Abstract
All CHMPs (charged multivesicular body proteins) reported to date have common features: they all contain approx. 200 amino acid residues, have coiled-coil regions and have a biased distribution of charged residues (basic N-terminal and acidic C-terminal halves). Yeast orthologues of CHMPs, including an ESCRT-III component Snf7, are required for the sorting of cargo proteins to intraluminal vesicles of multivesicular bodies. We have characterized a novel human ESCRT-III-related protein, designated CHMP7, which consists of 453 amino acid residues. CHMP7 contains an SNF7 domain and a distantly SNF7-related domain in its C-terminal half and N-terminal half respectively. Among the ten CHMP proteins classified previously in six subfamilies (CHMP1–CHMP6), the C-terminal SNF7 domain of CHMP7 is most similar to the SNF7 domain of CHMP6, which associates with CHMP4 proteins and EAP20, a component of ESCRT-II. Pull-down assays using lysates of HEK-293T (human embryonic kidney) cells that overexpressed Strep-tagged CHMP7 and GFP (green fluorescent protein)-fused CHMP4b (also named Shax1) revealed a positive interaction between the C-terminal half of CHMP7 and CHMP4b. However, interaction was not observed between CHMP7 and EAP20. Confocal fluorescence microscopic analyses revealed that FLAG–CHMP7 is distributed in HeLa cells diffusely throughout the cytoplasm, but with some accumulation, especially in the perinuclear area. The distribution of FLAG–CHMP7 was altered to a cytoplasmic punctate pattern by overexpression of either CHMP4b–GFP or GFP–Vps4BE235Q, a dominant-negative mutant of the AAA (ATPase associated with various cellular activities) Vps4B, and partially co-localized with them. Ubiquitinated proteins and endocytosed EGF accumulated in GFP–CHMP7-expressing cells. A dominant-negative effect of overexpressed GFP–CHMP7 was also observed in the release of virus-like particles from HEK-293T cells that transiently expressed the MLV (murine leukaemia virus) Gag protein. These results suggest that CHMP7, a novel CHMP4-associated ESCRT-III-related protein, functions in the endosomal sorting pathway.
Keywords: changed multivesicular body protein (CHMP), endosomal sorting complex required for transport (ESCRT), multivesicular body, retrovirus, SNF7
Abbreviations: AAA, ATPase associated with various cellular activities; AD, transcription-activation domain; Alix, ALG-2-interacting protein X; BD, DNA-binding domain; CaM, calmodulin; CC, coiled coil; CDD, conserved domain database; CHMP, charged multivesicular body protein; CHMP7NH, N-terminal half of CHMP7; CHMP7CH, C-terminal half of CHMP7; DMEM, Dulbecco's modified Eagle's medium; EEA1, early endosome antigen 1; EGF, epidermal growth factor; ESCRT, endosomal sorting complex required for transport; FBS, foetal bovine serum; GFP, green fluorescent protein; GST, glutathione S-transferase; HEK-293, human embryonic kidney; HRP, horseradish peroxidase; Lamp1, lysosome-associated membrane protein 1; mAb, monoclonal antibody; MLV, murine leukaemia virus; mRFP, monomeric red fluorescent protein; MVB, multivesicular body; Rh, tetramethylrhodamine; VLP, virus-like particle
INTRODUCTION
In eukaryotic cells, transmembrane proteins and their ligands that are destined for the lysosome are delivered from the plasma membrane and trans-Golgi network to intraluminal vesicles of the MVBs (multivesicular bodies). Protein complexes called ESCRT-I, -II and -III (endosomal sorting complex required for transport I, II and III) and their associated proteins are required for both the formation of MVB luminal vesicles and the sorting of ubiquitinated cargo proteins into these vesicles [1–4]. The ESCRT system is conserved from yeast to humans. ESCRT-III is the final major complex to be recruited to the endosomal membrane and disassembled by the AAA (ATPase associated with various cellular activities) called Vps4 (or SKD1 in mouse) for recycling. Yeast has four ESCRT-III components and two related proteins of similar sizes (∼220 amino acid residues) that are mutually similar in primary sequences. All six proteins (Vps2, Vps20, Vps24, Snf7/Vps32, Did2/Fti1 and Mos10/Vps60) have predicted CC (coiled coils) and are rich in basic and acidic residues in their N- and C-terminal halves respectively. Yeast genetic and biochemical studies have revealed that subcomplexes are formed between Vps2 and Vps24 and between Snf7 and Vps20 [5]. Deletion of each gene for ESCRT-III proteins, including Did2 and Vps60, causes abnormality in sorting of cargo molecules to vacuoles [5–7].
It has been reported that humans have a total of ten ESCRT-III-like proteins called CHMPs (charged MVB proteins), which can be subdivided into six subfamilies (CHMP1–CHMP6) that have identity with the six yeast proteins (CHMP1/Did2p, CHMP2/Vps2, CHMP3/Vps24, CHMP4/Snf7, CHMP5/Vps60 and CHMP6/Vps20). CHMP1 and CHMP2 each have two isoforms (CHMP1A, CHMP1B, CHMP2A and CHMP2B). CHMP4 has three isoforms [4,8–13], which are designated misleadingly in the published papers and DNA databases: Shax1/CHMP4b/CHMP4A, Shax2/CHMP4a/CHMP4B and Shax3/CHMP4c/CHMP4C. Since the approved symbol of the human Shax1 [Snf7 homologue associated with Alix (ALG-2-interacting protein X) 1] gene is designated CHMP4B (HUGO Gene Nomenclature Committee, http://www.gene.ucl.ac.uk/nomenclature/index.html), we use the nomenclature of CHMP4b and Shax1 in parallel to avoid further confusion in the names of CHMP4 proteins in the present paper. CHMPs associate with other ESCRTs either by direct physical interaction as demonstrated for CHMP6, a myristoylated protein, with EAP20 (an ESCRT-II component) [14], or by indirect interaction through Alix/AIP1 that bridges CHMP4 and TSG101 (an ESCRT-I component) [9–13]. Destruction of the mouse CHMP5 gene (Chmp5) by homologous recombination resulted in a phenotype of early embryonic lethality, reflecting defective late endosome function and dysregulation of multiple signal transductions, such as growth factor TGFβ (transforming growth factor β) and inflammatory cytokines via NF-κB (nuclear factor κB) [15]. Overexpression of CHMP proteins exhibited dominant-negative effects on the disappearance of endocytosed EGF (epidermal growth factor) [9,14], induced accumulation of cholesterol in the CHMP-localized compartments [16,17] and suppressed the release of retrovirus particles from infected host cells whose ESCRTs are utilized for virus budding [11–13]. Depletion of CHMP3 by siRNA (small interfering RNA) impaired transport of EGF receptor from early endosomes to lysosomes [18].
By searching for proteins that have sequences similar to those of CHMPs in the human DNA database, we have retrieved a cDNA sequence encoding a novel human CHMP family member designated CHMP7 (GenBank® accession number NM_152272). Although the DNA and its deduced amino acid sequences of CHMP7 are available in the database, no experimental results concerning biochemical and functional properties have been reported in any journals to date. In the present study, we investigated whether CHMP7 also plays roles in MVB sorting and displays properties similar to those of other characterized CHMP family members. We found that CHMP7 interacted with CHMP4b and that overexpression of GFP (green fluorescent protein)-fused CHMP7 caused impairment of transport of endocytosed EGF in the endosome–lysosome pathway. Moreover, the overexpressed CHMP7 proteins suppressed release of VLPs (virus-like particles) into culture medium in a co-transfection experiment. These results indicate that CHMP7 is a genuine member of the ESCRT-III-linked CHMP family.
MATERIALS AND METHODS
cDNA cloning and mammalian expression plasmids
A CHMP7 cDNA was cloned from a HEK-293 (human embryonic kidney 293) cell cDNA library by the PCR method using a pair of primers that were designed on the basis of a registered sequence (GenBank® accession number NM_152272, underlined) and an additional overhanging sequence for EcoRI digestion: 5′-GAGAATTCGTTCCGATGTGGTCCCCGGA-3′ (FP1) and 5′-CCGAATTCACAATGGCTTTAGAGTCGGTT-3′ (RP1). The PCR product was digested with EcoRI and first inserted into the EcoRI site of pBluescript II (Stratagene) and then subcloned further into the EcoRI sites of mammalian expression vectors pCMV-3xFLAG-A [9] and pEGFP-C2 (Clontech) respectively. The MCS (multiple cloning site) of the Strep-tag vector pEXPR-IBA105 (IBA GmbH) was modified into pEXPR-IBA105-A by inserting an oligonucleotide block (forward, 5′-GGAATTCGGGGATCCGATATCTCGAGGTACCA-3′; reverse, 5′-AGCTTGGTACCTCGAGATATCGGATCCCCGAATTCCGC-3′) between the SacII and HindIII sites before subcloning the EcoRI fragment of the CHMP7 cDNA. To express N- and C-terminal halves of CHMP7 (designated CHMP7NH and CHMP7CH respectively), cDNA fragments encoding amino acids 1–222 and 223–453 were amplified by PCR using forward and reverse primers containing additional EcoRI sites and a translation termination codon and were inserted into pEXPR-IBA105-A respectively. For expression of C-terminally GFP-fused CHMP7 (CHMP7–GFP), the CHMP7 termination codon was removed by PCR amplification with a forward primer (FP1) and reverse primer 5′-GCGGATCCAATGGCTTTAGAGTCGG-3′(RP2). The PCR product was inserted into the EcoRI/BamHI site of pSGG-GFP [14], a derivative of pEGFP-N1 (Clontech) containing a flexible Ser-Gly-Gly linker between the multiple cloning site and GFP-encoding sequence.
