Abstract
A thorough analysis, using MS, of aquaporins expressed in plant root PM (plasma membrane) was performed, with the objective of revealing novel post-translational regulations. Here we show that the N-terminal tail of PIP (PM intrinsic protein) aquaporins can exhibit multiple modifications and is differentially processed between members of the PIP1 and PIP2 subclasses. Thus the initiating methionine was acetylated or cleaved in native PIP1 and PIP2 isoforms respectively. In addition, several residues were detected to be methylated in PIP2 aquaporins. Lys3 and Glu6 of PIP2;1, one of the most abundant aquaporins in the PM, occurred as di- and mono-methylated residues respectively. Ectopic expression in Arabidopsis suspension cells of PIP2;1, either wild-type or with altered methylation sites, revealed an interplay between methylation at the two sites. Measurements of water transport in PM vesicles purified from these cells suggested that PIP2;1 methylation does not interfere with the aquaporin intrinsic water permeability. In conclusion, the present study identifies methylation as a novel post-translational modification of aquaporins, and even plant membrane proteins, and may represent a critical advance towards the identification of new regulatory mechanisms of membrane transport.
Keywords: aquaporin, methylation, N-terminal maturation, plasma membrane protein, post-translational modification, root
Abbreviations: AQP, aquaporin; 1meGlu6, monomethyl Glu6; 2meLys3, dimethyl Lys3; ESI, electrospray ionization; FW, fresh weight; MALDI–TOF, matrix-assisted laser-desorption ionization–time-of-flight; MS/MS, tandem MS; PG, pGreenII 0179 binary vector; PM, plasma membrane; PIP, PM intrinsic protein; PSD, post-source decay; T-DNA, transfer DNA; WT, wild-type
INTRODUCTION
Aquaporins define a large family of ubiquitous integral membrane proteins [1] which mediate the transport across membranes of water, small neutral solutes, and occasionally ions. In plants, aquaporins critically contribute to plant water homoeostasis during development and in response to challenging environmental conditions [2]. Most of the aquaporins consist of small (25–34 kDa) hydrophobic proteins assembling in tetramers. All aquaporin monomers share a typical organization with six transmembrane α-helices, the N- and C-terminus being located in the cytoplasm. The extra-membrane parts of the proteins, which include membrane-connecting loops and the N- and C-termini, are the most divergent regions among aquaporin homologues. A large body of data shows that these regions play a critical role in aquaporin expression and function and that these regulations are mediated in part by various post-translational modifications. For instance, the phosphorylation of serine residues at multiple sites was shown to modulate the gating of plant aquaporins [3–5] and the subcellular trafficking of animal aquaporins [6,7]. N-linked glycosylation of mammalian AQP2 (aquaporin-2) in extracellular loop C was recently shown to regulate cell-surface expression of the protein [8]. Intracellular redistribution of the plant vacuolar membrane aquaporin McTIP1;1 in response to osmotic stress also requires glycosylation [9]. Mammalian AQP1 undergoes ubiquitination in fibroblasts, a process which controls the differential stability of the protein between normal and osmotic stress conditions [10]. Finally, controlled proteolysis of the N- and C-terminal tails of AQP0 occurs in lens, but the functional significance of this processing remains as yet unknown [11–13].
Proteins are made of 20 main amino acids, but over 150 post-translationally modified amino acid forms have so far been reported. Thus a tremendous repertoire of modified proteins remains to be uncovered in both prokaryotic and eukaryotic organisms. For instance, protein methylation can occur as either N-methylation of residues such as lysine, arginine, histidine, alanine, proline, glutamine, phenylalanine, asparagine and methionine residues or carboxymethylation, i.e. O-methylesterification of glutamic acid and aspartic acid residues. S-Adenosylmethionine is used as a universal methyl donor and the extent of methylation varies from partial to complete. Whereas methylation (at glutamic acid residues) of bacterial chemotaxis receptors has been characterized as typically reversible [14], N-methylation of eukaryotic proteins has long been considered as irreversible. Yet, histone lysine demethylases have recently been identified [15,16]. Protein methylation affects various cell processes, including protein–protein and protein–nucleic acid interactions, chromatin remodelling and cellular signalling [17,18]. These processes mainly involve the methylation of soluble proteins. There have been only a few reports on methylation of eukaryotic transmembrane proteins and its role has remained elusive [19,20]. Recently, co-expression studies in Xenopus oocytes and mouse cortical collecting duct cells have shown that a novel protein O-methyltransferase can enhance the activity of an epithelial sodium channel, but the methylation site(s) was (were) not identified on the protein [21].
With the objective of revealing novel post-translational regulation of plant aquaporins, we have recently conducted a thorough analysis of the modification status of aquaporins expressed in root PM (plasma membrane) using MS. In a previous study, we have shown that the Arabidopsis root PM contains various PIP (PM intrinsic protein) aquaporin isoforms, including members of both the PIP1 and PIP2 subclasses [22]. Here, we show that the N-terminal tail of PIP aquaporins can exhibit multiple modifications and is differentially processed between the two PIP subclasses. More interestingly, we observed that several residues can be methylated in PIP2 aquaporins. Arabidopsis suspension cells were used to overexpress and characterize methylated WT (wild-type) PIP2;1 and mutants with altered methylation sites.
EXPERIMENTAL
Plant materials
Arabidopsis thaliana L. (Heynh.), ecotype Wassilewskija, plants were cultivated in hydroponic conditions as described in [22]. A. thaliana suspension cells, ecotype Columbia, were cultured at 24 °C under continuous light according to Gerbeau et al. [23].
Gene constructs
Mutated PIP2;1 cDNAs were constructed by using a sense primer that introduces a XhoI restriction site upstream of the start codon and contains the desired mutation, and an antisense primer, which introduces a XbaI restriction site downstream of the stop codon. The oligonucleotides used in the present study are described in Table 1. The resulting PCR products were digested with XbaI and XhoI and cloned into a pBlueScript derivative, downstream of a doubled CaMV35S promoter (CaMV70S) and upstream of the 3′-untranslated transcribed region of a nopaline synthase gene (nos). The resulting plasmid was then digested with both EcoRI and ClaI, and the cDNA-containing inserts were subcloned into the T-DNA (transfer DNA) region of a pGreenII 0179 binary vector (PG) (http://www.pgreen.ac.uk/). This vector contains an nptII gene for kanamycin selection in Escherichia coli and an aphIII gene for hygromycin selection in plants.
Table 1. Oligonucleotide primer sequences.
The primers were used for the mutation and cloning of WT and mutated forms of PIP2;1 in a pGreenII 0179 binary vector (PG), as described in the Experimental section. All primers are given in the 5′–3′ orientation. Cloning sites are indicated in boldface characters, mutated/inserted residues are indicated in italics and the start/stop codons are underlined.
| Primer name | Orientation | Sequence |
|---|---|---|
| K3A-XhoI | Sense | GACTCGAGATGGCAGCGGATGTGGAAGCCGTTCC |
| K3R-XhoI | Sense | CGACTCGAGATGGCAAGGAATGTGGAAGCCGTTCC |
| E6A-XhoI | Sense | GACTCGAGATGGCAAAGGATGTGGCAGCCGTTCC |
| PIP2;1-XhoI | Sense | GACTCGAGATGGCAAAGGATGTGGAAGCCGTTCC |
| PIP2;1-XbaI | Antisense | GATCTAGATTAGACGTTGGCAGCACTTC |
For ectopic expression of WT PIP2;1, we used a similar construct in pGreenII 0179. For water transport measurements, ectopic expression of WT-PIP2;1 was also obtained using pCAMBIA 1301 plasmid vector (Cambia, Canberra, Australia) carrying the PIP2;1 cDNA under the control of a CaMV35S promoter.
