Abstract
Despite certain structural and biochemical similarities, differences exist in the function of the NF-κB (nuclear factor κB) inhibitory proteins IκBα (inhibitory κBα) and IκBβ. The functional disparity arises in part from variance at the level of gene regulation, and in particular from the substantial induction of IκBα, but not IκBβ, gene expression post-NF-κB activation. In the present study, we probe the differential effects of IL (interleukin)-1β on induction of IκBα and perform the first characterization of the human IκBβ promoter. A consensus NF-κB-binding site, capable of binding NF-κB both in vitro and in vivo, is found in the IκBβ gene 5′ flanking region. However, the IκBβ promoter was not substantially activated by pro-inflammatory cytokines, such as IL-1β and tumour necrosis factor α, that are known to cause strong activation of NF-κB. Furthermore, in contrast with IκBα, NF-κB activation did not increase expression of endogenous IκBβ as assessed by analysis of mRNA and protein levels. Unlike κB-responsive promoters, IκBβ promoter-bound p65 inefficiently recruits RNA polymerase II, which stalls at the promoter. We present evidence that this stalling is likely due to the absence of transcription factor IIH engagement, a prerequisite for RNA polymerase II phosphorylation and transcriptional initiation. Differences in the conformation of promoter-bound NF-κB may underlie the variation in the ability to engage the basal transcriptional apparatus at the IκBβ and κB-responsive promoters. This accounts for the differential expression of IκB family members in response to NF-κB activation and furthers our understanding of the mechanisms involved in transcription factor activity and IκBβ gene regulation.
Keywords: chromatin immunoprecipitation (ChIP), cis-element, inhibitory κB (IκB), nuclear factor κB (NF-κB), RNA polymerase II, transcription
Abbreviations: ChIP, chromatin immunoprecipitation; CTD, C-terminal domain; DMEM, Dulbecco's modified Eagle's medium; DTT, dithiothreitol; EMSA, electrophoretic mobility-shift assay; FCS, foetal calf serum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HEK, human embryonic kidney; IκB, inhibitory κB; IL, interleukin; LTR, long terminal repeat; MCP, monocyte chemoattractant protein; NF-κB, nuclear factor κB; Pol II, polymerase II; RACE, rapid amplification of cDNA ends; RFP, red fluorescent protein; RHD, Rel homology domain; RLM-5′-RACE, RNA ligase-mediated 5′-RACE; TAD, transcriptional activation domain; TFIIH, transcription factor IIH; TNF, tumour necrosis factor; TSS, transcription start site
INTRODUCTION
The inducible expression of a wide range of immune and inflammatory response genes is controlled by the action of NF-κB (nuclear factor κB) [1]. NF-κB is a dimeric factor, comprised of members of the Rel family of proteins, some of which are transcriptionally active – p65, c-Rel (the product of the cellular homologue of the avian-recticuloendotheliosis-virus transforming gene) and RelB, while others lack TADs (transcriptional activation domains) and are synthesized as longer inactive precursors – p105 (p50) and p100 (p52) [2]. Rel proteins share a conserved 300-amino-acid N-terminal region known as the RHD (Rel homology domain) that contains regions responsible for dimerization, nuclear localization and site-specific DNA binding. The RHD is also responsible for mediating interactions with members of the IκB (inhibitory κB) family of proteins [3]. IκBα and IκBβ, the most extensively characterized members of this family, display numerous structural similarities. They contain a central ankyrin repeat domain responsible for association with NF-κB with 1:1 stoichiometry [4,5]. The interaction maintains NF-κB in an inactive state in unstimulated cells. In addition, IκBα and IκBβ contain specific N-terminal serine residues that are phosphorylated in response to NF-κB-inducing stimuli [6,7]. This tags the IκB protein for polyubiquitination and proteasome-mediated degradation, allowing NF-κB to enter the nucleus and bind to target promoters.
IκBα and IκBβ respond differently to the various inducers of NF-κB. Whereas IκBα is degraded by all stimuli known to activate NF-κB, IκBβ proteolysis only occurs in response to a subset, those causing long-term activation of the transcription factor [8]. As the IκBα gene is positively regulated by NF-κB, its expression is rapidly induced following initial NF-κB activation [9]. The newly synthesized protein can enter the nucleus, remove NF-κB from its target promoters and return it to the cytoplasm [10]. This inhibitory feedback accounts for the transient nature of the NF-κB response produced by certain stimuli. The importance of IκBα for regulating NF-κB was highlighted by studies on IκBα−/− mice, which display developmental defects postnatally, dying within 8 days. The haemopoietic tissue of these animals show increased levels of nuclear NF-κB and mRNAs of some NF-κB-responsive genes. Furthermore, in IκBα−/− embryonic fibroblasts, where signal-dependent NF-κB-activation is retained due to inhibition by IκBβ, nuclear NF-κB remains elevated in response to TNFα (tumour necrosis factor α) treatment relative to wild-type cells [11]. Interestingly, knock-in mice, in which the IκBα gene is replaced by the IκBβ gene under the control of the IκBα promoter, do not display postnatal developmental abnormalities and show an inducible NF-κB activation similar to wild-type animals [12]. This indicates that IκBβ can functionally compensate for IκBα, and suggests that it is the strong NF-κB-mediated up-regulation of IκBα expression that accounts for the main difference in IκBα and IκBβ activity. However, IκBβ is also likely to play unique roles in temporal regulation of NF-κB in some cell systems. We have previously shown that IL (interleukin)-1β causes sustained activation of NF-κB in human astrocytes and that this coincides with strong induction of IκBα but lack of resynthesis of IκBβ [13]. In the present study, we further probe the mechanisms underlying the differential induction of the two IκB forms by IL-1β in this system.
In contrast with IκBα, IκBβ expression in mouse is not strongly up-regulated by NF-κB, although it does contain a κB site in its promoter. Studies on the murine IκBβ promoter demonstrated that this κB site confers modest inducibility to NF-κB activation [14]. Such findings, coupled with our previous demonstration of the lack of resynthesis of IκBβ in response to IL-1β in human astrocytes, prompted us to perform the first characterization of the human IκBβ promoter and more significantly explore its regulation by NF-κB. In the present study, we clone the human IκBβ promoter and identify a putative κB site. We show that NF-κB can bind to this site in vitro, and more importantly that p65 binds to the IκBβ promoter in vivo in an IL-1β-dependent fashion. However, this binding is insufficient to promote up-regulation of IκBβ expression, as shown by the failure of NF-κB-activating stimuli to trigger substantial IκBβ promoter firing in transient transfections or to induce expression of endogenous IκBβ at the mRNA or protein level. We further show that this is due to the inability of DNA-bound p65 to efficiently recruit RNA Pol II (polymerase II) to the IκBβ promoter and also to the promoter-proximal stalling of RNA Pol II, with the resulting absence of transcriptional activity. This stalling appears to be due to the failure of p65 binding to augment recruitment of the general transcription factor TFIIH (transcription factor IIH), required for transcriptional initiation, to the IκBβ promoter, in contrast with its engagement at the κB-responsive IL-8 promoter. Given the critical role of IκB proteins in regulating NF-κB, and the importance of gene regulation in dictating IκB activity, probing the mechanisms underlying the differential expression of IκBα and IκBβ is of clear importance. The present study advances our understanding of those mechanisms, and offers an explanation for the failure of NF-κB to cause an induction of IκBβ expression.
MATERIALS AND METHODS
Materials
The human astrocytoma cell line, 1321N1, and HEK (human embryonic kidney)-293 cell line were obtained from the European Collection of Animal Cell Cultures (Salisbury, U.K.). DMEM (Dulbecco's modified Eagle's medium), FCS (foetal calf serum), penicillin/streptomycin and trypsin were from Gibco BRL. Human IL-1β and TNFα were purchased from R&D Systems Europe. The double-stranded oligonucleotide containing the HIV-LTR (long terminal repeat) κB site and T4 polynucleotide kinase were supplied by Promega. [γ-32P]ATP was purchased from Amersham International. PCR primers and oligonucleotides containing other κB site sequences were obtained from MWG Biotech. GeneJuice transfection reagent was from Novagen. Primers and probes for quantitative real-time PCR analysis of IκBβ and 18 S rRNA were supplied by Applied Biosystems. Rabbit polyclonal antibodies against IκBα (sc-203), IκBβ (sc-945), p50 (H-119), p65 (sc-372), c-Rel (sc-70X), RNA Pol II (sc-899) and p89 (sc-293) were obtained from Santa Cruz Biotechnology. Mouse monoclonal anti-β-actin antibody was from Sigma. Affinity-purified rabbit polyclonal antiserum against IκBϵ was a gift from Nancy Rice (NCI-Frederick Cancer Research and Development Center, Frederick, MD, U.S.A.).
