Abstract
Understanding the phylogeographic processes affecting endangered species is crucial both to interpreting their evolutionary history and to the establishment of conservation strategies. Lions provide a key opportunity to explore such processes; however, a lack of genetic diversity and shortage of suitable samples has until now hindered such investigation. We used mitochondrial control region DNA (mtDNA) sequences to investigate the phylogeographic history of modern lions, using samples from across their entire range. We find the sub-Saharan African lions are basal among modern lions, supporting a single African origin model of modern lion evolution, equivalent to the ‘recent African origin’ model of modern human evolution. We also find the greatest variety of mtDNA haplotypes in the centre of Africa, which may be due to the distribution of physical barriers and continental-scale habitat changes caused by Pleistocene glacial oscillations. Our results suggest that the modern lion may currently consist of three geographic populations on the basis of their recent evolutionary history: North African–Asian, southern African and middle African. Future conservation strategies should take these evolutionary subdivisions into consideration.
Keywords: bottleneck, human evolution, mitochondrial DNA, Panthera leo spelaea, palaeoenvironment, recent African origin model
1. Introduction
Fossil evidence suggests that the earliest lion-like cat appeared in East Africa during the Late Pliocene (5.0–1.8 Myr ago; Turner & Antón 1997). In a pattern broadly resembling that of humans, lions migrated out of Africa during the Middle Pleistocene (800–100 kyr ago) into Europe and Asia, eventually colonizing the entire Holarctic region (Stringer 2002; Yamaguchi et al. 2004). Lions were the most widespread large terrestrial mammals during the Late Pleistocene (100–10 kyr ago), with a range that included Africa, most of Eurasia and North America (Kurtén 1968; Turner & Antón 1997). Recent genetic studies have suggested that at least two distinct lineages of lion inhabited western Eurasia at the end of the Pleistocene: the Holarctic cave lion (Panthera leo spelaea), and the modern lion (P. l. ssp.; Burger et al. 2004).
Several models have been proposed to explain the diversification of lions. A multiregional origin model, based on the morphological analysis of lion remains from across its previous range, cites similarities in skull morphology between the middle Pleistocene (ca 500 kyr ago) European lion (P. l. fossilis) and modern North African–Asian lions, as evidence for long-term evolution in situ (Hemmer 1974). This interpretation has been challenged, however, by the discovery of a more recent population bottleneck of the modern lion (ca 74–203 kyr ago) that may have complicated the interpretation of morphological characteristics (O'Brien et al. 1987; Burger et al. 2004). It has recently been proposed that, following this bottleneck, a single population of lions replaced the older populations in Africa and southwestern Eurasia (Yamaguchi et al. 2004). This single origin replacement model of modern lion evolution provides an interesting parallel to the ‘recent African origin’ model of human evolution (in comparison to the ‘multiregional evolution’ model), in which modern Homo sapiens evolved in Africa ca 200 kyr ago and went on to replace hominids (e.g. the Neanderthals) elsewhere (Stringer 2002). However, as yet, no empirical study has investigated this issue in lions, in part due to the difficulty of accessing genetic material from all of the species' natural historic range.
Understanding the phylogeographic history of an endangered species is essential both for better understanding the evolutionary processes affecting the species and for developing conservation strategies (Crandall et al. 2000; Mace et al. 2003). Recent surveys of wild lion populations have estimated that as few as 18–47 000 wild lions currently live in Africa, and suggested that in addition to the already critically endangered Asiatic lion, the African lion may soon become endangered (Bauer & Van Der Merwe 2002; Chardonnet 2002). As lions have become increasingly confined to protected areas, current strategies for metapopulation management have included moving individuals over large distances between protected areas. Although little is known about the structure or history of the populations involved, these strategies have already been put into place on a small scale in southern Africa (Nowell & Jackson 1996; S. van der Merwe 2002, personal communication). The lack of objective consensus on intraspecific lion phylogeny is also complicating conservation efforts in other parts of Africa. For example, the population of lions living in west and central Africa could possibly be characterized as ‘critically endangered’; however, because their status as a separate lion subspecies (P. l. senegalensis) is unclear, the situation has not been officially recognized (Nowell & Jackson 1996; Bauer & Van Der Merwe 2002; Chardonnet 2002). A better understanding of the phylogeographic history of lions will, therefore, have immediate benefits not only for lion conservation ongoing in the field, but also for conservation legislation.