Biolistic transformation of suspension cells
Transformation of 5-day-old suspension cells was performed essentially as described in [24]. After bombardment, cells [1.5 g FW (fresh weight)] were transferred on to a culture medium containing 3 g/l Phytagel (Sigma) for 2 days and further transferred in 30 ml of liquid culture medium containing 50 mg/l hygromycin. When the percentage of dead cells reached 99%, the cells were resuspended in a fresh medium without hygromycin. The green calli, which eventually appeared after 5–30 days, were transferred in a medium containing 20 mg/l hygromycin and further amplified after 2 weeks in the presence of 40 mg/l hygromycin. Genomic DNA was extracted from suspension cells using the CTAB (cetyltrimethylammonium bromide) extraction procedure [25] and subjected to PCR to probe for T-DNA insertion.
Protein extraction
Total proteins were extracted from suspension cells (5–10 g FW) as described in [26], with the following modifications. The vacuum-dried pellet was resuspended in 0.1 ml/mg FW of a lysis buffer (9 M urea, 4% CHAPS, 0.5% Triton X-100 and 65 mM dithiothreitol) for 30 min under agitation. The extract was then centrifuged for 10 min at 10000 g and the supernatant containing total proteins was recovered. Protein concentration was measured using a modified Bradford procedure [22].
PM purification
A microsomal fraction was obtained from roots [22] and suspension cells [23]. In the latter case, cells were first homogenized with a Waring blender for 10 s and then fully disrupted in a cell disrupter (Constant System, Warwick, U.K.) at a pressure of 54 MPa. PM vesicles were purified by aqueous two-phase partitioning of a microsomal fraction, in a mixture of poly-(ethylene glycol) 3350/Dextran T-500, 6.4% (w/w) each in the presence of 5 mM KCl, as described in [22]. For both roots and suspension cells, the mean yield of PM extraction was 25 μg of protein/g FW. Extrinsic membrane proteins were stripped with a urea and NaOH treatment according to a previously described procedure [22].
Immunodetection
For Western blots, proteins were separated by SDS/PAGE on 11% acrylamide gels [22] and probed with a primary antibody raised against a 17-amino-acid C-terminal peptide of AtPIP2;1, as described in [22].
For ELISAs, serial 2-fold dilutions in a carbonate buffer (30 mM Na2CO3 and 60 mM NaHCO3, pH 9.5) of 2 μg of total protein extracts or 0.1 μg of PM proteins were loaded in duplicate on Maxisorp immunoplates (Nunc) overnight at 4 °C. The immunodetection was performed according to the manufacturer's instructions with 0.1% Tween 20 and 1% BSA when required. A 1:2000 dilution and a 1:2500 dilution of primary anti-PIP2;1 antibody and of secondary peroxidase-coupled anti-rabbit antibody respectively were successively applied for 2 h at 37 °C. A linear regression between the absorbance signal due to oxidized 2,2′-azinobis-(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt, as read with a multiplate reader (Victor; PerkinElmer), and the amount of total proteins was obtained for each sample and used for relative comparison between samples.
MS methods
Protein digestion and MALDI–TOF (matrix-assisted laser-desorption ionization–time-of-flight) MS analysis were performed as described in [22]. Post-translationally modified peptides were searched using FindMod software (http://us.expasy.org/tools/findmod/) [27], which systematically compares experimental peptide masses that did not match with native protein sequences with those obtained by addition of a putative modification. Mono-, di- and tri-methylations, and acetylation typically correspond to an increment in monoisotopic mass of 14.01, 28.03, 42.04 and 42.01 Da respectively.
PSD (post-source decay) was performed on a BiFlex III mass spectrometer. Peptide parent ions were isolated with a resolution M/ΔM≈100. A FAST™ procedure (Bruker, Bremen, Germany) was used to step scan the reflector tension and to reconstruct the daughter ion spectra; 200 spectra were summed for each reflector tension. An MS-BioTools™ software (Bruker) was used to confirm peptide sequence, and the final interpretation of spectra was done manually. ESI (electrospray ionization)–MS/MS (tandem MS) analyses were performed as described in [22].
Stopped-flow light scattering
Water transport measurements were performed at 20 °C essentially as previously described [23]. Briefly, membranes were diluted 100-fold into an equilibration solution which contained 20 mM KCl and 30 mM TrisHCl (pH 8.3). The shrinking of vesicles was initiated by mixing, in an SFM300 stopped-flow spectro-photometer (Biologic, Claix, France), the vesicles with an equal volume of the same buffer containing 500 mM mannitol. The subsequent kinetics of PM vesicles volume adjustment were followed by 90° light scattering at λex=510 nm. The exponential rate constant, kexp, fitted from the recorded data was used to estimate the osmotic water permeability of the vesicles [23]. It was assumed that, because they were prepared in parallel, membrane vesicles from independent cell lines had similar sizes.
RESULTS
The N-terminus of root PM aquaporins shows isoformspecific maturation
A PM fraction was purified from Arabidopsis roots by aqueous two-phase partitioning and enriched in hydrophobic proteins with a urea and NaOH treatment. In previous studies, the proteomic analysis of a major band at 28 kDa identified the presence of at least six PIP isoforms in this fraction (PIP1;1 and/or PIP1;2, PIP1;5, PIP2;1, PIP2;2, PIP2;4 and PIP2;7) [22,28]. These studies relied on the MS/MS sequencing of 12 tryptic peptides. Here, a more thorough analysis allowed us to identify additional tryptic peptides that cover new sequence regions of the previously identified PIP1 (Table 2) and PIP2 (Table 3). We also performed a systematic investigation of covalent modifications carried by the N-terminal tail of PIPs. For this, a FindMod software was used, which predicts from MALDI–TOF MS ‘fingerprints’, the presence of modifications based on the resulting mass shifts [27]. This study pointed to 11 new peptides that possibly map to the N-terminal region of PIPs and carry putative modifications, which were for most of them confirmed by MS/MS (Tables 2 and 3). In a more general context, FindMod predictions and subsequent sequence analyses allowed us to cover (determine the amino acid sequence by MS) a substantial part of the N-terminus of all PIPs. This corresponded to the entire N-terminal tail of PIP2;1 (Ala2–Arg39) and of PIP1;1 or PIP1;2 (Met1–Arg52). The N-termini of PIP2;2 and PIP2;4 were covered at 80%, whereas that of PIP2;7 was covered at 42% and those of PIP1;3 (or PIP1;4) and PIP1;5 were covered at 30% (Tables 2 and 3).
Table 2. Characterization by MALDI–TOF MS and ESI-MS/MS of N-terminal PIP1 peptides from a root PM extract.
The Table shows peptides that were identified from the 28 kDa band of a root PM fraction enriched in hydrophobic proteins [22] and that corresponded to putative PIP1 tryptic digests. The masses [M+H]+ of peptides are indicated in the first column. Masses in italics correspond to peptides that were described previously [22]. The corresponding sequence predicted by mass ‘fingerprinting’, the sequence determined by MS/MS analysis, the type of modification and the isoform name and position inside the sequence are indicated in the following columns. Peptides with modified residues are indicated in boldface characters. Conventions for post-translational modifications are as follows: acM1, acetyl Met1; 1me, monomethyl.
