Abstract
Because of the rise in atmospheric oxygen 2.3 billion years ago (Gya) and the subsequent changes in oceanic redox state over the last 2.3–1 Gya, trace metal bioavailability in marine environments has changed dramatically. Although theorized to have influenced the biological usage of metals leaving discernable genomic signals, a thorough and quantitative test of this hypothesis has been lacking. Using structural bioinformatics and whole-genome sequences, the Fe-, Zn-, Mn-, and Co-binding metallomes of 23 Archaea, 233 Bacteria, and 57 Eukarya were constructed. These metallomes reveal that the overall abundances of these metal-binding structures scale to proteome size as power laws with a unique set of slopes for each Superkingdom of Life. The differences in the power describing the abundances of Fe-, Mn-, Zn-, and Co-binding proteins in the proteomes of Prokaryotes and Eukaryotes are similar to the theorized changes in the abundances of these metals after the oxygenation of oceanic deep waters. This phenomenon suggests that Prokarya and Eukarya evolved in anoxic and oxic environments, respectively, a hypothesis further supported by structures and functions of Fe-binding proteins in each Superkingdom. Also observed is a proliferation in the diversity of Zn-binding protein structures involved in protein–DNA and protein–protein interactions within Eukarya, an event unlikely to occur in either an anoxic or euxinic environment where Zn concentrations would be vanishingly low. We hypothesize that these conserved trends are proteomic imprints of changes in trace metal bioavailability in the ancient ocean that highlight a major evolutionary shift in biological trace metal usage.
Keywords: bioinorganic chemistry, evolution, fold families, structural bioinformatics
The emergence of oxygenic photosynthesis is associated with major changes in global biogeochemistry and metabolism (1, 2). In particular, the rise in atmospheric oxygen ≈2.3 billion years ago (Gya) (3, 4) potentially led to the oxygenation of the entire ocean (5), whereas an alternative theory proposes that the deep ocean became euxinic (anoxic and sulfidic) ≈1.8 Gya (6, 7), before an oxygenation of deep waters ≈1 Gya (8). Putting aside for now when and where, these changes in the overall redox state of the ocean would dramatically influence trace metal chemistry and bioavailability, with an anoxic ocean being characterized by relatively high Fe, Mn, and Co but low Zn concentrations (9) (Fig. 4, which is published as supporting information on the PNAS web site). A euxinic ocean would have comparatively lower concentrations of all of these metals, particularly Zn (9) (Fig. 4). The oxygenation of oceanic deep waters would have dramatically increased Zn concentrations, with concomitant yet less severe decreases in Fe, Mn, and Co levels (9) (Fig. 4). As postulated by Williams and Frausto da Silva (10), these drastic shifts in metal bioavailability theoretically influenced the selection of trace elements for biological usage, leaving a record within the genomes and proteomes of extant organisms.
Protein structure has a remarkable level of redundancy, with a limited number of 3D folds describing all of life (11). Further, structure is retained over long evolutionary time scales, even when most sequence homology is lost, providing an excellent tool for this study. Already, the identification of domains within protein structures and the systematic and hierarchical classification of these domains have been used to study evolution (12). Within these hierarchical classifications reside fold superfamilies (FSF) and fold families (FF); a FSF contains structures believed to be evolutionarily related despite a lack of clear sequence similarity, whereas a FF contains structures with evident structural, functional, and sequence similarities (a FSF is composed of one or more FF). The gain or loss of a FSF or FF by an organism constitutes an important evolutionary event, either reducing or expanding the repertoire of functions available to that organism. Indeed, the presence or absence of FSFs in a proteome has been shown to discriminate species well enough to construct reasonable phylogenetic trees for all of life (13).
Here, we used structural bioinformatics to study the distribution of metal-binding protein structures within the proteomes of Archaea, Bacteria, and Eukarya, which to our knowledge has not been done before. The results suggest that ancient changes in trace metal geochemistry do indeed leave imprints observable within the genomes and proteomes of modern life and provide an important constraint on the evolution of Eukarya.
