Abstract
Tetrahydrobiopterin is an essential cofactor for aromatic amino acid hydroxylases, ether lipid oxidase and nitric oxide synthases. Its biosynthesis in mammals is regulated by the activity of the homodecameric enzyme GCH (GTP cyclohydrolase I; EC 3.5.4.16). In previous work, catalytically inactive human GCH splice variants differing from the wild-type enzyme within the last 20 C-terminal amino acids were identified. In the present study, we searched for a possible role of these splice variants. Gel filtration profiles of purified recombinant proteins showed that variant GCHs form high-molecular-mass oligomers similar to the wild-type enzyme. Co-expression of splice variants together with wild-type GCH in mammalian cells revealed that GCH levels were reduced in the presence of splice variants. Commensurate with these findings, the GCH activity obtained for wild-type enzyme was reduced 2.5-fold through co-expression with GCH splice variants. Western blots of native gels suggest that splice variants form decamers despite C-terminal truncation. Therefore one possible explanation for the effect of GCH splice variants could be that inactive variants are incorporated into GCH heterodecamers, decreasing the enzyme stability and activity.
Keywords: Chinese-hamster ovary cell (CHO cell), co-expression, enzyme stability, GTP cyclohydrolase I, splice variant, tetrahydrobiopterin
Abbreviations: CH1, GTP cyclohydrolase I type 1; ΔCH1, N-terminally truncated GTP cyclohydrolase I isoform 1; CHO, Chinese-hamster ovary; DRD, dopa-responsive dystonia; GCH, GTP cyclohydrolase I; GFRP, GCH feedback regulatory protein; H4-bip, (6R)-5,6,7,8-tetrahydro-L-biopterin, tetrahydrobiopterin; HRP, horseradish peroxidase
INTRODUCTION
H4-biopterin [tetrahydrobiopterin; (6R)-5,6,7,8-tetrahydro-L-biopterin] is an essential cofactor for biosynthesis of neurotransmitters by aromatic amino acid hydroxylases, ether lipid oxidase and nitric oxide synthases [1]. Its biosynthesis in mammals is regulated by the activity of the homodecameric enzyme GCH (GTP cyclohydrolase I). Two pentameric rings associate by intercalation of antiparallel helix pairs to give the toroidal GCH structure [2]. Intra-ring interactions result in the formation of a 20-stranded antiparallel β-barrel. Ten equivalent active sites are located at the periphery of the toroid, each formed at the interface of three adjacent subunits.
Mammalian GCH is inhibited by H4-biopterin and stimulated by phenylalanine through complex formation with the GFRP (GCH feedback regulatory protein) [3,4]. Two GFRP pentamers, which comprise β-propeller-like structures, cap the GCH decamer [5,6]. The allosteric effector molecules bind at the interface between GCH and GFRP. The biopterin-binding site is mainly formed by GCH, and conformational changes upon binding biopterin cause enzyme inhibition [6]. The phenylalanine-binding pocket, however, is formed mainly by GFRP, and phenylalanine binding stabilizes the active configuration of GCH [5]. In addition to this feedback regulation, GCH activity is modulated by phosphorylation [7,8], and gene transcription is induced by certain hormones and cytokines in a cell- and tissue-specific manner [9–11]. This is observed in cell types and tissues that otherwise express low GCH mRNA levels, whereas in liver and neuronal tissues GCH expression is constitutively high [10,11].
Mutations in the GCH gene cause hyperphenylalaninaemia and DRD (dopa-responsive dystonia), a variant form of Parkinsons disease [12]. A dominant-negative effect of mutant GCH on the wild-type enzyme was inferred from reduced enzyme activity observed in DRD patients and co-expression studies [13–15]. Instability and lower enzyme activity of chimaeric protein composed of wild-type and mutant subunits or reduced synthesis of wild-type GCH was suggested. Other workers proposed a reduction in the amount of enzyme, independent of mutation type, or a dominant-negative effect at either the translational or transcriptional level [16,17].
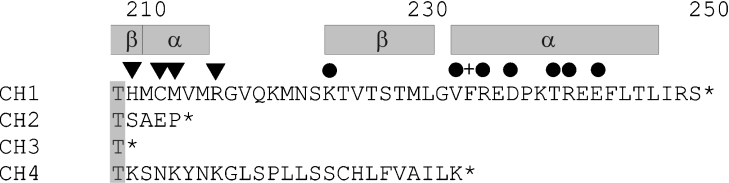
The mammalian GCH gene is organized in six exons containing introns of several kb of DNA [18], and multiple mRNA variants occur. Alternative splicing at the exon 5/6 boundary of the human gene gives GCH types 2 (cloned from liver) and 4 (from myelomonocytoma cells) and a premature stop yields type 3 (from liver) [19,20]. GCH mRNA variants were detected in various human cell types, and type 3 mRNA was present at significant levels relative to wild-type GCH in myelomonocytoma cells and dermal fibroblasts [19]. Although wild-type GCH is a 250-residue protein, the mRNA splice variants encode 213-residue (type 2), 209-residue (type 3) and 233-residue (type 4) proteins with alternative C-termini [19] (Figure 1). No catalytic activity has been determined for C-terminal splice variants [19,21,22] and expression of GCH type 2 in mammalian cells has been shown to decrease the level of wild-type enzyme [23].
Figure 1. C-termini of human GCH isoforms.
C-terminal sequences of GCH types 1–4 [19,20] are shown. Active-site residues (▼) and structural elements (α, helix; β, sheet) in wild-type GCH (type 1) are indicated according to the published structure (PDB 1FB1; [2]). Also marked are residues involved in binding phenylalanine (+) and biopterin (●) in the rat GCH–GFRP complex [5,6].
