Abstract
Rho GTPases (Rac, Rho, and Cdc42) play important roles in regulating cell function through their ability to coordinate the actin cytoskeleton, modulate the formation of signaling reactive oxidant species, and control gene transcription. Activation of Rho GTPase signaling pathways requires the regulated release of Rho GTPases from RhoGDI complexes, followed by their reuptake after membrane cycling. We show here that Src kinase binds and phosphorylates RhoGDI both in vitro and in vivo at Tyr156. Analysis of Rho GTPase–RhoGDI complexes using in vitro assays of complexation and in vivo by coimmunoprecipitation analysis indicates that Src-mediated phosphorylation of Tyr156 causes a dramatic decrease in the ability of RhoGDI to form a complex with RhoA, Rac1, or Cdc42. Phosphomimetic mutation of Tyr156→Glu results in the constitutive association of RhoGDIY156E with the plasma membrane and/or associated cortical actin. Substantial cortical localization of tyrosine-phosphorylated RhoGDI is also observed in fibroblasts expressing active Src, where it is most evident in podosomes and regions of membrane ruffling. Expression of membrane-localized RhoGDIY156E mutant is associated with enhanced cell spreading and membrane ruffling. These results suggest that Src-mediated RhoGDI phosphorylation is a novel physiological mechanism for regulating Rho GTPase cytosol membrane–cycling and activity.
INTRODUCTION
The Rho GTPases regulate cellular activities that include growth and differentiation, vesicular transport, production of reactive oxygen species (ROS), apoptosis, cell motility, and various other aspects of cytoskeletal dynamics and cell polarity (Van Aelst and Souza-Schorey, 1997; Bishop and Hall, 2000). Rho GTPases function as molecular switches in cell signaling, alternating between inactive GDP-bound states maintained as cytosolic complexes with GDP dissociation inhibitors (GDIs), and active GTP-bound states usually associated with membranes where effector targets reside. Complexation of Rho, Rac, or Cdc42 with GDIs inhibits GDP dissociation and localizes GTPases to the cytosol in inert forms unable to interact with GEFs (guanine nucleotide exchange factors), GAPs (GTPase activating proteins), or effector targets (Van Aelst and Souza-Schorey, 1997; DerMardirossian and Bokoch, 2005). GDIs maintain Rho GTPases as soluble cytosolic proteins by forming high-affinity complexes that mask the C-terminal geranylgeranyl membrane-targeting moiety within a hydrophobic pocket formed by the immunoglobulin-like domain of RhoGDI (Gosser et al., 1997; Keep et al., 1997; Hoffman et al., 2000). When Rho proteins are released from GDIs, they insert into the membrane lipid bilayer through their isoprenylated and polybasic C-terminal domains to be activated by membrane-associated GEFs, initiating the association with effector targets at the membrane. A reassociation with GDI, possibly associated with GTP hydrolysis, is postulated to induce recycling of the GTPase to the cytosol. Regulation of the cytosol-membrane cycling of the Rho GTPase by GDI thus serves a major role in controlling Rho GTPase activity and function. The signaling mechanisms and protein components that modulate this GTPase–RhoGDI regulatory cycle remain poorly defined.
RhoGDI appears to be a particularly critical regulator of Rho GTPase function in the kidney: In mice in which RhoGDI has been knocked out by homologous recombination, a major phenotype is death by renal failure (Togawa et al., 1999). The mice were characterized by age-related histological abnormalities of the kidney, including cystic dilation of the proximal and distal renal tubules, flattening and detachment of epithelial cells from the tubular basement membrane, glomerular sclerosis, disrupted glomerular podosomes, and increased infiltration of inflammatory cells. Renal function was progressively impaired, including decreases in creatinine clearance, polyuria and proteinuria, and renal failure leading to death. The knockout mouse data thus clearly indicate that RhoGDI acts as an important regulatory point for Rho GTPase activity in the kidney.
Signaling by Src family kinases also plays critical roles in renal function. Tyrosine phosphorylation of the podocyte transmembrane adhesion protein, nephrin, regulates downstream actin assembly necessary for normal functioning of the glomerular filtration apparatus (Benzing, 2004). Src is a well-known oncogene and plays important roles in regulating tumor cell invasion and metastasis (Martin, 2003). Src localizes to membrane ruffles and to podosomes in certain cancer cells. The latter are dynamic actin structures involved in extracellular matrix degradation during invasion (Abram et al., 2003; Martin, 2003). The action of Src is dependent on downstream activation of Rho GTPases, particularly Rac1 and RhoA (Urich et al., 1997; Martin, 2003; Berdeaux et al., 2004). Indeed, growth and metastasis of many forms of cancer are dependent on Rho GTPases, which are in equilibrium between active (GTP) and inactive (GDP) states (e.g., Jaffe and Hall, 2002). Perturbation of this equilibrium by dysregulation of GDIs, GEFs, or GAPs can therefore result in elevated cancer cell Rho GTPase activity. Both RhoGDI and D4GDI protein levels have been shown to be decreased in certain cancers, correlating with increased metastatic capability (Gildea et al., 2002; Jiang et al., 2003). Interestingly, proteomic analyses have identified Rho GDI (Unwin et al., 2005) and D4GDI (Rush et al., 2005) as tyrosine-phosphorylated proteins in cancer cells.
The regulation of the interaction of Rho GTPases with RhoGDI remains largely undefined, yet accumulating evidence suggests it is a complex process. Anionic lipids generated during cell signaling can modulate GTPase binding to GDIs (Chuang et al., 1993a; Ugolev et al., 2006). Several proteins have been reported to selectively induce dissociation of Rho GTPases from RhoGDI complexes, leading to activation (e.g., Yamashita and Tohyama, 2003). Additionally, a number of signaling cascades leading to the phosphorylation of either the Rho GTPase or to Rho GDI itself (DerMardirossian and Bokoch, 2005) have been shown to modulate RhoGDI–Rho GTPase complexation directly. Recently, we established that the phosphorylation of RhoGDI on two sites concurrently by p21-activated kinase 1 (Pak1) leads to a selective release of Rac from RhoGDI complexes and that this activity was necessary for Rac activation by growth factors (DerMardirossian et al., 2004).
Here we provide evidence that Src serves as a RhoGDI kinase in vitro and in vivo and demonstrate a specific effect of phosphorylation on Rho GTPase–RhoGDI association. Further, we show that this phosphorylation plays a unique role in modulating the persistent localization of RhoGDI to the plasma membrane and that tyrosine phosphorylation of RhoGDI by Src results in enhanced Rho GTPase cytoskeletal activity. Phosphorylation of RhoGDI by Src appears to represent a novel mechanism for the regulation of Rho GTPase–RhoGDI membrane cycling and signaling.