Results and Discussion
The Superfamily database (14, 15), derived from the Structural Classification of Proteins (SCOP) (16), provides an independent assessment of the presence and abundance of structural domains belonging to FSFs and FFs across a diverse set of species for which complete genome and translated proteome sequences are available. To extract the desired information from the Superfamily database, we manually annotated SCOP version 1.69 according to metal binding (Tables 3–6, which are published as supporting information on the PNAS web site). Both the raw structural data from the Protein Data Bank (PDB) (17) and the primary literature associated with each structure were used to identify covalently bound metals. Protein domains that bind a metal-containing cofactor (e.g., Co-containing B12 and Fe-containing heme) were considered metal binding. Here, an ambiguous FSF is defined as one in which the structures comprising that FSF bind different metals or contain a combination of both metal- and nonmetal-binding structures. Likewise, an ambiguous FF contains a mixture of metal- and nonmetal-binding structures or structures binding different metals. Approximately half of the metal-binding FSFs and 10% of the metal-binding FFs are ambiguous (Table 7, which is published as supporting information on the PNAS web site). Only unambiguous FFs were used for this study. When this hand-curated literature annotation of the SCOP is combined with the unannotated Superfamily data, frequencies of Fe-, Mn-, Co-, and Zn-binding structural domains in the proteomes of 23 Archaea, 233 Bacteria, and 57 Eukaryote species are obtained, providing the “metallome” based upon protein structure and the whole proteome of an organism. Although metals such as Cu, Mo, and Ni are biologically relevant, given the limited number of FFs that bind these metals (<0.3% of the average proteome), we have excluded them from our analysis of the overarching trends that are the focus of this paper. These metals and the issue of ambiguous FFs and FSFs may be addressed in subsequent work.
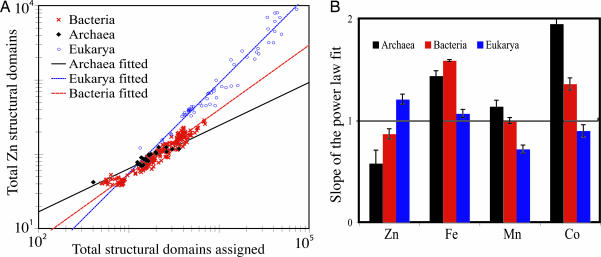
Using the metallome data, we observe that, although the percentage of a proteome that binds a given metal differs between the Superkingdoms (Fig. 5, which is published as supporting information on the PNAS web site), the abundances of metal-binding domains scale to proteome and genome size; therefore, stationary statistics like percentage are inappropriate for describing overall trends. Rather, the distributions of Fe-, Zn-, and Mn-binding domains within each Superkingdom conform to a power law (y = bxm) (Fig. 1 A, Table 1). Power law scaling to genome size has also been observed for functional categories of genes in Bacteria (18, 19); in contrast, the scaling category here describes an aspect of tertiary protein structure relatively independent of functional classification.
Fig. 1.
Power low scaling of metal-binding domains. (A) Log-log plot of the abundances of Zn-binding domains in Archaea (black ■), Bacteria (red x), and Eukarya (blue o) compared with the total number of structural domains in a proteome. Each point represents the number of metal-binding domains and the total number of assigned protein domains in a discrete proteome. The total number of structural domains annotated in a proteome scales linearly to both genome size and gene number (Fig. 6, which is published as supporting information on the PNAS web site). Also shown are the fitted power laws (black solid, Archaea; red dashed, Bacteria; blue dotted, Eukaryotes). (B) The power law slopes describing the abundances of Fe-, Zn-, Mn-, and Co/B12-binding structural domains in the proteomes of Archaea (black), Bacteria (red), and Eukarya (blue). The error bars denote 1 SD. The statistics for the quality of the power law fits are shown in Table 1. When the slopes of the curves are compared, they are significant as follows (A, Archaea; B, Bacteria; E, Eukaryote): Zn, all are significantly different at α = 0.5%. Fe, B vs. A and B vs. E α = 0.1%, A vs. E α = 5%. Mn, A vs. E, B vs. E α = 0.5%, A vs. B, not significantly different at α = 5%. Co, A vs. E α = 1%, B vs. E α = 5%, A vs. B not significantly different at α = 5%.
Table 1.
