Abstract
The mammalian host has a number of innate immune mechanisms designed to limit the spread of infection, yet many bacteria, including Salmonella, can cause systemic disease. Salmonella typhimurium-infected phagocytes traverse the gastrointestinal (GI) epithelium and enter the bloodstream within minutes after ingestion, thereby spreading throughout its host. Here, we provide a cellular and molecular basis for this phenomenon. We demonstrate that S. typhimurium manipulates the migratory properties of infected GI phagocytes with a type III secretion system. We show that one secreted effector, SrfH, interacts with the host protein TRIP6, a member of the zyxin family of adaptor proteins that regulate motility. SrfH promotes phagocyte motility in vitro and accelerates the systemic spread of infection away from the lumen of the intestine in the mouse. This is a previously uncharacterized mechanism by which an intracellular pathogen overcomes host defenses designed to immobilize infected cells.
Keywords: bacteremia, secreted effector, TRIP6, type 3 secretion, virulence
Numerous intracellular pathogens must breach epithelial barriers, navigate the lymphatic system, and travel considerable distances to reach their preferred sites of replication within a host. For enteric pathogens, in addition to the physical barrier that the gastrointestinal (GI) epithelium provides, microbes must also contend with a brush border, a thick mucus coat, an acidic environment, peristalsis, antimicrobial peptides, cell turnover, and endogenous flora. GI phagocytes can serve as vehicles for microbial dissemination into deeper tissue: they readily engulf microbes and can traverse epithelial barriers and inadvertently shield pathogens from other components of the immune system. However, when phagocytes internalize Gram-negative bacteria, their LPS binds Toll-like receptor 4, triggering the release of macrophage migration inhibition factor and other cytokines that strongly inhibits their motility (1, 2). Additionally, the best-characterized route for tissue phagocytes to reach the bloodstream is through the lymphatic system, and recirculation normally requires between 12 and 20 h (3–5). Thus, it is remarkable that Salmonella typhimurium- and Salmonella typhi-infected phagocytes can enter the bloodstream within as little as 15 min after ingestion (6, 7).
S. typhimurium penetrates to deeper tissue through two different pathways. In one route, the bacteria access systemic tissue by the lymphatic system and the Peyer's patches. In the second pathway, phagocytes are believed to carry intestinal bacteria directly into the bloodstream without passing through the Peyer's patches (6–10). After ingestion, bacteria reach systemic organs in <15 min in this pathway vs. ≈24 h by the lymphatic system (7). The bacteria are carried within CD18-expressing cells thought to be monocytes (11). Movement of bacteria-infected cells requires not only stimulation of cell motility but also inhibition of the inflammatory pathways that would normally block this motility (12). Thyroid receptor interacting protein 6 (TRIP6; ref. 13) is an adaptor protein that binds components of the Rac signaling pathway, critical for cell motility, and the NF-κB inflammatory pathways (14–18). Thus, Salmonella effector interation with TRIP6 might alter both the inflammatory response to infection as well as the motility of an infected cell, as described below.
Salmonella enterica virulence is mediated in part by the passage of proteins to the host through one or more type III secretion systems (19). Few of the type III secreted proteins have been characterized with respect to their mechanism of action. In this work, we describe how one such effector, SrfH, alters cell motility. This is a new and unexpected function for a type III secretion system, suggesting that Salmonella, and perhaps other intracellular pathogens, direct their course of infection by controlling the migratory properties of the host cells harboring them.
Results
SrfH Is a Secreted Type III Effector That Binds the LIM (Named for Lin-11, Isl-1, and Mec-3) Domains of TRIP6.
We recently identified srfH/sseI (SsrB regulated factor H/Salmonella secreted effector I), a chromosomal gene associated with SPI-2 whose expression is induced nearly 100-fold inside macrophages (20, 21). SrfH shares no homology with any database entries outside of its N-terminal sequence and possesses no conserved catalytic motifs, suggesting a unique function for the protein. We confirmed a report (22) that SrfH is secreted by SPI-2 across the vacuolar membrane after invasion (23); however, it plays no role in promoting intracellular proliferation rendering its function enigmatic (see supporting information, which is published on the PNAS web site; ref. 20).
To delineate SrfH's role within infected host cells, we used srfH as the “bait” in a yeast two-hybrid screen of a human cDNA library. Of approximately one million yeast transformants screened, only two contained host peptides that interacted significantly with SrfH, both of which contained LIM domain fragments of TRIP6 (Fig. 1A). Despite their similarities, SrfH does not interact with other members of this adaptor family (Fig. 1B).
Fig. 1.
