Abstract
The fungus Aspergillus niger is an industrial producer of pectin-degrading enzymes. The recent solving of the genomic sequence of A. niger allowed an inventory of the entire genome of the fungus for potential carbohydrate-degrading enzymes. By applying bioinformatics tools, 12 new genes, putatively encoding family 28 glycoside hydrolases, were identified. Seven of the newly discovered genes form a new gene group, which we show to encode exoacting pectinolytic glycoside hydrolases. This group includes four exo-polygalacturonan hydrolases (PGAX, PGXA, PGXB and PGXC) and three putative exo-rhamnogalacturonan hydrolases (RGXA, RGXB and RGXC). Biochemical identification using polygalacturonic acid and xylogalacturonan as substrates demonstrated that indeed PGXB and PGXC act as exo-polygalacturonases, whereas PGXA acts as an exo-xylogalacturonan hydrolase. The expression levels of all 21 genes were assessed by microarray analysis. The results from the present study demonstrate that exo-acting glycoside hydrolases play a prominent role in pectin degradation.
Keywords: Aspergillus niger, glycoside hydrolase, microarray, nano-LC-nanospray MS, pectin degradation, transcriptional profiling
Abbreviations: a-MHR-S, saponified apple modified ‘hairy’ regions; HG, homogalacturonan; HPAEC, high performance anion exchange chromatography; HPSEC, high performance size exclusion chromatography; MALDI-TOF MS, matrix-assisted laser-desorption ionization–time-of-flight MS; nano-RPLC/MS/MS, nano-reverse-phase liquid chromatography tandem MS; ORF, open reading frame; PG, polygalacturonan hydrolase; PGA, polygalacturonic acid; pnp-Rha, p-nitrophenyl-α-L-rhamnopyranoside; RG, rhamnogalacturonan; RGH, rhamnogalacturonan hydrolase; RT, reverse transcriptase; SBP, sugar beet pectin; XGA, xylogalacturonan; XGH, XGA hydrolase
INTRODUCTION
Pectin is a complex heteropolymer present in the middle lamella of the primary cell wall of plants. This biopolymer accounts for about one-third of the total cell wall material [1] and as such represents an important carbon source for bacteria and fungi. Pectin is composed of a number of distinct polysaccharides [2], such as HG (homogalacturonan), XGA (xylogalacturonan), RG (rhamnogalacturonan) I and RG II. RG carries short and long, linear and branched side chains that are built up from neutral sugars (e.g. arabinose and galactose).
Two pectin models exist, namely a pectin structure in which HG is an extension of RG I or vice versa and has neutral sugar side chains [3], and a pectin structure that is built from a backbone of RG I where HG and neutral sugars form the side chains [2].
HG, also known as the smooth region of pectin, is an unbranched polymer composed of (1,4)-linked α-D-galacturonic acid residues. XGA, RG I and RG II are part of the branched or ‘hairy’ regions of pectin [4]. In XGA, xylose is linked at O-3 of the galacturonyl residues of the HG backbone. RG I is composed of a backbone of alternating α-(1,2)-linked rhamnosyl and α-(1,4)-linked galactosyluronic acid residues [2]. The rhamnosyl residues are branched with O-4 attached neutral sugar side chains that can vary from a single galactose residue up to polymeric chains of glycosyl residues composed of arabinose and/or arabinose and galactose residues. RG II is comprised of a backbone of approximately nine α-(1,4)-linked D-galactosyluronic acid residues that carry four side chains, containing rare sugars, such as apiose and aceric acid [2].
The biodegradation of this complex and heterogeneous structure of pectin requires many different enzymatic activities. Exo- and endo-PGs (polygalacturonan hydrolases), pectin lyases and pectate lyases degrade HG. XGA can be degraded by endo-XGHs (XGA hydrolases) [5] and exo-PGs [6,7], whereas RGHs (RG hydrolases) [8] and RG lyases [9,10] degrade RG I. In addition, the complete enzymatic depolymerization of pectin requires the presence of different types of esterase activities [11].
The industrially used saprobic fungus Aspergillus niger is an excellent producer of pectinolytic enzymes which, unlike those produced by many other fungi of the same genus, enjoy a GRAS (generally recognized as safe) status. So far nine genes encoding pectinolytic glycoside hydrolases produced by A. niger have been cloned and the corresponding enzymes have been characterized in detail. The list includes seven different endo-PGs ([12] and references therein) and two RGHs [8], all belonging to family 28 of the general classification system of glycoside hydrolases [13].
The structures of two A. niger endo-PGs, PG I and PG II, have been solved [14,15]. This, together with the biochemical data obtained from site-directed mutagenesis of strictly conserved amino acids, allowed the identification of the residues involved in catalysis, substrate binding, substrate specificity and mode of action of PG II [16–18].
After the recent sequencing of the genome of A. niger, it became clear that only a part of the pectinase spectrum is currently explored. To obtain a complete inventory of pectinolytic glycoside hydrolases produced by A. niger, we applied bioinformatics and functional genomics tools. We were able to identify a total of 21 genes that belong to family 28 glycoside hydrolases. Their transcriptional levels were assessed using custom made Affymetrix gene chip DNA microarrays. Proteins from four newly discovered genes, encoding for novel exo-activities, have been overexpressed and their activities have been biochemically identified.
MATERIALS AND METHODS
Bioinformatics
Genome mining
The genome of A. niger strain CBS513.88, which is a natural derivative of strain NRRL3122, has been previously sequenced [19]. A list of accession numbers for currently available protein sequences belonging to family 28 glycoside hydrolases was obtained from the CAZy web-server at http://afmb.cnrs-mrs.fr/CAZY/index.html. The corresponding 309 protein sequences were retrieved from the Swiss-Prot database [21] at http://www.expasy.org/sprot/ and used to build a hidden Markov model profile using the HMMER package [22] from http://hmmer.wustl.edu/. The genome of A. niger was screened with the profile obtained using the WISE 2 package [23] from http://www.ebi.ac.uk/Wise2/.
Sequence analyses
Two-dimensional alignments of identified protein sequences were performed using the T-coffee program [24] and manually curated. Dendrograms and distance measurements were performed using the Mega 3 software package [25]. Sequences were compared using pairwise analysis algorithm. Distances only were computed. Gaps/Missing Data were calculated by the pairwise deletion algorithm. The substitution model used was amino p-distance with all substitutions included. Homogeneous pattern among lineages and uniform rates among sites were applied. Dendrograms were constructed using the Neighbour-Joining Method with the same distance parameters as described above. The nucleotide sequences reported in the current paper have been submitted to the DDBJ, EMBL and GenBank® Nucleotide Sequence Databases under the accession numbers DQ374422 to DQ374431, DQ417225 and DQ417226.
Verification of intron positions by RT (reverse transcriptase)-PCR
Total RNA was isolated from frozen ground mycelia using TRIZOL® reagent (Invitrogen) according to the manufacturer's instructions. RNA concentrations were estimated using a Nano-Drop® ND-1000 spectrophotometer. cDNAs of pgxC and rgxA were amplified by two step RT-PCR. In the first step, 1.5 μg of total RNA from 2 h transfer cultures on D-galacturonic acid and L-rhamnose respectively were used as a template in the reverse transcription reaction. Omniscript® RT (Qiagen) was used as described in the manufacturer's protocol. In the second step, RT-PCR was performed with 1 μl of product from the reverse transcription reaction using 2.5 units of SuperTaq Plus polymerase (Sphaero Q) under standard PCR conditions. Prior to sequencing, two overlapping fragments per mRNA were amplified with the following primers, for the amplification of the pgxC 5′ fragment: pgxCAFw 5′-CGTCATGTCTGTCTTCAAGG-3′ and pgxCBRv 5′-TGATA-CTGTCGGTGATCTGG-3′; for the amplification of the pgxC 3′ fragment: pgxCDFw 5′-CACTGCGGTGCAGAATATAG-3′ and pgxCCRv 5′-GCGTTCATGCAGATCACACT-3′; for the amplification of the rgxA 5′ fragment: rgxAAFw 5′-GGTATCGAGGTGAGCCAGGA-3′ and rgxABRv 5′-TATGGTCGGATCGGTACGTG-3′; for the amplification of the rgxA 3′ fragment: rgxADFw 5′-GGCTGTTCTTGTCGATTCCT-3′ and rgxACRv 5′-CAGCGACAGCTCACGAATTG-3′. RT-PCR products were gel purified with QIAquick® gel extraction kit (Qiagen) and outsourced to BaseClear Labservices (BaseClear B.V., Leiden, The Netherlands) for sequence determination.