The modern lion is customarily divided into eight subspecies, based on geographic range and differences in morphology (Hemmer 1974). Despite being one of the first felids whose phylogeny was investigated using molecular methods (O'Brien et al. 1987), there has as yet been no comprehensive genetic analysis of modern lion phylogeny. This may be in part due to difficulty in obtaining specimens from outside only a small fraction of their range. Indeed, all previously published lion sequences originate from seven countries that make up less than half of their natural range: Botswana, India, Kenya, Namibia, South Africa, Tanzania and Uganda (Janczewski et al. 1995; Burger et al. 2004; Dubach et al. 2005). Although lion specimens from outside these countries are accessible at museums, the possibility of ancient DNA (aDNA) analysis is restricted by the limited genetic variation observed in large felids (Culver et al. 2000; Eizirik et al. 2001; Uphyrinka et al. 2001), which makes it difficult to extract sufficient phylogenetic information from degraded ancient sequences.
In this paper, we attempt to overcome the lack of genetic variation in modern lions and consequential difficulties in working with museum specimens by focusing our analysis on the short highly variable region 1 (HVR1) of the mitochondrial DNA (mtDNA) control region, which is known to be the most variable segment of mtDNA in the genus Panthera (Jae-Heup et al. 2001). This allows us to extend the range of lion populations included in the analysis beyond the limits of previous analyses. We report an intraspecific phylogeny of the lion based on mtDNA control region sequences, covering not only its entire current range but also several extinct populations. We discuss the recent evolutionary history of the modern lion and implications for the conservation of this increasingly endangered species.
2. Material and methods
(a) Sampling and laboratory procedures
Small fragments of bone and dried tissues were sampled from museum specimens of lions of known origin (table 1). DNA extraction, polymerase chain reaction (PCR), sequencing and cloning were performed according to standard aDNA criteria (Shapiro et al. 2004) by following the procedures described elsewhere (Barnett et al. 2006).
Table 1.
Lion samples analysed in this study. (ID numbers correspond with those in figure 1. Sample sources are National Museums of Scotland, Edinburgh (I), National Museum of Natural History, Paris (II), National Natural History Museum, Leiden (III), Swedish Museum of Natural History, Stockholm (IV), Amathole Museum, King William's Town (V), Natural History Museum of Zimbabwe, Bulawayo (VI), Transvaal Museum, Pretoria (VII), American Museum of Natural History, New York (VIII), Palaeological Institute, Moscow (IX), University of Vienna, Vienna (X), University of Stuttgart, Stuttgart (XI) and Canadian Museum of Nature, Ottawa (XII). Abbreviations are as follows: CAR, Central African Republic; DRC, Democratic Republic of Congo; RSA, South Africa. Radiocarbon dating was performed at Oxford Radiocarbon Accelerator Unit (Oxford).)
| ID | sample type | origin | source | haplotype: museum ID (date) [comments] |
|---|---|---|---|---|
| modern lion | ||||
| 1 | frozen tissue | India | I | M9: [location: Gir Forest] |
| 2 | pelvis | India | II | M9: 1838-890 (31 Dec 1838) |
| 3 | skull | Iran | II | M10: 1962-2847 |
| 4 | skull | Iran | II | M10: 1962-2854 |
| 5 | dried skin | Tunisia | III | M11: specimen-c (1823) |
| 6 | vertebra | Algeria | II | M11: 1862-54 |
| 7 | vertebra | North Africa | II | M11: A-7912 (died 1839) |
| 8 | mandible | Barbary | IV | M11: A58:5287 (1831) |
| 9 | skull | Senegal | II | M4: A-1892 (1841) |
| 10 | skull | Senegal | II | M4: 1890-490 |
| 11 | skull | Burkina | II | M3: 1926-248 (died 23 Dec 1927) |
| 12 | skull | CAR | II | M6: 1996-2516 |
| 13 | skull | CAR | II | M6: 1996-2517 |
| 14 | vertebra | Sudan | II | M8: 1995-164 [location: Nubia] |
| 15 | skull | Ethiopia | II | M6: A-12942 |
| 16 | skull | Kenya | II | M2: 1962-2853 (1921) |
| 17 | skull | DRC | IV | M2: A59:5062 (1921) [location: s. of L. Edward] |
| 18 | mandible | DRC | IV | M2: A59:5066 (1921) [location: s. of L. Edward] |
| 19 | n.a. | Tanzania | GenBank | M2: Ple180-CL1 [location: Serengeti] |