Salmonella SrfH binds the adaptor protein TRIP6 in a yeast two-hybrid assay. (A) Yeast two-hybrid screen for SrfH interacting proteins. β-Gal assays were carried out on the indicated yeast cells. The level of β-gal expression is a measure of the strength of the binding (42). Fos + Jun are the c-Fos and c-Jun interacting partners used as a positive control. Laminin interaction is the negative nonspecific control. The TRIP6 fragments first identified contain the C-terminal LIM domains of TRIP6 starting from the amino acids indicated and thus contain either two or three LIM domains. Additional controls are shown. (B) SrfH interactions with LIM domain proteins. SrfH does not interact with any of the other zyxin family members in the yeast two-hybrid assay. Error bars in Figs. 1–6 represent the standard error of the mean.
To support the relevance of the SrfH/TRIP6 binding, we conducted additional experiments. Transient transfection of RAW264.7 cells with a hybrid SrfH-GFP protein shows localization to the membrane and to focal adhesions. When these same cells are stained with anti-TRIP6 antibody, they show >90% colocalization of the two proteins within macrophages (Fig. 2Upper). Transfection with a mammalian vector expressing GFP alone does not show colocalization with TRIP6 (Fig. 2 Lower). As shown in Fig. 2 Lower Left, staining with anti-TRIP6 antibody exhibits a characteristic punctate pattern that is diffusely spread across the cytoplasm, similar to staining observed in previous studies (17). After SrfH transfection, TRIP6 relocalizes to the cellular cytoskeleton (Fig. 2 Upper). It is noteworthy that cells transfected with a mammalian vector expressing SrfH show ruffling patterns consistent with those seen after stimulation of the Rac signaling pathway (Fig. 2; ref. 24), which is the major signal transduction pathway activated in cellular movement. In summary, the Salmonella secreted effector SrfH and mammalian adaptor protein TRIP6 interact; however, because there are >250 LIM-domain-containing proteins, we cannot rule out interaction with additional proteins. Further experiments that describe the nature of the TRIP6/SrfH interaction in detail will be published separately.
Fig. 2.
Evidence for SrfH/TRIP6 interaction. (Upper) Micrograph of RAW264.7 macrophages transfected with srfH-GFP and then analyzed to determine whether SrfH and TRIP6 colocalize. TRIP6 is stained with Texas red-labeled anti-TRIP6 antibody, whereas GFP is present as a translational fusion with SrfH. (Lower) Cells transfected with the same mammalian vector but expressing GFP alone and stained as in Upper for TRIP6.
SrfH Stimulates Phagocyte Migration.
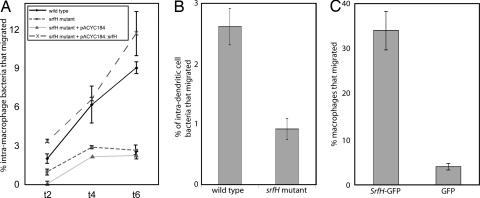
Given that TRIP6 is an adaptor in a pathway that can stimulate cellular motility and that knockdown as well as overproduction of TRIP6 alters cellular motility (16–18), we hypothesized that SrfH promotes the motility of infected phagocytes. To test this possibility, RAW264.7 macrophage-like cells and JAWS dendritic-like cells (both are CD18+) were infected with either wild-type S. typhimurium 14028s or an isogenic srfH mutant at a multiplicity of infection of 0.1 (21). After the initial infection period, all extracellular bacteria were removed by washing and treatment with gentamicin (25). The ability of the infected macrophages to chemotax was determined in a modified Boyden chamber (Chemotax System; Neuro Probe, Gaithersburg, MD). Boyden chambers are composed of two compartments separated by a hydrophobic filter that contains small holes. Macrophages are cultured in the top compartment, and various macrophage chemoattractants may be placed in the bottom compartment. The number of cfus reaching the bottom compartment was used to measure macrophage migration. The gentamicin-containing media in the bottom compartment were shown to be free of viable extracellular bacteria. We found that significantly more macrophages or dendritic cells infected with wild-type S. typhimurium migrated than these same cells types infected with an isogenic srfH mutant (Fig. 3A and B). Furthermore, SrfH expression in trans complemented a srfH mutant phenotype (Fig. 3A). Taken together with the results of the previous studies, these data indicated a role for SrfH in promoting the motility of infected cells, which may be a direct consequence of SrfH's interaction with TRIP6. However, these findings do not exclude the possibility that SrfH affects the motility of infected macrophages through an indirect mechanism requiring expression of additional effectors.
Fig. 3.