Cell culture
The human astrocytoma cell line, 1321N1, and HEK-293 cell line were cultured in DMEM supplemented with 100 units/ml of penicillin, 100 μg/ml streptomycin and 10% (v/v) FCS. Cells were maintained at 37 °C in a humidified atmosphere of 5% CO2. All cells were passaged using 1% (w/v) trypsin in PBS. Cytokine stimulation was performed on cells in a serum-containing medium at 37 °C for all experiments.
5′-RACE (rapid amplification of cDNA ends)
RNA was isolated from 1321N1 astrocytoma using TRI Reagent (Sigma). RLM-5′-RACE (RNA ligase-mediated 5′-RACE) was performed using the GeneRacer kit (Invitrogen) according to the manufacturer's instructions. Purified RACE products were TOPO-cloned into pCR2.1-TOPO (Invitrogen) and sequenced to identify the 5′-end of the transcripts.
EMSA (electrophoretic mobility-shift assay)
Double-stranded oligonucleotides containing the IκBβ, IκBα-κB1 and IκBα-κB2 NF-κB-binding sites were created by annealing the oligonucleotides 5′-AGTTGAGGGGAATTTCCCAGGC-3′, 5′-AGTTGAGGGAAATTCCCCAGGC-3′ and 5′-AGTTGAGGGGAAACCCCCAGGC-3′ to their respective complementary strands (putative κB sites are underlined). Annealing was performed by incubating 100 pmol of each strand in 10 mM Tris/HCl (pH 7.9) containing 2 mM MgCl2, 50 mM NaCl and 20 mM EDTA at 90 °C for 5 min. The incubation was cooled slowly to 50 °C, maintained at 50 °C for 5 min and then allowed to cool to room temperature (21 °C). Double-stranded oligonucleotides were labelled with [γ-32P]ATP (10 mCi/mmol) by T4 polynucleotide kinase [15]. In EMSA analysis, nuclear extracts (10 μg of protein, generated as described previously [16]) were incubated with 30000 d.p.m. of probe. Incubations were performed for 30 min at room temperature in 10 mM Tris/HCl buffer (pH 7.5) containing 100 mM NaCl, 1 mM EDTA, 5 mM DTT (dithiothreitol), 4% (w/v) glycerol, 4 μg of poly(dI-dC)·(dI-dC) and 1 mg/ml nuclease-free BSA. In the supershift analysis, polyclonal antibody (1 μg) against the NF-κB subunits p50, p65 or c-Rel, or non-immune IgG was added to the extracts and chilled for 30 min on ice prior to incubation with labelled oligonucleotide. In competition analysis, unlabelled oligonucleotide was added to the extracts and incubated for 20 min at room temperature (21 °C) in a total volume of 25 μl before probe addition. All incubations were subjected to electrophoresis on 4% (w/v) non-denaturing polyacrylamide gels, which were subsequently dried and autoradiographed.
Promoter-reporter constructs and cell transfections
The HIV-LTR κB-, β-κB- and α-κB1-luciferase constructs consisted of five consecutive copies of the HIV-LTR, IκBβ or IκBα-κB1 κB sites cloned into XhoI/HindIII sites in the pGL3-Basic reporter plasmid (Promega). To generate ‘β-Prom’, a 1 kb fragment of the IκBβ gene 5′ flanking region was amplified from 1321N1 genomic DNA by PCR using the GC-rich PCR system (Roche). The primers used were 5′-CCGCTCGAGCGGGATGTGGAACAAGTCCGTCTCTCTC-3′ and 5′-CCCAAGCTTG-GGTAAGGTTCACTCACGTGTCCCCATC-3′ (restriction sites and clamps underlined). This fragment was also cloned into XhoI/HindIII sites in the pGL3-Basic firefly luciferase plasmid (Promega). For transfections using these constructs, HEK-293 cells (2×105 cells/ml) were plated on to 24-well plates and allowed to grow for 24 h. Cells were then transfected using GeneJuice transfection reagent with constitutively expressed Renilla luciferase reporter construct (phRL-TK) (50 ng) and with promoter-firefly luciferase construct or empty pGL3-Basic vector (390 ng). Cells were allowed to recover overnight before treatment with IL-1β or TNFα (10 ng/ml) for 6 h. Cells treated with reporter lysis buffer (Promega) and extracts were assayed for firefly and Renilla luciferase activity using the luciferase assay system (Promega) and coelenterazine (Sigma; 1 μg/ml) respectively.
Quantitative real-time PCR analysis of IκBβ expression
1321N1 astrocytoma (2×105 cells/ml; 3 ml) were plated on to 6-well plates and grown for 48 h. Cells were treated with IL-1β (10 ng/ml) for various time periods. Cells were washed with PBS, and RNA was extracted using TRI Reagent (Sigma). After DNase I digestion, cDNA was generated from normalized RNA using Superscript II reverse transcriptase. Samples were assayed by quantitative real-time PCR for levels of IκBβ cDNA using the ABI Prism 7900HT thermal cycler. Reactions were performed using pre-validated primers and probes (Applied Biosystems).
p65 overexpression and Western immunoblotting
HEK-293 cells (2×105 cells/ml; 3 ml) were plated on to 6-well plates and allowed to grow for 24 h. Cells were transfected using GeneJuice transfection reagent with a p65 expression vector [p65–RFP (red fluorescent protein)] or with empty RFP vector (1 μg). After 24 h, medium was removed and cells were washed with ice-cold PBS. The cells were scraped into PBS (1 ml) and pelleted by centrifugation at 20000 g for 5 min at 4 °C. After discarding the supernatant, cells were resuspended in sample buffer [62.5 mM Tris/HCl, pH 6.8, 2% (w/v) SDS, 10% (w/v) glycerol, 50 mM DTT and 0.01% (w/v) Bromophenol Blue; 100 μl per sample] and chilled on ice for 10 min. Resuspended cells were then boiled for 5 min. Cell debris was pelleted by further centrifugation at 20000 g for 10 min. The supernatant containing the cell extract was removed to fresh Eppendorf tubes. Extracts were separated by SDS/PAGE using a 12% (w/v) resolving gel and proteins were transferred electrophoretically to nitrocellulose. Immunodetection of proteins was conducted as described previously [13]. Blots were blocked in TBS [Tris-buffered saline (20 mM Tris/HCl, pH 7.5, and 0.15 M NaCl)] and 5% (w/v) non-fat dried skimmed milk powder (Marvel). Incubations with primary antibodies against p65, IκBα, IκBβ, β-actin (all 1 μg/ml) or IκBϵ (1:2000 dilution) were for 2 h at room temperature. Horseradish-peroxidase-conjugated sheep anti-rabbit IgG and horseradish-peroxidase-conjugated goat anti-mouse IgG (Upstate Biotechnology) were used at concentrations of 1 and 0.5 μg/ml respectively. Immunoreactive bands were detected using the enhanced chemiluminescence detection system from Pierce according to the manufacturer's instructions.
ChIP (chromatin immunoprecipitation)
1321N1 astrocytoma cells were grown to confluence in 90 mm dishes and stimulated with or without IL-1β (10 ng/ml) for 1 h. ChIP assays were performed as described previously with some modifications [17]. Following stimulation, cells were cross-linked with 1% formaldehyde for 10 min at 37 °C. Isolated nuclei were subjected to seven 10 s sonication pulses from a Sanyo/MES Soniprep 150 at one-third of total power. Separate aliquots from each chromatin preparation were incubated overnight at 4 °C with 2 μg of anti-p65/Pol II antibody, or for 24 h with 8 μg of anti-p89 antibody, and with non-immune rabbit IgG antibody. An aliquot was also retained as an input sample to normalize PCR reactions and analyse shearing efficiency. The chromatin used had an average size of 750 bp. After reversion of cross-links by overnight incubation at 65 °C, DNA was extracted using the QIAquick purification kit (Qiagen) according to the manufacturer's instructions.
