Abstract
Four maternal systems are known to pattern the early Drosophila embryo. The key component of the anterior system is the homeodomain protein Bicoid (Bcd). Bcd needs the contribution of another anterior morphogen, Hunchback (Hb), to function properly: Bcd and Hb synergize to organize anterior development. A molecular mechanism for this synergy has been proposed to involve specific interactions of Bcd and Hb with TATA-binding protein-associated factors (TAFIIs) that are components of the general transcription machinery. Bcd contains three putative activation domains: a glutamine-rich region, which interacts in vitro with TAFII110; an alanine-rich domain, which targets TAFII60; and a C-terminal acidic region, which has an unknown role. We have generated flies carrying bcd transgenes lacking one or several of these domains to test their function in vivo. Surprisingly, a bcd transgene that lacks all three putative activation domains is able to rescue the bcdE1 null phenotype to viability. Moreover, the development of these embryos is not affected by the presence of dominant negative mutations in TAFII110 or TAFII60. This means that the interactions observed in vitro between Bcd and TAFII60 or TAFII110 aid transcriptional activation but are dispensable for normal development.
Keywords: Hunchback, morphogen, TAFII, transcription
In Drosophila, elaboration of the body plan is directed by maternally encoded information (1). Four maternal systems of genetic information are involved in setting up the pattern of the embryo. Patterning the anterior region requires input from both the terminal and anterior systems. The terminal system specifies the fate of the poles of the embryo by activating the uniformly distributed receptor tyrosine kinase Torso (Tor) at each end of the early embryo. Loss-of-function mutations in any of the genes implicated in the terminal system give rise to the loss of the anterior portion of the head and of all the structures posterior to and including the eighth abdominal segment (2–6).
The anterior region is organized by the bicoid (bcd) gene product, a homeodomain containing transcription factor (Bcd) (7, 8). bcd mRNA is synthesized during oogenesis and is transported into the egg, where it becomes localized at the anterior tip. This mRNA is translated after the egg is laid, and the protein diffuses away from the anterior tip in the syncytial environment of the early embryo, forming a concentration gradient (9). Bcd acts as a morphogen: it specifies distinct developmental fates by specifically activating zygotic target genes that respond to a series of Bcd concentration thresholds. In nuclei close to the anterior tip, high concentrations of Bcd activate head gap genes such as orthodenticle (10). At more posterior positions, lower concentrations of Bcd activate the thoracic gap gene hunchback (hb) (11–13). Finally, very low levels of Bcd are able to activate abdominal gap genes like knirps (14). In addition to its ability to transcriptionally activate a series of target genes, Bcd is also a repressor of posterior development by preventing translation of the ubiquitously distributed maternal caudal mRNA (15–17).
bcd mutant females produce embryos that lack all head and thoracic segments and some anterior abdominal segments and show duplication of the telson at the anterior end of the embryo (7). However, the remaining abdominal segments exhibit a normal anterior-posterior polarity that must be set up by other factors. Bcd is not the only morphogen active in the anterior region of the early fly embryo. A maternal gradient of the zinc-finger transcription factor Hb is also able to pattern the embryo (18–20). Therefore, not only does hb represent a zygotic gap gene, which is under the control of Bcd (11), but it also functions as a maternally provided morphogen. Translation of the homogeneously distributed maternal hb mRNA is blocked in the posterior of the embryo by nanos (21), thereby generating an anterior-posterior morphogenetic gradient of the Hb protein.
The two morphogens Bcd and Hb work together to organize proper anterior patterning of the fly embryo (22, 23). In the absence of both maternal and zygotic hb activities, bcd function is strongly affected. An artificial promoter that contains only strong Bcd binding sites is poorly activated in the anterior regions of the embryo. Addition of Hb sites dramatically increases the level of activation and enlarges the domain of reporter gene expression (23). Bcd and Hb synergize to fulfill their functions. The molecular mechanism underlying the synergy between Bcd and Hb has been elusive. Recently, biochemical studies of the interaction between Bcd, Hb, and the basal transcription factor complex TFIID led to a molecular model that could nicely explain how Bcd and Hb synergize in activating their target genes (24, 25).
Activation of transcription requires both the contribution of RNA polymerase II and the basal factors TFIIA, TFIIB, TFIIE, TFIIF, and TFIIH (reviewed in ref. 26). Moreover, the recruitment of the TFIID complex that contains the TATA-binding protein and the TATA-binding protein-associated factors (TAFIIs) is necessary for activated transcription. Most Drosophila TAFIIs have been cloned, among them TAFII60 (27) and TAFII110 (ref. 28; reviewed in ref. 29). It has been proposed that specific TAFIIs act as molecular bridges between specific activators and the general transcription machinery (30). However, different promoters might vary considerably in their requirement for specific TAFIIs for transcriptional activation (31–36).
Synergistic activation of transcription might result from two activators simultaneously contacting the TFIID complex through different adapter molecules, thus stabilizing the binding of TFIID to the promoter and the association of the remaining basal transcription factors (reviewed in ref. 26). Sauer et al. (24, 25) showed that Bcd interacts with TAFII60 and TAFII110 through two distinct domains, whereas Hb interacts only with TAFII60. They showed that transcriptional activation of a bcd target, the hb promoter, is synergistically enhanced in vitro by Bcd and Hb. However, this effect is observed only when both TAFII60 and TAFII110 are present. They suggested that the synergy observed in vivo is because of the corecruitment of TAFII110 and TAFII60 by Bcd and Hb, respectively.