The aim of the present study is to determine the potential role of splice variants in regulating GCH activity or accelerating its degradation through heteromeric interactions. Using bicistronic recombinant expression in mammalian cells, we investigated the properties of GCH splice variants and their influence on the wild-type enzyme activity and protein association state.
EXPERIMENTAL
pETDuetΔCH1 construction
Wild-type GCH [CH1 (GCH type 1)] was taken from a Bluescript SK− clone (Stratagene) of a cDNA library of stimulated THP-1 cells [19], cloned into pET-32a (Novagen) and then subcloned into multiple cloning site 1 of pETDuet-1 (Novagen) with deletion of amino acid residues 2–42 using NcoI/HindIII by standard techniques. PCR conditions were 2 min at 95 °C, 30 cycles of 30 s at 95 °C, 1 min at 58 °C and 1 min at 72 °C, followed by 7 min at 72 °C, with primers 5′-CGCCCATGGAGGCCAAGAGCGCGCAG-3′ and 5′-CTGAAGCTTGCTCAGCTCCTAATGAGAGTC-3′. All cloning steps were checked by sequencing of insert (Microsynth).
pET-29 vector construction
Plasmids for the GCH splice variants were created by PCR with oligo(dT)-primed cDNA, from THP-1 cells stimulated for 6–8 h with interferon-γ, as a template, and Herculase hot start polymerase (Stratagene). PCR conditions were 2 min at 95 °C, 35 cycles of 30 s at 94 °C, 1 min at 58 °C and 1 min at 72 °C, followed by 7 min at 72 °C, with the following primer combinations: chexp1B (forward primer) 5′-GCGGGACCATGGAGAAGG-3′ was combined with chexp2C 5′-TATCTAGAGCTATGGTTCTGCAGACGTT-GC-3′ for type 2, with primer chexp3 5′-TATCTAGATCACTTCTAGTGCACCATTATGACG-3′ for type 3 and with chexp2 5′-GCTCTAGATGTCTTCCACCGTCAGTTCATTC-3′ for type 4. PCR products were cloned using the Topo TA Cloning Vector (Invitrogen) and then subcloned using NotI/XbaI into pcDNA5TO (Invitrogen), and then into pET-29a with an N-terminal S-tag using KpnI/EcoRV with the In-Fusion PCR cloning kit (BD Biosciences) yielding ΔCH2s, ΔCH3s and ΔCH4s. PCR conditions were 2 min at 95 °C, 25 cycles of 30 s at 95 °C, 1 min at 58 °C and 1 min at 72 °C, followed by 7 min at 72 °C, using forward primer 5′-ACAGCCCAGATCTGGGTGAGGCCAAGAGCGCGCAG-3′ combined with reverse primers 5′-GAATTCGGATCCGATGGCTGATCAGCGGGTTTAAAC-3′ for pcDNA5TO templates (types 2 and 4) and 5′-GAATTCGGATCCGATTGCAGACTTACGTTG CTTCAAC-3′ for pCR-Topo template (type 3). All cloning steps were checked by sequencing of inserts.
Bacterial expression and protein purification
A culture of Tuner DE3 cells (Novagen) transformed with ΔCH1 or ΔCH2s–ΔCH4s plasmids was grown in LB (Luria–Bertani) medium containing kanamycin (50 μg/ml). Expression was induced by the addition of 0.5 mM isopropyl-β-D-thiogalactopyranoside (Serva), the culture was incubated for 6 h at 37 °C and harvested by centrifugation (5000 g for 20 min at 4 °C). The bacterial pellet was resuspended in BugBuster protein extraction reagent (Novagen) containing 0.2 mg·ml−1 lysozyme, 25 units·ml−1 Benzonase nuclease (Novagen) and protease inhibitor mix (Amersham Biosciences), incubated for 20 min at room temperature (25 °C) and centrifuged (16000 g for 20 min at 4 °C).
The extract was separated by anion-exchange chromatography at 4 °C on a column (2.5 cm×10 cm) of Q-Sepharose High Performance (Amersham Biosciences) equilibrated in 20 mM Bis-Tris propane HCl (pH 7), containing 0.02% NaN3. After washing the column with starting buffer, a linear gradient of 0–0.5 M NaCl over 500 ml was applied. The GCH-enriched fraction was identified by Western-blot analysis using GCH antisera [19] (to detect ΔCH1) or S-protein (to detect ΔCH2s, ΔCH3s and ΔCH4s). It was purified further by gel filtration at room temperature on a HiLoad Superdex 200 prep grade column (1.6 cm×60 cm; Amersham Biosciences) equilibrated in 50 mM potassium phosphate buffer (pH 7), containing 0.02% NaN3.
Protein purity was monitored by Tris/Tricine SDS/PAGE in the presence of the reducing agent 2-mercaptoethanol [24]. For estimation of molecular mass, low-range rainbow molecular-mass markers (Amersham Biosciences) were used. The identity and integrity of the purified ΔCH1 was confirmed by LCQ MS of protein desalted by reversed-phase HPLC. Samples were stored at 4 °C and concentrations were estimated by absorption at 280 nm.
pBud vector construction
GCH splice variants were subcloned into the EF-1α (elongation factor-1α) site of pBudCE4.1 using NotI/KpnI by standard techniques. In order to avoid inclusion of vector sequence in the protein N-terminus, these sequence parts were cut out by NotI/EcoRV digestions, filled with the Klenow fragment and religated. Wild-type GCH was inserted into the CMV (cytomegalovirus) cloning site of pBudCE4.1 (Invitrogen) using PstI/AccI restriction. S-tag was transferred from pET-32a (Novagen) as a short PCR product and inserted by restriction digest with NotI/EcoRV and ligation upstream of the N-terminus of the splice variants. PCR conditions were 2 min at 95 °C, 30 cycles of 30 s at 95 °C, 1 min at 64 °C and 1 min at 72 °C, followed by 7 min at 72 °C, with primers 5′-CTGCGGCCGCGCACCATCATCATCATCATTCTT-3′ and 5′-AGGATATCGCTGTCCATGTGCTGGCGTT-3′. Control vectors were created by cutting out the wild-type enzyme sequence with SbfI/PacI digestions, filled with the Klenow fragment and religated. All cloning steps were controlled by sequencing of inserts.