Construction of the expression vectors for FLAG–CHMP4b and CHMP4b–GFP was as described previously [9]. A cDNA expression construct of an ATPase-defective mutant of human Vps4B (Vps4BE235Q) has been described previously [19].
An mRFP1 (monomeric red fluorescent protein 1) cDNA derived from the prokaryotic expression vector, pRSET-B/mRFP1 [20] (a gift from Dr R.Y. Tsien, University of California at San Diego, San Diego, CA, U.S.A.), was amplified by the PCR method using a forward primer containing an additional NheI site and a Kozak consensus translation initiation site (5′-AATTGCTAGCGCTACCGGTCGCCACCATGGCCTCCTCCGAGGACGTC-3′) and a reverse primer containing an additional BsrGI site (5′-TACTTGTACACGGCGCCGGTGGAGTGGCGGC-3′). The PCR product was digested with NheI and BsrGI and then ligated with the larger fragment of NheI/BsrGI-digested pEGFP-C2 (Clontech) to construct a eukaryotic mRFP1 expression vector, pmRFP1-C2. A SKD1E235Q cDNA fragment was obtained from pGFP-SKD1E235Q [21] by digestion with EcoRI and was inserted into the EcoRI site of pmRFP1-C2 to construct pmRFP1-SKD1E235Q. A cDNA fragment encoding amino acids 1–222 of CHMP7 with additional EcoRI sites plus a stop codon was obtained using the PCR method and inserted into pEGFP-C2 (Clontech). The resultant plasmid was designated pEGFP-C2-CHMP7NH and was used to express GFP–CHMP7NH. To express C-terminally GFP-fused CHMP7CH (CHMP7CH–GFP), a cDNA encoding amino acids 223–453 was amplified by the PCR method and inserted into pSGG-GFP [14]. Then, an oligonucleotide block containing a Kozak consensus sequence and HindIII/EcoRI overhanging sites (5′-AGCTTGAGCCACCATGG-3′ and 5′-AATTCCATGGTGGCTCA-3′) was inserted into the upstream HindIII/EcoRI site.
Yeast two-hybrid assay
The Matchmaker™ Two-Hybrid System, including yeast strain AH109 and vectors, was obtained from Clontech. To construct expression vectors of the GAL4 BD (DNA-binding domain) fusion proteins (bait), full-length and truncated CHMP7 cDNAs were subcloned into the EcoRI site of pGBKT7. To construct the expression vectors for GAL4 AD (transcription-activation domain) fusion proteins (prey), each cDNA was subcloned into pGAD424 by the conventional gene-manipulation method. AD vectors of ESCRT and related proteins were constructed previously in the case of Alix, CHMP4a, CHMP4b, TSG101, Vps28, Vps37A [9,22] and newly constructed for CHMP1A, CHMP1B, CHMP2A, CHMP2B, CHMP3, CHMP4c, CHMP5, CHMP6, CHMP7, EAP20, EAP30, EAP45, Vps4A and Vps4B in the present study. Interaction was evaluated by growth on nutrient-dropout medium plates as described previously [23].
Antibodies
Mouse mAbs (monoclonal antibodies) were purchased from the following sources: anti-EEA1 (early endosome antigen 1) (clone 14) (Transduction Laboratories), anti-Lamp-1 (lysosome-associated membrane protein 1)/CD107a (H4A3) (Pharmingen), anti-ubiquitin (FK2) (MBL), anti-GFP (B2) (Santa Cruz Biotechnology) and anti-FLAG (M2) (Sigma). A goat anti-p30 (MLV Gag capsid protein) polyclonal antibody was a gift from Dr H. Amanuma (RIKEN, Wako, Japan). HRP (horseradish peroxidase)-conjugated anti-(Strep-tag II) was obtained from IBA GmbH. HRP-conjugated goat anti-mouse and donkey anti-goat IgG antibodies were obtained from Jackson Immunoresearch Laboratories and from Santa Cruz Biotechnology respectively. For indirect immunofluorescence analyses, Cy3-labelled goat anti-mouse IgG was obtained from Amersham Biosciences.
Cell culture and transfection
HEK-293T and HeLa cells were cultured in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% or 5% heat-inactivated FBS (foetal bovine serum), 100 units/ml penicillin and 100 μg/ml streptomycin at 37 °C under humidified air containing 5% CO2. At 24 h after HEK-293T cells had been seeded, the cells were transfected with the expression plasmid DNAs using the conventional calcium phosphate precipitation method and were cultured for 24 h. For fluorescence-microscopic analysis, HeLa cells were seeded on coverslips placed in 3-cm-diameter dishes, transfected using FuGENE6 (Roche Applied Science) and cultured for 24 h.
Strep-pull-down assay
At 24 h after transfection with expression vectors, HEK-293T cells were washed with PBS (137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4 and 1.5 mM KH2PO4, pH 7.3), and harvested cells were lysed in lysis buffer A (20 mM Hepes/NaOH, pH 7.4, 142.5 mM KCl, 1 mM EDTA, 1% Triton X-100 and 1% sodium deoxycholate) containing protease inhibitors {0.1 mM Pefabloc, 25 μg/ml leupeptin, 1 μM E-64 [trans-epoxysuccinyl-L-leucylamido-(4-guanidino)butane] and 1 μM pepstatin}. Supernatants after centrifugation at 10000 g for 10 min were incubated with Strep-tactin beads (IBA GmbH) for 1 h at 4 °C with gentle mixing. After the beads had been recovered by a low-speed centrifugation (600 g) for 3 min and washed three times with lysis buffer A, the bead-bound proteins (pull-down products) were subjected to SDS/PAGE followed by Western blotting using PVDF membranes (Immobilon-P; Millipore). The membranes were then blotted either with HRP-conjugated anti-Strep-tag II mAb or with anti-FLAG mAb followed by HRP-conjugated secondary antibody. Signals were detected using the chemiluminescence method using SuperSignal West Pico chemiluminescent substrate (Pierce).
Immunofluorescence microscopy
At 24 h after transfection, the transfected HeLa cells were washed three times with ice-cold PBS and fixed with 4% (w/v) paraformaldehyde in PBS for 20 min. The cells were permeabilized with 0.1% (w/v) Triton X-100 in PBS for 5 min and then blocked with 0.1% (w/v) gelatin in PBS (gelatin-PBS) for 30 min. After blocking, the cells were incubated with primary antibodies diluted with gelatin-PBS at 37 °C for 1 h. After the cells had been washed three times with gelatin-PBS, they were incubated with secondary antibodies diluted with gelatin-PBS. Then they were washed three times with gelatin-PBS and additionally with PBS twice. Finally, they were mounted with antifading solution as described previously [9] and observed under a confocal laser-scanning microscope, LSM PASCAL (Carl Zeiss).
EGF-uptake assay
EGF-uptake assay was performed essentially as described previously [9,14]. Briefly, the transfected HeLa cells grown on coverslips were incubated with serum-free medium (DMEM containing 10 mM Hepes/NaOH, pH 7.4, and 0.5 mg/ml BSA) for 1 h at 37 °C and then for 30 min at 4 °C. A small volume (50 μl) of 0.5 μg/ml Rh–EGF (tetramethylrhodamine–EGF) (Molecular Probes) solution in the serum-free medium was poured on to coverslips, and cells were incubated for 1 h at 4 °C to allow binding of Rh–EGF to cell-surface receptors. The cells were then washed with the serum-free medium, and Rh–EGF uptake was initiated by incubating in DMEM containing 10% FBS at 37 °C for the indicated times. Cells were then washed, fixed and observed under a confocal laser-scanning microscope.
