Abstract
HS (heparan sulfate) is essential for normal embryonic development. This requirement is due to the obligatory role for HS in the signalling pathways of many growth factors and morphogens that bind to sulfated domains in the HS polymer chain. The sulfation patterning of HS is determined by a complex interplay of Golgi-located N- and O-sulfotransferases which sulfate the heparan precursor and cell surface endosulfatases that selectively remove 6-O-sulfates from mature HS chains. In the present study we generated single or double knock-out mice for the two murine endosulfatases mSulf1 and mSulf2. Detailed structural analysis of HS from mSulf1−/− fibroblasts showed a striking increase in 6-O-sulfation, which was not seen in mSulf2−/− HS. Intriguingly, the level of 6-O-sulfation in the double mSulf1−/−/2−/− HS was significantly higher than that observed in the mSulf1−/− counterpart. These data imply that mSulf1 and mSulf2 are functionally co-operative. Unlike their avian orthologues, mammalian Sulf activities are not restricted to the highly sulfated S-domains of HS. Mitogenesis assays with FGF2 (fibroblast growth factor 2) revealed that Sulf activity decreases the activating potential of newly-synthesized HS, suggesting an important role for these enzymes in cell growth regulation in embryonic and adult tissues.
Keywords: endosulfatase, glycosaminoglycan, heparan sulfate, knock-out mice, signal transduction, sulfatase
Abbreviations: dp, degree of depolymerization; E, embryonic day; FGF, fibroblast growth factor; GlcA, glucuronic acid; GlcNAc, N-acetylglucosamine; GlcNS, N-sulfoglucosamine; HGF, hepatocyte growth factor; HS, heparan sulfate; IdoA, iduronic acid; m, murine; MEF, mouse embryonic fibroblast; NS, N-sulfation; 2S and 6S, 2-O- and 6-O-sulfation respectively; SAX-HPLC, strong-anion-exchange HPLC; UA, hexuronic acid; VEGF, vascular endothelial growth factor; ΔUA, Δ-4,5-unsaturated UA
INTRODUCTION
HS (heparan sulfate) proteoglycans are some of the most abundant proteins on the cell surface and within the extracellular matrix. The sulfate groups on the HS chains of these glycoconjugates form specific patterns which direct key aspects of cell growth, differentiation, adhesion and migration by regulating the interaction of growth factors with their cognate receptors and by modulating transport and diffusion of many growth and morphogenic factors [1]. For some ligands, such as members of the FGF (fibroblast growth factor) family, the interaction with specific sulfate patterns is essential for signalling [2,3]. Sulfate pattern regulation therefore plays a critical role during development and disease pathogenesis.
Within a mature HS chain (typically between 50 and 200 saccharides in length), long sequences of repeating GlcA–GlcNAc (glucuronic acid–N-acetylglucosamine) residues are separated by composite regions consisting of highly sulfated S-domains [predominantly IdoA(2S)–GlcNS (2-O-sulfated iduronic acid– N-sulfoglucosamine) sequences], with adjacent transition zones comprised of alternating GlcNS- and GlcNAc-containing disaccharides [4]. Both the S-domains and transition zones are further modified by 6S (6-O-sulfation) of glucosamine residues, and variations in the degree and patterning of 6S contribute significantly to the fine structural heterogeneity in HS. The structural diversity in HS is non-random, with regulation observed at the level of tissue type and developmental stage [5,6]. Within the specific domain types loss or gain of 6S alone is sufficient to alter the outcome of the ligand–HS interaction [7]. Dynamic changes in 6S correlate with modulated susceptibility to FGF signal transduction during neural development and tumour transformation [8,9].
The regulation of 6S patterning was thought to be the result solely of the action of the HS biosynthetic enzymes; however, the identification of the Sulfs, a family of HS 6-O-endosulfatases, has led to a reconsideration of this viewpoint. The first Sulf was discovered in a molecular cloning screen for Sonic hedgehog-responsive genes, which led to the characterization of QSulf1, a sulfatase regulating cellular differentiation through modulation of Wnt signalling in developing quail embryos [10]. Orthologues were identified in mammalian species [11]; however, many of the insights into Sulf activity have come from studies using QSulf1 and its recently characterized isoform QSulf2 [12–15]. The unique localization of the Sulfs at the cell surface and their endosulfatase activity, rather than the catabolic activity of lysosomal sulfatases, carried with it important implications for the novel hydrophilic domain that is inserted into the catalytic domain of the Sulfs. This highly and mainly positively charged domain has recently been characterized as a decisive functional element for both substrate recognition and cell surface localization [12]. The native substrate specificity of the Sulfs has, until now, been largely studied employing HS disaccharide analysis from QSulf over-expressing cellular systems or in vitro analysis [12,13]. Results from these experiments indicate a substrate specificity of QSulf1 and QSulf2 which is completely restricted to GlcA/IdoA(2S)–GlcNS(6S)-type disaccharides from the S-domains of HS [12–14].
Attention has now turned to the mammalian Sulf orthologues in mice and humans, as both Sulf1 and Sulf2 isoforms have been shown to be mis-regulated during mammalian tumorigenesis [16–18]. Investigations into the influence of Sulf activity on HS-binding factors active in tumorigenesis, such as VEGF (vascular endothelial growth factor), FGF, HGF (hepatocyte growth factor) and HB-EGF (heparin-binding epidermal-growth-factor-like growth factor) [19–21], have led to the hypothesis that the modulation of Sulf activity may be a target for therapeutic intervention.
In the present study we describe, for the first time, the characterization of HS produced by cells in which the genes for mSulf1, mSulf2 or both had been knocked out. We have combined detailed structural analysis of the HS with expression analyses, HS epitope characterization and growth factor activity studies. We conclude that mSulf1 and mSulf2 show functional co-operativity and that, although increased mSulf1 expression can compensate for loss of mSulf2 activity, mSulf2 is unable to fulfil the role of mSulf1. Importantly, the 6-O-desulfation catalysed by the mSulf enzymes was found to be extensive, and more widely distributed throughout the HS chain, than observed with the QSulf orthologues, with significant implications for the range of HS–ligand interactions affected. Finally, the use of a panel of ScFv (single chain fragment) antibodies to characterize mSulf-mediated changes in HS structure suggest a novel method for the in vivo detection of aberrant mSulf activity.
EXPERIMENTAL
Materials
Dulbecco's modified Eagle's medium and foetal calf serum were from Life Technologies. Heparinase I–III (Flavobacterium heparinum) were all from Grampian Enzymes (Orkney, U.K.). Chondroitinase ABC (Proteus vulgaris) and monoclonal antibodies against diphosphorylated ERK1/2 (extracellular-signal-regulated kinase 1/2) were from Sigma. The gels used were Sepharose CL-6B and DEAE-Sephacel (GE Healthcare) and Bio-Gels P-10 (fine grade) and P-2 (Bio-Rad). The ProPac PA-1 HPLC column was from Dionex (Camberley, U.K.). D-[6-3H]Glucosamine hydrochloride (20–45 Ci/mmol) and Na235SO4 (1050–1600 Ci/mmol) were from GE Healthcare and PerkinElmer Life Sciences respectively. Recombinant human FGF2 and HGF/SF (HGF/scatter factor) were from R&D Systems (Abingdon, Oxon., U.K.).
Generation of knock-out mice, genotyping and Northern analysis
By screening a genomic cosmid library of the 129Sv/Ola mouse (German Resource Centre for Genome Research, Berlin, Germany) with labelled cDNA fragments as probes, cosmids were identified that carried fragments of the mSulf1 (murine Sulf1) or mSulf2 gene. A 3.1 kb HindIII fragment containing exon 2 of mSulf1 and a 3.8 kb SalI/EcoRI fragment containing exon 1 of mSulf2 with approximately equal homology arms of 1.5–2.0 kb on both sides (Figures 1A and 1B) were sequenced, subcloned into pBluescriptII SK and used for construction of the targeting vectors. The neomycin-resistance cassette, a blunted XhoI/SalI fragment of the pMC1neo vector (Stratagene), was inserted into the BsaAI site of mSulf1 exon 2, as well as into the blunted BseRI site of mSulf2 exon 1 (Figures 1A and 1B), thereby disrupting the respective open reading frames and generating stop codons in all frames. NotI-linearized constructs were electroporated into 129Sv/Ola embryonic stem cells (cell line D3, provided by Peter Gruss, Department of Molecular Cell Biology, Max-Planck-Institute for Biophysical Chemistry, Goettingen, Germany). G418-resistant clones were selected and genotyped for specific recombination by Southern blotting. Positive ES clones were injected into C57BL/6 blastocysts to produce chimaeric mice. Male chimaeras were mated with C57BL/6 females, which led to germ-line transmission of the targeted alleles. From these, heterozygotes were intercrossed to generate wild-type and knock-out mice. Northern and Southern blotting was performed according to standard methods using PCR-generated probes (primers are listed in Supplementary Table 1 at http://www.BiochemJ.org/bj/400/bj4000063add.htm). The mSulf2 5′-external probe was a 420 bp EcoRI/HindIII fragment of EST clone IMAGp998G091033Q2 (German Resource Centre for Genome Research).
