
CaMKIIα and βe Were More Mobile within Cells than β and β′
To further probe into differential subcellular anchoring of the CaMKII isoforms and splice variants, we used FRAP of the mGFP fusion proteins expressed in Cos-7 cells (Figure 4). FRAP of the nonactin binding CaMKIIα and βe was indistinguishable from each other and relatively fast (Figure 4A) (k = 5.79 ± 0.26 and 5.12 ± 0.29 min−1, respectively, after curve fit to single exponential association). Fluorescence recovered almost completely (∼90%) within 2 min, indicating that α and βe are mobile and not persistently targeted to any subcellular structures within Cos-7 cells. FRAP of CaMKIIβ and β′ was also similar to each other, but significantly different from α and βe FRAP (Figure 4A). FRAP was slower (k = 1.55 ± 0.08 and 2.15 ± 0.09 min−1, respectively) and fluorescence recovered only partially (∼50%). This indicates that a significant pool of β and β′ remained persistently bound to F-actin and did not exchange in the unstimulated Cos-7 cells. Examples of βe and β′ FRAP images are shown in Figure 4A. In addition to bleaching βe in its typical evenly distributed localization (Figure 4A, first row), we also bleached βe enriched in actin-like structures (Figure 5A, second row), which is more typical for β or β′ localization (Figure 4A, third row). FRAP of βe was equally fast and complete in both localizations, indicating that any apparent actin-like localization of βe is different in nature from the more extensive actin association of β or β′.
Figure 4.
CaMKII variants differ in their motility within cells. FRAP of GFP-CaMKII was assessed in Cos-7 cells at 30°C. (A) Top, examples of GFP-CaMKII bleach and recovery in Cos-7 cells. Two examples are shown for βe: one bleach of its typical dispersed localization and one bleach is of an F-actin–like structure. Bottom, quantitative assessment of bleach recovery over time, with fluorescence before and immediately after bleach rescaled to 1 and 0, respectively. FRAP differed between cytosolic (α and βe) and actin-binding (β and β′) variants (p < 0.005 in two tailed t tests; n = 10 bleaches in 5 cells for each variant). (B) Apparent bleach efficiencies differed between cytosolic and actin-binding variants in live but not in fixed cells. *, not different from each other (p > 0.4), but different from all other measurements (p < 0.005); other data points do not differ from each other (p > 0.4; ANOVA) (n = 10). This suggests partial recovery even before the first picture could be taken (∼2 s after bleach). This indicates that the recoverable pool for the cytosolic variants is even greater than indicated by the rescaled measurements in A. Error bars show SEM.
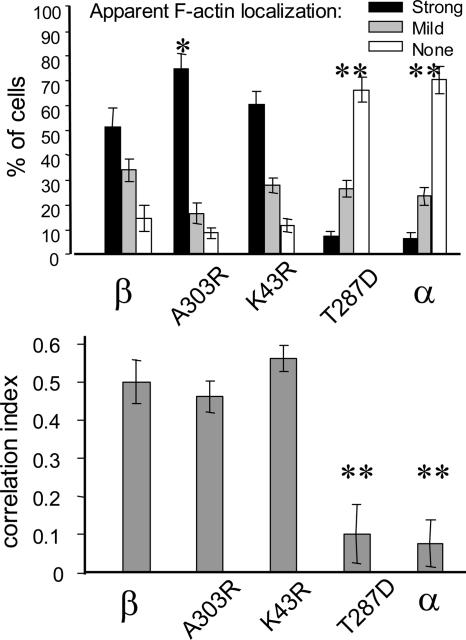
Figure 5.
F-actin localization of CaMKIIβ mutants. GFP-CaMKIIβ mutants A303R (CaM binding impaired), K43R (ATP binding impaired), and T287D (constitutively active) were tested for F-actin localization in Cos-7 cells by live scoring (n = 8) or correlation with phalloidin stain (n = 9) as described in Figure 3. CaMKIIβ T287D and CaMKIIα (**) showed significantly less F-actin localization than any other variant (p < 0.0005 in two-tailed t test), and they did not differ from each other. β A303R (*) showed slightly increased actin localization in the live scoring (p < 0.05 for the strong localization category).
Apparent bleach efficiencies for mGFP-CaMKIIα and βe in live cells were only about one-half of that seen for β and β′ (Figure 4B). This is probably because of significant diffusion of α and βe even during the time between the bleach and acquisition of the first image (∼2 s). Indeed, mGFP-CaMKIIα and β showed the same bleach efficiency in fixed cells, which was also undistinguishable from the apparent bleach efficiencies for β and β′ in live cells (Figure 4B). Thus, the recoverable pool of α and βe during FRAP is even higher than initially estimated from the rescaled fluorescence recovery shown in Figure 4A (∼95% instead of ∼90%). Notably, monomeric mGFP alone typically showed only minimal apparent bleach efficiency but in a much larger apparent bleach area (our unpublished data). Thus, FRAP of the soluble α and βe is probably resolvable in our experimental setup only because of relatively slow diffusion of the large mGFP-CaMKII holoenzymes (>900 kDa).