Statistics on the quality of the power law fits
| Superkingdom |
F values |
r2 values |
||||||
|---|---|---|---|---|---|---|---|---|
| Fe | Zn | Mn | Co | Fe | Zn | Mn | Co | |
| Archaea | 0.94 | 0.97 | 0.98 | 0.94 | 0.92 | 0.91 | 0.97 | 0.82 |
| Bacteria | 0.96 | 0.98 | 0.95 | 0.88 | 0.9 | 0.97 | 0.91 | 0.74 |
| Eukarya | 0.97 | 0.99 | 0.96 | 0.93 | 0.94 | 0.98 | 0.92 | 0.85 |
Both the F values, which describe the fraction of the variance in the data explained by the fitted curves, and the r2 values are shown.
Based upon the theory proposed by van Nimwegen (18), the observed power law slopes describe evolutionarily constant ratios for the size of a category of proteins relative to the size of the entire proteome. A slope of 1 indicates that the category is in equilibrium with proteome size, whereas a slope >1 indicates a preferential retainment of that category during increases in proteome size (and vice versa for a slope <1). Being empirically derived from a set of modern proteomes that resulted from different evolutionary trajectories, these ratios appear to be independent of the mechanism of proteome evolution (e.g., duplication, gene loss, horizontal gene transfer, and endosymbiotic events). Put another way, any event that changes the size and content of a proteome adheres to a given stoichiometry defined by power laws. Given this conserved behavior, the differences between the Superkingdoms are compelling. The power law slopes for Fe-, Mn-, and Co-/B12-binding FFs within the proteomes of Bacteria and Archaea are ≥1, but <1 for Eukarya (Fig. 1 B). This trend is reversed for the abundance of Zn-binding domains, with the Eukarya having a power law slope of >1 and the Prokaryotic proteomes exhibiting slopes ≤1 (Fig. 1 B). Notably, there appears to be an inflection point in the Eukarya between unicellular and multicellular organisms (Fig. 7, which is published as supporting information on the PNAS web site), but more complete proteomes are needed to robustly test this relationship.
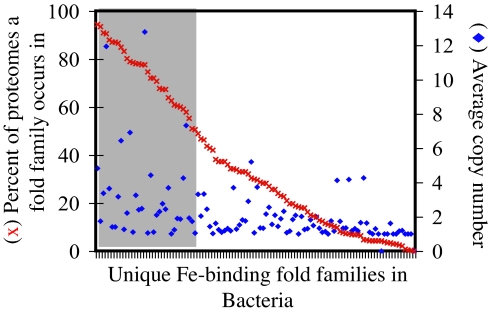
The observed scaling is not due to a core set of abundant metal-binding proteins (e.g., a metal-binding superfold) within a given Superkingdom; individual species have drawn broadly and diversely from the pool of available metalloproteins. That is, very few metal-binding domains are ubiquitous or are found within all of the proteomes of a Superkingdom, yet many are present in at least one proteome (e.g., see Fig. 2 for the case of Fe in Bacteria). Additionally, structural domains found in all or most of the proteomes are not necessarily more abundant in those proteomes than structural domains found in only a few proteomes (Fig. 2). Essentially, different organisms have different metal-binding domains, a logical extension of the results of Yang et al. (13), yet the total abundances of Fe-, Mn-, Zn-, or Co-binding domains within a proteome conform to fundamental constants defined by power laws.
Fig. 2.
Diversity and abundance of Fe-binding fold families in Bacteria. For each Fe-binding fold family (tick marks on x axis), the red × (left axis for scale) shows the percentage of proteomes in which it occurs, whereas the blue ♦ (right axis for scale) shows the average copy number in proteomes where it does occur. The shaded area highlights the number of fold families that occur in at least 50% of the Bacterial proteomes examined. Similar trends are observed for the other metals and Superkingdoms.
It appears that methodological limitations, including proteome coverage and sampling bias, do not contribute to the observed trends. According to Superfamily (14), on average 55% of Archaeal and Bacterial proteomes and 40% of Eukaryotic proteomes have fold families assigned. Although this coverage may seem limiting, results from the Protein Structure Initiative suggest that ≈90% of this unannotated space is comprised of variants of already discovered fold families (20), and that only 10% of the undiscovered fold families will actually be metal-binding (21). It appears that membrane proteins are similarly distributed, with the most abundant membrane folds being already described (22). Essentially, it seems unlikely that a new protein fold family will be discovered that is abundant enough to overly skew the observed results.