MATERIALS AND METHODS
Reagents
Cell culture medium, fetal bovine serum, and supplements were from Invitrogen (Carlsbad, CA). [γ-32P]ATP (specific activity 4500 mCi/mmol) was from ICN (Costa Mesa, CA). Plasmids for transfection were purified using the Qiagen Qiafilter system (Chatsworth, CA). The SrcY527F-transformed NIH-3T3 (Src3T3) cell line (Lock et al., 1998) was gift from Sara Courtneidge (The Burnham Institute, La Jolla, CA). The following reagents were purchased as indicated: ECL reagents from Pierce (Rockford, IL), protein G- or protein A-Sepharose beads from Amersham (Piscataway, NJ) and Repligen (Waltham, MA), respectively; recombinant human EGF from Fisher Scientific (Tustin, CA); recombinant Src (pp60c-src) Catalogue no. 14–117 was from Upstate Biotechnology (Charlottesville, VA); SU6656 Src inhibitor was from Calbiochem (572635; EMD Biosciences, Darmstadt, Germany); mouse glutathione S-transferase (GST) antibody, rabbit polyclonal RhoGDI, RhoA, Cdc42, c-Src, His, and monoclonal RhoA antibodies from Santa Cruz Biotechnology (Santa Cruz, CA; catalogue nos. Sc-138, Sc-320; Sc-179; Sc-87, Sc-18, Sc-803, and Sc-418, respectively); monoclonal anti-His from BabCO (Richmond, CA); monoclonal EGFP 3E6 antibody and Alexa568- or Alexa488-fluorochrome–conjugated secondary antibody and Alexa phalloidin were purchased from Molecular Probes (Eugene, OR); monoclonal Rac1 antibody 23A8 (05–389) and anti-phosphotyrosine antibody 4G10 (05–321) were from Upstate Biotechnology. 9E10 anti-myc antibody was prepared in house. Expression and purification of RhoGDI from Escherichia coli was as in Chuang et al. (1993b).
Plasmid DNA and Construction
All RhoGDI constructs (human RhoGDI Y156A, Y156E, and Y156F) were inserted into pCMV6 with a C-terminal His-epitope and mutants were prepared by site-directed mutagenesis using QuickChange kit (Stratagene, La Jolla, CA). Human RhoGDI wild type was cloned into pGEX4T3 vector at the BamHI/EcoRI site. Src plasmids (pcDNA3-c-Src wild-type, Y527F, and K295M) were generously provided by David Schlaepfer (TSRI).
Kinase Assays
In vitro kinase activity was determined as described in Knaus et al. (1995) using pure recombinant RhoGDI, immobilized on glutathione beads or not, as a substrate at 2 μg per reaction, and recombinant Src was used as the kinase at 1 μg per reaction. Reactions (30 min at 30°C) were started by the addition of ATP (final concentration: 20 μM of cold ATP and 0.5 μCi [γ-32P]ATP per reaction).
Cell Transfection and Western Blot
HeLa cells were maintained in DMEM (Sigma, St. Louis, MO) supplemented to 8% fetal bovine serum at 37°C and 5% CO2. HeLa cells were cultured on six-well plates and transfected with Src and/or His-tagged RhoGDI plasmids or with appropriate vector cDNA using Lipofectamine transfection reagent (Invitrogen).
For Western blot, whole cell extracts were prepared by washing cells grown on plates once with PBS before disruption in lysis buffer (25 mM Tris, pH 7.4, 5 mM MgCl2, 150 mM NaCl, 1 mM DTT, 1% NP40) supplemented with 1 mM leupeptin, 1 mM aprotinin, 1 mM sodium orthovanadate, and 1 mM PMSF. Lysates were clarified by centrifugation at 13,000 rpm, and the supernatant (whole cell lysate) was used for SDS-PAGE separation or immunoprecipitation. The immunoprecipitation assay was carried out as follows. Whole cell lysates containing the same amount of total protein were incubated with the appropriate antibody for overnight at 4°C. Twenty-five microliters of protein G-Sepharose beads was then added to the sample for another 1 h at 4°C. Beads were collected by centrifugation and washed four times with the lysis buffer. Proteins were eluted by boiling in 4× SDS sample buffer and subjected to SDS-PAGE and then Western blotting with the appropriate antibodies, as indicated in the figure legends.
Membrane and Cytosol Fractionation
Adherent cells grown in 10-cm dishes were treated with 1 mM vanadate for 1 h, washed once with cold 1× PBS, and extracted in ice-cold lysis buffer (100 mM Pipes, pH 7.3, 100 mM KCl, 3.5 mM MgCl2, 3 mM NaCl, 1 mM ATP supplemented with 1 mM PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin, and 1 mM vanadate) for 5 min. Cells were collected by scraping, and cell lysates were sonicated four times for 30 s. Homogenates were centrifuged at 10,000 rpm for 10 min to pellet nuclei and intact cells, and supernatants were then spun at 45,000 rpm at 4°C for 60 min in a refrigerated ultracentrifuge (TL100) to sediment plasma membranes. The cytosol-containing supernatant was removed, and the crude membrane pellet gently washed with lysis buffer. Membrane fractions were resuspended in lysis buffer, sonicated three times for 30 s, and then spun down at 45,000 rpm at 4°C for 60 min. Membrane and cytosol fractions were then assayed for total protein, and equal amounts were analyzed by Western blotting. The membrane fractions were verified to be free of contaminating cytosol by immunoblotting for the cytosolic marker Erk1/2.
For protein immunoprecipitation experiments, membrane fractions were solubilized with lysis buffer containing 1% NP40 for 1 h at 4°C and then centrifuged at 45,000 rpm at 4°C for 45 min. Individual proteins were immunoprecipitated using appropriate antibodies and analyzed by Western blotting, as indicated in the figure legends.
Membranes expressing GST-Rac1 were prepared from baculovirus-infected Sf9 cells as described by (Chuang et al., 1993b). Twenty micrograms of membrane protein containing 1.1 μg of GST-Rac1-GDP (determined by 35S-GTPγS binding) was incubated for 30 min at 4°C with either 1 μg of recombinant RhoGDIwt, RhoGDIY156E, RhoGDIY156F or without protein, as indicated in the figure. Membrane pellets and supernatants were collected by ultracentrifugation, and, in order to compare the capability of each RhoGDI protein to extract Rac1 from the membranes, each supernatant was analyzed by Western blot with GST antibody (1/1000 dilution).