Localization of CaMKIIβ Mutants
CaMKIIβ and several of its mutants were previously tested for their effect on dendritic arborization of hippocampal neurons in dissociated culture (Fink et al., 2003). We have now quantified the actin cytoskeletal localization of these CaMKIIβ mutants in Cos-7 cells (Figure 5). A303R (impaired for CaM binding) and K43R (impaired for ATP binding) localized to F-actin similarly as wild type. However, in the live cell scoring, A303R seemed to localize even stronger than wild type (p < 0.05 in two-tailed t test) (Figure 5). A303R does not disperse from F-actin upon Ca2+ signals (Shen and Meyer, 1999), likely because of impaired CaM binding; this seems to mildly enhance localization even in unstimulated cells. By contrast, the CaMKIIβ mutant T287D (constitutively active) did not show F-actin colocalization (Figure 5). T287D mimics autophosphorylation at T287, indicating that T287 autophosphorylation is involved in regulation of actin binding, either directly or by increasing affinity for Ca2+/CaM (Meyer et al., 1992). The T287D mutant had significantly lower effect on dendritic arborization than wild type (Fink et al., 2003), further supporting functional importance of cytoskeletal localization of CaMKIIβ.
F-Actin Phosphorylation by Basal Activity of Bound CaMKIIβ
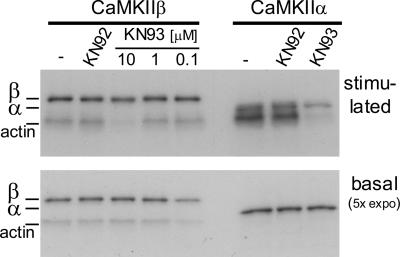
The CaM-binding impaired CaMKIIβ mutant A303R enhanced dendritic arborization in hippocampal neurons, whereas the ATP-binding impaired K43R mutant did not (Fink et al., 2003), indicating a possible role for CaM-independent kinase activity. Could F-actin binding directly enhance CaM-independent CaMKII activity? CaMKIIβ binding to F-actin did not increase phosphorylation of a substrate peptide, indicating that the activation state of the kinase was not changed (Fink et al., 2003). However, CaMKII can autophosphorylate at residues other than T286/T287, even by its basal activity (Colbran, 1993), that is, by activity without Ca2+/CaM stimulation or phosphorylation at T286/287 (Figure 6) (T286/T287 autophosphorylation requires binding of Ca2+/CaM to make T286/T287 accessible as a substrate, in addition to kinase activation; Hanson et al., 1994; Rich and Schulman, 1998). Thus, we hypothesized that basal activity of targeted CaMKIIβ could similarly suffice to phosphorylate other substrate proteins that are anchored in the right position. Indeed, purified CaMKIIβ phosphorylated F-actin in vitro even by nonstimulated basal activity, whereas the nonactin-binding CaMKIIα isoform did not (Figure 6). This is not because of differential substrate preference of the isoforms, because CaMKIIα did phosphorylate actin at least as well as CaMKIIβ when stimulated by Ca2+/CaM (Figure 6). The phosphorylated actin-sized band was only observed in presence of actin, but not when actin was omitted or substituted for BSA, or when CaMKIIβ was omitted (Supplemental Figure 3). The CaMKII inhibitor KN93, but not the inactive analogue KN92, inhibited Ca2+/CaM-stimulated actin phosphorylation by both CaMKIIα and β. Actin phosphorylation by basal activity of CaMKIIβ was not inhibited (Figure 6A), as expected, because KN93 is competitive with Ca2+/CaM (Sumi et al., 1991) and thus should not inhibit basal activity. Other inhibitors such as AIP or CaM-KIINtide are not competitive with Ca2+/CaM, but bind to CaMKII only in the activated and not in the basal state (Ishida et al., 1995; Chang et al., 2001). To further test actin phosphorylation by basal CaMKIIβ activity, mGFP-CaMKIIβ wild type and the ATP-binding impaired mutant K43R from extracts of transfected Cos-7 cells were used (Supplemental Figure 3). The K43R extract caused phosphorylation above background, indicating some residual kinase activity of this mutant; the alternative that actin bundling by CaMKIIβ activated another kinase seems less likely. Also, wild-type kinase extracts caused a higher degree of basal actin phosphorylation than both K43R extracts and extracts from mock-transfected cells (Supplemental Figure 3). These results strongly support phosphorylation by basal activity of targeted CaMKIIβ and give a possible explanation why the two activation-deficient CaMKIIβ mutants A303R (CaM-binding impaired) and K43R (ATP-binding impaired) have opposite effects on dendritic arborization (Fink et al., 2003).
Figure 6.
Purified CaMKIIβ, but not α, can phosphorylate actin in vitro even by nonstimulated basal activity. Both isoforms phosphorylate actin when stimulated by Ca2+/CaM. The CaM-competitive CaMKII inhibitor KN93, but not the inactive control compound KN92, inhibited stimulated kinase activity. Basal activity was not affected, as expected. KN concentration was 10 μM unless noted otherwise. Phosphorylation of actin and CaMKII autophosphorylation were detected by 32P incorporation and autoradiography.