To analyze the in vivo function of these interactions, we generated transgenic fly lines bearing deletion variants of bcd rescue constructs. Here, we show that the protein domains of Bcd that exhibit in vitro interaction with TAFII60 and TAFII110 are not essential in vivo. We also show that a bcd construct that lacks both domains is able to rescue bcd mutants to viability. Finally, even in this rescue situation, which should represent a highly sensitized system for detecting a function of TAFIIs, development of the embryo is not affected by mutations in TAFII110 or TAFII60. This suggests that the interactions observed in vitro between Bcd and TAFII60 or TAFII110 are not necessary for the function of bcd and its synergy with Hb in vivo.
MATERIALS AND METHODS
bcd Deletion Constructs.
To generate deletion constructs in the bcd coding region, we subcloned the 870-bp BspEI–NsiI fragment from pKSgenomicBCD (37) into pSL1180 (Pharmacia), from which part of the polylinker had been removed by a HpaI and SnaBI digest and religation. The fragment was then taken as a NotI–KpnI fragment and cloned into pBluescriptKS (Stratagene), generating pKSbcdBsp-Nsi, which was used in the following cloning steps. pBcdΔQ is the result of a HincII and partial BstUI digest and religation, thereby deleting amino acids 247–304. pBcdΔA and pBcdΔQA resulted from cloning the 550-bp NciI (Klenow filled-in) to NsiI fragment into a KasI (Klenow filled-in)- and NsiI- or HincII- and NsiI-treated vector backbone, thereby deleting amino acids 331–344 or 247–344, respectively. pBcdΔAC and pBcdΔQAC were generated by a KasI (Klenow filled-in) or HincII plus partial ScaI digest and religation, thereby deleting all amino acids C-terminal to amino acid 330 or 246, respectively. pBcdΔC and pBcdΔQC resulted from opening the vector pKSbcdBsp-Nsi with a BspEI and partial ScaI digest and cloning into it the BspEI to NciI (Klenow filled-in) fragment from pKSbcdBsp-Nsi (320 bp) or pBcdΔQ (150 bp), thereby deleting all amino acids C-terminal to amino acid 344. The in-frame fusions were verified by sequencing. The BspEI–NsiI fragments from pKSbcdBsp-Nsi and the deletion constructs were isolated and cloned back into pKSgenomicBCD, thereby replacing the wild-type BspEI–NsiI fragment. The complete genomic coding sequences were taken from these vectors and cloned as BamHI fragments into the BglII site of pCaSpeRBcdBglII. These bcd rescue constructs were then used to generate different transgenic fly lines (37).
In Vivo Rescue.
Two BcdWT, two BcdΔA, four BcdΔQ, five BcdΔC, three BcdΔQA, three BcdΔQC, two BcdΔAC, and six BcdΔQAC transgenic lines on the first or second chromosome were crossed into the background to the amorphic bcdE1 allele, and rescue of the bcd phenotype was assessed by cuticular preparations (38). For each construct, the line with the strongest rescue ability was chosen for further analysis. The presence of the bcdE1 homozygous mutation in the mothers was assessed by the ri and roe markers that flank bcd. The nature of the transgene was verified by genomic PCR amplification by using oligonucleotides flanking the deleted regions in Bcd: 5′ oligonucleotide, GTCACATGCACATGCAGTAT; and 3′ oligonucleotide, ACTCCCAAATCTCATCGATC. The sizes of the obtained bands for BcdWT, BcdΔA, BcdΔQ, BcdΔC, BcdΔAC, BcdΔQC, BcdΔQA, and BcdΔQAC were 624, 578, 449, 353, 307, 178, 330, and 58 bp, respectively (data not shown).
Recombination of TAFII Mutants onto bcd Mutant Chromosome.
The TAFII60YY dominant negative allele (39) was recombined onto the chromosome carrying the bcdE1 allele. We obtained the TAFII110ΔC dominant negative allele recombined on the bcdE1 chromosome from Gary Struhl (Columbia University, New York). The presence of the dominant alleles in the stocks was checked by using genomic PCR amplification, which confirmed the molecular nature of the two alleles used (TAFII110ΔC and TAFII60YY; G. Struhl, personal communication).
In Situ Hybridization.
Digoxigenin-labeled RNA probes for in situ hybridization were prepared as described (37). In situ hybridization on whole-mount embryos was performed as described originally (40), with an adaptation (M. Klingler, personal communication). Prehybridization and hybridization were performed at 70°C at pH 5. Embryos were mounted in methyl salicilate/Canada balsam (1:2) and photographed by using Nomarski optics.
RESULTS
In Vivo Function of the Putative Bcd Activation Domains.
Bcd contains three putative activation domains: a glutamine-rich (Q) domain, an alanine-rich (A) domain, and a C-terminal acidic region (C) (Fig. 1). The domains of interaction of Bcd with the TAFIIs have been mapped in vitro: TAFII110 binds to the Q domain, whereas TAFII60 binds to the A domain (25). The C-terminal region has been shown to be dispensable for bcd function in vivo, but it contributes to increase activation in yeast (41). Interestingly, the entire activation domain of Bcd can be functionally replaced by a generic activation domain (random acidic amphipathic helix B6) from Escherichia coli (37, 41–43).