Microscopy
For microscopic observations, HeLa cells were cultured in six-well plates on 35-mm-diameter coverslips overnight. To determine the localization of RhoGDI mutants, cells were transfected with 0.2 μg of His-tagged RhoGDI plasmids, as indicated, using Lipofectamine reagent. Cells were fixed in 4% paraformaldehyde 6 h after transfection and then permeabilized with 0.5% Triton X-100 in PBS for 10 min, followed by 3% BSA blocking buffer for 1 h. Fluorescence microscopy was performed using anti-His antibody (1/400 dilution), followed by incubation with Alexa568 fluorochrome-conjugated secondary antibody (1/500 dilution). Samples were analyzed on an epifluorescence microscope (Nikon, Melville, NY; Eclipse TE2000U; 60× or 90× oil immersion objective). To determine the localization of endogenous RhoGDI in SrcY527F-transformed NIH-3T3 cells, cells were fixed with 4% paraformaldehyde, and the cells were processed for immunofluorescence staining (antibody at 1/250 dilution) as described in the figure legends. To detect localization of endogenous Rac1 in HeLa cells expressing His-tagged RhoGDI Y156E, cells were fixed in 10% trichloroacetic acid for 15 min at 4°C. Staining was with Rac1 23A8 antibody at 1/100 dilution and His polyclonal antibody at 1/100 dilution. Images were processed using Metamorph software, version 6.1 (Universal Imaging, Downington, PA).
Confocal images were obtained on a Bio-Rad Radiance 2100 Rainbow laser scanning confocal microscope (LSCM; Zeiss, Thornwood, NY), attached to a Nikon TE2000-U microscope with a 60× oil objective. Colocalization analysis was performed using the software Image J (version 1.31). Colocalized data points were determined based on a Pearson's r ≥ 0.7 and are shown as white overlays (see Figure 5A, as indicated, and Figure 6).
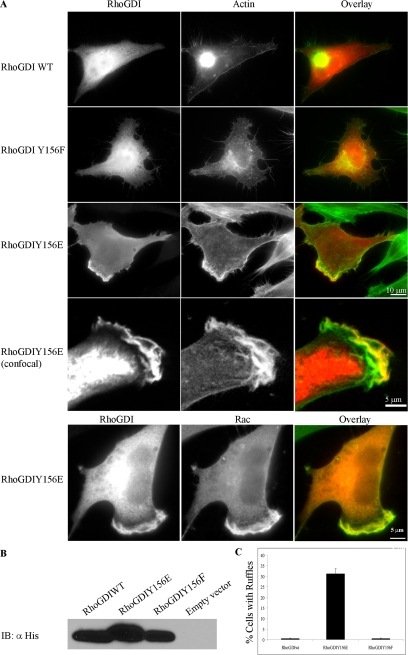
Figure 5.
Tyrosine–phosphorylated RhoGDI enhances and localizes to membrane ruffles. (A) HeLa cells were transfected with either His-tagged RhoGDI wt (WT), His-tagged RhoGDI Y156F (Y156F), or His-tagged RhoGDI Y156E (Y156E). At 6 h after transfection, the cells were fixed and stained for His tag (red) and F-actin (green). Arrowheads indicate ruffles prominently containing His-RhoGDI Y156E. Colocalization of RhoGDI Y156E with F-actin in membrane ruffles was verified by confocal microscopy (lower panels, as indicated), as in Materials and Methods. Colocalization of His-RhoGDI Y156E (red) with Rac1 (green) in membrane ruffles was also evident (lowermost panels) and was also confirmed by confocal microscopy (unpublished data). Bar, 10 μm or 5 μm (confocal), as indicated. Results shown are typical of more than three independent experiments. (B) RhoGDI immunoblots showing approximately equivalent levels of transfected RhoGDI expression in the cells used in A. (C) The percentage of cells expressing various RhoGDI mutants that exhibited a membrane ruffling phenotype was quantified. Results shown are from n = 272 cells in three separate experiments and are given as mean ± SE.
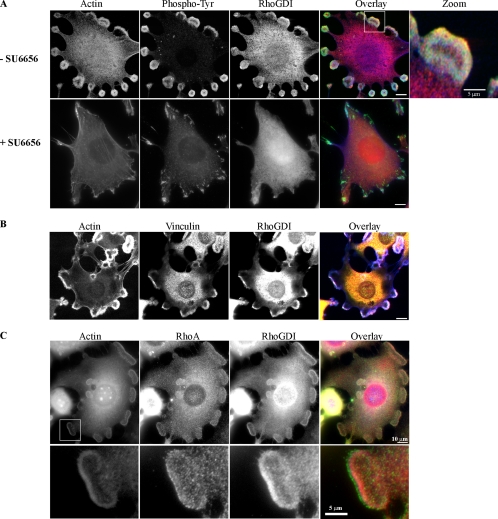
Figure 6.
RhoGDI localizes to Src-dependent rosettes in Src3T3 fibroblasts. SrcY527F-expressing NIH-3T3 cells were analyzed by immunofluorescence, as described in Materials and Methods. (A) Cells were either untreated (−SU6656) or treated with 10 μM SU6656 (+SU6656) overnight before analysis for localization of F-actin (blue; antibody at 1:10 dilution), phospho-tyrosine (green; antibody at 1:250 dilution), and RhoGDI (red; antibody at 1:250 dilution), as indicated. The rightmost panel is a threefold magnification of the boxed area indicated in the overlay. (B) Localization of F-actin (blue; antibody at 1:10 dilution), vinculin (green; antibody at 1:1000 dilution), and RhoGDI (red; antibody at 1:250 dilution), as indicated. (C) Localization of F-actin (blue; antibody at 1:10 dilution), RhoA (green; antibody at 1:100 dilution), and RhoGDI (red; antibody at 1:250 dilution), as indicated. In each case, the areas of colocalization for all three proteins are shown as white (Pearson's r ≥ 0.7). The lowermost panels are a threefold magnification of the boxed area indicated in the lower magnification actin panel.
RESULTS
Src Phosphorylates RhoGDI In Vitro
We had previously reported that RhoGDI was phosphorylated on tyrosine in lysates from human neutrophils stimulated with the chemoattractant, n-formyl-methionyl-leucine-phenyalanine (DerMardirossian et al., 2004). We observed that pure recombinant RhoGDI was extensively phosphorylated by recombinant Src kinase in vitro (Figure 1A). Phosphorylation by recombinant Src reached a stoichiometry approaching 1 mol/mol RhoGDI, as determined by filtration analysis, and occurred exclusively on tyrosine, as determined by phospho-amino acid analysis (unpublished data) and immunoblot with 4G10 anti-phosphotyrosine antibody. D4GDI was also an excellent substrate for Src and was phosphorylated nearly as well as RhoGDI (Figure 1A, bottom panel). The phosphorylation of RhoGDI by Src correlated with the ability of expressed Src to coimmunoprecipitate with RhoGDI (Figure 1B), indicating a relatively high-affinity physical interaction between these proteins.