Measurement of infected cell migration in a Boyden chamber. (A) Washed bacteria were used to infect RAW264.7 bacteria at a multiplicity of infection of 0.1–0.2, treated for 1 h with 100 μgm/ml gentamicin to kill extracellular bacteria, and placed in the top well of the Boyden chamber. At the times indicated, the cells in the bottom reservoir were lysed and the number of infected cells that migrated determined by plating and counting cfus. In the experiments shown, heat-treated Salmonella was used as a chemoattractant. Gentamicin (at 10 μg/ml) was used throughout the experiment to kill any released bacteria. Cell migration of infected RAW264.7 cells was stimulated by infection with srfH+ Salmonella but not srfH−. SrfH, supplied in trans, complemented a srfH mutation. (B) Similar experiments were carried out by using the JAWS dendritic cell line with very similar results. The 6-hr time point is shown. (C) SrfH alone stimulates cell motility. RAW264.7 macrophages were transfected with a mammalian vector expressing SrfH-GFP or GFP alone. Six hours later, cells were removed and the number of fluorescent cells in the bottom reservoir counted in the microscope.
To establish whether SrfH was sufficient for increased macrophage motility, we expressed SrfH within the macrophage cytosol in the absence of any other bacterial components and assayed for induction of macrophage motility. Separate populations of macrophages were transfected with either a plasmid expressing SrfH-GFP or a plasmid expressing GFP alone. More macrophages expressing SrfH-GFP migrated than the group expressing only GFP (Fig. 3C).
SrfH Has No Effect on Host-Cell Movement in the Absence of TRIP6.
We next assessed the degree to which the srfH phenotype depends on TRIP6 expression. It was established that TRIP6 levels could be greatly reduced when the corresponding transcripts were targeted with small interfering RNA (siRNA). Macrophages were transfected with either nonsilencing or silencing TRIP6 siRNA. Subsequently, the cells were infected with either the srfH mutant or the wild-type Salmonella strain. When TRIP6 expression was blocked with siRNA, macrophages harboring either type of bacteria migrated almost identically (Fig. 4B). In contrast, when macrophages were transfected with nonsilencing siRNA, SrfH conferred an ≈5-fold increase in macrophage motility, similar to what was observed in Fig. 3. Overall, the results of this experiment suggest that SrfH manipulates macrophage motility by interacting directly with the adaptor protein TRIP6. The finding that blocking TRIP6 expression resulted in increased cell motility is in agreement with others (18). We hypothesize that TRIP6 inhibits cell migration and that the binding of SrfH to TRIP6 blocks this inhibition.
Fig. 4.
Reducing the expression of TRIP6 by siRNA reduces the effect of SrfH on cell motility. (A) (Upper) Total proteins expressed by RAW264.7 treated with siRNA (Right) or an irrelevant siRNA (Left). (Lower) Western hybridization of the same gel probed with anti-TRIP6 antibody. TRIP6 expression was reduced by >95% with siRNA. (B) TRIP6 was required for SrfH to affect cell motility. Left-hand bars compare cell migration when RAW264.7 cells were treated with randomized control siRNA. Right-hand bars compare cell migration when RAW264.7 cells were treated with TRIP6 siRNA. RAW264.7 cells treated with TRIP6 siRNA show no srfH+/srfH− difference in cell migration.
S. typhimurium Travels Directly from the GI Tract to the Bloodstream Within CD18+ Phagocytes.
Because SrfH is able to promote macrophage movement in vitro, we hypothesized that in an infection model, SrfH expression would correlate with a rapid appearance of S. typhimurium-infected phagocytes in the bloodstream in a mouse infection (6, 7). During short time periods, S. typhimurium travels from the murine lumen of the intestine into the bloodstream within GI phagocytes (7). In CD18− mice, phagocytes cannot efficiently traverse the GI epithelium (7, 26, 27). CD18 is an integrin specific to immune system cells that facilitates leukocyte transmigration and is present on the professional phagocytic cells: neutrophils, macrophages, and dendritic cells. Ten CD18− mice were inoculated with 1 × 109 S. typhimurium by gavage. Thirty minutes postinfection, we did not recover any viable bacteria from their peripheral blood (Fig. 5A). This was not because of increased killing of intracellular bacteria, because CD18-deficient macrophages are less microbiocidal to S. typhimurium than are congenic control macrophages (7, 11). Thus, it was concluded that S. typhimurium travels from the GI tract to the bloodstream exclusively inside of CD18+ phagocytes in the short time frame used in this study. This finding allowed us to examine how various bacterial mutations may impact the migration of these cells.
Fig. 5.
