Abstract
Myosin-VI is a dimeric isoform of unconventional myosins. Single molecule experiments indicate that myosin-VI and myosin-V are processive molecular motors, but travel toward opposite ends of filamentous actin. Structural studies show several differences between myosin-V and VI, including a significant difference in the light-chain domain connecting the motor domains. Combining the measured kinetics of myosin-VI with the elasticity of the light chains, and the helical structure of F-actin, we compare and contrast the motility of myosin-VI with myosin-V. We show that the elastic properties of the light-chain domain control the stepping behavior of these motors. Simple models incorporating the motor elastic energy can quantitatively capture most of the observed data. Implications of our result for other processive motors are discussed.
INTRODUCTION
Besides muscle contraction, various isoforms of myosin are involved in cytoskeleton rearrangement, adhesion regulation, transport, and stereocilia repositioning (1–4). Single-molecule experiments show that myosin-V and myosin-VI are dimeric molecular motors that travel processively on actin filaments (1,5–15). Several lines of evidence suggest that these motors move via a hand-over-hand mechanism where the motor domains take regular steps of ∼30–36 nm (5–8,10–17). However, there are several differences between myosin-V and VI. While myosin-V processively travels toward the plus (barbed) end of F-actin, myosin-VI travels toward the minus (pointed) end (17–21). The step-size distribution of myosin-VI is also broader than myosin-V, indicating that myosin-VI takes irregular steps (10). In this article, we compare and contrast the biophysical properties of myosin-V and -VI using a unified mechanochemical model. We show that the elasticity of the light-chain domain and the helical geometry of the actin binding sites ultimately determine the preferred step size. Using a simplified kinetic scheme based on an earlier model of myosin-V (22), and a worm-like-chain (WLC) model for the light-chain domain, we compute the force-velocity relationship and the step-size distributions of myosin-VI. Implications of our result for other processive molecular motors are discussed.
Atomic structures of myosin-V and -VI monomers have been solved (19,23–26). The monomers contain a head (motor) domain, a converter, and a light-chain domain. Comparing the x-ray structures shows that there are three major structural differences distinguishing myosin-VI from myosin-V. First, there is an insert (∼50 residues) in the converter domain of myosin-VI, which reverses the swinging motion of the light-chain domain (1,3,4,13,18,19,27). Second, there is only one calmodulin-binding IQ motif in the light-chain domain of myosin-VI (1,3,4,19), instead of six in myosin-V (9,27–29), although the converter domain binds calmodulin as well (30). Third, the IQ motif is followed by a proximal tail region (∼83 residues) which appears to be an unfolded coiled-coil (10,11). In this article, we shall refer to the IQ motif and the proximal tail region combined as the light-chain domain of myosin-VI. Rock et al. (10,11) extensively examined the proximal tail region and concluded that it is flexible, and allows the motor domains to extend much further than a closed coiled-coil. Rock et al. (10,11) proposed that the light-chain domain acts as a soft spring, thus allowing the binding of 30–36 nm actin sites. These structural studies form the basis of the present model and ultimately explain the step-size distribution of myosin-VI.
Modeling of dimeric myosins largely followed two methodologies. Method One models the processive movement as a sequence of reactions characterized by reaction rate constants (13,31). To explain experimental data, fitting of rate constants is usually required. Method Two models the energetic landscape of the dimeric motor and use experimental kinetic data for the monomers to parameterize the model (22,32). Approximate structures of the motors are incorporated. Emphasis is placed on how the elastic energy of the motor complex modifies the rate constants. Regular steps and substeps are outcomes of the model (22). With the additional assumption that ADP release is gated by external force, the hand-over-hand mechanism emerges. Experiments on myosin monomers have shown that ADP release is indeed affected by load forces (6,9,33–35), although there is some controversy regarding which motor domain is rate-limited. Recently, we used Method Two to model the movement of myosin-V (22). This article follows the same methodology, but a slightly simpler kinetic scheme is used. Three-dimensional geometry of the motor binding sites and the elastic properties of the light-chain domains are incorporated. The computational results explain the broad step-size distributions of myosin-VI and the observed dwell times (13).
The general framework presented here is also applicable to other dimeric motors such as kinesin and dynein although the detailed parameterization depends on the biophysical properties of those molecules. Thus, the presentation focuses on the general framework first; the specific parameters are given in the Appendix.
METHODS
Motions of molecular motors can be understood within an energy landscape framework where the motor energy depends at least on two variables, E(ξ, s). The value ξ denotes the conformational variable: it describes the movement of protein structure as it responds to thermal fluctuations or other external forces. The value s denotes the chemical state of the proteins: it describes whether the motor domains are bound to actin and the nucleotide occupancy of the catalytic sites. Each motor domain can be in any of 10 possible states: the F-actin bound states are A.M.E, A.M.T*, A.M.T, A.M.DP, and A.M.D; the actin-free states are M.E, M.T*, M.T, M.DP, and M.D (36–38). Therefore, the total number of chemical states of the dimer is 100. For dimeric motors, ξ should reveal any possible structural asymmetry in the dimer. For the present treatment, we use four variables: ξ=(θ1, φ1, θ2, φ2), labeling the directions of the light chain in motors 1 and 2, respectively (see Fig. 1).
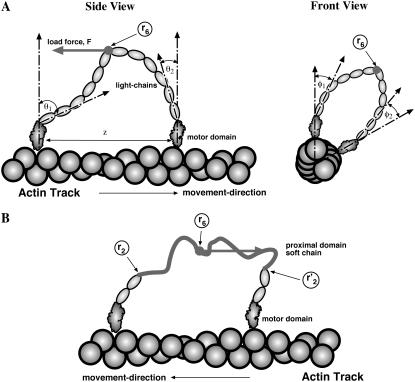
FIGURE 1.
A cartoon depiction of myosin-V and myosin-VI. (A) Conformations of myosin-V can be described by (θ1, φ1, θ2, φ2, z). The light-chain domains connecting the motor domains are described by a semiflexible rod model. The joint, r6, is free to bend and rotate. (B) Conformations of myosin-VI are described by in a similar manner with the same variables (not shown). The proximal domains between r2 and r6, and 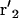 and r6, are soft. They are modeled by a worm-like-chain model.
and r6, are soft. They are modeled by a worm-like-chain model.
Experiments have not measured the myosin motor energy as a function of the conformational variables. However, a simple model can capture most of the important features of myosin-V and -VI. We write the dimer energy as
 |
(1) |
Here E0(θi, φi, si) is the monomer energy as a function of its conformation. This energy depends on the chemical state of the monomer, si, and contains information about the magnitude and direction of the power-stroke. The value El is the elastic energy of the light-chain domain linking the two motor domains. Since it is an elastic energy, it is not a function of the motor chemical states. However, it does depend on the forces coming from the motor monomers. More precisely, the motor domains provide boundary conditions, (θ1, φ1, θ2, φ2), on the possible conformations of the light chains. When both motors are bound, El also depends on the relative separation of the binding sites, z (see Fig. 1). When a single motor is bound, El is only a function of the bound motor conformation. Lastly, any externally applied load force F also affects the light-chain conformation.
Without performing any computations, one can see that Eq. 1 predicts an asymmetrical structure when s1=s2=A.M.D. In this state, both motor domains would like to have the same conformation and will tend to orient (θ1, φ1) and (θ2, φ2) in the same direction. However, El resists such geometries. Solving for a global optimal which is equivalent to mechanical equilibrium will reveal that (θ1, φ1) ≠ (θ2, φ2).