| 20 | skull | Tanzania | V | M1: 107.1 [location: s. Tanganiyka] |
| 21 | skull | Zambia | VI | M2: 5728 (1957) [location: Kafue National Park] |
| 22 | dried tissue | Zambia | V | M2: 15908 (1927) |
| 23 | skull | Zimbabwe | VI | M5: 29119 (1967) [location: Tsholotsho, sw. Zimbabwe] |
| 24 | skull | RSA (Cape) | V | M5: 15904 [location: King William's Town] |
| 25 | dried tissue | RSA | VI | M2: 1072 (1939) [location: s. Kalahari] |
| 26 | dried tissue | RSA | V | M2: 38248 [location: Kalahari Gemsbok National Park] |
| 27 | skull | Botswana | VI | M5: 63589 (1964) |
| 28 | dried tissue | Namibia | IV | M2: A58:1971 (1856) [location: Walvis Bay] |
| 29 | skull | Botswana | VI | M5: 63591 (1965) [location: Moremi, n. Botswana] |
| 30 | dried tissue | Namibia | V | M5: 18319 [location: Etosha Pan, n. Namibia] |
| 31 | skull | DRC | II | M7: 1961-2849 |
| 32 | phalange | Gabon | II | M5: 1960-3680 (1959) |
| Late Pleistocene Holarctic lion | ||||
| HL1 | tibia | Alaska | VIII | H13: FAM69167 (12 090±80 14C years old, OxA-13451) |
| HL2 | calcaneus | Siberia | IX | H13: 772-95 (46 200±1500 14C years old, OxA-13024) |
| HL3 | radius | Siberia | IX | H13: xDYa-84 (28 720±160 14C years old, OxA-12981) |
| HL4 | tibia | Austria | X | H14: GS-27 (49 900±1500 14C years old, OxA-13110) |
| HL5 | calcaneus | Germany | XI | H12: 9995.2/SIB01 (more than 48 100 14C years old, OxA-15354) |
| HL6 | humerus | Canada | XII | H13: 47294 (12 640±75 14C years old, OxA-10083) |
Strict criteria are required to confirm the authenticity of DNA sequences extracted from ancient specimens (Willerslev & Cooper 2005). As such, DNA extractions and PCR amplifications were performed with multiple negative controls. In addition, DNA sequences from extracts of 17/32 modern lion samples and 6/6 Late Pleistocene lions were amplified and sequenced at least twice, and at least one PCR product from each haplotype was cloned to evaluate DNA damage and investigate the presence of contaminating sequences. DNA extractions from nine museum specimens (ID numbers 2, 3, 4, 12, 13, 17, 27, 31 and 32; table 1) were performed twice in the laboratory in Oxford, and for seven specimens, DNA extraction, amplification and sequencing was independently replicated in a dedicated aDNA facility at University College London (ID numbers 8, 9, 20, 24, 32, HL1 and HL2; table 1). All intra- and inter-laboratory replications produced identical sequences.
Members of the Felidae are known to contain macrosatellites resulting from nuclear translocation of the mtDNA (numts; Lopez et al. 1997). Two cloned extracts produced a minority clone sequence similar to published numts, while all other extracts generated sequences that grouped closely with those obtained using purified mitochondria from modern tissue (Lopez et al. 1997; Cracraft et al. 1998; Jae-Heup et al. 2001). The high degree of divergence between the mitochondrial sequence and numt allowed us to identify and discard the minority clone numt sequences from further analysis.
(b) Data analysis
The sequences obtained were aligned by eye using the program Se-Al (Rambaut 1996). A cytosine repeat in three African lions (sample ID numbers 12, 13 and 15; table 1) could not be unambiguously scored even after multiple amplifications and cloning, due to slippage during the PCR, and this region was excluded from the analysis. A median-joining network was constructed using Network v. 4.1.0.3 (Bandelt et al. 1999) to depict phylogeographic patterns and potential ancestor–descendant relationships. Maximum parsimony bootstrap analysis (100 replicates, gaps as fifth state, TBR model) was implemented in PAUP* v. 4.0b10 (Swofford 2001) to show degree of statistical support for intraspecific groupings.
3. Results
Approximately 130 bp of HVR1 was amplified from 32 modern lion specimens representing most of its extant and historic range (table 1; figure 1). This corresponded to an addition of 12 countries of origin (Algeria, Burkina Faso, Central African Republic (CAR), Democratic Republic of Congo (DRC), Ethiopia, Gabon, Iran, Senegal, Sudan, Tunisia, Zambia and Zimbabwe) to those previously analysed (Burger et al. 2004; Dubach et al. 2005), vastly increasing the proportion of the lions' natural range now represented by mtDNA sequences.
Figure 1.
