Abstract
Cyclotides are a fascinating family of plant-derived peptides characterized by their head-to-tail cyclized backbone and knotted arrangement of three disulfide bonds. This conserved structural architecture, termed the CCK (cyclic cystine knot), is responsible for their exceptional resistance to thermal, chemical and enzymatic degradation. Cyclotides have a variety of biological activities, but their insecticidal activities suggest that their primary function is in plant defence. In the present study, we determined the cyclotide content of the sweet violet Viola odorata, a member of the Violaceae family. We identified 30 cyclotides from the aerial parts and roots of this plant, 13 of which are novel sequences. The new sequences provide information about the natural diversity of cyclotides and the role of particular residues in defining structure and function. As many of the biological activities of cyclotides appear to be associated with membrane interactions, we used haemolytic activity as a marker of bioactivity for a selection of the new cyclotides. The new cyclotides were tested for their ability to resist proteolysis by a range of enzymes and, in common with other cyclotides, were completely resistant to trypsin, pepsin and thermolysin. The results show that while biological activity varies with the sequence, the proteolytic stability of the framework does not, and appears to be an inherent feature of the cyclotide framework. The structure of one of the new cyclotides, cycloviolacin O14, was determined and shown to contain the CCK motif. This study confirms that cyclotides may be regarded as a natural combinatorial template that displays a variety of peptide epitopes most likely targeted to a range of plant pests and pathogens.
Keywords: cyclic cystine knot, cyclotide, cycloviolacin, kalata B1, sequence variation, Viola odorata
Abbreviations: CCK, cyclic cystine knot; CSI, chemical shift index; HSQC, heteronuclear single-quantum coherence; LC, liquid chromatography; MALDI–TOF, matrix-assisted laser-desorption ionization–time-of-flight; MS/MS, tandem MS; NOE, nuclear Overhauser effect; RBC, red blood cell; rmsd, root mean square deviation; RP-HPLC, reverse-phase HPLC; TEPA, tetraethylenepentamine
INTRODUCTION
Cyclotides are a recently characterized but rapidly growing family of head-to-tail macrocyclic plant-derived peptides. Typically containing 28–37 amino acids, they are defined by their unique knotted disulfide topology linking six conserved cysteine residues and their characteristic cyclic backbone [1–6]. These structural features, referred to as the CCK (cyclic cystine knot) motif [1,7,8] and illustrated in Figure 1, engender cyclotides with the remarkable thermal, chemical and enzymatic stability that has made them a source of interest [3]. Understanding the distribution and diversity of cyclotides has led to extensive efforts to discover new members of the family. To date, more than 80 cyclotides have been characterized from the plant families Rubiaceae, Violaceae and Cucurbitaceae, with a recent paper estimating that not only could individual plants contain up to 100 cyclotides, but that there may be in excess of 9000 different cyclotide sequences present in Nature [9]. Diversity of sequences appears to exist not just between species, but evidence is beginning to emerge that the suite of cyclotides produced within a species varies seasonally [10].
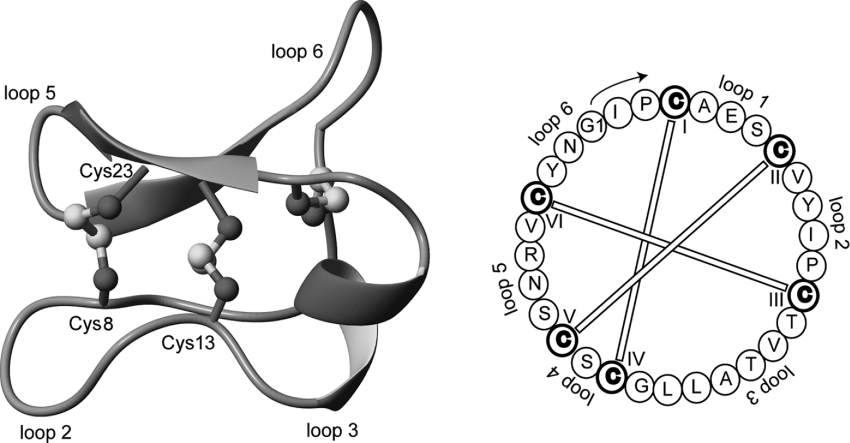
Figure 1. Structure (PDB ID: 1NBJ) and sequence of the cyclotide cycloviolacin O1 [1].
Cyclotides have a well-defined three-dimensional structure as a result of their interlocking disulfide bonds and cyclic peptide backbone. Selected backbone loops and cysteine residues are labelled on the structure to help orientation. By way of example, loop 1 corresponds to the segment between Cys4 and Cys8. The starting point for residue numbering used in the present paper is indicated on the sequence diagram to the right of the Figure and is based on the presumed processing point from the precursor protein [26].
Although the native function of cyclotides in plants is thought to be one of defence against insect pests and pathogens [11,12], a wide variety of biological activities are observed throughout the family. These include uterotonic, insecticidal, anti-HIV, antimicrobial, antineurotensive, cytotoxic and haemolytic activity [13–18]. In fact, the pharmacological utility of cyclotides has long been exploited, albeit unknowingly, by the African Lulua tribe where a boiled concoction of the plant Oldenlandia affinis was traditionally used to accelerate labour [19]. The active peptidic compound was structurally characterized in 1995 [20] and shown to contain the unusual features of a cyclic backbone and knotted arrangement of three disulfide bonds. This peptide, kalata B1, has since become the prototypic member of the cyclotide family. Although cyclotides are by far the largest known family of circular peptides, a range of other cyclic peptides from bacteria, plants and animals has been discovered over recent years [21,22] and a web-based database has been created to collate them [23].
It is convenient to discuss cyclotide sequences in terms of the backbone segments, or loops, between the six conserved cysteine residues that make the cystine knot. These segments, referred to as loops 1–6, are defined in Figure 1 for cycloviolacin O1, the first cyclotide discovered from Viola odorata [1]. Furthermore, cyclotides fall into one of two main structural subfamilies, Möbius and bracelet. Möbius cyclotides, the less common of the two subfamilies, contains a cis-proline residue in loop 5 that induces a local 180° backbone twist, whereas bracelet cyclotides contain a ring of all-trans peptide bonds linking their amino acids. Although the presence/absence of the cis-proline residue is the defining feature of the two subfamilies, they also tend to have high sequence homologies for several loops within subfamilies. To trace evolutionary relationships it is of interest to obtain more sequences to see if homology relationships are more broadly present.
The chemical, structural and biological properties of cyclotides suggest that they may be capable of overcoming the poor stability and susceptibility to proteolysis that restrict the utility of peptides in pharmaceutical and biotechnological applications. The potential for cyclotides to be used as frameworks/scaffolds for the incorporation of peptide epitopes to enhance their use as therapeutic agents is being explored [3,24]. Improvements in technologies for the production of peptides by chemical synthesis and transgenic methods have created an opportunity for the development and exploitation of peptides for a wide range of applications. To realize these applications, a better understanding of the natural diversity of cyclotides, the plasticity of the CCK motif, the nature of their stability and the practical requirements in the design and production of cyclotides is required.
The present paper reports 13 new cyclotide sequences from the plant V. odorata, practically doubling the number of cyclotides characterized from this species [1,25,26]. Haemolytic assays, incubation with proteolytic enzymes and solution-based NMR structural studies reveal that sequence variations greatly affect the haemolytic activity but do not induce significant changes in the global structure or stability of cyclotides. The findings reinforce the view that cyclotides may be thought of as natural combinatorial templates in which well-defined core structures are decorated with a diverse array of sequences. The combinatorial variety of these sequences and the large number of cyclotides expressed per plant presumably have evolved to allow plants to simultaneously target a range of pests or pathogens.
EXPERIMENTAL
Extraction and isolation of peptides from V. odorata
Above- and below-ground plant material from V. odorata was collected mid-morning during summer and winter from an outdoor field at the University of Queensland. Fresh plant material (∼1.5 kg) was ground and extracted with dichloromethane/methanol (1:1, v/v) overnight at room temperature (22 °C). The extract was partitioned with dichloromethane and water and the aqueous layer was concentrated on a rotary evaporator prior to freezedrying. The aqueous layer was then passed through a C18 flash column to remove impurities. Specifically, a C18/methanol slurry was made and poured into a column and then packed using compressed air and washed with Milli Q water. The aqueous extract was then loaded on to the column and eluted with increasing concentrations of solvent B (acetonitrile/water/trifluoro acetic acid; 9:1:0.005, by vol.). The eluent was concentrated on a rotary evaporator and freeze-dried in preparation for RP-HPLC (reverse-phase HPLC).
For preparative RP-HPLC, an Agilent 1100 series system with degasser, quad pump, variable wavelength detector and Phenomenex Jupiter column (C18; 250 mm×10 mm; 15 μm) was used. The sample was dissolved in 100% solvent A (water/trifluoro acetic acid; 10:0.005, v/v), loaded on to the column and eluted with a linear gradient from 20% solvent B to 80% solvent B over 70 min at a flow rate of 8 ml/min. RP-HPLC fractions were analysed on a Micromass LCT mass spectrometer equipped with a z-spray (nebulizer-assisted electrospray) atmospheric pressure ionization source to identify possible new cyclotides [1]. Samples (10 μl) were injected into a moving solvent (0.2 ml/min; 75% acetonitrile/0.1% formic acid), coupled directly with the ionization source via a fused silica capillary interface (50 mm internal diameter×50 cm length). Sample droplets were ionized with a positive capillary potential of 4 kV and entered the analyser through the z-spray source. The sample and extraction cone potentials were 30 and 15 V respectively. Data were acquired over the mass range m/z 400–2000.
To separate peptides with similar retention times under the above conditions, a TEPA (tetraethylenepentamine) buffer was substituted for solvents A and B to decrease peak tailing [27]. The TEPA buffer (water/H3PO4; 10:0.13, v/v, adjusted to pH 2.3 using triethylamine) was used to make buffers A and B [TEPA/acetonitrile in 9:1 and 2:3 (v/v) ratios respectively]. As the TEPA buffer resulted in the formation of salt adducts, the samples were analysed on a Voyager DE-STR MALDI–TOF (matrix-assisted laser-desorption ionization–time-of-flight) mass spectrometer. The samples were then desalted using the trifluoro acetic acid buffer (solvent B) on an analytical HPLC system and appropriate fractions were freeze-dried and prepared for MS/MS (tandem MS).