Cell culture and transfection
CHO (Chinese-hamster ovary)-K1 cells (American Type Culture Collection) were grown at 37 °C in a CO2 incubator with F-12K nutrient mixture (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (PAN Biotech). Cells (1×106) were grown for 24 h in 5 ml of growth medium, which was then replaced by Opti-MEM I medium (Invitrogen). The cells were transfected with 10 μg of endotoxin-free plasmid DNA (Qiagen) using 30 μl of Lipofectamine™ 2000 (Invitrogen) and the medium was replaced after 5 h. RNA was isolated from cells after 5 h using TRIzol® reagent (Invitrogen) according to the manufacturer's instructions. Controls were included where cells were treated with Lipofectamine™ but no plasmid DNA. Cells were harvested after 24 h using trypsin/EDTA solution (Sigma), then resuspended in Dulbecco's buffer (Serva) containing protease inhibitor mix (Amersham Biosciences), frozen in liquid N2 and thawed at 37 °C.
Quantitative PCR analysis
Total RNA isolated from CHO cells was reverse-transcribed into a first-strand cDNA using SuperScript II RNase H− reverse transcriptase (Invitrogen) and random hexamer primers. Quantitative PCR analysis using the Brilliant Core kit (Stratagene) was performed with real-time PCR (ABI Prism 7700 sequence detector; Applied Biosystems). Sequences for primers and probes specific for various GCH variants were as described previously [19], except for type 2 mRNA, which are 5′-CGCCTTACAAAACAAATTGCTGTA-3′ (sense), 5′TTCGCCCTTTATCTAGAGCTATGG3′ (antisense) and 5′-TTCTGCAGACGTTGCTTCAACCACTACC-3′ (probe).
Enzyme assay
GCH activity was assayed using a modification of the published method [25]. Low-molecular-mass compounds were removed from cell extracts by Sephadex G-25 chromatography (NAP-5 columns; Amersham Biosciences) and protein was eluted in 0.1 M Tris/HCl buffer (pH 7.8), containing 0.3 M KCl, 10% (v/v) glycerol and 2.5 mM EDTA. Samples were incubated with 2 mM GTP substrate (Sigma) for 90 min at 37 °C to generate 7,8-dihydroneopterin triphosphate. This was oxidized by iodine in HCl and cleaved by alkaline phosphatase (Sigma) to neopterin, which was detected by fluorescence after HPLC as described previously [25]. The protein content of eluates was determined by the Bradford assay [26] and activity was expressed as the amount of neopterin formed (pmol·mg−1·min−1).
Western blot analysis
Total protein (5 μg) was analysed by Tris/Tricine SDS/PAGE in the presence of the reducing agent 2-mercaptoethanol [24]. For native PAGE, 50–100 μg samples, in 60 mM Tris/HCl (pH 6.8), containing 25% glycerol and 0.1% (w/v) Bromophenol Blue, were analysed using 6% resolving and 4% stacking gels with a Tris/glycine buffer (25 mM Tris/HCl, pH 8.8, containing 190 mM glycine). Western blots were performed with standard procedures using PVDF membranes and ECL® Plus detection reagent (Amersham Biosciences). GCH antiserum was used as described previously [19], and S-protein–HRP (horseradish peroxidase) conjugate (Novagen) was diluted 1:1000 (SDS/PAGE) or 1:2000 (native PAGE) in Dulbecco's PBS containing 1% (w/v) BSA and 0.05% (v/v) Tween 20. Mouse anti-actin monoclonal antibody (Chemicon) was diluted 1:500 and was detected using a monoclonal anti-mouse IgG–HRP conjugate, diluted 1:50000. Blots were scanned (Typhoon Scanner) and bands from SDS/PAGE were quantified using ImageQuant TL software (Amersham Biosciences).
RESULTS
Gel-filtration chromatography of recombinant GCH splice variants
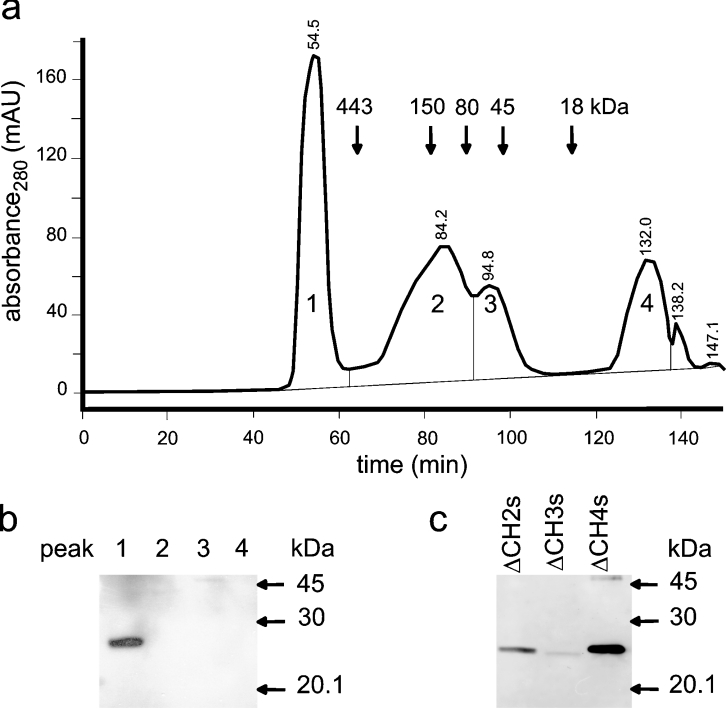
In order to characterize the biochemical properties of the inactive C-terminal splice variants, we investigated their gel-filtration profiles using proteins recombinantly expressed in Escherichia coli. Purification of full-length wild-type GCH is difficult due to proteolysis of the N-terminal segment [2]. These N-terminal residues are disordered in inhibitory and stimulatory complexes of mammalian GCH with GFRP and have no significant effect on enzyme activity, complex formation or inhibition by GFRP [2,5,6]. Recombinant expression of wild-type GCH lacking residues 2–42 [ΔCH1 (N-terminally truncated CH1)] yielded protein levels high enough for purification and for confirming its identity and integrity (molecular mass, 23476.7±2 Da) [2] by MS and CD spectra (results not shown). Since the splice variants studied here show no enzyme activity and are not stained with the GCH antiserum (see below), we added N-terminal S-tags to these reading frames. Although deletion of residues 2–42 in the splice variants did not increase their stability to a degree allowing for biophysical analysis, we could show that, similar to ΔCH1, the variant proteins eluted at high-molecular-mass by gel filtration. Figure 2(a) shows a typical elution profile for ΔCH2s after anion-exchange chromatography. Moreover, the predicted molecular masses for the S-tagged proteins (molecular mass: 21204 Da for ΔCH2s; 20820 Da for ΔCH3s; 23463 Da for ΔCH4s) collected from the high-molecular-mass peak by gel filtration were consistent with the migration of monomers during SDS/PAGE (Figure 2b).