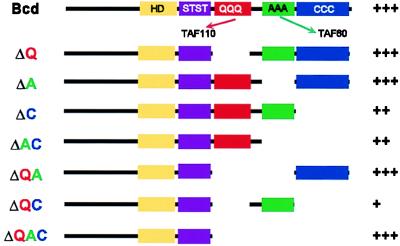
Figure 1.
Schematic representation of the Bcd deletion variants used in the bcd rescue experiment. For wild-type Bcd, the boxes represent identified domains in Bcd: HD, homeodomain; STST, S/T-rich region; QQQ, glutamine-rich domain that interacts with TAFII110 in vitro; AAA, alanine-rich domain that interacts with TAFII60 in vitro; CCC, C-terminal acidic region. Rescue of the bcdE1 phenotype by the different transgenes (assessed by cuticle preparations) is shown on the right. +++, rescue to viability with no cuticular defects; ++, thorax and most of the head structures are present; +, repression of the duplicated telson at the anterior end only.
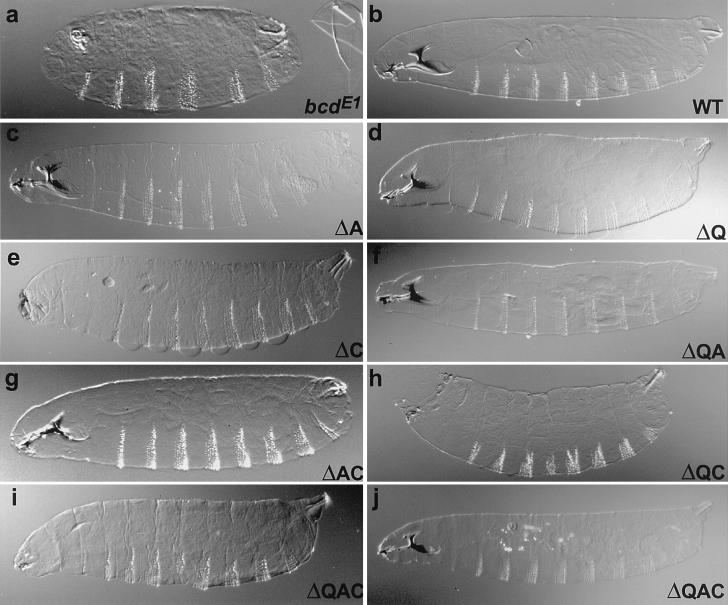
Because the corecruitment of TAFIIs could provide a powerful explanation to the synergistic effect observed between Bcd and Hb during early development, we generated bcd rescue constructs encoding proteins lacking one or several domains of interaction with the TAFIIs (Fig. 1). These constructs contain DNA encoding the wild-type or deletion variants of the Bcd protein in the context of the genomic bcd locus. We examined their ability to rescue the bcdE1 mutant phenotype. bcdE1 is a null allele that is truncated after the homeodomain and does not appear to produce a stable protein (12). Embryos from bcdE1 mothers lack all head and thorax segments and some abdominal segments. Instead, they have a second telson at the anterior end (Fig. 2a). A construct encoding a wild-type Bcd protein completely rescues the phenotype and behaves like the endogenous gene (Fig. 2b). Surprisingly, deletion of the A or Q (BcdΔA or BcdΔQ, respectively) domains or of both domains (BcdΔQA) does not abolish rescue activity (Fig. 2, c, d, and f). The BcdΔC transgene displays a hypomorphic bcd phenotype with only a very partial head but normal thorax and abdominal segments (Fig. 2e). The BcdΔAC construct shows an almost wild-type cuticular phenotype with only small head defects (Fig. 2g). The lines bearing the BcdΔQC deletion (Fig. 2h) exhibit very low rescue, with only the lack of a duplicated telson at the anterior. Although the lines lacking the C domain have a lower potential for rescue than the BcdΔQ, BcdΔA, and BcdΔQA lines, this is likely because of lower levels of expression rather than weaker activator proteins. Consistent with this interpretation, the domain of the hb staining is moved anteriorly in these lines (Fig. 3, d, f, and g). It has been argued that the position of the posterior border of hb indicates the amount of Bcd protein produced, whereas the intensity of staining within this domain reflects the activation potential of the Bcd deletion variant (41). In fact, two lines expressing a truncation construct lacking the C domain as well as both the Q and A domains (BcdΔQAC) have strong rescue activity: these lines exhibit rescue of the thorax and abdominal segments, loss of duplication of the telson at the anterior, and development of head structures (Fig. 2i). One of these two BcdΔQAC lines completely rescues the bcdE1 phenotype (Fig. 2j). Seventy-five percent of the embryos exhibit wild-type cuticles, and 30% of those are viable. This rescue is probably because of high-level expression of the transgene. Consistent with this, the posterior border of bcd target genes and the position of the cephalic furrow, both strong indicators of the slope of the Bcd protein gradient, are more posterior in this line than in wild-type embryos (Fig. 3h and data not shown). The ability of BcdΔQAC to activate transcription of target genes in vivo is consistent with Schneider cell cotransfection experiments in which the construct exhibits significant transcriptional activity (F.J., R. Sturny, F. Catala, and N. Dostatni, unpublished work). Because the phenotypic rescue of BcdΔQAC is not fully penetrant, the Q, A, and C domains do provide some contribution to the activity of Bcd but are not essential.
Figure 2.