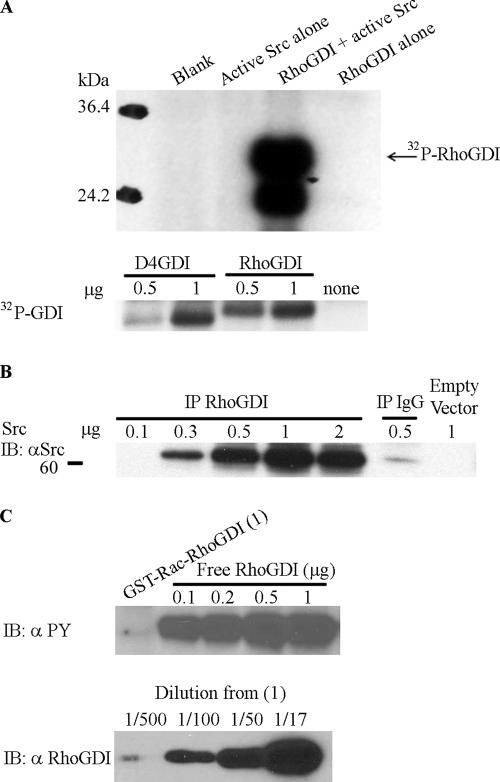
Figure 1.
Src phosphorylates uncomplexed RhoGDI at Tyrosine 156 in vitro. (A) Src phosphorylates RhoGDI directly. Recombinant RhoGDI (2 μg) purified from E. coli was subjected to in vitro kinase assay using recombinant active Src (1 μg), as in Materials and Methods (top panels). Phosphorylation of the indicated amounts of D4GDI versus RhoGDI were compared under the same conditions (bottom panels). (B) Interaction of His-tagged RhoGDIwt with overexpressed active Src. HeLa cells were cotransfected with His-tagged RhoGDI wt and different amounts of active Src Y527F cDNA, as indicated. RhoGDI was then immunoprecipitated and probed with Src antibody for coimmunoprecipitated Src proteins. The IgG IP control represents a nonspecific rabbit IgG pulldown in the presence of 0.5 μg Src plasmid DNA. (C) Src does not phosphorylate Rac-RhoGDI complex in vitro. (A) GST-Rac1–RhoGDI complex immobilized on glutathione beads and recombinant purified uncomplexed RhoGDI were subjected to an in vitro kinase assay using active recombinant Src, and then tyrosine phosphorylation was determined by blotting with mAb 4G10, as in Materials and Methods. The top panel shows that only free RhoGDI, but not RhoGDI complexed with Rac, is tyrosine phosphorylated by Src. The bottom panel shows a RhoGDI immunoblot of various dilutions (indicated in parentheses) of the GST-Rac1–RhoGDI complex used in the kinase assay of the top panel (undiluted = 1).
To evaluate whether the ability of Src to bind and phosphorylate RhoGDI was dependent on release of RhoGDI from complexes with Rho GTPases, we compared phosphorylation of free RhoGDI with that of the RhoGDI–Rac1 complex. As shown in Figure 1C, recombinant Src was unable to effectively phosphorylate RhoGDI presented as a preformed complex with GTPase. This is consistent with the location of the primary Src phosphorylation site (Tyr156; see below) within a region at the interface of the GTPase-binding site on RhoGDI.
Src Phosphorylates RhoGDI in Intact Cells
We expressed constitutively active SrcY527F in HeLa cells and examined phosphorylation of coexpressed RhoGDI. As shown in Figure 2, RhoGDI was phosphorylated on tyrosine upon expression of active Src. In contrast, inactive Src K295M did not phosphorylate RhoGDI, indicating that tyrosine phosphorylation of RhoGDI is dependent on the kinase activity of Src (unpublished data). These data establish that Src expressed in intact HeLa cells functions as a RhoGDI kinase. In contrast, the Src kinase family member Lck was unable to effectively phosphorylate RhoGDI (unpublished data).
Figure 2.
Src phosphorylates RhoGDI at Tyr156 in vivo. HeLa cells were cotransfected with active Src Y527F (or empty vector) and the indicated mutant versions of His-tagged RhoGDI. The top panel shows equal amounts of each construct were immunoprecipitated. Immunoblot of immunoprecipitated RhoGDI constructs with 4G10 phosphotyrosine antibody indicate that RhoGDI is primarily phosphorylated by Src at Tyrosine 156.
Evaluation of the primary amino acid sequence of RhoGDI indicated that Tyr27 and Tyr156 were contained in potential Src phosphorylation consensus motifs (EEIYGEFD/F; Songyang et al., 1995). To identify the exact site(s) of RhoGDI phosphorylation by Src, we mutated Tyr27 and Tyr156 to Phe or Ala, both individually and together, and then assessed phosphorylation of the mutant RhoGDIs when coexpressed with active Src. As shown in Figure 2, the mutation of Tyr156 eliminated the majority of RhoGDI phosphorylation by Src. There was a minor reduction in phosphorylation in the Y27A mutant, and mutation of both sites totally abrogated RhoGDI phosphorylation by Src (Figure 2). We conclude that Tyr156 is the major Src kinase phosphorylation site on RhoGDI. D4GDI also contains an equivalent tyrosine residue at aa 153, which appears to be the major site of Src-mediated phosphorylation in D4GDI.
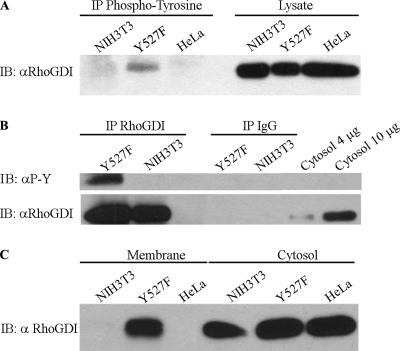
Endogenous RhoGDI in NIH-3T3 cells transformed by active SrcY527F (Src3T3 fibroblasts) was examined for tyrosine phosphorylation. As shown in Figure 3, A and B, we observed that RhoGDI was phosphorylated on tyrosine in the activated Src-expressing cells, but not in control NIH-3T3 cells. Interestingly, substantial amounts of the endogenous tyrosine-phosphorylated RhoGDI was detected in the membrane fraction (Figure 3C; see below). Phosphorylation was blocked using the Src inhibitors PP1 or SU6656 (Blake et al., 2000), consistent with this being a kinase-dependent process (unpublished data).
Figure 3.