Figure 2. Gel filtration of N-terminally truncated GCH splice variants.
Recombinant ΔCH2s purified using anion-exchange chromatography using Q-Sepharose was further separated by gel filtration on Superdex 200 (a). Elution positions of standard proteins are indicated by their apparent molecular masses in kDa: ferritin (443), alcohol dehydrogenase (150), transferrin (80), egg albumin (45) and myoglobin (18). S-tag detection on Western blots of peaks 1–4 showed that ΔCH2s eluted in the first peak (b). Also ΔCH3s and ΔCH4s collected from the high-molecular-mass peak migrated at the predicted monomer mass (see the Results section).
Co-expression of wild-type GCH and its splice variants
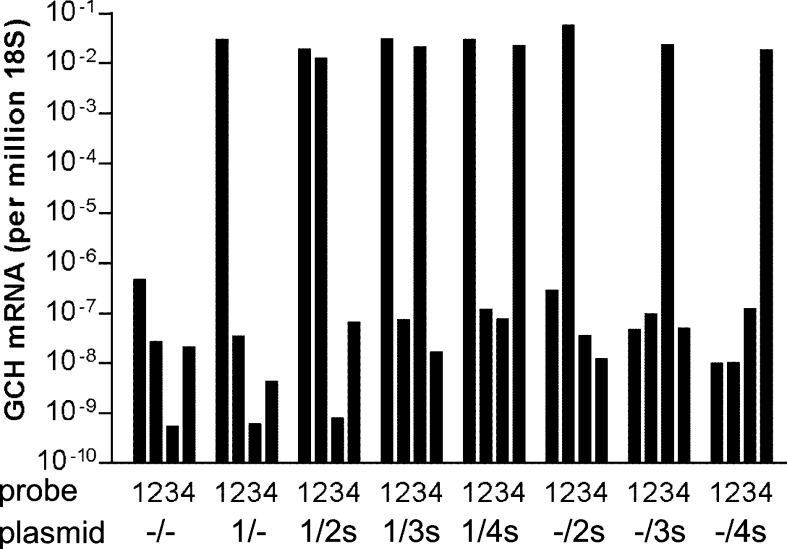
We then co-expressed inactive C-terminal splice variants of human GCH [19] together with the wild-type enzyme in mammalian cells to obtain some insight into their possible biological role. CHO cells were transfected with pBudCE4.1 vector constructs encoding genes for full-length GCH at the first cloning site and for variants at the second site (either with or without N-terminal S-tags). Use of pBud vectors ensures double transfection of two mRNAs within the same cell. Real-time PCR demonstrated that these bicistronic vectors achieved high mRNA expression levels, yielding comparable amounts for all isoforms irrespective of whether they were expressed alone or in combination with a second isoform (Figure 3).
Figure 3. Real-time PCR of GCH mRNA variants co-expressed in mammalian cells.
CHO cells were transfected with wild-type plus various variant GCH pBudCE4.1 plasmids and RNA was isolated 24 h thereafter for quantitative PCR analysis. 1, 2, 3 and 4, TaqMan probe specific for GCH type 1, 2, 3 and 4 respectively. –/–, empty vector; 1/–, only GCH type 1; 1/2s, 1/3s and 1/4s, GCH type 1 plus S-tagged GCH variants 2, 3 and 4 respectively; –/2s, –/3s and –/4s, S-tagged GCH variants without GCH type 1. Values are means for duplicate determinations of one of three similar experiments. GCH mRNA was below the detection limit in Lipofectamine™-treated control cells (not shown).
