Abstract
Valosin-containing protein (VCP; p97; cdc48 in yeast) is a hexameric ATPase of the AAA family (ATPases with multiple cellular activities) involved in multiple cellular functions, including degradation of proteins by the ubiquitin (Ub)–proteasome system (UPS). We examined the consequences of the reduction of VCP levels after RNA interference (RNAi) of VCP. A new stringent method of microarray analysis demonstrated that only four transcripts were nonspecifically affected by RNAi, whereas ∼30 transcripts were affected in response to reduced VCP levels in a sequence-independent manner. These transcripts encoded proteins involved in endoplasmic reticulum (ER) stress, apoptosis, and amino acid starvation. RNAi of VCP promoted the unfolded protein response, without eliciting a cytosolic stress response. RNAi of VCP inhibited the degradation of R-GFP (green fluorescent protein) and Ub-G76V-GFP, two cytoplasmic reporter proteins degraded by the UPS, and of α chain of the T-cell receptor, an established substrate of the ER-associated degradation (ERAD) pathway. Surprisingly, RNAi of VCP had no detectable effect on the degradation of two other ERAD substrates, α1-antitrypsin and δCD3. These results indicate that VCP is required for maintenance of normal ER structure and function and mediates the degradation of some proteins via the UPS, but is dispensable for the UPS-dependent degradation of some ERAD substrates.
INTRODUCTION
Valosin-containing protein VCP (p97; cdc48 in yeast) is a hexameric type II ATPase of the AAA family (ATPases with multiple cellular activities) that mediates disparate cellular functions, including endoplasmic reticulum-associated degradation (ERAD) via the ubiquitin–proteasome system (UPS) (Woodman, 2003; Dreveny et al., 2004; Wang et al., 2004; Bar-Nun, 2005; Halawani and Latterich, 2006). VCP interacts with at least 30 different cellular proteins, some of which may differentially mediate its functions. For example, VCP forms a complex with the Ufd1–Npl4 heterodimer that is essential for its role in ERAD, a process by which constituent and transient endoplasmic reticulum (ER) proteins are removed from the ER and degraded in the cytosol by the 26S proteasome. A general model for the role of VCPUfd1–Npl4 in ERAD involves binding of polyubiquitinated ERAD substrates at the cytoplasmic face of the ER membrane both before and after substrate ubiquitination, followed by a complete substrate extraction/dislocation and its transfer to the 26S proteasome (Meyer et al., 2000, 2002; Dai and Li, 2001; Rabinovich et al., 2002; Elkabetz et al., 2003; Ye et al., 2003). Substrate dislocation from the ER could result from mechanical stress transmitted to the bound substrate by conformational changes in VCP during cycles of VCP-catalyzed ATP hydrolysis (Zhang et al., 2000; Wang et al., 2004).
ERAD is a component of a coordinated cellular response to ER stress, termed the unfolded protein response (UPR) (Harding et al., 2002; Ma and Hendershot, 2002; Kostova and Wolf, 2003; Sitia and Braakman, 2003). UPR can be promoted by the buildup of unfolded proteins in the ER and constitutes a mechanism to reduce this burden. UPR acutely reduces translation of new proteins, followed by increased expression of chaperones to aid folding of existing proteins and enhanced elimination of proteins that cannot be refolded. In mammals, apoptosis is initiated if ER stress is not relieved. A critical UPR pathway is initiated by activation of IRE-1, an ER membrane endonuclease that splices XBP-1 mRNA (Yoshida et al., 2001). Translation of spliced XBP-1 mRNA promotes transcriptional activation of genes for UPR, including those required for ERAD (Sriburi et al., 2004).
VCP is required for the fusion of ER and Golgi membranes (Latterich et al., 1995; Rabouille et al., 1995; Patel et al., 1998). RNA interference (RNAi) of VCP in HeLa cells results in the formation of large intracellular vacuoles, likely derived from ER (Wojcik et al., 2004b). Thus, VCP seems to be required for normal ER function, whereas reduced VCP content seems to induce ER stress, perhaps as consequence of reduced constitutive ERAD and/or by disturbing the fusion of ER membranes. RNAi of VCP also caused a general increase in polyubiquitinated cellular proteins, indicative of impaired UPS function (Wojcik et al., 2004b). However, it is unclear whether this effect reflects the quantitative significance of ERAD to overall cellular protein degradation or whether VCP mediates UPS-dependent degradation of non-ERAD substrates, as shown previously for several individual proteins (Johnson et al., 1995; Ghislain et al., 1996; Dai et al., 1998; Dai and Li, 2001). To gain insight to these various issues, we have analyzed altered transcription profiles in mammalian cells subjected to RNAi of VCP and directly determined the role of VCP in UPR and in UPS-dependent degradation of specific ERAD and non-ERAD substrates. Our results demonstrate that VCP mediates multiple aspects of ER structure and function and multiple aspects of UPS function. Surprisingly, however, VCP is not required for the degradation of all ERAD substrates.
MATERIALS AND METHODS
Antibodies, Reagents, and Plasmids
Anti-hemagglutinin (HA) monoclonal antibody (mAb) was from Covance (Princeton, NJ), anti-ubiquitin mAb was from Santa Cruz Biotechnology (Santa Cruz, CA), anti-VCP mAb was from BD Biosciences (Franklin Lakes, NJ), anti-green fluorescent protein (GFP) mAb was from Roche (Alameda, CA), and anti-KDEL antibody detecting BiP and GRP94 as well as anti-hsp70 antibody were from Stressgen Biotechnologies (Victoria, British Columbia, Canada). The plasmids encoding ubiquitin (Ub)-R-GFP and Ub-G76V-GFP were derived from pEGFP-N1 (Dantuma et al., 2000), HA-δCD3 and HA-α chain of the T-cell receptor (αTCR) were on pcDNA 3.1 (Yang et al., 1998; Yu and Kopito, 1999), and α1-antirtrypsin Hong Kong mutant was on pCMV (Hosokawa et al., 2003). All the remaining reagents were from Sigma-Aldrich (St. Louis, MO).
Cell Culture and Establishment of Stable Cell Lines
HeLa cells were grown in Advanced DMEM (Invitrogen, Carlsbad, CA) supplemented with GlutaMAX, antibiotic/antimycotic solution, and 2% fetal bovine serum (Gemini Bioproducts, Woodland, CA). Plasmids used for transfection were sequenced using CEQ 2000XL DNA analysis system (Beckman Coulter, Fullerton, CA). Transfection was carried on using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). After transient transfection, HeLa cells were used for the production of stable cell lines by selection with Geneticin (Invitrogen). All clones that expressed a given protein showed accumulation of that protein after inhibition of the proteasome (our unpublished data). One clone from each group (UbG76V GFP, Ub-R-GFP, δCD3, and α1AT) with the highest basal expression level was selected for the study. None of these clones differed from the nontransfected or mock-transfected controls in morphology, growth characteristics, or time- and dose-dependent sensitivity to proteasome inhibitors (our unpublished data).
RNA Interference
Small interfering RNAs (siRNAs) were obtained by chemical synthesis using 2′-ACE chemistry (Hartsel et al., 2005) from Dharmacon RNA Technologies (Lafayette, CO). siRNAs were 2′ deprotected, desalted, purified by PAGE, and duplexed by the manufacturer. The mass of each siRNA was verified by matrix-assisted laser desorption ionization/time of flight mass spectrometry. After shipment in a dry form, the siRNAs were suspended in the 1× universal buffer (20 mM KCl, 6 mM HEPES-KOH, pH 7.5, and 0.2 mM MgCl2) at a 20 μM concentration, aliquoted, and frozen at −20°C for further use. Two siRNAs were obtained targeting VCP and one siRNA targeting enhanced green fluorescent protein (EGFP), a protein not found in HeLa cells. The first siRNA (positions 599–619 of human VCP mRNA; accession number NM_007126), called VCP-2, has been used previously, and it was originally selected from five different siRNAs based on its efficiency (Wojcik et al., 2004a, b). The second siRNA targeting VCP (positions 480–500), called VCP-6, was designed using Dharmacon's Web site siRNA design center (Reynolds et al., 2004). As control for nonspecific effects of RNAi, we have designed and used the siRNA targeting EGFP (positions 1101–1018 of CVU55763, preceded by AA). RNAi was performed by single Oligofectamine-mediated transfection (Invitrogen) as described previously (Wojcik et al., 2004a, b). Cells were collected 72 h after the transfection. HeLa cells mock transfected with EGFP siRNA served as a control.
Transmission Electron Microscopy
HeLa cells were grown on glass slides and submitted to RNAi of VCP by using either VCP-2 or VCP-6. Three days after RNAi, cells were fixed in 2% glutaraldehyde in a cacodylate buffer supplemented with 5 mM CaCl2, postfixed with OsO4 in the cacodylate buffer supplemented with CaCl2 and K4[Fe(CN)6], and then dehydrated with ethanol and acetone and embedded in LR White resin (Sigma-Aldrich). Resin blocks were cut, mounted on Formvar carbon-coated grids, counterstained with lead citrate and uranyl acetate, and observed in a Jeol JEM-100S electron microscope (Jeol, Tokyo, Japan).