| Measured mass (Da) [M+H]+ | Predicted peptide sequence | Determined peptide sequence | Modification | Isoform | Position in the sequence |
|---|---|---|---|---|---|
| 1017.55 | VGANKFPER | None | PIP1;1/PIP1;2/PIP1;3/PIP1;4 | 10–18 | |
| 1031.55 | VGANKFPER | 1me | PIP1;1/PIP1;2/PIP1;3/PIP1;4 | 10–18 | |
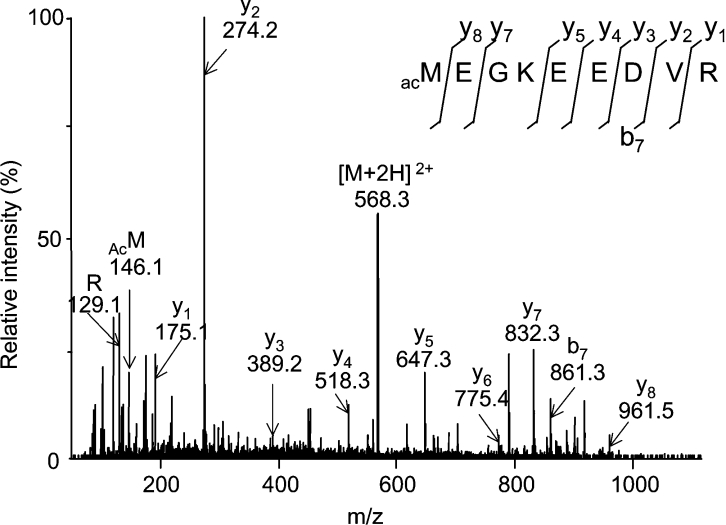
| 1134.52* | MEGKEEDVR+42 Da | MEGKEEDVR | acM1 | PIP1;1/PIP1;2/PIP1;3/PIP1;4 | 1–9 |
| 1537.75 | QPIGTSAQSDKDYK | None | PIP1;1/PIP1;2 | 19–32 | |
| 1638.80 | QPIGTSAQSTDKDYK | None | PIP1;4 | 19–33 | |
| 1660.82 | FPERQPIGTSAQSDK | None | PIP1;1/PIP1;2 | 15–29 | |
| 1960.97 | DYKEPPPAPFFEPGELK | Tag APFFE | None | PIP1;5 | 31–47 |
| 2313.14 | EPPPAPLFEPGELASWSFWR | None | PIP1;2 | 33–52 | |
| 2363.12 | EPPPAPFFEPGELSSWSFWR | EPPPAPFFEPGELSSWSFWR | None | PIP1;1 | 33–52 |
*Peptide, the ESI-MS/MS spectrum of which is shown in Figure 1.
Table 3. Characterization by MALDI–TOF MS and ESI-MS/MS of N-terminal PIP2 peptides from a root PM extract.
The Table shows putative PIP2 peptides that were identified from the 28 kDa band of a root PM fraction as described in Table 2. Peptides with modified residues are indicated in boldface characters. Conventions for post-translational modifications are as follows: 1me, monomethyl; 2me, dimethyl; 1meE6, monomethyl Glu6; 2meK3, dimethyl Lys3.
| Measured mass (Da) [M+H]+ | Predicted peptide sequence | Determined peptide sequence | Modification | Isoform | Position in the sequence |
|---|---|---|---|---|---|
| 886.46 | KWSFYR | None | PIP2;1/PIP2;6 | 34–39/33–38 | |
| 1234.56 | DVEGPEGFQTR | None | PIP2;2 | 4–14 | |
| 1340.65 | DLDVNESGPPAAR | None | PIP2;4 | 4–16 | |
| 1404.70 | DVEAVPGEGFQTR | DVEAVPGEGFQTR | None | PIP2;1 | 4–16 |
| 1418.70 | DVEAVPGEGFQTR+14 Da | 1me | PIP2;1 | 4–16 | |
| 1433.70 | AKDVEGPEGFQTR | Tag PEGFQTR | M1 cleaved | PIP2;2 | 2–14 |
| 1447.70 | AKDVEGPEGFQTR+14 Da | M1 cleaved+1me | PIP2;2 | 2–14 | |
| 1461.70 | AKDVEGPEGFQTR+28 Da | M1 cleaved+2me | PIP2;2 | 2–14 | |
| 1475.70* | AKDVEGPEGFQTR+42 Da | AKDVEGPEGFQTR | M1 cleaved+2meK3+1me | PIP2;2 | 2–14 |
| 1539.78 | AKDLDVNESGPPAAR | M1 cleaved | PIP2;4 | 2–16 | |
| 1603.80 | AKDVEAVPGEGFQTR | AKDVEGPEGFQTR | M1 cleaved | PIP2;1 | 2–16 |
| 1617.80 | AKDVEAVPGEGFQTR+14 Da | M1 cleaved+1me | PIP2;1 | 2–16 | |
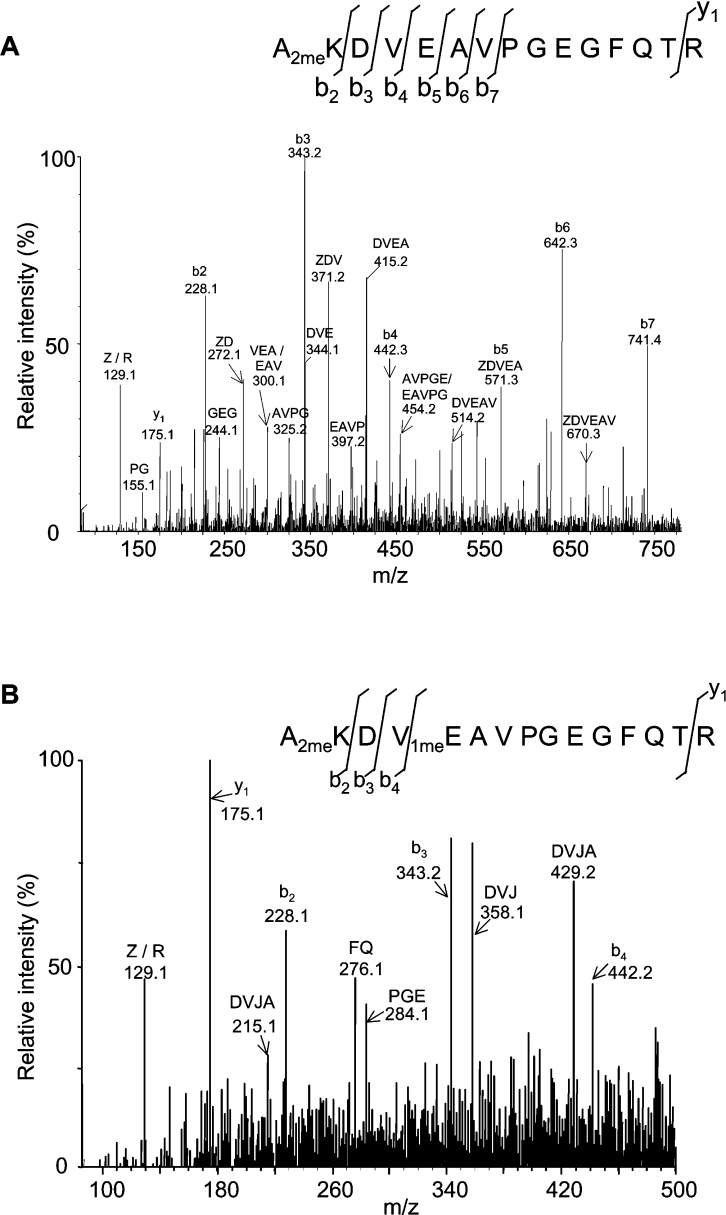
| 1631.80† | AKDVEAVPGEGFQTR+28 Da | AKDVEAVPGEGFQTR | M1 cleaved+2meK3 | PIP2;1 | 2–16 |
| 1645.80† | AKDVEAVPGEGFQTR+42 Da | AKDVEAVPGEGFQTR | M1 cleaved+2meK3+1meE6 | PIP2;1 | 2–16 |
| 1869.93 | DYVDPPPAPLLDMGELK | None | PIP2;7 | 16–32 | |
| 1872.90 | DYQDPPPAPFIDGAELK | DYQDPPPAPFI/LDGAELK | None | PIP2;1 | 17–33 |
| 2000.99 | DYQDPPPAPFIDGAELKK | DYQDPPPAPFI/LDGAEL/IKK | None | PIP2;1 | 17–34 |
| 2066.96 | DYKDPPPAPFFDMEELR | None | PIP2;4 | 17–33 | |
| 2096.94 | DYEDPPPTPFFDADELTK | DYEDPPPTPFFDADEL/ITK | None | PIP2;2 | 15–32 |
*The peptide sequence was deduced from PSD fragmentation. Lys3 was shown to be dimethylated (2meK3). The additional 14 Da mass increment indicates the presence of a methyl group that must be positioned within the following sequence DVEGPEGFQTR.