Endogenous RhoGDI is tyrosine-phosphorylated in Src3T3 cells and is found associated with the membrane fraction. (A) Whole cell lysates prepared from SrcY527F-expressing NIH-3T3 cells (Y527F) or control NIH-3T3 cells versus Hela cell controls were immunoblotted for RhoGDI (αRhoGDI) either in the whole lysate (right panels) or after immunoprecipitation with 4G10 phospho-tyrosine antibody, as in Materials and Methods. (B) Lysates from SrcY527F-expressing NIH-3T3 cells or control NIH-3T3 cells were immunoprecipitated with RhoGDI antibody (left lanes) or unspecific rabbit IgG (middle lanes) and then immunoblotted for RhoGDI and phospho-tyrosine. Four and 10 μg of total cytosol protein were loaded as controls (right lanes). (C) Membrane versus cytosol fractions were prepared from SrcY527F-expressing NIH-3T3 cells or control NIH-3T3 cells versus Hela cell controls as described in Materials and Methods, and then the fractions were immunoblotted for RhoGDI. Results shown here are representative of at least two experiments.
Phosphorylation of RhoGDI by Src Decreases Its Affinity for Rho GTPases
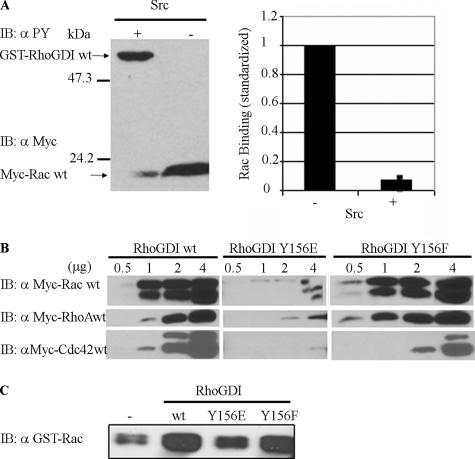
Because a primary function of RhoGDI is to bind Rho GTPases, we wanted to compare the GTPase-binding properties of wild-type RhoGDI versus Src-phosphorylated RhoGDI. To evaluate the effects of RhoGDI Tyr156 phosphorylation on Rho GTPase–RhoGDI complexes, we prepared GST-RhoGDI coupled to glutathione beads that we either phosphorylated by recombinant Src in vitro or subjected to kinase reaction conditions in the absence of Src. As shown in Figure 4A, there was a substantial reduction (>90%) in the ability of tyrosine-phosphorylated RhoGDI to bind overexpressed myc-Rac1 from added lysates. Similar reductions in complex formation were observed with RhoA and Cdc42 (unpublished data).
Figure 4.
Tyrosine-phosphorylated RhoGDI and a RhoGDI phosphomimetic mutant exhibit less affinity for Rac, Rho, and Cdc42. (A) Myc-tagged Rac1 was pulled down from cell lysates by recombinant GST-RhoGDI immobilized on glutathione beads either without (−) or with (+) in vitro phosphorylation by recombinant active Src (left panel), as in Materials and Methods. Pulldowns were immunoblotted for both phospho-tyrosine with 4G10 (αPY) and the presence of associated Rac1 with myc mAb 9E10 (αMyc). The results from three experiments were quantified (right panel) and are shown as mean ± SE. (B) Myc-tagged Rac1-, RhoA-, and Cdc42-expresing cell lysates were incubated with the indicated amounts (in micrograms) of recombinant GST-RhoGDI constructs immobilized on glutathione beads and then pulled down by centrifugation, washed, and analyzed. Substitution in RhoGDI of tyrosine 156 with glutamic acid (Y156E), but not with phenylalanine (Y156F), mimics the results observed with Src-phosphorylated RhoGDI in A by reducing complex formation with all three Rho GTPases at all levels of added GDI protein. (C) The ability of the indicated pure recombinant RhoGDI constructs to extract GST-Rac1 from Sf9 cell membranes was determined, as in Materials and Methods. There was a small amount of Rac1 that became soluble even in the absence of RhoGDI (shown as − control). Results shown above are typical of at least three independent experiments.
To verify this result, we generated recombinant RhoGDI in which Tyr156 was replaced with Glu as a phosphomimetic substitution versus Ala or Phe as nonphosphorylatable residues. Binding of Rac1, Cdc42, and RhoA were reduced by more than 90% in the RhoGDIY156E mutant compared with that of RhoGDIwt (Figure 4B). GTPase binding to the RhoGDIY156F/A mutants were comparable to wild-type (Figure 4B). These results support the in vitro phosphorylation data and indicate that the phosphorylation of Tyr156 by Src inhibits the binding of Rho GTPases to RhoGDI.
We examined endogenous Rac1 binding to RhoGDI in intact NIH-3T3 cells in which RhoGDI was phosphorylated by expression of active SrcY527F. However, we could detect no significant differences in the amount of Rac1 in precipitatable RhoGDI complexes in cells expressing active Src versus kinase dead Src or vector controls (unpublished data). We speculate that this is likely to be due to the inability of Src to effectively phosphorylate the preexisting RhoGDI-Rho GTPase complexes prevalent in unstimulated cells. No change in expression levels of endogenous Rac1 or RhoGDI was observed in cells expressing various Src constructs.
Activated Rho GTPases usually associate with the membrane to activate effectors and then must be recycled back to the cytosol as part of the deactivation process. To evaluate the effect of the reduced affinity of the tyrosine-phosphorylated RhoGDI on its ability to extract membrane-associated GTPases, we prepared membranes from cells overexpressing Rac1wt protein. Intact membranes were then incubated with purified RhoGDIwt, RhoGDIY156E, or RhoGDIY156F, and Rac1 extraction was determined. As is evident in Figure 4C, RhoGDIwt effectively extracted Rac1 from membranes, and the RhoGDIY156F control mutant was only slightly less effective. In contrast, the ability of RhoGDIY156E to extract Rac1 was dramatically reduced. Thus, the decreased affinity of Rho GTPase binding observed with RhoGDIY156E is also manifest in the inability of this tyrosine-phosphorylated form of RhoGDI to reextract membrane-bound Rac1 GTPase.