Cuticle preparations of embryos from females homozygous for the strong bcdE1 allele. (a) Without rescuing construct. (b) Full rescue by two copies of a wild-type rescue construct. (c–j) Rescue by two copies of the Bcd deletion construct as indicated on each panel. Embryos from bcdE1 mothers lack head, thorax, and some abdominal segments. Instead, they have a second telson (anal plates, tuft, spiracles, and filzkörper) at the anterior end (a). A construct encoding a wild-type Bcd protein rescues completely the phenotype and behaves like the endogenous gene (b). Deletion of the A (c) or Q (d) domains or of both domains (f) does not abolish rescue activity. The BcdΔC (e) transgene displays only a very partial head but shows a normal thorax and abdominal segments. The BcdΔAC (g) construct shows an almost wild-type cuticular phenotype with only small head defects. The line bearing the BcdΔQC (h) deletion exhibits only the repression of the development of a telson at the anterior end. Two lines BcdΔQAC have strong rescue activity: these lines exhibit rescue of the thorax and abdominal segments, loss of duplication of the telson at the anterior, and development of head structures (i). One of these two BcdΔQAC lines completely rescues the bcdE1 phenotype (j). In all panels, anterior is to the left and dorsal is up.
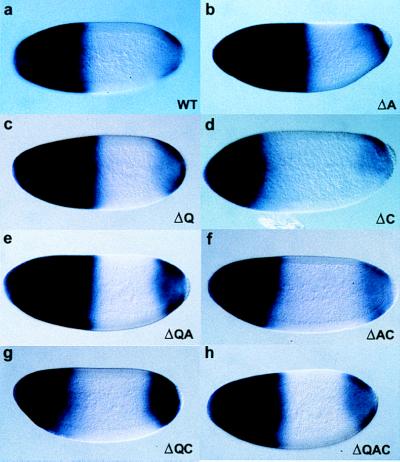
Figure 3.
hb RNA in situ hybridization of embryos from females homozygous for the strong bcdE1 allele and carrying: (a) two copies of a wild-type rescue construct; or (b–h) two copies of the Bcd deletion construct (representing the best rescuing lines), as indicated on each panel. In situ hybridization was performed with antisense probe for hb on whole-mount embryo preparations from the different transgenic lines containing Bcd deletion variants. The posterior boundary of the hb domain of expression is significantly moved anteriorly in embryos laid by females homozygous for the bcdE1 mutation and carrying two copies of the transgene: lines BcdΔC (d), BcdΔAC (f), and BcdΔQC (g). The BcdΔQ (c) and BcdΔQA (e) lines show normal amounts of hb expression within an enlarged domain. The BcdΔA line (b) exhibits a widely enlarged domain of hb transcription. The domain of expression mediated by the BcdΔQAC line that is able to rescue the bcdE1 phenotype to viability (h) appears slightly enlarged compared with wild-type Bcd.
Bcd-Dependent hb Activation.
In a wild-type embryo (Fig. 3a), hb is transcribed in a broad anterior domain as well as in a more restricted posterior domain. Transcription of the anterior domain of hb is under the control of Bcd and, therefore, is lost in embryos from bcdE1 mutant mothers (13). Whenever Bcd activity is altered, hb expression is also changed (see above). The posterior border of the anterior hb expression domain is, like the cephalic furrow, positioned in response to the amount of Bcd, whereas the level of expression within this domain reflects the activation potential of the respective Bcd variant (41). In embryos laid by females homozygous for the bcdE1 mutation and carrying two copies of the transgene BcdΔC, BcdΔAC, or BcdΔQC, the posterior boundary of hb expression is significantly moved toward the anterior (Fig. 3, d, f, and g). This is consistent with the fact that these lines only show partial rescue of the bcdE1 phenotype, which appears to be because of a lower level of expression of the transgenes. However, within the hb domain, hb is transcribed at normal levels, which indicates normal activity of these Bcd deletions. The BcdΔQ (Fig. 3c) and BcdΔQA (Fig. 3e) lines that fully rescue the bcdE1 phenotype show normal amounts of hb expression within a slightly enlarged domain. One of the BcdΔA lines (Fig. 3b) exhibits a widely enlarged domain of hb transcription, likely because of the very high level of transgene expression observed by immunostaining (data not shown). Another BcdΔA line shows a normal rescue pattern and exhibits a normal position of the cephalic furrow (data not shown). Finally, the domain of expression mediated by the BcdΔQAC (Fig. 3h) line that is able to rescue the bcdE1 phenotype to viability appears slightly enlarged compared to wild-type Bcd, indicating a high level of transgene expression. Thus, Bcd is able to activate its target gene hb, even in the absence of the protein domains that have been shown to interact in vitro with TAFII60 and TAFII110.
Requirement of TAFII Function for Bcd Activity.
TAFII110 and TAFII60 have been shown to mediate transcriptional activation in vitro (24), and mutations in the respective genes have been isolated in a dominant modifier screen in Drosophila (39). Mutations in TAFII60 and TAFII110 are homozygous embryonic lethal and cell lethal during eye development and oogenesis. However, viable dominant negative alleles of these two genes have allowed Sauer et al. (39) to investigate their effects on transcription. These mutations do not appear to cause a general reduction of transcription, but instead, they affect the expression of only a subset of genes (33, 39). The TAFII60YY allele encodes a protein that contains two tyrosines residues inserted at amino acid 207. The TAFII110ΔC allele lacks the C-terminal 126 amino acids of the protein. Although TAFII60 and TAFII110 have been shown to bind to both Bcd and TAFII250, the mutant proteins are defective for their interaction with TAFII250 but still retain their in vitro interaction with Bcd. Because these mutant proteins can still interact with the transcription activators but cannot mediate interaction with the transcription machinery, they can be considered dominant negative alleles.