Immunofluorescence Microscopy
HeLa cells were grown in Lab-Tek two-chamber slides (Nunc Nalgene, Naperville, IL). After 16-h treatment with 10 μg/ml tunicamycin or 5 μM brefeldin A (BFA) either alone or in combination with 10 μM MG132, cells were fixed with 2% formaldehyde in phosphate-buffered saline for 30 min, quenched in 50 mM NH4Cl, permeabilized in 0.1% Triton X-100, washed twice for 15 min each with Tris-buffered saline (TBS), pH 7.6, supplemented with 0.1% bovine serum albumin and 0.1% fish gelatin, and incubated with anti-polyubiquitin FK1 mAb (BIOMOL Research laboratories, Plymouth Meeting, PA) diluted in the same buffer containing Tween 20 for 2 h. After three 15-min washes in TBS with 0.1% bovine serum albumin and 0.1% fish gelatin, the cells were incubated with secondary rhodamine-conjugated anti-mouse F(ab′)2 fragment (Jackson ImmunoResearch Laboratories, West Grove, PA). Cells were then stained with 100 nM Yo-Pro1 iodide (Invitrogen). After two washes in TBS, cells were mounted using Gel/Mount (Biomeda, Foster City, CA). Slides were observed using the 60× Plan Apo objective of a Nikon Eclipse TE2000-U epifluorescence microscope. Images were acquired using the CoolSNAP ES charge-coupled device camera operated by the MetaMorph 6.3 software (Fryer Company, Cincinnati, OH).
RT-PCR
RNA was isolated using the modified method of Chomczynski (Chomczynski and Sacchi, 1987) from HeLa cells 72 h after transfection with two different siRNAs targeting VCP (VCP-2 and VCP-6) or from cells treated for 6 h with 10 μM MG132, 10 μg/ml tunicamycin, and 5 μM brefeldin A (all from Calbiochem, San Diego, CA). RT-PCR was performed with the OneStep kit (QIAGEN, Valencia, CA) by using the pairs of primers amplifying the genes of interest, as indicated in Table 1. For the XBP-1 transcript, the primers amplify the region that includes the 26-base pair deletion dependent on IRE-1 endonuclease activity (Yoshida et al., 2001). The number of cycles was adjusted to obtain a linear range of reaction products. Gels were scanned with the Kodak 4000MM Image Station (Eastman Kodak, Rochester, NY).
Table 1.
Primers used for semiquantitative RT-PCR
| Gene name | Accession no. | Forward primer | Reverse primer |
|---|---|---|---|
| XBP1 | AB076384 | CCTTGTAGTTGAGAACCAGG | GGGGCTTGGTATATATGTGG |
| VCP | NM_007126 | TGGAGTTCAAAGTGGTGGAAA | ATGGCAGGAGCATTCTTCTCA |
| β5 | BC057840 | GCTTCGAAATAAGGAACGCA | ATTGTCACTGGAGACTCGGAT |
| Rpt2 | NM_002802 | AACCAAACCTCAGCCACTTTC | TTAAGGCCATCAGACCAGCTT |
| GADD45 | NM_001924 | TTTTGCTGCGAGAACGACAT | ACTGGAACCCATTGATCCAT |
| BiP | X87949 | ACGTGGAATGACCCGTCTGT | ATGAAGTGTTCCATGACACGC |
| GDF15 | NM_004864 | AAGAACTCAGGACGGTGAATG | AAGAACTCAGGACGGTGAATG |
| ATF3 | NM_004024 | TCCTGGGTCACTGGTGTTTGA | TTCTTGTTTCGGCACTTTGC |
| CTH | NM_153742 | TTGCCCAGTTCCTGGAATCTA | TGCTGCCTTCAAAGCTTGAT |
| IL18 | NM_001562 | ATGGCTGCTGAACCAGTAGAA | AATAAATATGGTCCGGGGTG |
| Actin | NM_001101 | TTCCTTCCTGGGCATGGAGT | ATCCACATCTGCTGGAAGGT |
SDS-PAGE and Western Blotting
SDS-PAGE and Western blotting was conducted for indicated proteins as described previously (Wojcik et al., 2004b). Primary antibodies were detected using horseradish peroxidase-conjugated anti-mouse and anti-rabbit antibodies from Jackson ImmunoResearch Laboratories. Horseradish peroxidase was detected using the ECL Advance kit (GE Healthcare, Piscataway, NJ). Images were acquired with Kodak 4000MM Image Station. Densitometry was performed using Image Quant version 5.2 (GE Healthcare).
Preparation of RNA and Hybridization of Spotted Microarrays
Preliminary experiments established 72 h posttransfection as the optimal time for RNAi of VCP (Wojcik et al., 2004a, b) Therefore, total RNA was isolated 72 h after transfection with siRNA. We used the modified method of Chomczynski after lysis in TRIzol (Invitrogen) (Chomczynski and Sacchi, 1987). The quality of RNA was assessed by spectrophotometric analysis (measuring OD260/280 ratio) and by BioAnalyzer (Agilent Technologies, Palo Alto, CA). Samples were transferred to the Microarray Core Facility at University of Texas Southwestern Medical Center (Dallas, TX) where microarray experiments were performed using human 35k spotted oligonucleotide arrays version 3.0.2 (Operon Biotechnologies, Huntsville, AL). The samples were fluorescently labeled with either Cy3-dCTP or Cy5-dCTP (GE Healthcare). The labeled probes were mixed with preheated ASAP hybridization buffer (PerkinElmer Life and Analytical Sciences, Boston, MA), and hybridized to an oligo array according to the manufacturer's instruction. The slides were washed with SSC buffer from low to high stringency and scanned by GenePix scanner (Molecular Devices, Sunnyvale, CA) at 532 nm (Cy3) and 635 nm (Cy5). The Cy3 and Cy5 scans for each slide were superimposed, and the fluorescent ratio for each spot was obtained.
Design of Microarray Study
Transfection with any siRNA may induce certain sequence-independent effects, regardless of the lack or presence of specific effects (Sledz et al., 2003). To increase specificity of analysis, we used two independent siRNAs against VCP and one against, an irrelevant control target (EGFP), and compared results with those from cells treated with Oligofectamine only. RNA isolated from each experimental group was divided equally in two, with one-half of treated samples and one-half control samples labeled with Cy3 and Cy5, to randomize effects of the dye bias (Dobbin et al., 2003; Rosenzweig et al., 2004). To minimize other unknown variables, repeat experiments were performed on different days by using identical reagents. In total, we generated six microarray data sets for VCP knockdown by using VCP-2, four microarray data sets by using VCP-6, and four microarray data sets by using siRNA targeting EGPF.
Data Processing
The preliminary data analysis (flagging low-quality spots) was performed using Gene Pix 3.0 Prosoftware (Molecular Devices). Local Lowess normalization was done using Gene Traffic Duo software (Iobion Informatics, La Jolla, CA). Before further analysis, we rejected all spots flagged by Gene Traffic software. Further analysis was conducted using in-house custom C++ programs and Perl scripts developed for this project. We rejected all points with raw signal intensity <100 (in either red or green channel) and any point with sum of normalized green and red channel intensities smaller than 25 (to avoid meaningless high fold ratios). To make fold ratio distribution closer to Gaussian (normal), we converted fold ratios to logarithmic scale. After logarithm transformation, for each data set, we subtracted the average measurement from each probe measurement, thus setting the average log-ratio in each experiment to zero. We then performed principal component analysis, inspected the data, and observed that they did not reveal any structure. Because the data did not contain distinct clusters, and the distribution was close to normal, we examined outliers to identify transcripts of genes significantly changed during our experiments. We computed mean values and standard deviations of measurement for each experiment group (xexp and σexp) as well as a mean value and a SD of each transcript measurement during each knockdown (xtr and σtr). For ideal measurement, we would obtain outliers with 95% confidence by selecting as altered those transcripts that are more than σexp distant from the average during this experiment:
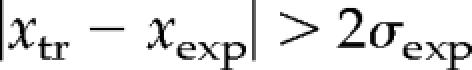 |
However, because the individual transcripts ratios contain errors of their own, even if the average ratio measured for a given transcripts, xtr, lies farther than 2σexp from the average result in this experiment, xexp, the measurement still may be not a true outlier.