Transcriptional profiling
Strains and growth conditions
Wild-type strain A. niger N400 (CBS 120.49) was used in all transcriptional profiling experiments.
For growth, 300 ml of minimal medium [26] (pH 6.0) containing 0.1% (w/v) yeast extract and Vishniac trace elements [27] with 2% (w/v) D-fructose as a sole carbon source was inoculated with 106 spores/ml and cultivated at 30 °C at 250 rev./min in an orbital shaker. After 18 h of incubation, mycelium were harvested on a Büchner funnel with nylon gauze, washed once with sterile 0.9% (w/v) NaCl and aliquots of 1.5 g (wet weight) of mycelium was transferred to 50 ml of minimal medium (pH 6.0) containing 0.1% (w/v) yeast extract, Vishniac trace element solution and 1% (w/v) of the various sole carbon sources: D-fructose (Merck), D-glucose (Merck), D-galacturonic acid (Fluka Chemica), L-rhamnose (ACROS Organics), D-xylose (Merck), D-sorbitol (Merck), PGA (polygalacturonic acid; United States Biochemical Corporation) and SBP (sugar beet pectin; GENU, Copenhagen). At 2, 4, 8 and 24 h after transfer, mycelium was harvested on a Büchner funnel with nylon gauze and immediately stored at −70 °C. The amount of monomeric sugars remaining in the culture fluid was assessed by standard HPLC techniques [7].
RNA manipulations and microarray processing
Before and during microarray processing RNA quality was verified by analysing aliquots with 1% TAE (Tris/acetate/EDTA) agarose gel electrophoresis and the Agilent Bioanalyzer ‘Lab-on-chip’ system (Agilent Technologies). mRNA levels were assessed using custom made ‘dsmM_ANIGERa_coll’ Affymetrix GeneChip® Microarrays kindly provided by DSM Food Specialties (Delft, The Netherlands).
Probe labelling, hybridization and scanning
Total RNA from mycelium samples were amplified, labelled and hybridized strictly following the Affymetrix protocols for ‘Eukaryotic Target Preparation’ and ‘Eukaryotic Target Hybridization’. For probe array wash and staining the ‘Antibody Amplification Washing and Staining Protocol’ was used. Probe arrays were scanned with an Agilent Technologies G2500A Gene Array Scanner at a pixel value of 3 μm and a wavelength of 570 nm. Raw intensity measurements and present/absent calls were derived using Microarray Suite Software version MAS5 (Affymetrix) after applying the ‘Mask all outliers’ algorithm. All chip data were scaled to an arbitrary target gene intensity of 500.
Cloning and overexpression of enzymes
PGXA, PGXB and RGXB were overexpressed in A. niger PlugBug® [28] and kindly supplied by DSM Food Specialities (Delft, The Netherlands).
Cloning and overexpression of PGXC
Molecular work was essentially carried out using standard techniques described previously [29]. All enzymes were used according to the instructions provided by the supplier. A. niger genomic DNA was isolated as described [30]. A PCR fragment containing the rgxC gene (1.5 kb) was amplified using primers PGXCFw 5′-TGGCATGGCAATTGGAGACC-3′ and PGXCRv 5′-GTGTGGCTTCCTGTGGATGG-3′. The obtained product was used as a template in a subsequent nested PCR reaction using primers PGXC3NsiIFw 5′-GCATCGTCATGCATGTCTTC-3′ and PGXCNotIRv 5′-TGGCGGCGGCTGGATGGCTTA-3′ for the introduction of restriction sites NsiI and NotI at the ATG start codon and 73 bp downstream of the stop codon of the pgxC gene respectively. Both PCR and nested PCR were performed using Pfu Turbo polymerase (Stratagene). The nested PCR product was digested with NsiI and NotI, gel-purified using the QIAquick® gel extraction kit (Qiagen) and ligated into plasmid pIM3710 [31], resulting in the pgxC overexpression vector pIM5150.
Transformation of A. niger NW188 (cspA1, pyrA6, leuA1, prtF28, goxC17) was performed as previously described [32] using 1 μg of pGW635 [33] and 20 μg of co-transforming pIM5150. Control transformation with 1 μg of pGW635 was performed in parallel. Determination of positive transformants and enzyme production were performed as described [34] with the following modifications: STREAMLINE SP and citrate buffer (pH 3.8) were used instead of Sephadex-DEAE and 10 mM piperazine/HCl. Following dialysis, further purification steps were performed as described below.
Enzyme purification and gel electrophoresis
PGXA, PGXB, PGXC and RGXB were purified by dialysis overnight at 4 °C against 50 mM NaOAc (pH 5.7) and subsequent size-exclusion chromatography was performed on a HiLoad 16/60 Superdex 75 column (Amersham Pharmacia Biotech) equilibrated with 50 mM NaOAc (pH 5.7). The elution was carried out with the same buffer at a flow rate of 1 ml/min, and fractions of 2 ml were collected. Protein homogeneity was determined by SDS/PAGE on a 8–25% polyacrylamide gradient gel under reducing conditions, using the PhastSystem following the manufacturer's protocol (Amersham Pharmacia Biotech). The gels were stained with Coomassie Brilliant Blue. Low molecular mass standard proteins (14.4–94 kDa) were used to determine the molecular mass of the purified enzymes.
Substrates
PGA and XGA
XGA-25 was prepared from gum tragacanth by treatment with alkali and TFA (trifluoroacetic acid) and had a xylose/galacturonic acid ratio of 1:4 [35]. PGA was from ICN Biomedials. As substrates for rhamnosidase activity, p-nitrophenyl-α-L-rhamnopyranoside (pnp-Rha; Sigma), naringin (Sigma) and hesperidin (Fluka) were used.
Amidated and methylated pectin
Three commercial pectins, A, B and E, extracted from lemon peels, were obtained from Degussa Texturant Systems SAS. These pectins were partially methyl-esterified with a degree of methyl-esterification of 74%, 72% and 30% respectively, where the distribution of methyl esters was blockwise for pectin A and random for pectin B [36].
Two other commercial pectins, O5 and O27, were obtained from Danisco. These pectins had a degree of amidation of 5% and 27% respectively. Prior to use, the methyl esters were removed from O5 and O27 by saponification [37]. Saponified amidated pectins are referred to as O5sap and O27sap.