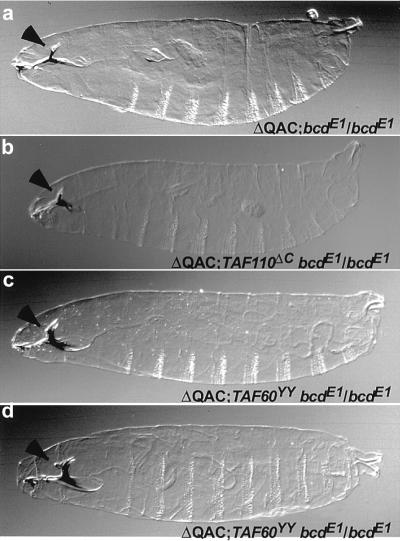
Although the BcdΔQAC truncation construct lacks the identified TAFII60 and TAFII110 interaction domains (25), it is still able to strongly activate the bcd target genes. It is possible that the BcdΔQAC truncation does not affect Bcd function because it can still interact with Hb, its partner involved in synergistic activation. Hb might still recruit TAFII60 to the promoter, even if Bcd cannot do it directly. If this is the case, the genetic rescue of bcdE1 by BcdΔQAC should be highly dependent on the interaction between Hb and TAFII60. To test this possibility, we examined the ability of BcdΔQAC to rescue a bcd mutant phenotype in the presence of either the dominant negative TAFII60YY, which is expected to interfere with activation by BcdΔQAC, or TAFII110ΔC, which should not interfere with the rescue. Typically, a single copy of the BcdΔQAC transgene is able to almost completely rescue the progeny of females homozygous mutant for bcdE1(Fig. 4a). Neither of the TAFII60YY nor TAFII110ΔC dominant negative mutations affect the rescue ability of the BcdΔQAC construct: the bcdE1 phenotype in the progeny of double mutant females TAFII60YY, bcdE1/bcdE1 or TAFII110ΔC, bcdE1/bcdE1 is rescued by the BcdΔQAC as well as that of the single bcdE1/bcdE1 mutant (Fig. 4, b and c). Rare rescue to wild type can be observed at the same frequency, independently of the presence (Fig. 4d) or absence of dominant negative TAFII mutations (data not shown).
Figure 4.
Cuticle preparations of embryos from females homozygous for the strong bcdE1 allele and rescued by one copy of the BcdΔQAC construct (best rescuing line). Arrowhead, dorsal bridge. (a) Wild type for TAFIIs. (b) In the presence of dominant negative TAFII110ΔC. (c and d) In the presence of dominant negative TAFII60YY. (a) A single copy of the BcdΔQAC transgene is able to almost completely rescue the amorphic mutant bcdE1, with only the dorsal bridge reduced. In b and c, the same rescue is obtained as in a, whereas in d, a rare rescue to wild type is shown.
Because the mutations identified in the genes encoding TAFII110 or TAFII60 have no direct or specific effect on Bcd activity, even in a highly sensitized situation, this suggests that the synergy between Bcd and Hb is not based on interactions with TAFIIs and must, therefore, be mediated by another, still elusive mechanism.
DISCUSSION
Are the TAFIIs Dispensable for in Vivo Transcriptional Activation by Bcd?
The mechanisms of transcriptional activation in eukaryotes have been studied extensively, and biochemical studies have established that in vitro mRNA synthesis by RNA polymerase II requires the regulated assembly of multiple protein complexes at promoters (45). Recent work points to the basal transcription factor complex TFIID as a key component in the response to transcription regulators (30, 46, 47). TFIID consists of the TATA binding protein and ∼10 other TATA binding protein-associated factors (TAFIIs). In the yeast Saccharomyces cerevisiae, the TFIID complex shares functional and structural similarities with higher eukaryotic TFIID. In particular, most yeast TAFIIs are homologous to higher eukaryotic TAFIIs (29, 48–50). Earlier reports have indicated that several TAFIIs are dispensable in yeast for in vivo transcriptional activation, despite being essential for growth (51, 52). More recently, it has been shown that a mutation in TAFII250 causes gene specific transcription defects (53). TAFII250 is in direct contact with TATA binding protein and is thought to function as an important scaffold for the TFIID complex by interacting with other TAFIIs such as TAFII110 and TAFII60 (54). However, there is extensive controversy as to the role TAFIIs play in vivo (31–36).