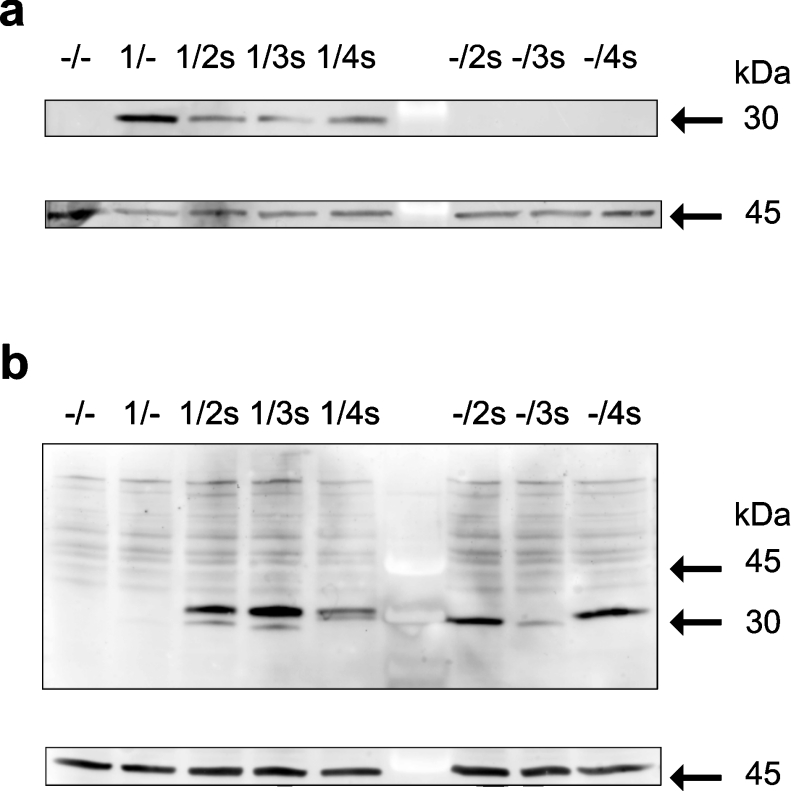
Western-blot analysis using a GCH antiserum revealed that co-expression with splice variants decreased GCH protein levels (Figure 4a). It also turned out that the GCH antiserum only stained the wild-type protein CH1 (molecular mass 27903 Da), but did not cross-react with the splice variants (Figure 4a) even though the proteins vary only within the last 20 amino acids of their C-terminus (see Figure 1). Detection of S-protein, however, clearly demonstrated expression of spliced proteins in isoform-only expressions as well as in co-expressions with wild-type CH1 showing bands at the appropriate size (Figure 4b). The calculated molecular mass for CH2s, CH3s and CH4s are 26493 Da, 26109 Da and 28752 Da respectively. As with the wild-type enzyme, these proteins appeared as slightly larger bands in SDS/PAGE.
Figure 4. SDS/PAGE and Western-blot analysis of extracts from mammalian cells co-expressing GCH splice variants.
GCH isoforms expressed in CHO cells transfected with the respective pBudCE4.1 plasmids are indicated above the lanes. Blots were analysed using a GCH antiserum [19] (a) or S-protein HRP conjugate (b). For a control, blots were probed also with an actin antiserum (lower panels of a and b). Results of one of three similar experiments are shown.
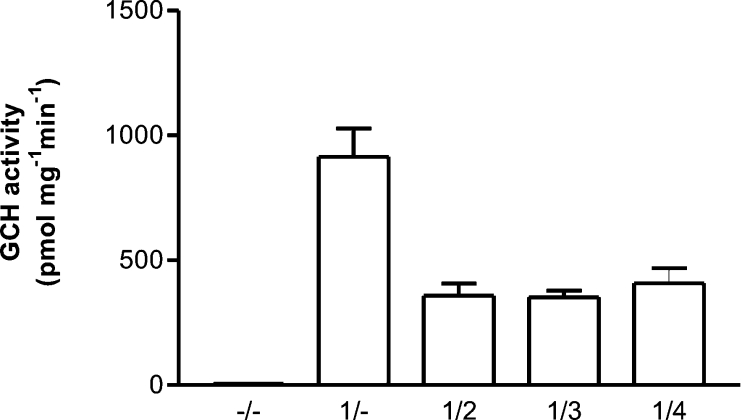
The effect of splice variants on wild-type GCH activity was investigated by assaying protein extracts from CHO cells co-expressing GCH isoforms. Enzyme activity data averaged from three transfection experiments are shown in Figure 5. The activity of 914±114 pmol·mg−1·min−1 for expression of wild-type enzyme is reduced 2.5-fold by co-expression with inactive C-terminal variants. For comparison, baseline levels were 3.83±0.95 pmol·mg−1·min−1 (empty vector) and 3.44±1.03 pmol·mg−1·min−1 (no plasmid; mean±S.D. of nine experiments). Control transfections with splice variants in the absence of wild-type enzyme did not exceed these baseline GCH activity levels (results not shown).
Figure 5. GCH activity in extracts from CHO cells co-expressing GCH splice variants.
CHO cells were transfected with various pBudCE4.1 plasmids expressing different GCH isoforms (–/–, empty vector; 1/–, GCH type 1; 1/2, 1/3 and 1/4, GCH type 1 plus variant proteins 2, 3 and 4 respectively). Enzyme activity was determined in cell extracts prepared 24 h after transfection. Values are means from three independent transfections ±S.D. Baseline levels were 3.83±0.95 (empty vector) and 3.44±1.03 pmol·mg−1·min−1.