Transitions between the chemical states are specified by rates  . In principle, the transition rates are functions of the conformational variables (θi, φi). Indeed, this dependence gives rise to gating, where one of the motor domains appears to release ADP faster than the other. Physically, gating arise from the dependence of the catalytic activity on enzyme conformation. In purified protein experiments with motor monomers, the measured rate constants correspond to reaction rates at monomer conformational equilibrium, (θ0(s), φ0(s)). In the present treatment, we do not include all the chemical states. Rather, a simplified kinetic scheme is used (see below). The simplifications do not adversely affect the results.
. In principle, the transition rates are functions of the conformational variables (θi, φi). Indeed, this dependence gives rise to gating, where one of the motor domains appears to release ADP faster than the other. Physically, gating arise from the dependence of the catalytic activity on enzyme conformation. In purified protein experiments with motor monomers, the measured rate constants correspond to reaction rates at monomer conformational equilibrium, (θ0(s), φ0(s)). In the present treatment, we do not include all the chemical states. Rather, a simplified kinetic scheme is used (see below). The simplifications do not adversely affect the results.
Given the energy landscape and the reaction rates, the dynamics of the dimer can be obtained from a Langevin equation or a Fokker-Planck equation (40). In most situations, conformational fluctuations are much faster than changes in the chemical state such as hydrolysis and binding to actin. Therefore, the reactions are not diffusion-limited and the waiting times of conformational changes can be neglected. The Fokker-Planck equation becomes equivalent to a master equation where the rate constants are functions of the motor conformation given by the overall energy (41). In the following subsections, we discuss the detailed specifications of the myosin-V and VI energy and rate constants.
Simplified kinetic model
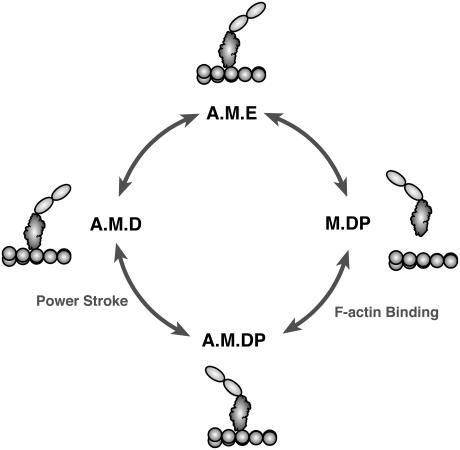
Our previous model of myosin-V included all 100 chemical states (22). However, it was found that each motor goes through essentially the same cycle of chemical states (shown as the red path in Fig. 2), although the phases of the cycles are different. In the present treatment, we simplify the problem by considering the rigor state (A.M.E), the actin-detached state (M.DP), the actin-attached state (A.M.DP), and the post-power-stroke state (A.M.D). Thus, the total number of states is reduced to 42=16. We will show that this simplification captures the dominant dynamical pathways of the motor, and can quantitatively explain the observed data.
FIGURE 2.
The simplified kinetic cycle in a myosin monomer. There are actually 10 possible chemical states. In the present treatment, this is reduced to four. In the motor dimer, both monomers can proceed through these states. The rate constants are regulated by the elastic energy, and are functions of the motor conformations.
As emphasized earlier, the transition rates between the chemical states depend on the motor conformation,  . More precisely, since the relaxation of motor conformation is rapid, rates depend on the dimer equilibrium conformation given by Eq. 1. For given occupancies of the catalytic sites, the dimer equilibrium conformations, (θi, φi), are obtained by solving
. More precisely, since the relaxation of motor conformation is rapid, rates depend on the dimer equilibrium conformation given by Eq. 1. For given occupancies of the catalytic sites, the dimer equilibrium conformations, (θi, φi), are obtained by solving
 |
(2) |
The dimer equilibrium conformation is different from the monomer equilibrium conformation, (θ0(si), φ0(si)). The rate constants at monomer equilibrium conformation have been measured for myosin-V and myosin-VI. As before, we use the measured data to parameterize 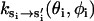 .
.
A.M.E → M.DP
This simplified step actually describes transitions through A.M.T*, A.M.T, M.T, and M.DP. The conformational dependence of ATP binding has not been measured. Since ATP binding depends on the openness of the binding pocket of A.M.E state, a dependence is expected. Conformational dependence of ATP binding in myosin-VI has been observed (39). We describe the dependence by a sigmoid function constructed using a combination of tanh functions. At the middle of the sigmoid, the rate constant depends exponentially on θi. We also assume that the rate is independent of φi. The function is
 |
(3) |
where  is the measured binding rate at conformational equilibrium. The function is designed so that
is the measured binding rate at conformational equilibrium. The function is designed so that  . For myosin-VI the parameters are Δ1=−7° and ε1=7. These parameters are not optimized, but are estimated to qualitatively reproduce experimental data. For myosin-V, the rate-limiting step is actually the loose to tight binding of ATP, A.M.T* → A.M.T. The overall rate is somewhat insensitive to θi.
. For myosin-VI the parameters are Δ1=−7° and ε1=7. These parameters are not optimized, but are estimated to qualitatively reproduce experimental data. For myosin-V, the rate-limiting step is actually the loose to tight binding of ATP, A.M.T* → A.M.T. The overall rate is somewhat insensitive to θi.
M.DP → A.M.DP
This step describes myosin binding to actin. Since we are modeling processive movement, binding of the free motor domain to actin occurs while the other motor domain is already bound. The binding step involves the diffusive search of the free motor domain, and in principle, all the available binding sites, including backward sites, are possible for binding. Note that this diffusive search is not free diffusion, because positioning the free motor domain at different sites results in different conformational energies. For myosin-V, the light-chain domain is relatively stiff. For myosin-VI, the light-chain domain is soft and flexible. Binding to F-actin is controlled by the elastic energy in the light-chain domains, given by Eq. 1. The conformational energy shifts the equilibrium between the bound and unbound states. We incorporate this change in the binding rate constant by writing  , where ΔE is the energy change before and after binding to F-actin,
, where ΔE is the energy change before and after binding to F-actin,
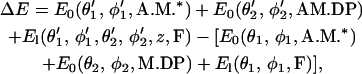 |
(4) |
where (θ′1,φ′1, θ′2, φ′2) are the equilibrium conformations of the dimer after both motor domains are bound to F-actin. Before binding to actin, only one of the light chains bears the load force and E1 is a function of (θ1, φ1) only. Because this energy difference is a function of z or the distance between the bound motor domains, the binding rates to the available sites are different. This leads to preferential binding to 36-nm site for myosin-V, and 30-nm site for myosin-VI.
Note that this specification of binding rate already contains the timescale associated with the diffusion of myosin monomer. This timescale is estimated by 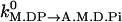 . The exact numerical dependence of the rate on ΔE is related to the energy landscape associated with this step. The form used here assumes that the transition state energy is increased by the change in elastic energy, ΔE. This form is also equivalent to a mean first-passage time analysis where the reaction rate is proportional to
. The exact numerical dependence of the rate on ΔE is related to the energy landscape associated with this step. The form used here assumes that the transition state energy is increased by the change in elastic energy, ΔE. This form is also equivalent to a mean first-passage time analysis where the reaction rate is proportional to 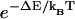 . Our earlier model used a general form
. Our earlier model used a general form 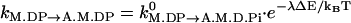 , where λ is between 0 and 1. The value λ=1 is consistent with the assumption that the energy of the transition state is dominated by the elastic energy of the light chains.
, where λ is between 0 and 1. The value λ=1 is consistent with the assumption that the energy of the transition state is dominated by the elastic energy of the light chains.
A.M.DP → A.M.D
This step describes phosphate release after hydrolysis. We assume it is independent of conformation: 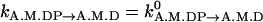 .
.