Because TAFIIs are supplied maternally and because homozygous TAFII mutants are cell lethal, it is, so far, impossible to create an early embryo that completely lacks TAFII function. Therefore, to test the in vivo relevance of the interaction between the Bcd activation domains and TAFII60 and TAFII110, we used alleles of TAFIIs that are considered dominant-negative because they can still interact with Bcd but not with TAFII250 (39). These mutations have been identified as suppressors of the rough eye phenotype induced by misexpression of an activated form of Ras1. This suppressor effect is likely because of the reduction of the sevenless promoter-mediated Ras1 expression by the TAFIIs mutations (39). Embryos derived from heterozygous mothers are loaded with both maternal wild type, and with the dominant negative mutant TAFIIs, we could show that the domains of interaction with the TAFIIs are dispensable for Bcd function in vivo, which suggests that Bcd does not use a specific interaction with TAFII60 or TAFII110 to activate its targets. Alternatively, it is possible that BcdΔQAC still contains cryptic TAFII interaction domains that have not been identified in in vitro experiments. However, if this was the case, BcdΔQAC should only weakly interact with TAFIIs and should, therefore, critically depend on these TAFIIs to function. However, even in the highly sensitized situation of a rescue by BcdΔQAC, a dose of neither wild-type TAFII60 nor wild-type TAFII110 is required. This shows that the interaction domains identified in vitro are not absolutely necessary for Bcd function in vivo, and that the requirement for TAFIIs observed in vitro is not observed in vivo. The interaction with TAFIIs might have accessory character only and contribute to transcriptional activation in a quantitative way, without being crucially important for its biological function.
Comparison Between the Bcd Truncation Constructs and bcd Alleles.
The extensive truncation of the BcdΔQAC protein shows that the putative activation domains are dispensable for proper development. Several bcd alleles have been characterized molecularly and have been placed into categories according to the extent of anterior pattern defects that they produce. Embryos from strong mutant alleles (bcdE1, bcdE2, and bcd33-5) show a complete lack head and thorax (9, 12), deletions, and fusions of anterior abdominal segments as well as a duplicated telson at the anterior. Generally, these mutations truncate the Bcd protein right after the homeodomain, they are more extensive than BcdΔQAC, and they might not produce a stable protein. A less extensive C-terminal deletion of Bcd is encoded by the bcdE5 allele, which belongs to the weakest class of alleles (with bcd111 and bcd2-13) (7). bcdE5 truncates the protein at amino acid 264 (12), which corresponds approximately to the BcdΔQAC deletion construct (truncation at amino acid 246). These weak alleles show only slight pattern defects in the anterior head region, lacking the labral derivatives (labrum and dorsal bridge), comparable to the defects produced by the weaker of the BcdΔQAC lines. It is likely that a slight increase of the amount of the bcdE5 protein would produce a near normal embryo.
Is the Tight Localization of the bcd mRNA Involved in the BcdΔQAC Rescue?
The ability of BcdΔQAC to fully rescue the bcdE1 phenotype is surprising with respect to previous reports by using injection of mRNA encoding truncation constructs of bcd (41). mRNA injections of a truncated construct similar to BcdΔQAC showed a low rescue potential, even at high concentrations: the construct only suppressed the formation of posterior structures at the anterior. It induced with high frequency thoracic structures and, more rarely, structures of the gnathal region of the head (38). The difference between the results obtained by mRNA injection or by using transgenic flies could be because of the lack of anterior localization of the injected mRNA. To carry out its morphogenetic function, a gradient of Bcd activity is established by the tight localization of the bcd mRNA to the anterior pole of the egg. This localization depends on the maternal genes exuperentia (exu), swallow (swa), and staufen (stau). Embryos from females homozygous mutant for exu, swa, or stau (2, 55) show head defects similar to those of embryos from females mutant for weak bcd alleles. The labrum is absent and the pharyngeal head skeleton is reduced. Strikingly, the swa, exu, and stau mutants, the weak bcd alleles, and the embryos rescued by injection of the bcd(1–264) mRNA, are all reminiscent of the torso phenotype: lack of dorsal bridge and labrum. This suggests that that Tor has a enhancing effect on Bcd activity, as has been reported for bcd target gene expression (56–58). The tight localization of the bcd mRNA might, thus, be required for Tor to enhance the function of the Bcd protein before it migrates.
Tor-Dependent Phosphorylation of Bcd.
It has been shown that Bcd is phosphorylated in response to Tor activity (44). The strong remaining activity in the BcdΔQAC protein that presents such an extensive deletion might be because of the presence of the S/T rich region located between the homeodomain and the Q, A, and C activation domains (Fig. 1). The S/T-rich sequence is the target of phosphorylations induced by the activity of the terminal Tor pathway: in embryos lacking Tor activity, Bcd phosphorylation is greatly diminished (44). Activation of the Tor signal transduction pathway leads to activation of a mitogen-activated protein kinase, a nuclear kinase that has been implicated in the phosphorylation of transcription factors. Most mitogen-activated protein phosphorylation sites of Bcd have been mapped in vitro to the S/T rich region. Site-directed mutagenesis indicates that most of the Bcd phosphorylations occur on these sites in vivo (F.J., R. Sturny, F. Catala, and N. Dostatni, unpublished work). The ability of the Tor pathway to create negative charges in this region might allow the generation of an acidic activation domain that compensates for the loss of the other Q, A, and C activation domains.
It has been shown that the Bcd phosphorylations mediate part of the Tor enhancing effect on Bcd activity (F.J., R. Sturny, F. Catala, and N. Dostatni, unpublished work). Hence, the rescue by BcdΔQAC could be explained by the fact that the S/T rich region is phosphorylated in vivo by the Tor cascade, thus creating an acidic activation domain on Bcd before it forms its concentration gradient. As long as the homeodomain and the S/T rich region are intact, good rescue is achieved, especially if the transgene is expressed at high levels and mRNA translation occurs in the region of Tor activity.