Reduction of peptides and MALDI-MS analysis
To approx. 6 nmol of peptide in 20 μl of 0.1 M ammonium bicarbonate (pH 8.0), 1 μl of 0.1 M TCEP was added and the solution was incubated at 65 °C for 10 min. Chemical reduction was confirmed by MALDI–TOF-MS after desalting using Ziptips (Millipore), which involved several washing steps followed by elution in 10 μl of 80% acetonitrile (v/v) containing 0.5% formic acid in water. The desalted samples were mixed in a 1:1 ratio (v/v) with matrix consisting of a saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile containing 0.5% aqueous formic acid. A Voyager DE-STR mass spectrometer was used to examine the samples: 200 shots per spectra were acquired in positive-ion reflector mode. The laser intensity was set to 2200, the accelerating voltage was set to 20000 V; the grid voltage set to 64% of the accelerating voltage and the delay time was 165 ns. The low mass gate was set to 500 Da. Data were collected between 500 and 5000 Da. Calibration was undertaken using a peptide mixture (MSCal1) obtained from Sigma–Aldrich.
Enzymatic digestion and nanospray MS/MS sequencing
To the reduced peptide (in 0.1 M ammonium bicarbonate, pH 8.0), trypsin, endoproteinase Glu-C, or a combination of both, was added to give a final peptide/enzyme ratio of 50:1 (w/v). The trypsin incubation was allowed to proceed for 1 h at 37 °C, the endoproteinase Glu-C digestion was over 3 h, whereas for the combined digestion trypsin was added for 1 h followed by endoproteinase Glu-C for a further 3 h. The digestions were quenched by the addition of an equal volume of aq. 0.5% formic acid and the solutions were desalted using Ziptips (Millipore). Samples were stored at 4 °C prior to analysis. The fragments resulting from the digestion were examined firstly by MALDI–TOF-MS followed by sequencing by nanospray MS/MS on an Applied Biosystems QStar mass spectrometer. A capillary voltage of 900 V was applied and spectra were acquired between m/z 60 and 2000 for both TOF spectra and product-ion spectra. The collision energy for peptide fragmentation was varied between 10 and 50 V, depending on the size and charge of the ion. The Analyst software program was used for data acquisition and processing. The MS/MS spectra were examined and the peptides were sequenced based on the presence of both b- and y-series of ions (N- and C-terminal fragments). Amino acid analysis was carried out on all peptides to confirm sequence composition (Australian Proteome Analysis Facility, Macquarie University, New South Wales, Australia). Chymotrypsin digests using the same conditions as for trypsin were also conducted to confirm the results and to identify the positions of leucine residues, as MS/MS alone cannot distinguish isoleucine from leucine. In the case of cycloviolacin O19, where isoleucine or leucine could not be distinguished using chymotrypsin digestion due to the presence of an adjacent proline, assignment of isoleucine/leucine was based on the results of amino acid analysis and by sequence homology with the co-eluting cycloviolacin O8.
LC (liquid chromatography)/MS analysis
A sample of the dried product after C18 flash column purification was used for LC/MS. This was dissolved in 10% solvent B and an aliquot of 100 μl was injected on the LC/MS. LC/MS analysis was performed on an Agilent 1100 Series HPLC system with Phenomenex Jupiter column (C18; 150 mm×2 mm; 5 μm; equipped with a SecurityGuard guard column) run at 200 μl/min. Peptides were eluted with a 1%/min gradient from 25 to 75% acetonitrile/0.1% formic acid (v/v). The eluent was fed directly (i.e. without a split system) into the ion spray source of a Micromass LCT time-of-flight mass spectrometer controlled by a PC running MassLynx version 3.5. Mass spectra were obtained in positive-ion mode over a range m/z 500–2000. While it is always possible that some cyclotides may elute prior to 25% acetonitrile/0.1% formic acid (v/v), the general hydrophobicity of cyclotides, one of the characteristics that expedites their separation from the residual plant material, typically means that they elute later than 30% acetonitrile/0.1% formic acid (v/v).
Haemolytic activity and stability against proteolytic degradation
Peptides were dissolved in water and serially diluted in PBS to give 20 μl test solutions in a 96-well U-bottomed microtitre plate (Nunc). Human type A RBCs (red blood cells) were washed with PBS and centrifuged at 1500 g for 60 s in a microcentrifuge several times until a clear supernatant was obtained. A 0.25% suspension of washed RBCs in PBS (100 μl) was added to the peptide solutions. The plate was incubated at 37 °C for 1 h and centrifuged at 150 g for 5 min. Aliquots of 100 μl were transferred to a 96-well flat-bottomed microtitre plate (Falcon) and the absorbance was measured at 405 nm with an automatic Multiskan Ascent plate reader (Labsystems). The amount of haemolysis was calculated as the percentage of maximum lysis (1% Triton X-100 control) after adjusting for minimum lysis (PBS control). Synthetic melittin (Sigma) was used for comparison. The haemolytic dose necessary to lyse 50% of the RBCs (HD50) was calculated using the regression constant from the linear portion of the haemolytic titration curve (Graphpad Prism software).
Enzymatic digestion assays were performed at a 50:1 substrate (peptide)/enzyme ratio (w/v) for trypsin, pepsin and thermolysin. A linear 34-amino-acid peptide corresponding to a truncated form of the bacterial chaperone Escherichia coli DnaK-(577–610) (DnaK-f) was utilized as a control peptide [28]. Kalata B1 was also included for comparison. For all assays, peptides were incubated with enzyme at 37 °C for up to 6 h [9]. Peptide solutions were prepared in the appropriate buffer as follows. Trypsin and thermolysin stocks were prepared in 100 mM ammonium bicarbonate buffer (pH 8) (containing 10 mM CaCl2 for thermolysin digests) and their incubation with the peptide was halted upon a 5 μl injection into 95 μl of 0.5% formic acid solution. Pepsin stock solution was prepared in 100 mM acetic acid/formic acid (1:1, v/v) at pH 2 and the reaction was halted by diluting into 100 mM ammonium bicarbonate buffer (pH 8). All digestion assay data were analysed by LC/MS chromatography on an Agilent 1100 series HPLC coupled with a QStar mass spectrometer (Applied Biosystems) equipped with an electrospray ionization source.
NMR sample analysis
Samples of cycloviolacin O14 were dissolved in 90% water/10% 2H2O (v/v) or 100% 2H2O to a concentration of 1.5 mM at pH 6.7 and 3.5. Spectra were recorded on Bruker ARX 500 or Bruker ARX 600 spectrometers at sample temperatures in the range 280–320 K. For resonance assignment and structure determination, a set of two-dimensional spectra, including double quantum-filtered COSY, TOCSY with a mixing period of 80 ms, ECOSY (exclusive COSY) and NOESY with mixing times of 100 and 200 ms, were recorded. HSQC (heteronuclear single-quantum coherence) spectra were recorded with the spectral widths in the 1H and 13C dimensions as 6010 and 12073 Hz respectively. All two-dimensional spectra were collected over 4096 data points in the f2 dimension and 512–600 increments in the f1 dimension and processed using XWINNMR (Bruker). Chemical shifts were referenced to internal sodium 2,2-dimethyl-2-silapentane-5-sulfonate.
Structure calculations
Interproton distance restraints for cycloviolacin O14 were derived from cross-peaks in a NOESY spectrum recorded with a mixing time of 200 ms. Cross-peaks were analysed and integrated using the program SPARKY [29] and initial structures were calculated within the DYANA package [30]. After an iterative process in which preliminary structures were used to resolve ambiguities, a set of 299 distance restraints, including 143 sequential, 77 medium and 79 long-range restraints, was derived for O14. Additionally, the spectral data yielded 14 backbone dihedral angle restraints based on 3JHNHα coupling constants derived from the splitting of the amide signals in the COSY and one-dimensional spectra and 4 φ1 dihedral angles based on 3JHαHβ coupling constants derived from an ECOSY spectrum. Five hydrogen bonds from amide exchange experiments were included in the calculations for the preliminary structures. Amide chemical shift temperature coefficients were also measured and these supported the slow exchange assignments. To determine the most likely disulfide connectivity, all 15 disulfide-bonding arrangements were tested and their target functions were compared. The pattern best fitting the restraint data was then selected and used for further structure refinement. After initial structure calculations using DYANA [30], sets of 50 structures were calculated using a torsion angle simulated annealing protocol within CNS 1.1 [31]. This protocol involves a high-temperature phase comprising 4000 steps of 0.015 ps of torsion angle dynamics, a cooling phase with 4000 steps of 0.015 ps torsion angle dynamics, and finally an energy minimization phase comprising 500 steps of Powell minimization. The resultant structures were subjected to further molecular dynamics and energy minimization in a water shell [32]. The refinement in explicit water involves the following steps: first heating to 500 K via steps of 100 K, each comprising 50 steps of 0.005 ps Cartesian dynamics; secondly, 2500 steps of 0.005 ps of Cartesian dynamics at 500 K before a cooling phase where the temperature is lowered in steps of 100 K, each comprising 2500 steps of 0.005 ps Cartesian dynamics; finally, the structures were minimized with 2000 steps of Powell minimization. Structures were analysed using the programs PROMOTIF [33] and PROCHECK-NMR [34].
Homology modelling of cycloviolacin O25
A model of the structure of cycloviolacin O25 was derived based on that of cycloviolacin O1 (PDB ID: 1NBJ) using the restraint-based modelling program MODELLER 8v1 (http://www.salilab.org/modeller). A set of 100 models was calculated for the sequence of O25, and the 20 lowest energy models were selected to represent the structure.