A further concern is that the observed results are biased by the available whole genomes. The Archaea sequenced are mostly thermophiles from anoxic environments (although this potentially provides a modern-day glimpse into ancient bioinorganic chemistry), whereas the sequenced Eukarya are almost entirely aerobic. The dataset does include the Eukaryotic anaerobic amitochondritic parasite Encephalitozoon cuniculi, which has metallomic features typical of aerobic Eukaryotes. In contrast, the analyzed Bacteria are from a broad array of environments and have a variety of oxygen tolerances and therefore can be used to gain a preliminary understanding of the influence of modern environment and metabolism on metallomic content. Surprisingly, within the Bacterial Superkingdom, differences in oxygen tolerance do not seem to influence the proteomic abundance of metal-binding domains (Fig. 8, which is published as supporting information on the PNAS web site). Instead, Phylum or Classes roughly group together (Fig. 9, which is published as supporting information on the PNAS web site), implying that the observed stoichiometries are vertically inherited. Future work will explore the causes of the nonsize-dependent variance within the data (<10%; see Table 1). Additionally, more genome and proteome sequences will allow for a continued updating of these results, with the proteomes of aerobic Archaea and anaerobic Eukaryotes providing key tests.
Accepting that the data are not limiting, the critical question remains as to the source of the Superkingdom level differences in the power law slopes. It seems reasonable to assume that the observed power law slopes are determined by selective pressure (23), and that trace metal bioavailability can produce such pressure (24). Hence, we hypothesize that the environmental bioavailability of trace elements during major periods of phylogenetic diversification shape the evolution of vertically inherited metal homeostasis systems that then continually influence the retention and loss of genetic material. The differences in the power law scaling for metal-binding structures within Prokaryotic and Eukaryotic proteomes are similar to the shifts in trace metal bioavailability caused by increasing oxygen, implying that Prokaryotic and Eukaryotic organisms diversified in anoxic and oxic environments, respectively. The proposed theory entails a closed feedback loop, whereby a biological phenomenon (cyanobacterial production of oxygen) incites a shift in trace metal geochemistry that in turn influences the evolution of bioinorganic chemistry. An alternative theory is that the observed differences between the Prokaryotes and Eukaryotes are due to an unknown but environmentally unrelated phenomenon. To address this possibility, we further examined the functions and structures of Zn- and Fe-binding proteins for environmentally consistent signals.
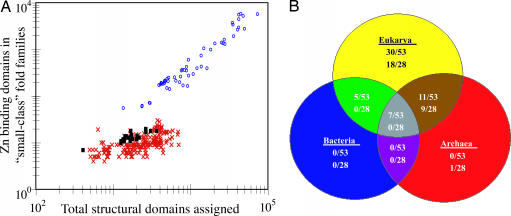
As stated, Eukaryotic and Prokaryotic proteomes show significant differences in the abundance of Zn-binding domains. These are wholly attributable to structures in the “small protein” structural class (Fig. 3A), typified by small Zn-binding domains such as Zn fingers and RING domains involved in protein–DNA/RNA interactions and protein–protein interactions, respectively. Eukaryotic proteomes also encode for a greater structural diversity of “small protein class” fold families that bind Zn (Fig. 3B), a noteworthy radiation in the diversity and usage of Zn within proteins, one that is predominantly structural. Most of the “small protein” class Zn-binding protein fold families are unique to Eukarya, although a significant subset is shared with Archaea (Fig. 3B). Because Zn concentrations would be vanishingly low in an anoxic or euxinic environment (ref. 9; Fig. 4), it seems unlikely that such a diversification in the biological usage of Zn could occur under such conditions.
Fig. 3.
The abundance and diversity of “small protein” class Zn-binding structures. (A) Log-log plot of the abundance of Zn-binding domains belonging to the “small protein” structural class in proteomes of Archaea, Bacteria, and Eukarya (symbols are the same as in Fig. 1). (B) The phylogenetic distribution of “small protein” class Zn-binding fold families. There are 53 distinct “small protein” class Zn-binding fold families that occur in at least one proteome, and the distribution of these is described by the top set of numbers in each set. The bottom numbers of each set detail distribution of fold families that occur in at least 50% of the proteomes of a Superkingdom (28 “small protein” class Zn-binding fold families occur in at least 50% of the proteomes of at least one Superkingdom). The lists of FFs in each category within the diagram are provided in Table 8, which is published as supporting information on the PNAS web site.