Linear RG oligosaccharides
a-MHR-S (saponified apple modified ‘hairy’ region) was prepared as described [38]. Enzymatic liquefaction of apples for the preparation of a-MHR-S was performed with a commercial enzyme preparation (Ultra-SP; Novozymes). This substrate was further treated with RGH and fractionated by HPSEC (high performance size exclusion chromatography) as described previously [39]. Fractions 45–60, which contained oligomeric products as determined by HPSEC analysis, were pooled and freeze-dried. As this material still contained some polymeric residue, further fractionation was performed by gel filtration on a Sephadex G50 column (GE Health Care), using Millipore water as the eluent at a flow rate of 1 ml/min. Fractions 89–100, which contained primarily oligosaccharides as determined by HPSEC analysis, were pooled and subsequently freeze-dried. This pool is referred to as a-MHR-S-fr1. Since the activity of a RG rhamnohydrolase and a RG galacturonohydrolase towards RG oligosaccharides is hindered by Gal side chains [40], a-MHR-S-fr1 was treated with β-galactosidase [41] for 19 h at 40 °C in 20 mM Bis-Tris (pH 5). The final protein concentration was 2.2 μg/ml. The enzyme was inactivated by heating for 10 min at 100 °C. Subsequently, β-galactosidase treated a-MHR-S-fr1 (a-MHR-S-fr1-g) was desalted on a Biogel P2 column (Bio-Rad) using Millipore water as the eluent at a flow rate of 1 ml/min. After desalting, a-MHR-S-fr1-g was analysed by MALDI-TOF (matrix-assisted laser-desorption ionization–time-of-flight) MS [39], for the presence of RG oligosaccharides. Four linear RG oligosaccharides were detected at m/z 539, m/z 685, m/z 861 and m/z 1007. Based on the mode of action of RGH towards RG, it was concluded that the produced RG oligosaccharides with a m/z at 685 and m/z at 1007 correspond to rhamnose–galacturonic acid–rhamnose–galacturonic acid and galacturonic acid–rhamnose–galacturonic acid–rhamnose–galacturonic acid respectively. The RG oligosaccharides at m/z 539 and at m/z 861 correspond to galacturonic acid–rhamnose–galacturonic acid and galacturonic acid–rhamnose–galacturonic acid–rhamnose–galacturonic acid. As these latter two oligosaccharides cannot be produced directly from RGH-treated RG it is assumed that these oligosaccharides are formed due to side activities in the RGH preparation, in particular RG rhamnosidase activity.
Enzyme activity measurements
PGXA, PGXB, PGXC and RGXB were analysed for their activity towards 0.25% (w/v) PGA, 0.25% (w/v) XGA-25, and 0.25% (w/v) of the pectins, encoded as A, B, C, O5sap and O27sap. Exo-PG from Aspergillus tubingensis [42] was included as a reference. The action of RGXB was also studied towards 0.25% (w/v) narigin, 0.25% (w/v) hespiridin, 0.02% (w/v) pnp-Rha and 0.5% (w/v) a-MHR-S-Fr1-g. For comparison, this latter substrate was also treated with RG galacturonohydrolase from Aspergillus aculeatus [43]. All digests were made in 50 mM NaOAc (pH 4.0) for 16 h at 30 °C. The final protein concentration was 1 μg/ml. Enzymes were inactivated by heating the reaction mixtures for 10 min at 100 °C. All digests were analysed by HPSEC and HPAEC (high performance anion exchange chromatography), as described previously [44], except for RG galacturonohydrolase or RGXB treated a-MHR-S-Fr1-g, which were analysed by MALDI-TOF MS as described previously [39]. Products released from XGA-25 and PGA after 1 h of incubation with PGXA, PGXB, PGXC or exo-PG were also analysed by HPAEC.
The specific activity of PGXA, PGXB, PGXC and exo-PG was determined towards 0.25% (w/v) PGA and 0.25% (w/v) XGA-25, as described previously [34]. Specific activity is in units/mg of enzyme, where 1 unit is defined as 1 μmol of reducing sugar released per min. The influence of pH on enzyme activity was determined as described previously [34].
Identification of the RGXC protein sequence by MS analysis
Tryptic digest of RG galacturonohydrolase from A. aculeatus
The protein concentration of RG galacturonohydrolase [43] was determined by the Bradford assay (Bio-Rad Laboratories) and the molecule subsequently digested with trypsin as described previously [45]. The tryptic digest was subsequently acidified to pH 3.0 by the addition of 10% formic acid. The sample was then analysed by nano-RPLC/MS/MS (nano-reverse-phase liquid chromatography tandem MS).
Nano-RPLC/MS/MS
Nano-RPLC/MS/MS analysis of the tryptic peptides was performed on an integrated Famos/Switchos/Ultimate 1D/2D nano-flow HPLC system (LC Packings, a division of Dionex), as described [46] with the following adaptations. The peptides were first trapped on a LC Packings C18 precolumn [300 μm i.d. (inner diameter)×5 mm, 100 Å (1 Å=0.1 nm)] and eluted after desalting onto a Pep Map C18 resolving column (3 μm, 100 Å, 75 μm i.d.×15 cm, LC Packings, Dionex) using the following elution profile: a linear gradient of increasing acetonitrile concentration in water (5–30%) over 40 min, isocratic at 30% for 5 min, a linear gradient (30–95%) over 30 min, isocratic at 95% for 5 min. All eluents contained 0.05% formic acid as the ion-pairing agent.
The electrospray needle was operated with a voltage differential of 3.0 kV, and the heated desolvation capillary was held at 180 °C. The specific mass-to-charge value of each peptide sequenced by MS/MS was excluded dynamically from reanalysis for 2 min.
Analysis of MS/MS and MS3 data
Data analysis of each raw tandem spectrum was performed as described previously [47] with adaptations. Processed tandem mass spectra were correlated with the various public protein databases and the A. niger protein database (DSM, Delft, The Netherlands) using the program Bio Works 3.1 (Thermo Finnigan). The allowed mass tolerance range between expected and observed masses for search was ±1.0 Da for MS peaks, and ±0.1 Da for MS/MS fragment ions. The correlation results were then filtered using the value of the cross-correlation score and the matched sequence for each spectrum. For singly charged peptides, spectra with a cross-correlation score to a tryptic peptide ≥1.5 were retained. For multiple-charged peptides, spectra with a cross-correlation to a tryptic peptide ≥2 were retained. All spectra with cross-correlation scores not meeting these criteria were eliminated from further consideration.
RESULTS AND DISCUSSION
Genome mining
Screening the genome sequence of A. niger with a hidden Markov model profile for family 28 glycoside hydrolases returned 21 significant alignments from which only nine corresponded to already known A. niger proteins. The start and the end of each alignment were extended to the nearest in frame start-codon and nearest stop-codon respectively. Further analysis of the Gene Wise output indicated possible frame-shifts in the sequences of pgxC and rgxA, therefore the full-length of the corresponding cDNAs were sequenced. Comparison of the obtained cDNA sequences with the predicted gene models demonstrated the presence of one frame-shift in the genomic sequence of rgxA and three frame-shifts in the genomic sequence of the pgxC. These corrected sequences were used for further analysis.