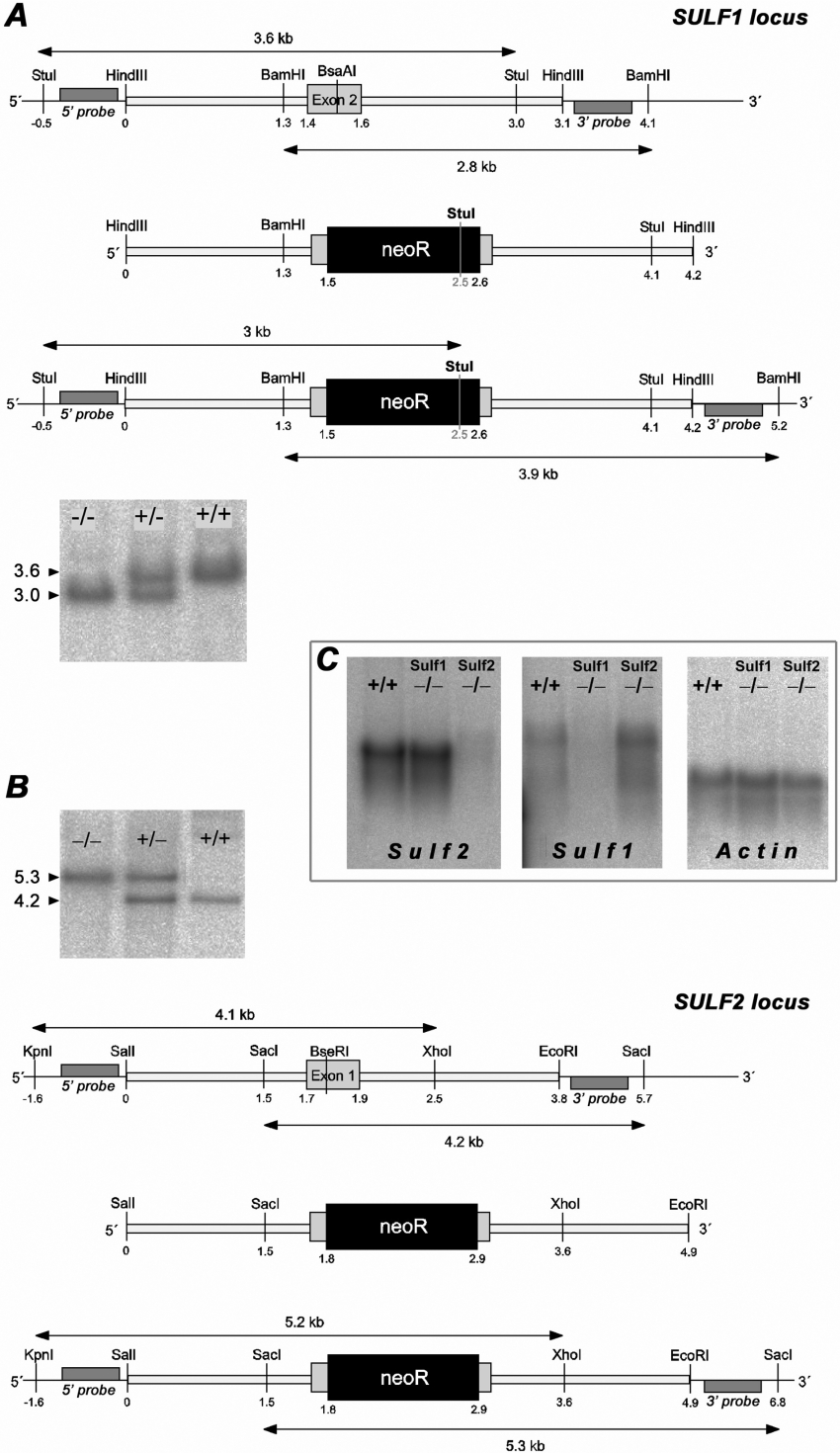
Figure 1. Targeted disruption of murine Sulf1 and Sulf2.
Representation of the murine Sulf1 (A) and Sulf2 (B) loci in the area of the targeted exon 2 (A) and exon 1 (B) respectively. The wild-type locus (top), the targeting vector (middle) and the recombined allele (bottom) are shown. Recombined alleles carry the neomycin-resistance cassette inserted into the BsaAI site of mSulf1 exon 2 (A) and the BseRI site of mSulf2 exon 1 (B) respectively. The location of the 5′ and 3′ external probes used for Southern hybridization and the relevant restriction sites and resulting fragments are indicated. Successful and specific germline targeting was confirmed by Southern blotting of StuI- or SacI-digested genomic DNA using the indicated external probes (see blots in A and B respectively). Results were confirmed by independent external probes detecting the indicated 3.9 kb fragment in BamHI-digested DNA (A) (Sulf1) and the 5.2 kb fragment in KpnI/XhoI-digested DNA (B) (Sulf2) (results not shown). (C) Northern blot analysis of total RNA (10 μg) from wild-type, mSulf1−/− and mSulf2−/− MEFs, using probes for mSulf2, mSulf1 and β-actin (control).
Preparation of HS from MEF (mouse embryonic fibroblast) cultures
Embryos [E (embryonic day) 12.5], obtained from mating mSulf1−/−, mSulf2−/− or mSulf1−/−/mSulf2−/− mice, were dissected, MEFs were isolated and the HS chains were radiolabelled and purified as described previously [22]. Primary MEFs from the mSulf1−/−/mSulf2−/− embryos could not be maintained in culture. We therefore prepared immortalized lines (mSulf1−/−/mSulf2−/− and wild-type control) by stable transfection of MEFs (E12.5) with the pMSSVLT vector containing an SV40 fragment including the large T gene [23], prior to isolation of HS chains.
Disaccharide composition analysis
3H-labelled HS chains (50000 c.p.m.) were digested with a combination of heparinases I, II and III, and the resulting disaccharides identified using SAX-HPLC (strong-anion-exchange HPLC) as described previously [22].
Low pH nitrous acid depolymerization
3H-labelled HS (100000 c.p.m.) was taken for low pH nitrous acid depolymerization [22]. Oligosaccharides produced were separated and the tetrasaccharides purified essentially as described by Viviano et al. [14]. Recovered tetrasaccharides were then separated by SAX-HPLC as described for the heparinase I-, II- and III-generated disaccharides.
Heparinase III digestion
3H-labelled HS (100000 c.p.m.) was treated with heparinase III essentially as described by Merry et al. [24]. Isolated sized fragments were pooled, desalted and further fractionated by SAX-HPLC. Disaccharides were analysed as described for heparinase I-, II- and III-generated disaccharides. For larger oligosaccharides, a single ProPac PA-1 SAX column was used with a linear gradient running from 0–0.75 M NaCl over 110 min. Peak identification was made by comparison with previously published profiles [24].