Standard PCR was performed using 1 μl (∼3% of total) template DNA, 500 nM primers and 0.2 unit of Taq DNA polymerase (Invitrogen) per 50 μl reaction. Quantitative real-time PCR reactions were performed in duplicate with 2 μl of template DNA, 50 nM primers and the SYBR Green Jumpstart Taq Readymix (Sigma) in a total volume of 20 μl, using the Mx3000P QPCR System (Stratagene). Dissociation curve analysis and gel electrophoresis of the final products confirmed that only the expected specific amplicon of correct size was generated for each target promoter. Real-time PCR data analysis was performed as described previously [18]. Results for each treatment are expressed as fold differences between DNA enrichment in the p65/Pol II/p89-ChIP sample relative to IgG-ChIP sample. The sequences of primers used are as follows: IκBα promoter 5′-GAAAGGACCGGCAGGTTGGCAAAC-3′ and 5′-GGGTCAGGCTCGGGGAATTTC-3′, IκBα gene 5′-GTAGGATCAGCCCTCATTTTGTTGC-3′ and 5′-CTGTTACATGTCACAGGATACCACTG-3′, IκBβ promoter 5′-GGAACGGCTAGAGAGTTGTAGTCC-3′ and 5′-CTTTGCCGGGAGTTCTGGAGCTTC-3′, IκBβ gene 5′-GATTCATTGCACGATGTCCAGTGTCTTC-3′ and 5′-CTACTGTATGCTAGGCCTTGTGACCAC-3′, IL-8 promoter 5′-GGAAGTGTGATGACTCAGGTTTGC-3′ and 5′-GATGGTTCCTTCCGGTGGTTTCTTC-3′, IL-8 gene 5′-CCAAGGGCCAAGAGAATATCCGAAC-3′ and 5′-CTTCCACATGTCCTCACAACATCAC-3′, IL-2 promoter 5′-CTTGCTCTTGTCCACCACAATATGC-3′ and 5′-CAAAGACTGACTGAATGGATGTAGGTG-3′, GAPDH (glyceraldehyde-3-phosphate dehydrogenase) promoter 5′-CTACTAGCGGTTTTACGGGCG-3′ and 5′-TCGAACAGGAGGAGCAGAGAGCGA-3′.
RESULTS
Identification of TSS (transcription start site) of the IκBβ gene
In order to study regulation of IκBβ transcription in human cells, we decided to characterize its promoter. However, it was initially necessary to identify the TSS of the IκBβ gene. We employed RLM-5′-RACE to identify the site. This approach identified nine independent TSSs upstream of the translation start codon of the IκBβ gene (Figure 1). 5′-RACE is a highly sensitive technique, and the ligation of an RNA oligonucleotide to the 5′-end of decapped RNA, and subsequent use of a primer specific for this 5′ oligonucleotide in conjunction with a gene-specific primer, ensures the amplification of only full-length transcripts. A range of TSSs, or ‘transcription window’, is not unusual in TATA-less, GC-rich promoters. However, the detection of numerous sites is in contrast with the murine IκBβ promoter, where a single major TSS was detected in an S1 nuclease protection assay [19].
Figure 1. Nucleotide sequence of the IκBβ gene 5′ flanking region.
A number of TSSs for the IκBβ gene were identified by 5′-RACE and are denoted by boldface upper-case letters. The most 5′ site identified is designated +1. The translation start codon is in boldface and is underlined. A putative κB site is located 5′ of the transcription window.
The murine IκBβ gene 5′ flanking region contains an NF-κB-binding site (κB site) 35 nucleotides upstream of the first TSS. Analysis of the equivalent human region and alignment with the murine promoter confirmed the presence of a consensus κB site in the human IκBβ gene 5′ flanking region (Figure 1). Between species there is variance at a single position – a change of C→T, from mouse to human, at the third nucleotide from the 3′-end of the decameric sequence. An interspecies comparison of 11 functional κB sites has previously shown total conservation despite a divergence in the sequence of surrounding regions, indicating the importance of the cis-element sequence in conferring responsiveness to the appropriate trans-acting factor [20]. Nevertheless, given that the human IκBβ promoter κB site retained the consensus sequence (5′-GGGRNNYYCC-3′), and that this site had been identified as a sequence capable of interacting with p65 and c-Rel in early studies on optimal NF-κB-binding sites [21], we decided to examine the functional relevance of this site and probed its ability to interact with activated NF-κB in an in vitro assay.
NF-κB binds to the IκBβ promoter κB site in an IL-1β-dependent manner in vitro
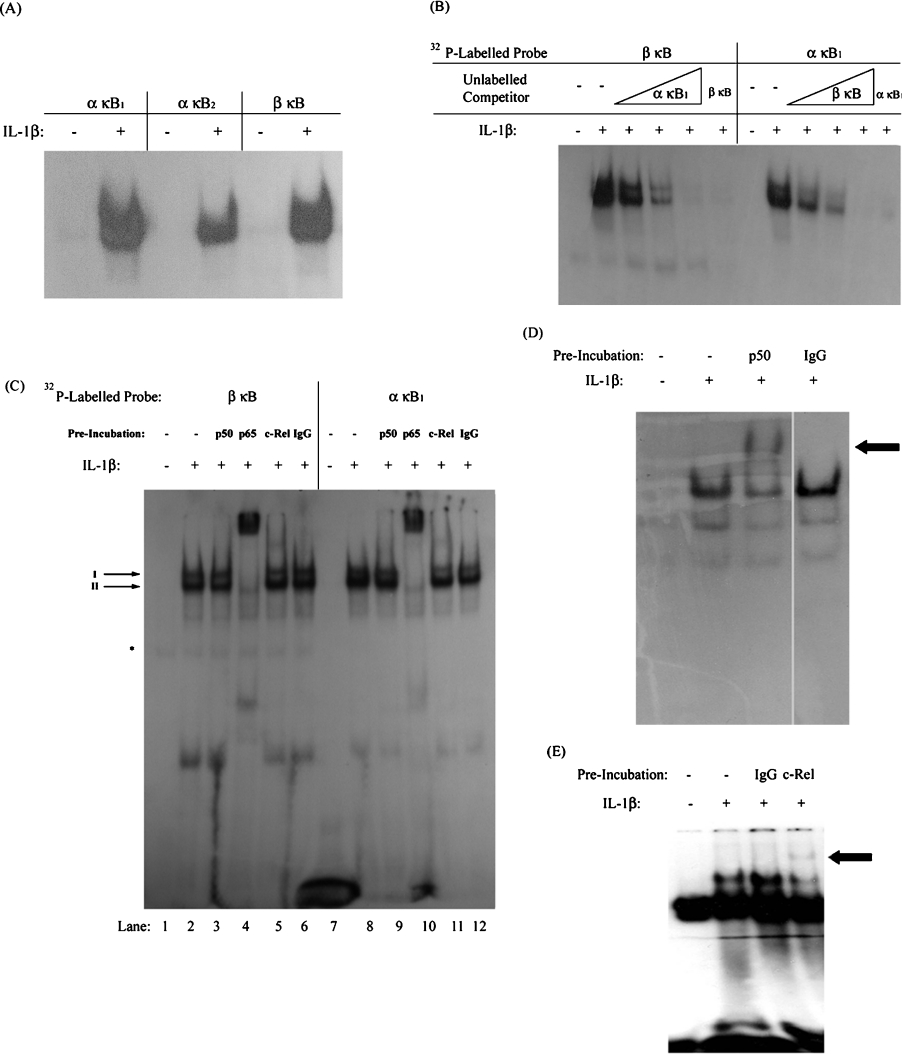
The binding of NF-κB to the IκBβ promoter κB site was initially characterized using an in vitro assay. A range of oligonucleotides containing single κB sites were radiolabelled for use in EMSA analysis. Two of these oligonucleotides contained known, functional κB sites from the IκBα promoter-designated α-κB1 and α-κB2 [22]. A third included the κB site from the IκBβ promoter – β-κB. Since we were especially interested in examining the differential effects of IL-1β on IκBα and IκBβ expression in human astrocytes, 1321N1 astrocytoma were used as an accepted model of the astrocytic cell system in which IL-1β is known to cause strong and sustained activation of NF-κB. Thus the probes were individually incubated with nuclear extracts from 1321N1 cells treated with or without IL-1β for 30 min. All three radiolabelled probes were found to bind activated NF-κB (Figure 2A). Given the ability of the β-κB site to bind NF-κB in vitro, additional EMSA analysis was conducted to compare the NF-κB binding to this site with that to the α-κB1 site. In a competition EMSA, effective competition of NF-κB binding with radiolabelled β-κB probe was achieved by pre-incubating nuclear extracts from cells stimulated with IL-1β for 30 min with increasing concentrations of unlabelled α-κB1 oligonucleotide (Figure 2B). Similarly, unlabelled β-κB oligonucleotide competed the binding of NF-κB to a radiolabelled α-κB1 probe. This indicates that the same form of NF-κB can bind to the β-κB site and the α-κB1 site in vitro. In addition, activated NF-κB appears to display similar affinity for both κB sites, as a similar concentration of one unlabelled oligonucleotide was required to eliminate binding to the other radiolabelled probe.
Figure 2. NF-κB binds to the κB site of the IκBβ promoter in vitro.
1321N1 astrocytoma were treated with or without IL-1β (10 ng/ml) for 30 min before examining NF-κB DNA-binding activity by EMSA. (A) Nuclear extracts (10 μg of protein) were incubated with radiolabelled oligonucleotides containing the IκBβ, IκBα-κB1 and IκBα-κB2 sites. (B) Nuclear extracts (10 μg of protein) were incubated with increasing amounts of unlabelled IκBα-κB1 oligonucleotide (0.07–1.75 pmol) or with an excess of unlabelled IκBβ-κB oligonucleotide (1.75 pmol) before addition of radiolabelled IκBβ-κB probe. Also, nuclear extracts were incubated with increasing amounts of unlabelled IκBβ-κB oligonucleotide (0.07–1.75 pmol) or with an excess of unlabelled IκBα-κB1 oligonucleotide (1.75 pmol) before adding radiolabelled IκBα-κB1 probe. (C) Nuclear extracts were pre-incubated with anti-p50, anti-p65, anti-c-Rel or non-immune (IgG) antibody. *Denotes an unidentified binding complex. (D, E) Nuclear extracts were pre-incubated with non-immune (IgG) and anti-p50 (D) or anti-c-Rel (E) antibody before addition of radiolabelled probe containing the HIV-LTR κB site. The arrows indicate the mobility of supershifted complexes.