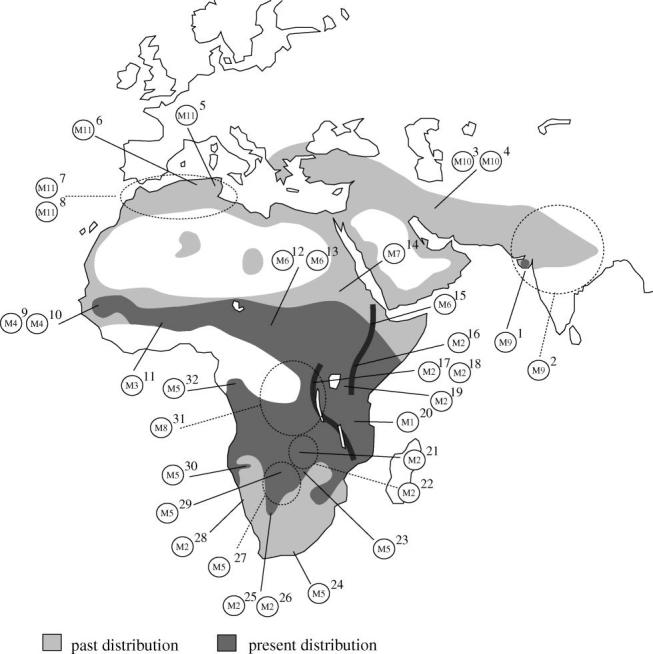
Map showing approximate sampling sites with mtDNA haplotypes: numbers in circles correspond to different haplotypes. The numbers superscripted beside the circles correspond with the ID numbers in table 1. If more than one sample originates from the same country (or region) and the exact sampling location is not known for every animal, a dashed circle and dashed line were used. The Great Rift Valley is shown as thick dark lines.
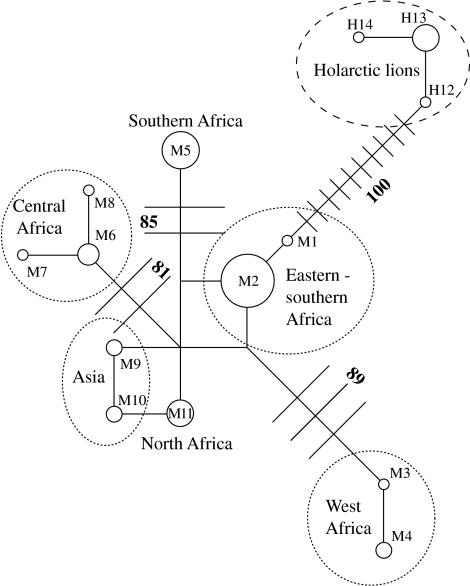
Eleven different haplotypes (M1–M11) were identified from the 32 modern lion specimens, defined by 13 variable sites in modern lions alone and 23 variable sites including extinct Holarctic lions (table 1). Two haplotypes (M2 and M5, figures 1 and 2) were widespread in eastern and southern Africa, but M5 was geographically restricted to the southwestern side of the Great Rift Valley (figure 1). The other nine haplotypes had more localized distributions (figure 1). Eight out of the 11 haplotypes were found in the lower latitudes of Africa between approximately 15° North and South and M9–M11 were found only in North Africa–Asia.
Figure 2.
Median-joining network for lion mtDNA haplotypes. Different numbers represent different haplotypes, corresponding to those in figure 1 and table 1. The area of the circle is proportional to the haplotype frequency, and the length of connecting lines to the distance between haplotypes, defined as the number of substitutions estimated by Network v. 4.1.0.3 (Bandelt et al. 1999). A place where lines meet without a circle indicates the possible ancestral state. Maximum parsimony bootstrap support for groupings is in bold.
A median-joining network of lion mtDNA sequences placed the sub-Saharan sequences basal to those of North Africa and Asia (figure 2) with haplotypes M1 and M2 (eastern–southern Africa) basal to all other sub-Saharan lions and most closely related to the extinct Pleistocene lion. The network also showed a relatively simple mtDNA structure within the North African–Asian population in comparison to that within the sub-Saharan population. Among the latter, closely related haplotypes were found in western (M3 and M4), northeastern (M6, M7 and M8), eastern–southern (M1 and M2) and southwestern (M5) Africa.