Expression of CaMKIIβ Variants during Development and in Different Tissues
RT-PCR analysis showed similar expression of the splice variants CaMKIIβ, β′, βe, and βe′ in frontal cortex of rat brain on day 2 after birth, whereas β dominated by day 14 and was even more prominent in adult brain (Figure 7A). This developmental switch is consistent with a previous report (Brocke et al., 1995). However, CaMKIIβe expression is not eliminated during development (Brocke et al., 1999; Supplemental Figure 2). Expression of CaMKIIβ variants was also detected in pancreatic islets and in skeletal muscle, but not in heart (Figure 7B). The major splice variant in skeletal muscle correspondend in length to CaMKIIβM (Bayer et al., 1998), whereas the major variant in islets corresponded to CaMKIIβ′. Identity of CaMKIIβ′ was confirmed by restriction digest (Figure 7C) and by direct sequencing of the RT-PCR product. Thus, skeletal muscle and islets expressed two different but F-actin–associated CaMKIIβ variants (cf. Figure 3). Additionally, small amounts of β and βe′ transcripts were identified in islets, based on the length of two minor RT-PCR products (our unpublished data).
Figure 7.
The actin-binding CaMKIIβ′ is the dominant splice variant in rat pancreatic islets. Products from RT-PCR with CaMKIIβ-specific primers flanking the variable region were separated by agarose gel electrophoresis. Grayscale-inverted images of ethidium bromide stains are shown. (A) Cortex and whole brain was analyzed at different days after birth (pn), as indicated. Assignment of CaMKIIβ splice variants to the bands was based on length and (Brocke et al. 1995). (B) CaMKIIβ splice variants were detected in brain, skeletal muscle, and pancreatic islets, but not in heart of adult rats. The variant detected in skeletal muscle corresponded in length to CaMKIIβM (and contained exon v1 sequences based on SacII digest; our unpublished data); the major variant detected in islets corresponded to CaMKIIβ′. (C) The RT-PCR product from rat pancreatic islets was digested with enzymes that cut in different variable exons (−, no enzyme; v1, SacII; v3, BamHI; and v4, MseI). Resistance only to MseI digest indicates lack of exon v4, corresponding to CaMKIIβ′. SacII and BamHI digests yielded band of similar length, as expected for β′ PCR amplificates (6 base pairs difference); the same digests of β amplificates would differ by 49 base pairs. Identity of the RT-PCR product as β′ was also confirmed by direct sequencing (our unpublished data).
DISCUSSION
The isoform specificity of CaMKIIβ function in enhancing dendritic arborization and synapse density is thought to be mediated by CaMKIIβ binding to F-actin, a property not shared by the α isoform (Fink et al., 2003). Consistent with this isoform-specific binding, the CaMKIIβ, but not the α isoform, assembled with F-actin into large complexes, including bundles of parallel actin filaments. Most importantly, alternative splicing of CaMKIIβ regulated its localization to F-actin within cells. CaMKIIβ and β′, but not βe, associated with the actin cytoskeleton, indicating that the sequence encoded by variable exon v1 is necessary for the interaction (Figure 2). Alternative splicing of exon v1 depends both on cell type and developmental stage.
CaMKIIβe is the dominant splice variant before birth; CaMKIIβ becomes dominant between day 4 and 13 of postnatal rat brain development, both in cortex and hippocampus (Brocke et al., 1995). Thus, alternative splicing of CaMKIIβ provides a developmental switch from a cytoplasmic to an actin cytoskeleton-associated isoform. The F-actin–binding CaMKIIβ enhances dendritic arborization and synapse formation (Fink et al., 2003), and the timing of the developmental switch to this splice variant correlates with extensive synapse formation. Notably, the βe variant dominates at a time when the other major brain isoform, CaMKIIα, is not yet significantly expressed (Burgin et al., 1990; Bayer et al., 1999). CaMKIIβe variant transcripts also were found also in mature rat brain. Thus, although there is a developmental switch in β splicing, expression of βe is not eliminated during development. Indeed, βe transcripts have been detected previously in mature hippocampal CA1 pyramidal neurons by single cell RT-PCR (Brocke et al., 1999). Four of 14 CaMKIIβ-positive cells expressed βe transcripts, indicating differentially regulated alternative splicing even among mature neurons of the same cell type (Brocke et al., 1999). Notably, splice variant expression seemed segregated, with only one cell expressing more than one β splice variant (Brocke et al., 1999). This segregation of expression may be functionally important, because CaMKIIβ coassembles also with nonactin binding isoforms and targets such heteromeric holoenzymes to the actin cytoskeleton (Shen et al., 1998). CaMKIIα can partially escape such coassembly and cotargeting, because its mRNA is targeted to dendrites (Burgin et al., 1990; Mayford et al., 1996); protein synthesis in a separate compartment then allows formation of homomeric holoenzymes. By contrast, coexpression of CaMKIIβ and βe would result in actin-targeted heteromers, which can be prevented by exclusive expression of only one variant within individual neurons. The mechanism for differential regulation of CaMKIIβ splicing even among individual neurons of the same cell type is presently unclear. However, regulation by synaptic stimulation is an intriguing possibility that may provide a novel form of long-lasting neuronal plasticity. Synaptic plasticity indeed involves F-actin dynamics (Kim and Lisman, 1999; Fischer et al., 2000; Krucker et al., 2000; Star et al., 2002; Penzes et al., 2003; Okamoto et al., 2004; for review, see Lamprecht and LeDoux, 2004), which may be differentially modulated by CaMKII isoforms with different actin association (Fink et al., 2003).