Real-time PCR
Primary wild-type, mSulf1−/− and mSulf2−/− MEFs, together with immortalized wild-type and mSulf1−/−/mSulf 2−/− MEFs, were washed in PBS then harvested. RNA was extracted using the RNeasy Protect mini-kit (Qiagen, catalogue 74124). Synthesis of cDNA from mRNA transcripts was performed using the following method: RNA (5 μg, denatured at 65 °C for 3 min and placed on ice), dNTP (250 μM), oligonucleotide dTs (5 μg; Promega), 1×AMV buffer (Promega) and AMV (avian myeloblastosis virus) reverse transcriptase (40 units; Promega) in a total volume of 200 μl was incubated at 42 °C for 1 h, followed by 98 °C for 5 min. Real-time PCR assays of the Sulf genes, together with housekeeping genes L19 and EEF1G, were designed using the Exiqon Mouse Universal ProbeLibrary system [25] (Roche Applied Science), with amplification primers obtained from MWG (Ebersberg, Germany). Amplicons were designed to be intronspanning, typically between 60–100 nt in length. Real-time PCR reaction mixtures were set up using SensiMix (dT) (Quantace) according to the manufacturer's instructions. Briefly, for a 10-μl reaction, 5 ng of cDNA, 0.1 μM of each forward and reverse primer, 0.1 μl of the appropriate mouse probe (Universal ProbeLibrary Set, Roche Applied Science), 5 μl of reaction buffer and water to make the total volume up to 10 μl were mixed in each well of a 384-well clear-optical reaction plate (ABgene). Each primer/cDNA set was set up in triplicate and the whole experiment was repeated three times. Primer/probe combinations are shown in Supplementary Table 2 (http://www.BiochemJ.org/bj/400/bj4000063add.htm). Experiments were performed on an ABI 7900 Real Time Sequence Detection System, using an Epmotion 5070 robot (Eppendorf, Hamburg, Germany) to set up the plates. Assays were tested for efficiency using reference RNA and a 10-fold dilution series; only assays with greater than 99% amplification efficiency were used for subsequent work. Data analysis was performed using the ΔΔCT method [26], normalizing against L19 and EEF1G to allow the fold change relative to wild-type to be calculated.
FACS analysis: 96-well-plate format
Cells were harvested using cell dissociation buffer (Gibco) and resuspended in PBS. Cells were distributed at 1×106 cells per well in a round-bottom 96-well plate and fixed in 1% formaldehyde at 4 °C for 15 min prior to staining. Cells were then washed in PBS and resuspended in 100 μl of FACS buffer (0.2% BSA and 0.1% sodium azide in PBS) containing one of the primary ScFv antibodies [HS4C3, RB4EA-12, AOB408, HS4E4, EV3C3 and MPB49V (control), periplasmic fractions 1/10], and incubated at 4 °C for 1 h. Cells were washed twice with 100 μl of PBS and resuspended in 100 μl of FACS buffer containing anti-mouse VSV antibody (1:1000; V-5507, Sigma), and incubated at 4 °C for 45 min. Cells were then washed twice in PBS. Tertiary antibody goat anti-mouse IgG-PE (1:100; P-852, Molecular Probes) was then added to 100 μl of FACS buffer and incubated at 4 °C for 45 min. Cells were then washed twice in PBS and transferred into a FACS tube (352054, Falcon). Control cells were incubated as above, without the primary antibody step. Cell fluorescence was measured in a FACScan (Becton Dickinson). Viable cells were gated using forward and side scatter, and the fluorescence of this population was measured.
Mitogenesis assay
A simple mitogenesis assay was established using wild-type and knock-out primary MEFs. Cells were plated at 25000 cells per well on a 24-well plate in 1 ml of serum-free Dulbecco's modified Eagle's medium. After incubation for 4 h at 37 °C, HGF or FGF2 was added at 10 ng/ml. Cells were subsequently incubated at 37 °C for 36 h before being counted manually under a light microscope in 15 individual fields per well. The average number of cells per well was calculated and the fold increase/decrease was determined by comparing the average number of cells per well in growth-factor-induced and untreated control cells.
RESULTS AND DISCUSSION
An essential function of HS is to provide a scaffold on the cell surface for mediating the interaction of signalling molecules, whereby the sulfation patterns of HS largely determine ligand-binding affinities. The endosulfatases Sulf1 and Sulf2 act as key players in sulfation-pattern remodelling. To investigate the role of these enzymes in vivo, mSulf1 and mSulf2 knock-out mice were generated.
Phenotypic characterization
mSulf1 and mSulf2 knock-out mice were generated by the classical approach, i.e. by insertion of a neomycin-resistance cassette into exon 2 of the murine Sulf1 gene and exon 1 of the murine Sulf2 gene, as shown in Figures 1(A) and 1(B). Northern analysis of mRNA from knock-out MEFs verified the absence of wild-type mSulf1 or mSulf2 transcripts (Figure 1C). Real-time PCR detected very small amounts of targeted, i.e. non-functional, transcripts (see below). mSulf1−/− mice showed no obvious abnormal phenotype. However, their mortality in the first month of life was slightly, but consistently, increased. mSulf2−/− mice show a small reduction in litter size and body mass. Their mortality within the first 6 weeks was increased. Death of the mSulf2−/− mice was usually associated with malformations, in particular of the brain (I. Kalus, M. Padva, R. D'Hooge, B. Salmen, C. Viebahn and T. Dierks, unpublished work). Mice that did not die appeared normal. Double knock-out animals were difficult to obtain. After many heterozygous matings the very few fertile mSulf1−/−/mSulf2−/− animals obtained were used for homozygous matings. In this way, we recently succeeded in obtaining five litters with an approx. 2-fold reduced size (4.4±0.9 animals). These animals typically have a very short life span and an obvious reduction in body mass. Double knock-out MEFs could be obtained from E12.5 embryos and are described below.
The knock-out approach is distinct from those which have previously investigated the function of the Sulf enzymes, in that we are interpreting Sulf activity from the effects of its loss, as opposed to the over-expression studies frequently used, or the in vitro recapitulations of activity using isolated enzyme fractions. We believe this strategy is more biologically significant, as the amount and subcellular localization of all the other components of the HS biosynthetic and modification machinery are maintained, and thus are only influenced by the loss of Sulf activity. Therefore the interplay between these factors and the Sulf enzymes is likely to be more representative of their true activity in vivo.
Molecular phenotype: effect on HS disaccharide composition and overall sulfation
As stated above, HS structure is tissue-specific; however, it has been demonstrated previously that the loss/mutation of HS biosynthetic enzymes in a knock-out model has a similar effect on all tissues [5,22]. We therefore chose MEFs as a convenient source of material for detailed structural analyses. Metabolically 3H/35S-radiolabelled HS was purified from primary wild-type, mSulf1−/− and mSulf2−/− MEFs, as well as from immortalized mSulf1−/−/mSulf2−/− and wild-type MEFs. Immortalization was necessary as it proved impossible to maintain the mSulf1−/−/2−/− MEFs in culture for long enough to allow the preparation of radiolabelled HS. HS from all cells was digested with a combination of heparinases I, II and III, and the resulting disaccharides resolved on a Bio-Gel P2 column. The disaccharides were then pooled and fractionated by SAX-HPLC (Table 1 and Supplementary Figure 1 at http://www.BiochemJ.org/bj/400/bj4000063add.htm). Significant increases were observed in all 6S disaccharide species in the mSulf1−/− HS compared with the wild-type HS (Table 1). No overall increase in 6S was observed in the mSulf2−/− HS, although there was a significant increase in the UA (hexuronic acid)–GlcNAc(6S) disaccharide unit known to be associated with transition zones (see below). The mSulf1−/−/mSulf2−/− HS showed large increases in all 6S disaccharides. Comparing the relative abundance of 6S disaccharides between single and double knock-out cells revealed a greater than 2-fold increase in S-domain-associated disaccharides, UA–GlcNS(6S) and UA(2S)–GlcNS(6S), in the double knock-out HS as compared with either single knock-out HS, thus indicating co-operativity between the two Sulf activities within the S-domains (Table 1).
Table 1. Disaccharide composition and overall sulfation of HS from cell extract fractions from knock-out mice.
| Composition (%) | |||||
|---|---|---|---|---|---|
| Disaccharide | WT | mSulf1−/− | mSulf2−/− | Immortalized WT | Immortalized mSulf1−/−/mSulf2−/− |
| UA–GlcNAc | 49.5 | 49.6 | 52.7 | 45.4 | 37.5 |
| UA–GlcNS | 21.5 | 20.0 | 22.2 | 23.5 | 20.3 |
| UA–GlcNAc(6S) | 5.9 | 11.1 | 7.7 | 7.5 | 12.6 |
| UA(2S)–GlcNAc | 2.7 | 1.7 | 1.3 | 1.4 | 1.0 |
| UA–GlcNS(6S) | 3.7 | 5.8 | 3.1 | 5.1 | 13.7 |
| UA(2S)–GlcNS | 10.9 | 4.5 | 8.8 | 10.4 | 1.8 |
| UA(2S)–GlcNS(6S) | 5.8 | 7.4 | 4.2 | 6.8 | 13.1 |
| Total NAc | 58.1 | 62.4 | 61.8 | 54.2 | 51.1 |
| Total NS | 41.9 | 37.6 | 38.2 | 45.8 | 48.9 |
| Total 2S | 19.3 | 13.5 | 14.3 | 18.5 | 15.9 |
| Total 6S | 15.4 | 24.2 | 15.0 | 19.4 | 39.3 |
Interestingly, the increase in 6S in the mSulf knock-out cells was accompanied by small, but statistically significant reductions in other sulfation events, particularly in the level of 2S (Table 1 and Supplementary Figure 1). It is therefore important to apply caution to the interpretation of experiments where loss (or gain) of Sulf activity influences HS–ligand interactions, as these may not necessarily be due to changes in 6S alone. The effect on NS (N-sulfation) and 2S may be due to the Sulf enzymes influencing the biosynthetic pathway. However, whether this occurs as a result of direct interaction between the Sulfs, the nascent HS chain and/or other processing enzymes in the Golgi remains to be determined. Alternatively, these changes in NS and 2S levels could result from downstream changes in HS biosynthetic enzyme expression levels as a consequence of changes in Sulf activity.