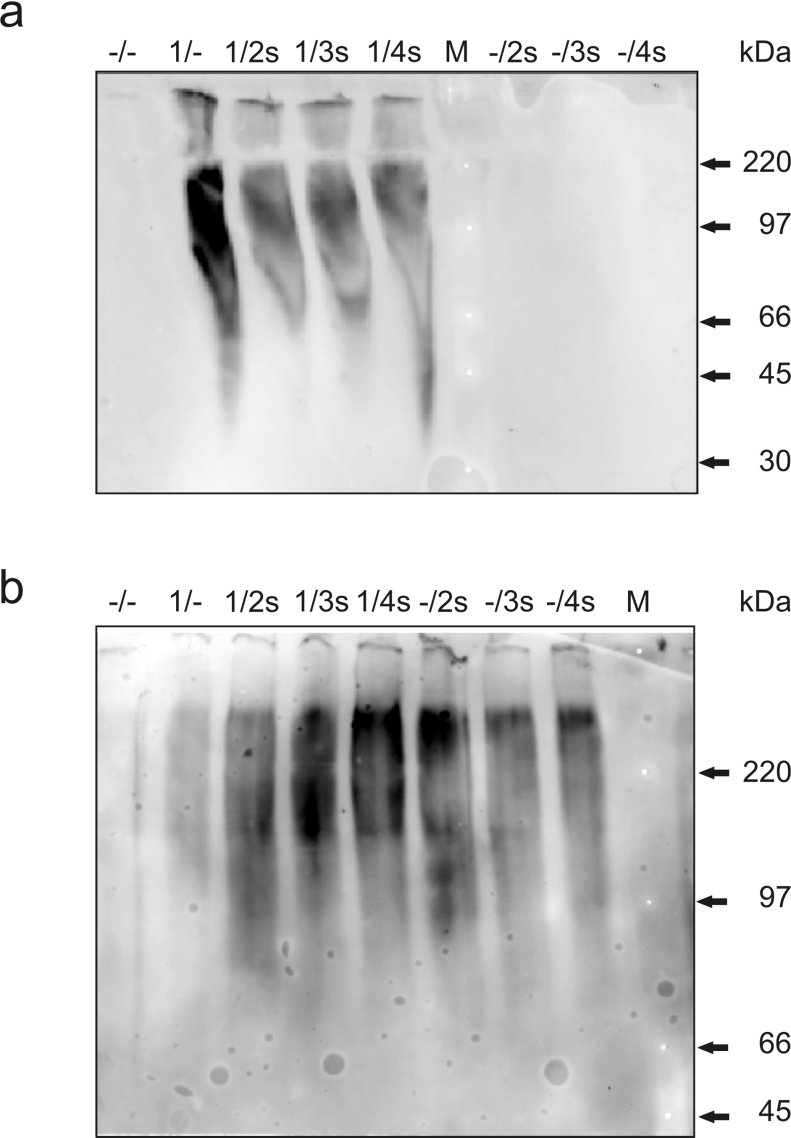
The association state of GCH isoforms was determined by native PAGE of protein extracts of transfected CHO cells. Co-expressions of CH1 with S-tagged isoforms (CH2s, CH3s and CH4s) were compared with expressions of CH1 or S-tagged isoforms alone. Separation of marker proteins in the range of 14300–220000 Da was achieved, although bands are broader than with denaturing gels. GCH antisera specifically detected CH1, which was present in higher levels in the absence of splice variants (Figure 6a). The migration of GCH is consistent with the formation of higher oligomers such as pentamers (140 kDa) or decamers (279 kDa) and clearly shows that monomers and dimers are not detectable under native conditions. Detection with S-protein revealed that C-terminally truncated isoforms also formed high-molecular-mass oligomers, both in the presence and absence of CH1 (Figure 6b).
Figure 6. Native PAGE and Western-blot analysis of extracts from CHO cells co-expressing GCH splice variants.
CHO cells were transfected with pBudCE4.1 plasmids expressing various GCH isoforms (–/–, empty vector; 1/–, CH1 alone; 1/2s, 1/3s and 1/4s, CH1 plus S-tagged CH2, CH3 and CH4 respectively; –/2s, –/3s and –/4s, S-tagged CH2–CH4 only). The migration of high-molecular-mass rainbow marker proteins is shown (M). Blots were analysed using GCH antiserum [19] (a) and S-protein HRP conjugate (b). Results of one of three similar experiments are shown.
DISCUSSION
Tissue-specific alternative splicing of the H4-biopterin-requiring enzyme tyrosine hydroxylase is proposed to allow differential regulation through dopamine binding and cAMP-dependent phosphorylation in Drosophila [27], but human isoforms show no effect of phosphorylation on dopa binding [28]. N-terminal splice variants of GCH with distinct expression patterns have been characterized in Drosophila [29]; however, they cannot be compared with mammalian GCH, which lacks the N-terminal extension present in Drosophila isoforms. Our work investigates the biological role of splice variants of the homodecameric enzyme GCH, which have alternative C-termini.
C-terminal residues in wild-type mammalian GCH contribute to structural elements, intra-ring contacts, catalysis and binding of substrate, GFRP and H4-biopterin (Figure 1a). The lack of the C-terminal β-strand in human GCH splice variants might be expected to interfere with association of the pentameric rings. However, as is shown in the present study by native gel electrophoresis and gel filtration of GCH isoforms expressed in bacterial and mammalian cells, variants assemble into high-molecular-mass oligomers despite C-terminal variation, even in the absence of wild-type GCH (Figures 2a and 6b). Although these results do not allow precise definition of the association state, it is clear that variants do not form monomers or dimers. By comparison with GCH controls, we can say that the splice variants associate very likely into decamers. Although these data do not prove heteromeric interactions, the ability of the C-terminal variants to self-associate suggests that hetero-oligomerization is possible.
Interestingly, the molecular chaperone Hsp90 (heat-shock protein 90) was recently shown to stabilize GCH mutations in DSD [30]. The absence of C-terminal structural elements in splice variants may cause general destabilization of the protein structure and increased susceptibility to proteolytic degradation. Therefore it is not surprising that purification of GCH splice variants expressed recombinantly did not yield the levels obtained for wild-type protein. Using Western-blot analysis of co-expressed GCH isoforms, we found that levels of wild-type enzyme were reduced by the presence of splice variants (Figure 4). The apparent inactivity of bacterially expressed GCH variants [19,22] can be attributed to the absence of active-site residues (His210, Cys212, Met213 and Arg216). Our finding that the enzyme activity of wild-type GCH is reduced 2.5-fold by co-expression with splice variants (Figure 5) suggests an effect through heteromeric interactions.