In addition to neurons, CaMKIIβ gene expression was found in pancreatic islets and in skeletal muscle, but not in heart. The dominant β transcript in skeletal muscle, CaMKIIβM, contains exon v1 (Bayer et al., 1998) and localized to the actin cytoskeleton in Cos-7 cells. However, the F-actin colocalization index for βM was lower than for β and β′. This may reflect additional targeting of βM to other subcellular locations, possibly mediated by the putative binding sites for SH3 domains present in the 12-kDa βM-specific insert (Urquidi and Ashcroft, 1995; Bayer et al., 1998).
The major CaMKIIβ transcript found in rat pancreatic islets was β′, which lacks exon v4 and was found to associate with the actin cytoskeleton. By contrast, previous studies in rat and human found predominant expression of CaMKIIβe′ (Rochlitz et al., 2000; Tabuchi et al., 2000), which additionally lacks exon v1 and did not associate with F-actin. This difference may be due to use of different rat strains (Sprague-Dawley versus Wistar), raising the possibility of variation also among individual humans. Notably, the two rat strains show some difference in their insulin secretion and response (Gaudreault et al., 2001; de Groot et al., 2004). CaMKII is thought to regulate insulin secretion and production (Wasmeier and Hutton, 1999; Bhatt et al., 2000; Tabuchi et al., 2000; Osterhoff et al., 2003), and it will be interesting to see whether and how these functions depend on actin association. CaMKIIδ may be the second major isoform in islets (Rochlitz et al., 2000; Osterhoff et al., 2003); however, it was not detected in all studies (Breen and Ashcroft, 1997; Tabuchi et al., 2000). CaMKIIδ variants do not contain sequences homologous to exon v1, but the CaMKIIδ association domain has been reported to mediate localization of a GFP-fusion protein to the actin cytoskeleton (Caran et al., 2001). However, it should be noted that actin localization of CaMKIIδ was rather mild, and, in contrast to the CaMKIIβ localization observed here, became apparent only after fixation of the cells (Caran et al., 2001).
CaMKIIβ bundled and cross-linked F-actin filaments. This suggests that CaMKIIβ holoenzymes can bind multiple F-actin filaments simultaneously. Inactive holoenzymes form disk-like structures composed of two stacked hexameric rings (Kolodziej et al., 2000; Hoelz et al., 2003; Rosenberg et al., 2005). Thus, at least two actin filaments should be able to bind to a holoenzyme without steric hindrance, one to each ring and at several possible angles to each other. A previous electron microscopic study detected binding of CaMKII to F-actin (Ohta et al., 1986), but it did not describe cross-linking of filaments. However, the CaMKII preparation used in the study likely contained mostly the α isoform (from a soluble, noncytoskeletal fraction of forebrain); small amounts of β isoform present may have been sufficient to mediate some binding, but not bundling or cross-linking.
CaMKIIβ dissociates from F-actin upon Ca2+/CaM stimulation (Fink et al., 2003; Figure 2). Thus, cross-linking of actin filaments by CaMKIIβ may play a role in Ca2+-regulated actin dynamics. However, such a possible direct structural function is presently unclear. Moreover, kinase activity is required for the morphogenic function of CaMKIIβ in enhancing dendritic arborization, because a kinase dead mutant (K43R) even had a dominant-negative effect (Fink et al., 2003). By contrast, a CaM binding-deficient mutant (A303R) still enhanced arborization (Fink et al., 2003), indicating a function of CaM-independent activity. Indeed, whereas both CaMKIIβ and α phosphorylated actin upon Ca2+/CaM stimulation in vitro, only the F-actin–binding CaMKIIβ phosphorylated actin even by its nonstimulated basal activity. Binding to F-actin does not increase CaMKIIβ activity toward a soluble peptide substrate (Fink et al., 2003), suggesting that targeting generated a high local substrate concentration that is sufficient to result in significant phosphorylation even by basal activity. Consistent with such interpretation, basal CaMKII activity is sufficient for autophosphorylation (at residues other than T286 in α or T287 in β) (Colbran, 1993). A previous study indicated that CaMKII can phosphorylate SynGAP also by basal activity when both proteins are bound to the adaptor protein MUPP1 (Krapivinsky et al., 2004). Thus, a function of basal kinase activity after targeting may underlie the different effects of the two activity- or activation-impaired mutants (Fink et al., 2003). Although the morphogenic effect of CaMKIIβ could involve direct phosphorylation of actin, it seems more likely that phosphorylation of other actin-bound regulatory proteins is required.
Previous studies showed that alternative splicing of CaMKIIβ modulates its Ca2+-spike frequency response (Bayer et al., 2002). The results presented here show that alternative splicing also regulated the association of CaMKIIβ with the actin cytoskeleton, which is thought to underlie the specific morphogenic role of CaMKIIβ in neurons (Fink et al., 2003). It will be interesting to elucidate how these two biochemical properties contribute to differential function of the CaMKIIβ splice variants, and how the alternative splice events are regulated.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Dot Dill and Francisco Ramirez (University of Colorado Health Sciences Center) for excellent technical assistance with the electron microscopy and islet preparations, respectively. We thank Dr. Mark Dell'Acqua (University of Colorado Health Sciences Center) and laboratory members, especially Drs. Jessica Gorski, Karen Smith, Eric Horne, and Matthew Pink for critical reading of the manuscript. We thank Drs. Tobias Meyer (University of California, Stanford) and Charles Fink (University of Wisconsin, Madison, WI) for sharing constructs. Purified actin was a kind gift from Dr. R. Rock. We thank Dr. Mike Bradshaw (Roche, Palo Alto, CA) for discussions and help with CaMKII purification. The research was supported by a Pilot and Feasibility Award P30 DK57516 from the National Institutes of Health Diabetes and Endocrine Research Center at University of Colorado Health Sciences Center, by National Institutes of Health Grant DK-070735, and by American Heart Association Grant SDG 0430196N.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-03-0252) on August 23, 2006.