Comparison of A. niger glycoside hydrolases
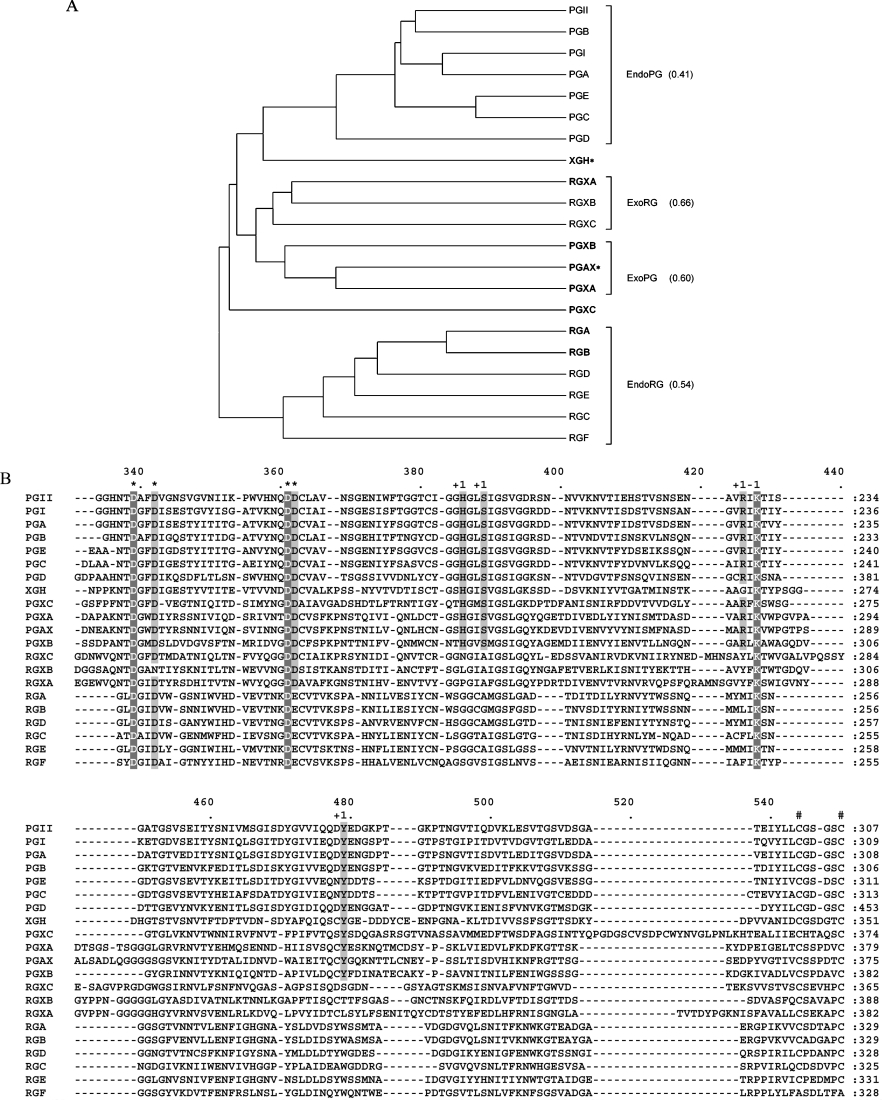
The protein sequence alignment of the complete A. niger family 28 glycoside hydrolases and the corresponding sequence distance dendrogram are presented in Figure 1. Based on sequence similarity three major groups are observed (Figure 1A). The first group is composed of all previously known endo-PGs of A. niger. No newly identified sequences are added to this group. The second group contains six newly discovered ORFs (open reading frames) of A. niger of which one designated PGAX is a very close homologue of A. tubingensis exo-PG [42] with 99% amino acid identity. Based on sequence similarity, the six ORFs can be divided into two sub-groups (Figure 1A) each comprising three proteins. PGAX forms a subgroup with PGXA and PGXB, therefore it is very likely that PGXA and PGXB are also exo-PGs. RGXA, RGXB and RGXC form the second distinct subgroup and evidence is presented that they are similar to RG galacturonohydrolase [43] (see below). The conservation of intron positions within ORFs of both sub-groups further underpins this subdivision. The most deviating sequence from all 21 identified proteins is PGXC. Also, there are no introns in the encoding gene pgxC to support its classification. However, exo-PG activity was determined for the encoded enzyme (see below). The last group includes two previously characterized endo-acting A. niger proteins, RGHA and RGHB, as well as four putative enzymes which we propose to be endo-RGHs. Finally XGH, the strongly conserved homologue (97% amino acid identity) of XGH of A. tubingensis [5] did not group together with any of the other sequences.
Figure 1. Comparative sequence analysis of A. niger family 28 glycoside hydrolases.
(A) Dendrogram of A. niger family 28 glycoside hydrolases. The average pairwise distance within the different functional groups of A. niger hydrolases is indicated in brackets. Biochemically identified enzymes are in bold. *A. niger enzyme amino acid sequences that are greater than or equal to 97% identical to that of a previously characterized A. tubingensis enzyme. (B) Region of the multiple alignment of A. niger family 28 glycoside hydrolases. *Columns containing catalytic residues; #conserved cysteine bridge; +1, columns containing residues involved in substrate binding at subsite +1; −1, columns containing residues involved in substrate binding at subsite −1. The complete multiple alignment of A. niger family 28 glycoside hydrolases is provided as Supplementary Figure 1 (http://www.BiochemJ.org/bj/400/bj4000043add.htm).
Detailed inspection of the amino acid conservation between all 21 sequences revealed that only one of the four disulfide bridges between columns 544 and 550 of the multiple alignment (Figure 1B), demonstrated by the protein structures of PG II of A. niger [14] and RGA of A. aculeatus [48], is well conserved within family 28 glycoside hydrolases of A. niger. The other three cysteine bridges are well conserved in the groups of endo-PGs and endo-RGHs but not in the exo-hydrolase group. Inspection of the catalytic residues revealed that all previously identified catalytic aspartate residues are conserved within the A. niger family except for Asp362 (Figure 1B). In the group of RGHs, Asp362 is replaced by glutamate which is characteristic for this type of enzymatic activity [48]. The substrate-binding lysine residue at alignment position 428 is 100% conserved, whereas the second substrate-binding residue Arg426 is present in all endo-PG sequences and in only four of the seven proteins from the exo-group, PGAX, PGXA, PGXB and PGXC. Armand et al. [16] have demonstrated by site-directed mutagenesis that the corresponding arginine residue of A. niger PG II (Arg256) is essential for the proper orientation of the substrate in the catalytic cleft and mutations of this residue dramatically affect the substrate affinity of the enzyme. Interestingly, other amino acids shown to be involved in substrate binding at subsite +1, His386 and Ser389, are conserved within the same subset of sequences. The same is true for the Tyr479, which in PG II is involved in the stabilization of the substrate at subsite +1 [16,49]. The partial conservation of amino acids responsible for substrate specificity and binding within the exo-hydrolase group further supports the suggestion that only PGXA, PGXB, PGXC and PGAX have exo-PG activity.
Identification of RGXC as a close homologue of A. aculeatus RG galacturonohydrolase
A RG galacturonohydrolase activity, specifically removing terminal galacturonosyl residues from RG I, has been characterized in the commercial mixture Pectinex Ultra SP produced by A. aculeatus [43]. In order to identify a possible A. niger homologue of this enzyme, a tryptic digest of the A. aculeatus protein was analysed by MS. The obtained peptide masses were used to search the inferred A. niger proteome. At least six peptides perfectly matched the sequence of RGXC, resulting in a total of 29% overall coverage of the protein (results not shown). Based on sequence similarity, RGXC is grouped together with RGXA and RGXB indicating that there are three candidate genes encoding proteins with exo-RGH activity.