Salmonella septicemia is observed 30 min after oral infection of the mouse. (A) The number of bacteria recovered by cardiac puncture 30 min after oral infection with 1 × 109 bacteria of the strains indicated. The first two bars show that five times more wild-type S. typhimurium was recovered than in an isogenic srfH mutant. The results shown are for BALB/c mice, and the error bars represent the standard error of the mean. The third bar shows the number of bacteria recovered for SrfH expression in trans from a multicopy plasmid. The y axis is discontinuous, so that the phenotypes could be displayed on the same graph. The fourth bar represents bacteria recovered for a SPI-2 type III secretion mutant (ssaK::cm). As a control, mice were inoculated with E. coli K12 at 10 times the dose used with S. typhimurium, and no bacteria were recovered from the bloodstream. The last bar shows that no wild-type bacteria were recovered from CD18− mice inoculated with 1 × 109 bacteria. (B) The number of bacteria recovered from the spleen and liver 24 h after oral infection with either a srfH mutant or an arbitrary MudJ insertion that does not affect virulence (srfJ; ref. 41). Wild-type S. typhimurium 14028 gave the same numbers as the srfJ mutation that is shown. A MudJ insertion is displayed based on concern that bacteriophage Mu might alter the phenotype of Salmonella.
SrfH Accelerates Entry of S. typhimurium to the Bloodstream.
To determine whether SrfH accelerates the rate at which infected phagocytes travel from the lumen of the intestine into the bloodstream, we introduced either wild-type S. typhimurium or the srfH mutant into mice by oral gavage (1 × 109). Thirty minutes after inoculation, an average of five times more wild-type S. typhimurium than srfH mutants were recovered from the bloodstream (Fig. 5A). When the srfH mutation was complemented by SrfH expression in trans from a multicopy plasmid, the complemented mutant showed an even higher level of bacteria than that seen with wild-type S. typhimurium (Fig. 5A). Each of these experiments was carried out with groups of five mice on seven separate occasions. In a dissemination assay with a Salmonella strain defective in all SPI-2-dependent type III secretion (ssaK::cm), no mutant bacteria were recovered in 18 of 25 mice intragastrically infected with the ssaK mutant. Thus, the phenotype of a complete knockout of the SPI-2 type III secretion systems is much stronger than the srfH mutation alone, which suggests that additional effectors are used during the course of infection in the mouse.
To establish whether the accelerated bacteremia resulted in accelerated colonization of internal organs, we assayed the number of bacteria in the spleen and liver, preferred sites for Salmonella replication, 24 h after oral inoculation. At 24 h, SrfH increased the rate at which S. typhimurium colonized both the liver and spleen by an order of magnitude (Fig. 5B). The rapidity with which mouse infection was observed was surprising given the fact that previous studies suggested that expression of SPI-II and its effectors is delayed for hours in vitro (see ref. 28 for discussion). This conundrum has been resolved by direct assays for SPI-2 expression within ligated mouse ileal loops, which demonstrated rapid expression in the gut (28). In fact, constitutive expression of SrfH did result in increased bacteremia after gavage. Thus, SrfH expression may limit the level of infection (Fig. 5A). In summary, after Salmonella typhimurium 14028 oral infection of mice, at least one organism in one million inoculated traveled to the bloodstream within minutes.
As further verification that the bacteria are present within cells in the bloodstream, we performed FACS analysis of infected white blood cells isolated from Salmonella::GFP orally infected mice (Fig. 6). Most of the bacteria were present within monocytes (GR-1+/CD11b+), corroborating earlier observations (11), and the number of infected cells closely paralleled the number of cfu recovered in the previous experiment (Fig. 5A).
Fig. 6.
Blood from mice orally infected with 1 × 109 cfu of GFP expressing Salmonella analyzed by FACS. GFP fluorescence vs. side scatter is shown in the scatter plots. Mice were inoculated IG with the Salmonella strain indicated above each image, and the percentage of GFP positive cells found in the bloodstream 30 min postinoculation is indicated in the upper right corner of each plot. Results are shown for pooled blood from three mice. An LSRII (Becton Dickinson) FACS analysis machine equipped with a 488-nm laser was used to detect GFP-expressing bacteria in infected peripheral blood cells. FACS data were analyzed by using FlowJo software (Tree Star).
Bacteria Are Not Killed in Transit to the Bloodstream.