†Peptide, the ESI-MS/MS spectrum of which is shown in Figure 2.
The peptide with a mass [M+H]+ of 1134.52 Da (Table 2) corresponded to the perfectly conserved N-terminus of the PIP1;1, PIP1;2, PIP1;3 and PIP1;4 isoforms, with the initiating methionine being acetylated (Table 2; Figure 1). The corresponding peptide in PIP1;5, whether acetylated or not, was too small to fall into the detection range of the MALDI instrument.
Figure 1. Representative sequence analysis of an N-terminal PIP1 tryptic peptide from root PM.
A Met1–Arg9 peptide of PIP1 was analysed by ESI–MS/MS and both N-terminal (b-type) and C-terminal (y-type) fragments were used to interpret the spectra. Note that PIP1;1, PIP1;2, PIP1;3 and PIP1;4 isoforms share a perfectly conserved Met1–Arg9 sequence. Analysis was performed on the [M+2H]2+ species of 567.76 Da that corresponds to the [M+H]+ species of 1134.52 Da, as described in Table 2. The whole sequence can be deduced from the spectrum. AcM, acetylated methionine.
MS/MS further established that, by contrast with PIP1s, excision of the initiating methionine occurred in PIP2;1 and PIP2;2 (Table 3, Figure 2). PIP2;4 was also found in the PM fraction [22] and the MALDI detection of a peptide with a mass [M+H]+ of 1539.78 Da strongly suggested that the initiating methionine was also cleaved (Table 3). One peptide specific for PIP2;7 was also detected in the PM fraction (Table 3), but the N-terminal sequence next to the initiating methionine was not covered.
Figure 2. Representative sequence analysis of N-terminal PIP2;1 tryptic peptides from root PM.
Ala2–Arg16 peptides of PIP2;1 were analysed by ESI–MS/MS, and N-terminal (b-type), C-terminal (y-type) and internal fragments were used to interpret the spectra. Fragmented peptides are indicated according to either b and y series, or by their presumed sequence in the case of internal fragments. (A) Analysis was performed on the [M+2H]2+ species of 816.40 Da that corresponds to the [M+H]+ species of 1631.80 Da, as described in Table 3. The whole sequence can be deduced from the spectrum. The initiating methionine is cleaved. Lys3 is dimethylated (2meK). Z=K+28 Da. (B) Analysis was performed on the [M+3H]3+ species of 549.27 Da that corresponds to the [M+H]+ species of 1645.80 Da, as described in Table 3. The whole sequence can be deduced from the spectrum. The initiating methionine is cleaved. Lys3 is dimethylated (2meK), whereas Glu6 is monomethylated (1meE). Z=K+28 Da; J=E+14 Da.
In summary, the initiating methionine was co-translationally processed in all PIP isoforms studied. Whereas it was Nα-acetylated in members of the PIP1 subclass, methionine was cleaved in PIP2 isoforms.
Modification of aquaporins by methylation
‘Fingerprint’ analysis of the tryptic digest also revealed putative PIP2;1 and PIP2;2 peptides with mass increments of approx. 14, 28 and 42 Da (Tables 3 and 4). These can be attributed to methylation, dimethylation, and trimethylation or acetylation respectively. The fragmentation by ESI–MS/MS of a PIP2;1 peptide with a mass [M+H]+ of 1631.80 Da induced N-terminal (b-type), C-terminal (y-type) and internal fragments (Figure 2A). Analysis of these fragments revealed that Lys3 was dimethylated [2meLys3, (dimethyl Lys3)] (Tables 3 and 4, Figure 2A). Note that this modification can be unambiguously identified, due to the high resolution of MS. The fragmentation of a PIP2;1 peptide with a mass [M+H]+ of 1645.80 Da revealed that in addition to being dimethylated at Lys3, the peptide was monomethylated at Glu6 [1meGlu6 (monomethyl Glu6)] (Tables 3 and 4; Figure 2B). Sequence analysis of a homologous N-terminal peptide from PIP2;2 with [M+H]+=1475.70 Da showed that in this isoform also, Lys3 was dimethylated (results not shown). An additional monomethylation site was detected, but could not be unambiguously positioned (Table 3). Because of the presence of a trypsin miscleavage after Lys3, the N-terminal peptides of PIP2;1 and PIP2;2 with the highest signal were identified as Ala2–Arg16 and Ala2-Arg14 peptides respectively. However, trypsin cleavage occurred occasionally and the Asp4–Arg16 and the Asp4–Arg14 peptides of PIP2;1 and PIP2;2 respectively could also be detected (Table 3). Overall, the N-terminal extremity (Ala2–Arg16) of PIP2;1 occurred in the PM fraction in four distinct molecular forms (Tables 3 and 4): unmodified, dimethylated on Lys3 (2meLys3), monomethylated on Glu6 (1meGlu6) and trimethylated (2meLys31meGlu6).
Table 4. Characterization by MS of N-terminal peptides of WT and mutated forms of PIP2;1.
The first column refers to the form of PIP2;1, either WT or mutated. The second column refers to the measured peptide mass. The indicated peptides were identified after trypsin digestion of the proteins. They correspond to all identified forms of the 2–16 N-terminal peptide. Note that, because of an inefficient trypsin cleavage after Lys3, the Asp4–Arg16 peptide represents, with respect to Ala2–Arg16, a minor digestion product almost undetectable in the MALDI–TOF ‘fingerprint’. The predicted sequence is indicated in the third column. The last column refers to the relative abundance (±S.E.M.) in PM fractions from either roots or suspension cells. n refers to the number of MS spectra from at least two independent transformed cell lines or plant cultures. acA, acetyl alanine; 1meE, monomethyl glutamic acid; 2meK, dimethyl lysine.
| Abundance (%) | ||||
|---|---|---|---|---|
| Form expressed | Measured peptide mass (Da) | Predicted sequence | Roots | Suspension cells |
| WT | 1603.80* | AKDVEAVPGEGFQTR | 64±9 | 49±27 |
| 1617.80 | AKDV1meEAVPGEGFQTR | 3±2 | 17±3 | |
| 1631.80* | A2meKDVEAVPGEGFQTR | 7±4 | 9±6 | |
| 1645.80* | A2meKDV1meEAVPGEGFQTR | 26±6 (n=9) | 25±15 (n=4) | |
| E6A | 1545.80* | AKDVAAVPGEGFQTR | 100 (n=2) | |
| K3A | 1546.75 | AADVEAVPGEGFQTR | 13±5 | |
| 1588.75* | acAADVEAVPGEGFQTR | 82±1 | ||
| 1602.75* | acA1meADVEAVPGEGFQTR | 4±4 (n=3) | ||
| K3R | 1631.81 | ARDVEAVPGEGFQTR | 94±6 | |
| 1645.81† | ARDVEAVPGEGFQTR+1me | 4±4 | ||
| 1673.81* | acARDVEAVPGEGFQTR | 2±4 (n=4) | ||
*Sequence determined by MS/MS.
†The position of the methyl group was not determined.