Acknowledgments
We are very grateful to Terry Turner, who was extremely helpful with technical assistance during injection and fly stock maintenance. We acknowledge the Desplan laboratory for its constant support during this work as well as Franck Sauer and David Wassarman for generously providing the TAFII mutants. Gary Struhl generated the TAFII110ΔC, bcdE1 chromosome and, through constant discussions, had much input into this project. We also thank Nathalie Dostatni for important comments on the experiments and Matthieu Schaeffer for his help as a summer student. V.S. thanks Christine Dambly-Chaudière for her constant support and encouragement. V.S. and F.J. were supported by fellowships from the French “Ministère de la Recherche,” and E.A.W. was supported by fellowships from European Molecular Biology Organization and Human Frontier Science Program Organization.
ABBREVIATION
- TAFII
TATA-binding protein-associated factor
References
- 1.Nusslein-Volhard C, Frohnhofer H G, Lehmann R. Science. 1987;238:1675–1681. doi: 10.1126/science.3686007. [DOI] [PubMed] [Google Scholar]
- 2.Schüpbach T, Wieschaus E. Roux’s Arch Dev Biol. 1986;195:302–317. doi: 10.1007/BF00376063. [DOI] [PubMed] [Google Scholar]
- 3.Stevens L M, Frohnhofer H G, Klingler M, Nusslein-Volhard C. Nature (London) 1990;346:660–663. doi: 10.1038/346660a0. [DOI] [PubMed] [Google Scholar]
- 4.Savant-Bhonsale S, Montell D J. Genes Dev. 1993;7:2548–2555. doi: 10.1101/gad.7.12b.2548. [DOI] [PubMed] [Google Scholar]
- 5.Martin J R, Raibaud A, Ollo R. Nature (London) 1994;367:741–745. doi: 10.1038/367741a0. [DOI] [PubMed] [Google Scholar]
- 6.Casanova J, Furriols M, McCormick C A, Struhl G. Genes Dev. 1995;9:2539–2544. doi: 10.1101/gad.9.20.2539. [DOI] [PubMed] [Google Scholar]
- 7.Frohnhofer H G, Nusslein-Volhard C. Nature (London) 1986;324:120–125. [Google Scholar]
- 8.Berleth T, Burri M, Thoma G, Bopp D, Richstein S, Frigerio G, Noll M, Nusslein-Volhard C. EMBO J. 1988;7:1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Driever W, Nusslein-Volhard C. Cell. 1988;54:83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- 10.Gao Q, Finkelstein R. Development (Cambridge, UK) 1998;125:4185–4193. doi: 10.1242/dev.125.21.4185. [DOI] [PubMed] [Google Scholar]
- 11.Driever W, Nusslein-Volhard C. Nature (London) 1989;337:138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- 12.Struhl G, Struhl K, Macdonald P M. Cell. 1989;57:1259–1273. doi: 10.1016/0092-8674(89)90062-7. [DOI] [PubMed] [Google Scholar]
- 13.Tautz D. Nature (London) 1988;332:281–284. doi: 10.1038/332281a0. [DOI] [PubMed] [Google Scholar]
- 14.Rivera-Pomar R, Lu X, Perrimon N, Taubert H, Jackle H. Nature (London) 1995;376:253–256. doi: 10.1038/376253a0. [DOI] [PubMed] [Google Scholar]
- 15.Dubnau J, Struhl G. Nature (London) 1996;379:694–699. doi: 10.1038/379694a0. [DOI] [PubMed] [Google Scholar]
- 16.Rivera-Pomar R, Niessing D, Schmidt-Ott U, Gehring W J, Jackle H. Nature (London) 1996;379:746–749. doi: 10.1038/379746a0. [DOI] [PubMed] [Google Scholar]
- 17.Chan S K, Struhl G. Nature (London) 1997;388:634. doi: 10.1038/41692. [DOI] [PubMed] [Google Scholar]
- 18.Hulskamp M, Pfeifle C, Tautz D. Nature (London) 1990;346:577–580. doi: 10.1038/346577a0. [DOI] [PubMed] [Google Scholar]
- 19.Struhl G, Johnston P, Lawrence P A. Cell. 1992;69:237–249. doi: 10.1016/0092-8674(92)90405-2. [DOI] [PubMed] [Google Scholar]
- 20.Schulz C, Tautz D. Development (Cambridge, UK) 1994;120:3043–3049. doi: 10.1242/dev.120.10.3043. [DOI] [PubMed] [Google Scholar]
- 21.Gavis E R, Lehmann R. Cell. 1992;71:301–313. doi: 10.1016/0092-8674(92)90358-j. [DOI] [PubMed] [Google Scholar]
- 22.Small S, Kraut R, Hoey T, Warrior R, Levine M. Genes Dev. 1991;5:827–839. doi: 10.1101/gad.5.5.827. [DOI] [PubMed] [Google Scholar]
- 23.Simpson-Brose M, Treisman J, Desplan C. Cell. 1994;78:855–865. doi: 10.1016/s0092-8674(94)90622-x. [DOI] [PubMed] [Google Scholar]
- 24.Sauer F, Hansen S K, Tjian R. Science. 1995;270:1783–1788. doi: 10.1126/science.270.5243.1783. [DOI] [PubMed] [Google Scholar]
- 25.Sauer F, Hansen S K, Tjian R. Science. 1995;270:1825–1828. doi: 10.1126/science.270.5243.1825. [DOI] [PubMed] [Google Scholar]