One concern we had at the outset was that the reduced numbers of mutant bacteria that reach the bloodstream was a consequence of bacterial death in transit and not a difference in infected cell motility. To investigate this possibility, we developed a more sensitive PCR assay for the presence of live or dead bacteria. The assay is based on incorporating a high copy number plasmid into the bacteria to provide a PCR template that is not readily degraded and can be easily detected. In our assay, we purified plasmid DNA from infected mouse blood, relying on its structure as a closed circular molecule, and then used the purified plasmid DNA to template PCR. In control experiments, we could detect even a single heat-killed bacterium in 1 ml of blood (supporting information). No bacteria were recovered by plating or PCR in dissemination experiments with Escherichia coli K12, a nonpathogenic relative of S. typhimurium. To determine whether ssaK mutant bacteria were being killed during the short time period of this assay, we repeated the more sensitive PCR experiments. We could not detect live or dead bacteria within the bloodstreams of 10 additional mice that were inoculated with 1 × 1010 SPI-2 secretion mutant bacteria, 10 times the original inoculum used. Therefore, the earlier recovery of a few bloodstream bacteria in 7 of 24 mice inoculated with the type III secretion systems structural mutant was likely the result of abrasion by the gavage needle. To summarize, we show that phagocytes do not kill ssaK mutants in transit (supporting information). Because SPI-2 genes mediate neither GI tract survival (supporting information) nor adherence and uptake by phagocytic cells (supporting information), the results imply that SPI-2 secretes additional effectors that manipulate cellular motility.
Discussion
Our results show that SrfH is responsible for accelerating the movement of infected CD18+ cells from the lumen of the intestine to the liver and spleen, which are the preferred sites for Salmonella replication. Partial experimental support for a direct interaction between SrfH and TRIP6 is provided but will be considered in detail in a later publication. Results from the Boyden chamber assays show that SrfH expression in infected cells is both necessary and sufficient for an increase in macrophage and dendritic cell motility. Most important, infection of mice with wild-type Salmonella, but not the srfH mutant, resulted in the presence of infected cells in the bloodstream within minutes. These bacteria are carried exclusively within CD18+ cells: in mice lacking these cells, bacteremia takes place by a different route and is delayed many hours. SrfH does not affect bacterial adherence, cell invasion, survival within the intestine, or replication within macrophages, dendritic cells, or epithelial cells (supporting information and ref. 21). Our data support the hypothesis that SrfH alters the motility of infected cells by its interaction with TRIP6.
TRIP6 Functions in both Inflammatory and Cell Motility Pathways.
TRIP6 is an adaptor that interacts with components of inflammatory pathways as well as the Rac pathway for cell motility (15–18, 24). Professional phagocytic cells chemotax toward infecting microbes, limiting the spread of infection by destroying the invading microbe and initiating the adaptive immune response. They are directed along a concentration gradient of bacterial products but stop at the site of infection. This must require a complex interaction between the Toll-like receptor pathways and the Rac cell motility pathway (18). In the macrophage-like cell we have studied, reducing expression of TRIP6 stimulates motility of the infected cells, thus TRIP6 must be acting as a brake to inhibit infected cells from disseminating, and SrfH, in effect, removes the brake (our results; ref. 18).
Antigen Sampling and Bacterial Infection.
Numerous intracellular pathogens can traverse epithelial barriers and disseminate within hosts with surprising speed. Accordingly, it has long been speculated that phagocytes might inadvertently spread such pathogens as part of a host-specified antigen-sampling pathway (7, 29). However, our results suggest that S. typhimurium uses the SPI-2 encoded type III secretion systems to stimulate infected GI phagocytes to carry them across the epithelium. This allows Salmonella to bypass the lymphatic system and penetrate the bloodstream, thereby accelerating the colonization of internal organs. This study provides one of the first descriptions of a virulence mechanism in which a pathogen directly alters the motility of infected host cells for its own benefit.
A Common Virulence Strategy.
The manipulation and subversion of host cell motility is a virulence strategy likely to be used by other intracellular pathogens aside from S. typhimurium. Because such pathogens must traverse the host integuments and move within the host to reach their preferred site(s) of replication, the manipulation and subversion of host cell motility is a virulence strategy likely to be used by other intracellular pathogens aside from S. typhimurium. Thus, it is not surprising that Yersinia enterocolitica and Vaccinia virus alter the motility of infected cells (8–10, 30). For these pathogens, the proteins responsible have not yet been identified. However, such observations suggest that numerous infectious microbes subvert phagocytes as vehicles for their own intrahost dissemination. Thus, it is surprising that srfH shows a very limited distribution within S. enterica given the fact that many strains of Salmonella have the ability to disseminate rapidly within mice and humans and to reach the bloodstream within minutes after oral infection (6). It is therefore likely that there will be additional genes identified encoding proteins with similar function but not necessarily sequence homology.