RESULTS
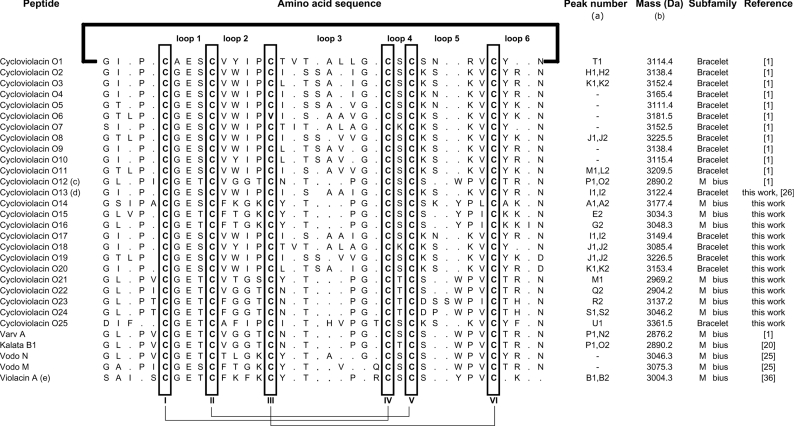
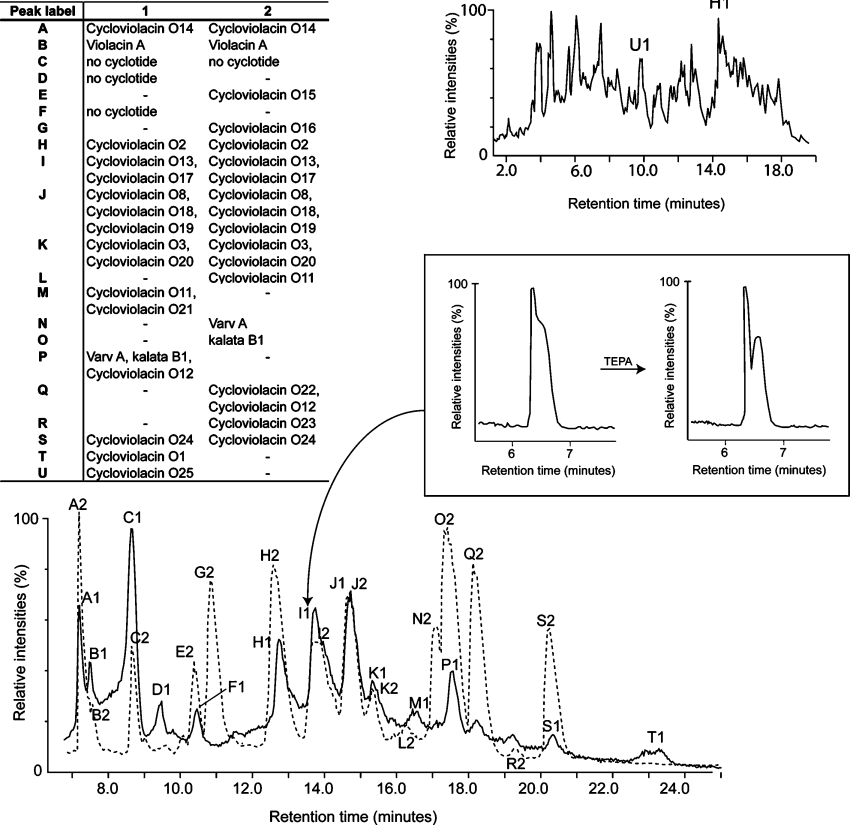
To further our understanding of the natural diversity of cyclotides and the implications of sequence variation on structure, function and stability, we set out to characterize the suite of cyclotides expressed in the plant V. odorata. Cyclotides were isolated from winter and summer extractions using RP-HPLC, monitored by UV and MS. Peptides were then sequenced by MS/MS and their composition was confirmed with quantitative amino acid analyses. From two seasonally distinct extractions, we characterized six new bracelet and seven new Möbius cyclotides, effectively doubling the number of known cyclotides in this plant species. These peptide sequences are summarized in Table 1 where they are labelled according to their elution time in RP-HPLC. Figure 2 presents the summer and winter LC/MS profiles and illustrates the extent of seasonal cyclotide expression variation. It was beyond the scope of the current project to quantify all of the factors that might contribute to variations in the seasonal or temporal expression of cyclotides; the two-season sampling was made simply to increase the potential number of cyclotides detected. Figure 2 shows that there are substantial differences in the peptide profile for the above ground and below ground parts of the plant. Only two cyclotides were identified in roots in the crude extract but at least three other cyclotides are present in roots based on a previous report [35].
Table 1. Cyclotides from V. odorata.
(a) Cyclotides in this Table are referenced to the LC/MS profile in Figure 2. (b) Masses are reported as monoisotopic mass. Average masses are typically 2 Da higher than the monoisotopic mass unless the total mass is greater than 3.5 kDa in which case the average mass may be 3 Da higher. (c) Cycloviolacin O12 is also named varv E [37]. (d) The previously reported cDNA clone Voc3 [26] encodes the peptide discovered here and named cycloviolacin O13. (e) Denotes a linear cyclotide derivative.
Figure 2. LC/MS profiles of summer (solid line) and winter (dotted line) cyclotide extractions from V. odorata.
To account for any minor variations in buffer and column conditions, the LC/MS profiles were aligned based on the most hydrophilic and hydrophobic cyclotides that are present in both extracts. The LC/MS profile at the upper right is for plant roots from the summer extract. LC/MS profiles are displayed as total ion chromatograms. The expanded RP-HPLC trace (boxed) illustrates the improved separation of peak I1 following the use of a TEPA buffer. This improved resolution allowed cycloviolacin O13 and O17 to be separated and sequenced. Peaks C, D and F had masses outside the expected range for cyclotides (<700 Da) and were not further investigated.
Separation, isolation and characterization of cycloviolacins O13–O25
LC/MS analysis of crude extracts from aerial parts and roots of V. odorata indicated the possible existence, based on retention times, of 30 cyclotides of which 13 had novel masses. Separation and isolation of these involved RP-HPLC purification and MS analysis. The above ground extractions yielded approx. 1.5 g of cyclotides (combined total mass) per kg of wet plant material, which is comparable with cyclotide yields previously reported [13]. The use of a TEPA buffer in RP-HPLC purification aided in separating overlapping peaks by suppressing chromatographic tailing, thus helping to identify peptides not clearly visible in the original LC/MS profile. The value of this procedure is evident in the inset of Figure 2, where improved peak separation led to the isolation of the novel peptides cycloviolacins O13 and O17. The compositions of the new sequences were confirmed with quantitative amino acid analysis and the sequences were named according to the nomenclature scheme reported previously [1]. The known sequences included cycloviolacins O1–O12, the recently reported linear cyclotide derivative violacin A [36], and several cyclotides previously reported in other plants of the Violaceae and Rubiaceae families, including the prototypic cyclotide kalata B1.
Analysis of novel sequences
As seen in Table 1, the cyclotides in V. odorata exhibit extensive variations in the composition and size of their loops. The six cysteine residues are absolutely conserved and presumably contribute to the preservation of the CCK motif throughout the 30 sequences. Throughout the cyclotide suite, loop 1 shows the least variation and the glutamic acid residue in this loop is absolutely conserved. Loop 4 is similarly highly conserved, always comprising just a single residue, namely a serine, threonine or lysine. Although loops 2, 3, 5 and 6 show extensive variations in their composition, there are highly conserved elements within them, including the preference towards an asparagine (or occasionally aspartic acid) residue at the putative cyclization point [11,26,36] in loop 6.
The 13 new sequences, combined with the previously known sequences, provide new information on the composition of cyclotide subfamilies. The seven novel Möbius cyclotides characterized take the total count of Möbius cyclotides in V. odorata to 13, making it the richest known natural source of cyclotides from this subfamily. Previously, of the 17 characterized peptides expressed in V. odorata, only six were from the Möbius subfamily. Although Möbius cyclotides are not more difficult to isolate and characterize, they are less common among other species examined so far (less than 25% of total number of cyclotides reported from all species). In addition, some Möbius cyclotides such as cycloviolacins O14–O16 have a higher than average number of cationic side chains and are thus more hydrophilic than their bracelet counterparts. This puts them outside of the standard LC/MS elution range (35–50% solvent B) for cyclotide extraction that was arbitrarily set in early cyclotide discovery programmes [1,37]. Cycloviolacin O14 is the most hydrophilic and positively charged (containing four lysine residues) cyclotide discovered in V. odorata and has the earliest elution time on RP-HPLC (Figure 2). Interestingly, in addition to having a very similar elution time, it has a similar loop 2 sequence to the recently reported linear cyclotide violacin A [36].
As cyclotides are thought to be important in defence against plant pests/pathogens, and most have been isolated so far from aerial plant parts, it is reasonable to assume that root-specific cyclotides would have atypical amino acid sequences, reflecting the different pests, growth requirements and environmental stresses of roots relative to aerial tissues. Although only two cyclotides, cycloviolacins O2 and O25, were characterized from roots, O25 is substantially different from the other cyclotides reported from V. odorata. Aside from being the earliest eluting bracelet cyclotide from V. odorata, O25 is also the only cyclotide from any species to contain an Asn-Asp dipeptide sequence in loop 6. Single asparagine or aspartic residues in loop 6 are thought to be required for cyclization of cyclotides via an asparaginyl endopeptidase [11,26,36], but until now both residues have not been seen adjacent to one another in an individual cyclotide. Furthermore, O25 is the only cyclotide to contain three phenylalanine residues. Homology modelling of O25 suggested that these residues (Phe3, Phe10 and Phe30) are solvent-exposed (see Supplementary Figure at http://www.BiochemJ.org/bj/400/bj4000001add.htm), which supports a previous study that suggested that hydrophobic residues of cyclotides are generally solvent-exposed [38].