Transcriptional profiling
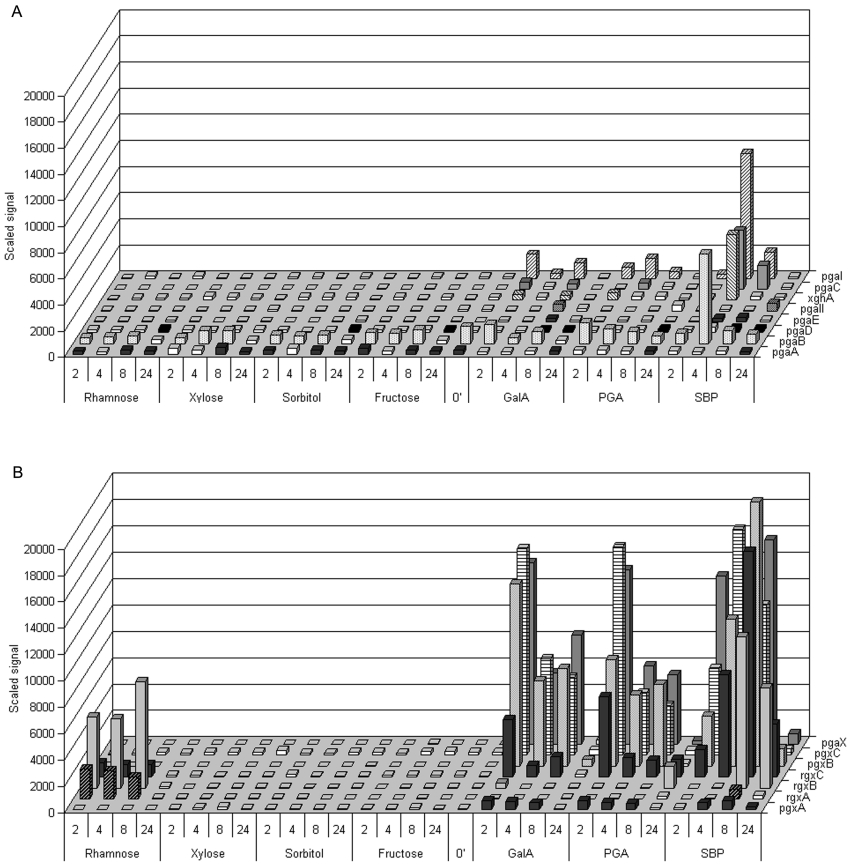
The complex substrates SBP and PGA were chosen as primary carbon sources to investigate the expression of selected pectinolytic genes. Galacturonic acid, rhamnose and xylose represent the most abundant sugar residues present in HG, RG and XGA and were used to assess the effects on gene expression caused by simple well-defined carbon sources. Fructose, as a strong repressor of the expression of genes that are under carbon catabolite regulation, and sorbitol, as a non-inducing sugar-like alcohol which does not affect the carbon catabolite regulation mechanisms, were chosen as control substrates. After transfer from 18 h pre-culture on 2% fructose, mycelium was sampled at four time points during 24 h of growth on the different substrates. The corresponding total RNA samples were used in microarray experiments to monitor the mRNA levels of family 28 glycoside hydrolase encoding genes (Figure 2).
Figure 2. Transcriptional levels of A. niger glycoside hydrolases: 24 h time course on different carbon sources.
(A) Endo-PGs and endo-XGH. (B) Exo-acting hydrolases. Minor axis intervals, time points of harvesting after transfer from fructose (in h); 0′–18 h preculture on 2% (w/v) fructose; White colour indicates absent call. The numeric values and calls from the obtained expression data are provided as Supplementary Table 1 (http://www.BiochemJ.org/bj/400/bj4000043add.htm).
During growth on non-pectic carbon sources, namely fructose and sorbitol, none of the genes encoding for enzymes from the exo-hydrolase group were transcribed. In contrast, transfer from fructose to galacturonic acid and PGA caused a strong induction peak in the first 2 h after transfer for pgaX, pgxB and pgxC followed by a decrease to relatively moderate expression levels during the next 8 h and further reduction of the signal to background levels 24 h after transfer, coinciding with the depletion of the carbon source. Although less strongly induced, rgxC displayed a similar transcriptional profile on galacturonic acid and PGA. This gene was also moderately expressed, together with rgxA and rgxB, after transfer to rhamnose (Figure 2B). Interestingly pgxA showed only low expression levels on these carbon sources.
Transfer to SBP caused a more complex transcriptional pattern. pgaX was quickly induced and remained highly expressed reaching a maximum at 8 h after transfer. pgxC mRNA levels reached a maximum at 4 h after transfer. pgxB and rgxC levels gradually increased and reached a maximum 8 h after transfer. The signals obtained for rgxC were much higher on SBP compared with rhamnose or galacturonic acid as the sole carbon source. SBP caused a similar effect on the induction of the rgxB gene. rgxA mRNA was hardly detected at 8 h after transfer to SBP.
In contrast with the strong induction observed for genes from the exo-group, the transfer to galacturonic acid and PGA did not cause a strong effect on the expression of endo-PGs. SBP moderately induced pgaI, pgaB and pgaC 4 h after transfer, coinciding with the slight accumulation of extracellular free galacturonic acid (results not shown).
pgaA and pgaB have been reported to be constitutively expressed [50], which is in agreement with the data obtained in the present study (Figure 2A). Yet a slight increase of pgaB mRNA was observed coinciding with the depletion of fructose during the time course of incubation, suggesting that, although weak, pgaB is under carbon catabolite regulation control. In contrast, when pectic substrates were used, the signal of pgaB decreased in time to reach a background level at 24 h (Figure 2A). Instead, low levels of pgaD mRNA were detected. PGD, the protein encoded by the pgaD gene, has been proposed to be a cell wall- attached oligogalacturonase with high tolerance towards methyl-esterified pectins [12]. HPLC measurements (results not shown) demonstrated that at the time points of expression of pgaD, free sugars are being depleted from the culture fluid, which, together with its functional properties, suggests that the expression of pgaD may in part be due to carbon starvation. Still, during incubation on galacturonic acid and SBP, pgaD mRNA could be detected slightly earlier, 8 h after incubation.
Despite many efforts, including several genetic screens [51], no specific transcription factors regulating the expression of pectinolytic glycoside hydrolases have been identified so far. The strong discrepancy between the general expression profiles observed for the exo- and the endo-hydrolase encoding genes suggests separate or loosely linked regulation mechanisms for both groups. Exo-PGs are well-induced by galacturonic acid and the observed expression levels on pectin, although slightly more elevated, do not differ much from the levels observed on its monomeric substitute. Unexpectedly, galacturonic acid and PGA as a sole carbon source appeared to be only weak inducers for endo-PGs, whereas SBP caused profoundly higher expression of endo-PGs. In addition, the expression pattern of all studied genes on SBP has a much more complex character compared with monomeric sugars, which suggests that a multi-factorial regulation system, not just galacturonic acid, is responsible for the induction of pectinolytic genes.
Biochemical identification of PGXA, PGXB, PGXC and RGXB
Purified PGXA, PGXB, PGXC and RGXB appeared as a single band on SDS/PAGE (results not shown) and had apparent molecular masses of 78 kDa, 67 kDa, 79 kDa and 82 kDa respectively (Table 1). The difference between the measured and calculated molecular masses of these enzymes is most probably caused by a high degree of glycosylation, which has also been demonstrated previously for the exo-PG from A. tubingensis [42].
Table 1. Properties of PGXA, PGXB, PGXC and exo-PG.
Specific activity of the enzymes were determined towards 0.25% (w/v) PGA and 0.25% (w/v) XGA in 50 mM NaOAc (pH 4.0). The optimum pH for each enzyme was determined towards 0.25% (w/v) PGA in McIlvaine buffers (over a pH range of 2.5–7.5). n.d., not determined.
| Specific activity (units/mg) | |||||
|---|---|---|---|---|---|
| Enzyme | Molecular mass (kDa) | Calculated molecular mass (kDa) | PGA | XGA | pH optimum towards PGA |
| PGXA | 78 | 47 | 7 | 25 | 3.5–4.0 |
| PGXB | 67 | 48 | 242 | 33 | 4.0–4.5 |
| PGXC | 79 | 45 | 223 | 202 | 3.5–4.0 |
| Exo-PG | 781 | 47 | 230 | 17 | 4.21 |
| RGXB | 82 | 50 | n.d. | n.d. | n.d. |
1 As determined by Kester et al. [42].
The action of PGXA, PGXB, PGXC and RGXB was studied towards PGA, XGA-25 and pectins with various degrees of methylation or amidation. As determined by HPSEC, only PGA and XGA-25 were degraded by PGXA, PGXB and PGXC. Similar results were observed for exo-PG, which indicates that PGXA, PGXB and PGXC have an exo-PG activity. RGXB was not active towards these galacturonic acid-containing substrates, indicating that this enzyme is not a PG. To investigate whether RGXB exhibits exo-RGH activity, a mixture of RG oligosaccharides was treated with RGXB and analysed by MALDI-TOF MS. These oligosaccharides were also treated with exo-acting RG galacturonohydrolase from A. aculeatus [43] for comparison. As determined by MALDI-TOF MS (results not shown), RG oligosaccharides galacturonic acid–rhamnose–galacturonic acid–rhamnose–galacturonic acid and galacturonic acid–rhamnose–galacturonic acid, at m/z 861 and m/z 539 respectively, disappeared upon treatment with RG galacturonohydrolase, which implies the expected removal of the non-reducing galacturonic acid residue from these oligosaccharides [43]. The enzyme RGXB did not degrade these RG oligosaccharides. Also the RG oligosaccharides rhamnose–galacturonic acid–rhamnose–galacturonic acid–rhamnose–galacturonic acid at m/z 1007 and rhamnose–galacturonic acid–rhamnose–galacturonic acid at m/z 685 respectively were not attacked (results not shown). From these results it was concluded that RGXB does not act towards these RG oligosaccharides from the non-reducing end nor from the reducing end. However, this is still unclear for RG oligosaccharides with a rhamnose residue at the reducing end.