The immediate consequence of the presence of intracellular bacteria is to block the motility of infected cells by release of cytokines such as migration inhibition factor (MIF; refs. 31 and 32). MIF is also proinflammatory and a pivotal regulator of innate immunity (reviewed in ref. 33). It follows that a mechanism to overcome this component of the innate immune system must be an essential virulence determinant of pathogens that cause systemic infection. The underlying microbial machinery may provide a new generation of antimicrobial targets as well as tools for delineating the molecular mechanisms responsible for cell movement.
Methods
Bacterial Strains, Constructs, and Media.
S. typhimurium 14028s and derivatives as well as cultured RAW264.7 macrophages from American Type Culture Collection were used throughout this study. Bacteria and tissue culture cells were grown as described (34). An SPI-2 structural mutant was created by disrupting the chromosomal ssaK allele, as described (21). For in vitro motility assays, complemented derivatives of 14028s srfH::MudJ (MJW704; ref. 21) were generated. The srfH ORF and 65 bp of upstream sequence were PCR amplified, cloned into pWKS30 (35), and electroporated into MJW704. For transfection experiments, srfH was cloned into the eukaryotic expression vectors, pEGFP-N1.
General Methods.
All molecular biology and genetic manipulations were performed with established protocols (36, 37). Transfections were performed with Effectene for mammalian vectors and with HiPerFect for siRNA (Qiagen, Valencia, CA). Higher concentrations than specified by the manufacturer were necessary for macrophage transfection but did not appear to affect the viability of the transfected cells. Intramacrophage survival/growth assays were performed at an multiplicity of infection of ≤1 (38). TRIP6 was identified as the binding partner for SrfH with the Hybrid Hunter yeast two-hybrid kit (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions, using a commercial prey bank derived from HeLa cells (Invitrogen). β-Gal assays were performed on cultures grown overnight with rigorous shaking at 30°C according to the assay conditions provided with the Invitrogen kit.
Microscopy.
Samples were fixed in 4% formaldehyde, permeabilized, and stained with a native TRIP6 antibody and an anti-mouse secondary conjugated to Texas red (Molecular Probes, Eugene, OR). SrfH-GFP 3D colocalization analysis was performed with the Applied Precision (Marlborough, U.K.) Deltavision image restoration system. Deconvolution using the iterative constrained algorithm of Sedat and coworkers (39) and image processing were performed on an SGI Octane workstation. Colocalization analysis was performed with the CoolLocalizer algorithm (CytoLight, Ann Arbor, MI), on sets of 99 Z sections captured from representative macrophages expressing both GFP and Texas red.
In Vitro Motility Assays.
Macrophages were infected with bacteria grown to saturation and washed three times with PBS at a multiplicity of infection of 0.1–0.2. Bacteria were centrifuged onto the monolayers and incubated for 1 hr. Infected monolayers were then washed four times, for 5 min each wash. The extracellular bacteria were killed with a 1-hr incubation in DMEM supplemented with 100 μg/ml gentamicin (34). The monolayers were washed as before and collected in 1.0 ml of DMEM containing 10 μg/ml gentamicin and 50 μl of this suspension added to the top of a 12-μM pore chemotaxis plate from the Chemotax system (Neuro Probe, Gaithersburg, MD). In some experiments, heat-treated S. typhimurium (90°C for 10 min) from a saturated overnight culture (diluted 1,000-fold in DMEM augmented with 10 μg/ml gentamicin) was used as a chemoattractant (30 μl of this mixture in the bottom reservoir). Counting macrophages with a hemocytometer as well as counting bacteria after macrophage lysis determined migration. Transfection studies were performed similarly, except background migration was determined with mock-transfected macrophages and subtracted from the values presented.
Animal Experiments.
Four-week-old female BALB/c mice were used in all animal experiments except for those concerning CD18. For these, 8-week-old female C57BL/6J Itgb2 and C57BL/6J Itgb2tm1/Bay mice were used (27). Mice were orally inoculated by gavage with 1 × 109 or 1 × 1010 bacteria. Thirty minutes after inoculation, mice were anesthetized and peripheral blood obtained by cardiac puncture. Host cells were lysed with 1% Triton and bacteria recovered on MacConkey agar plates. Bacteria were confirmed to be S. typhimurium by their growth and appearance on MacConkey agar, and sensitivity to bacteriophage P22 and, where appropriate, by resistance to antibiotics. Bacterial survival within the GI tract and the ability of bacterial strains to colonize the spleen and liver were performed as described (40).