Stability against proteolytic degradation and haemolytic activity
A selection of the new cyclotides (cycloviolacin O13, O14, O16 and O24) was tested for their stability against proteolytic degradation by pepsin, trypsin and thermolysin to investigate how tolerant the characteristic proteolytic resistance of the cyclotide framework is to sequence variation. The peptides were primarily selected based on their abundance, but also to cover a wide range of biophysical properties such as hydrophobicity, as judged by their elution times on RP-HPLC. Kalata B1 was included for comparison, as was a linear control peptide. No degradation by proteases was observed for any of the cyclotides tested following 6 h of incubation with enzymes, while the linear control peptide was completely degraded in less than 5 min (results not shown).
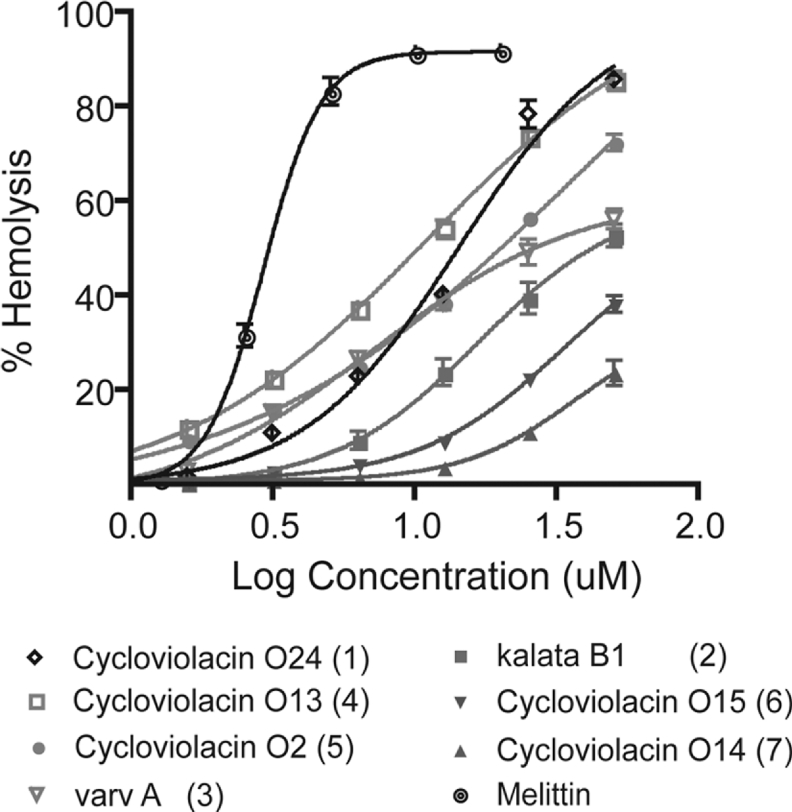
The insecticidal activity of cyclotides has been proposed to result from damage to membranes within the insect gut [11] and various other activities of cyclotides have similarly been associated with membrane interactions. Accordingly, in the present study, we used haemolytic activity as a marker for bioactivity for a selection of the new cyclotides. Figure 3 shows that the haemolytic activity of the various sequences does vary considerably. At a concentration of 25 μM, a more than 6-fold difference exists between the most haemolytic cyclotide, cycloviolacin O24 (∼75% haemolysis), and the least haemolytic cyclotide, cycloviolacin O14 (∼11% haemolysis). The sensitivity of haemolytic activity to variations in the peptide sequence is evident when comparing cycloviolacins O2 and O13. Here, the only sequence deviation is a single residue replacement of a serine in loop 3 of O2 (HD50∼36 μM) to an alanine in the homologous position in O13 (HD50∼11 μM). The loss of this single hydroxy group changes the HD50 more than 3-fold
Figure 3. Haemolytic activities of cyclotide natural variants from V. odorata relative to the well-known synthetic haemolytic agent melittin.
A hydrophobicity ranking, based on RP-HPLC retention time (a ranking of 1 corresponds to the most hydrophobic cyclotide), is included in parentheses.
1H-NMR resonance assignments and secondary structure of cycloviolacin O14
It was of interest to determine the structure of cycloviolacin O14 since it differs from other cyclotides structurally determined so far in a number of ways. First, it exhibits the high stability typical of cyclotides but has the lowest observed haemolytic activity of the new cyclotides. Secondly, it is the most hydrophilic of any reported cyclotide from V. odorata. Finally, it has a proline residue at the second last position of loop 5, a motif that has been proposed to be the signature of the Möbius subfamily of cyclotides, assuming that the proline adopts a cis conformation. However, O14 has five residues in loop 5, not four as is usually the case, and no cyclotide structure has been reported to determine whether the cis-proline conformation exists in the five-residue loop. Including the new cyclotides reported here, only nine Möbius cyclotides have more than four residues in loop 5 and no structures of these have been reported, so O14 provides a test case to determine whether the cis-proline is dependent on loop size.
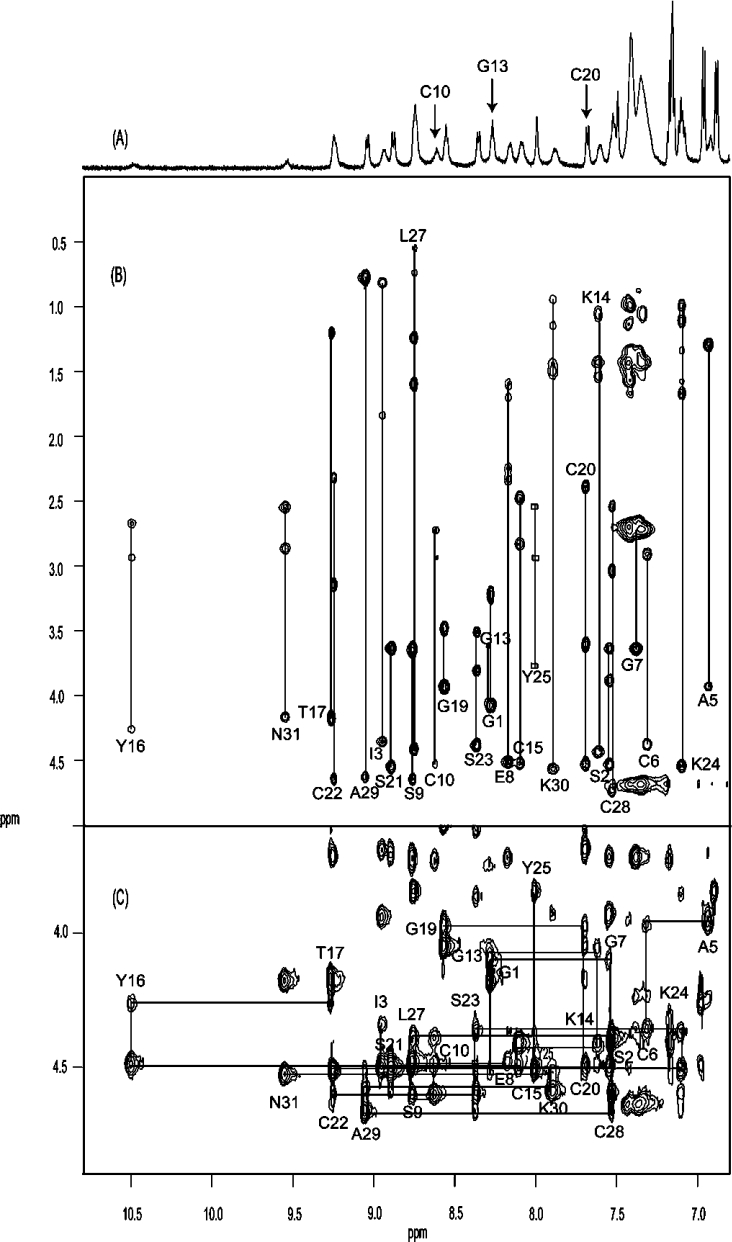
Accordingly, NMR spectra of cycloviolacin O14 were recorded to determine its three-dimensional structure. In the first instance, it was of interest to confirm that it contains a CCK motif characteristic of cyclotides. As illustrated in Figures 4(A) and 4(B), the peaks in the one-dimensional spectrum and the two-dimensional total correlation spectrum (TOCSY) of O14 are well dispersed in the amide region, and the individual amino acids spin systems were readily identified. These were assigned to specific amino acid residues in the sequence using two-dimensional NOESY. The ‘fingerprint’ region of the NOESY spectrum in Figure 4(C) shows the expected series of αHi-NH(i+1) sequential connectivities and is consistent with the proposed sequence of the peptide. The presence of strong αH(i−1)-δHi NOE (nuclear Overhauser effect) signals for Pro4 and Pro18 and strong αH(i−1)-αHi NOE signals for Pro26 confirmed that the amide bonds preceding these residues are in the trans, trans and cis conformations respectively. A 13C-1H HSQC spectrum recorded at natural abundance supported these assignments. In general, in the case of a trans-proline peptide bond conformation, the 13C chemical shift difference between proline Cβ and Cγ is approx. 5 p.p.m., while a cis-proline shows a significantly greater chemical shift dispersion of approx. 10 p.p.m. [39]. The Cβ and Cγ shift difference in cycloviolacin O14 is approx. 2 p.p.m. for the trans-proline Pro4 and Pro18 and approx. 8 p.p.m. for the cis-proline Pro26. The presence of the cis peptide bond preceding the proline in loop 5 (Pro26) confirms the generality of the Möbius subfamily nomenclature for cyclotides [1].
Figure 4. (A) 600 MHz one-dimensional, (B) TOCSY and (C) NOESY NMR spectra for cycloviolacin O14.
All spectra were recorded at 290 K in 90% water/10% 2H2O at pH 3.5. (B) Amino acid spin systems are labelled. Note the absence of signals for residues Phe11 and Lys12 and the broadened signals for Cys10 and Gly13 (arrowed in the one-dimensional spectrum). The peaks for Tyr25 are boxed, as there were no observable TOCSY signals for this residue. The ‘fingerprint’ region of the NOESY spectrum (C) shows the sequential connectivities between the residues.