Release of VLPs
An EF1α promoter-driven MLV (murine leukaemia virus) Gag–Pol expression plasmid, pGag-pol-IRES-bsr [24], a gift from Dr Kitamura (University of Tokyo, Tokyo, Japan), was modified to remove the Pol sequence as follows: an MluI site was introduced before the termination codon of the Gag sequence by site-directed mutagenesis and the MluI/NcoI fragment containing the Pol sequence was replaced with an oligonucleotide block (5′-CGCGTCGATATCCGATTACAAGGATGACGACGATAAGTAGC-3′; 5′-GGCCGCTACTTATCGTCGTCATCCTTGTAATCGGATATCGA-3′). Although the resultant plasmid, pGag-FL, encodes a FLAG-sequence at the C-terminus of Gag, the sensitivity and signal-to-noise ratio to detect the expressed protein were greater by using an anti-Gag polyclonal antibody than by using an anti-FLAG mAb (results not shown). At 24 h after transfection of HEK-293T cells with the MLV Gag expression plasmid pGag-FL, culture media were collected and centrifuged at 1000 g for 3 min to remove cell debris, and the supernatants were centrifuged further at 50000 rev./min for 30 min at 4 °C using a Beckman TLA 100.2 rotor. The pellets were washed by suspending with PBS and re-centrifuged under the same conditions to collect VLPs released from cells. For the analyses of intracellular Gag proteins, cells remaining after removing the culture medium were washed with PBS, lysed and subjected to Western blotting.
RESULTS
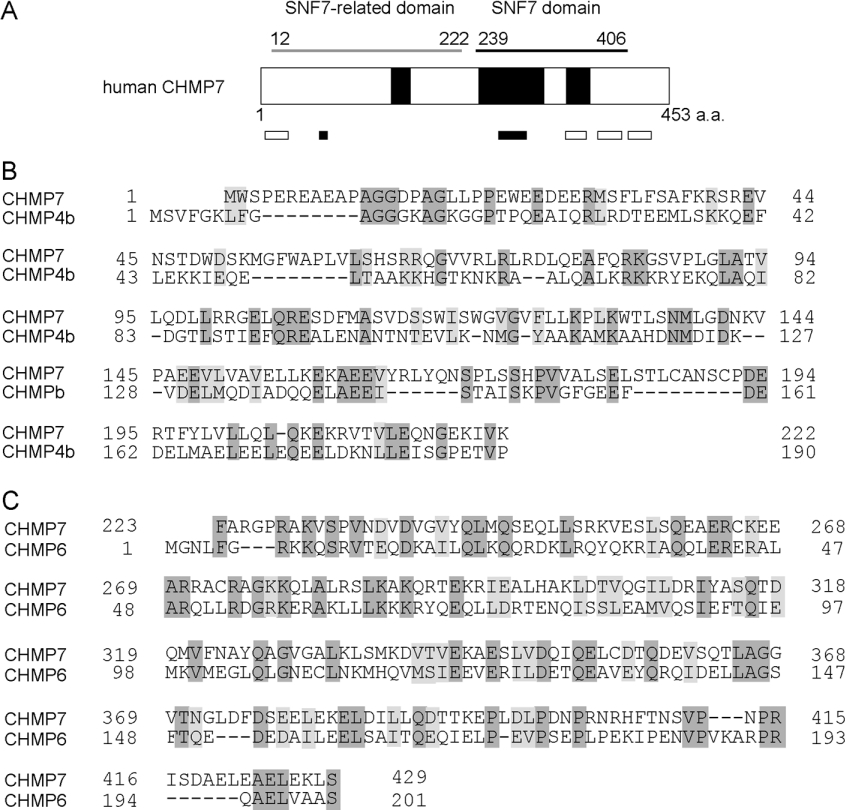
The human CHMP7 cDNA encodes a longer translated region than do other human CHMP family members (CHMP1A, 196 amino acids; CHMP1B, 196 amino acids; CHMP2A, 222 amino acids; CHMP2B, 213 amino acids; CHMP3, 222 amino acids; CHMP4a/Shax2, 222 amino acids; CHMP4b/Shax1, 224 amino acids; CHMP4c/Shax3, 233 amino acids; CHMP5, 219 amino acids; CHMP6, 201 amino acids; CHMP7, 453 amino acids). A BLAST search revealed the presence of an SNF7 domain [CDD (conserved domain database) (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) accession number pfam03357] in its C-terminal half (Figure 1A). CHMP7 contains predicted CC regions (residues 146–166, 243–314 and 340–365) and clusters of basic and acidic residues (basic: residues 65–73 and 264–294; acidic: residues 5–29, 338–360, 374–400 and 418–432). Among the human CHMP family members, the sequence of CHMP7CH is most similar to that of CHMP6 (CHMP7, amino acids 223–429 compared with CHMP6, amino acids 5–201, amino acid identity: 25.2%) (Figure 1C). Although a BLAST search of CHMP7NH did not result in the presence of any conserved domain in CDD, a weak identity was found between CHMP7NH and CHMP4b using a program of pairwise alignment algorithms, EMBOSS-Align (http://www.ebi.ac.uk/emboss/align/index.html) (CHMP7, amino acids 1–222; CHMP4b, amino acids 1–190, amino acid identity: 21.4%) (Figure 1B). CHMP7NH is distantly related to CHMPs and was assigned as an outgroup in the phylogenetic tree (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/400/bj4000023add.htm). CHMP7 orthologues are found from lower to higher eukaryotes (see Supplementary Figure S2 at http://www.BiochemJ.org/bj/400/bj4000023add.htm).
Figure 1. Schematic representation of human CHMP7 and comparison with other CHMP family proteins.
(A) Domain structure of CHMP7. CHMP7 contains an SNF7 domain (CDD accession number pfam03357) which is found in a group of Snf7 homologues, including ESCRT-III components. The N-terminal half also has a sequence related to the SNF7 domain, indicated by a grey bar (SNF7-related domain). CCs predicted by using a program on the Internet (http://www.ch.embnet.org/software/COILS_form.html; window 21, score above 0.2), are indicated by large closed boxes. Clusters of basic and acidic residues are indicated by small closed and open boxes respectively. Amino acid sequence alignments between CHMP7NH (amino acids 1–222) and CHMP4b (B) and between CHMP7CH (amino acids 223–429) and CHMP6 (C). Alignment was performed by using a pairwise alignment program, EMBOSS-Align. Identical and similar residues are shaded dark grey and light grey.
Analysis of interaction between CHMP7 and ESCRT-related proteins
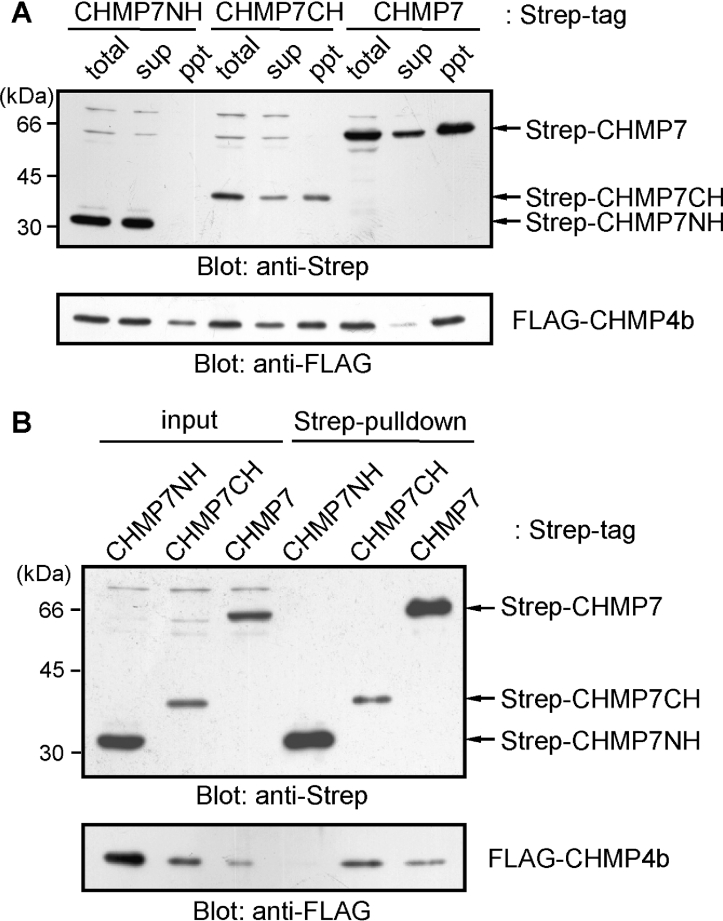
We reported previously that CHMP6, a myristoylated ESCRT-III component, interacts directly with CHMP4b as well as EAP20, a component of ESCRT-II [14]. First, we investigated whether CHMP7, having an SNF7 domain most similar to that of CHMP6, also associates with CHMP4b. HEK-293T cells were co-transfected with the expression vector of FLAG–CHMP4b and each vector of Strep-tagged CHMP7 protein of full-length (Strep–CHMP7), N-terminal half (Strep–CHMP7NH) or C-terminal half (Strep–CHMP7CH). Recovery of each Strep-tagged protein in the supernatant of the cell lysate was poor for Strep–CHMP7 and Strep–CHMP7CH, probably due to co-precipitation with FLAG–CHMP4b to a detergent-insoluble fraction (Figure 2A). The cleared lysates were incubated with Strep-tactin beads, and the proteins bound to the beads (pull-down products) were analysed by Western blotting. FLAG–CHMP4b was pulled-down with Strep–CHMP7 and Strep–CHMP7CH, but not with Strep–CHMP7NH (Figure 2B). As described above, although total expression levels of FLAG–CHMP4b were similar irrespective of co-expressing CHMP7 constructs, recovery of FLAG–CHMP4b in the supernatant fraction (designated ‘sup’ in Figure 2A and ‘input’ in Figure 2B) was significantly reduced by co-expression with Strep–CHMP7CH or Strep–CHMP7 when compared with Strep–CHMP7NH. Next, we investigated whether CHMP7 interacts with EAP20 by co-expressing epitope-tagged proteins in HEK-293T cells. FLAG-tagged EAP20 was not co-immunoprecipitated with GFP-fused CHMP7 proteins under the conditions in which interaction was observed between EAP20 and CHMP6 (see Supplementary Figure S3 at http://www.BiochemJ.org/bj/400/bj4000023add.htm).