To further characterize NF-κB binding to the β-κB site, nuclear extracts from IL-1β-treated cells were pre-incubated with antisera against the Rel proteins p50, p65 and c-Rel prior to addition of radiolabelled β-κB probe and subsequent electrophoresis. As shown above, IL-1β induces the activation of NF-κB, observed here as two major complexes of similar electrophoretic mobility (Figure 2C, lane 2). These IL-1β-induced complexes were unaffected by pre-incubation with the non-immune IgG isotype control (lane 6). Complexes I and II were supershifted by preincubation with anti-p65 antiserum, with complex I disappearing and complex II substantially reduced in intensity (lane 4). Preincubation with anti-c-Rel antiserum also caused the appearance of a faint supershifted complex, although this was not accompanied by a significant decrease in the level of any IL-1β-induced NF-κB–DNA complex (lane 5). This indicates that p65 was contained in both of the IL-1β-induced NF-κB complexes that bound to the β-κB site, while a complex containing c-Rel most likely constitutes a fraction of either complex I or II. No supershift was observed upon pre-incubation with anti-p50 antibody (lane 3). The ability of both the anti-p50 and anti-c-Rel antibodies to cause a supershift of NF-κB–DNA complexes was confirmed using a radiolabelled probe containing the HIV-LTR κB site and nuclear extracts from 1321N1 astrocytoma stimulated in the presence and absence of IL-1β (Figures 2D and 2E).
Interestingly, an identical profile of supershifted complexes was observed when EMSAs were performed with radiolabelled α-κB1 following pre-incubation with anti-p65 or anti-c-Rel antisera (Figure 2C, lanes 7–12). It therefore appears that despite the difference in κB site sequence, the α-κB1 and β-κB sites bind the same range of NF-κB dimers in vitro. The Rel subunits p65 and c-Rel harbour substantial transactivation potential, arising from the presence of C-terminal TADs [23,24]. This suggests that transcriptionally active complexes of NF-κB bind similarly to the κB site of the IκBα and IκBβ promoters and that any differences in IL-1β-induced expression of IκBα and IκBβ are not due to the differential binding of NF-κB to the two promoters. However, while the EMSA procedure offers information on the ability of NF-κB to bind to oligonucleotides in vitro, transcription factor interaction with cis-elements in the intact cell depends on chromatin structure and promoter accessibility. Therefore we next investigated whether NF-κB could bind to the IκBα and IκBβ promoters in vivo.
p65 binds to the IκBβ promoter in vivo
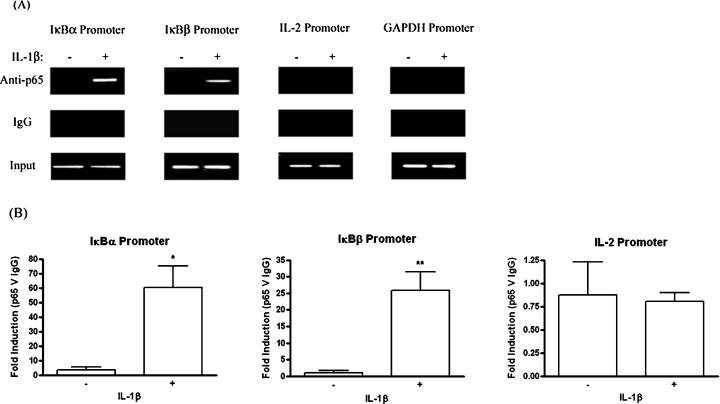
ChIP studies were employed to examine whether IL-1β-mediated NF-κB activation in human astrocytes led to the association of p65 with the IκBβ promoter in vivo. As NF-κB is known to mediate the induction of IκBα following activation [9], binding to the IκBα promoter was used as a positive control for this assay. 1321N1 astrocytoma were stimulated with or without IL-1β for 1 h prior to immunoprecipitation of nuclear lysates with anti-p65 antisera. Intriguingly, analysis of immunoprecipitated DNA by PCR indicated that p65 was present at both the IκBα and IκBβ promoters 1 h post-IL-1β stimulation (Figure 3A). This was corroborated by real-time PCR analysis of immunoprecipitated DNA, where statistically significant levels of p65 binding to both promoters was recorded in response to IL-1β (Figure 3B). The specificity of IκBα and IκBβ promoter enrichment was confirmed by the absence of GAPDH promoter in p65 immunoprecipitates. In addition, p65 binding was not detected at the IL-2 promoter by standard or real-time PCR of immunoprecipitated DNA. The IL-2 gene is regulated by NF-κB, but is only expressed in T-cells. Therefore the appearance of p65 at a promoter in response to IL-1β is not a general phenomenon at all promoters containing κB sites, but rather is specific to a subset, including the IκBβ promoter. As IL-1β stimulation induced the binding of NF-κB to the κB site of the IκBβ promoter in vitro, and resulted in p65 engagement of the IκBβ promoter in vivo, we next assessed the ability of the human IκBβ κB site to confer inducibility on IL-1β in functional assays.
Figure 3. IL-1β induces p65 binding to the IκBβ gene promoter in vivo.
1321N1 astrocytoma were treated with or without IL-1β (10 ng/ml) for 1 h. Nuclear lysates were generated and immunoprecipitated with anti-p65 or control IgG antibodies. Immunoprecipitated DNA was analysed for IκBα, IκBβ, IL-2 and GAPDH promoters by PCR and gel electrophoresis (A) and IκBα, IκBβ and IL-2 promoters by quantitative real-time PCR (B). Data from real-time PCR are expressed as fold differences between DNA enrichment in the anti-p65 ChIP sample relative to IgG ChIP sample and are displayed as means±S.E.M. for three to five independent experiments. *P<0.05, **P<0.01 versus unstimulated control (unpaired t test).
The β-κB site confers low inducibility on IL-1β in transient transfection assays
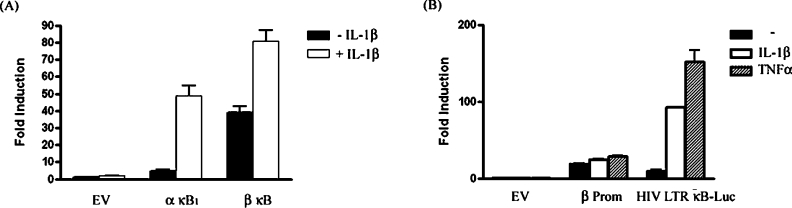
Promoter–reporter constructs containing five consecutive β-κB or α-κB1 sites upstream of a luciferase reporter gene (β-κB–luciferase and α-κB1–luciferase) were generated as described in the Materials and methods section. HEK-293 cells transfected with one of these constructs or empty pGL3-Basic vector were treated with or without IL-1β for 6 h before assessing reporter gene activity. IL-1β stimulation caused a 10-fold induction of α-κB1–luciferase expression. In contrast, only 2-fold induction of β-κB–luciferase expression was observed upon IL-1β treatment (Figure 4A). Together with the EMSA analysis, these data suggest that while both the β-κB and α-κB1 sites bind identical forms of NF-κB, stronger transactivation is seen from α-κB1-bound NF-κB. The reason for the high basal level of β-κB–luciferase activity relative to α-κB1–luciferase is currently unknown. However, EMSA analysis detected an unidentified complex binding to the β-κB, but not α-κB1, probe, which was unaffected by IL-1β treatment or pre-incubation with any anti-Rel subunit antisera (Figure 2C, denoted by ‘*’). While sequence analysis did not reveal the presence of any known cis-elements within the β-κB site that were absent from the α-κB1 site, it is possible that this complex represents a factor involved in stimulating basal levels of reporter gene activity.
Figure 4. NF-κB causes strong activation of the IκBα promoter but weak activation of the IκBβ promoter in 1321N1 cells.
(A) HEK-293 cells were co-transfected with α-κB1–luciferase, β-κB–luciferase or empty pGL3-Basic vector (EV), as well as phRL-TK (constitutively expressed Renilla luciferase). After overnight recovery, cells were stimulated in the presence or absence of IL-1β (10 ng/ml) for 6 h. (B) HEK-293 cells were co-transfected with β-Prom, HIV-LTR κB-luc or empty pGL3-Basic vector (EV), and phRL-TK. After overnight recovery, cells were stimulated in the presence or absence of IL-1β or TNFα (10 ng/ml) for 6 h. Cell lysates were assayed for firefly and Renilla luciferase. Data are presented relative to untreated cells transfected with empty vector. Results represent means±S.E.M. for three independent experiments.