A feature of interest noted during the assignment process was that the signals for Phe11 and Lys12 were absent from the one-dimensional, TOCSY and NOESY spectra, presumably as a result of severe broadening. The homologous positions in other cyclotide structures occur in a turn region of loop 2 and the residues involved have not been reported to have broad peaks [40]. However, similar broadening was noted in the linear cyclotide derivative violacin A [36]. The latter peptide has a similar combination of loop 2/loop 5 residues to cycloviolacin O14 and is the only other reported cyclotide to have a lysine residue in the second position of loop 2 (Lys12). The exchange broadening of the NMR signals of residues Phe11 and Lys12 appears to be highly localized, as signals from other residues are not broadened.
Following dissolution of the peptide in 2H2O, five slowly exchanging amide protons were detected, namely for Cys20, Ser21, Leu27, Cys28 and Ala29, suggesting their likely involvement in intramolecular hydrogen bonds. Amide chemical-shift temperature coefficients over the range 280–320 K for these residues were relatively small (δ/T=−1.13, −1.83, −2.33, −4.9 and −1.1 p.p.b./K respectively), supporting their likely involvement in hydrogen-bonding. The number of slowly exchanging amides is less than is typically seen in other cyclotides, suggesting that O14 may be more flexible than other cyclotides characterized so far. The severe broadening of signals for Phe11 and Lys12 also supports this suggestion.
Trends in the measured chemical shifts provided a useful first insight into the secondary structure of cycloviolacin O14. CSI (chemical-shift index) values for the backbone α protons were determined using the measured chemical shifts and random coil values. Most of the residues have chemical shifts that differ from random coil values by more than 0.1 p.p.m. and hence have CSI values of ±1, suggesting that O14 is a structured peptide. Successive negative CSI values for residues Ile3-Cys6 and successive positive values for residues Cys20-Ser21 and Cys28-Ala29 suggested the presence of an α-helix and β-strands respectively. This was confirmed following full three-dimensional structure determination.
Structure determination
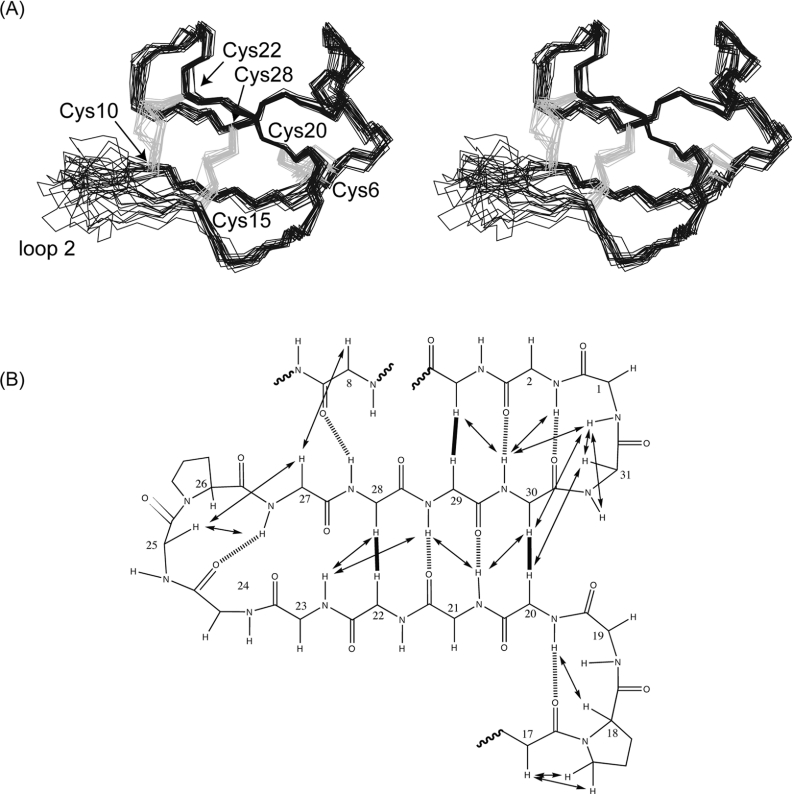
Solution structures for cycloviolacin O14 were determined by simulated annealing calculations incorporating experimental distances and dihedral angle restraints, with refinement using the explicit inclusion of solvent molecules. The connectivity of the disulfide bonds was deduced based on preliminary structure calculations in which energy functions were evaluated for all 15 possible disulfide connectivities (see Supplementary Table at http://www.BiochemJ.org/bj/400/bj4000001add.htm). The lowest energy solution fitted the connectivity pattern of known cyclotides [38,41], i.e. I–IV, II–V and III–VI, corresponding to Cys6-Cys20, Cys10-Cys22 and Cys15-Cys28. Following structure refinement, it was determined that in addition to the slowly exchanging amides, residues Glu8 and Lys30 were also hydrogen-bonded. This conclusion was based on the relative orientation of their amide NH bonds and the insensitivity of their chemical shifts to changes in temperature (δ/T=−0.77 and 0.3 p.p.b./K respectively). Of the final 50 calculated structures, the 20 lowest energy structures consistent with experimental data were chosen to represent the family of O14 solution structures. A summary of the structural statistics for the family is given in Table 2. The ensemble of NMR structures and a diagram highlighting the hydrogen bonds and important NOEs are shown in Figure 5. The structural ensemble is generally very well defined apart from some disorder in loop 2.
Table 2. Geometric and energetic statistics for NMR structures of cycloviolacin O14.
The values are given as means±S.D. for the ensemble of the 20 final solution structures. Cdih, constrained dihedral angle.
| Energies (kcal/mol) | |
| Overall | −1003.17±26.97 |
| Bonds | 6.21±0.87 |
| Angles | 32.98±7.46 |
| Improper | 5.13±1.38 |
| van der Waals | −77.01±8.29 |
| NOE | 7.26±3.15 |
| Cdih | 0.53±0.25 |
| Dihedral | 120.35±8.29 |
| Electrostatic | −1098.00±25.43 |
| rmsd | |
| Bond (Å) | 0.00377±0.00026 |
| Angle (deg) | 0.521±0.057 |
| Improper (deg) | 0.387±0.050 |
| NOE (Å) | 0.0213±0.0046 |
| Cdih (deg) | 0.713±0.184 |
| Pairwise rmsd (Å) | |
| Mean global backbone (1–9, 15–31) | 0.55±0.14 |
| Mean global heavy (1–9, 15–31) | 1.34±0.31 |
| Experimental data | |
| Distance restraints | 299 |
| Dihedral restraints | 18 |
| NOE violations >0.2 Å | 0 |
| Dihedral violations >2.0° | 0 |
| Ramachandran statistics for residues 1–9 and 15–31 | |
| Most favoured (%) | 81.3 |
| Additionally allowed (%) | 18.7 |
| Generously allowed (%) | 0 |
| Disallowed (%) | 0 |
Figure 5. Structural representations of cycloviolacin O14.
(A) Stereo-view of the 20 lowest energy structures of cycloviolacin O14 superimposed over the backbone atoms (N, C and Cα). The backbone is in black with the disulfide bonds in grey. Loop 2 of O14 (residues 10–14) is disordered due to peak broadening and a lack of restraint data in this region. (B) Schematic diagram of the secondary structure of cycloviolacin O14 showing the inter-strand NOEs as continuous arrows or thick lines and hydrogen bonds as broken lines. Sequential NOEs are omitted for clarity. Hydrogen bonds were inferred from slow-exchange data, sensitivity of amide chemical shifts to changes in temperature and preliminary structure calculations, as described in the text.
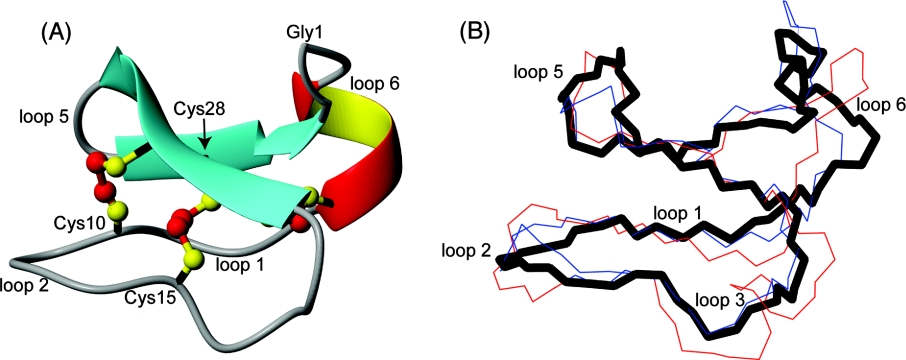
Analysis of the family of structures using the program PROMOTIF shows that the main elements of secondary structure are an α-helix (residues 3–6), a β-sheet (with the stands comprising residues 20–24 and 27–31) and two β-turns (residues 17–18 and 25–26). There is also a β-bulge involving residues 22–24 and 27–28, occluding these residues from regular hydrogen-bonding interactions. A ribbon representation of the structure is shown in Figure 6(A). The small helical segment in loop 6 is unique among cyclotides and may be explained by the extended loop 6 of O14 (longer than 81 of the 83 reported cyclotides). The structures are in good agreement with the experimental restraint data, with no distance violations exceeding 0.2 Å (1 Å=0.1 nm) and no dihedral angle violations exceeding 2°. The precision of the family of structures is slightly lower than seen for other cyclotides, presumably as a result of the disorder of loop 2 due to a relative lack of NOE data for Cys10, Phe11, Lys12 and Gly13.
Figure 6. Structural comparison of cycloviolacin O14 with other cyclotides.
(A) Ribbon representation of the mean structure of cycloviolacin O14. Cysteine residues and loops are labelled. (B) O14 (black thick line) is labelled and overlaid with kalata B1 (blue, PDB ID: 1NB1) and cycloviolacin O1 (red, PDB ID: 1NBJ). There is preservation of the defining structural features (the CCK motif) despite the sequence variations that cause changes in cytotoxic activity.
Despite the disorder in loop 2 and the presence of the α-helix in loop 6, cycloviolacin O14 superimposes well over the cystine knot motif of the prototypic cyclotide kalata B1 (also from the Möbius subfamily) and cycloviolacin O1 (from the bracelet subfamily), with backbone rmsd (root mean square deviation) values of 0.67 and 0.63 Å respectively. These superimpositions are shown in Figure 6(B).