For example, if σtr = 5σexp, the accuracy of the transcript log-ratio measurement is clearly not sufficient to determine whether it is altered significantly. To take into account a varying accuracy of individual transcript log-ratio measurements, we used a conservative criterion of defining a transcript as up- or down-regulated only if its log-ratio together with its 95% confidence interval (corresponding to ca. 2σ) lies in the outlier region:
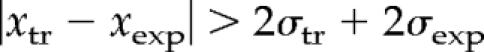 |
Thus, our method eliminates transcripts with high fold ratios, which may have been caused by high experimental error. Because we performed at least four replicates of each experiment, we also used information about variation between experiments to judge the quality and reproducibility of the results. We define a transcript as up-regulated if its significance (i.e., 1 minus probability) of being up- or down-regulated is <0.05. The significance is computed as follows:
where erf is the error function. The probability of a gene being down-regulated is computed analogously. To define transcripts that remain unchanged during knockdown, we assumed the measured transcript log-ratio, together with its 95% confidence interval, lies within 95% confidence interval of the measured mean log-ratio during this experiment. The significance is computed as follows:
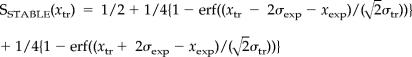 |
RESULTS
RNAi Causes Few Nonspecific Effects on the Transcriptome Level
To gain insight into cellular roles of VCP, we determined the effects of decreased levels of VCP on the gene expression profile of HeLa cells subjected to RNAi of VCP. To discriminate between specific and off-target effects, we used two different siRNAs against VCP and a control siRNA against EGFP that does not match any sequence in the human genome (Jackson et al., 2003). Expression profiles of cells transfected with all three siRNAs were compared with control cells treated with the transfection reagent alone. Transcription profiles were analyzed using the strict criteria outlined in Materials and Methods. From 34,993 spots on the microarrays, 9637 produced high-quality data in all 12 hybridizations and were therefore analyzed further. The complete results of the microarray experiments have been deposited in the public Arrayexpress database (http://www.ebi.ac.uk/arrayexpress, accession no. E-MEXP-817). Transcripts that were up- or down-regulated after RNAi of VCP with both siRNAs with at least 95% confidence level and were not significantly up- or down-regulated after siRNA of EGFP were considered specific for the VCP knockdown. In contrast, transcripts that were altered after transfection with siRNAs for both VCP and EGFP were considered nonspecific, probably reflecting a sequence-independent cellular response to siRNAs. Only one transcript was down-regulated and three transcripts were up-regulated after transfection by each of these three siRNAs (Table 2). These results indicate that siRNA transfection per se causes remarkably few nonspecific changes in the gene expression profile. This conclusion, however, is limited by the single time point of our analysis. Thus, it is possible that some transcripts were altered at earlier times but returned to normal levels after 72 h.
Table 2.
Transcripts nonspecifically up- or down-regulated after transfection with any tested siRNA
| Name | Id | Fold | Sign. | Description | Localization |
|---|---|---|---|---|---|
| SERPINE2 | NM_006216 | 2.9 | 4.6 × 10−6 | Protease nexin 1 | EC |
| NRG1 | NM_013959 | 1.9 | 4.5 × 10−6 | Neuregulin 1 | PM |
| IDS | NM_000202 | 2.1 | 3.0 × 10−10 | Iduronate 2 sulfatase | L |
| CABLES1 | NM_138375 | −1.5 | 0.010 | Cdk5 and Abl enzyme substrate | N, membranes |
Name, gene name; Id, systematic id (RefSeq for mRNA or GenBank accession no.); Fold, average fold change for all VCP knockdown experiments; Sign., significance (as explained in Materials and Methods) that a given transcript is more than 1.96 σ down-regulated and stable during control knockdown; PM, plasma membrane; N, nucleus; L, lysosomes; and EC, extracellular, secreted.
Trancripts up- or down-regulated by RNAi of VCP and control EGFP. Transfection of HeLa cells with siRNAs was conducted as described in Materials and Methods.
RNAi of VCP Down-Regulates Expression of a Limited Number of Transcripts
RNAi of VCP resulted in a 2.2-fold down-regulation of the VCP transcript, verifying the effectiveness of RNAi against this target (Table 3). Six other transcripts were down-regulated in response to RNAi of VCP. Two of the down-regulated sequences encode unknown proteins, whereas the others encode proteins that do not seem to be functionally related. Two are receptors located at the plasma membrane, whereas two are cytosolic proteins. One cytosolic protein contains an F-box and therefore is a putative component of an SCF-type ubiquitin ligase complex. To further analyze the specificity of these effects, we compared the sequences of each down-regulated transcript with the sequences of each VCP siRNA. Partial matching of both siRNAs with the transcripts was detected (Table 4), raising the question of whether down-regulation of these transcripts might result from off-target effects (Jackson et al., 2003). Additional analysis of possible off-target sequences by using the Web engine http://rnai.cs.unm.edu/offTarget (Qiu et al., 2005) revealed 11 possible off-target transcripts for the VCP-2 siRNA with an off-target score ≤35, but only one off-target transcript for the VCP-6 siRNA with an off-target score ≤35. This difference may be due to the different algorithms used to design these siRNAs (Elbashir et al., 2001; Reynolds et al., 2004). Nevertheless, none of the possible off-target sequences identified by this search corresponded to the transcripts down-regulated by both VCP siRNAs. Moreover, none of the predicted 11 off-target transcripts for VCP-2 was found among the eight transcripts down-regulated by VCP-2 alone (our unpublished data). These results suggest that RNAi of VCP results in decreased expression of a minimal number of genes. Most if not all down-regulated transcripts presented in Table 3 are specifically down-regulated in response to low cellular VCP levels.
Table 3.
Transcripts specifically down-regulated after RNAi of VCP with two different siRNAs
| Name | Id | Fold | Sign. | Description | Localization |
|---|---|---|---|---|---|
| — | XM_378620 | −2.3 | 1.9 × 10–5 | Hypothetical protein, predicted by PSORTII to be mitochondrial (60.9%), nuclear (34.8%), or ER (4.3%) | ? |
| VCP | NM_007126 | −2.2 | 0.02 | Valosin-containing protein | C, N |
| — | AK091343 | −1.7 | 0.010 | Hypothetical protein, predicted by PSORTII to be nuclear (47.8%) or mitochondrial (34.8%) | ? |
| ADRA2A | NM_000681 | −1.5 | 0.03 | α-2A adrenergic receptor | PM |
| PAFAH1B2 | BC001774 | −1.5 | 0.010 | β subunit of platelet-activating factor acetylhydrolase, isoform Ib | C |
| VIPR2 | NM_003382 | −1.4 | 4.5 × 10-3 | Vasoactive intestinal polypeptide receptor 2 | PM |
| FBP5 | NM_012177 | −1.4 | 0.05 | F-box only protein 5 | C, N |
Name, gene name; Id, systematic id (RefSeq for mRNA or GenBank accession no.); Fold, average fold change for all VCP knockdown experiments; Sign., significance (as explained in Materials and Methods) that a given transcript is more than 1.96 σ down-regulated and stable during control knockdown; PM, plasma membrane; C, cytoplasm; N, nucleus; L, lysosomes; EC, extracellular, secreted; and ?, unknown.
Transcripts specifically down-regulated by RNAi of VCP.
Table 4.
Alignment of the two different siRNAs targeting VCP with transcripts specifically down-regulated after RNAi of VCP
| XM_378620 | TCTCGTGTTTGATTATATCCTG | XM_378620 | TTACCATA-TGATACAGCCTA |
| VCP2 siRNA | TGTAGGGTATGATGACAT—TG | VCP6 siRNA | TAACCTTCGTG-TAC-GCCTA |
| AK091343 | TGTAGCCTA–ATGA-ATTG | AK091343 | TAACCTAGCAGTACTCC-A |
| VCP2 siRNA | TGTAGGGTATGATGACATTG | VCP6 siRNA | TAACCTTCGTGTACGCCTA |
| ADRA2A | -GTTGAG-ATCATGTCATTG | ADRA2A | TCCCCTTCTTCTTCACCTA |
| VCP2 siRNA | TGTAGGGTATGATGACATTG | VCP6 siRNA | TAACCTTCGTGTACGCCTA |
| PAFAH1B2 | TGTGGTGTATGA-G-CATTG | PAFA | TATTCCTTCGCCCACGCATT |
| VCP2 siRNA | TGTAGGGTATGATGACATTG | VCP6 siRNA | TAA-CCTTCGTGTACGCCTA |
| VIPR2 | TGTAGGGTTTG–GACA—G | VIPR2 | TCACCTTGGTTTGCAAAACCCAT |
| VCP2 siRNA | TGTAGGGTATGATGACATTG | VCP6 siRNA | TAACCTTCGTGT—–ACGCCT |
| FBP5 | -GTAAAACCTGATGACATTG | FBP5 | TCAACTTC-TGGA-GTCTA |
| VCP6 siRNA | TAACCTTCGTGTACGCCTA | VCP2 siRNA | TGTAGGGTATGATGACATTG |
Alignment of the two different siRNAs targeting VCP (denoted VCP-2 and VCP-6 siRNAs) with down-regulated transcripts.