In order to determine whether the modest IL-1β-mediated inducibility seen with the β-κB-luciferase construct was retained within the context of the intact promoter, we next cloned the human IκBβ 5′-flanking region. The IκBβ gene had previously been assigned to chromosome band 19q13 by fluorescence in situ hybridization [25]. A search of the NCBI database using the IκBβ cDNA sequence identified a contig (CTC-360G5) containing the IκBβ gene. An approx. 1 kb fragment of the 5′-flanking region was amplified by PCR and cloned into the pGL3-Basic vector upstream of the luciferase gene as described in the Materials and methods section. HEK-293 cells transfected with the IκBβ-promoter-reporter (β-Prom) or empty pGL3-Basic vector were stimulated in the presence or absence of IL-1β or TNFα. β-Prom displayed a high level of basal activity, approx. 20-fold over empty vector (Figure 4B). Treatment with IL-1β and TNFα, another pro-inflammatory cytokine known to activate NF-κB, caused a 1.3- and 1.5-fold induction of β-Prom activity respectively. In order to verify the responsiveness of HEK-293 cells to these cytokines, cells were transfected with a promoter-reporter construct containing a luciferase gene immediately downstream of five tandemly repeated κB sites from the HIV LTR (HIV-LTR κB-luc). IL-1β and TNF induced a 9- and 13-fold induction of reporter gene activity respectively.
Thus transient transfections using β-κB–luciferase and the IκBβ-promoter-reporter indicated that NF-κB activation does not lead to substantial transactivation at the IκBβ promoter. As these constructs constitute artificial extrachromosomal templates, we also examined the effect of NF-κB activity on endogenous IκBβ expression at the level of both mRNA and protein.
Activation of NF-κB does not induce endogenous IκBβ gene expression
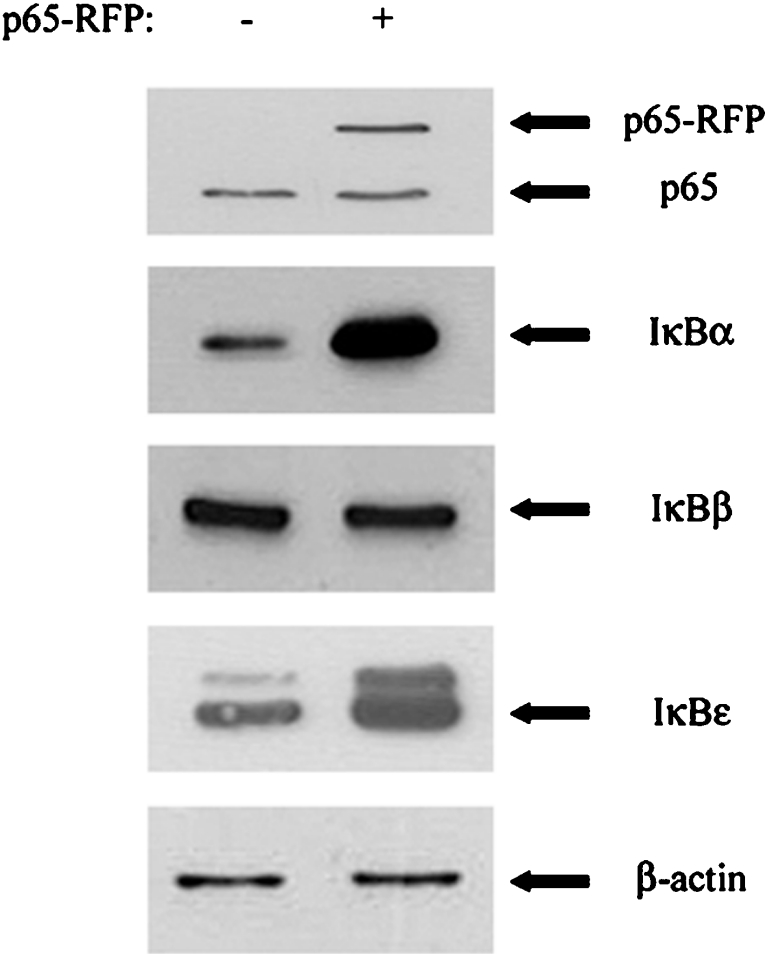
To examine whether IκBβ gene expression was induced by NF-κB at the transcriptional level, 1321N1 astrocytoma were treated with or without IL-1β, a strong activator of NF-κB, for various time periods. mRNA levels were then quantified by real-time PCR. No significant up-regulation of IκBβ mRNA was recorded following IL-1β stimulation for 3, 8 or 24 h (results not shown). This confirms our earlier Northern-blot analysis of IκBβ transcript levels in response to activators of NF-κB [8,13]. As a positive control, IL-8 mRNA levels were also monitored by quantitative real-time PCR, with a robust up-regulation of mRNA expression observed in response to IL-1β (results not shown). The effect of NF-κB activation on IκBβ protein levels was also assessed. p65 was overexpressed in HEK-293 cells, before monitoring levels of IκBα, IκBβ and another IκB family member, IκBϵ, by Western immunoblotting (Figure 5). The expression of IκBα and IκBϵ is known to be up-regulated by NF-κB [13], and this was reflected by increased levels of these proteins detected in cells expressing the p65–RFP fusion protein. However, levels of IκBβ were equivalent in cells transfected with and without the p65 expression vector. Together, these data demonstrate that expression of endogenous IκBβ is not increased by activation of NF-κB, despite the binding of p65 to the IκBβ promoter in an IL-1β-dependent manner.
Figure 5. NF-κB causes strong induction of endogenous IκBα but fails to up-regulate endogenous IκBβ gene expression in HEK-293 cells.
HEK-293 cells were transfected with a plasmid expressing a p65–RFP fusion protein or empty RFP vector. Cell lysates were generated after 24 h and subjected to SDS/PAGE and Western immunoblotting to measure levels of p65, IκBα, IκBβ, IκBϵ and β-actin. These results are representative of three independent experiments.
As shown above, p65 was found to be present at the IκBβ promoter following 1 h stimulation with IL-1β. The IκBβ mRNA 3′-UTR (untranslated region) does not contain an AU-rich sequence. This indicates that the transcript is not subject to rapid turnover and, were mRNA levels to be up-regulated, this would most likely be detectable at 3 h. In addition, previous Northern blots performed by this group have demonstrated the absence of an IκBβ mRNA induction after 1 h IL-1β treatment [13]. The question thus remained as to how binding of p65 at the IκBβ promoter failed to lead to transcriptional activation. To address this issue, we returned to ChIP analysis of the IκBβ promoter, monitoring the recruitment of a critical component of the transcriptional machinery, RNA Pol II.
p65 binding to the IκBβ promoter results in relatively inefficient recruitment and stalling of RNA Pol II at the promoter
Transcriptional activators operate by binding to a gene promoter and increasing the rate of transcriptional initiation. Following site-specific DNA binding, p65 can recruit components of the basal transcription apparatus either directly or indirectly, via co-activators such as CBP [CREB (cAMP-response-element-binding)-binding protein]/p300 [26–28]. Following initiation of transcription, RNA Pol II moves along the gene in the elongation phase. The presence or absence of RNA Pol II at a gene promoter, and within the gene at sites distant from the TSS, provides a picture of the transcriptional status of that gene. We chose three target genes in which to monitor RNA Pol II activity, IκBα, IκBβ and IL-8. p65 binds to the promoters of all three targets in an IL-1β-dependent manner (Figure 3 and [16]).