Effect on N-sulfate distribution and transition zones
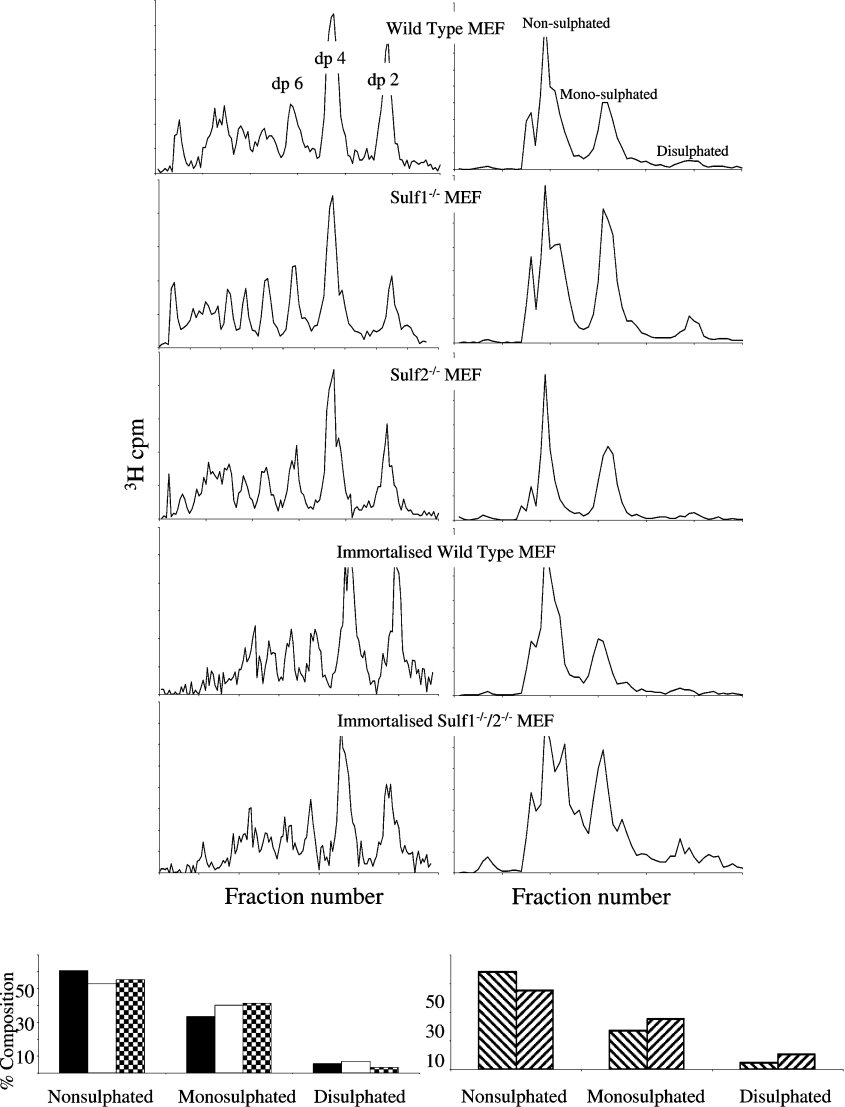
Previous analysis in vitro using QSulfs indicated that these enzymes preferentially remove 6S from the tri-sulfated IdoA(2S)–GlcNS(6S) disaccharide, and have no activity outside of the highly sulfated S-domains [14]. In order to analyse the domain specificity of the mammalian Sulfs, low pH nitrous acid and heparinase III depolymerization were used to isolate and analyse the respective S-domains, non-sulfated domains and transition zones which make up the HS chain [14,24]. HS from all cell populations was cleaved at GlcNS residues using low pH nitrous acid [27]. We compared the profiles of the resulting oligosaccharides on a Bio-Gel P10 column to determine the proportion of GlcNS-containing disaccharides within the chains (Figure 2), and to additionally provide information on domain organization (see Figure 3A). Regions of contiguous N-sulfated disaccharides (S domains) will be depolymerized to disaccharides (dp2; where dp is the degree of depolymerization), with regions of alternating GlcNS and GlcNAc residues (transition zones) generating tetrasaccharides (dp4). All profiles (Figure 2, left-hand panels), were characteristic of a typical HS structure with N-sulfated residues occurring predominantly in S-domain clusters or in UA–GlcNS–UA–GlcNAc-type transition zones. Minimal differences in overall macromolecular organization were visible between wild-type and mSulf knock-out cells. However, preparative fractionation using Bio-Gel P-10 and SAX-HPLC of low pH nitrous acid-resistant tetrasaccharides revealed important differences in sulfate composition of the transition zones (Figure 2, right-hand panels). The transition zone-derived tetrasaccharides separated into non-, mono- and di-sulfated species, with the variation primarily due to the addition of 6S, as these regions typically contain little 2S [6]. By comparing the contribution of each of these species, we demonstrated that there is a significant increase in 6S in the transition zones of all mSulf knock-out HS, predominantly observed as an increase in mono-sulfated tetrasaccharides, whereas in the mSulf1−/−/mSulf2−/− HS there was a significant increase in both mono- and di-sulfated species (Figure 2, lower panels).
Figure 2. Low pH nitrous acid depolymerization of cell extract HS.
Following treatment with low pH nitrous acid, HS was fractionated by Bio-Gel P-10 (left-hand panels) to separate size-defined oligosaccharides (dp2, dp4 etc.). The proportion of counts in each sized peak was found to be very similar between the samples: dp2=14–15%, dp4 28–30% (primary cells) and dp2=22%, dp4=31–32% (immortalized cells). Tetrasaccharides (from UA–GlcNAc–UA–GlcNS regions of the chains) were further fractionated by SAX-HPLC (right-hand panels) into di-, mono- and non-sulfated species. These were then quantified (lower panels) for the primary MEFs: wild-type (black bars), mSulf1−/− (white bars) and mSulf2−/− (checked bars); and the immortalized MEF lines: wild-type (downwards diagonal bars) and mSulfI−/−/mSulf2−/− (upwards diagonal bars). All profiles are representative of at least three separate fractionations.