Indeed, GCH mutants associated with disease are proposed to cause a dominant-negative effect by sequestering functional wild-type enzyme in inactive oligomers [13–15]. Our co-expression studies suggest that splice variants exert their effect by incorporation in heterodecamers. However, it is unclear whether the decreased enzyme activity results directly from active site disruption and loss of positive co-operativity in GCH heterodecamers. The observation of correspondingly low GCH levels in co-expression studies suggests an indirect effect, whereby heteromerization causes destabilization and degradation resulting in lower levels of active enzyme. It should be kept in mind that by using the bicistronic pBudCE4.1 vector, equal expression of wild-type and variant proteins was achieved in the present study, whereas under physiological conditions the amount of variant protein(s) is substantially lower than that of wild-type GCH (see below). However, even low levels of variant protein can be physiologically significant if they effect destabilization of wild-type enzyme through heteromerization.
Splice variants were proposed to regulate GCH by reducing its basal activity, allowing a bigger amplitude of induction [23]. GCH variant mRNAs are present in immune-responsive cells (THP-1 and human dermal fibroblasts) pre-stimulation only at basal levels [19]. However, their increased levels, corresponding to 20% of the wild-type GCH mRNA, in stimulated cells may be a mechanism for non-specific regulation of GCH through rapid degradation. Indeed, sub-stoichiometric amounts of a naturally occurring truncated splice variant of transcription factor E3 cause a dominant-negative effect [31]. The absence of residues involved in GFRP and H4-biopterin binding in the splice variants may serve to uncouple GCH from feedback regulation.
In summary, we have demonstrated the ability of GCH splice variants to form high-molecular-mass oligomers. Co-expressions of GCH splice variants with wild-type enzyme resulted in reduced levels of GCH protein and enzyme activity. One possible explanation of how GCH splice variants interfere with wild-type GCH could be that inactive variants are incorporated into heterodecamers, decreasing the enzyme stability and activity.
Acknowledgments
We thank Petra Loitzl and Renate Kaus for their excellent technical assistance, and Bettina Sarg for performing MS. This work was supported by the Austrian Science Fund (FWF Project P16059).
References
- 1.Thony B., Auerbach G., Blau N. Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem. J. 2000;347:1–16. [PMC free article] [PubMed] [Google Scholar]
- 2.Auerbach G., Herrmann A., Bracher A., Bader G., Gutlich M., Fischer M., Neukamm M., Garrido-Franco M., Richardson J., Nar H., et al. Zinc plays a key role in human and bacterial GTP cyclohydrolase I. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13567–13572. doi: 10.1073/pnas.240463497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harada T., Kagamiyama H., Hatakeyama K. Feedback regulation mechanisms for the control of GTP cyclohydrolase I activity. Science. 1993;260:1507–1510. doi: 10.1126/science.8502995. [DOI] [PubMed] [Google Scholar]
- 4.Milstien S., Jaffe H., Kowlessur D., Bonner T. I. Purification and cloning of the GTP cyclohydrolase I feedback regulatory protein, GFRP. J. Biol. Chem. 1996;271:19743–19751. doi: 10.1074/jbc.271.33.19743. [DOI] [PubMed] [Google Scholar]
- 5.Maita N., Okada K., Hatakeyama K., Hakoshima T. Crystal structure of the stimulatory complex of GTP cyclohydrolase I and its feedback regulatory protein GFRP. Proc. Natl. Acad. Sci. U.S.A. 2002;99:1212–1217. doi: 10.1073/pnas.022646999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maita N., Hatakeyama K., Okada K., Hakoshima T. Structural basis of biopterin-induced inhibition of GTP cyclohydrolase I by GFRP, its feedback regulatory protein. J. Biol. Chem. 2004;279:51534–51540. doi: 10.1074/jbc.M409440200. [DOI] [PubMed] [Google Scholar]
- 7.Hesslinger C., Kremmer E., Hultner L., Ueffing M., Ziegler I. Phosphorylation of GTP cyclohydrolase I and modulation of its activity in rodent mast cells. GTP cyclohydrolase I hyperphosphorylation is coupled to high affinity IgE receptor signaling and involves protein kinase C. J. Biol. Chem. 1998;273:21616–21622. doi: 10.1074/jbc.273.34.21616. [DOI] [PubMed] [Google Scholar]
- 8.Lapize C., Pluss C., Werner E. R., Huwiler A., Pfeilschifter J. Protein kinase C phosphorylates and activates GTP cyclohydrolase I in rat renal mesangial cells. Biochem. Biophys. Res. Commun. 1998;251:802–805. doi: 10.1006/bbrc.1998.9552. [DOI] [PubMed] [Google Scholar]
- 9.Nichol C. A., Smith G. K., Duch D. S. Biosynthesis and metabolism of tetrahydrobiopterin and molybdopterin. Annu. Rev. Biochem. 1985;54:729–764. doi: 10.1146/annurev.bi.54.070185.003501. [DOI] [PubMed] [Google Scholar]
- 10.Werner-Felmayer G., Golderer G., Werner E. R. Tetrahydrobiopterin biosynthesis, utilization and pharmacological effects. Curr. Drug Metab. 2002;3:159–173. doi: 10.2174/1389200024605073. [DOI] [PubMed] [Google Scholar]