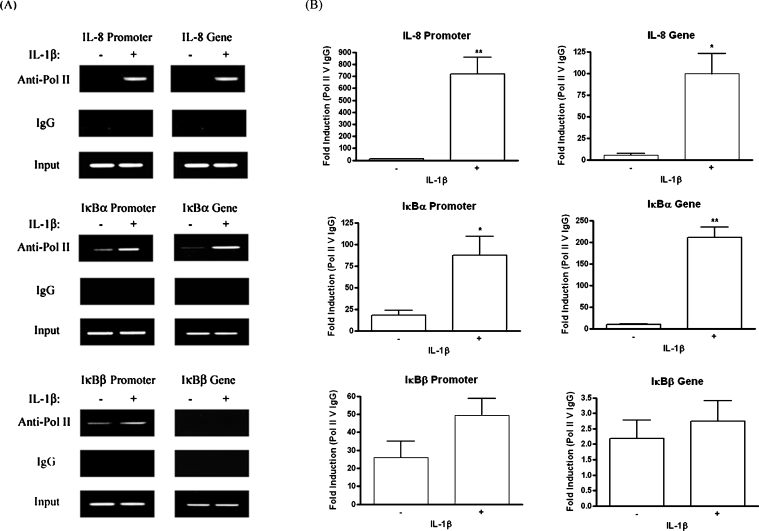
As for p65-ChIP analysis, 1321N1 astrocytoma cells were treated with or without IL-1β for 1 h. Following anti-Pol II immunoprecipitation of nuclear lysates, immunoprecipitated DNA was analysed for the presence of target sequences by standard and quantitative real-time PCR (Figure 6). For each gene, amplification of two target sequences was performed. The first was located in the promoter, just upstream of the TSS. The second was at least 2000 bp downstream from the TSS. As the average size of sonicated chromatin was 750 bp, this second target was considered to be sufficiently far from the TSS to avoid false positive results, otherwise attainable by the immunoprecipitation of RNA Pol II stalled at the upstream promoter. While absent from the IL-8 promoter in the basal state, RNA Pol II binding occurred following IL-1β stimulation. Furthermore, the enzyme was detected downstream in the IL-8 gene, in response to IL-1β, demonstrating the effectiveness of Pol II-ChIP as a method to study IL-1β-induced changes in transcriptional activity at target genes. Some RNA Pol II was present at the IκBα promoter in the basal state, but an increase in binding was observed in response to IL-1β, and this increase was deemed statistically significant by analysis of quantitative real-time PCR results. In addition, examination of a downstream region of the IκBα gene showed that active transcription is in progress 1 h post-IL-1β stimulation.
Figure 6. Stimulation of 1321N1 cells with IL-1β results in relatively inefficient recruitment and stalling of RNA Pol II at the IκBβ promoter.
1321N1 astrocytoma were stimulated in the presence or absence of IL-1β (10 ng/ml) for 1 h. Nuclear lysates were generated and immunoprecipitated with anti-Pol II or control IgG antibodies. Immunoprecipitated DNA was analysed for IL-8, IκBα and IκBβ promoter and gene sequences by PCR and gel electrophoresis (A) and by quantitative real-time PCR analysis (B). Data from real-time PCR are expressed as fold differences between DNA enrichment in the anti-Pol II ChIP sample relative to IgG ChIP sample and are displayed as means±S.E.M. for three independent experiments. *P<0.05, **P<0.01 versus unstimulated control (unpaired t-test).
As shown earlier, IL-1β induces binding of p65 to the IκBβ promoter in vivo, but this is not accompanied by an up-regulation of gene expression. Pol II ChIP showed that RNA Pol II is found at the IκBβ promoter in unstimulated cells. IL-1β treatment tended to cause some further recruitment of the enzyme. While quantitative real-time PCR indicated that the increase was approx. 2-fold, it was not found to be statistically significant at P<0.05. No RNA Pol II was detected downstream within the IκBβ gene sequence in the presence or absence of IL-1β. This indicates that while IL-1β induces the binding of NF-κB to the IκBβ promoter, p65 is relatively inefficient at recruiting RNA Pol II to the TSS, and furthermore, the recruited polymerase stalls at the promoter.
p65 binding to the IκBβ promoter is not followed by TFIIH recruitment
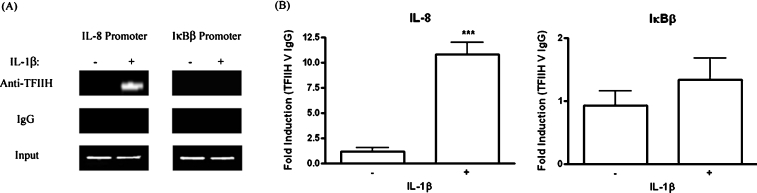
In the stepwise assembly model of transcriptional initiation, preinitiation complex formation is completed by the loading of TFIIH [29]. This multi-subunit general transcription factor possesses a DNA helicase and a kinase activity, the latter of which phosphorylates the evolutionarily conserved heptapeptide repeat in the CTD (C-terminal domain) of RNA Pol II. This modification appears necessary for transcriptional initiation, possibly by inducing a conformational change that allows the polymerase to disengage from the pre-initiation complex [30]. In order to probe the mechanism underlying the failure of IκBβ promoter-bound RNA Pol II to initiate transcription, ChIP assays were performed to assess the genomic location of p89, the largest TFIIH subunit, following IL-1β treatment of 1321N1 astrocytoma. Standard and real-time PCRs of p89-immunoprecipitated DNA demonstrated that IL-1β caused loading of p89 on the IL-8 promoter, with real-time PCR recording a 9-fold induction of binding. However, no binding was detectable at the IκBβ promoter (Figures 7A and 7B). This strongly suggests that the absence of IκBβ promoter firing in response to NF-κB activation stems from the failure of DNA-bound p65 to efficiently recruit RNA Pol II and to directly or indirectly engage TFIIH, with a resulting lack of RNA Pol II CTD phosphorylation and promoter clearance. This is a highly credible mechanism underlying the non-inducibility of the IκBβ gene by NF-κB, and thus IL-1β, in human astrocytes.
Figure 7. TFIIH is not recruited to the IκBβ promoter in response to IL-1 stimulation of 1321N1 cells.
1321N1 astrocytoma were stimulated in the presence or absence of IL-1β (10 ng/ml) for 1 h. Nuclear lysates were generated and immunoprecipitated with anti-p89 or control IgG antibodies. Immunoprecipitated DNA was analysed for IL-8 and IκBβ promoter sequences by PCR and gel electrophoresis (A) and by quantitative real-time PCR analysis (B). Data from real-time PCR are expressed as fold differences between DNA enrichment in the anti-p89 ChIP sample relative to IgG ChIP sample and are displayed as means±S.E.M. for three or four independent experiments. ***P<0.001 versus unstimulated control (unpaired t test).
DISCUSSION
The activity of transcription factors must be tightly marshalled to avoid inappropriate gene expression. IκB proteins are charged with regulating NF-κB activity. A notable disparity in the regulation of IκBα and IκBβ expression is the strong induction of the former, but not the latter, in response to NF-κB activation. As functional compensation by IκBβ did not occur in IκBα−/− mice, it has been proposed that it is the up-regulation of IκBα, but not IκBβ, following activation of NF-κB that underlies the difference in activity of the two IκBs. In a study of the murine IκBβ promoter, Budde et al. [14] identified a κB site upstream of the TSS and demonstrated that NF-κB could bind to the site and cause modest transcriptional activation. The human IκBβ promoter has a κB site in a similar position. We examined the human IκBβ κB site at the level of NF-κB binding, and assessed the functional consequences of NF-κB activation on IκBβ promoter activity. We were especially interested in understanding the role of NF-κB in the differential induction of IκBα and IκBβ by IL-1β in human astrocytes, a situation that leads to sustained activation of NF-κB and likely chronic neuroinflammation.
EMSA analysis demonstrated that NF-κB, activated in response to IL-1β, could interact with the IκBβ-κB site in addition to two functionally competent κB sites from the IκBα promoter. The consistent electrophoretic mobility of the NF-κB–DNA complexes detected with different probes indicated that a similar form of NF-κB was mediating binding in each case. This likelihood was strengthened through competition EMSAs, where the NF-κB complex was found to bind to both the β-κB and α-κB1 probes with equal affinity. Further support came from the immunodetection of the same Rel subunits in NF-κB complexes bound to both sites in vitro. p65 was found to be contained in all IL-1β-induced NF-κB complexes, with the presence of c-Rel also apparent, although to a lesser extent. Thus EMSA analysis showed that a form of NF-κB with substantial transactivation potential can bind to the IκBβ promoter κB site in vitro. ChIP analysis was performed to investigate whether this interaction had any relevance in living cells. p65 was chosen as the target Rel subunit since it had been identified in the NF-κB complexes as the predominant subunit to interact with both the β-κB and α-κB1 probes in vitro. Furthermore, its presence at a promoter could point towards an up-regulation of transcriptional activity. Interestingly, like the IκBα promoter, p65 was detected at the IκBβ promoter in an IL-1β-dependent manner.