As RGXB was able to degrade pnp-Rha (results not shown), we have termed this enzyme a pnp-rhamnohydrolase. In contrast to the pnp-rhamnohydrolase from A. aculeatus [40], RGXB did not hydrolyse hespiridin or naringin.
The enzymes PGXA, PGXB and PGXC were optimally active towards PGA at pH 3.5–4.0, pH 4.0–4.5 and pH 3.5–4.0 respectively (Table 1). pH optima for exo-PGs close to this value have been previously reported by several investigators [42,52,53].
The specific activities of PGXA, PGXB, PGXC towards PGA and XGA-25 are presented in Table 1. Also the specific activity of A. tubingensis exo-PG towards these polymers is included in Table 1 for comparison. PGXB, PGXC and exo-PG have a similar activity towards PGA. The specific activity of PGXB and exo-PG towards XGA were less than towards PGA, which shows that these enzymes have a higher preference for PGA. However, PGXC has no preference as it was equally active on both substrates. Compared with the other enzymes, PGXA has a significantly lower specific activity towards PGA and it is the only enzyme that has a higher specific activity towards XGA-25 than towards PGA. This was also demonstrated by HPAEC analysis (see below).
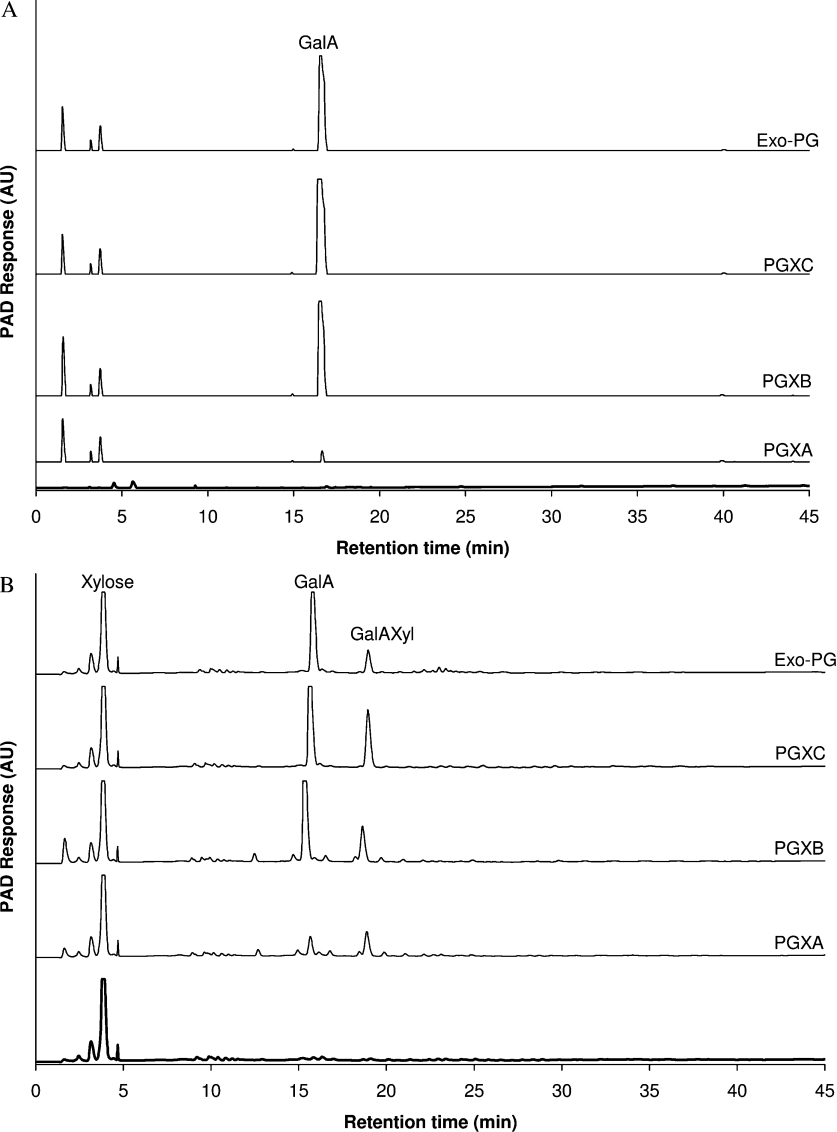
As determined by HPSEC analysis (results not shown), PGA and XGA-25 were degraded by PGXA, PGXB and PGXC respectively, without a dramatic decrease in the molecular mass of these polymers. These results were comparable with those obtained for exo-PG and imply that PGXA, PGXB and PGXC degrade both polymers in an exo-fashion. This is further substantiated by analysis of the products by HPAEC. PGXA, PGXB and PGXC predominantly produce galacturonic acid from PGA, and a mixture of galacturonic acid and the disaccharide galacturonic acid–xylose from XGA-25 at the early stage of the reaction (i.e. after 1 h of incubation, see Figure 3), as well as after prolonged incubation (results not shown). These results were similar to those observed for exo-PG, thus again confirming the exolytic mode of action of these enzymes as predicted from the sequence alignment.
Figure 3. HPAEC of (A) PGA and (B) XGA hydrolysis after treatment for 1 h with PGXA, PGXB, PGXC or exo-PG.
Spectra of untreated PGA and XGA are in bold. PAD, pulsed amperomatric detection. AU, arbitrary units.
A detailed inspection of the HPAEC results demonstrates that PGXA acts differently towards PGA and XGA-25 than PGXB, PGXC and exo-PG. As shown in Figure 3(A), the production of galacturonic acid from PGA (after 1 h of incubation) was relatively low for PGXA when compared with PGXB, PGXC and exo-PG. These results are also in accordance with the specific activities of these enzymes towards PGA (Table 1). As illustrated in Figure 3(B), compared with the other enzymes, PGXA also produced significantly more galacturonic acid–xylose in ratio to galacturonic acid from XGA-25, showing that PGXA prefers to act towards xylosylated galacturonic acid residues of the substrate XGA-25. It can be concluded that PGXA is primarily behaving as an exo-XGH (and not an exo-PG), but is also able to degrade PGA.
CONCLUSIONS
The present paper provides for the first time an overview of the entire set of pectinolytic glycoside hydrolases encoded in the genome of A. niger.
In addition to the already well-studied gene group of seven endo-PGs, we were able to extend the number of endo-RGHs to six. Furthermore, a new gene group comprising seven genes encoding for exo-acting pectinolytic glycoside hydrolases was identified. In addition, one gene encoding for a pectinolytic glycoside hydrolase was identified to be a XGH.
Subsequent sequence analysis of the new gene group in combination with transcriptional data allowed the prediction of two subgroups of enzyme activities. Four genes were assessed as encoding the exo-PGs, PGAX, PGXA, PGXB, PGXC and three genes as encoding the exo-RGHs, RGXA, RGXB and RGXC. PGAX encoded by the A. niger gene pgaX is almost identical with the previously described exo-PG of A. tubingensis.