DISCUSSION
The present study reports the characterization of 13 novel cyclotides from the European sweet violet V. odorata and shows that amino acid sequence variations affect the cytotoxic activity but not the proteolytic stability of cyclotides. The NMR structure determined for one of the new cyclotides, cycloviolacin O14, is well defined and contains the CCK motif that is characteristic of the cyclotide family together with the loop 5 cis-proline that defines the Möbius subfamily. Comparison of this structure with previously reported cyclotide structures reveals that sequence variations elicit only minimal structural perturbations. The diversity of the sequences discovered and the sensitivity of cytotoxic activity to these sequence variations with minimal effect on structure and proteolytic stability confirm the concept that cyclotides appear to be a natural combinatorial template that displays a wide range of peptide epitopes targeted to different pests or pathogens. These findings also further validate the potential applicability of cyclotides as templates in drug design and development programmes.
The cyclotides have several defining characteristics that can be exploited when extracting them from plants. Their typical masses of 2.8–4 kDa, late elution times and resistance to chemical, thermal and enzymatic degradation help to distinguish them from other plant proteins and facilitate their isolation and purification. However, the multitude of cyclotides present in V. odorata means that there is a high probability of co-elution, thereby potentially creating separation difficulties during RP-HPLC. A common cause of co-elution due to peak tailing in RP-HPLC is the secondary retention that occurs when an ion-exchange interaction takes place between a positively charged solute (amine) and an acidic silanol on the surface of the silica stationary phase support particles. To overcome this problem, a TEPA buffer was used in the present study. TEPA interacts strongly with silanols, acting as a competing amine to the mobile phase, thereby inhibiting silanol–amine interactions and thus improving peak shape and elution. Using this approach, we were able to separate cyclotides that would otherwise have been inseparable.
The 30 cyclotide sequences reported here from V. odorata show extensive variations in the size and composition of their loops. Figure 7 summarizes the sequence variations and also highlights regions of conservation. In general, it would be expected that those residues that are highly conserved are likely to play a role in maintaining the structural integrity of the framework, while those that vary might be associated with adaptation to target different pests. Residues involved in the biosynthetic cyclization would also be expected to be conserved. The residues that are conserved throughout the cyclotide suite are the cysteine residues, a glutamic acid residue in loop 1, a hydroxyl-containing residue in the third position in loop 3 and a hydrophobic residue in the final position in loop 5. NMR structural characterization of O14 shows that these residues have roles in structural stabilization through hydrogen-bonding or hydrophobic interactions. The asparagine/aspartic residue in loop 6 is also conserved and has been implicated as a crucial feature in the in-planta cyclization of cyclotides [11,26,36,38]. Overall, as the conserved residues account for only one-third of the scaffold, cyclotide-expressing plants have many possibilities for evolving the remainder of the sequence to optimize the required biological activities.
Figure 7. Cyclotides from the (A) bracelet and (B) Möbius subfamilies from V. odorata.
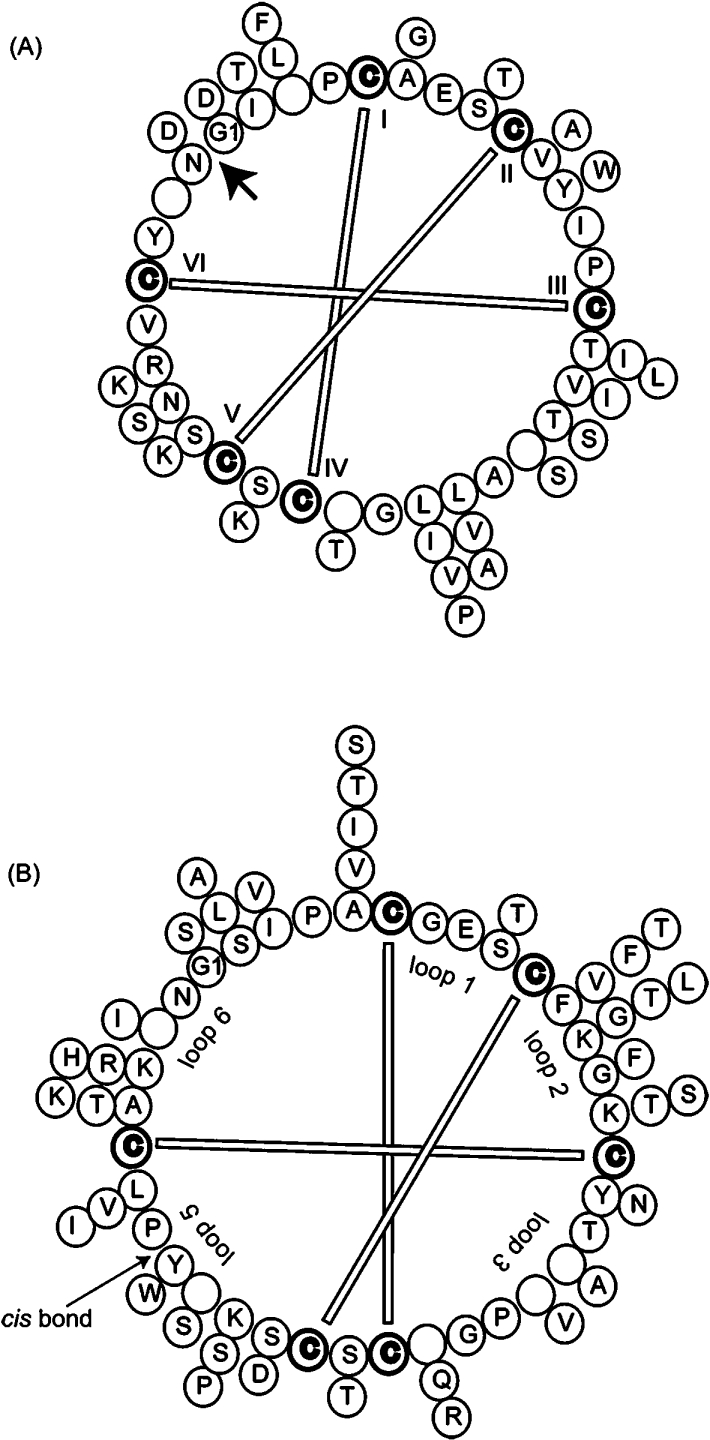
The bracelet and Möbius subfamilies are represented by cycloviolacin O1 and O14 respectively and their backbone sequences are indicated in the inner ‘backbone wheel’. Empty circles represent points where insertions are found in other cyclotides compared and the variations for each position are given by additional amino acids outside the ‘backbone wheel’. The more abundant an amino acid is at a particular position, the closer it is to the representative backbone. Disulfide bonds and loops are represented and the protein precursor processing point is also indicated (heavy arrow in A). The cis-proline bond that defines the Möbius subfamily is also indicated.
In the current study, haemolytic assays were used as a measure of biological activity, as a previous study suggested a possible link with insecticidal activity against Helicoverpa insect species [11]. Haemolytic activity provides a convenient marker that is reflective of the membrane interactions [42] typical of many peptide defence molecules and, given the number of cyclotides expressed per plant, activities against a wide range of pests and pathogens is anticipated. Our study showed that minor variations in sequence caused significant changes in haemolytic activity, confirming the importance of sequence variations in membrane interactions.
Why are so many cyclotide variants produced by individual plants? Some degree of targeting of specific cyclotides to particular pests seems likely, but it would also seem unlikely that each plant would face an array of different pests similar in number to its cyclotide suite. Both in the current study and in previous work [10], seasonal variation in cyclotide expression of V. odorata has been noted. These variations may reflect seasonal variations in pest populations. It is interesting to note that in the Australian native violet Viola hederaceae, minimal seasonal variation in cyclotide expression has been reported [10]. This may indicate a propensity for V. hederaceae to grow in more stable climates than V. odorata and thus have to manage smaller changes in pest populations. An alternative explanation for the large number of cyclotides produced is that diversity and redundancy in cyclotide expression may assist in combating the development of resistance in particular pests over time. It is interesting to note that despite the vast numbers of cyclotides produced by plants, some individual cyclotides, such as kalata B1, are present in plants as diverse as V. odorata and O. affinis, from the Violaceae and Rubiaceae families respectively. Even more interesting is that fact that despite the identical peptide sequences of kalata B1 in the two plants the genes corresponding to the same peptide are quite different in the two species. In O. affinis kalata B1 derives from the Oak1 gene, which encodes a single mature cyclotide domain, whereas in V. odorata kalata B1 derives from the second of three cyclotide domains in the Vok1 gene [26].
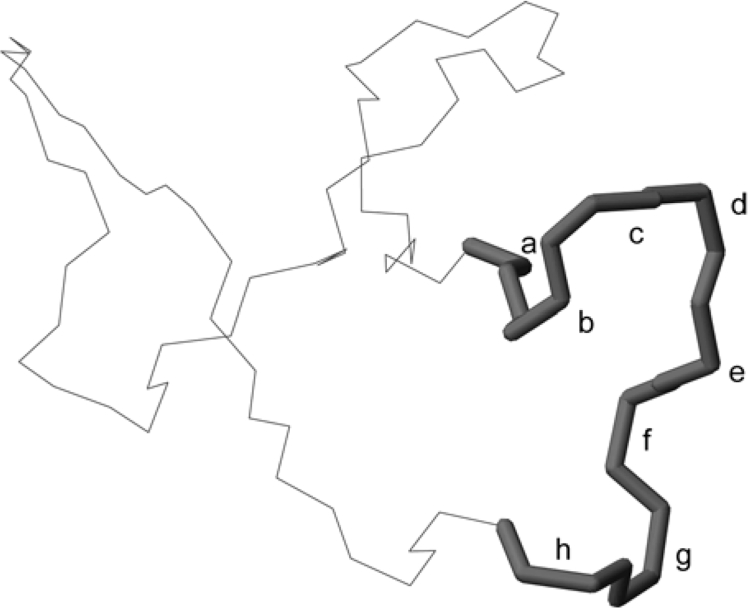
The different genes for kalata B1 in different plants suggests that there is redundancy in the way in which cyclic proteins can be produced but, as mentioned earlier, an asparagine/aspartic residue in loop 6 appears to be crucial in the cyclization mechanism that produces mature cyclotides from their precursor proteins. Sequence variations of other residues in this loop are thus of particular interest, with one of the new cyclotides, cycloviolacin O25, having some substitutions not seen previously in cyclotides. A summary of the loop 6 sequences of cycloviolacin O25 and other cyclotides and the functional/structural roles of these residues is presented in Table 3 and Figure 8. Of particular interest is the presence of an aspartic acid residue following the conserved asparagine residue implicated in the cyclization process. In all other cyclotides discovered to date, the adjacent residue is a glycine or serine, with it previously being assumed that the cyclization process requires a small amino acid at this position. The discovery of cycloviolacin O25 suggests that the cyclization mechanism is more tolerant to sequence changes than has previously been thought. Cycloviolacin O25 also differs from other cyclotides in having multiple hydrophobic residues (Tyr29, Phe30 and Phe3) in loop 6.