4. Discussion
(a) Validation of results
Although our results are based on only a small ca 130 bp fragment of HVR1, the number of variable sites identified (13 among modern lions, 23 if the extinct Holarctic lion is included) are of the same order of magnitude as previous studies based on much larger sequence fragments. Dubach et al. (2005) found 26 variable sites between modern eastern–southern African lions in 2360 bp of cytochrome b and NADH5, and Burger et al. (2004) found ca 50 variable sites between modern and extinct Holarctic lions in 1051 bp of cytochrome b. Additionally, our results are consistent with previously determined phylogeographic patterns in Eastern–Southern African lions (Burger et al. 2004; Dubach et al. 2005), in which two major clades were identified: one confined to the southwest of the Great Rift Valley (corresponding to our haplotype-M5), and the other in eastern and southern Africa (corresponding to our haplotype-M2). This suggests that the short HVR1 is useful for deducing the general phylogeographic pattern across the lions range, although finer resolution will require more variable sites.
(b) Origin of the modern lion
Our results show that the sub-Saharan African lions are phylogenetically basal to all modern lions, when compared to extinct Pleistocene lions (figure 2). Additionally, sub-Saharan African lions have a much broader range of mtDNA variation than other modern lions, despite the limited number of variable sites identified by this analysis. This suggests a relatively recent dispersal from sub-Saharan Africa into North Africa–Asia. Given the position of haplotypes M1 and M2 in the network, the centre of origin for modern lions is probably eastern–southern Africa. This may reflect either the evolutionary focal point for modern lions (cf. Turner & Antón 1997), or a more recent post-bottleneck refugium (cf. Burger et al. 2004).
Severe anthropogenic reduction and fragmentation of lion populations in North Africa–Asia in the last few hundred years (Yamaguchi & Haddane 2002) may have artificially reduced the mtDNA diversity of the regions' lion populations leaving only haplotypes closely related to each other. However, this does not explain why only closely related haplotypes were left among the lion populations scattered in the vast geographic area between North Africa and India (figure 1), if the area had possessed many distinct haplotypes as does the middle part of Africa. The data, therefore, support the single origin replacement model of modern lion evolution (Yamaguchi et al. 2004), as opposed to the multiregional model (Hemmer 1974).
(c) Current lion diversity
The results suggest that the modern lion currently consists of populations (or units) diagnosable through mtDNA, which are sorted into several coherent phylogeographic groups including North African–Asian, West African, eastern Sahel (steppe/savannah areas immediately south of the Sahara), eastern–southern African and southwestern African types (figures 1 and 2). Bootstrap values give reasonable support for most of these divisions. This phylogeographic pattern may reflect the Sahara Desert, dense equatorial rain forests and the Great Rift Valley all acting as natural physical barriers. The Great Rift Valley has previously been implicated as a barrier to lion dispersal (Burger et al. 2004; Dubach et al. 2005). Furthermore, similar phylogeographic patterns are detected in other large African mammals whose habitat preferences are comparable to that of the lion, including hartebeest (Alcelaphus buselaphus), topi (Danaliscus lunatus), wildebeest (Connochaetes taurinus), roan antelope (Hippotragus equinus) and warthog (Phacochoerus africanus) (Arctander et al. 1999; Muwanika et al. 2003; Alpers et al. 2004). The distribution pattern of physical barriers within Africa (see figure 1) may also explain the larger number of haplotypes (eight) currently found in the low latitudes (15° N–15° S), when compared to southern Africa (two haplotypes), in spite of these regions being of similar size and having been sampled equally.
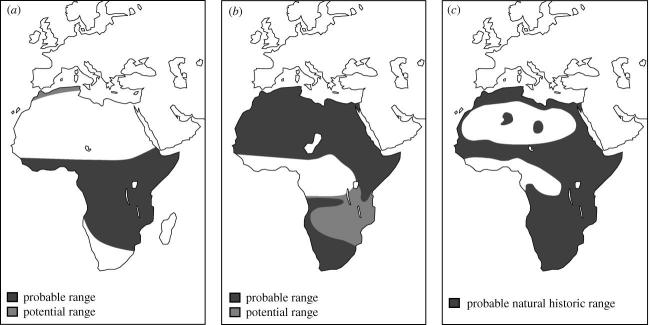
While some of our results are explained by the presence of physical barriers to dispersal, this does not explain the geographic clustering of all of the observed haplotypes (e.g. West African and eastern Sahel populations). Such phylogeographic patterns without physical barriers may represent an intermediate state of a spontaneous diffusion process after the removal of barriers (Hewitt 2000, 2004). The timing of the lion population bottleneck ca 74–203 kyr ago suggests that the modern lion has experienced the last two glacial–interglacial cycles with corresponding environmental changes, which are characterized in Africa by inversely related expansions and contractions of mid latitude deserts and low latitude forests (Mannion 1999; Hewitt 2000; Burger et al. 2004; De Vivo & Carmignotto 2004). Lions generally do not occur in either dry desert or dense forest (Nowell & Jackson 1996), suggesting the widespread super aridity during the last glacial maximum in both northern and southern Africa and in mid latitude southwestern Eurasia (De Menocal 2004; De Vivo & Carmignotto 2004) have had a detrimental impact on the regions' lion populations (figure 3a). On the other hand, it has been suggested that less dense habitats existed in low latitude Africa even during the Early Holocene wetter period (De Menocal 2004; De Vivo & Carmignotto 2004) when widespread forests likely fragmented lion populations in the region (figure 3b). As forests receded after the Early Holocene wetter period (De Vivo & Carmignotto 2004), surviving lion populations could have re-colonized the area, as well as lions invading from both north and south, contributing to the mtDNA haplotype mosaic currently observed in low latitude Africa.