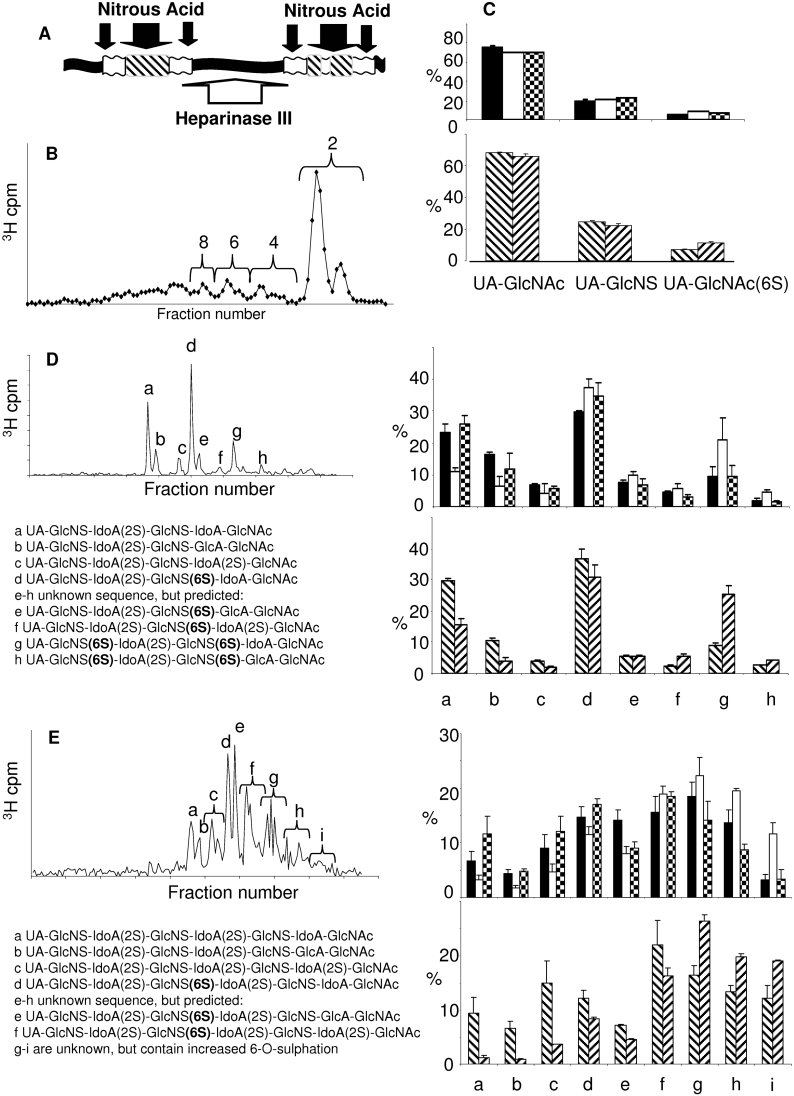
Figure 3. Sulfated domain analysis of cell extract HS.
HS was digested with heparinase III. (A) Site of action of heparinase III (white arrow) and low pH nitrous acid (black arrows). Non-N-sulfated domains (black box), transition zone NS/NAc (white box) and S-domains (striped box) are indicated. (B) Typical HS profile following heparinase III digestion and separation on a Bio-Gel P-10 column. The di-, tetra-, hexa- and octa-saccharide fractions indicated were pooled and fractionated further using SAX-HPLC. Three disaccharide species were identified (C), eight hexasaccharide species (D) and a complex range of octasaccharide species, some of which were pooled as indicated (E). Many of the oligosaccharides could be identified by comparison of elution position with that of known standards and where possible, by disaccharide analysis. Data from at least three profiles were quantified to generate the histograms shown. Primary MEFs: wild-type (black bars), mSulf1−/− (white bars) and mSulf2−/− (checked bars). Immortalized MEF lines: wild-type (downwards diagonal bars) and mSulf1−/−/mSulf2−/− (upwards diagonal bars).
Effect on S-domain organization and composition
Heparinase III excises the S-domains in HS by specific scission of the non-sulfated regions and transition zones (see Figure 3A) [24]. Following heparinase III digestion and subsequent fractionation of the resultant oligosaccharides by Bio-Gel P-10 (Figure 3B), we observed that, similar to the nitrous acid fractionation, the overall chain macromolecular organization was conserved between the HS types (results not shown). Each of the size-defined peaks was isolated separately and re-fractionated using SAX-HPLC. For tetrasaccharides and longer oligosaccharides, we took advantage of a previous study in which heparinase III digestion was used to isolate S-domains from NIH 3T3 fibroblast HS prior to sequence analysis [24]. By comparing the profiles generated in the present study with those from the NIH 3T3 fibroblast HS [24], we were able to identify confidently many of the oligosaccharides generated. Where possible, peak assignments were confirmed by isolation and disaccharide-compositional analysis.
Disaccharide analysis
The disaccharide peak generated by heparinase III contained three species corresponding to UA–GlcNAc, UA–GlcNS and UA–GlcNAc(6S). These disaccharides are typically restricted to the non-sulfated and transition regions of the HS chain, confirming heparinase III enzyme specificity. In comparison with wild-type HS, a significant 2-fold increase in UA–GlcNAc(6S) residues (p≤0.01) was seen in mSulf1−/− material (Figure 3C and Table 2). Compensatory decreases in the non-6-O-sulfated disaccharide UA–GlcNAc were observed, balancing this increase in UA–GlcNAc(6S). A similar decrease in UA–GlcNAc residues was also seen in mSulf2−/− material, although only a 40% increase in 6-O-sulfated UA–GlcNAc(6S) residues was apparent. mSulf1−/−/mSulf2−/− material also showed a significant (60%) increase in UA–GlcNAc(6S) when compared with the wild-type control. Thus, as verified by the independent nitrous acid-mediated domain analysis (see above), both mSulf1 and to a lesser extent mSulf2 are active 6-O-endosulfatases within transition domains marked by GlcNAc(6S).
Table 2. Relative amounts of heparinase III-generated disaccharides.
| Relative amount (%) | |||||
|---|---|---|---|---|---|
| Disaccharide | WT | mSulf1−/− | mSulf2−/− | Immortalized WT | Immortalized mSulf1−/−/mSulf2−/− |
| UA–GlcNAc | 75.5 | 69.7 | 70.5 | 68.1 | 66.1 |
| UA–GlcNS | 19.9 | 21.4 | 23.0 | 24.7 | 22.4 |
| UA–GlcNAc(6S) | 4.7 | 9.0 | 6.5 | 7.2 | 11.5 |
Tetrasaccharide analysis
Only one major tetrasaccharide species was visible in all MEF HS types following separation by SAX-HPLC (results not shown). This tetrasaccharide corresponds to ΔUA–GlcNS–IdoA(2S)–GlcNAc (where ΔUA is Δ-4,5-unsaturated UA), and was confirmed by disaccharide analysis.
Hexasaccharide analysis
Eight major peaks were detected in all heparinase III-derived hexasaccharides (dp6s) (Figure 3D, peaks a–h). These species fell into two groups, those that were 6S-deficient (6a–6c) and those with 6S (6d–6h). Peaks a–d have been sequenced previously [24], and are identified as shown in Figure 3(D). Hexasaccharide peaks e–h have an unknown sequence, however, the peaks form a repeating pattern, with a, b and c anticipated to have the same relationship to each other as d, e and f, etc. We could therefore predict the sequences of these species. Compared with wild-type, mSulf1−/− cell extract HS contains fewer lower-sulfated sequences (peaks a–c), whereas hexasaccharides containing one or more 6-O-sulfated glucosamine residue are enriched (peaks d–h) (Figure 3D). A 53% decrease in peak a was seen in mSulf1−/− HS, consistent with a 26% and 27% increase in peaks d and g, which are modifications of a, to contain either one or two 6S glucosamine residues respectively. The mSulf2−/− profiles indicate little difference in the sulfated regions compared with wild-type, where the only consistent difference was an 18% increase in peak d, containing one 6-O-sulfated glucosamine residue. Comparison of wild-type and mSulf1−/−/mSulf2−/− hexasaccharides extended the trend established in the mSulf1−/− material, showing a marked increase in highly sulfated hexasaccharide species. These data are thus comparable with the compositional disaccharide analysis whereby distinct differences were detected between the various MEF cells in UA–GlcNS(6S) and UA(2S)-GlcNS(6S) residues contained within the S-domains (Table 1).
Octasaccharide analysis
As observed previously [24], the S-domain SAX-HPLC profiles became more complex with increasing length. Numerous species of octasaccharide (dp8) were visible (Figure 3E, peaks a–i). Some of these sequences have been previously characterized [24], however, the oligosaccharide peaks labelled g–i are of unknown composition, and are likely to contain more than one octasaccharide species per peak, an observation supported by subsequent analytical work [28]. mSulf1−/− material contained clearly reduced levels of species a–c, representing S-domain regions devoid of 6S (Figure 3E). However, mSulf1−/− material was enriched in species which elute at higher salt concentrations from the column, most noticeably in octasaccharides f–i, with a 40% total increase in these saccharides compared with wild-type. Again, the trends observed for mSulf1−/− material were amplified when comparing immortalized wild-type and mSulf1−/−/mSulf2−/− octasaccharides (Figure 3E).