REFERENCES
- Bayer K. U., De Koninck P., Leonard A. S., Hell J. W., Schulman H. Interaction with the NMDA receptor locks CaMKII in an active conformation. Nature. 2001;411:801–805. doi: 10.1038/35081080. [DOI] [PubMed] [Google Scholar]
- Bayer K. U., De Koninck P., Schulman H. Alternative splicing modulates the frequency-dependent response of CaMKII to Ca(2+) oscillations. EMBO J. 2002;21:3590–3597. doi: 10.1093/emboj/cdf360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer K. U., Harbers K., Schulman H. alphaKAP is an anchoring protein for a novel CaM kinase II isoform in skeletal muscle. EMBO J. 1998;17:5598–5605. doi: 10.1093/emboj/17.19.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer K. U., LeBel E., McDonald G., O'Leary H., Schulman H., De Koninck P. Transition from reversible to persistent binding of CaMKII to postsynaptic sites and NR2B. J. Neurosci. 2006;26:1164–1174. doi: 10.1523/JNEUROSCI.3116-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer K. U., Lohler J., Schulman H., Harbers K. Developmental expression of the CaM kinase II isoforms: ubiquitous gamma- and delta-CaM kinase II are the early isoforms and most abundant in the developing nervous system. Brain Res. Mol. Brain Res. 1999;70:147–154. doi: 10.1016/s0169-328x(99)00131-x. [DOI] [PubMed] [Google Scholar]
- Bhatt H. S., Conner B. P., Prasanna G., Yorio T., Easom R. A. Dependence of insulin secretion from permeabilized pancreatic beta-cells on the activation of Ca(2+)/calmodulin-dependent protein kinase II A re-evaluation of inhibitor studies. Biochem Pharmacol. 2000;60:1655–1663. doi: 10.1016/s0006-2952(00)00483-4. [DOI] [PubMed] [Google Scholar]
- Bradshaw J. M., Hudmon A., Schulman H. Chemical quenched flow kinetic studies indicate an intraholoenzyme autophosphorylation mechanism for Ca2+/calmodulin-dependent protein kinase II. J. Biol. Chem. 2002;277:20991–20998. doi: 10.1074/jbc.M202154200. [DOI] [PubMed] [Google Scholar]
- Breen M. A., Ashcroft S. J. Human islets of Langerhans express multiple isoforms of calcium/calmodulin-dependent protein kinase II. Biochem. Biophys. Res. Commun. 1997;236:473–478. doi: 10.1006/bbrc.1997.6871. [DOI] [PubMed] [Google Scholar]
- Brocke L., Chiang L. W., Wagner P. D., Schulman H. Functional implications of the subunit composition of neuronal CaM kinase II. J. Biol. Chem. 1999;274:22713–22722. doi: 10.1074/jbc.274.32.22713. [DOI] [PubMed] [Google Scholar]
- Brocke L., Srinivasan M., Schulman H. Developmental and regional expression of multifunctional Ca2+/calmodulin-dependent protein kinase isoforms in rat brain. J. Neurosci. 1995;15:6797–6808. doi: 10.1523/JNEUROSCI.15-10-06797.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin K. E., Waxham M. N., Rickling S., Westgate S. A., Mobley W. C., Kelly P. T. In situ hybridization histochemistry of Ca2+/calmodulin-dependent protein kinase in developing rat brain. J. Neurosci. 1990;10:1788–1798. doi: 10.1523/JNEUROSCI.10-06-01788.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caran N., Johnson L. D., Jenkins K. J., Tombes R. M. Cytosolic targeting domains of gamma and delta calmodulin-dependent protein kinase II. J. Biol. Chem. 2001;276:42514–42519. doi: 10.1074/jbc.M103013200. [DOI] [PubMed] [Google Scholar]
- Chang B. H., Mukherji S., Soderling T. R. Calcium/calmodulin-dependent protein kinase II inhibitor protein: localization of isoforms in rat brain. Neuroscience. 2001;102:767–777. doi: 10.1016/s0306-4522(00)00520-0. [DOI] [PubMed] [Google Scholar]
- Colbran R. J. Inactivation of Ca2+/calmodulin-dependent protein kinase II by basal autophosphorylation. J. Biol. Chem. 1993;268:7163–7170. [PubMed] [Google Scholar]
- Colbran R. J., Brown A. M. Calcium/calmodulin-dependent protein kinase II and synaptic plasticity. Curr. Opin. Neurobiol. 2004;14:318–327. doi: 10.1016/j.conb.2004.05.008. [DOI] [PubMed] [Google Scholar]
- de Groot M., de Haan B. J., Keizer P. P., Schuurs T. A., van Schilfgaarde R., Leuvenink H. G. Rat islet isolation yield and function are donor strain dependent. Lab. Anim. 2004;38:200–206. doi: 10.1258/002367704322968885. [DOI] [PubMed] [Google Scholar]
- De Koninck P., Schulman H. Sensitivity of CaM kinase II to the frequency of Ca2+ oscillations. Science. 1998;279:227–230. doi: 10.1126/science.279.5348.227. [DOI] [PubMed] [Google Scholar]