A.M.D → A.M.E
This step describes ADP release, which has been shown to be a sensitive function of conformation. Following our earlier model, we use a combination of tanh functions, similar to the ATP binding step,
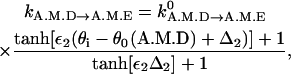 |
(5) |
where for myosin-VI, Δ2=0.5° and ε2=100; for myosin-V, Δ2=15° and ε2=6. At the preferred conformation θ0(A.M.D), the rate equals to the equilibrium ADP release rate 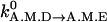 .
.
In these specifications of reaction rates, the out-of-plane angle φ does not influence the rate constants. In reality, myosin-V mutants with shorter light chains process more quickly than wild-type, suggesting that φ has an influence on the rate (27). We have not considered a more complicated rate function.
The reverse reaction rates are all obtained from the detailed balance condition. Since the total energy of the dimer is known via Eq. 1, the reverse reaction rates are given by, for instance,
 |
(6) |
where (θ′1, φ′1, θ′2, φ′2) is the equilibrium conformation for the state (s′1, s2). Given our specifications of the forward rates, the reverse rates are unambiguously defined. We see that the overall motor energy changes (or regulates) the reaction cycle in each motor domain. This regulation introduces a phase difference in the two reaction cycles and ultimately explains motor processivity. The implementation of these rate constants in a master equation model is described in more detail in Appendix B.
Motor energy
In this section, we describe how we compute the overall elastic energy of the motor. This energy is important for obtaining the dimer equilibrium configuration for each state and the rate constants. First, the energy of the motor monomers is given by E0(θi, φi, si). A simple but sufficient way to model E0 is using harmonic functions,
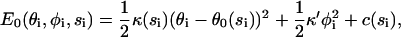 |
(7) |
where κ(si), κ′ are the moduli of in-plane (along the actin filament) and off-plane bendings. In our model, these parameters are fitted to obtain results consistent with experimental data; the numerical values are summarized in Table 1. However, as we emphasized earlier, the flexibility of the motor domains is crucial for obtaining an asymmetrical conformation when ADP is in both motor domains. These parameters may be measured from experiments or computed from molecular dynamics simulations. The value θ0(si) indicates the equilibrium conformations of the myosin monomer. Here, we assume at the equilibrium conformation, φi = 0. The equilibrium conformation of the monomer is a function of its chemical state. Due to structural differences in the converter domain, the equilibrium conformations of myosin-V are also different from myosin-VI. The values we have used are given in Table 2 in the Appendix. The last constant in Eq. 7, c(si), is not fitted. This constant represents the energy difference between monomer equilibrium conformations and can be obtained from monomer kinetics data (36,38) via detailed balance conditions:
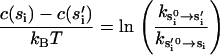 |
(8) |
The remaining energy is the elastic energy of the light-chain domain, which has different morphologies in myosin-V and myosin-VI. In myosin-V, the light-chain domain is an α-helix, decorated by six IQ motifs where calmodulins can bind. Since calmodulins protect the helix from water, the domain is expected to be folded and stable. In a separate study, it was shown that bending persistence length of an α-helix is ∼100 nm (45). With the bound calmodulins, the light-chain domain is probably stiffer. Our earlier treatment considered only in-plane bending explicitly. Here, we develop the elastic model more fully. In myosin-VI, the light-chain domain has one IQ motif but the converter domain also binds calmodulin (30). The remaining proximal region of the light chain (∼83 residues) is mostly unfolded and extended (11,13). Taking these structural facts into account, we use two different forms of El.
TABLE 1.
Miscellaneous parameters used in the model
| Symbol | Description |
|---|---|
| R | Radius of actin filament, 5.5 nm. |
| Δz | Size of actin monomer, 5.5 nm. |
| lm | Length of the myosin motor domain, 6 nm. |
| lp1 | Bending and twist persistence length of myosin-V light-chains (lp1=150 nm, lp2=400 nm, lp3=400 nm). |
| α | Bending persistence length of myosin-VI proximal tail, 0.9 nm. |
| Δs | Length of a single IQ motif, 5.0 nm. |
| L | Length of myosin V IQ motifs, 27 nm. |
| Length of myosin VI converter plus IQ motif, 10 nm. | |
| l | Total length of one proximal tail, 29 nm. |
TABLE 2.
Parameters used to specify myosin-V monomer energy E0(θi, φi, si)
| si | Identity | κ(si) (kBT) | θ0(si) (degrees) | κ′(si) (kBT) | c(si)(kBT) |
|---|---|---|---|---|---|
| 1 | A.M.E | 20.0 | 10.0 | 100.0 | 25(0.0) |
| 2 | M.DP | 16.0 | −50.0 | 100.0 | 16.1 |
| 3 | A.M.DP | 16.0 | −50.0 | 100.0 | 10.3 |
| 4 | A.M.D | 20.0 | 15.0 | 100.0 | −2.4 |
The constants c(si) are obtained for [ATP] = 1 mM, [ADP] = 1 μM, and [Pi] = 1 mM. At other conditions, c(si) will change.
For myosin-V, we treat the light-chain domains as two elastic rods with a completely free joint in between. The elastic energy can be written as
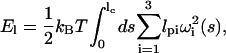 |
(9) |
where lc is the light-chain contour length, ωi is the ith torsion angle, and i ranges from 1 to 3. The values lp1 and lp2 are the bending persistence lengths in two principal bending directions. The value lp3 is the twist persistence length. If lp1=lp2, then the rod is isotropic. For myosin-V, the persistence length of the light-chain domain is unknown. Here, we find that an anisotropic rod model is necessary; the values are summarized in Table 1. Since there are six IQ motifs, we compute the light-chain energy using a discrete form of Eq. 9,
 |
(10) |
where Δs is the segment size and N– 1 indicates that there are N − 1 joints along the rod and N=12. (Note that since the joint between the light-chain domains is free, lp1 for the 6th joint is zero. There is no bending or twisting energy for this joint.) For each segment, we define a local frame (e1(n), e2(n), e3(n)), specifying the orientation of the nth segment in three-dimensional space. The skewed symmetric rotation matrix connecting the adjacent segments is defined as
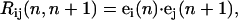 |
(11) |
where Rij is an element of the rotation matrix R(n, n+1). This allows us to define the torsional angles ωi(n) using the relations
 |
(12) |
 |
(13) |
where εjk is the permutation symbol. The three-dimensional position of the nth segment is obtained by
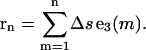 |
(14) |
If there is an external force applied at the joint between the light-chain domains, then the energy becomes
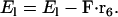 |
(15) |
We assume F is parallel to F-actin, although any F can be modeled.
Given the elastic constants (lpi), the light-chain energy is a function of its boundary conditions. The boundary conditions are determined by the orientation of the first segment and last segment, (θ1, φ1, θ2, φ2), and the location of the second motor domain with respect to the first one. We treat the actin filament as a helical rod, and there is a one-to-one mapping between the interhead distance z and the location of the binding sites. Details of this relationship are given in Appendix B.