Ectopic expression of PIP2;1 in suspension cells
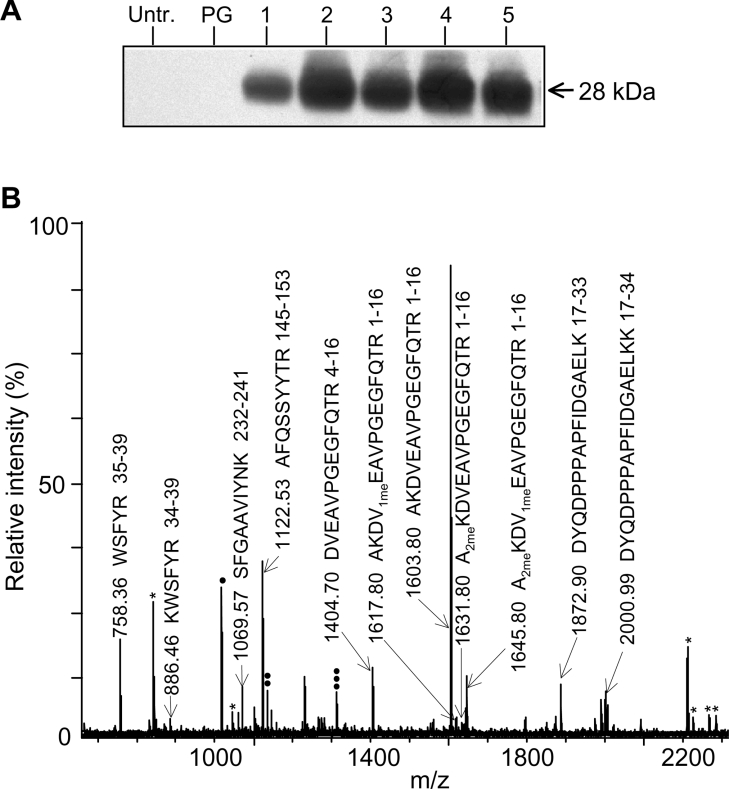
PIP2;1 is one of the most abundant PIPs in the Arabidopsis root PM [22] and supposedly plays a central role in water uptake by roots. By contrast with root cells, Arabidopsis suspension cells show a low endogenous expression level of PIP aquaporins ([23]; V. Santoni, S. Prak and L. Verdoucq, unpublished work). With the prospect of studying N-terminal methylation of PIP2;1 in a simplified system, we evaluated the ability of these cells to express this aquaporin ectopically. Biolistic transformation using a CaMV70S::PIP2;1 expression cassette yielded seven independent transformed cell lines. Western-blot analysis of total protein extracts using an anti-PIP2;1 peptide antibody revealed a significant overexpression of PIP2 aquaporins in these cells, as compared with untransformed cells or cells transformed with an empty vector (PG) (Figure 3A). The 28 kDa band of a PM extract from cells overexpressing PIP2;1 was further analysed by MS. Of the approx. 20 most abundant peptides of the spectrum, 11 were shown to derive from PIP2;1 (Figure 3B). Further MS/MS analysis of the PIP2;1 peptides revealed a modification status similar to that of endogenous PIP2;1 expressed in the root PM (Table 4): the initiating Met was cleaved, and Lys3 and Glu6 were either native or present in dimethylated and monomethylated forms respectively.
Figure 3. Ectopic expression of PIP2;1 in suspension cells.
(A) Western-blot analysis, using an anti-PIP2;1 peptide antibody, of total protein extracts from independent cell lines, either untransformed (Untr.) or transformed with an empty vector (PG) or with a CaMV70S::PIP2;1 construct (lines 1–5). Equal amounts of proteins (5 μg) were loaded on to each lane. (B) MALDI–TOF analysis of the trypsin-cleaved 28 kDa band from PM of suspension cells overexpressing PIP2;1. Peptides derived from PIP2;1 are described by their monoisotopic mass, sequence and position inside the protein sequence. Three peptides were assigned to endogenous PIP aquaporins: (●), [M+H]+=1017.55 Da, attributed to sequence VGANKFPER of PIP1 isoforms; (●●), [M+H]+=1134.52 Da, attributed to sequence acMEGKEEDVR of PIP1;1/PIP1;2/PIP1;3/PIP1;4 isoforms; (●●●), [M+H]+=1312.65 Da, attributed to sequence SFGAAVIYNNEK of PIP2;4/PIP2;7/PIP2;8 isoforms. The trypsin autolysis peptides are also indicated (*). 1meE, monomethylated glutamic acid; 2meK: dimethylated lysine.
A semi-quantitative MS characterization of the methylation status of PIP2;1 was performed, based on the peak intensity of the detected peptides assuming equal ionization efficiencies for peptides with identical primary sequences (Table 4). In both roots and suspension cells, the fully unmodified form was the most abundant. The proportion of peptides containing 1meGlu6 was slightly more abundant in suspension cells (42%) than in roots (29%), as indicated by the addition of their respective peak intensities in the MALDI spectrum (Table 4). By contrast, the proportion of peptides carrying 2meLys3 was similar in suspension cells (34%) and root cells (33%) (Table 4). Thus suspension and root cells appear to have comparable PIP2;1 methylation patterns, and the former cells can be used as an expression system to further investigate N-terminal PIP2;1 methylation.
Modification profile of PIP2;1 mutants with altered methylation sites
With the prospect of dissecting the respective roles of PIP2;1 methylation at positions 3 and 6, we introduced single changes at these positions, the corresponding residues being in particular replaced with Ala (E6A-PIP2;1 and K3A-PIP2;1 mutants). The resulting mutants were then overexpressed in Arabidopsis suspension cells, and their modification profile in purified PM was characterized as described above for WT-PIP2;1 (Table 4).
PM of cells expressing the E6A-PIP2;1 mutant revealed a unique N-terminal peptide. As in the WT form, the initiating methionine was cleaved (Table 4). In addition, the peptide carried no post-translational modification. Thus substitution of alanine for Glu6 resulted in a complete lack of methylation not only at position 6, but also at position 3 (Table 4).
All N-terminal peptides of K3A-PIP2;1 typically showed methionine excision but with a 42 Da mass increment in 86% of the peptides, which was due to an additional co-translational Nα-terminal acetylation [29] (Table 4). MS/MS also showed a residual monomethylation on 4% of the Ala2–Arg16 K3A-PIP2;1 peptide. Curiously, methylation occurred on Ala3 and not on the non-mutated methylation site at position 6 (Table 4). Thus the K3A mutation was unable to fully prevent methylation at this position, whereas it strongly interfered with methylation of the adjacent site. The complex alteration of the PIP2;1 N-terminus modification pattern induced by the K3A mutation prompted us to investigate the effects of a more conservative substitution of Lys3 by an arginine residue. Similar to WT-PIP2;1, the PIP2;1-K3R mutant showed a typical methionine excision (Table 4). We also observed monomethylation and acetylation, but on very minor fractions (2–4%) of the N-terminal peptides (Table 4). Thus the K3R mutation resulted in an almost complete lack of methylations at position 3 and 6. Overall, these and the data obtained with the E6A-PIP2;1 and K3A-PIP2;1 mutants show mutual interactions of methylation at the two sites.