Figure 3.
Estimated shift of the natural distribution range of the modern lion in the last ca 20 000 years: (a) at the last glacial maximum ca 20 000 years ago, (b) during the Early Holocene wetter period ca 10 000–4000 years ago and (c) present.
The foregoing arguments suggest that over its natural range (figure 3c) the modern lion may be evolutionarily subdivided into three geographic populations: North African–Asian, southern African and middle African, determined by expansions and contractions of mid latitude deserts and low latitude forests. The former two are characterized by relatively simple mtDNA haplotype structure, and the latter by a diverse haplotype mosaic. The North African–Asian population is separated from the sub-Saharan African populations by the vast Sahara Desert, and its skull is morphologically distinguishable from those of sub-Saharan lions (Hemmer 1974), suggesting its distinct taxonomic status.
(d) Implications for conservation
Although identifying appropriate management units (MU) is crucial for conservation, the concept of unit is scale-dependent, and ultimately depends on the agreed definition of the unit (Moritz 1994; Crandall et al. 2000). The modern lion consists of a handful of populations diagnosable with mtDNA (scale-1), which are sorted into five coherent phylogeographic groups (scale-2), which in turn are further sorted into three geographic groups based on the deduced recent (evolutionary) history (scale-3). The groups at the scale-2 level may be comparable to both evolutionary significant units and MU as defined by Moritz (1994), while some of the scale-1 groups may qualify as MU too. Although such units may warrant separate management, the deduced recent (evolutionary) history of the lion also makes it appropriate to keep a gradual evolutionary diffusion (or split) among populations in the long term. Conservationists need to design management plans choosing corresponding temporal and spatial scales—smaller scale units may be managed separately in the short term while they all may be incorporated into the management of larger scale units in the long term. To facilitate long-term lion conservation and conserve the perpetual process of evolution, research is needed into the dispersal patterns of each sex, including frequency, speed and distance after adjustment for physical barriers such as high mountains and large rivers (Eizirik et al. 2001).
Alternatively, although zoos probably cannot conserve long-term evolutionary processes, they may be able to contribute by keeping as many viable diagnosable units as possible in accordance with the concept of the tree of life (Mace et al. 2003). In this context, it would be appropriate for captive breeding programmes to distinguish minor MU characterized by different genetics, morphology, or simply geographic regions separated by natural barriers or distances. According to the International Species Information System (http://www.isis.org/abstracts/abs.asp), there are currently 29 lions registered in captivity as P. l. bleyenberghi (from Angola and Zimbabwe) and similarly 100 as P. l. krugeri (South Africa), eight as P. l. massaicus (Uganda and Kenya), 23 as P. l. nubicus (Tanzania), 115 as P. l. persica (India) and 13 as P. l. senegalensis (Senegal to Cameroon), in addition to ca 970 lions whose origins are uncertain. Considering the diagnosable units suggested by the results, lions from West Africa and eastern Sahel appear to be underrepresented in captivity for preservation of the diversity of the species. Since lions in these regions may be critically endangered in the wild (Bauer & Van Der Merwe 2002; Chardonnet 2002), establishing ex situ breeding programmes is a conservation priority.
Acknowledgments
We thank C. Claude, W. Cotteril, J. Cuisin, B. Fernholm, O. Grönwall, C. Harington, F. Kigozi, A. Kitchener, G. Lenglet, D. MacFadyen, D. Nagel, F. Renoud, D. Robineau, A. Sher, C. Smeenk, M. Tranier, P. J. H. van Bree, L. Werdelin, R. Ziegler and V. Ziswiler for access to samples. We also thank B. Shapiro, L. Werdelin and S. Y. W. Ho, as well as three anonymous referees for insightful comments on the manuscript. Particular thanks are also due to Tom Higham and the NERC ORADS program. Financial support was provided by NERC (R.B., I.B. and A.C.), BBSRC (R.B.), and the Leverhulme (A.C.) and Wellcome (I.B. and A.C.) Trusts.