For myosin-VI, the elastic energy of the IQ motifs can be modeled in the same way as Eq. 9, except that the contour length is much shorter. In between the IQ motifs is the soft proximal tail region. We model the total elastic energy of the light-chain region as
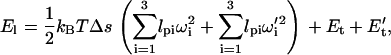 |
(16) |
where the first term is the elastic energy in the first and last set of IQ motifs. Since there are two calmodulins in each light-chain domain, there are two joints, described by torsion angles ωi and ω′i. The remaining part is the proximal tail energies, Et and E′t, which are given by a WLC description. Since the position of the IQ motifs are known, we define x = |r2 − r6|, where r2 and r6 are the end of the IQ motif and the joint between the domains (see Fig. 1). The value x is the end-to-end distance of the WLC. The elastic energy of one of the segment of the proximal tail is then
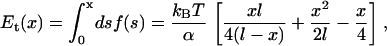 |
(17) |
where f(s) is the worm-like chain force-extension curve,
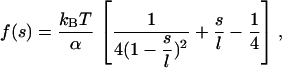 |
(18) |
where l and α are the overall contour length and the persistence length of the proximal tail, respectively (42–44). The energy of the other segment of the proximal tail, E′t, has a similar form, except the end-to-end distance is defined as x=|r′2 − r6|. Note that Et is essentially a quadratic function of extension, x. Again, an external load will add a term to the overall energy in the same fashion as Eq. 15. The light-chain energy and the positions of r2, r′2, and r6 are obtained by force balance (mechanical equilibrium). The light-chain energy again depends on the boundary conditions and interhead separation, and applied force (θ1, φ1, θ2, φ2, z, F). Having computed the light-chain energy and the total energy, the equilibrium conformations of the dimer can be computed by minimizing the total energy via Eq. 2. This allows us to define rate constants and their dependences on external forces and z.
RESULTS
Motor energy and conformation as a function of binding site
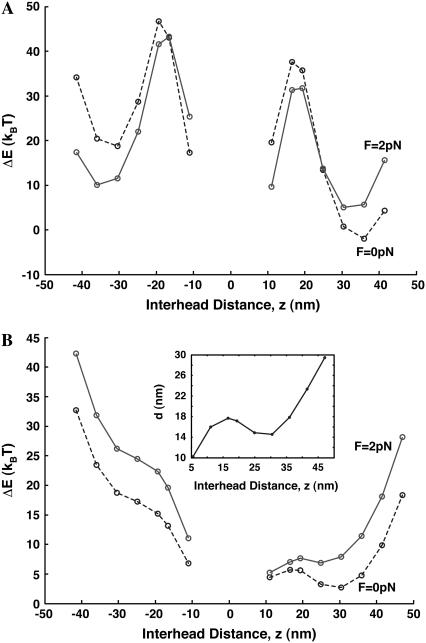
Binding of the free motor domain in our model occurs at (A.M.D, M.DP) → (A.M.D, A.M.DP). The rate constant for this step determines the step size, and the rate constant is regulated by the ΔE, given in Eq. 4. Fig. 3 shows ΔE as function of binding site for myosin-V and -VI under different load force conditions. Since there are approximately two possible binding sites for each z, we show the lower energy binding site only. For myosin-V, there is a low energy conformation at the 13th binding site (z ∼ 36 nm) and a local minimum at the−13th binding site (z ∼−36 nm). This implies that the binding rate to F-actin is fastest for z=36 nm. For myosin-VI, there is a much shallower well at z=30 nm, corresponding to the 12th site. Therefore, the probabilities of binding to 25 nm and 36 nm are comparable to binding to z = 30 nm. However, backward binding to z =−30 disappears and the lowest energy backward site is z=10 nm. This ensures that the net movement direction of myosin-VI is forward.
FIGURE 3.
The elastic energy difference of the motor dimer, ΔE, given by Eq. 4. The dashed line is for F=0.0 pN and the red solid line is for F=2 pN. (A) Myosin-V shows large variations in ΔE and a low energy for z=36 nm. This implies that the forward 36-nm step is preferred. The −36 nm step is also possible, and becomes almost equally probable as the 36-nm step when F=2 pN. (B) Myosin-VI has a softer connection between motor domains and therefore ΔE shows a smaller variation. The 31-nm step is the preferred binding site. The value−10 nm is the preferred backward step. When the load force is 2 pN, the preferred binding site shifts slightly to 25 nm. In the inset, we show the equilibrium extension of the proximal tail region, d=|r2 − r′2|, as function of interhead separation. The behavior of d largely explains the low energy conformation at z=30 nm. According to our model, binding sites at ±5 nm and ±10 nm are also low in energy and therefore small steps in these sites are possible.
The observed behavior of ΔE for myosin-VI can be explained by the helical structure of F-actin. The extension between the IQ motifs, |r2 − r2|, is not a linear function of z. This is shown in the inset in Fig. 3 b. Even though the 30-nm binding site is quite far in z, binding sites such as 24 nm and 18 nm have a longer extension.
Loading at the joint, r6, between the light-chain domains changes motor energy. For myosin-V, z-positions of the favorable sites remain the same as the load force is increased. However, the energies of 36 nm and−36 nm sites become similar, suggesting that the probabilities of binding to those sites become comparable at F=2 pN. At this load force, the dimer can step either forward or backward, leading to no net motion. The net binding rate at F=2 pN is also exponentially smaller by a factor of ∼e−7. For myosin-VI, the favorable forward binding position shifts by 5 nm under load force >1.0 pN. This has been observed in experiments (13).
An interesting feature in Fig. 3 is that ±5 nm and ±10 nm binding sites are also favorable binding sites. Whether this is actually occurring in experiments is controversial. Yanagida and co-workers (46) have observed 5-nm steps in myosin-V fixed to an AFM tip. Our model uses very simple descriptions of the proximal domain. For instance, for small extensions, WLC is not correct and factors such as the finite size of the chains and excluded volume come into play. Thus, the computed energies of ±5 nm and ±10 nm sites should be regarded as estimates.
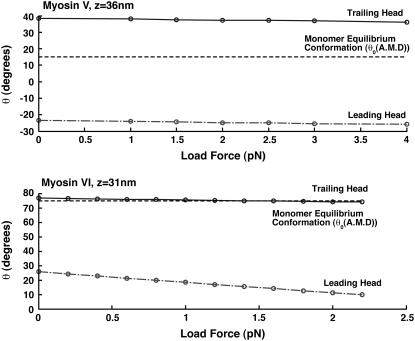
After binding to F-actin and phosphate release, the dimer remains for a long period in the (A.M.D, A.M.D) state. In this state, both motor domains are waiting to release ADP. Fig. 4 shows the equilibrium conformations of the dimer, (θ1, φ1, θ2, φ2), as a function of the applied load. We see that the leading motor domain has a different geometry than the trailing motor domain, indicating that the ADP release rate is different. The trailing head releases ADP faster and therefore the dimer processes forward.
FIGURE 4.
The equilibrium conformations of myosin-V and myosin-VI dimer. Here, the equilibrium (θ1, θ2) of the dimer are plotted for the (A.M.D, A.M.D) state. The monomer equilibrium conformation is the dashed line. For myosin-V, the conformations are fairly independent of the load force. For myosin-VI, most of the conformational difference occurs in the leading head.
The elastic model also allows us to explicitly compute the forces transmitted between the motor domains. These results can be used in conjunction with the measurement of force-dependent kinetic in single monomers. Since the conformations of the motor domain is defined by (θi, φi), transmitted stress is also a torque. For instance, the torque in the θ1 direction is computed by
 |
(19) |
where the derivative is taken at the dimer equilibrium conformation. The forces acting on the motor domain can also be computed by taking the derivative of the energy with respect to the motor domain position. For myosin-VI, since the IQ motifs are stiffer than the proximal tail, the torque is ∼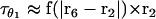 , where the magnitude of the extension force |f| is given by Eq. 18. Our model predicts that the leading head and trailing head experience dramatically different forces and torques, especially when their separation is large. For example, |f| for the leading and trailing heads is ∼5 pN at zero load force and z = 30 nm. However, as the load force increases to 2 pN, the force on the trailing head decreases to ∼3 pN while the force on the leading head remains essentially constant. These different forces and torques ultimately lead to very different kinetics.
, where the magnitude of the extension force |f| is given by Eq. 18. Our model predicts that the leading head and trailing head experience dramatically different forces and torques, especially when their separation is large. For example, |f| for the leading and trailing heads is ∼5 pN at zero load force and z = 30 nm. However, as the load force increases to 2 pN, the force on the trailing head decreases to ∼3 pN while the force on the leading head remains essentially constant. These different forces and torques ultimately lead to very different kinetics.