RNAi of VCP Up-Regulates Expression of Multiple Transcripts
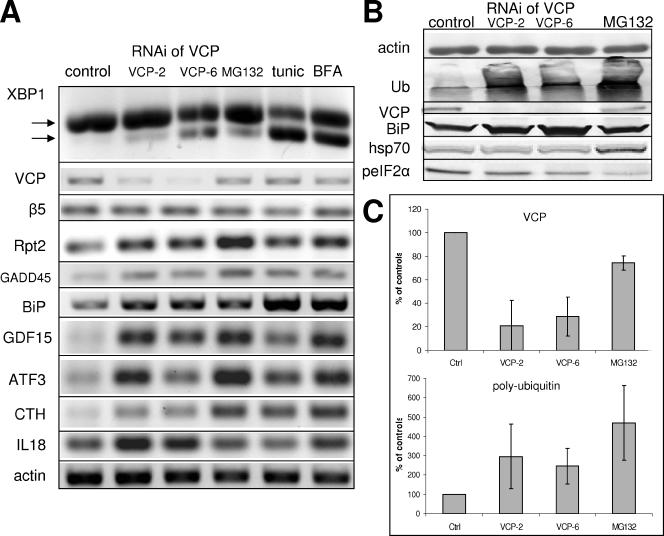
RNAi of VCP specifically up-regulated transcripts of 28 genes (Table 5). The up-regulation of three transcripts, HERP, INSIG2, and SAT, was detected independently by two different oligonucleotides present in the microarray. In contrast to the identified down-regulated genes, many up-regulated transcripts encode functionally or structurally related proteins. For example, 12 transcripts (46%) encode proteins that are either secreted or reside in compartments of the secretory pathway. Moreover, many of these proteins are known to be up-regulated by ER stress and/or may participate in the unfolded protein response. Four other cytosolic proteins associate with the cytoplasmic face of cellular membranes. Ten transcripts are known to be involved in various forms of cellular stress, including ER stress and oxidative stress, and five transcripts encode proteins involved in apoptosis. Interestingly, none of the up-regulated transcripts is known to physically interact with VCP and with the exception of one putative ubiquitin-specific hydrolase, none is a recognized component of the UPS. We performed semiquantitative RT-PCR for selected transcripts to verify their up-regulation by RNAi of VCP (Figure 1A). The mRNA levels of GADD45, GDF15, ATF3, CTH, and IL18 were increased by each siRNA against VCP (Figure 1A). These results are in excellent accord with the microarray data and strongly support the conclusion that RNAi of VCP up-regulates expression of these transcripts. We have also performed semiquantitative RT-PCR for selected ER stress-related transcripts that were not detected by our screen, in particular BiP, the β5 subunit of the 20S proteasome, and the S4/Rpt2 subunit of the PA700. Although mRNA levels of β5 were not altered, those of BiP and S4/Rpt2 were increased. These transcripts are present in our microarray data set, but they did not pass our strict criteria for significant change. BiP was rejected because one of four control microarrays yielded a flagged spot, even though it was significantly increased (2.3-fold, significance of 1.3 × 10−3) in other RNAi experiments. S4/Rpt2 (NM_002802) showed a consistent pattern of expression characteristic of specifically up-regulated genes, but the low magnitude of change (average 1.4-fold difference) did not meet our criteria. Several other AAA protein subunits of the 26S proteasome showed similar changes (see original data).
Table 5.
Transcripts specifically up-regulated after RNAi of VCP with two different siRNAs
| Name | Id | Fold | Sign. | Description | Localization |
|---|---|---|---|---|---|
| GDF15 | NM_004864 | 4.6 | 0.050 | Growth differentiation factor 15 (macrophage inhibitory cytokine 1, MIC1) | EC |
| VLDLR | NM_003383 | 3.3 | 2.4 × 10−6 | Very low density lipoprotein receptor | PM |
| ATF3 | NM_004024 | 3.1 | 1.4 × 10−4 | Activation transcription factor 3, involved in stress responses | N |
| HRK | NM_003806 | 2.9 | 0.046 | Harakiri, BCL2 interacting protein, involved in apoptosis | C, membrane-associated |
| CTH | NM_153742 | 2.7 | 0.020 | Cystathionase, enzyme of transsulfuration pathway, increased in oxidative stress | C |
| SSAT | NM_002970 | 2.6 | 4.7 × 10−5 | Diamine acetyltransferase/Spermidine/spermine N(1)-acetyltransferase | C |
| Myosin VB | L29143 | 2.5 | 2.6 × 10−6 | Myosin VB mRNA | C, membrane-associated |
| TGFβ2 | NM_003238 | 2.4 | 0.035 | Transforming growth factor, β2 | EC |
| USP53 | AB037771 | 2.3 | 0.049 | Ubiquitin-specific protease 53, according to PSORTII nuclear (73.9%) | ? |
| GADD45 | NM_001924 | 2.2 | 0.002 | Growth arrest and DNA damage-inducible/DDIT1 | N |
| WIPI1, Atg18 | NM_017983 | 2.2 | 6.0 × 10−6 | WD repeat domain, phosphoinositide interacting, involved in autophagy | Autophagosome |
| IL18 | NM_001562 | 2.2 | 0.031 | interleukin 18 | EC |
| HERP, MIF1 | NM_014685 | 2.1 | 1.3 × 10−4 | Homocysteine-responsive ER-resident Ub like | ER |
| — | NP_060840 | 2.1 | 0.001 | Hypothetical protein NP_060840, according to PSORTII transmembrane (100%) | ? |
| CHST11 | NM_018413 | 2.0 | 0.003 | carbohydrate (chondroitin 4) sulfotransferase 11 | Golgi |
| BLVRP | NM_000713 | 1.9 | 0.025 | Biliverdin reductase B/Flavin reductase | C |
| EPHA2/ECK | NM_004431 | 1.9 | 0.035 | Ephrin type-A receptor 2 | PM |
| IER3 | NM_003897 | 1.9 | 0.047 | Immediate early response 3 (IER3), involved in apoptosis | N (PML bodies) |
| SLC2A1, GLUT1 | NM_006516 | 1.8 | 0.011 | Solute carrier family 2, facilitiated glucose transporter member 1 | PM |
| PKCA, PRKCA | NM_002737 | 1.8 | 0.003 | Protein kinase C, α type | C, membrane-associated |
| ICAM2 | NM_000873 | 1.8 | 0.038 | intercellular adhesion molecule 2 | PM |
| — | D87454 | 1.8 | 0.003 | KIAA0265, has 3 Kelch motifs, according to PSORTII cytoplasmic (73.9%) | ? |
| MAX | NM_145114 | 1.8 | 0.013 | MYC associated factor X (MAX), transcript variant 4, transcription factor, binds ATF3 and GADD45 promoters | N |
| INSIG2 | NM_016133 | 1.8 | 0.048 | Insulin induced gene 2, block processing of SREBP by binding SCAP | ER |
| EMP1 | NM_001423 | 1.7 | 0.034 | Epithelial membrane protein 1 | PM |
| WARS | NM_004184 | 1.7 | 0.002 | Tryptophanyl-tRNA synthetase | C |
| GPNMB | NM_002510 | 1.6 | 0.036 | Glycoprotein (transmembrane) nmb transcript variant 2 | PM |
Name, gene name; Id, systematic id (RefSeq for mRNA or GenBank accession no.); Fold, average fold change for all VCP knockdown experiments; Sign., significance (as explained in Materials and Methods) that a given transcript is more than 1.96 σ down-regulated and stable during control knockdown; PM, plasma membrane; C, cytoplasm; N, nucleus; L, lysosomes; EC, extracellular, secreted; and ?, unknown.
Transcripts specifically up-regulated by RNAi of VCP.
Figure 1.
RNAi of VCP induces ER stress and induction of UPR. HeLa cells were subjected to RNAi of VCP by using either VCP-2 or VCP-6 siRNA as described in Materials and Methods. (A) RT-PCR was performed for the indicated messages from cells subjected to RNAi and from cells treated for 6 h with 10 μM MG132, 10 μg/ml tunicamycin, and 5 μM brefeldin A. Arrows show spliced and unspliced transcripts of XBP-1. (B) Western blotting was performed for the indicated proteins from cells subjected to RNAi against VCP and from cells treated with 10 μM MG132 for 6 h. (C) Effect of RNAi on levels of VCP and polyubiquitinated proteins. HeLa cells were subjected to RNAi of VCP with the indicated siRNAs or treated with 10 μM MG132 for 6 h. Cell lysates were Western blotted for VCP (top) or polyubiquitin (bottom). Blots were quantified, and data are expressed as a percentage of control values. Results represent means ± SEM from five independent experiments.