Overall, the analyses of heparinase III-generated hexa- and octa-saccharides demonstrated that there is a clear increase in 6S in mSulf1−/− and mSulf1−/−/mSulf2−/− material observed in all sizes of S-domain studied. mSulf2−/− HS, however, shows an increase in 6S almost exclusively within the non-sulfated and transition zones. Thus the mSulf2 knock-out primarily becomes manifest outside the S-domains. Importantly, this observation contrasts with previous studies on the avian Sulf1 and Sulf2 orthologues where activity was restricted to within the S-domains. Specific analysis of the longer sulfated S-domains released by heparinase III as hexa- (dp6) and octa-saccharides (dp8) was made possible by incorporation of data from previous studies, in which we have sequenced many of these sub-species [24]. This demonstrated clear differences in the effect of each mSulf knock-out on S-domain sulfation patterning. As shown in Figures 3(D) and 3(E), mSulf1−/− HS contained a greater proportion of dp6s with either one (6d–f) or two (6g–h) 6S groups; a corresponding decrease in the non-6S species (6a–c) was observed. There was also some indication that mSulf1 activity may be favoured on adjacent GlcNS(6S) residues, as the increase in species 6g containing this pattern, was greater than that seen in 6d, containing only one GlcNS(6S) residue. mSulf1−/−/mSulf2−/− HS-derived S-domain oligosaccharides were similar to, but more extreme than, those seen in the mSulf1−/− HS. This detailed domain-specific HS analysis provides the molecular basis needed to gain insight into the effect of dynamic changes in 6S on the biological phenotypes of the mSulf knock-out mice, which are currently being characterized.
Regulation underlying 6S patterning
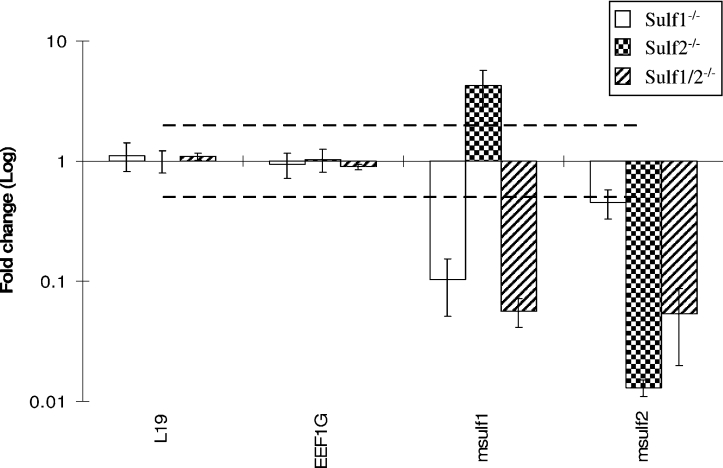
We were interested in a possible co-regulation of mSulf1 and mSulf2 at the level of gene expression. Real-time PCR analysis identified an up-regulation of mSulf1 expression in the mSulf2−/− MEFs (Figure 4, mSulf1 increased by 4.3-fold in mSulf2−/− cells), which was confirmed by Northern analysis (Figure 1C). This compensatory increase in mSulf1 in mSulf2−/− MEFs is in good agreement with our disaccharide data where mSulf2−/− material showed a surprising decrease in S-domain-related 6S disaccharides [UA–GlcNS(6S) and UA(2S)-GlcNS(6S)], and where the effect of the mSulf1−/−/mSulf2−/− double knock-out was in excess of that suggested by either single knock-out alone.
Figure 4. Real-time PCR analysis of the expression of mSulf enzymes in knock-out MEFs.
An average of the two house-keeping genes L19 and EEF1G was used to normalize the cDNAs of each sample relative to each other. The fold change for mSulf1−/− and mSulf2−/− MEFs was calculated relative to the primary MEF wild-type control. Values for mSulf1−/−/mSulf2−/− MEFs were calculated relative to an immortalized wild-type control. A fold change of greater than 2 was judged to be significant (broken line). mSulf1−/− (open bars), mSulf2−/− (checked bars) and mSulf1−/−/mSulf2−/− (upwards diagonal bars). mSulf1 expression was reduced 10-fold in the mSulf1−/− cells, increased 4.3-fold in the mSulf2−/− cells and decreased 18-fold in the mSulf1−/−/mSulf2−/− cells. mSulf2 expression was decreased 2.2-fold in the mSulf1−/− cells, decreased 77-fold in the mSulf2−/− cells and decreased 19-fold in the mSulf1−/−/mSulf2−/− cells. Results are expressed as the means±S.D.
This differential activity of mSulf1 and mSulf2 in the mSulf knock-out MEFs may have important biological implications. Indeed, it is important to note that although no mouse 6-O-sulfotransferase knock-outs have yet been published, in vitro experiments suggest that individual isoforms are non-specific and complementary in their sites of action [29,30]. Therefore the predominant control of 6S patterning, as seen by binding ligands, is probably due to mSulf activity. How mSulf activity is dynamically coupled to generate specific sulfation patterns is likely to be critical for understanding how changes in HS structure are generated to direct cell signalling events during development and disease.
mSulf activity alters surface epitopes recognized by HS-specific antibodies
Methods to quickly and non-destructively analyse cell-surface HS sulfation patterns could significantly improve our ability to screen for sulfation-state changes involved in developmental processes and disease. To approach this issue we have recently developed a method that is based on the use of a panel of phage-display-derived antibodies which have specificity for a variety of saccharide patterns within HS. The ability of these antibodies to recognize specific native HS structures (some of which have recently been defined) can provide a ‘fingerprint’ of HS epitope expression on a cell [31]. When this panel of antibodies was screened against immortalized knock-out and wild-type cells, we found that for some of the antibodies there was little change in binding, for others the binding was decreased in the knock-outs, and for one antibody, RB4EA-12, binding was increased, particularly for the mSulf1−/−/mSulf2−/− line (Figure 5). This differential binding behaviour supports biochemical analyses, as RB4EA-12 has been shown previously to have a specific requirement for 6S [32,33]. Furthermore, antibodies EV3C3 and HS4E4, which showed reduced binding to mSulf knock-out MEFs, are known to be inhibited by 6S [32]. Therefore, in keeping with the documented complexity of Sulf activity on HS-ligand binding, we found that, depending on the antibody used, loss of mSulf activity had a negative effect (i.e. less binding), no effect or a positive effect on antibody binding (Figure 5), demonstrating the potential for differential regulation of ligand-binding epitopes within HS by altered mSulf activity. This further indicates that mSulf activity can simultaneously influence multiple ligand-binding events, probably through induced changes in HS structure [34]. In view of the paucity of detailed three-dimensional information on HS motifs, this panel of antibodies provides a useful screening tool for evaluating these changes.
Figure 5. HS epitope-specific ScFv antibodies show altered cell binding characteristics for mSulf knock-out MEFs.
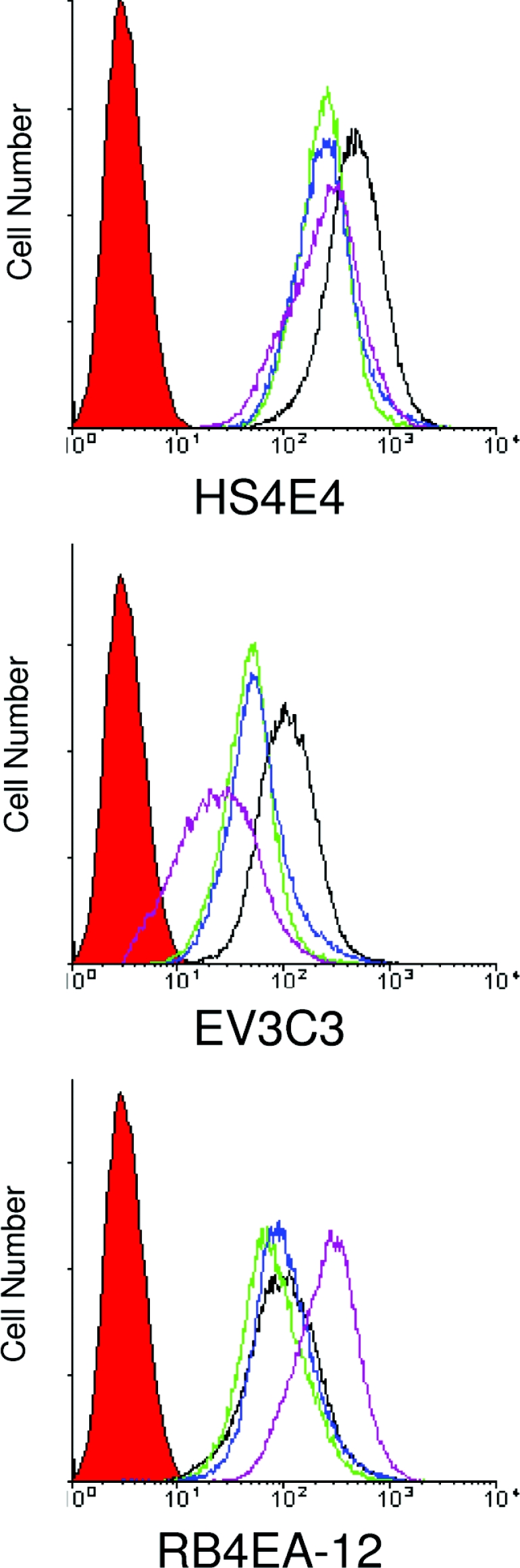
Immortalized wild-type (black line), mSulf1−/− (green line), mSulf2−/− (blue line) and mSulf1−/−/mSulf2−/− (purple line) MEFs were incubated with a panel of HS sequence-specific ScFv antibodies. These were then analysed by FACS to demonstrate a shift in binding profile for each of the MEFs with the various antibodies. The profiles shown for three of the antibodies, as indicated, are representative of at least three different experiments. Control profiles (using secondary antibody alone) are shown as the filled red profiles.