Biochemical characterization of the three proposed exo-PGs, PGXA, PGXB and PGXC, demonstrated that indeed all three enzymes were active towards HG in an exo-fashion. Yet, although the biochemical properties of PGXB do not differ much from those of the exo-PG of A. tubingensis, the data obtained for PGXA strongly indicate that this enzyme is a novel exo-XGH. Interestingly, PGXC appears to be an enzyme with reduced substrate specificity and remains unaffected in its activity when galacturonic acid residues of the substrate are substituted with xylose.
Additionally, the MS analysis of a tryptic digest of the RG galacturonohydrolase produced by A. aculeatus, a protein for which no gene sequence is known, allowed the identification of its A. niger homologue RGXC.
Although RGXB showed some activity towards pnp-Rha we were not able to identify the natural substrate of this enzyme. Additional analysis is required to investigate the possible function of RGXA and RGXB. Yet, both enzymes possess the catalytic residues and the proper spacing required for the inverting mechanism characteristic for enzymes from family 28 glycoside hydrolases. The expression of the genes encoding these two enzymes on rhamnose and SBP suggests their involvement in the degradation of pectin regions that contain rhamnose, most probably RG I.
The generally higher expression levels observed for genes encoding exo-acting glycoside hydrolases compared with those of genes encoding endo-activities, the strong initial rates of the reactions catalysed by them and the fact that A. niger is able to utilize galacturonic acid in its monomeric form suggest that exo-activities play a much more important physiological role than expected previously during pectin degradation.
Online data
Acknowledgments
This research was supported by Senter IOP, The Netherlands (IGE01021). We would like to thank Esther Tichon (Laboratory of Microbiology, Wageningen University, Wageningen, The Netherlands) for cloning the pgxC gene. We also thank Margaret Bosveld (Laboratory of Food Chemistry, Wageningen University, Wageningen, The Netherlands) for the preparation of linear RG oligosaccharides.
References
- 1.Carpita N. C., Gibeaut D. M. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 2.Vincken J.-P., Schols H. A., Oomen R. J. F. J., Beldman G., Visser R. G. F., Voragen A. G. J. Pectin – The hairy thing. In: Voragen A. G. J., Schols H., Visser R., editors. Advances in Pectin and Pectinase Research. Dordrecht: Kluwer Academic Publishers; 2003. pp. 47–59. [Google Scholar]
- 3.Voragen A. G. J., Pilnik W., Thibault J.-F., Axelos M. A. V., Renard C. M. G. C. Pectins. In: Stephen A. M., editor. Food Polysaccharides. New York-Basel-Hing Kong: Marcel Dekker, Inc; 1995. pp. 287–339. [Google Scholar]
- 4.Schols H. A., Voragen A. G. J. Complex pectins: structure elucidation using enzymes. In: Visser J., Voragen A. G. J., editors. Pectins and Pectinases. Amsterdam: Elsevier Science B.V.; 1996. pp. 3–19. [Google Scholar]
- 5.Van der Vlugt-Bergmans C. J. B., Meeuwsen P. J. A., Voragen A. G. J., van Ooyen A. J. J. Endo-xylogalacturonan hydrolase, a novel pectinolytic enzyme. Appl. Environ. Microbiol. 2000;66:36–41. doi: 10.1128/aem.66.1.36-41.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beldman G., van den Broek L. A. M., Schols H. A., Searle-van Leeuwen M. J. F., van Laere K. M. J., Voragen A. G. J. An exogalacturonase from Aspergillus aculeatus able to degrade xylogalacturonan. Biotechnol. Lett. 1996;18:707–712. [Google Scholar]
- 7.Kester H. C., Benen J. A., Visser J. The exopolygalacturonase from Aspergillus tubingensis is also active on xylogalacturonan. Biotechnol. Appl. Biochem. 1999;30:53–57. [PubMed] [Google Scholar]
- 8.Suykerbuyk M. E., Kester H. C., Schaap P. J., Stam H., Musters W., Visser J. Cloning and characterization of two rhamnogalacturonan hydrolase genes from Aspergillus niger. Appl. Environ. Microbiol. 1997;63:2507–2515. doi: 10.1128/aem.63.7.2507-2515.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kofod L. V., Kauppinen S., Christgau S., Andersen L. N., Heldt-Hansen H. P., Dorreich K., Dalboge H. Cloning and characterization of two structurally and functionally divergent rhamnogalacturonases from Aspergillus aculeatus. J. Biol. Chem. 1994;269:29182–29189. [PubMed] [Google Scholar]
- 10.Azadi P., O'Neill M. A., Bergmann C., Darvill A. G., Albersheim P. The backbone of the pectic polysaccharide rhamnogalacturonan I is cleaved by an endohydrolase and an endolyase. Glycobiology. 1995;5:783–789. doi: 10.1093/glycob/5.8.783. [DOI] [PubMed] [Google Scholar]
- 11.de Vries R. P., Visser J. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol. Mol. Biol. Rev. 2001;65:497–522. doi: 10.1128/MMBR.65.4.497-522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parenicova L., Kester H. C., Benen J. A., Visser J. Characterization of a novel endopolygalacturonase from Aspergillus niger with unique kinetic properties. FEBS Lett. 2000;467:333–336. doi: 10.1016/s0014-5793(00)01173-x. [DOI] [PubMed] [Google Scholar]
- 13.Henrissat B., Davies G. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struc. Biol. 1997;7:637–644. doi: 10.1016/s0959-440x(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 14.van Santen Y., Benen J. A. E., Schroter K.-H., Kalk K. H., Armand S., Visser J., Dijkstra B. W. 1.68-A crystal structure of endopolygalacturonase II from Aspergillus niger and identification of active site residues by site-directed mutagenesis. J. Biol. Chem. 1999;274:30474–30480. doi: 10.1074/jbc.274.43.30474. [DOI] [PubMed] [Google Scholar]
- 15.van Pouderoyen G., Snijder H. J., Benen J. A. E., Dijkstra B. W. Structural insights into the processivity of endopolygalacturonase I from Aspergillus niger. FEBS Lett. 2003;554:462–466. doi: 10.1016/s0014-5793(03)01221-3. [DOI] [PubMed] [Google Scholar]
- 16.Armand S., Wagemaker M. J. M., Sanchez-Torres P., Kester H. C. M., van Santen Y., Dijkstra B. W., Visser J., Benen J. A. E. The active site topology of Aspergillus niger endopolygalacturonase II as studied by site-directed mutagenesis. J. Biol. Chem. 2000;275:691–696. doi: 10.1074/jbc.275.1.691. [DOI] [PubMed] [Google Scholar]
- 17.Pages S., Heijne W. H., Kester H. C., Visser J., Benen J. A. Subsite mapping of Aspergillus niger endopolygalacturonase II by site-directed mutagenesis. J. Biol. Chem. 2000;275:29348–29353. doi: 10.1074/jbc.M910112199. [DOI] [PubMed] [Google Scholar]
- 18.Pages S., Kester H. C., Visser J., Benen J. A. Changing a single amino acid residue switches processive and non-processive behavior of Aspergillus niger endopolygalacturonase I and II. J. Biol. Chem. 2001;276:33652–33656. doi: 10.1074/jbc.M105770200. [DOI] [PubMed] [Google Scholar]
- 19.Groot G., Peij N. V., Ooyen A. V. Exploring and exploiting the Aspergillus niger genome; Annual Meeting of the Society for Industrial Microbiology, Philadelphia, PA, U.S.A. August 2002; 2002. [Google Scholar]
- 20. Reference deleted.