Force-velocity relations and dwell times
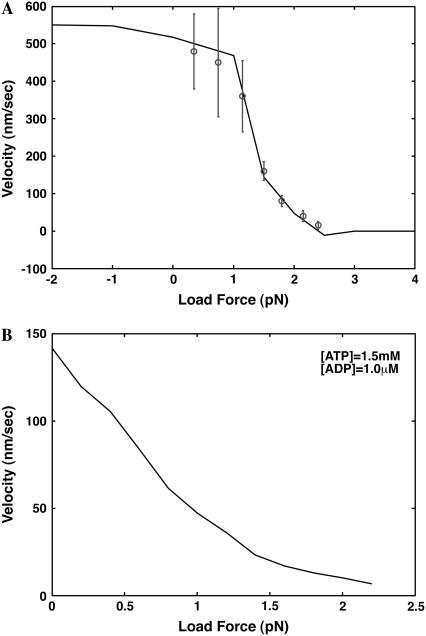
Fig. 5 shows the force-velocity curve for myosin-V and myosin-VI. The myosin-V prediction is shown together with the experimental data under saturating [ATP]. Under low force (<1.0 pN), stepping velocity is not sensitive to the applied force, and is within the reasonable range of 400 nm/s ∼ 500 nm/s. Between ∼1.0 pN and 1.5 pN, velocity drops dramatically to ∼100 nm/s. And for forces >1.5 pN, velocity slowly approaches to zero. The stall force from our model is ∼2.5 pN. Beyond the stall force, there is a small negative velocity until ∼3 pN.
FIGURE 5.
Computed force-velocity curves of myosin-V and myosin-VI. (A) The myosin-V result is compared with the experimental measurements of Uemura et al. (7). (B) Myosin-VI force-velocity relationship has a different behavior. Instead of a plateau in the velocity, the speed decreases monotonically with increasing force. This results from competition between ADP rebinding and ATP binding, which depend differently on the load force.
The behavior of the force-velocity curve can be rationalized by examining the rate-limiting steps. Under low load force, the rate-limiting step is the trailing head ADP release ∼15 s−1. As the force increases beyond 1.0 pN, F-actin binding rate becomes comparable with ADP release, and both contribute to decreasing velocity. When the load force is >1.5 pN, F-actin binding becomes rate-limiting and the movement slows and stalls. Here, backward binding is equally probable as well.
The force-velocity curve for myosin-VI has a different behavior. While the maximum velocity at zero load force is comparable to myosin-V, our model predicts that under certain nucleotide conditions (ADP added), the velocity decreases with increasing load, with no plateau regions. The predicted stall force is, however, quite high, near 2.2 pN. The lack of a plateau region is explained by the competition between ADP rebinding and ATP binding at the A.M.E state (13). ADP rebinding rate is affected by the load and becomes comparable to ATP binding rate. This slows down the motor. Stall is reached when binding to actin becomes rate-limiting.
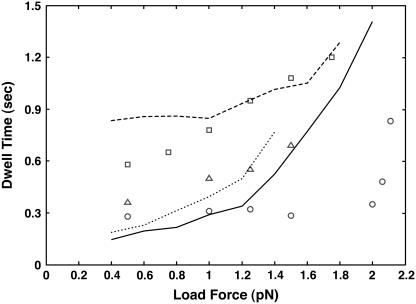
From the stochastic trajectories, it is possible to obtain the statistics of dwell times between steps. Indeed, the force velocity relation is approximately the average step size divided by the average dwell time. Note that dwell times between steps is not equivalent to dwell times between states since the motor can be in several states before stepping. Dwell times of myosin-VI have been measured. Our model results are shown in comparison with the experiments in Fig. 6. The experimental result shows a sharp change in the dwell time at high [ATP], high load force, and no [ADP]. The model result shows a much slower change. The explanation of this result has to rely on a more sophisticated model of the light-chain elastic energy. The WLC model can only be an approximation to the actual mechanical behavior of the proximal tail. WLC also assumes that the contour length is infinite when compared to the persistence length, and gives zero extension at no load force. This is clearly unrealistic. Nevertheless, the model qualitatively captures the observed dwell times at higher [ADP].
FIGURE 6.
The dwell time before taking a step as a function of the load force is shown for [ATP]=2 mM, [ADP]=0 μM (solid line and circles); [ATP]=1.5 mM, [ADP]=1 μM (dotted line and triangles); and [ATP]=100 μM, [ADP]=0 μM (dashed line and squares). The symbols are measurements from Altman et al. (13). The model results are in reasonable agreement, although for high [ATP] and no [ADP], the agreement is poor. This is a limitation of the WLC model.
At higher [ADP] concentrations, due to competition between ADP rebinding and ATP binding, the velocity becomes a smoother function of the load force. This is also consistent with the force-velocity curve.
Step-size distributions
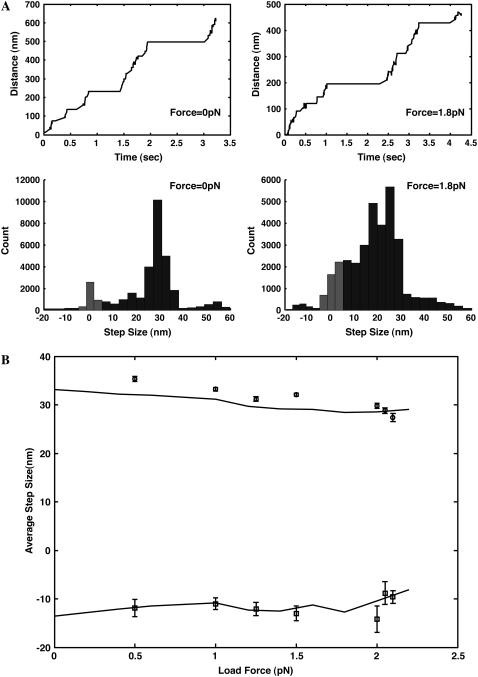
Our model, combined with a kinetic Monte Carlo scheme, allows us to compute trajectories of dimer stepping. The computational procedure gives continuous time results whereas most experimental apparatus has a finite time resolution of ∼10−3–10−6 s. Therefore, we perform some averaging with an interval of δt=10−5 s (windowing). If both motor domains are bound to F-actin for >δt=10 ms, we define the difference between neighboring average positions as the step size. Fig. 7 shows the distributions for myosin-VI.
FIGURE 7.
(A) Trajectories and step-size distributions of myosin-VI under different load conditions. The ATP concentration is 2 mM and no ADP is present. Model results have essentially infinite positional resolution. Therefore, very small steps (5–10 nm) are resolved and shown in the step-size distributions as light shaded bars. In an experiment, these steps may be indistinguishable from stationary positions. (B) The average step size for the forward and backward steps are shown as a function of the load force. The experimental results (symbols) are taken from Altman et al. (13).
Step-size distributions of myosin V are essentially unchanged from our earlier model. There is a large peak at 36 nm for all load forces. This is a consequence of Fig. 3. Backward steps and substeps are also seen. The relative probability of these steps would be functions of the load force. The substeps have been explained before. Our current model is consistent with the earlier predictions.