Figure 2. Interaction between CHMP7 and CHMP4b.
(A) HEK-293T cells were co-transfected with plasmids that express FLAG–CHMP4b and Strep–CHMP7, Strep-CHMP7NH or Strep-CHMP7CH. After 24 h, the cells were lysed and separated into supernatants (sup) and pellets (ppt) by centrifugation. Expressed proteins were detected by Western blotting using anti-Strep mAb (upper panel) and anti-FLAG mAb (lower panel). (B) The supernatants (input) were incubated with Strep-tactin beads, and the proteins bound to the beads (pull-down) were analysed by Western blotting.
To explore further the possibility of protein–protein interactions that might occur between CHMP7 and ESCRT-related proteins, we performed a yeast two-hybrid assay. Since the expression of GAL4 BD fused with a full-length construct of CHMP7 exhibited growth suppression of the yeast transformant, we expressed the N-terminal half (CHMP7NH, amino acids 1–222) and the C-terminal half (CHMP7CH, amino acids 223–453) separately. We tested a GAL4 AD fused with TSG101, Vps28, Vps37A, EAP20, EAP30, EAP45, CHMP1A, CHMP1B, CHMP2A, CHMP2B, CHMP3, CHMP4a, CHMP4b, CHMP4c, CHMP6, CHMP7, Vps4A, Vps4B or Alix. Growth of colonies on selection agar plates was observed only when CHMP7CH fusion protein (BD–CHMP7CH) and three AD–CHMP4 isoforms were used as bait and prey respectively. No significant differences were observed in the growth rates among the yeast transformants that co-expressed BD–CHMP7CH and AD–CHMP4a, AD–CHMP4b or AD–CHMP4c on selection agar plates (results not shown), suggesting similar abilities of CHMP4 isoforms to bind CHMP7. No colonies appeared when BD–CHMP7NH was tested in any case (results not shown). CHMP5 exhibited toxicity to the growth of yeast transformant under our experimental conditions and interaction could not be tested.
Subcellular distribution of FLAG- and GFP-tagged CHMP7
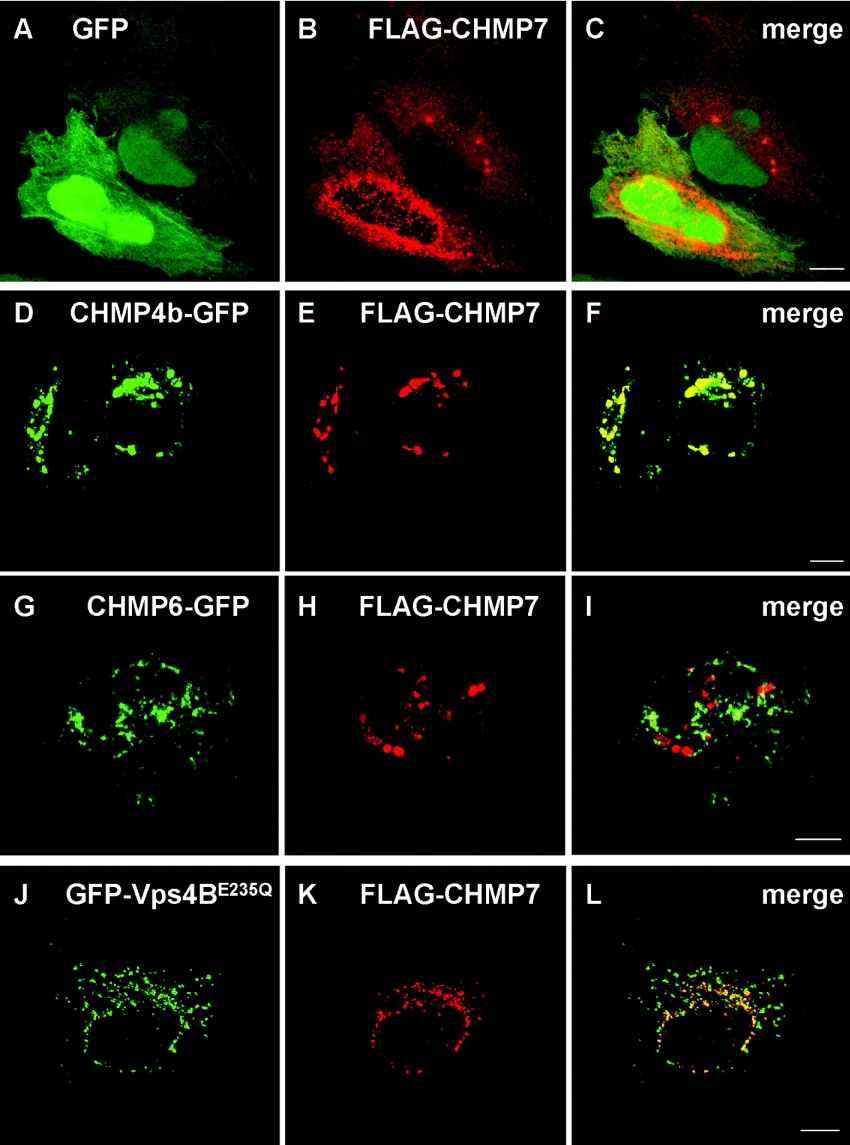
To investigate the subcellular distribution of CHMP7, HeLa cells expressing FLAG–CHMP7 were subjected to immunofluorescence microscopic analysis using anti-FLAG mAb (M2) and Cy3-labelled anti-mouse IgG anitibody. FLAG–CHMP7 co-expressed with GFP showed diffuse cytoplasmic staining and a punctate pattern, especially in the perinuclear area (Figures 3A, GFP, 3B, FLAG–CHMP7, and 3C, merged image), and the distribution pattern was similar to that of FLAG–CHMP7 expressed alone (results not shown). Co-expression with CHMP4b–GFP caused accumulation of FLAG–CHMP7 in the perinuclear area in a punctate pattern, and both overexpressed proteins showed a considerable overlap in the distribution (Figures 3D–3F), supporting their physical interaction in vivo (Figure 2). We also examined the effect of CHMP6–GFP expression on the distribution of FLAG–CHMP7. No effect was observed at a low CHMP6–GFP expression level (results not shown), but the distribution of FLAG–CHMP7 was significantly altered at a higher expression level of CHMP6–GFP (Figures 3G–3I). However, co-localization of the two CHMP proteins was limited compared with the case of CHMP4b–GFP and FLAG–CHMP7. Protein molecules involved in endosomal sorting are known to accumulate on endosomal membranes in cells expressing a dominant-negative mutant allele of the AAA Vps4/SKD1 [8,9,16,22,25–28]. Fluorescent signals of overexpressed GFP-fused dominant-negative Vps4B (GFP–Vps4BE235Q) exhibited a punctate pattern in the perinuclear area and partly merged with those of FLAG–CHMP7 (Figures 3J–3L).
Figure 3. Subcellular localization of CHMP7.
HeLa cells were co-transfected with plasmids encoding FLAG–CHMP7 and GFP (A–C), CHMP4b–GFP (D–F), CHMP6–GFP (G–I) or GFP–Vps4BE235Q (J–L). After 24 h, the cells were fixed and subjected to immunofluorescence confocal microscopic analysis by direct visualization of GFP or GFP-fusion proteins (A, D, G and J; green) and by staining with anti-FLAG mAb (M2) and Cy3-labelled goat anti-mouse IgG antibody (B, E, H and K; red). Their merged images are shown in (C, F, I and L) respectively. Scale bars, 10 μm.