Table 3. Functional and structural roles of amino acids in loop 6 of cyclotides from V. odorata.
| Positiona | Residuesb | Incidencec (%) | Functional/structural comments |
|---|---|---|---|
| a | Cys† | 100 | Flanking cysteine at start of loop. Part of the cystine knot [20]. |
| b | Tyr†, Thr, Lys*, Ala* | 62 | Typically a hydrogen bond donor or acceptor [38] stabilizing the cyclotide structure. |
| c | Arg, Lys, His*, Phe*† | 48 | Typically a solvent-exposed positively charged residue [38]. |
| d | Asn†, Asp | 90 | Functional role in the cyclization mechanism [26,36,38,47,48]. |
| e | Gly, Ser, Asp*† | 90 | Typically, a small and uncharged amino acid involved in the cyclization mechanism [47] as the N-terminal residue that forms a bond with the C-terminal asparagine/aspartic residue |
| f | Ile†, Leu, Thr*, Ser* | 41 | Typically, a hydrophobic residue believed to be involved in haemolytic activity [38]. |
| g | Pro, Val*, Ser*, Phe*† | 93 | Typically, turn-inducing and involved in hydrophobic interactions with residues in loops 5 and 6 [38]. |
| h | Cys† | 100 | Final cysteine of loop 6. Part of the cystine knot [20]. |
aPosition in loop 6 as outlined on the structure of cycloviolacin O14 (PDB ID: 2GJ0) in Figure 8.
bThe residues expressed in V. odorata cyclotides at these positions. Residues shown in boldface are the most common in this position; *indicates residues seen in fewer than 20% of cyclotides, and †indicates residue in cycloviolacin O25.
cIncidence of the most common residue.
Figure 8. A trace of the backbone structure of cycloviolacin O14 (PDB ID: 2GJ0).
Loop 6 of cycloviolacin O14 is emboldened and the amino acid positions are labelled a–h for reference with Table 3.
Why do plants produce cyclic proteins if this presumably involves extra energy expenditure to activate the machinery necessary for cyclization? The current study shows that the new cyclotides are completely resistant to enzyme hydrolysis and thus are exceptionally stable proteinaceous molecules. It appears that there are stability advantages to having a cyclic backbone that outweigh any potential disadvantages. Since the stability is independent of the sequence in the natural variants examined so far, it appears to be an intrinsic property of the CCK motif.
The presence of the CCK motif in the new cyclotides was confirmed by determining the structure of cycloviolacin O14 in solution using NMR methods. It was chosen as an example that is most different in properties than the other cyclotides, being the most hydrophilic, with the rationale being that if it adopts the CCK motif then the other new cyclotides that have more similar sequences to previously characterized cyclotides are also likely to adopt the motif. The structural study also unequivocally showed the presence of a cis-proline peptide bond in loop 5, confirming that the presence of a proline residue in this loop is a valid determinant for defining the Möbius subfamily of cyclotides, irrespective of loop size. Finally, NMR structure determination of O14 showed that loop 2 is disordered and appears to access multiple conformations. In all but one cyclotide for which structures have been determined, this loop is well defined. The exception is the linear cyclotide derivative violacin A, which elutes close to O14 on RP-HPLC (see Figure 2) and has some sequence similarities in loop 2 to O25. Specifically, while all reported loop 2 sequences are preferentially composed of hydrophobic residues, violacin A and O14 are the only examples to contain lysine residues in both the second and final positions of this loop. The replacement of hydrophobic residues in this loop appears to lead to the disruption of favourable interactions with the adjacent loop 5, resulting in flexibility. The flexibility appears to be greater in O14 than in violacin A, judging by the increased broadening of loop 2 signals in O14 due to a smaller hydrophobic patch in O14 relative to violacin A. Phe12 in violacin A is replaced with Gly13 in O14, leaving only Phe11 to interact with Tyr25 and Phe26 of loop 5.
The apparent use by plants of cyclotides as a natural combinatorial template to display various bioactivities, together with their high stability [28] and amenity to chemical synthesis [43], has led to suggestions of their use in protein engineering and drug design applications [3,24]. It was therefore of interest to examine the natural ‘plasticity’ of the CCK framework as assessed by sequence variations of the new cyclotides reported here. Each additional residue that a loop can hold greatly increases the number of possible epitopes available for insertion into the cyclotide framework. Consequently, it was important to identify the natural upper limits of the loop sizes. From this study, loops 1, 2 and 4 appear to be largely fixed at three, four and one residue respectively, while loops 3, 5 and 6 can take up to six to eight residues. In the first instance, epitopes for drug development may be limited to approximately eight residues.
One factor that needs to be considered in drug design applications in which foreign epitopes are grafted onto the cyclotide framework is that any residual activity of the framework itself should be minimized. Previous studies have shown that anti-HIV [44], anti-bacterial [17] and haemolytic [45] activities of natural cyclotides are broadly correlated with hydrophobicity. The 2-fold difference in haemolytic activity between violacin A [36] and cycloviolacin O14 supports the proposed link between cyclotide hydrophobicity and haemolytic activity [9,36]. Violacin A was hypothesized to have haemolytic activity associated with a hydrophobic patch formed by residues from loops 2 and 5 [36], and this hydrophobic patch is reduced in O14. Thus, while O14 has an equivalent number of charged residues and hence has only a slight decrease in hydrophobicity, the reduction of the concentrated patch of hydrophobic residues leads to a 2-fold decrease in IC50 haemolytic activity. The glutamic acid residue in loop 1 is absolutely conserved throughout the suite of cyclotides reported in the present study and a recent report has identified this residue as essential for the maintaining cytotoxic activity of cyclotides [46].
In summary, this work highlights the extensive sequence diversity of cyclotides expressed in V. odorata. Although amino acid sequence variations considerably influenced the activity of the new cyclotides, only minimal changes to the structure and proteolytic stability of the cyclotide framework occurred, owing to the stabilizing influence of the conserved CCK motif. The study reinforces the suggestion that cyclotides have a conserved scaffold on to which a combinatorial array of sequences is displayed for host defence purposes. The new sequence variations observed in loop 6 suggest that the cyclization mechanism is even more robust than previously anticipated and that further diversity in cyclotide sequences can be expected.
Online data
Acknowledgments
D. C. I. is supported by a University of Queensland Graduate School scholarship and an NHMRC (National Heart and Medical Research Council) scholarship. This work was supported in part by a grant from the ARC (Australian Research Council) and in part by the NHMRC. D. J. C. is an ARC Professorial Fellow. We thank Dr Norelle Daly (Institute for Molecular Bioscience, University of Queensland, Queensland, Australia) for her assistance with NMR data analysis.