The functions and structures of the prevalent Fe-binding domains in each Superkingdom also are consistent with the evolution of Eukarya in an oxic environment. Fe-binding FFs were characterized according to the mode of Fe binding (Fe-S, heme, or direct amino acid), and the abundances of these binding forms were quantified for each Superkingdom (Table 2). Archaeal and Bacterial metallomes have significantly more Fe-S proteins and fewer heme proteins than the Eukaryotic metallomes (Table 2). Both the observed Fe-S clusters and hemes function in e− transfer reactions, but Fe-S clusters are oxygen-sensitive and have more negative reduction potentials than heme-based Fe proteins such as cytochromes (25). The proteomes of aerobic Bacteria also contain fewer Fe-S clusters and more hemes than anaerobic Bacteria (Table 9, which is published as supporting information on the PNAS web site), suggesting that the actual repertoire of metalloproteins within the constrained totals may partially reflect physiological adaptations in addition to evolutionary history. Functionally, the most abundant Fe-binding domains in Archaea and Bacteria are involved in electron transfer, vitamin/cofactor biosynthesis, or dissolved gas sensing, with most of the catalyzed reactions excluding oxygen (Table 2). In contrast, the prevalent Eukaryotic Fe-binding domains catalyze a wide variety of mostly oxygen-dependent interactions, with the abundance of the hypoxia induction factor proteins being particularly telling (Table 2). Note that some of the “non-O2” domains do participate in O2-dependent pathways, but that they do not actually contact oxygen. Because structural protein domains can make diverse combinations (26), we feel that the direct interaction with O2 is more pertinent to the issue at hand than a downstream or upstream interaction.
Table 2.
The function and structure of abundant Fe-binding domains in each Superkingdom
| Superkingdom | Fold families | Percentage | Fe binding | O2 | Overall percentage of Fe bound by |
||
|---|---|---|---|---|---|---|---|
| Fe-S | Heme | Amino | |||||
| Eukarya | Cytochrome P450 | 0.44 ± 0.48 | Heme | Yes | |||
| Cytochrome c3-like | 0.13 ± 0.3 | Heme | No | ||||
| Cytochrome b5 | 0.12 ± 0.09 | Heme | No | ||||
| Purple acid phosphatase | 0.11 ± 0.08 | Amino | No | 21 ± 9 | 47 ± 19 | 32 ± 12 | |
| Penicillin synthase-like | 0.07 ± 0.1 | Amino | Yes | ||||
| Hypoxia-inducible factor | 0.07 ± 0.04 | Amino | Yes | ||||
| Di-heme elbow motif | 0.06 ± 0.01 | Heme | No | ||||
| Archaea | 4Fe-4S ferredoxins | 1.80 ± 0.7 | Fe-S | No | |||
| MoCo biosynthesis proteins | 1.60 ± 0.3 | Fe-S | No | ||||
| Heme-binding PAS domain | 1.10 ± 1.0 | Heme | * | ||||
| HemN | 0.80 ± 0.20 | Fe-S | No | 68 ± 12 | 13 ± 14 | 19 ± 6 | |
| α helical ferrodoxin | 0.60 ± 0.16 | Fe-S | No | ||||
| Biotin synthase | 0.55 ± 0.1 | Fe-S | No | ||||
| ROO N-terminal domain-like | 0.5 ± 0.1 | Amino | No | ||||
| Bacteria | High potential iron protein | 0.38 ± 0.25 | Fe-S | No | |||
| Heme-binding PAS domain | 0.3 ± 0.4 | Heme | * | ||||
| MoCo biosynthesis proteins | 0.21 ± 0.15 | Fe-S | No | ||||
| HemN | 0.2 ± 0.15 | Fe-S | No | 47 ± 11 | 22 ± 12 | 31 ± 16 | |
| 4Fe-4S ferredoxins | 0.2 ± 0.2 | Fe-S | No | ||||
| Cytochrome c | 0.14 ± 0.2 | Heme | No | ||||
| α helical ferrodoxin | 0.12 ± 0.09 | Fe-S | No | ||||
The seven most abundant fold families in each Superkingdom are listed, along with the average percentage of a proteome they comprise, the mode of Fe binding, and whether oxygen is directly involved in the catalyzed reactions. The final three columns show the overall percentage of Fe-S, heme, or amino acid binding in the Fe-binding proteome for each Superkingdom.
*Some, but not all, PAS domains actually sense oxygen.