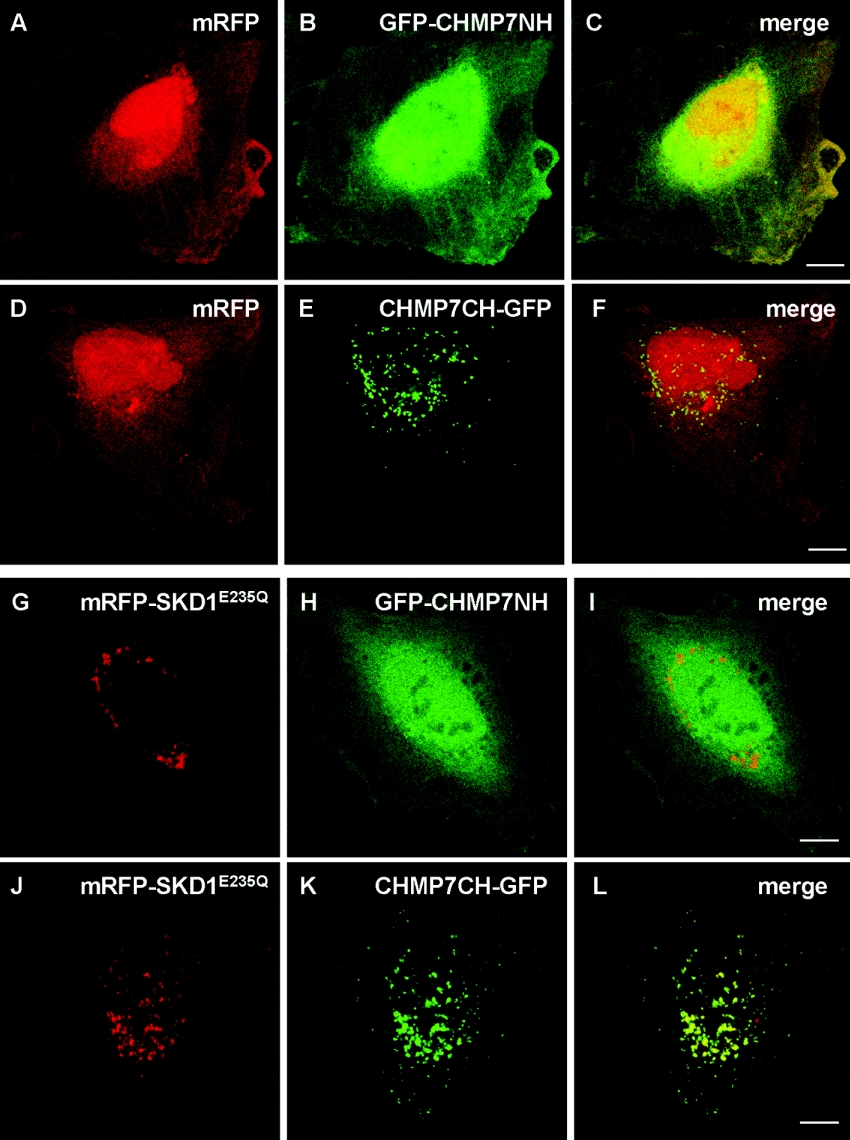
Next, we investigated which domain of CHMP7 was responsible for the punctate distribution. We constructed expression plasmids pGFP-CHNP7NH and pCHMP7CH-GFP, which expressed GFP fused with an SNF7-related domain (N-terminal half) or an SNF7 domain (C-terminal half) of CHMP7 respectively. As shown in Figure 4, while CHMP7CH–GFP exhibited a punctate pattern in the perinuclear area (panel E), GFP–CHMP7NH exhibited a diffuse pattern similar to that of mRFP (panels A–C). In contrast with mRFP (panel A), however, the fluorescent signals of GFP–CHMP7NH are more intense in the perinuclear area than in the nucleus (panel B). The distribution of GFP–CHMP7NH was not significantly influenced by co-expression with mRFP–SKD1E235Q (SKD1: murine Vps4B) (panels G–I), whereas CHMP7CH–GFP co-localized well with mRFP–SKD1E235Q.
Figure 4. Punctate distribution of CHMP7CH and no effect of Vps4/BSKD1E235Q overexpression on subcellular distribution of CHMP7NH.
HeLa cells were co-transfected with plasmids encoding either mRFP (A–F) or a dominant-negative mutant of AAA Vps4B/SKD1 fused with mRFP (mRFP–SKD1E235Q) (G–L) together with each GFP-fused CHMP7 construct: (A–C and (G–I), GFP–CHMP7NH; (D–F and J–L), CHMP7CH–GFP. After 24 h, the cells were fixed and subjected to fluorescence confocal microscopic analysis by direct visualization of mRFP (A, D, G and J) and GFP (B, E, H and K). Merged images are shown in panels C, F, I and L. Scale bars, 10 μm.
Localization of GFP–CHMP7
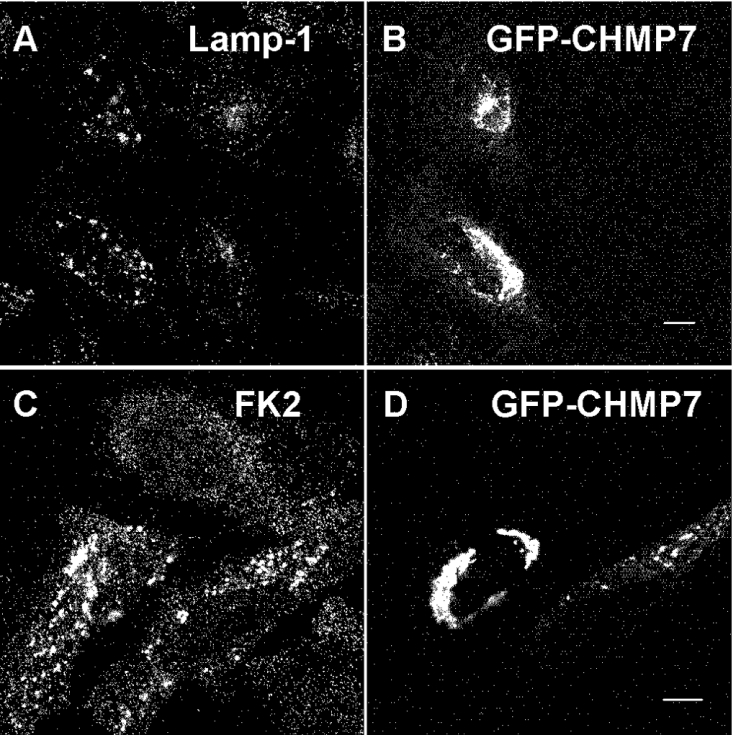
To investigate further the subcellular localization of CHMP7, we expressed GFP–CHMP7 in HeLa cells and immunostained the cells with mAbs of endosomal markers. While overexpressed GFP–CHMP7 did not co-localize with EEA1 (a marker for early endosomes) or affect its distribution (results not shown), punctate dots of Lamp-1 (a marker for late endosomes and lysosomes) became slightly enlarged in comparison with those in untransfected cells (Figures 5A and 5B).
Figure 5. Effects of GFP–CHMP7 overexpression on the distribution of Lamp-1 and ubiquitinated proteins.
HeLa cells overexpressing GFP–CHMP7 were fixed and subjected to immunofluorescence confocal microscopic analysis by staining with either anti-Lamp-1 (A) or anti-ubiquitin mAb (FK2) (C) and direct visualization of GFP–CHMP7 (B and D). Their merged images in colour are shown in Supplementary Figure S4 at http://www.BiochemJ.org/bj/400/bj4000000add.htm. Lamp-1 or ubiquitinated proteins, red; GFP–CHMP7, green. Scale bars, 10 μm.
Mono-ubiquitination of cell-surface receptors at multiple sites is known to function as a signal for efficient sorting of endocytosed proteins to internal vesicles of MVBs and sequential degradation in lysosomes [29]. To determine whether the distribution of ubiquitinated proteins was influenced by CHMP7 overexpression, we performed immunostaining of GFP–CHMP7-expressing HeLa cells using an anti-ubiquitin mAb (FK2) that recognizes mono- and poly-ubiquitinated proteins but not free mono-ubiquitin [30,31]. Compared with the untransfected cells, intensities of the fluorescent signals of ubiquitinated proteins became stronger and accumulated throughout the cytoplasm (Figures 5C and 5D). However, co-localization of ubiquitinated proteins with GFP–CHMP7 was not observed (see Supplementary Figure S4 at http://www.BiochemJ.org/bj/400/bj4000023add.htm for the merged image).