- Easom R. A. CaM kinase II: a protein kinase with extraordinary talents germane to insulin exocytosis. Diabetes. 1999;48:675–684. doi: 10.2337/diabetes.48.4.675. [DOI] [PubMed] [Google Scholar]
- Erondu N. E., Kennedy M. B. Regional distribution of type II Ca2+/calmodulin-dependent protein kinase in rat brain. J. Neurosci. 1985;5:3270–3277. doi: 10.1523/JNEUROSCI.05-12-03270.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink C. C., Bayer K. U., Myers J. W., Ferrell J. E., Jr, Schulman H., Meyer T. Selective regulation of neurite extension and synapse formation by the beta but not the alpha isoform of CaMKII. Neuron. 2003;39:283–297. doi: 10.1016/s0896-6273(03)00428-8. [DOI] [PubMed] [Google Scholar]
- Fischer M., Kaech S., Wagner U., Brinkhaus H., Matus A. Glutamate receptors regulate actin-based plasticity in dendritic spines. Nat. Neurosci. 2000;3:887–894. doi: 10.1038/78791. [DOI] [PubMed] [Google Scholar]
- Gaudreault N., Santure M., Pitre M., Nadeau A., Marette A., Bachelard H. Effects of insulin on regional blood flow and glucose uptake in Wistar and Sprague-Dawley rats. Metabolism. 2001;50:65–73. doi: 10.1053/meta.2001.18569. [DOI] [PubMed] [Google Scholar]
- Giese K. P., Fedorov N. B., Filipkowski R. K., Silva A. J. Autophosphorylation at Thr286 of the alpha calcium-calmodulin kinase II in LTP and learning. Science. 1998;279:870–873. doi: 10.1126/science.279.5352.870. [DOI] [PubMed] [Google Scholar]
- Griffith L. C. Calcium/calmodulin-dependent protein kinase II: an unforgettable kinase. J. Neurosci. 2004;24:8391–8393. doi: 10.1523/JNEUROSCI.2888-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hain J., Onoue H., Mayrleitner M., Fleischer S., Schindler H. Phosphorylation modulates the function of the calcium release channel of sarcoplasmic reticulum from cardiac muscle. J. Biol. Chem. 1995;270:2074–2081. doi: 10.1074/jbc.270.5.2074. [DOI] [PubMed] [Google Scholar]
- Hanson P. I., Meyer T., Stryer L., Schulman H. Dual role of calmodulin in autophosphorylation of multifunctional CaM kinase may underlie decoding of calcium signals. Neuron. 1994;12:943–956. doi: 10.1016/0896-6273(94)90306-9. [DOI] [PubMed] [Google Scholar]
- Heist E. K., Srinivasan M., Schulman H. Phosphorylation at the nuclear localization signal of Ca2+/calmodulin-dependent protein kinase II blocks its nuclear targeting. J. Biol. Chem. 1998;273:19763–19771. doi: 10.1074/jbc.273.31.19763. [DOI] [PubMed] [Google Scholar]
- Hoelz A., Nairn A. C., Kuriyan J. Crystal structure of a tetradecameric assembly of the association domain of Ca2+/calmodulin-dependent kinase II. Mol. Cell. 2003;11:1241–1251. doi: 10.1016/s1097-2765(03)00171-0. [DOI] [PubMed] [Google Scholar]
- Hudmon A., Schulman H. Neuronal CA2+/calmodulin-dependent protein kinase II: the role of structure and autoregulation in cellular function. Annu. Rev. Biochem. 2002;71:473–510. doi: 10.1146/annurev.biochem.71.110601.135410. [DOI] [PubMed] [Google Scholar]
- Huntley G. W., Benson D. L., Colman D. R. Structural remodeling of the synapse in response to physiological activity. Cell. 2002;108:1–4. doi: 10.1016/s0092-8674(01)00631-6. [DOI] [PubMed] [Google Scholar]
- Ishida A., Kameshita I., Okuno S., Kitani T., Fujisawa H. A novel highly specific and potent inhibitor of calmodulin-dependent protein kinase II. Biochem. Biophys. Res. Commun. 1995;212:806–812. doi: 10.1006/bbrc.1995.2040. [DOI] [PubMed] [Google Scholar]
- Kim C. H., Lisman J. E. A role of actin filament in synaptic transmission and long-term potentiation. J. Neurosci. 1999;19:4314–4324. doi: 10.1523/JNEUROSCI.19-11-04314.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej S. J., Hudmon A., Waxham M. N., Stoops J. K. Three-dimensional reconstructions of calcium/calmodulin-dependent (CaM) kinase IIα and truncated CaM kinase IIα reveal a unique organization for its structural core and functional domains. J. Biol. Chem. 2000;275:14354–14359. doi: 10.1074/jbc.275.19.14354. [DOI] [PubMed] [Google Scholar]