Evidence from the chemical fossil record and phylogenetic studies also corroborates the idea that the Prokaryotic and Eukaryotic Superkingdoms arose separately in anoxic and oxic environments, respectively. The Bacterial and Archaeal Superkingdoms certainly had undergone extensive diversification before the emergence of oxygenic photosynthesis (27). Eukaryotic-specific lipid biomarkers (which notably require oxygen to synthesize) have been found in rocks from 2.7 Gya (28), whereas molecular clock studies date the early diversification events of Eukarya to the late Proterozoic (0.9–1.2 Gya; ref. 29). The deep ocean was potentially anoxic or euxinic during both of these periods (30); these data have been used by some to argue that Eukarya evolved and diversified in anaerobic environments (31). This contention is contrary to the theory proposed by Anbar and Knoll that low Cu and Mo bioavailability in a euxinic ocean limited Eukaryotic diversification (30). Our results support the latter hypothesis that oxygen-induced changes in trace metal bioavailability occurred before the diversification of Eukarya and implicate Zn as another relevant metal. The fossil record indicates early evolutionary radiations of Eukarya likely occurred in shallow coastal environments (32, 33), where a combination of high oxygen concentrations and a terrestrial supply of trace metals may have increased the bioavailability of Zn, Cu, and Mo. Note that, although oxic microenvironments may have existed in the surface ocean since the advent of oxygenic photosynthesis, the supply of trace metals to these microenvironments would have been unchanged until the oxygenation of deep waters, in contrast to coastal environments.
The idea that the rise in oxygen affected the usage of trace metals was originally proposed by Williams and Frausto da Silva (10), and a few studies have used sequence-based methods to study the coevolution of biology and geochemistry. Morgan et al. (34) found that Eukarya have a higher diversity and abundance of Ca+2-binding protein sequence families. Zerkle et al. (35) examined the distributions of ORFs annotated as known metal-binding proteins within the genomes of Prokarya, finding differences based upon metabolism and phylogeny. The analysis conducted here expands on these theories and efforts. Within a proteome, the abundances of metal-binding domains conform to a stoichiometry defined by evolutionary constants despite the wide diversity of physiologies and environments of the analyzed organisms. Further, these constants exhibit Superkingdom-specific behavior consistent with development within anoxic vs. oxic environments. It must be noted that the observed proteomic stoichiometries likely do not define the physiological metal requirements of a specific organism. Single metalloproteins can constitute a large portion of an organism's metal usage. However, whole proteomes are less susceptible to gene acquisition events or evolutionarily recent ecological or physiological adaptations, such as those observed in coastal and open ocean cyanobacteria (36). Hence, we feel that the whole-proteome patterns observed represent a broader and more durable view into the ancient environment of Earth than physiological quotas.
Methods
Data Sources.
SCOP (16) provides a hierarchical classification of all protein domains published in the PDB (17). SCOP Version 1.69 has sorted 70,800 domains into 945 defined folds that are assigned to 1,539 superfamilies and further subdivided into 2,845 families. The Superfamily database (14, 15) was the source of all domain assignments; Release 1.69 covers 313 complete genomes (23 Archaea, 233 Bacteria, and 57 Eukaryota). The Superfamily database, using a hybrid approach of a hidden Markov model searching protocol and subsequent pairwise comparisons (15), uses a probability cutoff of E = 2 × 10−2 for identifying likely members of a group; it also provides a confidence level (in the form of an E value) for every candidate identified. As was done by Yang et al. (13), a more stringent E value cutoff of 10−4 was used for the domain assignments here.
Annotation of SCOP per Metal Binding.
Each FSF in the SCOP database was manually examined for structures containing a covalently bound inorganic ion. This objective requires examining the FF, fold domains, and specific example structures within each FSF. For a FSF or FF to be considered metal-binding, only one of the representative structures has to contain a bound metal. If all of the representative structures in a FSF or FF bind the same metal, the family is considered unambiguous; only these FFs were used for this study.