Previous studies have suggested that NF-κB activation has, at best, a modest effect on IκBβ expression. Nevertheless, given the binding of p65 to the IκBβ promoter induced by IL-1β, we examined the effect of NF-κB activation on IκBβ promoter activity in transient transfection assays, where it was shown to confer only a low level of inducibility. The effect of NF-κB activation on endogenous IκBβ gene expression was then assessed at the level of both mRNA and protein, where no up-regulation was observed. This indicated that the binding of p65 to the IκBβ promoter was insufficient to cause transcriptional activation of the gene.
p65 contains two independent TADs in its C-terminal portion [31]. Through these TADs, p65 can interact with co-activators and components of the basal transcription apparatus [27,28]. p65 has in fact been shown to interact directly with general transcription factors, thereby increasing the rate at which transcription is initiated at the relevant promoter [26]. ChIP studies with anti-RNA Pol II allowed us to monitor the recruitment of the enzyme to the IκBβ, IκBα and IL-8 promoters. In addition, by selecting a second target sequence downstream of the promoter, within which to probe for the presence of RNA Pol II, it was possible to ascertain whether the gene was being actively transcribed after 1 h IL-1β. IL-8 is known to be strongly induced by IL-1β, while not being expressed in basal state. Indeed, a major recruitment of RNA Pol II to the IL-8 promoter was recorded in response to IL-1β. As the enzyme was also detected within the IL-8 gene sequence in an IL-1β-dependent manner, this appeared to be followed by transcriptional activity. While RNA Pol II was detected at the IκBα promoter in unstimulated cells, further enzyme was recruited in response to IL-1β and, as for IL-8, RNA Pol II-mediated transcription resulted. RNA Pol II was also detected at the IκBβ promoter in the basal state. While IL-1β appeared to cause some further engagement, no RNA Pol II was detected downstream within the IκBβ gene sequence, in the presence or absence of IL-1β stimulation. It should be noted that the target sequences in both the IκBα and IL-8 genes were within exons, while the IκBβ gene target was within intronic DNA. It has been suggested that RNA Pol II accumulates at exons during transcription, making coding DNA a preferable target in Pol II ChIP [32,33]. However, technical difficulties precluded the use of a sequence within the sufficiently large exons 3 or 5 in the IκBβ gene. The target sequence used was just 5′ of exon 2. As the average chromatin size was 750 bp, it was expected that Pol II-immunoprecipitated DNA would have been enriched for this sequence, were the enzyme present in exon 2 during transcription.
p65 can therefore bind to the IκBβ promoter in response to IL-1β, although this does not result in efficient recruitment of RNA Pol II. In addition, the modest levels of recruited polymerase appear to stall at the promoter. As promoter clearance requires the phosphorylation of Ser5 of the RNA polymerase II CTD heptapeptide sequence by the Cdk7 subunit of TFIIH [30,34], we finally investigated whether this general transcription factor was recruited to the IκBβ promoter post-IL-1β stimulation. However, unlike the κB-responsive IL-8 promoter, p65 binding to the IκBβ promoter was not followed by loading of TFIIH. A mechanism may thus be proposed whereby p65 binding to the IκBβ promoter causes a slight recruitment of RNA Pol II but fails to facilitate TFIIH loading, with a resulting absence of transcriptional initiation and gene expression.
The failure of IκBβ-promoter-bound p65 to interact efficiently with the basal transcription apparatus, either directly or indirectly through co-activators, may occur for various reasons. NF-κB can bind to a range of κB site sequences and it has been proposed that the nucleotide sequence of a κB site dictates the conformation of the bound NF-κB dimer. This allosteric role for DNA has a precedent in the binding characteristics of the nuclear receptor protein family [35]. The NF-κB conformation induced by the κB site has functional consequences. An alteration of the site sequence can impact transcriptional activation mediated by bound NF-κB, most likely by modifying the ability of NF-κB to interact with other trans-acting factors or with co-activators [20,36]. Thus an induced conformation that does not favour recruitment of coactivators or basal transcription factors is one possible explanation for the failure of IκBβ promoter-bound p65 to activate transcription. However, an identical decameric κB site supports transactivation in the MIP (macrophage inflammatory protein)-2 and MCP (monocyte chemoattractant protein)-1 promoters [37,38]. Furthermore, in the case of the MCP-1 promoter, p65–p65 homodimers and p65–c-Rel heterodimers, the Rel subunits found to bind to the IκBβ promoter κB site in vitro, display the ability to activate transcription through this site [37]. However, it should be noted that the sequence flanking the κB site also influences the conformation of NF-κB–DNA complexes, as the structure of DNA is sequence-dependent [39–41]. As the flanking sequences differ between MCP-1and IκBβ, this could lead to the same NF-κB complex binding to identical decameric κB site sequences, but assuming different conformations due to the divergence in κB site DNA structure imposed by surrounding bases.
The characterization of many promoters in which NF-κB is known to play an important role in transcriptional activation has revealed the requirement for numerous other trans-acting factors and co-activators, combining to form a gene-specific enhanceosome [42,43]. p65 binding to the IκBβ promoter may provide just one part of the combinatorial complex necessary for gene expression. Other signals may be provided in a stimulus- and cell-type-specific manner. In addition, post-translational modifications of p65 have been shown to regulate interactions with co-activators. Such modifications are stimulus-dependent and may even occur in a promoter-specific manner, highlighting the complexities of gene regulation at the level of the promoter [44,45].
The failure of NF-κB activation to elicit a substantial increase in IκBβ gene expression has previously been demonstrated [8,13]. Here, the model is advanced to show that NF-κB can in fact bind to the IκBβ promoter, but that this does not trigger efficient general transcription factor or RNA Pol II recruitment or promoter firing. The present findings also provide a potential mechanistic understanding of the basis for sustained activation of NF-κB in some systems. We have previously shown that IL-1β causes sustained activation of NF-κB in human astrocytes and this is likely to play a key role in promoting chronic inflammation in the brain [46]. Such sustained activation coincides with strong induction of IκBα, but lack of resynthesis of IκBβ, suggesting the latter to be a key regulator of NF-κB in this system. We now define the molecular basis of the differential effects of IL-1β on IκBα and IκBβ expression in human astrocytes, in that IL-1β fails to stimulate efficient recruitment or processing of the basal transcriptional machinery at the IκBβ promoter. The delineation of mechanisms that increase such an efficiency may be of great value in up-regulating IκBβ expression and controlling chronic activation of NF-κB in the brain.
Acknowledgments
This work was supported by funding from the Health Research Board of Ireland and Science Foundation Ireland.
References
- 1.Kopp E. B., Ghosh S. NF-κB and rel proteins in innate immunity. Adv. Immunol. 1995;58:1–27. doi: 10.1016/s0065-2776(08)60618-5. [DOI] [PubMed] [Google Scholar]
- 2.Thanos D., Maniatis T. NF-κB: a lesson in family values. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 3.Nolan G. P., Ghosh S., Liou H. C., Tempst P., Baltimore D. DNA binding and IκB inhibition of the cloned p65 subunit of NF-κB, a rel-related polypeptide. Cell. 1991;64:961–969. doi: 10.1016/0092-8674(91)90320-x. [DOI] [PubMed] [Google Scholar]
- 4.Baeuerle P. A., Baltimore D. IκB: a specific inhibitor of the NF-κB transcription factor. Science. 1988;242:540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- 5.Verma I. M., Stevenson J. K., Schwarz E. M., Van Antwerp D., Miyamoto S. Rel/NF-κB/IκB family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 6.Brockman J. A., Scherer D. C., McKinsey T. A., Hall S. M., Qi X., Lee W. Y., Ballard D. W. Coupling of a signal response domain in IκBα to multiple pathways for NF-κB activation. Mol. Cell. Biol. 1995;15:2809–2818. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiDonato J. A., Hayakawa M., Rothwarf D. M., Zandi E., Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 8.Thompson J. E., Phillips R. J., Erdjument-Bromage H., Tempst P., Ghosh S. IκB-β regulates the persistent response in a biphasic activation of NF-κB. Cell. 1995;80:573–582. doi: 10.1016/0092-8674(95)90511-1. [DOI] [PubMed] [Google Scholar]