Tyrosine-phosphorylated RhoGDI Localizes to Areas of Membrane Ruffling and to Podosome Rosettes in Src-transformed Fibroblasts
As shown in Figure 3C, a large amount of RhoGDI was found associated with the isolated membrane fraction in SrcY527F-expressing NIH-3T3 cells. When we examined the localization of His-tagged RhoGDI in HeLa cells expressing RhoGDIY156E by immunofluorescence, we observed that in contrast to wild-type RhoGDI, which was almost entirely localized within the cytosol, a large fraction of RhoGDIY156E was associated with areas of membrane ruffling (Figure 5A). Membrane association was not observed with the control mutant, RhoGDIY156F (Figure 5A) or RhoGDIY156A (unpublished data). Specific association of the phosphomimetic RhoGDI, but not RhoGDIwt, with the plasma membrane was confirmed biochemically by immunoblot analysis of isolated membrane versus cytosol fractions. Confocal microscopy confirmed the presence of RhoGDIY156E in membrane ruffles where it partially colocalized with F-actin (Figure 5A, lower two panels) and Rac1 GTPase (Figure 5A, lowermost panels). Equivalent amounts of each RhoGDI construct were expressed (Figure 5B). Similar membrane localization was observed when RhoGDIY156E was expressed in normal NIH-3T3 cells, indicating that this was not a cell type–specific phenomenon.
We also examined the localization of endogenous RhoGDI in SrcY527F-expressing NIH-3T3 fibroblasts (Figure 6). These cells are characterized by the formation of abundant podosomes, which group to form the characteristic “rosettes” of active Src-expressing cells. These Src-dependent, actin-rich structures are often localized to the ventral surface (Abram et al., 2003). Podosomes are involved in driving locomotion and invasion of Src-transformed cells (Chen, 1989) and are also found in invasive human cancers, osteoclasts, and macrophages (Teti et al., 1991; Bowden et al., 1999). Endogenous RhoGDI was found to localize strongly within rosettes, where it colocalized with phosphotyrosine and F-actin (Figure 6A) and the marker vinculin (Figure 6B). RhoA was closely associated with RhoGDI in rosettes (Figure 6C): a region of RhoA at the very edge of the rosette was adjacent to an internal region that contained partially colocalizing RhoGDI and RhoA. On treatment of these cells with the selective Src inhibitor SU6656 or with PP1, podosomes were induced to disassemble, accompanied by loss of RhoGDI immunostaining of rosettes. Under these conditions, RhoGDI is no longer tyrosine phosphorylated.
Tyrosine-phosphorylated RhoGDI No Longer Effectively Inhibits Rho GTPase Activity In Vivo
Expression of RhoGDI in cells inhibits the activities of endogenous Rho GTPases because of the ability of RhoGDI to rebind and extract these GTPases from membrane locations where effector regulation takes place. However, we observed that HeLa cells expressing RhoGDIY156E were more spread than cells expressing RhoGDIwt or RhoGDIY156F/A. In addition, these cells exhibited a higher fraction of cells with extensive membrane ruffles and active protrusions in a wounded monolayer (Figure 5A; quantified in Figure 5C). These results suggest that RhoGDI in the phosphorylated state is less effective at suppressing Rho GTPase activity and are consistent with the reduced ability of tyrosine-phosphorylated RhoGDI to bind Rho GTPases observed in vitro (Figure 4). In confirmation of these differences in cytoskeletal phenotype, cells expressing RhoGDIY156E exhibited motile responses that were comparable to vector-transfected control cells, whereas cells expressing RhoGDI wt or RhoGDIY156A/F had a markedly reduced level of cell motility (unpublished data). Thus, phosphorylation of Tyr156 in RhoGDI abrogates the ability of RhoGDI to rebind and inhibit activated Rho GTPases both in vitro and in intact cells.
DISCUSSION
We describe here that the Src family kinase member, pp60Src is able to efficiently phosphorylate RhoGDI on Tyr156, both in vitro and in vivo. The equivalent tyrosine residue is conserved in the RhoGDI homolog D4GDI, and we observe that Src effectively phosphorylates D4GDI as well (Figure 1A). Consistent with our findings, RhoGDI was recently identified as a putative Src substrate in a screen involving chemical rescue of an overexpressed mutant form of Src (Qiao et al., 2006). We observed that phosphorylation of RhoGDI by Src does not occur with preformed Rho GTPase–RhoGDI complexes, but requires RhoGDI to be in the uncomplexed state. This observation is consistent with the location of Tyr156 within an acidic region at the interface of the binding site on RhoGDI for Rho GTPases, as described in several structural analyses of GTPase–GDI complexes (Gosser et al., 1997; Keep et al., 1997; Hoffman et al., 2000). As a consequence of phosphorylation at Tyr156, we show that binding of RhoGDI to Rho GTPases (RhoA, Rac1, and Cdc42) is substantially reduced, both in vitro and in vivo. We speculate that phosphorylation of Tyr156 in this critical interface between the two proteins acts to disrupt Rho GTPase binding, either through steric hindrance or via an induced conformational change.
Our data suggest the following model of RhoGDI regulation by phosphorylation at Tyr156: On cell activation by appropriate stimuli, RhoGDI becomes dissociated from bound Rho GTPases during the GTPase activation process. We show that this dissociation is initiated at specific sites associated with localized Rho GTPase activity (e.g., sites of membrane ruffling, podosomes). Indeed, Dransart et al. (2004) recently identified mutations in RhoGDI within the GTPase-binding interface (D45 and D185) that induce RhoGDI localization to membrane ruffles and protrusions and colocalization with Cdc42 GTPase. RhoGDI has also been observed to be enriched in pseudopods isolated from an MDCK-derived cell line (Jia et al., 2005).
The activation of Src by these same (or other) activating stimuli would phosphorylate the free RhoGDI released at these membrane interaction sites, where the RhoGDI normally transiently interacts with a membrane acceptor/binding protein and/or regulatory partner. Our data indicate that phosphorylation at Tyr156 would serve to 1) prevent the interaction and rebinding of membrane-associated Rho GTPases with RhoGDI, thereby prolonging the period of Rho GTPase activation and 2) interrupt the steady state recycling function of RhoGDI, thereby “delaying” the normally transient localization of RhoGDI at the membrane and revealing these transient steps as they become rate limiting. Presumably, this membrane-associated RhoGDI would be available, and positioned, to rebind Rho GTPases upon dephosphorylation of Tyr156. It will be of interest to identify the relevant phosphatase involved in regulating GTPase rebinding.
In support of this model, we observed that RhoGDIY156E and endogenous Src-phosphorylated RhoGDI localized to the plasma membrane at sites associated with Rho GTPases and cytoskeletal activity (Figures 5 and 6). In the former case it was membrane ruffles, whereas in the latter, it was in podosome rosettes, the major actin structures in Src-transformed fibroblasts. Podosome formation is known to be Rho GTPase–dependent (Chellaiah et al., 2000; Chellaiah, 2005; Osiak et al., 2005), and we found that RhoGDI was closely associated with RhoA in the rosettes (Figure 6C). RhoGDIY156E-expressing cells exhibited increased cell spreading, membrane ruffling, and motility compared with the suppression of these responses by wild-type RhoGDI or RhoGDIY156F. This is likely due to the inability of the phosphomimetic mutant to effectively rebind and recycle active Rho GTPases. Alternatively, this phenotype could result from competition of the phosphomimetic RhoGDIY156E with endogenous RhoGDI for membrane-binding sites. Although less likely, we cannot rule out that this phenotype is induced directly by the action of membrane-associated RhoGDIY156E itself.