Automated annotations of SCOP in this fashion are inhibited by two factors: (i) Some structures are crystallized with nonnative metals, and (ii) some PDB data files are less thorough in the description of binding mode and domain. The manual examination procedures were simplistic; the accompanying PDB file was examined for an inorganic ion and covalent binding of that ion. Some PDB files provide metal-binding information (i.e., heme, amino acid, and specific binding residues), and whether the metal is native or simply part of the crystallization buffer. In the cases where this was not clear, the primary literature citation was examined. Attention was also paid to which structural domains actually bind the metal. For example, there are numerous distinct FSFs and FFs that contain domains of cytochrome c oxidase in the SCOP, and each entry states “complexed with cdl, chd, cu, cua, dmu, hea, mg, na, pek, pgv, psc, tgl, unx, zn,” yet only a select few are Cu-, Fe-, Mg-, or Zn-binding. The FFs that unambiguously bind Fe, Zn, Mn, or Co are shown in Tables 3–6.
Data Management and Analysis.
Matrices were constructed, with each row representing a distinct species and each column representing the abundances of a specific metal-binding FF in each species. For the power law distributions in Fig. 1, the FFs in a given proteome that unambiguously bind a metal were summed and plotted against the total number of domains assigned to that proteome (the sum of all FF assignments for a proteome). For Table 2, the matrices were normalized by dividing the abundances of each FF within a species by the total number of structural domains assigned to that species' proteome. These internal percentages were then averaged over the entire Superkingdom.
Power law fits were determined in Matlab (Mathworks, Natick, MA). The data were log-transformed, and the linear fit was found by using a geometric mean least-squares fitting technique. Groupings of points were compared by using ANCOVA (the aoctool function in Matlab). Slopes were compared by using the multicompare function. Power law fit qualities (F values; Table 1) were determined by using the method of van Nimwegen (18). Briefly, the data were log-transformed. The distance from each point (defined by xi, yi) to the center of the scatter (Dc) is determined by Dc = √ ((xi − mean of all x)2 + (yi − mean of all y)2). Then the distance of each point to the fitted line (Dl) is determined by Dl = √((yi –mxi − b)2/(m2 +1)), where m and b equal the power law slope and intercept. Then the fraction of the variance explained by the data is given by F = 1 − (Σ(Dl)2)/(Σ(Dc)2).
Supplementary Material
Acknowledgments
We thank K. N. Chang, A. J. Lucas, R. J. P. Williams, S. Veretnik, and anonymous reviewers for constructive comments and suggestions. P.E.B. was supported by National Institutes of Health Grant GM63208, and C.L.D. is grateful for funding from a National Defense Science and Engineering Graduate Research Fellowship and the National Science Foundation/Department of Energy-supported Princeton Center for Bioinorganic Chemistry.
Abbreviations
- FSF
fold superfamily
- FF
fold family
- SCOP
structural classification of proteins
- PDB
Protein Data Bank
- Gya
billion years ago.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Kopp RE, Kirschvink JL, Hilburn IA, Nash CZ. Proc Natl Acad Sci USA. 2005;102:11131–11136. doi: 10.1073/pnas.0504878102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raymond J, Segre D. Science. 2006;311:1764–1767. doi: 10.1126/science.1118439. [DOI] [PubMed] [Google Scholar]
- 3.Bekker A, Holland HD, Wang PL, Rumble D, Stein HJ, Hannah JL, Coetzee LL, Beukes NJ. Nature. 2004;427:117–120. doi: 10.1038/nature02260. [DOI] [PubMed] [Google Scholar]
- 4.Farquhar J, Bao H, Thiemens M. Science. 2000;289:756–758. doi: 10.1126/science.289.5480.756. [DOI] [PubMed] [Google Scholar]
- 5.Holland HD. The Chemical Evolution of the Atmosphere and Oceans. Princeton: Princeton Univ Press; 1984. [Google Scholar]