- 9.Sun S. C., Ganchi P. A., Ballard D. W., Greene W. C. NF-κB controls expression of inhibitor IκBα: evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- 10.Arenzana-Seisdedos F., Turpin P., Rodriguez M., Thomas D., Hay R. T., Virelizier J. L., Dargemont C. Nuclear localization of IκBα promotes active transport of NF-κB from the nucleus to the cytoplasm. J. Cell Sci. 1997;110:369–378. doi: 10.1242/jcs.110.3.369. [DOI] [PubMed] [Google Scholar]
- 11.Beg A. A., Sha W. C., Bronson R. T., Baltimore D. Constitutive NF-κB activation, enhanced granulopoiesis, and neonatal lethality in IκBα-deficient mice. Genes Dev. 1995;9:2736–2746. doi: 10.1101/gad.9.22.2736. [DOI] [PubMed] [Google Scholar]
- 12.Cheng J. D., Ryseck R. P., Attar R. M., Dambach D., Bravo R. Functional redundancy of the nuclear factor κB inhibitors IκBα and IκBβ. J. Exp. Med. 1998;188:1055–1062. doi: 10.1084/jem.188.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourke E., Kennedy E. J., Moynagh P. N. Loss of IκB-β is associated with prolonged NF-κB activity in human glial cells. J. Biol. Chem. 2000;275:39996–40002. doi: 10.1074/jbc.M007693200. [DOI] [PubMed] [Google Scholar]
- 14.Budde L. M., Wu C., Tilman C., Douglas I., Ghosh S. Regulation of IκBβ expression in testis. Mol. Biol. Cell. 2002;13:4179–4194. doi: 10.1091/mbc.01-07-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J., Fritsch E. F., Maniatis T. 2nd edn. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 16.Curran N. M., Griffin B. D., O'Toole D., Brady K. J., Fitzgerald S. N., Moynagh P. N. The synthetic cannabinoid R(+)WIN 55,212-2 inhibits the interleukin-1 signaling pathway in human astrocytes in a cannabinoid receptor-independent manner. J. Biol. Chem. 2005;280:35797–35806. doi: 10.1074/jbc.M507959200. [DOI] [PubMed] [Google Scholar]
- 17.Saccani S., Pantano S., Natoli G. p38-dependent marking of inflammatory genes for increased NF-κB recruitment. Nat. Immunol. 2002;3:69–75. doi: 10.1038/ni748. [DOI] [PubMed] [Google Scholar]
- 18.Chakrabarti S. K., James J. C., Mirmira R. G. Quantitative assessment of gene targeting in vitro and in vivo by the pancreatic transcription factor, Pdx1: importance of chromatin structure in directing promoter binding. J. Biol. Chem. 2002;277:13286–13293. doi: 10.1074/jbc.M111857200. [DOI] [PubMed] [Google Scholar]
- 19.Budde L. M., Ghosh S. Cloning and characterization of the gene encoding mouse IκBβ. Gene. 2000;247:279–286. doi: 10.1016/s0378-1119(00)00051-2. [DOI] [PubMed] [Google Scholar]
- 20.Leung T. H., Hoffmann A., Baltimore D. One nucleotide in a κB site can determine cofactor specificity for NF-κB dimers. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Kunsch C., Ruben S. M., Rosen C. A. Selection of optimal κB/Rel DNA-binding motifs: interaction of both subunits of NF-κB with DNA is required for transcriptional activation. Mol. Cell. Biol. 1992;12:4412–4421. doi: 10.1128/mcb.12.10.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Bail O., Schmidt-Ullrich R., Israel A. Promoter analysis of the gene encoding the IκB-α/MAD3 inhibitor of NF-κB: positive regulation by members of the rel/NF-κB family. EMBO J. 1993;12:5043–5049. doi: 10.1002/j.1460-2075.1993.tb06197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore P. A., Ruben S. M., Rosen C. A. Conservation of transcriptional activation functions of the NF-κB p50 and p65 subunits in mammalian cells and Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:1666–1674. doi: 10.1128/mcb.13.3.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bull P., Morley K. L., Hoekstra M. F., Hunter T., Verma I. M. The mouse c-rel protein has an N-terminal regulatory domain and a C-terminal transcriptional transactivation domain. Mol. Cell. Biol. 1990;10:5473–5485. doi: 10.1128/mcb.10.10.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamoto T., Ono T., Hori M., Yang J. P., Tetsuka T., Kawabe T., Sonta S. Assignment of the IκB-β gene NFKBIB to human chromosome band 19q13.1 by in situ hybridization. Cytogenet. Cell Genet. 1998;82:105–106. doi: 10.1159/000015077. [DOI] [PubMed] [Google Scholar]
- 26.Schmitz M. L., Stelzer G., Altmann H., Meisterernst M., Baeuerle P. A. Interaction of the COOH-terminal transactivation domain of p65 NF-κB with TATA-binding protein, transcription factor IIB, and coactivators. J. Biol. Chem. 1995;270:7219–7226. doi: 10.1074/jbc.270.13.7219. [DOI] [PubMed] [Google Scholar]
- 27.Gerritsen M. E., Williams A. J., Neish A. S., Moore S., Shi Y., Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc. Natl. Acad. Sci. U.S.A. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kee B. L., Arias J., Montminy M. R. Adaptor-mediated recruitment of RNA polymerase II to a signal-dependent activator. J. Biol. Chem. 1996;271:2373–2375. doi: 10.1074/jbc.271.5.2373. [DOI] [PubMed] [Google Scholar]
- 29.Zawel L., Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu. Rev. Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- 30.Cismowski M. J., Laff G. M., Solomon M. J., Reed S. I. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol. Cell. Biol. 1995;15:2983–2992. doi: 10.1128/mcb.15.6.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitz M. L., dos Santos Silva M. A., Baeuerle P. A. Transactivation domain 2 (TA2) of p65 NF-κB: similarity to TA1 and phorbol ester-stimulated activity and phosphorylation in intact cells. J. Biol. Chem. 1995;270:15576–15584. doi: 10.1074/jbc.270.26.15576. [DOI] [PubMed] [Google Scholar]
- 32.Sandoval J., Rodriguez J. L., Tur G., Serviddio G., Pereda J., Boukaba A., Sastre J., Torres L., Franco L., Lopez-Rodas G. RNAPol-ChIP: a novel application of chromatin immunoprecipitation to the analysis of real-time gene transcription. Nucleic Acids Res. 2004;32:e88. doi: 10.1093/nar/gnh091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brodsky A. S., Meyer C. A., Swinburne I. A., Hall G., Keenan B. J., Liu X. S., Fox E. A., Silver P. A. Genomic mapping of RNA polymerase II reveals sites of co-transcriptional regulation in human cells. Genome Biol. 2005;6:R64. doi: 10.1186/gb-2005-6-8-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trigon S., Serizawa H., Conaway J. W., Conaway R. C., Jackson S. P., Morange M. Characterization of the residues phosphorylated in vitro by different C-terminal domain kinases. J. Biol. Chem. 1998;273:6769–6775. doi: 10.1074/jbc.273.12.6769. [DOI] [PubMed] [Google Scholar]
- 35.Rastinejad F. Retinoid X receptor and its partners in the nuclear receptor family. Curr. Opin. Struct. Biol. 2001;11:33–38. doi: 10.1016/s0959-440x(00)00165-2. [DOI] [PubMed] [Google Scholar]
- 36.Chen-Park F. E., Huang D. B., Noro B., Thanos D., Ghosh G. The κB DNA sequence from the HIV long terminal repeat functions as an allosteric regulator of HIV transcription. J. Biol. Chem. 2002;277:24701–24708. doi: 10.1074/jbc.M200007200. [DOI] [PubMed] [Google Scholar]
- 37.Ueda A., Ishigatsubo Y., Okubo T., Yoshimura T. Transcriptional regulation of the human monocyte chemoattractant protein-1 gene: cooperation of two NF-κB sites and NF-κB/Rel subunit specificity. J. Biol. Chem. 1997;272:31092–31099. doi: 10.1074/jbc.272.49.31092. [DOI] [PubMed] [Google Scholar]
- 38.Widmer U., Manogue K. R., Cerami A., Sherry B. Genomic cloning and promoter analysis of macrophage inflammatory protein (MIP)-2, MIP-1α, and MIP-1β, members of the chemokine superfamily of proinflammatory cytokines. J. Immunol. 1993;150:4996–5012. [PubMed] [Google Scholar]
- 39.Tisne C., Delepierre M., Hartmann B. How NF-κB can be attracted by its cognate DNA. J. Mol. Biol. 1999;293:139–150. doi: 10.1006/jmbi.1999.3157. [DOI] [PubMed] [Google Scholar]
- 40.Tisne C., Hantz E., Hartmann B., Delepierre M. Solution structure of a non-palindromic 16 base-pair DNA related to the HIV-1 κB site: evidence for BI–BII equilibrium inducing a global dynamic curvature of the duplex. J. Mol. Biol. 1998;279:127–142. doi: 10.1006/jmbi.1998.1757. [DOI] [PubMed] [Google Scholar]
- 41.Tisne C., Hartmann B., Delepierre M. NF-κB binding mechanism: a nuclear magnetic resonance and modeling study of a GGG→CTC mutation. Biochemistry. 1999;38:3883–3894. doi: 10.1021/bi982402d. [DOI] [PubMed] [Google Scholar]
- 42.Hoffmann Multiple control of interleukin-8 gene expression. J. Leukocyte Biol. 2002;72:847–855. [PubMed] [Google Scholar]
- 43.Maniatis T., Falvo J. V., Kim T. H., Kim T. K., Lin C. H., Parekh B. S., Wathelet M. G. Structure and function of the interferon-β enhanceosome. Cold Spring Harbor Symp. Quant. Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 44.Steinbrecher K. A., Wilson W., III, Cogswell P. C., Baldwin A. S. Glycogen synthase kinase 3β functions to specify gene-specific, NF-κB-dependent transcription. Mol. Cell. Biol. 2005;25:8444–8455. doi: 10.1128/MCB.25.19.8444-8455.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong H., May M. J., Jimi E., Ghosh S. The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol. Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- 46.Griffin B. D., Moynagh P. N. Persistent interleukin-1β signaling causes long term activation of NF-κB in a promoter-specific manner in human glial cells. J. Biol. Chem. 2006;281:10316–10326. doi: 10.1074/jbc.M509973200. [DOI] [PubMed] [Google Scholar]