Footnotes
Sequences are available in GenBank with accession numbers: DQ248045–DQ248055, DQ459397–DQ459402.
References
- Alpers D.L, van Vuuren J, Arctander P, Robinson T.J. Population genetics of the roan antelope (Hippotragus equinus) with suggestions for conservation. Mol. Ecol. 2004;13:1771–1784. doi: 10.1111/j.1365-294X.2004.02204.x. doi:10.1111/j.1365-294X.2004.02204.x [DOI] [PubMed] [Google Scholar]
- Arctander P, Johansen C, Coutellec-Vreto M. Phylogeography of three closely related African bovids (tribe Alcelaphini) Mol. Biol. Evol. 1999;16:1724–1739. doi: 10.1093/oxfordjournals.molbev.a026085. [DOI] [PubMed] [Google Scholar]
- Bandelt H.J, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Barnett, R., Yamaguchi, N., Barnes, I. & Cooper. A. 2006 Lost populations and preserving genetic diversity in the lion Panthera leo: implications for its ex situ conservation. Conserv. Genet Available online 1 March, 2006.
- Bauer H, Van Der Merwe S. The African lion database. Cat News. 2002;36:41–53. [Google Scholar]
- Burger J, et al. Molecular phylogeny of the extinct cave lion Panthera leo spelaea. Mol. Phylogenet. Evol. 2004;30:841–849. doi: 10.1016/j.ympev.2003.07.020. doi:10.1016/j.ympev.2003.07.020 [DOI] [PubMed] [Google Scholar]
- Chardonnet P. International Foundation for the Conservation of Wildlife; Paris, France: 2002. Conservation of African lion. [Google Scholar]
- Cracraft J, Feinstein J, Vaughn J, Helm-Bychowski K. Sorting out tigers (Panthera tigris): mitochondrial sequences, nuclear inserts, systematics, and conservation genetics. Anim. Conserv. 1998;1:139–150. doi:10.1111/j.1469-1795.1998.tb00021.x [Google Scholar]
- Crandall K.A, Bininda-Emonds O.R.P, Mace G.M, Wayne R.K. Considering evolutionary processes in conservation biology. Trends Ecol. Evol. 2000;15:290–295. doi: 10.1016/s0169-5347(00)01876-0. doi:10.1016/S0169-5347(00)01876-0 [DOI] [PubMed] [Google Scholar]
- Culver M, Johnson W.E, Pecon-Slattery J, O'Brien S.J. Genomic ancestry of the American puma (Puma concolor) J. Hered. 2000;91:186–197. doi: 10.1093/jhered/91.3.186. doi:10.1093/jhered/91.3.186 [DOI] [PubMed] [Google Scholar]
- De Menocal P.B. African climate change and faunal evolution during the Pliocene–Pleistocene. Earth Planet. Sci. Lett. 2004;220:3–24. doi:10.1016/S0012-821X(04)00003-2 [Google Scholar]
- De Vivo M, Carmignotto A.P. Holocene vegetation change and the mammal faunas of South America and Africa. J. Biogeogr. 2004;31:943–957. doi:10.1111/j.1365-2699.2004.01068.x [Google Scholar]
- Dubach J, Patterson B.D, Briggs M.B, Venzke K, Flamand J, Stander P, Scheepers L, Kays R.W. Molecular genetic variation across the southern and eastern geographic ranges of the African lion Panthera leo. Conserv. Genet. 2005;6:15–24. doi:10.1007/s10592-004-7729-6 [Google Scholar]
- Eizirik E, Kim J.H, Menotti-Raymond M, Crawshaw P.G, Jr, O'Brien S.J, Johnson W.E. Phylogeography, population history and conservation genetics of jaguar (Panthera onca, Mammalia, Felidae) Mol. Ecol. 2001;10:65–79. doi: 10.1046/j.1365-294x.2001.01144.x. doi:10.1046/j.1365-294X.2001.01144.x [DOI] [PubMed] [Google Scholar]
- Hemmer H. Untersuchungen zur stammesgeschichte der Pantherkatzen (Pantherinae). Teil III. Zur artgeschichte des Löwen Panthera (Panthera) leo (Linnaeus 1758) Veröff. Zool. Staatssamml. München. 1974;17:167–280. [Google Scholar]
- Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. doi:10.1038/35016000 [DOI] [PubMed] [Google Scholar]