- 6.Canfield DE, Teske A. Nature. 1996;382:127–132. doi: 10.1038/382127a0. [DOI] [PubMed] [Google Scholar]
- 7.Arnold GL, Anbar AD, Barling J, Lyons TW. Science. 2004;304:87–90. doi: 10.1126/science.1091785. [DOI] [PubMed] [Google Scholar]
- 8.Canfield DE. Nature. 1998;396:450–453. [Google Scholar]
- 9.Saito MA, Sigman DM, Morel FMM. Inorg Chim Acta. 2003;356:308–318. [Google Scholar]
- 10.Williams RJP, Frausto da Silva JJR. The Chemistry of Evolution: The Development of Our Ecosystem. Amsterdam: Elsevier; 2006. [Google Scholar]
- 11.Chothia C. Nature. 1992;357:543–544. doi: 10.1038/357543a0. [DOI] [PubMed] [Google Scholar]
- 12.Koonin EV, Wolf YI, Karev GP. Nature. 2002;420:218–223. doi: 10.1038/nature01256. [DOI] [PubMed] [Google Scholar]
- 13.Yang S, Doolittle RF, Bourne PE. Proc Natl Acad Sci USA. 2005;102:373–378. doi: 10.1073/pnas.0408810102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gough J, Karplus K, Hughey R, Chothia C. J Mol Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 15.Gough J. Nucleic Acids Res. 2006;34:3625–3633. doi: 10.1093/nar/gkl484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murzin AG, Brenner SE, Hubbard T, Chothia C. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 17.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Nimwegen E. Koonin EV, Wolf YI, Karev GP. Power Laws, Scale-Free Networks, and Genome Biology. Austin, TX: Eurekah; 2006. [Google Scholar]
- 19.Konstantinidis KT, Tiedje JM. Proc Natl Acad Sci USA. 2004;101:3160–3165. doi: 10.1073/pnas.0308653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandonia J, Brenner SE. Science. 2006;311:347–351. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- 21.Shi W, Zhan C, Ignatov A, Manjasetty BA, Marinkovic N, Sullivan M, Huang R, Chance MR. Structure (Cambridge, UK) 2005;13:1473–1486. doi: 10.1016/j.str.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 22.Oberai A, Ihm Y, Kim S, Bowie JU. Protein Sci. 2006;15:1723–1734. doi: 10.1110/ps.062109706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Nimwegen E. Trends Genet. 2003;19:479–484. doi: 10.1016/S0168-9525(03)00203-8. [DOI] [PubMed] [Google Scholar]
- 24.Williams RJP, Frausto da Silva JJR. J Theor Biol. 2003;220:323–343. doi: 10.1006/jtbi.2003.3152. [DOI] [PubMed] [Google Scholar]
- 25.Lippard SJ, Berg JM. Principles of Bioinorganic Chemistry. Mill Valley, CA: University Science Books; 1994. [Google Scholar]
- 26.Apic G, Gough J, Teichmann SA. J Mol Biol. 2001;310:311. doi: 10.1006/jmbi.2001.4776. [DOI] [PubMed] [Google Scholar]
- 27.Xiong J, Fischer WM, Inoue K, Nakamura M, Bauer CE. Science. 2000;289:1724–1730. doi: 10.1126/science.289.5485.1724. [DOI] [PubMed] [Google Scholar]
- 28.Brocks JJ, Logan GA, Buick R, Summons RE. Science. 1999;285:1033–1036. doi: 10.1126/science.285.5430.1033. [DOI] [PubMed] [Google Scholar]
- 29.Douzery EJP, Snell EA, Bapteste E, Delsuc F, Philippe H. Proc Natl Acad Sci USA. 2004;101:15386–15391. doi: 10.1073/pnas.0403984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anbar AD, Knoll AH. Science. 2002;297:1137–1142. doi: 10.1126/science.1069651. [DOI] [PubMed] [Google Scholar]
- 31.Theissen U, Hoffmeister M, Grieshaber M, Martin W. Mol Biol Evol. 2003;20:1564–1574. doi: 10.1093/molbev/msg174. [DOI] [PubMed] [Google Scholar]
- 32.Javaux EJ, Knoll AH, Walter MR. Nature. 2001;412:66–69. doi: 10.1038/35083562. [DOI] [PubMed] [Google Scholar]
- 33.Knoll AH, Javaux EJ, Hewitt D, Cohen P. Philos Trans R Soc. 2006;361:1023–1038. doi: 10.1098/rstb.2006.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morgan RO, Martin-Almedina S, Iglesias JM, Gonzalez-Florez MI, Fernandez MP. Biochim Biophys Acta. 2004;1742:133–140. doi: 10.1016/j.bbamcr.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Zerkle AL, House CH, Brantley SL. Am J Sci. 2005;305:467–502. [Google Scholar]
- 36.Palenik B, Ren Q, Dupont CL, Myers GS, Heidelberg JF, Badger JH, Madupu R, Nelson WC, Brinkac LM, Dodson RJ, et al. Proc Natl Acad Sci USA. 2006;103:13555–13559. doi: 10.1073/pnas.0602963103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.