RNAi of VCP Induces the UPR
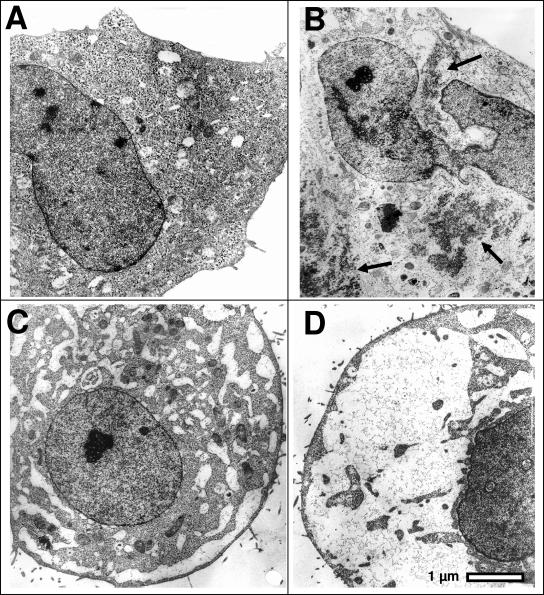
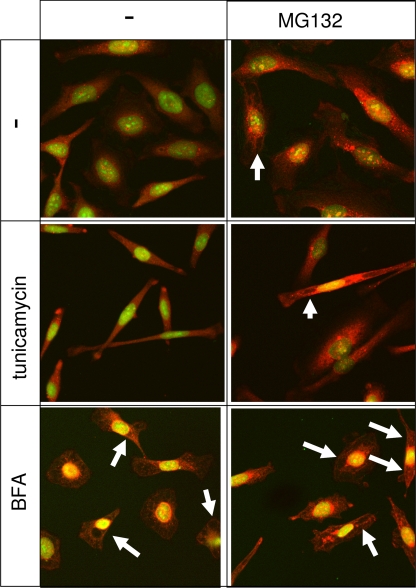
VCP has an established but incompletely defined role in various aspects of ERAD (Bar-Nun, 2005; Romisch, 2005). However, the role of VCP in ERAD has been studied more extensively in yeast than in mammals. We previously demonstrated that RNAi of VCP in HeLa cells induces an accumulation of polyubiquitinated proteins and promotes extensive cellular vacuolization due to swelling of the endoplasmic reticulum (Wojcik et al., 2004b). Transmission electron microscopy of HeLa cells subjected to RNAi of VCP confirms and extends the latter finding, demonstrating the presence of distended vacuole-like membrane-limited compartments probably corresponding to swollen ER cisternae identified previously by the immunofluorescent labeling (Figure 2, C and D). Surprisingly, the lumen of the distended cisternae has a very low electron density, suggesting an osmotic mechanism of swelling rather than one caused by an accumulation of protein aggregates within the ER lumen as a consequence of inhibited ERAD. In contrast, proteasome inhibition with MG132 induces the formation of electron-dense cytosolic aggregates (Figure 2B) as observed previously (Wojcik et al., 1996). Nevertheless, the dramatic alterations in cell morphology caused by RNAi of VCP are probably associated with induction of ER stress and promotion of the UPR. Interestingly, vacuolization is not caused by all inducers of ER stress. For example, brefeldin A promotes vacuolization, whereas tunicamycin does not (Figure 3). These data suggest that the induction of UPR by RNAi of VCP may reflect a BFA-sensitive function of VCP in membrane fusion rather than an effect of VCP on accumulation with unfolded proteins.
Figure 2.
RNAi of VCP promotes vacuolization of cells. HeLa cells were transfected with siRNAs directed against EGFP (control) or VCP as described in Materials and Methods, or treated with MG132 for 6 h before processing for transmission electron microscopy. (A) Control cells. (B) MG132-treated cells. Arrows show aggregation of high-electron-density cytosolic material. (C) RNAi of VCP by using VCP-2 siRNA. (D) RNAi of VCP by using VCP-6 siRNA.
Figure 3.
Pharmacological induction of ER stress is not always associated with cell vacuolization. Immunofluorescence images of HeLa cells labeled by the FK1 anti-ubiquitin antibody (red) and the nuclear dye YoPro iodide (green). Cells were submitted to a 16 h treatment with 5 μM BFA and 10 μg/ml tunicamycin, either alone or in combination with 10 μM MG132. Although both MG132 and BFA induce cell vacuolization, probably by distension of ER cistaernae (arrows), tunicamycin induces an elongated cellular phenotype without inducing vacuoles, which can be induced by a cotreatment with MG132.
To determine whether RNAi of VCP specifically induced UPR, we assayed IRE-1–dependent splicing of mRNA encoding the transcription factor XBP-1, a diagnostic feature of UPR (Yoshida et al., 2001). RNAi of VCP by each siRNA reduced VCP protein levels by more than 85% and consistently induced splicing of XBP-1 mRNA (Figure 1A). Although the magnitude of RNAi-induced XBP-1 splicing was somewhat variable in different experiments, it was similar to that promoted by the proteasome inhibitor MG132, but not as great as that caused by established inducers of ER stress such as tunicamycin or brefeldin A. RNAi of VCP caused a twofold increase in the levels of polyubiquitinated proteins, and increased levels of BiP, an ER chaperone whose expression increases as part of the UPR, but it had no effect on levels of cytosolic hsp70, which was increased by MG132 (Figure 1B). RNAi of VCP had no effect on the levels of phosphorylated eukaryotic initiation factor 2α (eIF2α), which was decreased by the treatment with MG132. Phosphorylation of eIF2α is a downstream event in multiple stress signaling pathways, including ER stress and UPR activation (Wek et al., 2006). The decrease in eIF2a phosphorylation after proteasome inhibition may be a consequence of reduced degradation of GADD34, a known phosphatase of eIF2a, or of a direct inhibition of PERK (Nawrocki et al., 2005). These results directly demonstrate that reduction of VCP levels by RNAi induces some, but not all features of UPR, without inducing a cytosolic stress response. In contrast, proteasome inhibition or induction of ER stress by either tunicamycin or BFA did not induce up-regulation of VCP mRNA or protein (Figure 1A).
RNAi of VCP Differentially Affects Degradation of Various UPS Substrates
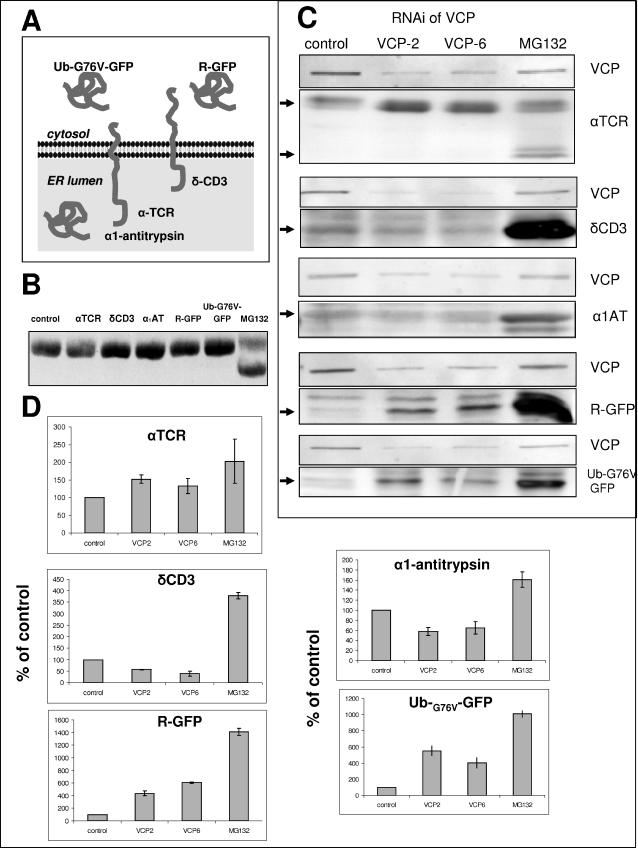
VCP is implicated in the degradation of both cytosolic and ER proteins (Dai et al., 1998; Dai and Li, 2001; Ye et al., 2001). To further examine the role of VCP in the degradation of various cellular proteins, we engineered HeLa cell lines that stably express five established substrates of the UPS: Ub-G76V-GFP, a cytosolic substrate of the ubiquitin-fusion degradation (UFD) pathway (Johnson et al., 1995; Dantuma et al., 2000); R-GFP, a rapidly degraded cytosolic substrate of the N-end rule pathway (Bachmair et al., 1986; Dantuma et al., 2000); αTCR and δCD3, two different ER transmembrane subunits of the T-cell receptor subject to ERAD (Yu et al., 1997; Yang et al., 1998; Yu and Kopito, 1999; Tiwari and Weissman, 2001); and α1-antitrypsin Hong Kong mutant, a misfolded lumenal ER protein subject to ERAD (Hosokawa et al., 2001; Hosokawa et al., 2003) (Figure 4A). Treatment of each cell line with MG132 caused time-dependent accumulation of the respective protein, thereby confirming the role of the UPS in its degradation (Figure 4). Because overexpression of misfolded proteins in the ER may promote ER stress and constitutive UPR, we examined the status of XBP-1 splicing in each cell line. XBP-1 splicing did not differ significantly in any cell line compared with the parental nontransfected HeLa cells (Figure 4B). Moreover, these cells had normal morphology and growth characteristics (our unpublished data). Thus, chronic expression of transfected proteins did not seem to induce features of ER stress.