Effect of GFP–CHMP7 overexpression on the endosome–lysosome degradation pathway
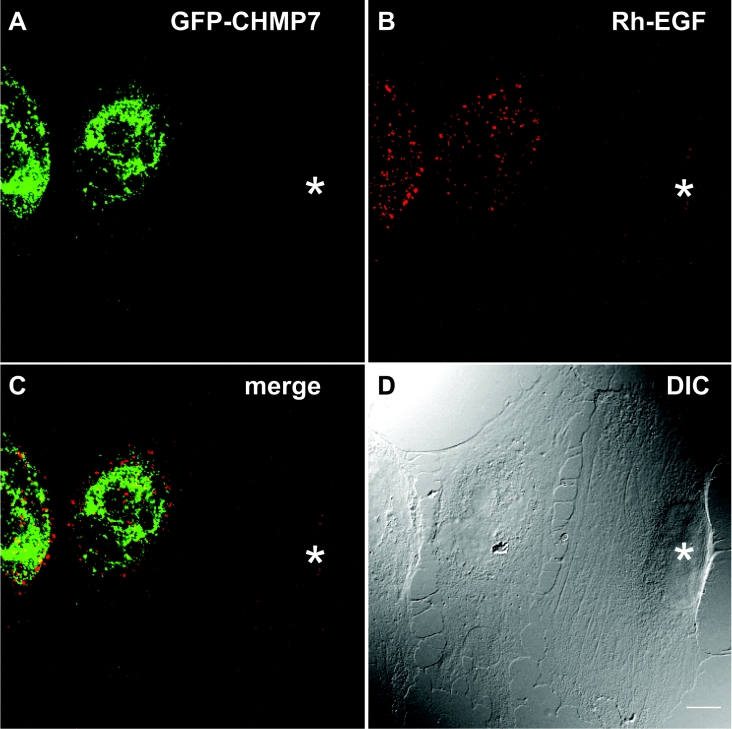
The accumulation of ubiquitinated proteins in cells overexpressing CHMP7 suggests that overexpression of CHMP7 may disturb the endosome–lysosome degradation pathway. To provide evidence supporting this speculation, we monitored the fate of endocytosed EGF. HeLa cells transfected with an expression vector of GFP–CHMP7 were incubated with Rh–EGF at 4 °C for 1 h, washed, and then allowed to take up EGF at 37 °C for 10 or 60 min. At 10 min, endocytosed Rh–EGF showed a punctate pattern similar to that of endosomes, and the intensity of the signals of Rh–EGF was reduced in some populations of GFP–CHMP7-expressing cells probably due to reduction in the numbers or binding abilities of cell-surface EGF receptors (results not shown, see below for discussion). At 60 min, most of the fluorescent signals of endocytosed Rh–EGF were no longer detectable in the untransfected cells, whereas they remained in the GFP–CHMP7-expressing cells, but did not co-localize with GFP–CHMP7, as shown in Figure 6.
Figure 6. Effects of GFP–CHMP7 overexpression on the fate of endocytosed EGF.
HeLa cells overexpressing GFP–CHMP7 were incubated in the presence of Rh–EGF at 4 °C for 1 h, washed, and then incubated at 37 °C for 1 h. The cells were fixed and subjected to immunofluorescence confocal microscopic analysis. (A) GFP–CHMP7; (B) Rh–EGF; (C) merged image of (A) and (B); (D) DIC (differential interference contrast) microscopic image. The asterisk indicates an untransfected cell. Scale bar, 10 μm.
Effect of overexpressed GFP-fusion proteins on the release of VLPs
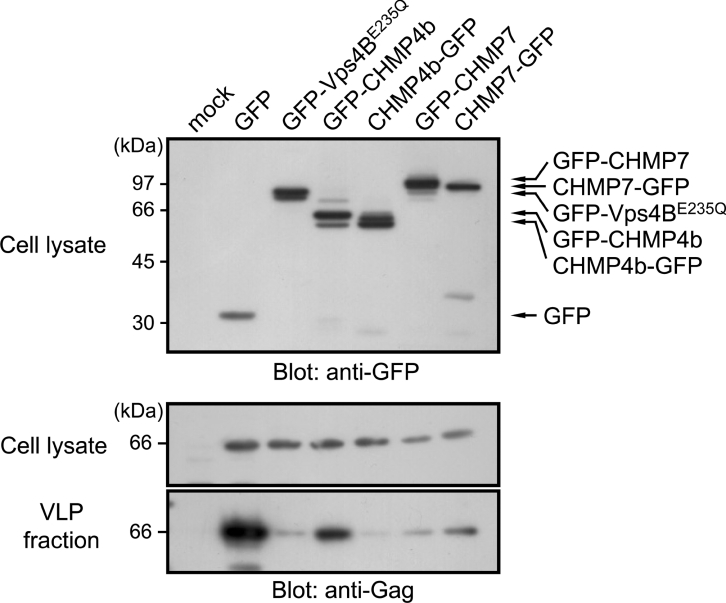
Enveloped RNA viruses are known to usurp cellular MVB-sorting machineries to release viral particles via budding from infected host cells [32,33]. Retroviral Gag proteins recruit TSG101, Alix/AIP1 or ubiquitin ligase Nedd4 by exposing specific motifs for interaction. Factors to be recruited depend on viruses, and MLV Gag has a unique ability to interact with all three factors [34]. In model experiments using cultured cells and transfection of proviral expression plasmids, overexpression of ESCRT-III proteins fused with GFP or other fluorescent proteins exhibited dominant-negative effects and suppressed the release of retroviruses to different degrees, depending on CHMP family members and tagged proteins [11–13]. Without Env proteins, retroviral Gag proteins alone can form VLPs and exit from cells [35]. By constructing an expression vector of MLV Gag full-length precursor protein (Gag-FL) that lacked both sequences encoding retrovirus protease and polymerase, we assessed the effects of overexpression of GFP-fused CHMP7 on VLP release from transfected HEK-293T cells. At 24 h after transfection, aliquots of cell lysates (0.4% of total culture) and VLP fractions (8.6% of total culture) prepared from culture medium (100000 g pellets) were subjected to Western blotting using anti-Gag antibody. As shown in Figure 7, a specific band corresponding to Gag-FL was detected in both the cell lysate and the VLP fraction (lane 1, mock transfection; lane 2, co-transfection with GFP-expressing plasmid). Co-expression of Gag-FL with GFP–Vps4BE235Q dramatically reduced the release of VLPs into the culture medium (lane 3). The amount of Gag-FL detected in the cell lysate was comparable with control cells, indicating a negligible effect of GFP–Vps4BE235Q on Gag-FL expression itself. Co-expression with GFP-fused CHMP4b suppressed VLP release and its effect was greater by C-terminally tagged protein (CHMP4b–GFP) (lane 5) than N-terminally tagged protein (GFP–CHMP4b) (lane 4). Co-expression of GFP-fused CHMP7 also suppressed release of VLPs at similar levels irrespective of N-terminal or C-terminal tagging (lanes 6 and 7). An inhibitory effect of overexpressed CHMP7 on Gag release was also observed using a Gag–Pol construct, in which Gag was processed into fragments by the viral protease encoded between Gag and Pol (results not shown).
Figure 7. Effects of overexpression of GFP-fused CHMP7 on the release of VLPs from HEK-293T cells.
HEK-293T cells were transfected with plasmids encoding the indicated GFP-fusion proteins and full-length MLV Gag (Gag-FL). At 24 h after transfection, the culture media were collected, VLPs released into the media were recovered by ultracentrifugation, and the pellets were washed and analysed as a VLP fraction. Cells remaining after removing the culture medium were washed with PBS, lysed and subjected to Western blotting using anti-Gag antibody and anti-GFP antibody. Molecular masses are given in kDa.
DISCUSSION
Mammalian orthologues of ESCRT-III components and their related proteins are collectively called CHMPs [8]. There are six CHMP subfamilies (CHMP1–CHMP6), each of which has single or multiple members, resulting in ten different proteins in humans [4]. Recently, a cDNA sequence of CHMP7 has been deposited in DNA databases as a newly assigned member of the CHMP family based on similarity of the deduced amino acid sequence to those of other members. Since no other information was available regarding this new member, we characterized CHMP7 and evaluated its membership. Like other members, as recently revealed in the three-dimensional structure of CHMP3 [36], CHMP7 contains a predicted CC region and a biased distribution of basic and acidic residues in the SNF7 domain (Figure 1A). The SNF7 domain in the C-terminal half of CHMP7, CHMP7CH, is most similar to that of CHMP6 among all CHMP family members (Figure 1C, and Supplementary Figure S1 at http://www.BiochemJ.org/bj/400/bj4000023add.htm).