- 11.Werner E. R., Gorren A. C., Heller R., Werner-Felmayer G., Mayer B. Tetrahydrobiopterin and nitric oxide: mechanistic and pharmacological aspects. Exp. Biol. Med. 2003;228:1291–1302. doi: 10.1177/153537020322801108. [DOI] [PubMed] [Google Scholar]
- 12.Thony B., Blau N. Mutations in the GTP cyclohydrolase I and 6-pyruvoyl-tetrahydropterin synthase genes. Hum. Mutat. 1997;10:11–20. doi: 10.1002/(SICI)1098-1004(1997)10:1<11::AID-HUMU2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 13.Hirano M., Tamaru Y., Nagai Y., Ito H., Imai T., Ueno S. Exon skipping caused by a base substitution at a splice site in the GTP cyclohydrolase I gene in a Japanese family with hereditary progressive dystonia dopa responsive dystonia. Biochem. Biophys. Res. Commun. 1995;213:645–651. doi: 10.1006/bbrc.1995.2180. [DOI] [PubMed] [Google Scholar]
- 14.Hirano M., Tamaru Y., Ito H., Matsumoto S., Imai T., Ueno S. Mutant GTP cyclohydrolase I mRNA levels contribute to dopa-responsive dystonia onset. Ann. Neurol. 1996;40:796–798. doi: 10.1002/ana.410400517. [DOI] [PubMed] [Google Scholar]
- 15.Hirano M., Yanagihara T., Ueno S. Dominant negative effect of GTP cyclohydrolase I mutations in dopa-responsive hereditary progressive dystonia. Ann. Neurol. 1998;44:365–371. doi: 10.1002/ana.410440312. [DOI] [PubMed] [Google Scholar]
- 16.Ichinose H., Ohye T., Takahashi E., Seki N., Hori T., Segawa M., Nomura Y., Endo K., Tanaka H., Tsuji S., Nagatsu T. Hereditary progressive dystonia with marked diurnal fluctuation caused by mutations in the GTP cyclohydrolase I gene. Nat. Genet. 1994;8:236–242. doi: 10.1038/ng1194-236. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki T., Ohye T., Inagaki H., Nagatsu T., Ichinose H. Characterization of wild-type and mutants of recombinant human GTP cyclohydrolase I: relationship to etiology of dopa-responsive dystonia. J. Neurochem. 1999;73:2510–2516. doi: 10.1046/j.1471-4159.1999.0732510.x. [DOI] [PubMed] [Google Scholar]
- 18.Ichinose H., Ohye T., Matsuda Y., Hori T., Blau N., Burlina A., Rouse B., Matalon R., Fujita K., Nagatsu T. Characterization of mouse and human GTP cyclohydrolase I genes. Mutations in patients with GTP cyclohydrolase I deficiency. J. Biol. Chem. 1995;270:10062–10071. doi: 10.1074/jbc.270.17.10062. [DOI] [PubMed] [Google Scholar]
- 19.Golderer G., Werner E. R., Heufler C., Strohmaier W., Grobner P., Werner-Felmayer G. GTP cyclohydrolase I mRNA: novel splice variants in the slime mould Physarum polycephalum and in human monocytes (THP-1) indicate conservation of mRNA processing. Biochem. J. 2001;355:499–507. doi: 10.1042/0264-6021:3550499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Togari A., Ichinose H., Matsumoto S., Fujita K., Nagatsu T. Multiple mRNA forms of human GTP cyclohydrolase I. Biochem. Biophys. Res. Commun. 1992;187:359–365. doi: 10.1016/s0006-291x(05)81501-3. [DOI] [PubMed] [Google Scholar]
- 21.Hirano M., Imaiso Y., Ueno S. Differential splicing of the GTP cyclohydrolase I RNA in dopa-responsive dystonia. Biochem. Biophys. Res. Commun. 1997;234:316–319. doi: 10.1006/bbrc.1997.6632. [DOI] [PubMed] [Google Scholar]
- 22.Gutlich M., Jaeger E., Rucknagel K. P., Werner T., Rodl W., Ziegler I., Bacher A. Human GTP cyclohydrolase I: only one out of three cDNA isoforms gives rise to the active enzyme. Biochem. J. 1994;302:215–221. doi: 10.1042/bj3020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwu W. L., Yeh H. Y., Fang S. W., Chiang H. S., Chiou Y. W., Lee Y. M. Regulation of GTP cyclohydrolase I by alternative splicing in mononuclear cells. Biochem. Biophys. Res. Commun. 2003;306:937–942. doi: 10.1016/s0006-291x(03)01091-x. [DOI] [PubMed] [Google Scholar]
- 24.Schagger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 25.Werner E. R., Wachter H., Werner-Felmayer G. Determination of tetrahydrobiopterin biosynthetic activities by high-performance liquid chromatography with fluorescence detection. Methods Enzymol. 1997;281:53–61. doi: 10.1016/s0076-6879(97)81008-7. [DOI] [PubMed] [Google Scholar]
- 26.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 27.Vie A., Cigna M., Toci R., Birman S. Differential regulation of Drosophila tyrosine hydroxylase isoforms by dopamine binding and cAMP-dependent phosphorylation. J. Biol. Chem. 1999;274:16788–16795. doi: 10.1074/jbc.274.24.16788. [DOI] [PubMed] [Google Scholar]
- 28.Sura G. R., Daubner S. C., Fitzpatrick P. F. Effects of phosphorylation by protein kinase A on binding of catecholamines to the human tyrosine hydroxylase isoforms. J. Neurochem. 2004;90:970–978. doi: 10.1111/j.1471-4159.2004.02566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLean J. R., Krishnakumar S., O'Donnell J. M. Multiple mRNAs from the Punch locus of Drosophila melanogaster encode isoforms of GTP cyclohydrolase I with distinct N-terminal domains. J. Biol. Chem. 1993;268:27191–27197. [PubMed] [Google Scholar]
- 30.Hwu W. L., Lu M. Y., Hwa K. Y., Fan S. W., Lee Y. M. Molecular chaperones affect GTP cyclohydrolase I mutations in dopa-responsive dystonia. Ann. Neurol. 2004;55:875–878. doi: 10.1002/ana.20122. [DOI] [PubMed] [Google Scholar]
- 31.Roman C., Cohn L., Calame K. A dominant negative form of transcription activator mTFE3 created by differential splicing. Science. 1991;254:94–97. doi: 10.1126/science.1840705. [DOI] [PubMed] [Google Scholar]