Figure 4.
Differential effects of RNAi of VCP on the degradation of UPS substrates. (A) Schematic representation of five UPS substrates used in this study. (B) Overexpression of UPS substrates does not induce UPR in stable cell lines as assessed by XBP-1 splicing. (C) HeLa cells stably
Each cell line was subjected to RNAi of VCP with two different siRNAs (Figure 4C). RNAi of VCP increased levels of polyubiquitinated cellular proteins and promoted UPR in each cell line, similarly to the effects in nonengineered cells (our unpublished data), but it had divergent effects on the degradation of the respective expressed proteins. RNAi of VCP caused a four- to sixfold accumulation of the cytosolic proteins R-GFP and UbG76VGFP. Analogous results with different UFD and N-end rule substrates have been reported previously in yeast expressing mutant VCP homolog (Cdc48) (Bachmair et al., 1986; Johnson et al., 1995; Ghislain et al., 1996). RNAi of VCP also inhibited degradation of αTCR, in accord with the established role of VCP in ERAD (Figure 4C). Proteasome inhibition with MG132 caused selective accumulation of a faster migrating (∼29-kDa) form of αTCR, probably corresponding to the deglycosylated form, as described previously (Yu et al., 1997; Huppa and Ploegh, 1997; Yang et al., 1998; Yu and Kopito, 1999). In contrast, RNAi of VCP caused accumulation of the slower migrating (∼38-kDa) glycosylated αTCR. These results suggest that MG132 inhibited degradation of αTCR after its extraction from the ER, whereas RNAi of VCP inhibited degradation of αTCR by preventing its extraction from the ER. In surprising contrast to αTCR, RNAi of VCP did not affect the levels of two other ERAD substrates, α1-antitrypsin and δCD3. These results support the conclusion that VCP mediates proteolysis in multiple cellular compartments and suggest that certain established ERAD substrates can be processed by VCP-independent mechanisms.
DISCUSSION
VCP (aka p97 or Cdc48 in yeast) is an AAA ATPase that has been implicated in numerous and diverse cellular functions (Woodman, 2003; Dreveny et al., 2004; Wang et al., 2004; Bar-Nun, 2005; Halawani and Latterich, 2006). Despite considerable work, a comprehensive view of its biological role(s) remains elusive. To gain insight into the cellular roles of VCP, we have conducted a microarray analysis to identify transcripts altered in response to RNAi of VCP in HeLa cells. We analyzed the data by a stringent method designed to reduce spurious positives that devalue some microarray results. This analysis demanded comparable effects from each of two different siRNAs in each of four different experiments to designate a transcript as altered. Although these stringent requirements probably resulted in elimination of some authentic responses (e.g., BiP and some proteasome subunits; see below), it produced a tractable list of affected transcripts. The altered expression of multiple selected transcripts was verified by independent methodology.
Our microarray analysis identified ∼30 genes whose expression is altered by RNAi of VCP. Although these proteins have diverse functions and many would not necessarily have been anticipated to emerge in this screen, most have plausible connections to known or suspected functions of VCP, including ERAD and ER stress. For example, the most up-regulated transcript is growth differentiation factor 15 (GDF15), up-regulated 4.6×; GDF15 is induced in different tissues after multiple types of chemical and physical injury, including oxidative stress and heat shock (Hsiao et al., 2000; Zimmers et al., 2005). Several proteins involved in cholesterol homeostasis also were up-regulated, including the very low density lipoprotein receptor and insulin-induced protein 2. Up-regulation of genes involved in cholesterol uptake and cholesterol biosynthetic pathway have been shown to be induced by ER stress (Werstuck et al., 2001). Activating transcription factor 3, a transcription factor activated by various stress stimuli, including ER stress and proteasome inhibition (Wek et al., 2006), was up-regulated 3.1-fold. Homocysteine-responsive ER-resident ubiquitin like protein, a transmembrane ER ubiquitin-like protein shown previously to be involved in ERAD (van Laar et al., 2001; Hori et al., 2004), was up-regulated more than twofold. Contrary to other reports (Sledz et al., 2003), we have not observed an interferon-like response to introduction of siRNAs; only four transcripts out of the 34,993 analyzed by the microarrays were affected by RNAi in a sequence-independent manner, thereby validating the specificity of the VCP knockdown.
The most extensively studied function of VCP involves ERAD, a process by which resident and transient ER proteins can be selectively or constitutively degraded in the cytoplasm by the UPS (Tsai et al., 2002; Meusser et al., 2005; Romisch, 2005; Bar-Nun, 2005). VCP in conjunction with Ufd1 and Npl4 may couple ATP hydrolysis and polyubiquitin chain binding properties to power extraction of proteins from the ER for delivery to and degradation by the 26S proteasome. Because this process probably involves multiple proteins whose functions are mechanistically linked, we were somewhat surprised not to identify many components of the UPS as altered transcripts. USP53, a poorly-characterized ubiquitin-specific protease, was the only specifically up-regulated UPS component, whereas Emi1, an F-box protein and thus a putative E3 ubiquitin ligase, was the only down-regulated UPS component. Emi1 is a known regulator of progression through mitosis (Reimann et al., 2001). Therefore, its down-regulation may be involved in severe mitotic abnormalities caused by RNAi of VCP (Wojcik et al., 2004b). Down-regulation of the β subunit of acetylhydrolase, a protein involved in control of intracellular microtubule-dependent motility (Arai, 2002), may be related to the previously described defect in the formation of aggresomes that are linked to altered UPS function (Wojcik et al., 2004b). Our analysis may have failed to detect some UPS components, such as the subunits of the PA700(19S) proteasome regulatory complex, whose altered expression fell short of our strict criteria. Alteration of PA700 content may be particularly significant because PA700 seems to have some similar functions as those attributed to VCP during ERAD (see below).
Previous work from many investigators has indicated a role for VCP in other aspects of ER and Golgi function, including mediation of homotypic membrane fusion (Latterich et al., 1995; Patel et al., 1998; Rabouille et al., 1998). Here, we provide direct evidence that RNAi of VCP not only induces UPR but also alters cellular ultrastructure. We also observed an up-regulation of several genes involved in ER stress. Despite the lack of increased eIF2α phosphorylation (Wek et al., 2006), three genes involved in the response of cells to protein starvation are induced: the biosynthetic enzyme tryptophanyl-tRNA synthetase; Atg18 required for autophagy; and spermidine/spermine N1-acetyltransferase, a rate-limiting enzyme in the catabolic pathway of polyamine metabolism. Several up-regulated transcripts are different gene products involved in apoptosis, a process induced by VCP knockdown (Wojcik et al., 2004b). Proapoptotic transcripts included GADD45A, Harakiri, EPHA2, and MAX. Oxidative stress also triggers UPR through a novel pathway involving inactivation of the VCP ATPase activity by the oxidative modification of Cys522 (Noguchi et al., 2005). Thus, RNAi of VCP may mimic the cellular effects of oxidative inactivation of VCP. In fact, we observed up-regulation of transcripts known to be induced by oxidative stress, such as SSAT (Chopra and Wallace, 1998), cytosolic flavin reductase (Sedlak and Snyder, 2004), and CTH (cystathionin-γ-lyase) (Ishii et al., 2004). These effects may result directly from decreased VCP levels or may be secondary to oxidative stress resulting from accumulation of misfolded ERAD substrates (Haynes et al., 2004).
VCP was required for degradation by the UPS of two cytoplasmic proteins, R-GFP and Ub-G76V-GFP. These results are in accord with findings in yeast, where VCP homolog (Cdc48) has been shown to be required for the degradation of substrates of the N-end rule and UFD pathways (Johnson et al., 1995; Ghislain et al., 1996). RNAi of VCP also inhibited degradation of a prototypical ERAD substrate, αTCR. Accumulation of the fully glycosylated form of αTCR indicates that lack of VCP delays its extraction from the ER membrane. In contrast, we failed to detect altered degradation of two other ERAD substrates, the lumenal α1-antitrypsin and the transmembrane δ-CD3 glycoproteins. The reason for the different sensitivity of these ERAD substrates to VCP depletion is unclear. It is possible that the localization (lumenal α1-antitrypsin versus transmembrane αTCR and δCD3) of proteins, the size of the domain in each compartment (short cytosolic portion in αTCR and large cytosolic portion in δCD3), and the nature of polyubiquitin chain linkages determine functional interactions with VCP. Previous work demonstrated that when cells are treated with proteasome inhibitors, at least some αTCR is exported to and accumulates in the cytosol, whereas δCD3 remains ER associated (Yang et al., 1998; Tiwari and Weissman, 2001). Regardless, the current results suggest that VCP is not required for retrotranslocation of all ERAD substrates. Although it has been recognized that ERAD proceeds by different pathways depending upon the localization of misfolded domains (Taxis et al., 2003; Vashist and Ng, 2004), it is assumed that different pathways of proteasome-dependent ERAD converge at a common step requiring VCP (Bar-Nun, 2005). Our results suggest that even that step may not be common to all ERAD, and therefore they are in accord with suggestions of others (Romisch, 2005). Emerging evidence demonstrates that certain ERAD substrates can be removed from the ER directly by PA700 (19S), the regulatory cap of the 26S proteasome (Lee et al., 2004). This VCP-independent process may involve binding of PA700 directly the Sec61 retrotranslocation channel (Kalies et al., 2005). PA700 contains a heterohexameric ring of AAA ATPases and therefore may share important functional features of VCP in ERAD (Glickman et al., 1998; DeMartino and Slaughter, 1999; Zhang et al., 2000). Moreover, retrotranslocation of the cholera toxin A1 chain, which hijacks the retrotranslocation pathway, does not require active VCP (Kothe et al., 2005). Thus, the role of VCP in ERAD is considerably more complex than envisioned by established models (Tsai et al., 2002; Bar-Nun, 2005). Our results may reflect such complexity whereby VCP functions as a partition for the fate of polyubiquitinated proteins (Halawani and Latterich, 2006). Thus, VCP may promote degradation of certain proteins but deubiquitination and salvage of others. The molecular basis for these distinctions will require additional work.