Intrinsic water transport activity of PIP2;1 mutants with altered methylation sites
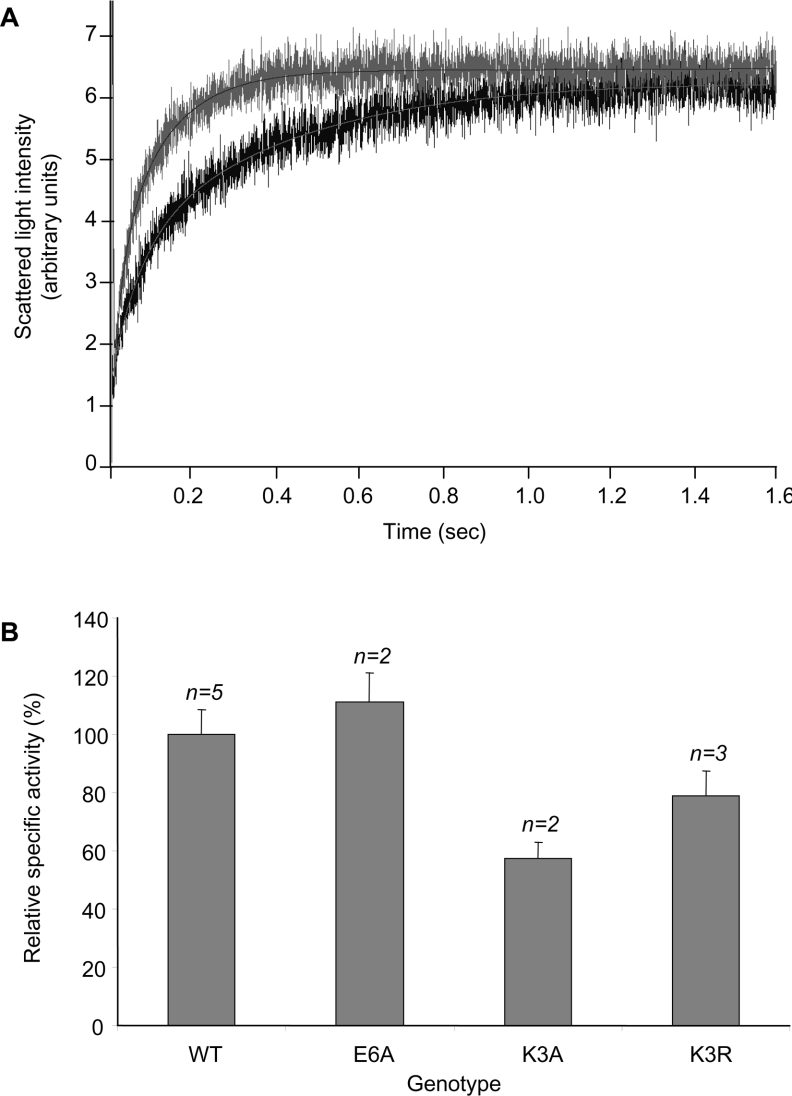
Methylation of PIP2;1 N-terminus may modulate the intrinsic water transport activity of the aquaporin. To explore this possibility, we investigated the water transport activity of the WT and mutant PIP2;1 forms. PM vesicles were purified from at least two representative cell lines of each of the PG, WT-PIP2;1, E6A-PIP2;1, K3A-PIP2;1 and K3R-PIP2;1 genotypes, and water transport in these cells was characterized by means of stopped-flow spectrophotometry. Figure 4(A) shows typical kinetic changes in light scattering as induced by a sudden hyperosmotic challenge on purified vesicles from PG and WT-PIP2;1 genotypes. The exponential rate constant (kexp) of these changes is indicative of the membrane osmotic water permeability. Membranes purified from WT-PIP2;1 and all other transformed cell lines investigated showed a water transport activity that was about 2-fold higher than in membranes from PG cells (Figure 4A; results not shown). These results suggest that both WT and mutant PIP2;1 can be expressed as active forms in the PM of suspension cells. The relationship between the abundance of PIP2;1 forms in purified PM (as assayed by ELISA) and the water permeability of these membranes was used to evaluate the intrinsic transport activity of each form. Figure 4(B) shows that PIP2;1-E6A and PIP2;1-K3R mutants had an intrinsic water transport activity similar to that of PIP2;1-WT. By contrast, the PIP2;1-K3A mutant showed a tendency towards a smaller activity (Figure 4B). Altogether, the results show that mutation of PIP2;1 at Lys3 or Glu6 and/or reduced methylation had no major effect on PIP2;1 water transport activity.
Figure 4. Relationship between water transport activity and PIP2 abundance in purified PM vesicles.
(A) Representative time course of stopped-flow scattered light intensity following imposition of an inwardly directed osmotic gradient on PM vesicles purified from PG (bottom trace) and WT-PIP2;1 (top trace) cells. The data were fitted with a single exponential function with a rate constant of kexp=5.90 and 9.39 s−1 for PG and WT-PIP2;1 cells respectively. (B) PM vesicles were prepared from at least two representative cell lines of each genotype, i.e. from cells containing an empty vector (PG) or overexpressing the WT, E6A, K3A or K3R form of PIP2;1. PM vesicles were probed for their water transport activity by stopped-flow measurements (kexp) and for PIP2 abundance by an ELISA. For each cell line, water transport activity was calculated as the ratio of kexp to PIP2. The specific activity of overexpressed PIP2;1 was deduced by substitution of the mean water transport activity measured in PG cells and was expressed as percentage of the specific activity of WT form. Each measurement was performed in triplicate from the indicated number (n) of independent cell lines.
DISCUSSION
Because of their relatively high abundance, aquaporins provide a unique model to address general aspects of membrane protein expression and regulation. In the present study, we have made use of the power and resolution of MS techniques to reveal, in the natural context of plant root membranes, novel co- and post-translational modifications of PM aquaporins. In roots, distinct maturation profiles of the initiating Met, through acetylation or excision, were exhibited by native members of the PIP1 and PIP2 subclasses respectively. In suspension cells, WT-PIP2;1 and its K3A, K3R and E6A derivatives also showed N-terminal methionine excision. Excision of the N-terminal methionine has been extensively studied in soluble proteins [30,31] and to a much lesser extent in membrane proteins. This process was shown to depend on the nature of penultimate residues, and occurs in the presence of residues with side chains of reduced steric hindrance [30,31]. In agreement with this rule, the three Arabidopsis PIP2s studied (PIP2;1, PIP2;2 and PIP2;4), which exhibit an alanine residue at position 2, all had their methionine excised, whereas PIP1s, which have a perfectly conserved glutamic acid residue, did not. N-terminal acetylation, which has been thoroughly studied in soluble mammalian and yeast proteins, depends on N-terminal sequences [29]. For instance, of 64 eukaryotic proteins examined, which similar to PIP1s exhibited a Met-Glu (or a Met-Asp) N-terminal end, all were acetylated, as were PIP1s [29]. Similar analyses suggested that, although the presence of a serine or alanine residue at the N-terminus as in PIP2s may permit acetylation, the presence of an adjacent basic residue usually prevents this process. Accordingly, WT-PIP2;1 and the K3R-PIP2;1 mutant were not or almost not acetylated (Table 4). By contrast, the substitution of alanine for Lys3 yielded a K3A-PIP2;1 mutant with very significant N-terminal acetylation. Curiously, we found that a minor proportion (∼13%) of the mutant was devoid of acetylation (Table 4). This form could reflect an incomplete modification or be indicative of an as yet unknown deacetylase activity.
In the present study, we also observed that PIP2;1 and PIP2;2 isoforms carried methylated residues, a discrete but less commonly described post-translational modification. Lys3 and Glu6 of PIP2;1, although they existed in their native form, occasionally carried dimethyl (Lys3) or monomethyl (Glu6) moieties (Tables 3 and 4). The methylation of lysine residues has been observed in soluble proteins and recently described in a few mammalian membrane-intrinsic or membrane-associated proteins, including subunit c of mitochondrial ATP synthase [19] or cytochrome c [20]. Also, a proteomic analysis of rat brain membranes uncovered 15 proteins that possibly carried mono-, di- or tri-methylated lysine [32], suggesting that lysine modification might be common in membrane proteins.
Glutamic acid methylesterification is by far less common than lysine methylation, and its observation in PIP2s represents a novel modification for eukaryotic membrane proteins. In bacteria, chemotaxis receptors undergo reversible methylation at glutamic acid residues, a process that modulates their signalling output [33–35]. We were aware that artefactual carboxymethylation of proteins can take place, when these ones are simultaneously treated by trichloroacetic acid and alcohols (methanol), as during gel Coomassie Blue staining [36]. These treatments were, however, carefully avoided in the present study.
Additional analysis indicated that alanine can also be methylated in the K3A-PIP2;1 mutant (Table 4). In that case, we assume that methylation occurs at the peptide bond. Such N-methylation has been described in several biologically active natural peptides and was shown to improve their lipophilicity and proteolytic stability [37]. Its role in membrane proteins is to our knowledge as yet unknown.
The significance of observations made on PIP2;1 N-terminal tail surely goes beyond the context of the present study, since putative methylated residues could be identified in other PIPs such as PIP1;1 (Table 2) and other PM proteins (V. Santoni and L. Verdoucq, unpublished work). These are, to our knowledge, the first observed methylations in aquaporins and even plant membrane proteins.