For myosin-VI, broader step-size distributions are observed. Under low load forces, the peak in the distribution is at ∼31 nm. Increasing the force decreases the step size and for force >1.5 pN, the peak step size becomes 26 nm. A smaller peak at ∼0–10 nm is also visible in the distribution. The distribution is also broader. Since the free energy differences between the sites are not large, the leading head could attach and detach from unfavorable sites many times before it finally finds a more favorable binding site. The relative energy of Fig. 3 determines the width of the step-size distribution in Fig. 7. Repeated attachment and detachment in the presence of ATP was suggested by de la Cruz et al. (47). The backward steps of myosin-VI only occurs at ∼5−10 nm. At higher load forces, the peak at ∼0−10 nm is due to favorable binding energy to those sites. In experiments, depending on the spatial resolution of the apparatus, binding to these sites is difficult to observe. Here, steps due to these binding events are marked with gray bars.
Larger steps (>50 nm) are observed for both motors. These only arise after windowing with a time interval δt. Rapid successive steps appear as a single step if there is finite time resolution.
DISCUSSION AND CONCLUSIONS
We have developed a unified mechanochemical model to explain the processivity of dimeric myosin motors. The helical structure of F-actin and the mechanical properties of the light-chain domains are crucial for regulating processivity. In myosin-V, the light-chain domains are relatively stiff, resulting in regular steps of 36 nm. When load forces are applied, the regular step size does not change. In myosin-VI, the light-chain domains, which contain a soft proximal tail region, are floppy and act as entropic springs. The amount of extension between the IQ motifs determines the stepping behavior. Experimentally measured rate constants are used to parameterize the model. Postulates of how the rate constants depend on the motor conformation are made.
Stall force is a key measurable for motor proteins, and our model provides an explanation for the observed stall force. Ultimately, two factors are important. First, ΔE of Eq. 4 controls the probability of binding to the available sites. When a load force is applied, the backward step eventually becomes as probable as the forward step and the motor stalls. Second, with increasing load force, ΔE also rises, implying that the absolute rate of binding to F-actin also slows. Indeed, ΔE can be interpreted as the transition state energy of binding to F-actin and eventually, the barrier becomes prohibitively high for binding. From an efficiency standpoint, the second factor does not waste ATP near stall where the first factor leads to futile hydrolysis of ATP. Our model shows that the second factor is probably dominant for myosin-V and VI near stall.
We note that the mechanical model introduced here is simple and only approximates the complex protein elasticity, although the model appears to capture most of the experimental observations. According to the model, in addition to the favorable binding sites at 36 nm and 30 nm, sites at ±5 nm and ±10 nm are also probable (Fig. 3). While there is no concrete experimental evidence in favor of this observation, the resolution of experiments is such that ruling out these small steps is difficult. We note that our model lacks contributions from the excluded volume interaction between the light chains. These interactions will raise the energy of ±5 nm and ±10 nm sites and render them less probable. Another possibility is that binding to F-actin alters the structure of actin at ±5 nm and ±10 nm sites, and thus the second head cannot bind to these sites. At present, our model cannot rule out stepping to those sites (our earlier treatment used a more approximate treatment of out-of-plane bending and did not predict binding to ±5 nm and ±10 nm sites) and further investigation of this issue is interesting.
Other models on dimeric motors are mostly based on some proposed kinetic schemes. These models assume distinct configurations, corresponding to a unique combination of chemical state and conformation on the energy landscape. Transition rates between configurations are typically postulated. In our model, each chemical state can have a continuum of conformations, characterized by variables (θ1, φ1, θ2, φ2). The energy landscape is constructed using simple elastic models. The transition rates between chemical states also depend on conformation which reflects experimental observation. As such, the kinetic models can be considered as subsets of models such as ours. We note that the number of parameters in our model is not necessarily larger than other models, but different biophysical measurements such as bending elasticity of protein subunits are needed to establish the parameters.
The basic model framework introduced here is equally applicable to other dimeric processive motors such as kinesin and dynein, although for these systems, the details of the model are likely to be more complex. Kinesin and dynein are microtubule motors, implying that the number of accessible sites for the free head is potentially much larger than that of myosin-V and VI. Single-molecule experiments show that kinesin seems to process on a single protofilament (48). From our model, we see that the preferred binding site is ultimately related to the elasticity of the connection between motor domains. In kinesin, the mechanical properties of the neck-linker are likely to be complex. In dynein, the connection between the microtubule binding domains containing the hexameric AAA domain is bulky (49). The interaction between the AAA domains is significant. Phenomenological models of dynein have shown that the step size depends on load force (50,51). The full explanation of dimeric processive motor proteins will depend on further understandings of the elasticity of protein domains.
Acknowledgments
The authors acknowledge helpful discussions with Dr. James Sellers, Dr. Peter Knight, and Dr. Claudia Veigel.
This work has been supported by the National Science Foundation through NSF grants Nos. CHE-0514749 and CHE-0547041.
APPENDIX A: SUMMARY OF PARAMETERS
The numerical values of the parameters used in specifying the dimer energy is given in Tables 1–3. In this model, κ and κ′, θ0(si), lpi, and α are estimated to explain the experimental data. Other parameters in Table 1 are not fitted.
TABLE 3.
Parameters used to specify myosin-VI monomer energy E0(θi, φi, si)
| s1 | Identity | κ(si) (kBT) | θ0(si) (degrees) | κ′(si) (kBT) | c(si)(kBT) |
|---|---|---|---|---|---|
| 1 | A.M.E | 8.5 | 90.0 | 50.0 | 25(0.0) |
| 2 | M.DP | 5.0 | −10.0 | 50.0 | 16.1 |
| 3 | A.M.DP | 5.0 | −10.0 | 50.0 | 12.7 |
| 4 | A.M.D | 10.0 | 75.0 | 50.0 | −2.4 |
The value c(si) is unavailable for M.DP. We have used the same value as myosin-V.
Table 4 shows the monomer equilibrium reaction rate constants, 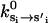 . These values are taken from kinetic data of de la Cruz et al. (36,38). Note for A.M.E → M.DP, the rate is limited by ATP binding in myosin-VI, but for myosin-V, this rate is limited by A.M.T* → A.M.T. We have used the reaction rate from the limiting steps since most reported data are for saturating [ATP]. This also implies that for myosin-V, this step is not sensitive to the motor conformation.
. These values are taken from kinetic data of de la Cruz et al. (36,38). Note for A.M.E → M.DP, the rate is limited by ATP binding in myosin-VI, but for myosin-V, this rate is limited by A.M.T* → A.M.T. We have used the reaction rate from the limiting steps since most reported data are for saturating [ATP]. This also implies that for myosin-V, this step is not sensitive to the motor conformation.
TABLE 4.
Kinetic rate constants at monomer conformation equilibrium
| Myosin V | Myosin VI | |
|---|---|---|
| A.M.E → M.DP | ∼300 s−1 | 0.018 μM−1 s−1 |
| A.M.E → A.M.D | 12.6 μM−1 s−1 | 0.3 μM−1 s−1 |
| M.DP → A.M.DP | 4700 s−1 | 5000 s−1 |
| A.M.DP → M.DP | 14.4 s−1 | 200 s−1 |
| A.M.DP → A.M.D | 250 s−1 | 90 s−1 |
| A.M.D → A.M.E | 15 s−1 | ∼4 s−1 |
| M.DP → A.M.E | ∼0 s−1 | ∼0 s−1 |
| A.M.D → A.M.DP | ∼7.7 × 10−4 mM−1s−1 | ∼7.7 × 10−4 mM−1 s−1 |
Several rate constants for myosin-VI are unavailable; in these cases, myosin-V values are used as estimates.
Several constants, e.g., c(si) for M.DP state and the rate constant for A.M.D → A.M.DP, are unavailable for myosin-VI. Most of the results are reported for [Pi] = 0.0 mM. Therefore, these values are not important for the reported results. In the tables, myosin-V values are substituted for myosin-VI when appropriate.