PCR and RT-PCR.
pBluescript, which has a copy number of >300 per cell, was electroporated into the various strains to serve as a sensitive target that would allow us to detect the presence of dead bacteria in the bloodstream. The PCR primers are 5′-CAAGGCGAGTTACATGATCCC and 5′-ACTGCGGCCAACTTACTTCTG. On two independent occasions, five mice for each strain tested were infected with ≈1 × 1010 bacteria and, 30 min later, total blood was collected and combined. Any plasmid DNA present was purified with plasmid DNA isolation columns (Qiagen), and the total eluates were used as templates in standard PCRs. All strains were tested side by side, with the same primers.
siRNA Studies.
Macrophages were transfected with the TRIP6-specific siRNA construct shown in Fig. 4 or a randomized sequence containing the same base composition (Qiagen), washed 24 h later, and then incubated for an additional 24 h. At this time, the macrophages were infected with bacteria, and the motility assay was performed as previously described.
FACS Analysis.
Mice were inoculated IG with 108 bacteria. Thirty minutes after inoculation, mice were anesthetized, and cardiac punctures were performed. Blood samples from three mice were pooled, red blood cells lysed, and white blood cells collected by using a lympholyte gradient (Cedarlane Laboratories, Burlington, NC) and passed through a 70-μm cell filter (Falcon, Colorado Springs, CO). Cells were resuspended in PBS containing 2% FBS and 0.1% sodium azide, then analyzed by FACS. A LSRII (Becton Dickinson, Franklin Lakes, NJ) FACS machine equipped with a 488-nm laser was used to detect GFP fluorescence (>500,000 cells were analyzed per sample using FlowJo software; Tree Star, Ashland, OR).
Supplementary Material
Acknowledgments
We are indebted to Dr. Kim Saunders for invaluable help with animal experiments. We are grateful to Dr. Mary Beckerle and Dr. Richard Klemke for providing us with encouragement and insight, and to Dr. Mary Beckerle for providing the anti-TRIP6 antibody. We are also indebted to Rebecca Tempel and Jean Gustin for help with this manuscript.
Abbreviations
- GI
gastrointestinal
- siRNA
small intestinal RNA
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Calandra T, Bucala R. Crit Rev Immunol. 1997;17:77–88. doi: 10.1615/critrevimmunol.v17.i1.30. [DOI] [PubMed] [Google Scholar]
- 2.Hermanowski-Vosatka A, Mundt SS, Ayala JM, Goyal S, Hanlon WA, Czerwinski RM, Wright SD, Whitman CP. Biochemistry. 1999;38:12841–12849. doi: 10.1021/bi991352p. [DOI] [PubMed] [Google Scholar]
- 3.MacPherson GG, Jenkins CD, Stein MJ, Edwards C. J Immunol. 1995;154:1317–1322. [PubMed] [Google Scholar]
- 4.Rothkotter HJ, Pabst R, Bailey M. Vet Immunol Immunopathol. 1999;72:157–165. doi: 10.1016/s0165-2427(99)00128-2. [DOI] [PubMed] [Google Scholar]
- 5.Westermann J, Puskas Z, Pabst R. Scand J Immunol. 1988;28:203–210. doi: 10.1111/j.1365-3083.1988.tb02432.x. [DOI] [PubMed] [Google Scholar]
- 6.Gerichter CB. J Hygiene. 1960;58:307–319. doi: 10.1017/s0022172400038420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vazquez-Torres A, Jones-Carson J, Baumler AJ, Falkow S, Valdivia R, Brown W, Le M, Berggren R, Parks WT, Fang FC. Nature. 1999;401:804–808. doi: 10.1038/44593. [DOI] [PubMed] [Google Scholar]
- 8.Autenrieth IB, Kempf V, Sprinz T, Preger S, Schnell A. Infect Immun. 1996;64:1357–1368. doi: 10.1128/iai.64.4.1357-1368.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marra A, Isberg RR. Infect Immun. 1997;65:3412–3421. doi: 10.1128/iai.65.8.3412-3421.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pepe JC, Miller VL. Proc Natl Acad Sci USA. 1993;90:6473–6477. doi: 10.1073/pnas.90.14.6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vazquez-Torres A, Fang FC. Curr Opin Microbiol. 2000;3:54–59. doi: 10.1016/s1369-5274(99)00051-x. [DOI] [PubMed] [Google Scholar]
- 12.Stupack DG, Cho SY, Klemke RL. Immuol Res. 2000;21:83–88. doi: 10.1385/IR:21:2-3:83. [DOI] [PubMed] [Google Scholar]