Methylation of PIP2;1 was examined in detail since this aquaporin is one of the most abundantly expressed aquaporins in plants. Although MALDI–TOF analyses do not allow an absolute quantification of the modified versus unmodified forms of a peptide, their relative peak intensities can be indicative of their relative abundance. Such analyses revealed an interplay between adjacent methylations of PIP2;1 at Lys3 and Glu6 (Table 4). In WT-PIP2;1, the apparent degree of methylation at one position was enhanced by methylation at the other site. For instance, 1meGlu6 was found in 26 and 73% of peptides carrying unmodified Lys3 or 2meLys3 respectively. Conversely, 2meLys3 was found in 15 and 59% of peptides carrying unmodified Glu6 or 1meGlu6 respectively. We also observed that a PIP2;1 mutant with a single E6A substitution did not display any methylation on Lys3 (Table 4). Conversely, the single K3A or K3R substitutions and possibly the altered N-terminal maturation of the former mutant protein prevented any methylation of Glu6 (Table 4). Such interactions between modifications at adjacent sites are reminiscent of observations made in histones. In histone H3 for instance, methylation of Lys4 prevents methylation of Lys9 (for a review, see [38]). Our findings also provide primary information on the specificity of protein methyltransferases involved in modifying the PIP2;1 N-terminal end. They conform to the idea that these enzymes have local recognition sequences that encompass a few residues [39]. Enzymes involved in lysine and glutamic acid methylation of PIP2;1 are very likely distinct in nature, and both remain to be identified.
In recent years, the significance of histone methylation in epigenetic control of gene expression has been extensively studied [40], and the effects of methylation on the activity or the subcellular trafficking of a few other soluble proteins have been elucidated [39,41–43]. Although recent data point to a role of methylation in controlling the activity of an epithelial sodium channel [21], the functional significance of specific methylation events on eukaryotic membrane proteins remains to be elucidated. PIP aquaporins can be gated by phosphorylation of their cytosolic loop B or C-terminal tail [4] or protonation of their cytosolic loop D [44]. To investigate whether methylation of PIP2;1 N-terminal tail plays a similar role, we coupled water transport measurements with aquaporin immunoquantification in PM vesicles expressing WT or mutant PIP2;1 forms. The E6A-PIP2;1 and K3R-PIP2;1 mutants, which both are not methylated at Lys3 and Glu6 (Table 4), exhibited a water transport activity similar to that of WT-PIP2;1. These results suggest that methylation at Lys3 and Glu6 does not interfere with intrinsic water permeability. By contrast, the K3A-PIP2;1 showed a tendency to a lower water transport activity. We suppose that the non-conservative K3A substitution and/or the resulting alteration of the protein maturation profile, including an abnormal Nα-acetylation, may result in a partially inactive protein.
As an alternative approach for investigating the functional significance of aquaporin methylation, we also searched for physiological conditions that would induce changes in the modification profile of root PM aquaporins. We observed that salinity that down-regulates root water transport by inhibiting aquaporins [45] does not alter their methylation profile (S. Prak, C. Maurel and V. Santoni, unpublished work). Since methylation of PIP2;1 seems not to control the gating of this aquaporin, we now investigate the role of this modification in the protein stability and subcellular localization.
In conclusion, the present study identifies methylation as a new post-translational modification of aquaporins, and plant membrane proteins in general, and points to interactions between adjacent methylated sites. Although the functional role of this modification remains as yet unclear, our work may represent a critical advance in identifying new regulatory mechanisms of membrane water transport.
Acknowledgments
We are grateful to Dr Carmela Giglione, Dr Thierry Meinnel (both of the Institut des Sciences du Végétal, Centre National de la Recherche Scientifique, Gif-sur-Yvette Cédex, France) and Dr Hervé Sentenac (Biochimie et Physiologie Moléculaires des Plantes, Agro.M-CNRS-INRA-UM2, Montpellier, France) for critical reading of this paper before its submission. Some MS/MS analyses were performed in the Plateau de Protéomique Fonctionelle, IGF UPR 2580 CNRS, of the Montpellier Languedoc-Roussillon Genopole®. This work was supported in part by Genoplante (AF 2001093).
References
- 1.Zardoya R., Villalba S. A phylogenetic framework for the aquaporin family in eukaryotes. J. Mol. Evol. 2001;52:391–404. doi: 10.1007/s002390010169. [DOI] [PubMed] [Google Scholar]
- 2.Luu D.-T., Maurel C. Aquaporins in a challenging environment: molecular gears for adjusting plant water status. Plant Cell Environ. 2005;28:85–96. [Google Scholar]
- 3.Tornroth-Horsefield S., Wang Y., Hedfalk K., Johanson U., Karlsson M., Tajkhorshid E., Neutze R., Kjellbom P. Structural mechanism of plant aquaporin gating. Nature. 2006;439:688–694. doi: 10.1038/nature04316. [DOI] [PubMed] [Google Scholar]
- 4.Johansson I., Karlsson M., Shukla V. K., Chrispeels M. J., Larsson C., Kjellbom P. Water transport activity of the plasma membrane aquaporin PM28A is regulated by phosphorylation. Plant Cell. 1998;10:451–459. doi: 10.1105/tpc.10.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guenther J. F., Chanmanivone N., Galetovic M. P., Wallace I. S., Cobb J. A., Roberts D. M. Phosphorylation of soybean nodulin 26 on serine 262 enhances water permeability and is regulated developmentally and by osmotic signals. Plant Cell. 2003;15:981–991. doi: 10.1105/tpc.009787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamsteeg E.-J., Heijnen I., van Os C. H., Deen P. M. T. The subcellular localization of an aquaporin-2 tetramer depends on the stoichiometry of phosphorylated and non phosphorylated monomers. J. Cell Biol. 2000;151:919–929. doi: 10.1083/jcb.151.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Balkom B. W. M., Savelkoul P. J., Markovich D., Hofman E., Nielsen S., van der Sluijs P., Brown D., Deen P. M. T. The role of putative phosphorylation sites in the targeting and shuttling of the aquaporin-2 water channel. J. Biol. Chem. 2002;277:41473–41479. doi: 10.1074/jbc.M207525200. [DOI] [PubMed] [Google Scholar]
- 8.Hendricks G., Koudijs M., van Balkom B. W. M., Oorschot V., Klumperman J., Deen P. M. T., van der Sluijs P. Glycosylation is important for cell surface expression of the water channel aquaporin-2 but is not essential for tetramerization in the endoplasmic reticulum. J. Biol. Chem. 2004;279:2975–2983. doi: 10.1074/jbc.M310767200. [DOI] [PubMed] [Google Scholar]
- 9.Vera-Estrella R., Barkla B. J., Bonhert H. J., Pantoja O. Novel regulation of aquaporins during osmotic stress. Plant Physiol. 2004;135:2318–2329. doi: 10.1104/pp.104.044891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leitch V., Agre P., King L. S. Altered ubiquitination and stability of aquaporin-1 in hypertonic stress. Proc. Natl. Acad. Sci. U.S.A. 2001;98:2894–2898. doi: 10.1073/pnas.041616498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ball L. E., Little M., Nowak M. W., Garland D. L., Crouch R. K., Schey K. L. Water permeability of C-terminally truncated aquaporin 0 (AQP0 1–243) observed in the aging human lens. Invest. Ophthalmol. Visual Sci. 2003;44:4820–4828. doi: 10.1167/iovs.02-1317. [DOI] [PubMed] [Google Scholar]