- Krapivinsky G., Medina I., Krapivinsky L., Gapon S., Clapham D. E. SynGAP-MUPP1-CaMKII synaptic complexes regulate p38 MAP kinase activity and NMDA receptor-dependent synaptic AMPA receptor potentiation. Neuron. 2004;43:563–574. doi: 10.1016/j.neuron.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Krucker T., Siggins G. R., Halpain S. Dynamic actin filaments are required for stable long-term potentiation (LTP) in area CA1 of the hippocampus. Proc. Natl. Acad. Sci. USA. 2000;97:6856–6861. doi: 10.1073/pnas.100139797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht R., LeDoux J. Structural plasticity and memory. Nat. Rev. Neurosci. 2004;5:45–54. doi: 10.1038/nrn1301. [DOI] [PubMed] [Google Scholar]
- Lantsman K., Tombes R. M. CaMK-II oligomerization potential determined using CFP/YFP FRET. Biochim. Biophys. Acta. 2005;1746:45–54. doi: 10.1016/j.bbamcr.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Lisman J., Schulman H., Cline H. The molecular basis of CaMKII function in synaptic and behavioural memory. Nat. Rev. Neurosci. 2002;3:175–190. doi: 10.1038/nrn753. [DOI] [PubMed] [Google Scholar]
- Lisman J. E., McIntyre C. C. Synaptic plasticity: a molecular memory switch. Curr. Biol. 2001;11:R788–R791. doi: 10.1016/s0960-9822(01)00472-9. [DOI] [PubMed] [Google Scholar]
- Malenka R. C., Nicoll R. A. Long-term potentiation–a decade of progress? Science. 1999;285:1870–1874. doi: 10.1126/science.285.5435.1870. [DOI] [PubMed] [Google Scholar]
- Malinow R., Schulman H., Tsien R. W. Inhibition of postsynaptic PKC or CaMKII blocks induction but not expression of LTP. Science. 1989;245:862–866. doi: 10.1126/science.2549638. [DOI] [PubMed] [Google Scholar]
- Mayford M., Baranes D., Podsypanina K., Kandel E. R. The 3′-untranslated region of CaMKII alpha is a cis-acting signal for the localization and translation of mRNA in dendrites. Proc. Natl. Acad. Sci. USA. 1996;93:13250–13255. doi: 10.1073/pnas.93.23.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee A. W., Bredt D. S. Assembly and plasticity of the glutamatergic postsynaptic specialization. Curr. Opin. Neurobiol. 2003;13:111–118. doi: 10.1016/s0959-4388(03)00008-4. [DOI] [PubMed] [Google Scholar]
- Meyer T., Hanson P. I., Stryer L., Schulman H. Calmodulin trapping by calcium-calmodulin-dependent protein kinase. Science. 1992;256:1199–1202. doi: 10.1126/science.256.5060.1199. [DOI] [PubMed] [Google Scholar]
- Morris E. P., Torok K. Oligomeric structure of alpha-calmodulin-dependent protein kinase II. J. Mol. Biol. 2001;308:1–8. doi: 10.1006/jmbi.2001.4584. [DOI] [PubMed] [Google Scholar]
- Ohta Y., Nishida E., Sakai H. Type II Ca2+/calmodulin-dependent protein kinase binds to actin filaments in a calmodulin-sensitive manner. FEBS Lett. 1986;208:423–426. doi: 10.1016/0014-5793(86)81061-4. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Nagai T., Miyawaki A., Hayashi Y. Rapid and persistent modulation of actin dynamics regulates postsynaptic reorganization underlying bidirectional plasticity. Nat. Neurosci. 2004;7:1104–1112. doi: 10.1038/nn1311. [DOI] [PubMed] [Google Scholar]
- Osterhoff M., Mohlig M., Schwanstecher M., Seufert J., Ortmann J., Schatz H., Pfeiffer A. F. Ca2+/calmodulin-dependent protein kinase II delta2 regulates gene expression of insulin in INS-1 rat insulinoma cells. Cell Calcium. 2003;33:175–184. doi: 10.1016/s0143-4160(02)00227-0. [DOI] [PubMed] [Google Scholar]
- Penzes P., Beeser A., Chernoff J., Schiller M. R., Eipper B. A., Mains R. E., Huganir R. L. Rapid induction of dendritic spine morphogenesis by trans-synaptic ephrinB-EphB receptor activation of the Rho-GEF kalirin. Neuron. 2003;37:263–274. doi: 10.1016/s0896-6273(02)01168-6. [DOI] [PubMed] [Google Scholar]
- Rich R. C., Schulman H. Substrate-directed function of calmodulin in autophosphorylation of Ca2+/calmodulin-dependent protein kinase II. J. Biol. Chem. 1998;273:28424–28429. doi: 10.1074/jbc.273.43.28424. [DOI] [PubMed] [Google Scholar]
- Rochlitz H., Voigt A., Lankat-Buttgereit B., Goke B., Heimberg H., Nauck M. A., Schiemann U., Schatz H., Pfeiffer A. F. Cloning and quantitative determination of the human Ca2+/calmodulin-dependent protein kinase II (CaMK II) isoforms in human beta cells. Diabetologia. 2000;43:465–473. doi: 10.1007/s001250051330. [DOI] [PubMed] [Google Scholar]
- Rosenberg O. S., Deindl S., Sung R. J., Nairn A. C., Kuriyan J. Structure of the autoinhibited kinase domain of CaMKII and SAXS analysis of the holoenzyme. Cell. 2005;123:849–860. doi: 10.1016/j.cell.2005.10.029. [DOI] [PubMed] [Google Scholar]