Loss of mSulf activity increases the mitogenic response of MEFs
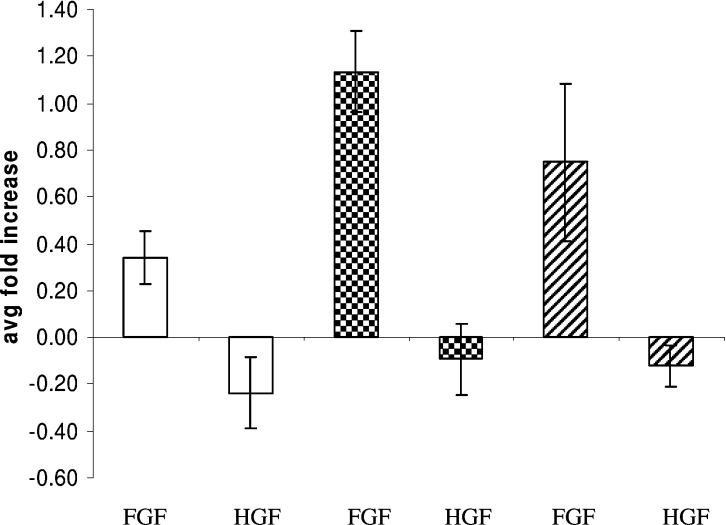
The antibody study above suggested that ligand binding may be affected by mSulf activity. However, the regulatory activity of HS for its many interacting factors is more complex than can be explained by binding affinity alone [24], and, indeed, in some instances high-affinity binding is inhibitory for activity. mSulf activity has previously been linked with the activity of a number of HS-dependent growth factors and morphogens, including the multi-functional FGF2 [15]. We therefore employed a simple proliferation assay using primary MEFs to ascertain if there was any differential activity between the mSulf mutant lines. MEF cell cultures have previously been shown to be suitable for these studies [22]. Results demonstrated a significant increase in proliferation of both single mSulf knock-out cells, particularly for mSulf1−/− in which FGF2 induced a 3-fold increase in proliferation compared with wild-type (Figure 6). No mitogenic response was observed with HGF in any of the MEFs tested, in agreement with other studies (C. L. R. Merry, unpublished work).
Figure 6. Loss of mSulf activity increases the mitogenic response of MEFs to FGF2.
Primary MEFs were allowed to quiesce in serum-free medium prior to induction with either FGF2 or HGF. mSulf1−/− (white bars) and mSulf2−/− (checked bars) cells showed a markedly increased mitogenic response to FGF2 as compared with wild-type (upwards diagonal bars). Exposure to HGF resulted in no mitogenic response in all MEFs. Results are expressed as the means (‘avg’)±S.D.
CONCLUSIONS
In conclusion, loss of mSulf1 or mSulf2 activity in MEFs is associated with increases in HS 6S. Notably, the domain-specific pattern of this increase, its influence on other sulfation activity and the potential effect this has on ligand-binding patterns (as revealed by reactivity with antibodies) are different for the two enzymes. Additionally, the present study provides strong evidence to suggest that mSulf1 and mSulf2 act co-operatively and are the major regulators of 6S patterns within HS (Figure 7). The focus of future studies will be the analysis of the knock-out mice themselves, particularly to characterize the ability of the mSulfs to regulate specific developmental patterning events and tumorigenesis in vivo. Based on this characterization of mSulf knock-out MEFs, we would predict many HS–ligand interactions will be affected by the structural alterations, particularly for the mSulf1−/−/mSulf2−/− mouse.
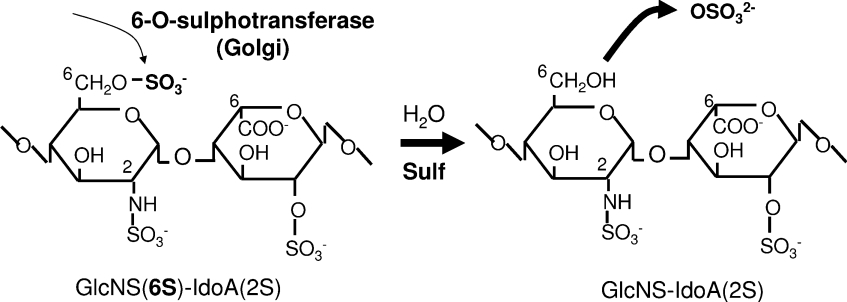
Figure 7. Sulfs are major regulators of 6S within HS.
The addition of 6S to the nascent HS chain within the Golgi is mediated by a family of 6-O-sulfotransferases. Our data suggest that these modifications are fairly extensive throughout the chain (leading to approx. 40% of disaccharides containing 6S groups). It is the activity of the mSulf enzymes that reduces this level to the approx. 15%, which is found in HS prepared from tissues and which finalizes the complex sulfate patterning observed on mature HS chains.
Online data
Acknowledgments
We would like to thank Nicole Tasch, Ellen Eckermann-Felkl, Martina Balleininger and Annegret Schneemann for excellent technical assistance, Peter Gruss for providing embryonic stem cells, and Paul Saftig for his advice in knock-out mouse generation. This work was supported by the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie, Cancer Research UK and the British Council UK–Netherlands Partnership Programme in Science.