APPENDIX B: COMPUTATION OF LIGHT-CHAIN ENERGY IN MYOSIN-V AND MYOSIN-VI
From the geometry of F-actin binding sites, the positions of the protofilaments can be written as two helices. This implies that for each binding site on each protofilament, we can write the approximate position of the converter domain (joint between motor domain and light-chain domain) of a bound motor domain as
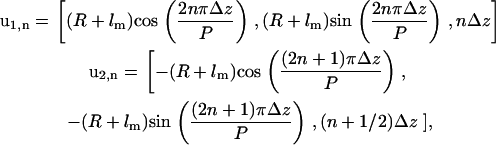 |
(B1) |
where u1, n and u2, n are the positions of bound myosin on site n of protofilament 1 and 2, respectively. R is the radius of F-actin and lm is the approximate length of the myosin motor domain. P=72 nm is the helical pitch of F-actin and Δz=72/13 ≈ 5.5 nm is the size of actin monomer. The value ui, n defines the initial position of the light-chain domain and (θi, φi) defines its orientation. The total energy of the bound dimer is described in the main text. To find the equilibrium (θi, φi), two methodologies are used: Method One assumes that the light-chain domains and the proximal tail region are in mechanical equilibrium; with this assumption, the equilibrium conformation of the dimer can be obtained by force balance, or minimizing the overall energy. Method Two makes no assumptions. A Monte Carlo simulation is carried out for (θ1, φ1, θ2, φ2) and all the degrees of freedom of the light-chain domain. From this simulation, the most probable conformation is called the equilibrium conformation. The energy of the dimer is given by umbrella sampling of the free energy as a function of interhead separation. These methods yield essentially the same results. The results shown here are obtained from Method One.
APPENDIX C: SOLUTION OF THE MODEL
Given the total dimer energy equation (Eq. 1), and the transition rates between the states, the dynamics of the dimer can be computed using a Fokker-Planck equation. This was carried out in our previous model of myosin-V. However, conformational relaxation is much faster than transitions between the chemical states. Under these conditions, the Fokker-Planck equation can be simplified to a kinetic master equation. Therefore, we simplify the description and define states on the energy surface using si and the equilibrium conformations (θi, φi). For given chemical states (s1, s2), interhead distance z (z = 0 for single head-bound state), and the load force F, the equilibrium configuration is uniquely defined. A kinetic master equation of the form
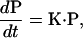 |
(C1) |
where P(t) is the vector whose ith element is the probability of being in the ith state. There is a one-to-one mapping between i and (s1, s2) and the conformation (θ1, φ1, θ2, φ2, z, F). The matrix of transition rates, K, are defined by the rate constants specified in the main text, and depend on the dimer equilibrium conformation. The monomer equilibrium rates used to parameterize the model are summarized in Table 2. Since for the ith state, the equilibrium conformation of the dimer is known and the total energy is known, this unambiguously defines K. The total number of elements of P is >4 × 4 = 16. If ∼13 possible binding sites are included, the number of elements is 13 × 3 × 3 + 2 × 3.
The steady-state velocity of the motor dimer can be computed by setting the left-hand side of Eq. C1 to zero. Alternatively, a stochastic trajectory can be generated starting from Eq. C1. The algorithm is based on the method of Bortz et al. (52), and was described earlier. Many trajectories are computed to obtain the average speed, step-size distributions, and dwell times.
References
- 1.Sellers, J. R. 2000. Myosins: a diverse superfamily. Biochim. Biophys. Acta. 1496:3–22. [DOI] [PubMed] [Google Scholar]
- 2.Langford, G. M. 2002. Myosin-V, a versatile motor for short-range vesicle transport. Traffic. 3:859–865. [DOI] [PubMed] [Google Scholar]
- 3.Buss, F., G. Spudich, and J. Kendrick-Jones. 2004. Myosin VI: cellular functions and motor properties. Annu. Rev. Cell Dev. Biol. 20:649–676. [DOI] [PubMed] [Google Scholar]
- 4.Roberts, R., I. Lister, S. Schmitz, M. Walker, C. Veigel, J. Trinick, F. Buss, and J. Kendrick-Jones. 2004. Myosin VI: cellular functions and motor properties. Philos. Trans. R. Soc. B. 359:1931–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehta, A. D., R. S. Rock, M. Rief, J. A. Spudich, M. S. Mooseker, and R. E. Cheney. 1999. Myosin-V is a processive actin-based motor. Nature. 400:590–593. [DOI] [PubMed] [Google Scholar]
- 6.Veigel, C., F. Wang, M. Bartoo, J. Sellers, and J. Molloy. 2001. The gated gait of the processive molecular motor, myosin V. Nat. Cell Biol. 4:59–65. [DOI] [PubMed] [Google Scholar]
- 7.Uemura, S., H. Higuchi, A. O. Olivares, E. M. de la Cruz, and S. Ishiwata. 2004. Mechanochemical coupling of two substeps in a single myosin V motor. Nat. Struct. Mol. Biol. 11:877–883. [DOI] [PubMed] [Google Scholar]
- 8.Rief, M., R. Rock, A. Mehta, M. Mooseker, R. Cheney, and J. Spudich. 2000. Myosin-V stepping kinetics: a molecular model for processivity. Proc. Natl. Acad. Sci. USA. 97:9482–9486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veigel, C., S. Schmitz, F. Wang, and J. R. Sellers. 2005. Load-dependent kinetics of myosin-V can explain its high processivity. Nat. Cell Biol. 7:861–869. [DOI] [PubMed] [Google Scholar]
- 10.Rock, R. S., S. E. Rice, A. L. Wells, T. J. Prcall, J. A. Spudich, and H. L. Sweeney. 2001. Myosin VI is a processive motor with a large step size. Proc. Natl. Acad. Sci. USA. 98:13655–13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rock, R. S., B. Ramamurthy, A. R. Dunn, S. Beccafico, B. R. Rami, C. Morris, B. J. Spink, C. Franzini-Armstrong, J. A. Spudich, and H. L. Sweeney. 2005. A flexible domain is essential for the large step size and processivity of myosin VI. Mol. Cell. 17:603–609. [DOI] [PubMed] [Google Scholar]
- 12.Ali, M. Y., K. Homma, A. H. Iwane, K. Adachi, H. Itoh, K. Kinosita, Jr., T. Yanagida, and M. Ikebe. 2004. Unconstrained steps of myosin VI appear longest among known molecular motors. Biophys. J. 86:3804–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altman, D., H. L. Sweeney, and J. A. Spudich. 2004. The mechanism of myosin VI translocation and its load-induced anchoring. Cell. 116:737–749. [DOI] [PubMed] [Google Scholar]
- 14.Nishikawa, S., K. Homma, Y. Komori, M. Iwaki, T. Wazawa, A. H. Iwane, J. Saito, R. Ikebe, E. Katayama, T. Yanagida, and M. Ikebe. 2002. Class VI myosin moves processively along actin filaments backward with large steps. Biochem. Biophys. Res. Commun. 290:311–317. [DOI] [PubMed] [Google Scholar]
- 15.Yildiz, A., H. Park, D. Safer, Z. Yang, L. Chen, P. R. Selvin, and H. L. Sweeney. 2004. Myosin VI steps via a hand-over-hand mechanism with its lever arm undergoing fluctuations when attached to actin. J. Biol. Chem. 279:37223–37226. [DOI] [PubMed] [Google Scholar]