- 12.Ball L. E., Garland D. L., Crouch R. K., Schey K. L. Post-translational modifications of aquaporin-0 (AQP0) in the normal human lens: spatial and temporal occurrence. Biochemistry. 2004;43:9856–9865. doi: 10.1021/bi0496034. [DOI] [PubMed] [Google Scholar]
- 13.Schey K. L., Fowler J. G., Shearer T., David L. Modifications to rat lens major intrinsic protein in selenite-induced cataract. Invest. Ophthalmol. Visual Sci. 1999;40:657–667. [PubMed] [Google Scholar]
- 14.Kim C., Jackson M., Lux R., Khan S. Determinants of chemotactic signal amplification in Escherichia coli. J. Mol. Biol. 2001;301:119–135. doi: 10.1006/jmbi.2000.4389. [DOI] [PubMed] [Google Scholar]
- 15.Tsukada Y., Fang J., Erdjument-Bromage H., Warren M. E., Borchers C. H., Tempst P., Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 16.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., Casero R. A., Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 17.McBride A. E., Silver P. A. State of the arg: protein methylation at arginine comes of age. Cell. 2001;106:5–8. doi: 10.1016/s0092-8674(01)00423-8. [DOI] [PubMed] [Google Scholar]
- 18.Aletta J. M., Cimato T. R., Ettinger M. J. Protein methylation: a signal event in post-translational modification. Trends Biochem. Sci. 1998;23:89–91. doi: 10.1016/s0968-0004(98)01185-2. [DOI] [PubMed] [Google Scholar]
- 19.Chen R., Fearnley I. M., Palmer D. N., Walker J. E. Lysine 43 is trimethylated in subunit c from bovine mitochondrial ATP synthase and in storage bodies associated with Batten disease. J. Biol. Chem. 2004;279:21883–21887. doi: 10.1074/jbc.M402074200. [DOI] [PubMed] [Google Scholar]
- 20.Kluck R. M., Ellerby L. M., Ellerby H. M., Naiem S., Yaffe M. P., Margoliash E., Bredesen D., Mauk A. G., Sherman F., Newmeyer D. D. Determinants of cytochrome c pro-apoptotic activity. The role of lysine 72 trimethylation. J. Biol. Chem. 2000;275:16127–16133. doi: 10.1074/jbc.275.21.16127. [DOI] [PubMed] [Google Scholar]
- 21.Edinger R. S., Yospin J., Perry C., Kleyman T. R., Johnson J. P. Regulation of epithelial Na+ channels (ENaC) by methylation: a novel methyltransferase stimulates ENaC activity. J. Biol. Chem. 2006;281:9110–9117. doi: 10.1074/jbc.M509232200. [DOI] [PubMed] [Google Scholar]
- 22.Santoni V., Vinh J., Pflieger D., Sommerer N., Maurel C. A proteomic study reveals novel insights into the diversity of aquaporin forms expressed in the plasma membrane of plant roots. Biochem. J. 2003;373:289–296. doi: 10.1042/BJ20030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerbeau P., Amodeo G., Henzler T., Santoni V., Ripoche P., Maurel C. The water permeability of Arabidopsis plasma membrane is regulated by divalent cations and pH. Plant J. 2002;30:71–81. doi: 10.1046/j.1365-313x.2002.01268.x. [DOI] [PubMed] [Google Scholar]
- 24.Savino G., Briat J.-F., Lobréaux S. Inhibition of the iron-induced ZmFer1 maize ferritin gene expression by antioxidants and serine/threonine phosphatase inhibitors. J. Biol. Chem. 1997;272:33319–33326. doi: 10.1074/jbc.272.52.33319. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J., Fritsch E. F., Maniatis T. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 26.Santoni V., Bellini C., Caboche M. Use of two-dimensional protein-pattern analysis for the characterization of Arabidopsis thaliana mutants. Planta. 1994;192:557–566. [Google Scholar]
- 27.Wilkins M. R., Gasteiger E., Gooley A. A., Herbert B. R., Molloy M. P., Binz P. A., Ou K., Sanchez J.-C., Bairoch A., Williams K. L., Hochstrasser D. F. High-throughput mass spectrometric discovery of protein post-translational modifications. J. Mol. Biol. 1999;289:645–657. doi: 10.1006/jmbi.1999.2794. [DOI] [PubMed] [Google Scholar]
- 28.Javot H., Lauvergeat V., Santoni V., Martin F., Güclü J., Vinh J., Heyes J., Franck K. I., Schäffner A. R., Bouchez D., Maurel C. Role for a single aquaporin isoform in root water uptake. Plant Cell. 2003;15:509–522. doi: 10.1105/tpc.008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poledova B., Sherman F. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J. Mol. Biol. 2003;325:595–622. doi: 10.1016/s0022-2836(02)01269-x. [DOI] [PubMed] [Google Scholar]
- 30.Moerschell R. P., Hosokawa Y., Tsunasawa S., Sherman F. The specificities of yeast methionine aminopeptidase and acetylation of amino-terminal methionine in vivo. J. Biol. Chem. 1990;265:19638–19643. [PubMed] [Google Scholar]
- 31.Giglione C., Boularot A., Meinnel T. Protein N-terminal methionine excision. Cell Mol. Life Sci. 2004;61:1455–1474. doi: 10.1007/s00018-004-3466-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu C. C., MacCoss M. J., Howell S. A., Yates J. R. A method for the comprehensive proteomic analysis of membrane proteins. Nat. Biotech. 2003;21:532–538. doi: 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- 33.Springer W. R., Koshland D. E. J. Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc. Natl. Acad. Sci. U.S.A. 1977;74:533–537. doi: 10.1073/pnas.74.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simms S. A., Subbaramaiah K. The kinetic mechanism of S-adenosyl-L-methionine: glutamylmethyltransferase from Salmonella typhimurium. J. Biol. Chem. 1991;266:12741–12746. [PubMed] [Google Scholar]
- 35.Djordjevic S., Goudreau P. N., Xu Q., Stock A. M., West A. H. Structural basis for methylesterase CheB regulation by a phosphorylation-activated domain. Proc. Natl. Acad. Sci. U.S.A. 1998;95:1381–1386. doi: 10.1073/pnas.95.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haebel S., Albrecht T., Sparbier K., Walden P., Körner R., Steup M. Electrophoresis-related protein modification: alkylation of carboxy residues revealed by mass spectrometry. Electrophoresis. 1998;19:679–686. doi: 10.1002/elps.1150190513. [DOI] [PubMed] [Google Scholar]
- 37.Haviv F., Fitzpatrick T. D., Swenson R. E., Nichols C. J., Mort N. A., Bush E. N., Diaz G., Bammert G., Nguyen A., Rhutasel N. S., et al. Effect of N-methyl substitution of the peptide bonds in luteinizing hormone-releasing hormone agonists. J. Med. Chem. 1993;36:363–369. doi: 10.1021/jm00055a007. [DOI] [PubMed] [Google Scholar]
- 38.Turner B. M. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- 39.Chuikov S., Kurash J. K., Wilson J. R., Xiao B., Justin N., Ivanov G. S., McKinney K., Tempst P., Prives C., Gamblin S. J., et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 40.Kouzarides T. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 41.Green D. M., Marfatia K. A., Crafton E. B., Zhang X., Cheng X., Corbett A. H. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 2002;277:7752–7760. doi: 10.1074/jbc.M110053200. [DOI] [PubMed] [Google Scholar]
- 42.Liang X., Lu Y., Wilkes M., Neubert T. A., Resh M. D. The N-terminal SH4 region of the Src family kinase fyn is modified by methylation and heterogeneous fatty acylation. J. Biol. Chem. 2004;279:8133–8139. doi: 10.1074/jbc.M311180200. [DOI] [PubMed] [Google Scholar]
- 43.Smith W. A., Schurter B. T., Wong-Staal F., David M. Arginine methylation of RNA helicase A determines its subcellular localization. J. Biol. Chem. 2004;279:22795–22798. doi: 10.1074/jbc.C300512200. [DOI] [PubMed] [Google Scholar]
- 44.Tournaire-Roux C., Sutka M., Javot H., Gout E., Gerbeau P., Luu D.-T., Bligny R., Maurel C. Early effects of salinity on water transport in Arabidopsis roots. Molecular and cellular features of aquaporin expression. Nature. 2003;425:393–397. doi: 10.1038/nature01853. [DOI] [PubMed] [Google Scholar]
- 45.Boursiac Y., Chen S., Luu D.-T., Sorieul M., van den Dries N., Maurel C. Gating of aquaporins by cytosolic pH regulates root water transport during anoxic stress. Plant Physiol. 2005;139:790–805. doi: 10.1104/pp.105.065029. [DOI] [PMC free article] [PubMed] [Google Scholar]