References
- 1.Bernfield M., Gotte M., Park P. W., Reizes O., Fitzgerald M. L., Lincecum J., Zako M. Functions of cell surface heparin sulphate proteoglycans. Annu. Rev. Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 2.Rapraeger A. C., Krufka A., Olwin B. B. Requirement of heparin sulphate for bFGF-mediated fibroblast growth and myoblast differentiation. Science. 1991;252:1705–1708. doi: 10.1126/science.1646484. [DOI] [PubMed] [Google Scholar]
- 3.Yayon A., Klagsbrun M., Esko J. D., Leder P., Ornitz D. M. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–848. doi: 10.1016/0092-8674(91)90512-w. [DOI] [PubMed] [Google Scholar]
- 4.Esko J. D., Lindahl U. Molecular diversity of heparin sulfate. J. Clin. Invest. 2001;108:169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ledin J., Staatz W., Li J. P., Gotte M., Selleck S., Kjellen L., Spillmann D. Heparan sulfate structure in mice with genetically modified heparan sulfate production. J. Biol. Chem. 2004;279:42732–42741. doi: 10.1074/jbc.M405382200. [DOI] [PubMed] [Google Scholar]
- 6.Maccarana M., Sakura Y., Tawada A., Yoshida K., Lindahl U. Domain structure of heparan sulfates from bovine organs. J. Biol. Chem. 1996;271:17804–17810. doi: 10.1074/jbc.271.30.17804. [DOI] [PubMed] [Google Scholar]
- 7.Pye D. A., Vives R. R., Hyde P., Gallagher J. T. Regulation of FGF-1 mitogenic activity by heparan sulfate oligosaccharides is dependent on specific structural features: differential requirements for the modulation of FGF-1 and FGF-2. Glycobiology. 2000;10:1183–1192. doi: 10.1093/glycob/10.11.1183. [DOI] [PubMed] [Google Scholar]
- 8.Brickman Y. G., Ford M. D., Gallagher J. T., Nurcombe V., Bartlett P. F., Turnbull J. E. Structural modification of fibroblast growth factor-binding heparin sulphate at a determinative stage of neural development. J. Biol. Chem. 1998;273:4350–4359. doi: 10.1074/jbc.273.8.4350. [DOI] [PubMed] [Google Scholar]
- 9.Jayson G. C., Vives C., Paraskeva C., Schofield K., Coutts J., Fleetwood A., Gallagher J. T. Coordinated modulation of the fibroblast growth factor dual receptor mechanism during transformation from human colon adenoma to carcinoma. Int. J. Cancer. 1999;82:298–304. doi: 10.1002/(sici)1097-0215(19990719)82:2<298::aid-ijc23>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 10.Dhoot G. K., Gustafsson M. K., Ai X., Sun W., Standiford D. M., Emerson C. P., Jr Regulation of Wnt signalling and embryo patterning by an extracellular sulfatase. Science. 2001;293:1663–1666. doi: 10.1126/science.293.5535.1663. [DOI] [PubMed] [Google Scholar]
- 11.Morimoto-Tomita M., Uchimura K., Werb Z., Hemmerich S., Rosen S. D. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J. Biol. Chem. 2002;277:49175–49185. doi: 10.1074/jbc.M205131200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ai X., Do A. T., Kusche-Gullberg M., Lindahl U., Lu K., Emerson C. P., Jr Substrate specificity and domain functions of extracellular heparan sulfate 6-O-endosulfatases, QSulf1 and QSulf2. J. Biol. Chem. 2006;281:4969–4976. doi: 10.1074/jbc.M511902200. [DOI] [PubMed] [Google Scholar]
- 13.Ai X., Do A. T., Lozynska O., Kusche-Gullberg M., Lindahl U., Emerson C. P., Jr QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling. J. Cell Biol. 2003;162:341–351. doi: 10.1083/jcb.200212083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Viviano B. L., Paine-Saunders S., Gasiunas N., Gallagher J., Saunders S. Domain-specific modification of heparan sulfate by QSulf1 modulates the binding of the bone morphogenetic protein antagonist noggin. J. Biol. Chem. 2004;279:5604–5611. doi: 10.1074/jbc.M310691200. [DOI] [PubMed] [Google Scholar]
- 15.Wang S., Ai X., Freeman S. D., Pownall M. E., Lu Q., Kessler D. S., Emerson C. P., Jr QSulf1, a heparan sulfate 6-O-endosulfatase, inhibits fibroblast growth factor signaling in mesoderm induction and angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4833–4838. doi: 10.1073/pnas.0401028101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai J., Chien J., Staub J., Avula R., Greene E. L., Matthews T. A., Smith D. I., Kaufmann S. H., Roberts L. R., Shridhar V. Loss of HSulf-1 up regulates heparin-binding growth factor signaling in cancer. J. Biol. Chem. 2003;278:23107–23117. doi: 10.1074/jbc.M302203200. [DOI] [PubMed] [Google Scholar]
- 17.Li J., Kleeff J., Abiatari I., Kayed H., Giese N. A., Felix K., Giese T., Buchler M. W., Friess H. Enhanced levels of Hsulf-1 interfere with heparin-binding growth factor signaling in pancreatic cancer. Mol. Cancer. 2005;4:14. doi: 10.1186/1476-4598-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morimoto-Tomita M., Uchimura K., Bistrup A., Lum D. H., Egeblad M., Boudreau N., Werb Z., Rosen S. D. Sulf-2, a proangiogenic heparan sulfate endosulfatase, is up regulated in breast cancer. Neoplasia. 2005;7:1001–1010. doi: 10.1593/neo.05496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai J. P., Chien J., Strome S. E., Staub J., Montoya D. P., Greene E. L., Smith D. I., Roberts L. R., Shridhar V. HSulf-1 modulates HGF-mediated tumor cell invasion and signaling in head and neck squamous carcinoma. Oncogene. 2004;23:1439–1447. doi: 10.1038/sj.onc.1207258. [DOI] [PubMed] [Google Scholar]
- 20.Lai J. P., Chien J. R., Moser D. R., Staub J. K., Aderca I., Montoya D. P., Matthews T. A., Nagorney D. M., Cunningham J. M., Smith D. I., et al. HSulf-1 sulfatase promotes apoptosis of hepatocellular cancer cells by decreasing heparin-binding growth factor signaling. Gastroenterology. 2004;126:231–248. doi: 10.1053/j.gastro.2003.09.043. [DOI] [PubMed] [Google Scholar]
- 21.Uchimura K., Morimoto-Tomita M., Bistrup A., Li J., Lyon M., Gallagher J., Werb Z., Rosen S. D. HSulf-2, an extracellular endoglucosamine-6-sulfatase, selectively mobilizes heparin-bound growth factors and chemokines: effects on VEGF, FGF-1, and SDF-1. BMC Biochem. 2006;7:2. doi: 10.1186/1471-2091-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merry C. L., Bullock S. L., Swan D. C., Backen A. C., Lyon M., Beddington R. S., Wilson V. A., Gallagher J. T. The molecular phenotype of heparan sulfate in the Hs2st−/− mutant mouse J. Biol. Chem. 2001;276:35429–35434. doi: 10.1074/jbc.M100379200. [DOI] [PubMed] [Google Scholar]
- 23.Schuermann M. An expression vector system for stable expression of oncogenes. Nucleic Acids Res. 1990;18:4945–4946. doi: 10.1093/nar/18.16.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merry C. L. R., Lyon M., Deakin J. A., Hopwood J. J., Gallagher J. T. Highly sensitive sequencing of the sulfated domains of heparan sulphate. J. Biol. Chem. 1999;274:18455–18462. doi: 10.1074/jbc.274.26.18455. [DOI] [PubMed] [Google Scholar]
- 25.Mouritzen P., Nielsen P. S., Jacobsen N., Noerholm M., Lomholt C., Pfundheller H. M., Ramsing N. B., Kauppinen S., Tolstrup N. The probe library: expression profiling of 99% of all human genes using only 90 dual labeled real time PCR probes. BioTechniques. 2004;37:492–495. [Google Scholar]
- 26.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Shively J. E., Conrad H. E. Formation of anhydrosugars in the chemical depolymerization of heparin. Biochemistry. 1976;15:3932–3942. doi: 10.1021/bi00663a005. [DOI] [PubMed] [Google Scholar]
- 28.Robinson C. J., Mulloy B., Gallagher J. T., Stringer S. E. VEGF165-binding sites within heparan sulfate encompass two highly sulfated domains and can be liberated by K5 lyase. J. Biol. Chem. 2006;281:1731–1740. doi: 10.1074/jbc.M510760200. [DOI] [PubMed] [Google Scholar]
- 29.Do A. T., Smeds E., Spillmann D., Kusche-Gullberg M. Overexpression of heparan sulfate 6-O-sulfotransferases in human embryonic kidney 293 cells results in increased N-acetylglucosaminyl 6-O-sulfation. J. Biol. Chem. 2005;9:5348–5356. doi: 10.1074/jbc.M509584200. [DOI] [PubMed] [Google Scholar]
- 30.Smeds E., Habuchi H., Do A. T., Hjertson E., Grundberg H., Kimata K., Lindahl U., Kusche-Gullberg M. Substrate specificities of mouse heparan sulphate glucosaminyl 6-O-sulphotransferases. Biochem. J. 2003;372:371–380. doi: 10.1042/BJ20021666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Kuppevelt T. H., Dennissen M. A., van Venrooij W. J., Hoet R. M., Veerkamp J. H. Generation and application of type-specific anti-heparan sulfate antibodies using phage display technology. Further evidence for heparan sulfate heterogeneity in the kidney. J. Biol. Chem. 1998;273:12960–12966. doi: 10.1074/jbc.273.21.12960. [DOI] [PubMed] [Google Scholar]
- 32.Dennissen M. A., Jenniskens G. J., Pieffers M., Versteeg E. M., Petitou M., Veerkamp J. H., van Kuppevelt T. H. Large, tissue-regulated domain diversity of heparan sulfates demonstrated by phage display antibodies. J. Biol. Chem. 2002;277:10982–10986. doi: 10.1074/jbc.M104852200. [DOI] [PubMed] [Google Scholar]
- 33.Jenniskens G. J., Oosterhof A., Brandwijk R., Veerkamp J. H., van Kuppevelt T. H. Heparan sulfate heterogeneity in skeletal muscle basal lamina: demonstration by phage display-derived antibodies. J. Neurosci. 2000;20:4099–4111. doi: 10.1523/JNEUROSCI.20-11-04099.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raman R., Sasisekharan V., Sasisekharan R. Structural insights into biological roles of protein–glycosaminoglycan interaction. Chem. Biol. 2005;12:267–277. doi: 10.1016/j.chembiol.2004.11.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.