- 21.Bairoch A., Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucl. Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durbin R., Eddy S., Krogh A., Mitchison G. A tutorial introduction to hidden Markov models and other probabilistic modelling approaches in computational sequence analysis. Cambridge University Press; 1998. Biological sequence analysis: probabilistic models of proteins and nucleic acids. [Google Scholar]
- 23.Birney E., Clamp M., Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Notredame C., Higgins D. G., Heringa J. T-coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 25.Kumar S., Tamura K., Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 26.Pontecorvo G., Roper J. A., Hemmons L. M., Macdonald K. D., Bufton A. W. J. The genetics of Aspergillus nidulans. Adv. Genet. 1953;5:141. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 27.Vishniac W., Santer M. The thiobacilli. Bacteriol. Rev. 1957;21:195–213. doi: 10.1128/br.21.3.195-213.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Dijck P. W. M. Made by genetic engineering: a series of articles to promote a better understanding of the use of genetic engineering. J. Biotechnol. 1999;67:77. [Google Scholar]
- 29.Sambrook J., Fritsch E. F., Maniatis T. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 30.de Graaff L., van den Broek H., Visser J. Isolation and transformation of the pyruvate kinase gene of Aspergillus nidulans. Curr. Genet. 1988;13:315–321. doi: 10.1007/BF00424425. [DOI] [PubMed] [Google Scholar]
- 31.Benen J. A. E., Kester H. C. M., Visser J. Kinetic characterization of Aspergillus niger N400 endopolygalacturonases I, II and C. Eur. J. Biochem. 1999;259:577–585. doi: 10.1046/j.1432-1327.1999.00080.x. [DOI] [PubMed] [Google Scholar]
- 32.Kusters-van Someren M. A., Harmsen J. A., Kester H. C., Visser J. Structure of the Aspergillus niger pelA gene and its expression in Aspergillus niger and Aspergillus nidulans. Curr. Genet. 1991;20:293. doi: 10.1007/BF00318518. [DOI] [PubMed] [Google Scholar]
- 33.Goosen T., Bloemheuvel G., Gysler C., de Bie D. A., van den Broek H. W., Swart K. Transformation of Aspergillus niger using the homologous orotidine-5′-phosphate-decarboxylase gene. Curr. Genet. 1987;11:499. doi: 10.1007/BF00384612. [DOI] [PubMed] [Google Scholar]
- 34.Parenicova L., Benen J. A., Kester H. C., Visser J. pgaE encodes a fourth member of the endopolygalacturonase gene family from Aspergillus niger. Eur. J. Biochem. 1998;251:72–80. doi: 10.1046/j.1432-1327.1998.2510072.x. [DOI] [PubMed] [Google Scholar]
- 35.Beldman G., Vincken J.-P., Meeuwsen P. J. A., Herweijer M., Voragen A. G. J. Degradation of differently substituted xylogalacturonans by endoxylogalacturonan hydrolase and endopolygalacturonases. Biocatal. Biotransform. 2003;21:189–198. [Google Scholar]
- 36.Guillotin S. E., Bakx E. J., Boulenguer P., Mazoyer J., Schols H. A., Voragen A. G. J. Populations having different GalA blocks characteristics are present in commercial pectins which are chemically similar but have different functionalities. Carbohydr. Polym. 2005;60:391. [Google Scholar]
- 37.Guillotin S. E. Ph.D. Thesis. Wageningen, The Netherlands: Wageningen University; 2005. Studies on the intra- and intermolecular distributions of substituents in commercial pectins; p. 155. [Google Scholar]
- 38.Schols H. A., Posthumus M. A., Voragen A. G. J. Hairy (ramified) regions of pectins. Part I. Structural features of hairy regions of pectins isolated from apple juice produced by the liquefaction process. Carbohydr. Res. 1990;206:117–129. [Google Scholar]
- 39.Zandleven J., Beldman G., Bosveld M., Benen J., Voragen A. Enzymatic degradation studies of xylogalacturonans from apple and potato. using XGH Carbohydr. Polym. 2006 in the press. [Google Scholar]
- 40.Mutter M., Beldman G., Schols H. A., Voragen A. G. J. Rhamnogalacturonan α-L-rhamnopyranosylhydrolase. A novel enzyme specific for the terminal non-reducing rhamnosyl unit in rhamnogalacturonan regions of pectin. Plant Physiol. 1994;106:241–250. doi: 10.1104/pp.106.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van de Vis J. W. Ph.D. Thesis. Wageningen, The Netherlands: Wageningen University; 1994. Characterization and mode of action of enzymes degrading galactan structures of arabinegalactans; p. 161. [Google Scholar]
- 42.Kester H. C., Kusters-van Someren M. A., Muller Y., Visser J. Primary structure and characterization of an exopolygalacturonase from Aspergillus tubingensis. Eur. J. Biochem. 1996;240:738–746. doi: 10.1111/j.1432-1033.1996.0738h.x. [DOI] [PubMed] [Google Scholar]
- 43.Mutter M., Beldman G., Pitson S. M., Schols H. A., Voragen A. G. J. Rhamnogalacturonan α-D-galactopyranosyluronohydrolase. An enzyme that specifically removes the terminal nonreducing galacturonosyl residue in rhamnogalacturonan regions of pectin. Plant Physiol. 1998;117:153–163. doi: 10.1104/pp.117.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zandleven J., Beldman G., Bosveld M., Benen J., Voragen A. Mode of action of xylogalacturonan hydrolase towards xylogalacturonan and xylogalacturonan oligosaccharides. Biochem. J. 2005;387:719–725. doi: 10.1042/BJ20041583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao J., Opiteck G. J., Friedrichs M. S., Dongre A. R., Hefta S. A. Changes in the protein expression of yeast as a function of carbon source. J. Proteome Res. 2003;2:643–649. doi: 10.1021/pr034038x. [DOI] [PubMed] [Google Scholar]
- 46.VerBerkmoes N. C., Hervey W. J., Shah M., Land M., Hauser L., Larimer F. W., Van Berkel G. J., Goeringer D. E. Evaluation of ‘shotgun’ proteomics for identification of biological threat agents in complex environmental matrixes: experimental simulations. Anal. Chem. 2005;77:923–932. doi: 10.1021/ac049127n. [DOI] [PubMed] [Google Scholar]
- 47.Link A. J., Eng J., Schieltz D. M., Carmack E., Mize G. J., Morris D. R., Garvik B. M., Yates J. R. Direct analysis of protein complexes using mass spectrometry. Nat. Biotech. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 48.Petersen T. N., Kauppinen S., Larsen S. The crystal structure of rhamnogalacturonase A from Aspergillus aculeatus: a right-handed parallel β-helix. Structure. 1997;5:533. doi: 10.1016/s0969-2126(97)00209-8. [DOI] [PubMed] [Google Scholar]
- 49.Cho S., Lee S., Shin W., Cho S. W., Lee S., Shin W. The X-ray structure of Aspergillus aculeatus polygalacturonase and a modeled structure of the polygalacturonase-octagalacturonate complex. J. Mol. Biol. 2001;311:863–878. doi: 10.1006/jmbi.2001.4919. [DOI] [PubMed] [Google Scholar]
- 50.Parenicova L., Benen J. K., Kester H. C., Visser J. pgaA and pgaB encode two constitutively expressed endopolygalacturonases of Aspergillus niger. Biochem. J. 2000;345:637–644. [PMC free article] [PubMed] [Google Scholar]
- 51.Parenicova L. Ph.D. Thesis. Wageningen, The Netherlands: Wageningen University; 2000. Pectinases of Aspergillus niger: a molecular and biochemical characterisation; p. 200. [Google Scholar]
- 52.Hara T., Lim J. Y., Ueda S. Purification and some properties of exopolygalacturonase from Aspergillus niger cultured in the medium containing satsuma mandarin peel. J. Jpn. Soc. Food Sci. Technol. 1984;31:581–586. [Google Scholar]
- 53.Sakamoto T., Bonnin E., Quemener B., Thibault J.-F. Purification and characterisation of two exo-polygalacturonases from Aspergillus niger able to degrade xylogalacturonan and acetylated homogalacturonan. Biochim. Biophys. Acta. 2002;1572:10–18. doi: 10.1016/s0304-4165(02)00277-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.