- Hewitt G.M. Genetic consequences of climatic oscillations in the quaternary. Phil. Trans. R. Soc. B. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. doi:10.1098/rstb.2003.1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jae-Heup K, Eizirik E, O'Brien S.J, Johnson W.E. Structure and patterns of sequence variation in the mitochondrial DNA control region of the great cats. Mitochondrion. 2001;1:279–292. doi: 10.1016/s1567-7249(01)00027-7. doi:10.1016/S1567-7249(01)00027-7 [DOI] [PubMed] [Google Scholar]
- Janczewski D.N, Modi W.S, Stephens J.C, O'Brien S.J. Molecular evolution of mitochondrial 12S RNA and cytochrome b sequences in the Pantherine lineage of Felidae. Mol. Biol. Evol. 1995;12:690–707. doi: 10.1093/oxfordjournals.molbev.a040232. [DOI] [PubMed] [Google Scholar]
- Kurtén B. Weidenfield and Nicolson; London, UK: 1968. Pleistocene mammals of Europe. [Google Scholar]
- Lopez J.V, Culver M, Stephens J.C, Johnson W.E, O'Brien S.J. Rates of nuclear and cytoplasmic mitochondrial DNA sequence in mammals. Mol. Ecol. Evol. 1997;14:277–286. doi: 10.1093/oxfordjournals.molbev.a025763. [DOI] [PubMed] [Google Scholar]
- Mace G.M, Gittleman J.L, Purvis A. Preserving the tree of life. Science. 2003;300:1707–1709. doi: 10.1126/science.1085510. doi:10.1126/science.1085510 [DOI] [PubMed] [Google Scholar]
- Mannion A.M. Routledge; London, UK: 1999. Natural environmental change. [Google Scholar]
- Moritz C. Applications of mitochondrial-DNA analysis in conservation—a critical-review. Mol. Ecol. 1994;3:401–411. [Google Scholar]
- Muwanika V.B, Nyakaana S, Siegismund H.R, Arctander P. Phylogeography and population structure of the common warthog (Phacochoerus africanus) inferred from variation in mitochondrial DNA sequences and microsatellite loci. Heredity. 2003;91:361–372. doi: 10.1038/sj.hdy.6800341. doi:10.1038/sj.hdy.6800341 [DOI] [PubMed] [Google Scholar]
- Nowell K, Jackson P. IUCN/SSC Cat Specialist Group; Gland, Switzerland: 1996. Wild Cats. [Google Scholar]
- O'Brien S.J, Martenson J.S, Packer C, Herbst L, De Vos V, Joslin P, Ott-Joslin J, Wildt D.E, Bush M. Biochemical genetic variation in geographic isolates of African and Asiatic lions. Natl Geogr. Res. 1987;3:114–124. [Google Scholar]
- Rambaut, A. 1996 Se-Al: sequence alignment editor. University of Oxford, UK. Available at http://evolve.zoo.ox.ac.uk/
- Shapiro B, et al. Rise and fall of the Beringian steppe bison. Science. 2004;306:1561–1565. doi: 10.1126/science.1101074. doi:10.1126/science.1101074 [DOI] [PubMed] [Google Scholar]
- Stringer C. Modern human origins: progress and prospects. Phil. Trans. R. Soc. B. 2002;357:563–579. doi: 10.1098/rstb.2001.1057. doi:10.1098/rstb.2001.1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, MA: 2001. PAUP*4.0b10. Phylogenetic analysis using parsimony and other methods 4.0b10. [Google Scholar]
- Turner A, Antón M. Columbia University Press; New York, NY: 1997. The big cats and their fossil relatives. [Google Scholar]
- Uphyrkina O, Johnson W.E, Quigley H, Miquelle D, Marker L, Bush M, O'Brien S.J. Phylogenetics, genome diversity and origin of modern leopard, Panthera pardus. Mol. Ecol. 2001;10:2617–2633. doi: 10.1046/j.0962-1083.2001.01350.x. doi:10.1046/j.0962-1083.2001.01350.x [DOI] [PubMed] [Google Scholar]
- Willerslev E, Cooper A. Ancient DNA. Proc. R. Soc. B. 2005;272:3–16. doi: 10.1098/rspb.2004.2813. doi:10.1098/rspb.2004.2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Haddane B. The north African barbary lion and the Atlas lion project. Int. Zoo News. 2002;49:465–481. [Google Scholar]
- Yamaguchi N, Cooper A, Werdelin L, Macdonald D.W. Evolution of the mane and group-living in the lion (Panthera leo): a review. J. Zool. Lond. 2004;263:329–342. [Google Scholar]