There is increasing evidence for roles of phosphorylation in regulating the interactions of Rho GTPases with GDIs (reviewed in DerMardirossian and Bokoch, 2005). DerMardirossian et al. (2004) recently described the binding and phosphorylation of RhoGDI, both in vitro and in vivo, by Pak1, a downstream effector of Rac and Cdc42. This phosphorylation was shown to occur on two sites (Ser101 and Ser174) in RhoGDI on the external surface of the hydrophobic cleft in which the GTPase prenyl group binds. Both of these sites lie adjacent to hydrophobic residues that directly line the RhoGDI geranylgeranyl-binding pocket. Phosphorylation of these two sites resulted in the selective release of Rac1, but not RhoA, from the GDI complex, leading to its subsequent activation by exchange factors. The phosphorylation of RhoGDI by Pak1 might serve as a positive feed-forward mechanism to account for sustained Rac activation during processes such as cell motility.
On the basis of our current results, as well as previous work describing phosphorylation-based regulation of GTPase–GDI complexes, we suggest that the action of distinct kinases acting on specific sites in Rho GTPases or RhoGDI itself may be an important means of controlling Rho GTPase signaling in response to diverse receptor-mediated stimuli. Phosphorylation provides a flexible, yet simple mechanism for coordinating Rho GTPase action in response to cell activation through various growth factors, hormones, and extracellular matrix molecules. RhoGDI kinases may act in concert with regulatory phosphatases, the formation of lipid mediators, and potentially other covalent modifications to control the specificity and coordinate the dynamics of Rho GTPase action in response to multiple extracellular signals.
In summary, we have described for the first time an endogenous regulatory pathway controlling the ability of RhoGDI to bind and recycle Rho GTPases from their sites of action at the membrane/membrane cytoskeleton. Our results support those of Dransart et al. (2004), which showed that RhoGDI mutations that exhibit impaired GTPase complex stability become membrane localized. The effect of Src, and perhaps other Src-family kinases, to enhance and/or prolong Rho GTPase activity by modulating membrane-to-cytosol recycling may play significant roles in cellular regulation. In particular, the process of cell transformation and invasion induced by oncogenic Src may be potentiated by the prolonged periods of Rho GTPase activity resulting from tyrosine phosphorylation of RhoGDI and/or D4GDI. It will be of interest to investigate whether the connection between GDI protein levels and metastatic capability relates to the observed tyrosine phosphorylation of RhoGDI and D4GDI detected by proteomic analysis. Additional studies of Src-mediated phosphorylation of RhoGDI are likely to have profound implications for the abnormal activity of Rho GTPases in numerous disease states, including cancer, neurological disorders, inflammation, and various cardiovascular and kidney diseases.
ACKNOWLEDGMENTS
We acknowledge the excellent technical assistance of B. Bohl and editorial assistance from Judith Preston. Dr. Perihan Nalbant provided assistance with confocal microscopy. We thank Dr. Sara Courtneidge (The Burnham Institute) for advice and for generously providing SrcY527F-transformed NIH-3T3 (Src3T3) cells and Dr. David Schlaepfer (TSRI) for Src constructs. We acknowledge the support of Dr. Richard Ulevitch. This is publication number 18267 from The Scripps Research Institute.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-06-0533) on August 30, 2006.
REFERENCES
- Abram C. L., Seals D. F., Pass I., Salinsky D., Maurer L., Roth T. M., Courtneidge S. A. The adaptor protein fish associates with members of the ADAMs family and localizes to podosomes of Src-transformed cells. J. Biol. Chem. 2003;278:16844–16851. doi: 10.1074/jbc.M300267200. [DOI] [PubMed] [Google Scholar]
- Benzing T. Signaling at the slit diaphragm. J. Am. Soc. Nephrol. 2004;15:1382–1391. doi: 10.1097/01.asn.0000130167.30769.55. [DOI] [PubMed] [Google Scholar]
- Berdeaux R. L., Diaz B., Kim L., Martin G. S. Active Rho is localized to podosomes induced by oncogenic Src and is required for their assembly and function. J. Cell Biol. 2004;166:317–323. doi: 10.1083/jcb.200312168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A. L., Hall A. Rho GTPases and their effector proteins. Biochem. J. 2000;348:241–255. [PMC free article] [PubMed] [Google Scholar]
- Blake R. A., Broome M. A., Liu X., Wu J., Gishizky M., Sun L., Courtneidge S. A. SU6656, a selective src family kinase inhibitor, used to probe growth factor signaling. Mol. Cell. Biol. 2000;20:9018–9027. doi: 10.1128/mcb.20.23.9018-9027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden E. T., Barth M., Thomas D., Glazer R. I., Mueller S. C. An invasion-related complex of cortactin, paxillin and PKCmu associates with invadopodia at sites of extracellular matrix degradation. Oncogene. 1999;18:4440–4449. doi: 10.1038/sj.onc.1202827. [DOI] [PubMed] [Google Scholar]
- Chellaiah M. A. Regulation of actin ring formation by rho GTPases in osteoclasts. J. Biol. Chem. 2005;280:32930–32943. doi: 10.1074/jbc.M500154200. [DOI] [PubMed] [Google Scholar]
- Chellaiah M. A., Soga N., Swanson S., McAllister S., Alvarez U., Wang D., Dowdy S. F., Hruska K. A. Rho-A is critical for osteoclast podosome organization, motility, and bone resorption. J. Biol. Chem. 2000;275:11993–12002. doi: 10.1074/jbc.275.16.11993. [DOI] [PubMed] [Google Scholar]
- Chen W. T. Proteolytic activity of specialized surface protrusions formed at rosette contact sites of transformed cells. J. Exp. Zool. 1989;251:167–185. doi: 10.1002/jez.1402510206. [DOI] [PubMed] [Google Scholar]