- 16.Balci, H., T. Ha, H. L. Sweeney, and P. R. Selvin. 2005. Interhead distance measurements in myosin VI via SHRImP support a simplified hand-over-hand model. Biophys. J. 89:413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yildiz, A., and P. R. Selvin. 2005. Fluorescence imaging with one nanometer accuracy: application to molecular motors. Acc. Chem. Res. 38:574–582. [DOI] [PubMed] [Google Scholar]
- 18.Schliwa, M. 1999. Myosin steps backwards. Nature. 401:431–432. [DOI] [PubMed] [Google Scholar]
- 19.Menetrey, J., A. Bahloul, A. L. Wells, C. M. Yengo, C. A. Morris, H. L. Sweeney, and A. Houdusse. 2005. The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature. 435:779–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wells, A. L., A. W. Lin, L. Q. Chen, D. Safer, S. M. Cain, T. Hasson, B. O. Carragher, R. A. Milligan, and H. L. Sweeney. 1999. Myosin VI is an actin-based motor that moves backwards. Nature. 401:505–508. [DOI] [PubMed] [Google Scholar]
- 21.Homma, K., M. Yoshimura, J. Saito, R. Ikebe, and M. Ikebe. 2001. The core of the motor domain determines the direction of myosin movement. Nature. 412:831–834. [DOI] [PubMed] [Google Scholar]
- 22.Lan, G., and S. X. Sun. 2005. Dynamics of myosin-V processivity. Biophys. J. 88:999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houdusse, A., V. N. Kalabokis, D. Himmel, A. G. Szent-Gyorgyi, and C. Cohen. 1999. Atomic structure of scallop myosin subfragment S1 complexed with MgADP: a novel conformation of the myosin head. Cell. 97:459–470. [DOI] [PubMed] [Google Scholar]
- 24.Liu, J., D. W. Taylor, E. B. Krementsova, K. M. Trybus, and K. A. Taylor. 2006. Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography. Nature. Advance online publication. [DOI] [PubMed]
- 25.Coureux, P. D., A. L. Wells, J. Menetrey, C. M. Yengo, C. Morris, H. L. Sweeney, and A. Houdusse. 2003. A structural state of the myosin V motor without bound nucleotide. Nature. 425:419–423. [DOI] [PubMed] [Google Scholar]
- 26.Coureux, P. D., H. L. Sweeney, and A. Houdusse. 2004. Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J. 23:4527–4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purcell, T. J., C. Morris, J. A. Spudich, and H. L. Sweeney. 2002. Role of the lever arm in the processive stepping of myosin V. Proc. Natl. Acad. Sci. USA. 99:14159–14164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Espindola, F. S., D. M. Suter, L. B. Partata, T. Cao, J. S. Wolenski, R. E. Cheney, S. M. King, and M. S. Mooseker. 2000. The light chain composition of chicken brain myosin-Va: calmodulin, myosin-II essential light chains, and 8-kDa dynein light chain/PIN. Cell Motil. Cytoskeleton. 47:269–281. [DOI] [PubMed] [Google Scholar]
- 29.Cheney, R. E., M. K. O'Shea, J. E. Heuser, M. V. Coelho, J. S. Wolenski, E. M. Espreafico, P. Forscher, R. E. Larson, and M. S. Mooseker. 1993. Brain myosin-V is a two-headed unconventional myosin with motor activity. Cell. 75:13–23. [DOI] [PubMed] [Google Scholar]
- 30.Bahloul, A., G. Chevreux, A. L. Wells, D. Martin, J. Nolt, Z. Yang, L. Q. Chen, N. Potier, A. Van Dorsselaer, S. Rosenfeld, A. Houdusse, and H. L. Sweeney. 2004. The unique insert in myosin VI is a structural calcium-calmodulin binding site. Proc. Natl. Acad. Sci. USA. 101:4787–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolomeisky, A., and M. E. Fisher. 2003. A simple kinetic model describes the processivity of myosin-V. Biophys. J. 84:1642–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vilfan, A. 2005. Elastic lever-arm model for myosin V. Biophys. J. 88:3792–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sellers, J. R., and C. Veigel. 2006. Walking with myosin V. Curr. Opin. Cell Biol. 18:68–73. [DOI] [PubMed] [Google Scholar]
- 34.Olivares, A. O., and E. M. de la Cruz. 2005. Holding the reins on myosin V. Proc. Natl. Acad. Sci. USA. 102:13719–13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purcell, T. J., H. L. Sweeney, and J. A. Spudich. 2005. A force-dependent state controls the coordination of processive myosin V. Proc. Natl. Acad. Sci. USA. 102:13873–13878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de la Cruz, E. M., A. L. Wells, S. S. Rosenfeld, E. M. Ostap, and H. L. Sweeney. 1999. The kinetic mechanism of myosin V. Proc. Natl. Acad. Sci. USA. 96:13726–13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de La Cruz, E. M., H. L. Sweeney, and E. M. Ostap. 2000. ADP inhibition of myosin V ATPase Activity. Biophys. J. 79:1524–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de La Cruz, E. M., E. M. Ostap, and H. L. Sweeney. 2001. Kinetic mechanism and regulation of myosin-VI. J. Biol. Chem. 276:32373–32381. [DOI] [PubMed] [Google Scholar]
- 39.Robblee, J. P., A. O. Olivares, and E. M. de la Cruz. 2004. Mechanism of nucleotide binding to actomyosin VI: evidence for allosteric head-head communication. J. Biol. Chem. 279:38608–38617. [DOI] [PubMed] [Google Scholar]
- 40.Bustamante, C., D. Keller, and G. Oster. 2001. The physics of molecular motors. Acc. Chem. Res. 34:412–420. [DOI] [PubMed] [Google Scholar]
- 41.Xing, J., H. Wang, and G. Oster. 2005. From continuum Fokker-Planck models to discrete kinetic models. Biophys. J. 89:1551–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bustamante, C., J. F. Marko, E. D. Siggia, and S. Smith. 1994. Entropic elasticity of λ-phage DNA. Science. 265:1599–1600. [DOI] [PubMed] [Google Scholar]
- 43.Marko, J. F., and E. D. Siggia. 1995. Stretching DNA. Macromolecules. 28:8759–8770. [Google Scholar]
- 44.Schwaiger, I., A. Kardinal, M. Schleicher, A. A. Noegel, and M. Rief. 2004. A mechanical unfolding intermediate in an actin-crosslinking protein. Nat. Struct. Mol. Biol. 11:81–85. [DOI] [PubMed] [Google Scholar]
- 45.Choe, S., and S. X. Sun. 2005. The elasticity of α-helices. J. Chem. Phys. 122:244912. [DOI] [PubMed] [Google Scholar]
- 46.Kitamura, K., M. Tokunaga, A. H. Iwane, and T. Yanagida. 1999. A single myosin head moves along an actin filament with regular steps of 5.3 nanometres. Nature. 397:129–134. [DOI] [PubMed] [Google Scholar]
- 47.de la Cruz, E. M., and E. M. Ostap. 2004. Relating biochemistry and function in the myosin superfamily. Curr. Opin. Cell Biol. 16:61–67. [DOI] [PubMed] [Google Scholar]
- 48.Ray, S., E. Meyhofer, R. A. Milligan, and J. Howard. 1993. Kinesin follows the microtubule's protofilament axis. J. Cell Biol. 121:1083–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakato, M., and S. M. King. 2004. Design and regulation of the AAA+ microtubule motor dynein. J. Struct. Biol. 146:58–71. [DOI] [PubMed] [Google Scholar]
- 50.Singh, M. P., R. Mallik, S. P. Gross, and C. C. Yu. 2005. Monte Carlo modeling of single-molecule cytoplasmic dynein. Proc. Natl. Acad. Sci. USA. 102:12059–12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao, Y. Q. 2006. A simple theoretical model explains dynein's response to load. Biophys. J. 90:811–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bortz, A., M. Kalos, and J. Lebowitz. 1975. A new algorithm for Monte Carlo simulation of Ising spin systems. J. Comput. Phys. 17:10–18. [Google Scholar]