CHMP7 is a unique member in that it is twice as large than other members due to a longer N-terminal region. Orthologues of CHMP7 are found from lower to higher eukaryotes, including budding yeast YJL049W (Supplementary Figure S2 at http://www.BiochemJ.org/bj/400/bj4000023add.htm). YJL049W is assigned as a non-essential gene of unknown function (http://www.yeastgenome.org/). The N-terminal half contains a sequence that is weakly similar to CHMP4b (Figure 1B). The amino acid sequence conservation rate of CHMP7/Yjl049wp between humans and budding yeast was found to be lower in the N-terminal half (identity, 17.0%; similarity, 32.4%, gaps, 27.0%, score, 69.0) than in the C-terminal half (identity, 21.7%; similarity, 41.8%, gaps, 27.0%, score, 102.5) using a program of pairwise alignment algorithms, EMBOSS-Align (http://www.ebi.ac.uk/emboss/align/index.html). Notably, the conservation rates are both lower than the rate between human CHMP6 and yeast Vps20 (identity, 26.2%; similarity, 50.7%, gaps, 15.7%, score, 247.0). This suggests that the CHMP7 gene was created by duplication of the ancestral SNF7 gene during evolution and evolved rapidly to acquire specific functions.
Individual CHMP proteins have been shown to interact with other ESCRT and non-ESCRT proteins by yeast two-hybrid assays, pull-down assays or co-immunoprecipitation assays [11–13,37,38]. We demonstrated previously that CHMP6 binds directly to CHMP4b and to the ESCRT-II component EAP20 [14]. CHMP6 and its yeast orthologue Vps20 are myristoylated proteins and are thus proposed to play a role as ESCRT-II acceptors of ESCRT-III on endosomal membranes [4,14,39]. While the C-terminal half of CHMP7 (designated CHMP7CH) associates with CHMP4b (Figure 2), no interaction was detected between CHMP7 and EAP20 (Supplementary Figure S3 at http://www.BiochemJ.org/bj/400/bj4000023add.htm). Using a yeast two-hybrid assay using the N-terminal half and C-terminal half of CHMP7 as bait, only CHMP4 isoforms displayed positive interaction with the C-terminal half of CHMP7 among the ESCRT-related proteins (results not shown). Recently, degradation of endocytosed EGF and virally ubiquitinated MHC class I was reported to be independent of mammalian ESCRT-II [40]. Linking of ESCRT-I and ESCRT-III is provided by interaction between each component (TSG101 and CHMP4s) and Alix/AIP1 [9–13], where the TSG101–Alix interaction is facilitated by their calcium-dependent binding partner ALG-2 ([22], and F. Ichioka, H. Shibata and M. Maki, unpublished work). Interestingly, although yeast Bro1 (Alix orthologue) does not associate with Vps23 (TSG101 orthologue), yeast Vps20 (CHMP6 orthologue) associates with the ESCRT-I component Vps28 [37,41]. Regardless of the sequence similarity of CHMP7CH to CHMP6, CHMP7CH did not show such interaction in our yeast two-hybrid assay (results not shown). Although regulatory roles of CHMP7 in the MVB sorting system remain to be established, CHMP7 might exert its function via interaction with unknown factors that associate directly or indirectly with the upstream ESCRTs.
By searching for a motif in the Eukaryotic Linear Motif resource (http://elm.eu.org/), the N-terminal half was predicted to have two putative CaM (calmodulin)-binding IQ motifs (amino acids 89–108 and 198–217). The IQ motif is one of several CaM-binding consensus sequences and is widely distributed in both myosins and non-myosins [42]. To determine whether CHMP7 associates with CaM, we prepared recombinant CaM protein fused with GST (glutathione S-transferase) (GST–CaM) and performed a GST-pull-down assay using FLAG–CHMP7 expressed in mammalian cells, but we did not detect either Ca2+-dependent or -independent binding (results not shown). We also carried out a co-immunoprecipitation assay using FLAG–CaM and GFP–CHMP7, but failed again to detect interaction between CaM and CHMP7. Binding of CaM to CHMP7, if any, seems to be too weak to be detected under our experimental conditions, because the predicted IQ motifs are not perfectly matched with the consensus sequence (http://calcium.uhnres.utoronto.ca/ctdb/flash.htm). The N-terminal half contains a sequence, 150-LVAVE, that was predicted be a potential binding site for clathrin. Association of CHMP7 with clathrin remains to be clarified.
We analysed the subcellular localization of CHMP7 using confocal laser-scanning microscopy. Exogenously expressed CHMP7 exhibited a punctate pattern in the perinuclear area, but did not show co-localization with various organelle markers such as EEA1 (a marker for early endosomes) (results not shown) and Lamp-1 (a marker for late endosomes and lysosomes) (Figure 5). We also tested MVB marker antibodies of CD63 and lysobisphosphatidic acid, but no co-localization was observed (results not shown). This result is in contrast with the distribution of CHMP4b and CHMP6, which showed similar punctate patterns but exhibited partial co-localization with endosomal markers [9,14]. The distributions of the N- and C-terminal halves of CHMP7 fused with GFP were diffuse throughout the cytoplasm and punctate in the perinuclear area respectively (Figures 4B and 4E). Moreover, the distribution of GFP–CHMP7NH was not affected by co-overexpression with a dominant-negative construct of Vps4B/SKD1 (Figures 4G–4I). These results indicate that the C-terminal half dominates the distribution of the full-length CHMP7 and that the N-terminal half, regardless of having a weak similarity to the SNF7 domain, lacks the biochemical properties common to CHMPs. The degree of accumulation of CHMP7 in the perinuclear area seems to depend on expression levels of CHMP7 tagged with either GFP or FLAG (results not shown). Since overexpressed protein may be distributed differently from endogenous protein, we attempted to analyse the subcellular distribution of CHMP7 using a polyclonal antibody against this recombinant protein. Unfortunately, regardless of affinity-purification, the specificity of the antibody obtained was not good enough to be used for immunofluorescence microscopic analysis. Further studies are required to determine the subcellular distribution of endogenous CHMP7.
ESCRTs play important roles in the sorting of ubiquitinated cargos into intraluminal vesicles in endosomes [1–4]. The sorting system appears to be impaired by overexpression of GFP-fused CHMP7 in HeLa cells, as demonstrated by accumulation of ubiquitinated proteins in the perinuclear area (Figures 5C and 5D) and inhibition of the disappearance of endocytosed Rh–EGF (Figure 6). Previously, we observed similar inhibition of disappearance of endocytosed Rh–EGF by overexpession of GFP-fused CHMP4b and CHMP6 [9,14]. In contrast with our previous results, at 5–6 h after uptake of Rh–EGF, however, fluorescent signals decreased near to a background level and differences between GFP–CHMP7-expressing and non-expressing cells were not clear (results not shown). Compared with CHMP4b and CHMP6, the dominant-negative potency of overexpressed GFP-fused CHMP7 seems to be weaker regarding MVB sorting of EGF receptor. This may be related to a lesser degree of co-localization between ubiquitinated proteins and GFP–CHMP7 than between ubiquitinated proteins and CHMP4b–GFP or CHMP6–GFP (Figures 5C and 5D) [9,14].
Since overexpressed CHMP7 co-localized with GFP-fused CHMP4b and Vps4BE235Q (Figure 3), the impairment of the normal sorting system may be due to sequestration of endogenous CHMP4 and Vps4 proteins. Alternatively, they may paralyse the function indirectly by causing accumulation of other ESCRT proteins on endosomes and gumming up the endosomal system. Dominant-negative effects of GFP-fused CHMP proteins were also observed for inhibition of release of VLPs composed of MLV Gag protein into culture medium (Figure 7). In the case of CHMP4b, the C-terminally GFP-tagged construct had a greater inhibitory effect than did the N-terminally GFP-tagged construct. In contrast, the inhibition rate was not significantly different between GFP–CHMP7 and CHMP7–GFP. The amounts of Gag-FL recovered from the lysates of CHMP7-overexpressing cells were slightly less than those of either control GFP or other constructs. Overexpression of CHMP7 may reduce translation efficiency, which may explain reduction in the numbers of cell-surface EGF receptors (results not shown).
In the present study, we demonstrated that CHMP7 possesses properties similar to those of other CHMP proteins, based on structural features and functions of overexpressed protein as described above. We conclude that CHMP7 is a genuine member of the ESCRT-III protein family. The unique N-terminal region may interact with other factors and exert a specific biological function(s). Studies to isolate proteins that interact with CHMP7 are in progress.
Online data
Acknowledgments
We are grateful to Y. Terasawa and E. Takaya for their work related to this study and to Dr K. Hitomi for his encouraging suggestions. We thank Dr H. Amanuma for providing an anti-Gag antibody. We also thank Dr T. Kitamura, Dr S. Ghosh, Dr T. Yoshimori, Dr Y. Ono, Dr M. Takahashi, and Dr R. Y. Tsien for providing us with the variety of cDNAs used in this study. This work was supported by a Grant-in-Aid for Scientific Research B (to M. M.), a Grant-in-Aid for Young Scientists B (to H. S.), and Fellowships for Young Scientists (K. K. and C. Y.) from JSPS (Japan Society for the Promotion of Science), and by Industrial Technology Research Grant Program (04A01042) from NEDO (New Energy and Industrial Technology Development Organization) (to H. S.).