- 26.Buratowski S. Science. 1995;270:1773–1774. doi: 10.1126/science.270.5243.1773. [DOI] [PubMed] [Google Scholar]
- 27.Weinzierl R O, Ruppert S, Dynlacht B D, Tanese N, Tjian R. EMBO J. 1993;12:5303–5309. doi: 10.1002/j.1460-2075.1993.tb06226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoey T, Weinzierl R O, Gill G, Chen J L, Dynlacht B D, Tjian R. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 29.Tansey W P, Herr W. Cell. 1997;88:729–732. doi: 10.1016/s0092-8674(00)81916-9. [DOI] [PubMed] [Google Scholar]
- 30.Verrijzer C P, Tjian R. Trends Biochem Sci. 1996;9:338–342. [PubMed] [Google Scholar]
- 31.Hahn S. Cell. 1998;95:579–582. doi: 10.1016/s0092-8674(00)81625-6. [DOI] [PubMed] [Google Scholar]
- 32.Grant P A, Workman J L. Nature (London) 1998;396:410–411. doi: 10.1038/24723. [DOI] [PubMed] [Google Scholar]
- 33.Zhou J, Zwicker J, Szymanski P, Levine M, Tjian R. Proc Natl Acad Sci USA. 1998;95:13483–13488. doi: 10.1073/pnas.95.23.13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Apone L M, Virbasius C A, Holstege F C, Wang J, Young R A, Green M R. Mol Cell. 1998;2:653–661. doi: 10.1016/s1097-2765(00)80163-x. [DOI] [PubMed] [Google Scholar]
- 35.Michel B, Komarnitsky P, Buratowski S. Mol Cell. 1998;2:663–673. doi: 10.1016/s1097-2765(00)80164-1. [DOI] [PubMed] [Google Scholar]
- 36.Moqtaderi Z, Keaveney M, Struhl K. Mol Cell. 1998;2:675–682. doi: 10.1016/s1097-2765(00)80165-3. [DOI] [PubMed] [Google Scholar]
- 37.Bellaiche Y, Bandyopadhyay R, Desplan C, Dostatni N. Development (Cambridge, UK) 1996;122:3499–3508. doi: 10.1242/dev.122.11.3499. [DOI] [PubMed] [Google Scholar]
- 38.Nusslein-Volhard C, Wieschaus E, Kluding H. Roux’s Arch Dev Biol. 1984;193:267–282. doi: 10.1007/BF00848156. [DOI] [PubMed] [Google Scholar]
- 39.Sauer F, Wassarman D A, Rubin G M, Tjian R. Cell. 1996;87:1271–1284. doi: 10.1016/s0092-8674(00)81822-x. [DOI] [PubMed] [Google Scholar]
- 40.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 41.Driever W, Ma J, Nusslein-Volhard C, Ptashne M. Nature (London) 1989;342:149–154. doi: 10.1038/342149a0. [DOI] [PubMed] [Google Scholar]
- 42.Ma J, Ptashne M. Cell. 1987;51:113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- 43.Driever W. In: The Development of Drosophila melanogaster. Bate M, Martinez-Arias A, editors. I. 1993. pp. 301–324. [Google Scholar]
- 44.Ronchi E, Treisman J, Dostatni N, Struhl G, Desplan C. Cell. 1993;74:347–355. doi: 10.1016/0092-8674(93)90425-p. [DOI] [PubMed] [Google Scholar]
- 45.Roeder R G. Trends Biochem Sci. 1996;9:327–335. [PubMed] [Google Scholar]
- 46.Goodrich J A, Tjian R. Curr Opin Cell Biol. 1994;6:403–409. doi: 10.1016/0955-0674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 47.Burley S K, Roeder R G. Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 48.Reese J C, Apone L, Walker S S, Griffin L A, Green M R. Nature (London) 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- 49.Poon D, Bai Y, Campbell A M, Bjorklund S, Kim Y J, Zhou S, Kornberg R D, Weil P A. Proc Natl Acad Sci USA. 1995;92:8224–8228. doi: 10.1073/pnas.92.18.8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moqtaderi Z, Yale J D, Struhl K, Buratowski S. Proc Natl Acad Sci USA. 1996;93:14654–14658. doi: 10.1073/pnas.93.25.14654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. Nature (London) 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 52.Walker S S, Reese J C, Apone L M, Green M R. Nature (London) 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 53.Wang E H, Zou S, Tjian R. Genes Dev. 1997;11:2658–2669. doi: 10.1101/gad.11.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen J L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 55.Stephenson E C, Chao Y-C, Fackenthal J D. Genes Dev. 1988;2:1655–1665. doi: 10.1101/gad.2.12a.1655. [DOI] [PubMed] [Google Scholar]
- 56.Grossniklaus U, Cadigan K M, Gehring W J. Development (Cambridge, UK) 1994;120:3155–3171. doi: 10.1242/dev.120.11.3155. [DOI] [PubMed] [Google Scholar]
- 57.Wimmer E A, Simpson-Brose M, Cohen S M, Desplan C, Jackle H. Mech Dev. 1995;53:235–245. doi: 10.1016/0925-4773(95)00439-8. [DOI] [PubMed] [Google Scholar]
- 58.Gao Q, Wang Y, Finkelstein R. Mech Dev. 1996;56:3–15. doi: 10.1016/0925-4773(96)00504-7. [DOI] [PubMed] [Google Scholar]