References
- 1.Craik D. J., Daly N. L., Bond T., Waine C. Plant cyclotides: a unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J. Mol. Biol. 1999;294:1327–1336. doi: 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- 2.Craik D. J., Daly N. L. Oxidative folding of the cystine knot motif in cyclotide proteins. Protein Pept. Lett. 2005;12:147–152. doi: 10.2174/0929866053005863. [DOI] [PubMed] [Google Scholar]
- 3.Craik D. J., Simonsen S., Daly N. L. The cyclotides: novel macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discov. Dev. 2002;5:251–260. [PubMed] [Google Scholar]
- 4.Göransson U., Svangard E., Claeson P., Bohlin L. Novel strategies for isolation and characterization of cyclotides: the discovery of bioactive macrocyclic plant polypeptides in the Violaceae. Curr. Opin. Drug Discov. Dev. 2004;5:317–329. doi: 10.2174/1389203043379495. [DOI] [PubMed] [Google Scholar]
- 5.Gustafson K. R., McKee T. C., Bokesch H. R. Anti-HIV cyclotides. Curr. Protein Pept. Sci. 2004;5:331–340. doi: 10.2174/1389203043379468. [DOI] [PubMed] [Google Scholar]
- 6.Craik D. J., Daly N. L., Mulvenna J., Plan M. R., Trabi M. Discovery, structure and biological activities of the cyclotides. Curr. Protein Pept. Sci. 2004;5:297–315. doi: 10.2174/1389203043379512. [DOI] [PubMed] [Google Scholar]
- 7.Craik D. J. Plant cyclotides: circular, knotted peptide toxins. Toxicon. 2001;39:1809–1813. doi: 10.1016/s0041-0101(01)00129-5. [DOI] [PubMed] [Google Scholar]
- 8.Daly N. L., Koltay A., Gustafson K. R., Boyd M. R., Casas-Finet J. R., Craik D. J. Solution structure by NMR of circulin A: a macrocyclic knotted peptide having anti-HIV activity. J. Mol. Biol. 1999;285:333–345. doi: 10.1006/jmbi.1998.2276. [DOI] [PubMed] [Google Scholar]
- 9.Simonsen S., Sando L., Ireland D. C., Colgrave M. L., Craik D. J. A continent of plant defense peptide diversity: cyclotides in Australian hybanthus (Violaceae) Plant Cell. 2005;17:3176–3189. doi: 10.1105/tpc.105.034678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trabi M., Svangard E., Herrmann A., Göransson U., Claeson P., Craik D. J., Bohlin L. Variations in cyclotide expression in viola species. J. Nat. Prod. 2004;67:806–810. doi: 10.1021/np034068e. [DOI] [PubMed] [Google Scholar]
- 11.Jennings C., West J., Waine C., Craik D. J., Anderson M. Biosynthesis and insecticidal properties of plant cyclotides: the cyclic knotted proteins from Oldenlandia affinis. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10614–10619. doi: 10.1073/pnas.191366898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jennings C., Rosengren K. J., Daly N. L., Plan M. R., Stevens J., Scanlon M. J., Waine C., Norman D. G., Anderson M. A., Craik D. J. Isolation, solution structure, and insecticidal activity of kalata B2, a circular protein with a twist: do Möbius strips exist in Nature? Biochemistry. 2005;44:851–860. doi: 10.1021/bi047837h. [DOI] [PubMed] [Google Scholar]
- 13.Gran L. Oxytoxic principles of Oldenlandia affinis. Lloydia. 1973;36:174–178. [PubMed] [Google Scholar]
- 14.Gustafson K. R., Sowder R. C., Henderson L. E., Parsons I. C., Kashman Y., Cardellina J. H., McMahon J. B., Buckheit R. W. J., Pannell J. K., Boyd M. R. Circulins A and B. Novel human immunodeficiency virus (HIV)-inhibitory macrocyclic peptides from the tropical tree Chassalia parvifolia. J. Am. Chem. Soc. 1994;116:9337–9338. [Google Scholar]
- 15.Lindholm P., Göransson U., Johansson S., Claeson P., Gulbo J., Larsson R., Bohlin L., Backlund A. Cyclotides: a novel type of cytotoxic agents. Mol. Cancer Ther. 2002;1:365–369. [PubMed] [Google Scholar]
- 16.Schöpke T., Hasan Agha M. I., Kraft R., Otto A., Hiller K. Hämolytisch aktive komponenten aus Viola tricolor L. und Viola arvensis Murray. Sci. Pharmaceut. 1993;61:145–153. [Google Scholar]
- 17.Tam J. P., Lu Y. A., Yang J. L., Chiu K. W. An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8913–8918. doi: 10.1073/pnas.96.16.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Witherup K. M., Bogusky M. J., Anderson P. S., Ramjit H., Ransom R. W., Wood T., Sardana M. Cyclopsychotride A, a biologically active, 31-residue cyclic peptide isolated from Psychotria longipes. J. Nat. Prod. 1994;57:1619–1625. doi: 10.1021/np50114a002. [DOI] [PubMed] [Google Scholar]
- 19.Gran L. An oxytoxic principle found in Oldenlandia affinis DC. Medd. Nor. Farm. Selsk. 1970;32:173–138. [Google Scholar]
- 20.Saether O., Craik D. J., Campbell I. D., Sletten K., Juul J., Norman D. G. Elucidation of the primary and three-dimensional structure of the uteronic polypeptide kalata B1. Biochemistry. 1995;34:4147–4158. doi: 10.1021/bi00013a002. [DOI] [PubMed] [Google Scholar]
- 21.Trabi M., Craik D. J. Circular proteins – no end in sight. Trends Biochem. Sci. 2002;27:132–138. doi: 10.1016/s0968-0004(02)02057-1. [DOI] [PubMed] [Google Scholar]
- 22.Craik D. J. Seamless proteins tie up their loose ends. Science. 2006;311:1563–1564. doi: 10.1126/science.1125248. [DOI] [PubMed] [Google Scholar]
- 23.Mulvenna J., Wang C., Craik D. J. Cybase – a circular protein database. Nucleic Acids Res. 2006;24:D192–D194. doi: 10.1093/nar/gkj005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Craik D. J., Cemazar M., Daly N. L. The cyclotide and related macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discov. Dev. 2006;9:251–260. [PubMed] [Google Scholar]
- 25.Svangard E., Göransson U., Smith D., Verma C., Backlund A., Bohlin L., Claeson P. Primary and 3-D modelled structures of two cyclotides from Viola odorata. Phytochemistry. 2003;64:135–142. doi: 10.1016/s0031-9422(03)00218-8. [DOI] [PubMed] [Google Scholar]
- 26.Dutton J. L., Renda R. F., Waine C., Clark R. J., Daly N. L., Jennings C., Anderson M., Craik D. J. Conserved structural and sequence elements implicated in the processing of gene-encoded circular proteins. J. Biol. Chem. 2004;279:46858–46867. doi: 10.1074/jbc.M407421200. [DOI] [PubMed] [Google Scholar]
- 27.Pennington M. W., Dunn B. M. Totowa, NJ: Humana Press; 1994. Peptide Synthesis Protocols. [Google Scholar]
- 28.Colgrave M. L., Craik D. J. Thermal, Chemical and enzymatic stability of the cyclotide kalata B1: the importance of the cyclic cystine knot. Biochemistry. 2004;43:5965–5975. doi: 10.1021/bi049711q. [DOI] [PubMed] [Google Scholar]
- 29.Goddard T. D., Kneller D. G. San Francisco, CA: University of California; 1989. Sparky 3 Manual. [Google Scholar]
- 30.Güntert P., Mumenthaler C., Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 31.Brünger A. T., Adams P. D., Rice L. M. New applications of simulated annealing in X-ray crystallography and solution NMR. Structure (Camb.) 1997;5:325–336. doi: 10.1016/s0969-2126(97)00190-1. [DOI] [PubMed] [Google Scholar]
- 32.Linge J. P., Nilges M. Influence of non-bonded parameters on the quality of NMR structures: a new force field for NMR structure calculation. J. Biomol. NMR. 1999;13:51–59. doi: 10.1023/a:1008365802830. [DOI] [PubMed] [Google Scholar]
- 33.Hutchinson E. G., Thornton J. M. PROMOTIF: a program to identify and analyse structural motifs in proteins. Protein Sci. 1996;5:212–220. doi: 10.1002/pro.5560050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laskowski R. A., Rullmann J. A., MacArthur M. W., Kaptein R., Thornton J. M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 35.Göransson U., Broussalis A. M., Claeson P. Expression of Viola cyclotides by liquid chromatography-mass spectrometry and tandem mass spectrometry sequencing of intercysteine loops after introduction of charges and cleavage sites by aminoethylation. Anal. Biochem. 2003;318:107–117. doi: 10.1016/s0003-2697(03)00114-3. [DOI] [PubMed] [Google Scholar]
- 36.Ireland D. C., Colgrave M. L., Nguyencong P., Daly N. L., Craik D. J. Discovery and characterization of a linear cyclotide from Viola odorata: implications for the processing of circular proteins. J. Mol. Biol. 2006;357:1522–1535. doi: 10.1016/j.jmb.2006.01.051. [DOI] [PubMed] [Google Scholar]
- 37.Göransson U., Luijendijk T., Johansson S., Bohlin L., Claeson P. Seven novel macrocyclic polypeptides from Viola arvensis. J. Nat. Prod. 1999;62:283–286. doi: 10.1021/np9803878. [DOI] [PubMed] [Google Scholar]
- 38.Rosengren K. J., Daly N. L., Plan M. R., Waine C., Craik D. J. Twists, knots, and rings in proteins. J. Biol. Chem. 2003;278:8606–8616. doi: 10.1074/jbc.M211147200. [DOI] [PubMed] [Google Scholar]
- 39.Hill J. M., Alewood P. F., Craik D. J. Three-dimensional solution structure of u-conotoxin GIIIB, a specific blocker of skeletal muscle sodium channels. Biochemistry. 1996;35:8824–8835. doi: 10.1021/bi960073o. [DOI] [PubMed] [Google Scholar]
- 40.Craik D. J., Cemazar M., Wang C. K. L., Daly N. L. The cyclotide family of circular mini-proteins: Nature's combinatorial peptide template. Biopolymers. 2006;84:250–266. doi: 10.1002/bip.20451. [DOI] [PubMed] [Google Scholar]
- 41.Göransson U., Craik D. J. Disulfide mapping of the cyclotide kalata B1. J. Biol. Chem. 2003;278:48188–48196. doi: 10.1074/jbc.M308771200. [DOI] [PubMed] [Google Scholar]
- 42.Kamimori H., Hall K., Craik D. J., Aguilar M. I. Studies on the membrane interactions of the cyclotides kalata B1 and kalata B6 on model membrane systems by surface plasmon resonance. Anal. Biochem. 2005;337:149–153. doi: 10.1016/j.ab.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 43.Daly N. L., Love S., Alewood P. F., Craik D. J. Chemical synthesis and folding pathways of large cyclic polypeptides: studies of the cystine knot polypeptide kalata B1. Biochemistry. 1999;38:10606–10614. doi: 10.1021/bi990605b. [DOI] [PubMed] [Google Scholar]
- 44.Daly N. L., Gustafson K. R., Craik D. J. The role of the cyclic peptide backbone in the anti-HIV activity of the cyclotide kalata B1. FEBS Lett. 2004;574:69–72. doi: 10.1016/j.febslet.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 45.Chen B., Colgrave M. L., Wang C., Craik D. J. Cycloviolacin H4, a hydrophobic cyclotide from Viola hederaceae. J. Nat. Prod. 2006;69:23–28. doi: 10.1021/np050317i. [DOI] [PubMed] [Google Scholar]
- 46.Herrmann A., Svangard E., Claeson P., Gullbo J., Bohlin L., Göransson U. Key role of glutamic acid for the cytotoxic activity of the cyclotide cycloviolacin O2. Cell. Mol. Life Sci. 2006;63:235–245. doi: 10.1007/s00018-005-5486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daly N. L., Clark R. J., Plan M. R., Craik D. J. Kalata B8, a novel antiviral circular protein, exhibits conformation flexibility in the cystine knot motif. Biochem. J. 2006;393:619–626. doi: 10.1042/BJ20051371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mulvenna J., Sando L., Craik D. J. Processing of a 22 kDa precursor protein to produce the circular protein tricyclon A. Structure (Camb.) 2005;13:691–701. doi: 10.1016/j.str.2005.02.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.