ACKNOWLEDGMENTS
We acknowledge the generous gifts of pEGFP-N1-Ub-G76V-GFP and pEGFP-N1-Ub-R-GFP plasmids from Dr. Maria Masucci (Karolinska Institutet, Stockholm, Sweden), pcDNA3.1-HA-δCD3 from Allan Weissman (National Institutes of Health, Bethesda, MD), pCDNA3.1-HA-α-TCR from Dr. Ron Kopito (Stanford University, Stanford, CA), and pCMV-α1-antitrypsin Hong Kong from Dr. Nobuko Hosokawa (Kyoto University, Kyoto, Japan). This work was supported by American Heart Association, Texas Affiliate Grant 0365148Y (to C.W.), Biomedical Research Grant from Indiana University School of Medicine 22–812-57 (to C.W.), American Cancer Society Grant IRG-84-002-22 (to C.W.), National Institutes of Health Grant DK-46181 (to G.N.D.), and the Welch Foundation (to G.N.D.). D.N. is on temporary leave from Department of Immunology, Medical University of Warsaw, Warsaw, Poland.
Abbreviations used:
- AAA
ATPases with multiple cellular activities
- BFA
brefeldin A
- ERAD
endoplasmic reticulum-associated degradation
- GFP
green fluorescent protein
- R-GFP
GFP with the N-terminal Met replaced by Arg
- siRNA
small interfering RNA
- αTCR
α chain of the T-cell receptor
- Ub-G76V-GFP
GFP with a noncleavable, mutated (Gly76Val) Ub attached to its N terminus
- UFD
ubiquitin-fusion degradation
- UPR
unfolded protein response
- UPS
ubiquitin-proteasome system
- VCP
valosin-containing protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-05-0432) on August 16, 2006.
REFERENCES
- Arai H. Platelet-activating factor acetylhydrolase. Prostaglandins Other Lipid Mediat. 2002;68–69:83–94. doi: 10.1016/s0090-6980(02)00023-0. [DOI] [PubMed] [Google Scholar]
- Bachmair A., Finley D., Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Bar-Nun S. The role of p97/Cdc48p in endoplasmic reticulum-associated degradation: from the immune system to yeast. Curr. Top. Microbiol. Immunol. 2005;300:95–125. doi: 10.1007/3-540-28007-3_5. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Chopra S., Wallace H. M. Induction of spermidine/spermine N1-acetyltransferase in human cancer cells in response to increased production of reactive oxygen species. Biochem. Pharmacol. 1998;55:1119–1123. doi: 10.1016/s0006-2952(97)00601-1. [DOI] [PubMed] [Google Scholar]
- Dai R. M., Chen E., Longo D. L., Gorbea C. M., Li C. C. Involvement of valosin-containing protein, an ATPase Co-purified with IκBα and 26 S proteasome, in ubiquitin-proteasome-mediated degradation of IκBα. J. Biol. Chem. 1998;273:3562–3573. doi: 10.1074/jbc.273.6.3562. [DOI] [PubMed] [Google Scholar]
- Dai R. M., Li C. C. Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat. Cell Biol. 2001;3:740–744. doi: 10.1038/35087056. [DOI] [PubMed] [Google Scholar]
- Dantuma N. P., Lindsten K., Glas R., Jellne M., Masucci M. G. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat. Biotechnol. 2000;18:538–543. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- DeMartino G. N., Slaughter C. A. The proteasome, a novel protease regulated by multiple mechanisms. J. Biol. Chem. 1999;274:22123–22126. doi: 10.1074/jbc.274.32.22123. [DOI] [PubMed] [Google Scholar]
- Dobbin K., Shih J. H., Simon R. Statistical design of reverse dye microarrays. Bioinformatics. 2003;19:803–810. doi: 10.1093/bioinformatics/btg076. [DOI] [PubMed] [Google Scholar]
- Dreveny I., Pye V. E., Beuron F., Briggs L. C., Isaacson R. L., Matthews S. J., McKeown C., Yuan X., Zhang X., Freemont P. S. p97 and close encounters of every kind: a brief review. Biochem. Soc. Trans. 2004;32:715–720. doi: 10.1042/BST0320715. [DOI] [PubMed] [Google Scholar]
- Elbashir S. M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elkabetz Y., Shapira I., Rabinovich E., Bar-Nun S. Distinct steps in dislocation of luminal ERAD substrates: roles of ER-bound p97/Cdc48p and proteasome. J. Biol. Chem. 2003;279:3980–3989. doi: 10.1074/jbc.M309938200. [DOI] [PubMed] [Google Scholar]
- Ghislain M., Dohmen R. J., Levy F., Varshavsky A. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 1996;15:4884–4899. [PMC free article] [PubMed] [Google Scholar]
- Glickman M. H., Rubin D. M., Coux O., Wefes I., Pfeifer G., Cjeka Z., Baumeister W., Fried V. A., Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- Halawani D., Latterich M. p97, the cell's molecular purgatory? Mol. Cell. 2006;22:713–717. doi: 10.1016/j.molcel.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Harding H. P., Calfon M., Urano F., Novoa I., Ron D. Transcriptional and translational control in the Mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 2002;18:575–599. doi: 10.1146/annurev.cellbio.18.011402.160624. [DOI] [PubMed] [Google Scholar]
- Hartsel S. A., Kitchen D. E., Scaringe S. A., Marshall W. S. RNA oligonucleotide synthesis via 5′-silyl-2′-orthoester chemistry. Methods Mol. Biol. 2005;288:33–50. doi: 10.1385/1-59259-823-4:033. [DOI] [PubMed] [Google Scholar]
- Haynes C. M., Titus E. A., Cooper A. A. Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol. Cell. 2004;15:767–776. doi: 10.1016/j.molcel.2004.08.025. [DOI] [PubMed] [Google Scholar]
- Hori O., et al. Role of Herp in the endoplasmic reticulum stress response. Genes Cells. 2004;9:457–469. doi: 10.1111/j.1356-9597.2004.00735.x. [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Tremblay L. O., You Z., Herscovics A., Wada I., Nagata K. Enhancement of endoplasmic reticulum (ER) degradation of misfolded Null Hong Kong α1-antitrypsin by human ER mannosidase I. J. Biol. Chem. 2003;278:26287–26294. doi: 10.1074/jbc.M303395200. [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Wada I., Hasegawa K., Yorihuzi T., Tremblay L. O., Herscovics A., Nagata K. A novel ER alpha-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2001;2:415–422. doi: 10.1093/embo-reports/kve084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao E. C., Koniaris L. G., Zimmers-Koniaris T., Sebald S. M., Huynh T. V., Lee S. J. Characterization of growth-differentiation factor 15, a transforming growth factor beta superfamily member induced following liver injury. Mol. Cell. Biol. 2000;20:3742–3751. doi: 10.1128/mcb.20.10.3742-3751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppa J. B., Ploegh H. L. The alpha chain of the T cell antigen receptor is degraded in the cytosol. Immunity. 1997;7:113–122. doi: 10.1016/s1074-7613(00)80514-2. [DOI] [PubMed] [Google Scholar]
- Ishii I., Akahoshi N., Yu X. N., Kobayashi Y., Namekata K., Komaki G., Kimura H. Murine cystathionine gamma-lyase: complete cDNA and genomic sequences, promoter activity, tissue distribution and developmental expression. Biochem. J. 2004;381:113–123. doi: 10.1042/BJ20040243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. L., Bartz S. R., Schelter J., Kobayashi S. V., Burchard J., Mao M., Li B., Cavet G., Linsley P. S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Johnson E. S., Ma P. C., Ota I. M., Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- Kalies K. U., Allan S., Sergeyenko T., Kroger H., Romisch K. The protein translocation channel binds proteasomes to the endoplasmic reticulum membrane. EMBO J. 2005;24:2284–2293. doi: 10.1038/sj.emboj.7600731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostova Z., Wolf D. H. For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 2003;22:2309–2317. doi: 10.1093/emboj/cdg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe M., Ye Y., Wagner J. S., De Luca H. E., Kern E., Rapoport T. A., Lencer W. I. Role of p97 AAA-ATPase in the retrotranslocation of the cholera toxin A1 chain, a non-ubiquitinated substrate. J. Biol. Chem. 2005;280:28127–28132. doi: 10.1074/jbc.M503138200. [DOI] [PubMed] [Google Scholar]
- Latterich M., Frohlich K. U., Schekman R. Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell. 1995;82:885–893. doi: 10.1016/0092-8674(95)90268-6. [DOI] [PubMed] [Google Scholar]