- Rosenberg O. S., Deindl S., Comolli L. R., Hoelz A., Downing K. H., Nairn A. C., Kuriyan J. Oligomerization states of the association domain and the holoenyzme of Ca/CaM kinase II. FEBS J. 2006;273:682–694. doi: 10.1111/j.1742-4658.2005.05088.x. [DOI] [PubMed] [Google Scholar]
- Segal M. Dendritic spines and long-term plasticity. Nat. Rev. Neurosci. 2005;6:277–284. doi: 10.1038/nrn1649. [DOI] [PubMed] [Google Scholar]
- Shen K., Meyer T. Dynamic control of CaMKII translocation and localization in hippocampal neurons by NMDA receptor stimulation. Science. 1999;284:162–166. doi: 10.1126/science.284.5411.162. [DOI] [PubMed] [Google Scholar]
- Shen K., Teruel M. N., Subramanian K., Meyer T. CaMKIIbeta functions as an F-actin targeting module that localizes CaMKIIalpha/beta heterooligomers to dendritic spines. Neuron. 1998;21:593–606. doi: 10.1016/s0896-6273(00)80569-3. [DOI] [PubMed] [Google Scholar]
- Silva A. J., Stevens C. F., Tonegawa S., Wang Y. Deficient hippocampal long-term potentiation in alpha-calcium-calmodulin kinase II mutant mice. Science. 1992;257:201–206. doi: 10.1126/science.1378648. [DOI] [PubMed] [Google Scholar]
- Singla S. I., Hudmon A., Goldberg J. M., Smith J. L., Schulman H. Molecular characterization of calmodulin trapping by calcium/calmodulin-dependent protein kinase II. J. Biol. Chem. 2001;276:29353–29360. doi: 10.1074/jbc.M101744200. [DOI] [PubMed] [Google Scholar]
- Soderling T. R., Chang B., Brickey D. Cellular signaling through multifunctional Ca2+/calmodulin-dependent protein kinase II. J. Biol. Chem. 2001;276:3719–3722. doi: 10.1074/jbc.R000013200. [DOI] [PubMed] [Google Scholar]
- Srinivasan M., Edman C. F., Schulman H. Alternative splicing introduces a nuclear localization signal that targets multifunctional CaM kinase to the nucleus. J. Cell Biol. 1994;126:839–852. doi: 10.1083/jcb.126.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star E. N., Kwiatkowski D. J., Murthy V. N. Rapid turnover of actin in dendritic spines and its regulation by activity. Nat. Neurosci. 2002;5:239–246. doi: 10.1038/nn811. [DOI] [PubMed] [Google Scholar]
- Sumi M., Kiuchi K., Ishikawa T., Ishii A., Hagiwara M., Nagatsu T., Hidaka H. The newly synthesized selective Ca2+/calmodulin dependent protein kinase II inhibitor KN-93 reduces dopamine contents in PC12h cells. Biochem. Biophys. Res. Commun. 1991;181:968–975. doi: 10.1016/0006-291x(91)92031-e. [DOI] [PubMed] [Google Scholar]
- Tabuchi H., Yamamoto H., Matsumoto K., Ebihara K., Takeuchi Y., Fukunaga K., Hiraoka H., Sasaki Y., Shichiri M., Miyamoto E. Regulation of insulin secretion by overexpression of Ca2+/calmodulin-dependent protein kinase II in insulinoma MIN6 cells. Endocrinology. 2000;141:2350–2360. doi: 10.1210/endo.141.7.7553. [DOI] [PubMed] [Google Scholar]
- Tobimatsu T., Fujisawa H. Tissue-specific expression of four types of rat calmodulin-dependent protein kinase II mRNAs. J. Biol. Chem. 1989;264:17907–17912. [PubMed] [Google Scholar]
- Tombes R. M., Faison M. O., Turbeville J. M. Organization and evolution of multifunctional Ca(2+)/CaM-dependent protein kinase genes. Gene. 2003;322:17–31. doi: 10.1016/j.gene.2003.08.023. [DOI] [PubMed] [Google Scholar]
- Urquidi V., Ashcroft S. J. A novel pancreatic beta-cell isoform of calcium/calmodulin-dependent protein kinase II (beta 3 isoform) contains a proline-rich tandem repeat in the association domain. FEBS Lett. 1995;358:23–26. doi: 10.1016/0014-5793(94)01381-a. [DOI] [PubMed] [Google Scholar]
- Wang J., Best P. M. Inactivation of the sarcoplasmic reticulum calcium channel by protein kinase. Nature. 1992;359:739–741. doi: 10.1038/359739a0. [DOI] [PubMed] [Google Scholar]
- Wasmeier C, Hutton J. C. Secretagogue-dependent phosphorylation of phogrin, an insulin granule membrane protein tyrosine phosphatase homologue. Biochem. J. 1999;341:563–569. [PMC free article] [PubMed] [Google Scholar]
- Xu A., Hawkins C., Narayanan N. Phosphorylation and activation of the Ca(2+)-pumping ATPase of cardiac sarcoplasmic reticulum by Ca2+/calmodulin-dependent protein kinase. J. Biol. Chem. 1993;268:8394–8397. [PubMed] [Google Scholar]
- Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.