References
- 1.Katzmann D. J., Odorizzi G., Emr S. D. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 2.Gruenberg J., Stenmark H. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- 3.Babst M. A protein's final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- 4.Hurley J. H., Emr S. D. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babst M., Katzmann D. J., Estepa-Sabal E. J., Meerloo T., Emr S. D. Escrt-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 6.Amerik A. Y., Nowak J., Swaminathan S., Hochstrasser M. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell. 2000;11:3365–3380. doi: 10.1091/mbc.11.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kranz A., Kinner A., Kolling R. A family of small coiled-coil-forming proteins functioning at the late endosome in yeast. Mol. Biol. Cell. 2001;12:711–723. doi: 10.1091/mbc.12.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howard T. L., Stauffer D. R., Degnin C. R., Hollenberg S. M. CHMP1 functions as a member of a newly defined family of vesicle trafficking proteins. J. Cell Sci. 2001;114:2395–2404. doi: 10.1242/jcs.114.13.2395. [DOI] [PubMed] [Google Scholar]
- 9.Katoh K., Shibata H., Suzuki H., Nara A., Ishidoh K., Kominami E., Yoshimori T., Maki M. The ALG-2-interacting protein Alix associates with CHMP4b, a human homologue of yeast Snf7 that is involved in multivesicular body sorting. J. Biol. Chem. 2003;278:39104–39113. doi: 10.1074/jbc.M301604200. [DOI] [PubMed] [Google Scholar]
- 10.Katoh K., Shibata H., Hatta K., Maki M. CHMP4b is a major binding partner of the ALG-2-interacting protein Alix among the three CHMP4 isoforms. Arch. Biochem. Biophys. 2004;421:159–165. doi: 10.1016/j.abb.2003.09.038. [DOI] [PubMed] [Google Scholar]
- 11.von Schwedler U. K., Stuchell M., Muller B., Ward D. M., Chung H. Y., Morita E., Wang H. E., Davis T., He G. P., Cimbora D. M., et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 12.Strack B., Calistri A., Craig S., Popova E., Gottlinger H. G. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 13.Martin-Serrano J., Yarovoy A., Perez-Caballero D., Bieniasz P. D. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yorikawa C., Shibata H., Waguri S., Hatta K., Horii M., Katoh K., Kobayashi T., Uchiyama Y., Maki M. Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting. Biochem J. 2005;387:17–26. doi: 10.1042/BJ20041227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shim J. H., Xiao C., Hayden M. S., Lee K. Y., Trombetta E. S., Pypaert M., Nara A., Yoshimori T., Wilm B., Erdjument-Bromage H., et al. CHMP5 is essential for late endosome function and down-regulation of receptor signaling during mouse embryogenesis. J. Cell Biol. 2006;172:1045–1056. doi: 10.1083/jcb.200509041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peck J. W., Bowden E. T., Burbelo P. D. Structure and function of human Vps20 and Snf7 proteins. Biochem. J. 2004;377:693–700. doi: 10.1042/BJ20031347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reid E., Connell J., Edwards T. L., Duley S., Brown S. E., Sanderson C. M. The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum. Mol. Genet. 2005;14:19–38. doi: 10.1093/hmg/ddi003. [DOI] [PubMed] [Google Scholar]
- 18.Bache K. G., Stuffers S., Malerod L., Slagsvold T., Raiborg C., Lechardeur D., Walchli S., Lukacs G. L., Brech A., Stenmark H. The ESCRT-III subunit hVps24 is required for degradation but not silencing of the epidermal growth factor receptor. Mol. Biol. Cell. 2006;17:2513–2523. doi: 10.1091/mbc.E05-10-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urata S., Noda T., Kawaoka Y., Yokosawa H., Yasuda J. Cellular factors required for Lassa virus budding. J. Virol. 2006;80:4191–4195. doi: 10.1128/JVI.80.8.4191-4195.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell R. E., Tour O., Palmer A. E., Steinbach P. A., Baird G. S., Zacharias D. A., Tsien R. Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshimori T., Yamagata F., Yamamoto A., Mizushima N., Kabeya Y., Nara A., Miwako I., Ohashi M., Ohsumi M., Ohsumi Y. The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol. Biol. Cell. 2000;11:747–763. doi: 10.1091/mbc.11.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh K., Suzuki H., Terasawa Y., Mizuno T., Yasuda J., Shibata H., Maki M. The penta-EF-hand protein ALG-2 interacts directly with the ESCRT-I component TSG101, and Ca2+-dependently co-localizes to aberrant endosomes with dominant-negative AAA ATPase SKD1/Vps4B. Biochem. J. 2005;391:677–685. doi: 10.1042/BJ20050398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satoh H., Shibata H., Nakano Y., Kitaura Y., Maki M. ALG-2 interacts with the amino-terminal domain of annexin XI in a Ca2+-dependent manner. Biochem. Biophys. Res. Commun. 2002;291:1166–1172. doi: 10.1006/bbrc.2002.6600. [DOI] [PubMed] [Google Scholar]
- 24.Morita S., Kojima T., Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 25.Babst M., Wendland B., Estepa E. J., Emr S. D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bishop N., Woodman P. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol. Biol. Cell. 2000;11:227–239. doi: 10.1091/mbc.11.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujita H., Yamanaka M., Imamura K., Tanaka Y., Nara A., Yoshimori T., Yokota S., Himeno M. A dominant negative form of the AAA ATPase SKD1/VPS4 impairs membrane trafficking out of endosomal/lysosomal compartments: class E vps phenotype in mammalian cells. J. Cell Sci. 2003;116:401–414. doi: 10.1242/jcs.00213. [DOI] [PubMed] [Google Scholar]
- 28.Lin Y., Kimpler L. A., Naismith T. V., Lauer J. M., Hanson P. I. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J. Biol. Chem. 2005;280:12799–12809. doi: 10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- 29.Haglund K., Di Fiore P. P., Dikic I. Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem. Sci. 2003;28:598–603. doi: 10.1016/j.tibs.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Usuba T., Ishibashi Y., Okawa Y., Hirakawa T., Takada K., Ohkawa K. Purification and identification of monoubiquitin-phosphoglycerate mutase B complex from human colorectal cancer tissues. Int. J. Cancer. 2001;94:662–668. doi: 10.1002/ijc.1524. [DOI] [PubMed] [Google Scholar]
- 31.Haglund K., Sigismund S., Polo S., Szymkiewicz I., Di Fiore P. P., Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 2003;5:461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- 32.Morita E., Sundquist W. I. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 33.Demirov D. G., Freed E. O. Retrovirus budding. Virus Res. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 34.Segura-Morales C., Pescia C., Chatellard-Causse C., Sadoul R., Bertrand E., Basyuk E. Tsg101 and Alix interact with murine leukemia virus Gag and cooperate with Nedd4 ubiquitin ligases during budding. J. Biol. Chem. 2005;280:27004–27012. doi: 10.1074/jbc.M413735200. [DOI] [PubMed] [Google Scholar]
- 35.Resh M. D. Intracellular trafficking of HIV-1 Gag: how Gag interacts with cell membranes and makes viral particles. AIDS Rev. 2005;7:84–91. [PubMed] [Google Scholar]
- 36.Muziol T., Pineda-Molina E., Ravelli R. B., Zamborlini A., Usami Y., Göttlinger H., Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 37.Bowers K., Lottridge J., Helliwell S. B., Goldthwaite L. M., Luzio J. P., Stevens T. H. Protein–protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic. 2004;5:194–210. doi: 10.1111/j.1600-0854.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- 38.Tsang H. T., Connell J. W., Brown S. E., Thompson A., Reid E., Sanderson C. M. A systematic analysis of human CHMP protein interactions: additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics. 2006;88:333–346. doi: 10.1016/j.ygeno.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 39.Teo H., Perisic O., Gonzalez B., Williams R. L. ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev. Cell. 2004;7:559–569. doi: 10.1016/j.devcel.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 40.Bowers K., Piper S. C., Edeling M. A., Gray S. R., Owen D. J., Lehner P. J., Luzio J. P. Degradation of endocytosed epidermal growth factor and virally ubiquitinated major histocompatibility complex class I is independent of mammalian ESCRTII. J. Biol. Chem. 2006;281:5094–5105. doi: 10.1074/jbc.M508632200. [DOI] [PubMed] [Google Scholar]
- 41.Pineda-Molina E., Belrhali H., Piefer A. J., Akula I., Bates P., Weissenhorn W. The crystal structure of the C-terminal domain of Vps28 reveals a conserved surface required for Vps20 recruitment. Traffic. 2006;7:1007–1016. doi: 10.1111/j.1600-0854.2006.00440.x. [DOI] [PubMed] [Google Scholar]
- 42.Bähler M., Rhoads A. Calmodulin signaling via the IQ motif. FEBS Lett. 2002;20:107–113. doi: 10.1016/s0014-5793(01)03239-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.