- Lee R. J., Liu C. W., Harty C., McCracken A. A., Latterich M., Romisch K., DeMartino G. N., Thomas P. J., Brodsky J. L. Uncoupling retro-translocation and degradation in the ER-associated degradation of a soluble protein. EMBO J. 2004;23:2206–2215. doi: 10.1038/sj.emboj.7600232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Hendershot L. M. The mammalian endoplasmic reticulum as a sensor for cellular stress. Cell Stress. Chaperones. 2002;7:222–229. doi: 10.1379/1466-1268(2002)007<0222:tmeraa>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meusser B., Hirsch C., Jarosch E., Sommer T. ERAD: the long road to destruction. Nat. Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Meyer H. H., Shorter J. G., Seemann J., Pappin D., Warren G. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 2000;19:2181–2192. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H. H., Wang Y., Warren G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 2002;21:5645–5652. doi: 10.1093/emboj/cdf579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki S. T., Carew J. S., Dunner K., Jr, Boise L. H., Chiao P. J., Huang P., Abbruzzese J. L., McConkey D. J. Bortezomib inhibits PKR-like endoplasmic reticulum (ER) kinase and induces apoptosis via ER stress in human pancreatic cancer cells. Cancer Res. 2005;65:11510–11519. doi: 10.1158/0008-5472.CAN-05-2394. [DOI] [PubMed] [Google Scholar]
- Noguchi M., Takata T., Kimura Y., Manno A., Murakami K., Koike M., Ohizumi H., Hori S., Kakizuka A. ATPase activity of p97/Valosin-containing protein is regulated by oxidative modification of the evolutionally conserved 522nd cysteine residue in Walker A motif. J. Biol. Chem. 2005;280:41332–41341. doi: 10.1074/jbc.M509700200. [DOI] [PubMed] [Google Scholar]
- Patel S. K., Indig F. E., Olivieri N., Levine N. D., Latterich M. Organelle membrane fusion: a novel function for the syntaxin homolog Ufe1p in ER membrane fusion. Cell. 1998;92:611–620. doi: 10.1016/s0092-8674(00)81129-0. [DOI] [PubMed] [Google Scholar]
- Qiu S., Adema C. M., Lane T. A computational study of off-target effects of RNA interference. Nucleic Acids Res. 2005;33:1834–1847. doi: 10.1093/nar/gki324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich E., Kerem A., Frohlich K. U., Diamant N., Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C., Kondo H., Newman R., Hui N., Freemont P., Warren G. Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell. 1998;92:603–610. doi: 10.1016/s0092-8674(00)81128-9. [DOI] [PubMed] [Google Scholar]
- Rabouille C., Levine T. P., Peters J. M., Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- Reimann J. D., Freed E., Hsu J. Y., Kramer E. R., Peters J. M., Jackson P. K. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell. 2001;105:645–655. doi: 10.1016/s0092-8674(01)00361-0. [DOI] [PubMed] [Google Scholar]
- Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W. S., Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Romisch K. Endoplasmic reticulum-associated degradation. Annu. Rev. Cell Dev. Biol. 2005;21:435–456. doi: 10.1146/annurev.cellbio.21.012704.133250. [DOI] [PubMed] [Google Scholar]
- Rosenzweig B. A., Pine P. S., Domon O. E., Morris S. M., Chen J. J., Sistare F. D. Dye bias correction in dual-labeled cDNA microarray gene expression measurements. Environ. Health Perspect. 2004;112:480–487. doi: 10.1289/ehp.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlak T. W., Snyder S. H. Bilirubin benefits: cellular protection by a biliverdin reductase antioxidant cycle. Pediatrics. 2004;113:1776–1782. doi: 10.1542/peds.113.6.1776. [DOI] [PubMed] [Google Scholar]
- Sitia R., Braakman I. Quality control in the endoplasmic reticulum protein factory. Nature. 2003;426:891–894. doi: 10.1038/nature02262. [DOI] [PubMed] [Google Scholar]
- Sledz C. A., Holko M., de Veer M. J., Silverman R. H., Williams B. R. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- Sriburi R., Jackowski S., Mori K., Brewer J. W. XBP 1, a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J. Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis C., Hitt R., Park S. H., Deak P. M., Kostova Z., Wolf D. H. Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J. Biol. Chem. 2003;278:35903–35913. doi: 10.1074/jbc.M301080200. [DOI] [PubMed] [Google Scholar]
- Tiwari S., Weissman A. M. Endoplasmic reticulum (ER)-associated degradation of T cell receptor subunits. Involvement of ER-associated ubiquitin-conjugating enzymes (E2s) J. Biol. Chem. 2001;276:16193–16200. doi: 10.1074/jbc.M007640200. [DOI] [PubMed] [Google Scholar]
- Tsai B., Ye Y., Rapoport T. A. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell Biol. 2002;3:246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- van Laar T., van der Eb A. J., Terleth C. Mif 1, a missing link between the unfolded protein response pathway and ER-associated protein degradation? Curr. Protein Pept. Sci. 2001;2:169–190. doi: 10.2174/1389203013381189. [DOI] [PubMed] [Google Scholar]
- Vashist S., Ng D. T. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Song C., Li C. C. Molecular perspectives on p97-VCP: progress in understanding its structure and diverse biological functions. J. Struct. Biol. 2004;146:44–57. doi: 10.1016/j.jsb.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Wek R. C., Jiang H. Y., Anthony T. G. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- Werstuck G. H., et al. Homocysteine-induced endoplasmic reticulum stress causes dysregulation of the cholesterol and triglyceride biosynthetic pathways. J. Clin. Invest. 2001;107:1263–1273. doi: 10.1172/JCI11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik C., Fabunmi R., DeMartino G. N. Modulation of gene expression by RNAi. In: Fennell J. P., Baker A. H., editors. Hypertension. Methods and Protocols. Totowa, NJ: Humana Press; 2004a. pp. 381–394. [Google Scholar]
- Wojcik C., Schroeter D., Wilk S., Lamprecht J., Paweletz N. Ubiquitin-mediated proteolysis centers in HeLa cells: indication from studies of an inhibitor of the chymotrypsin-like activity of the proteasome. Eur. J. Cell Biol. 1996;71:311–318. [PubMed] [Google Scholar]
- Wojcik C., Yano M., DeMartino G. N. RNA interference of valosin-containing protein (VCP/p97) reveals multiple cellular roles linked to ubiquitin/proteasome-dependent proteolysis. J. Cell Sci. 2004b;117:281–292. doi: 10.1242/jcs.00841. [DOI] [PubMed] [Google Scholar]
- Woodman P. G. p97, a protein coping with multiple identities. J. Cell Sci. 2003;116:4283–4290. doi: 10.1242/jcs.00817. [DOI] [PubMed] [Google Scholar]
- Yang M., Omura S., Bonifacino J. S., Weissman A. M. Novel aspects of degradation of T cell receptor subunits from the endoplasmic reticulum (ER) in T cells: importance of oligosaccharide processing, ubiquitination, and proteasome-dependent removal from ER membranes. J. Exp. Med. 1998;187:835–846. doi: 10.1084/jem.187.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Meyer H. H., Rapoport T. A. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Ye Y., Meyer H. H., Rapoport T. A. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Yu H., Kaung G., Kobayashi S., Kopito R. R. Cytosolic degradation of T-cell receptor alpha chains by the proteasome. J. Biol. Chem. 1997;272:20800–20804. doi: 10.1074/jbc.272.33.20800. [DOI] [PubMed] [Google Scholar]
- Yu H., Kopito R. R. The role of multiubiquitination in dislocation and degradation of the alpha subunit of the T cell antigen receptor. J. Biol. Chem. 1999;274:36852–36858. doi: 10.1074/jbc.274.52.36852. [DOI] [PubMed] [Google Scholar]
- Zhang X., et al. Structure of the AAA ATPase p97. Mol. Cell. 2000;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- Zimmers T. A., Jin X., Hsiao E. C., McGrath S. A., Esquela A. F., Koniaris L. G. Growth differentiation factor-15/macrophage inhibitory cytokine-1 induction after kidney and lung injury. Shock. 2005;23:543–548. [PubMed] [Google Scholar]