- 13.Lee JW, Choi H-S, Gyurist J, Brent R, Moore DD. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 14.Kain KH, Klemke RL. J Biol Chem. 2001;276:16185–16192. doi: 10.1074/jbc.M100095200. [DOI] [PubMed] [Google Scholar]
- 15.Kassel O, Schneider S, Heilbock C, Litfin M, Gottlicher M, Herrlich P. Genes Dev. 2004;18:2518–2528. doi: 10.1101/gad.322404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai YJ, Chen CS, Lin WC, Lin FT. Mol Cell Biol. 2005;25:5859–5868. doi: 10.1128/MCB.25.14.5859-5868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, Lai YJ, Lin WC, Lin FT. J Biol Chem. 2004;279:10459–10468. doi: 10.1074/jbc.M311891200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yi J, Kloeker S, Jensen CC, Bockholt S, Honda H, Hirai H, Beckerle MC. J Biol Chem. 2002;277:9580–9589. doi: 10.1074/jbc.M106922200. [DOI] [PubMed] [Google Scholar]
- 19.Hueck CJ. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao EA, Miller SI. Proc Natl Acad Sci USA. 2000;97:7539–7544. doi: 10.1073/pnas.97.13.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Worley MJ, Ching KH, Heffron F. Mol Microbiol. 2000;36:749–761. doi: 10.1046/j.1365-2958.2000.01902.x. [DOI] [PubMed] [Google Scholar]
- 22.Miao EA, Brittnacher M, Haraga A, Jeng RL, Welch MD, Miller SI. Mol Microbiol. 2003;48:401–415. doi: 10.1046/j.1365-2958.2003.t01-1-03456.x. [DOI] [PubMed] [Google Scholar]
- 23.Geddes K, Worley M, Niemann G, Heffron F. Infect Immun. 2005;73:6260–6271. doi: 10.1128/IAI.73.10.6260-6271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klemke RL, Leng J, Molander R, Brooks PC, Vuori K, Cheresh DA. J Cell Biol. 1998;140:961–972. doi: 10.1083/jcb.140.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fields PI, Swanson RV, Haidaris CG, Heffron F. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li XC, Miyasaka M, Issekutz TB. J Immunol. 1998;161:6258–6264. [PubMed] [Google Scholar]
- 27.Wilson RW, Ballantyne CM, Smith CW, Montgomery C, Bradley A, O'Brien WE, Beaudet AL. J Immunol. 1993;151:1571–1578. [PubMed] [Google Scholar]
- 28.Brown NF, Vallance BA, Coombes BK, Valdez Y, Coburn BA, Finlay BB. PLoS Pathog. 2005;1:e32. doi: 10.1371/journal.ppat.0010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl J-P, Ricciardi-Castagnoli P. Nat Immunol. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 30.Sanderson CM, Way M, Smith GL. J Virol. 1998;72:1235–1243. doi: 10.1128/jvi.72.2.1235-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lue H, Kleemann R, Calandra T, Roger T, Bernhagen J. Microbes Infect. 2002;4:449–460. doi: 10.1016/s1286-4579(02)01560-5. [DOI] [PubMed] [Google Scholar]
- 32.Stavitsky AB, Xianli J. Cell Immunol. 2002;217:95–104. doi: 10.1016/s0008-8749(02)00516-6. [DOI] [PubMed] [Google Scholar]
- 33.Roger T, Froidevaux C, Martin C, Calandra T. J Endotoxin Res. 2003;9:119–123. doi: 10.1179/096805103125001513. [DOI] [PubMed] [Google Scholar]
- 34.van der Velden AW, Lindgren SW, Worley MJ, Heffron F. Infect Immun. 2000;68:5702–5709. doi: 10.1128/iai.68.10.5702-5709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang RF, Kushner SR. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 36.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 2005. [Google Scholar]
- 37.Maloy SR, Stewart VJ, Taylor RK. Genetic Analysis of Pathogenic Bacteria. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 38.Worley MJ, Stojiljkovic I, Heffron F. Mol Microbiol. 1998;29:1471–1480. doi: 10.1046/j.1365-2958.1998.01030.x. [DOI] [PubMed] [Google Scholar]
- 39.Hanser BM, Gustafsson MG, Agard DA, Sedat JW. J Microsc. 2004;216:32–48. doi: 10.1111/j.0022-2720.2004.01393.x. [DOI] [PubMed] [Google Scholar]
- 40.van der Velden AW, Baumler AJ, Tsolis RM, Heffron F. Infect Immun. 1998;66:2803–2808. doi: 10.1128/iai.66.6.2803-2808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beuzon CR, Meresse S, Unsworth KE, Ruiz-Albert J, Garvis S, Waterman SR, Ryder TA, Boucrot E, Holden DW. EMBO J. 2000;19:3235–3249. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Estojak J, Brent R, Golemis EA. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.