- Chuang T. H., Bohl B. P., Bokoch G. M. Biologically-active lipids are regulators of Rac-GDI complexation. J. Biol. Chem. 1993a;268:26206–26211. [PubMed] [Google Scholar]
- Chuang T. H., Xu X. M., Knaus U. G., Hart M. J., Bokoch G. M. GDP dissociation inhibitor prevents intrinsic and GTPase activating protein-stimulated GTP hydrolysis by the Rac GTP-binding protein. J. Biol. Chem. 1993b;268:775–778. [PubMed] [Google Scholar]
- DerMardirossian C., Bokoch G. M. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–363. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- DerMardirossian C., Schnelzer A., Bokoch G. M. Phosphorylation of RhoGDI by Pak1 mediates dissociation of Rac GTPase. Mol. Cell. 2004;15:117–127. doi: 10.1016/j.molcel.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Dransart E., Morin A., Cherfils J., Olofsson B. Uncoupling of inhibitory and shuttling functions of rhoGDIs. J. Biol. Chem. 2004;280:4674–4683. doi: 10.1074/jbc.M409741200. [DOI] [PubMed] [Google Scholar]
- Gildea J. J., Seraj M. J., Oxford G., Harding M. A., Hampton G. M., Moskaluk C. A., Frierson H. F., Conaway M. R., Theodorescu D. RhoGDI2 is an invasion and metastasis suppressor gene in human cancer. Cancer Res. 2002;62:6418–6423. [PubMed] [Google Scholar]
- Gosser Y. Q., Nomanbhoy T. K., Aghazadeh B., Manor D., Combs C., Cerione R. A., Rosen M. K. C-terminal binding domain of Rho GDP-dissociation inhibitor directs N-terminal inhibitory peptide to GTPases. Nature. 1997;387:814–819. doi: 10.1038/42961. [DOI] [PubMed] [Google Scholar]
- Hoffman G. R., Nassar N., Cerione R. A. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell. 2000;100:345–356. doi: 10.1016/s0092-8674(00)80670-4. [DOI] [PubMed] [Google Scholar]
- Jaffe A. B., Hall A. Rho GTPases in transformation and metastasis. Adv. Cancer Res. 2002;84:57–80. doi: 10.1016/s0065-230x(02)84003-9. [DOI] [PubMed] [Google Scholar]
- Jia Z., Barbier L., Stuart H., Amraei M., Pelech S., Dennis J. W., Metalnikov P., O'Donnell P., Nabi I. R. Tumor cell pseudopodial protrusions. Localized signaling domains coordinating cytoskeleton remodeling, cell adhesion, glycolysis, RNA translocation, and protein translation. J. Biol. Chem. 2005;280:30564–30573. doi: 10.1074/jbc.M501754200. [DOI] [PubMed] [Google Scholar]
- Jiang W. G., Watkins G., Lane J., Cunnick G. H., Douglas-Jones A., Mokbel K., Mansel R. E. Prognostic value of Rho GTPases and Rho guanine nucleotide dissociation inhibitors in human breast cancers. Clin. Cancer Res. 2003;9:6432–6440. [PubMed] [Google Scholar]
- Keep N. H., Barnes M., Barsukov I., Badii R., Lian L. Y., Segal A. W., Moody P. C., Roberts G. C. A modulator of Rho family G proteins, RhoGDI, binds these G proteins via an immunoglobulin-like domain and a flexible N-terminal arm. Structure. 1997;5:623–633. doi: 10.1016/s0969-2126(97)00218-9. [DOI] [PubMed] [Google Scholar]
- Knaus U. G., Morris S., Dong H. J., Chernoff J., Bokoch G. M. Regulation of human leukocyte p21-activated kinases through G protein–coupled receptors. Science. 1995;269:221–223. doi: 10.1126/science.7618083. [DOI] [PubMed] [Google Scholar]
- Lock P., Abram C. L., Gibson T., Courtneidge S. A. A new method for isolating tyrosine kinase substrates used to identify fish, an SH3 and PX domain-containing protein, and Src substrate. EMBO J. 1998;17:4346–4357. doi: 10.1093/emboj/17.15.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. S. Cell signaling and cancer. Cancer Cell. 2003;4:167–174. doi: 10.1016/s1535-6108(03)00216-2. [DOI] [PubMed] [Google Scholar]
- Osiak A. E., Zenner G., Linder S. Subconfluent endothelial cells form podosomes downstream of cytokine and RhoGTPase signaling. Exp. Cell Res. 2005;307:342–353. doi: 10.1016/j.yexcr.2005.03.035. [DOI] [PubMed] [Google Scholar]
- Qiao Y., Molina H., Pandey A., Zhang J., Cole P. A. Chemical rescue of a mutant enzyme in living cells. Science. 2006;311:1293–1297. doi: 10.1126/science.1122224. [DOI] [PubMed] [Google Scholar]
- Rush J., Moritz A., Lee K. A., Guo A., Goss V. L., Spek E. J., Zhang H., Zha X. M., Polakiewicz R. D., Comb M. J. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- Songyang Z., et al. Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- Teti A., Marchisio P. C., Zallone A. Z. Clear zone in osteoclast function: role of podosomes in regulation of bone-resorbing activity. Am. J. Physiol. 1991;261:C1–C7. doi: 10.1152/ajpcell.1991.261.1.C1. [DOI] [PubMed] [Google Scholar]
- Togawa A., et al. Progressive impairment of kidneys and reproductive organs in mice lacking Rho GDIalpha. Oncogene. 1999;18:5373–5380. doi: 10.1038/sj.onc.1202921. [DOI] [PubMed] [Google Scholar]
- Ugolev Y., Molshanski-Mor S., Weinbaum C., Pick E. Liposomes comprising anionic but not neutral phospholipids cause dissociation of [Rac(1 or 2)-RhoGDI] complexes and support amphiphile-independent NADPH oxidase activation by such complexes. J. Biol. Chem. 2006;281:19204–19219. doi: 10.1074/jbc.M600042200. [DOI] [PubMed] [Google Scholar]
- Unwin R. D., Sternberg D. W., Lu Y., Pierce A., Gilliland D. G., Whetton A. D. Global effects of BCR/ABL and TEL/PDGFRbeta expression on the proteome and phosphoproteome: identification of the Rho pathway as a target of BCR/ABL. J. Biol. Chem. 2005;280:6316–6326. doi: 10.1074/jbc.M410598200. [DOI] [PubMed] [Google Scholar]
- Urich M., Senften M., Shaw P. E., Ballmer-Hofer K. A role for the small GTPase Rac in polyomavirus middle-T antigen-mediated activation of the serum response element and in cell transformation. Oncogene. 1997;14:1235–1241. doi: 10.1038/sj.onc.1200982. [DOI] [PubMed] [Google Scholar]
- Van Aelst L., Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- Yamashita T., Tohyama M. The p75 receptor acts as a displacement factor that releases Rho from Rho-GDI. Nat. Neurosci. 2003;6:461–467. doi: 10.1038/nn1045. [DOI] [PubMed] [Google Scholar]